ABSTRACT.
The role of microbial coinfection in the pathogenesis of pneumonia in children is not well known. The aim of this work was to describe the prevalence of microorganism co-detection in nasopharyngeal samples (NPS) of pneumonia cases and control subjects and to study the potential association between nasopharyngeal microorganism co-detection and pneumonia. A case-control study was carried out from 2010 to 2014 in nine study sites located in low- or middle-income countries. The data from 888 children under 5 years of age with pneumonia (cases) and 870 children under 5 without pneumonia (controls) were analyzed. Nasopharyngeal samples were collected; reverse transcription polymerase chain reaction (RT-PCR) enabled the detection of five bacteria and 19 viruses. Multiple, mixed-effects logistic regression modeling was undertaken to evaluate the association between microorganism co-detection and pneumonia. A single Streptococcus pneumoniae colonization was observed in 15.2% of the controls and 10.1% of the cases (P = 0.001), whereas S. pneumoniae and a single virus co-detection was observed in 33.3% of the cases and in 14.6% of the controls (P < 0.001). Co-detections with rhinovirus, respiratory syncytial virus, parainfluenza virus, human metapneumovirus, and influenza virus were more frequent in the cases compared with the controls (P < 0.001) and were significantly associated with pneumonia in multiple regression analysis. The proportion of single virus detection without bacterial co-detection was not different between cases and controls (13.6% versus 11.3%, P = 0.13). This study suggests that coinfection of S. pneumoniae and certain viruses may play a role in the pathophysiology of pneumonia in children.
INTRODUCTION
Pneumonia caused 13.3% of 5.05 million deaths in 2019 according to the Global Burden of Diseases Study 2019, and it remains the leading cause of mortality from infectious disease in children under 5 years of age. 1 The burden of the different pathogens associated with pneumonia has substantially changed in the last few years, mainly through the broad implementation of the pneumococcal conjugate vaccine (PCV) and Haemophilus influenzae type B vaccination policies. The number of bacterial pneumonias could have decreased in favor of viral pneumonia. Several studies have been carried out in low- and middle-income countries, for example, the Pneumonia Etiology Research of Child Health (PERCH) study run between 2011 and 2014. In this study, authors estimated that viruses accounted for 61.4% of pneumonia causes and bacteria (excluding mycobacteria) for 27.3%. 2 Bacterial infections can be more severe than viral infections, which are often confined to the upper respiratory tract. However, severe viral infections of the lower respiratory tract and viral pneumonia can also occur, in particular, in infants under 2 years of age. 3, 4
Nowadays, the pathophysiology of pneumonia is well explained. It is also known that viruses interact with the bacteria of the respiratory microbiome. 5, 6 Occurrences of bacterial and viral coinfection have been frequently reported in the literature. 7 However, the prevalence of microorganism coinfection in pneumonia, and their association with the disease in children is not well known, particularly in low- and middle-income countries. 8 Moreover, it is commonly admitted that different viral infections (e.g., influenza) predispose the host to secondary bacterial pneumonia 9 – 11; but conversely, it has also been suggested that bacterial colonization plays a role in virus-associated pneumonia. 12, 13
Between 2010 and 2014, the Mérieux Foundation, Global Approach for Biological Research, Infectious Diseases and Epidemics in Low-Income Countries (the GABRIEL Network) carried out a large, prospective, multicenter case-control study. 14 Its primary objectives were to identify the causative agents involved in the onset of pneumonia in children under 5 in low- and middle-income countries and to assess individual and microbial factors associated with the risk of pneumonia. 15 The microorganisms most frequently attributable to pneumonia were Streptococcus pneumoniae (adjusted population attributable fraction: 42.2%), followed by respiratory syncytial virus (RSV) (18.2%) and rhinovirus (RV) (11.2%). 16 The present study focused on a secondary objective of the GABRIEL Pneumonia study. The aim of the present work was to describe the prevalence of microorganism co-detection in nasopharyngeal samples (NPS) of pneumonia cases and control subjects, and to study the association of various detections and/or co-detections of microorganisms with pneumonia.
MATERIALS AND METHODS
Study sites and design.
The GABRIEL Network pneumonia study was a large, multicenter case-control study carried out between 2010 and 2014 in nine sites in eight low- or middle-income countries: Cambodia, China, Haiti, India (two sites), Madagascar, Mali, Mongolia, and Paraguay (see Supplemental material). The study protocol and sites have been previously described in detail. 15
Definitions of cases and controls.
The eligible cases were children from 2 to 60 months of age admitted for suspected pneumonia. Eligible cases were identified on admission by study clinicians at each participating site. Incident cases were included if they complied with protocol definitions and met all inclusion criteria. Eligible cases were included in the study on the basis of clinical features of pneumonia following WHO criteria: 1) cough and/or dyspnea, and 2) tachypnea, as delineated by the WHO, and 3) first symptoms appearing within the last 14 days, and 4) radiological confirmation of pneumonia on chest x-ray, as defined per WHO guidelines. 17 Pneumonia was considered hypoxemic when oxygen saturation was < 90%. According to the WHO, hypoxemia is one of the criteria defining severe pneumonia. 17 Controls were children 1) without symptoms of respiratory illness/upper respiratory tract infection, and 2) between 2 and 60 months of age, and 3) hospitalized for surgery or attending routine outpatient appointment at hospital site (see Supplemental material). For both cases and controls, their parents or legal guardians gave informed consent before inclusion.
Data sources and quality control.
Demographic characteristics, medical history, vital signs, clinical symptoms and biological parameters on admission were recorded prospectively for each subject on a standardized data collection form. Data quality was monitored and evaluated by the Foundation Mérieux study conduct team including the Emerging Pathogens Laboratory (Lyon, France).
Biological samples.
Specimens were collected within 48 hours after hospitalization. Nasopharyngeal samples were collected from all pneumonia case and control subjects. Nasopharyngeal samples enabled the identification of viruses and bacteria by reverse-transcription polymerase chain reaction (RT-PCR) assay with a panel of five bacteria and 19 viruses (Fast-track Diagnostic Respiratory Pathogens 21 PLUS, Esch-sur-Alzette, Luxembourg): S. pneumoniae, Staphylococcus aureus, H. influenzae type B, Mycoplasma pneumoniae, Chlamydophila pneumoniae, influenza virus (IV) A, A H1N1 and B, coronavirus (CoV) 229E, OC43, NL63 and HKU1, parainfluenza virus (PIV) 1, 2, 3 and 4, human metapneumovirus (HMPV) A and B, RV, RSV A and B, adenovirus (AdV), enterovirus (EV), parechovirus (HPeV), and bocavirus (BoV). 18 The specimen collection was performed by trained clinical staff, nurses, and the principal investigator of each site. The samples of 1–2 mL were aliquoted, analyzed on-site, and stored at −80°C. A centralized, blinded PCR respiratory quality control panel was provided to all sites to ensure procedure validation on-site before the specimens were processed locally.
Statistical analysis.
Descriptive analysis of the included and excluded populations was performed. Continuous variables were reported as median and interquartile range (IQR) with comparisons by Mann–Whitney U test. Categorical variables were computed as number of individuals and proportions with χ2 or Fisher’s exact test as appropriate for comparison. Calculated proportions of microorganism detection represented mean occurrence over the study period. They were reported per 100 subjects with their 95% CI.
Multiple, mixed-effects logistic regression modeling was undertaken to evaluate the association between microorganism co-detection and pneumonia, with center as the random effect. Models were adjusted for gender, age, other microorganisms detected and time period (year). Age was stratified in three groups: 2–11 months, 12–23 months, and 24–60 months. Interactions between microorganisms were tested for each different couple of microorganisms of interest. Multiple, mixed-effects logistic regression modeling was also undertaken to evaluate the association between microorganism co-detection and hypoxemic pneumonia, with center as the random effect. For the regression analyses, site-stratified regressions were also undertaken as sensitivity analyses. All tests were two-tailed, with P < 0.05 considered as statistically significant. Statistical analysis was undertaken with Stata Version 13.0 (College Station, TX).
Inclusion process and subject characteristics.
The inclusion process is described in Supplemental Figure 1; 2,247 subjects were enrolled. Of those, 489 (21.8%, N = 280 [24.0%] cases, N = 171 [16.4%] controls) were excluded owing to missing data and/or because they did not meet inclusion criteria. By study site, the percentage of excluded subjects ranged from 0.5% of all assessed subjects in Paraguay to 46.4% in Madagascar. A description of the excluded subjects is presented in Supplemental Table 1. Among the 1,758 included subjects, there were 888 (50.5%) cases and 870 (49.5%) controls, 1,024 (58.4%) children were male and the median age was 16 months (IQR 9–30 months). Subject characteristics are shown in Supplemental Tables 2 and 3. 19
Ethics.
All study documents were submitted to and approved by the institutional Research Ethics Committee of each site.
RESULTS
Overall detection of viruses and bacteria.
Overall detection of microorganisms in NPS (in combination or alone) are described in Supplemental Table 4. Human metapneumovirus, RSV, PIV, and IV were more frequently detected in pneumonia cases compared with controls (P < 0.001 for each). Streptococcus pneumoniae was more frequently detected in pneumonia cases compared with controls (P < 0.001), whereas S. aureus was more frequently detected in controls compared with cases (P = 0.003). The median number of bacteria detected was 1, IQR [0–1] in cases and 1, IQR [0–1] in controls, P = 0.84. The median number of viruses detected was 1, IQR [1–1] in cases and 0, IQR [0–1] in controls, P < 0.001.
Detection of viruses.
A single virus was detected in the NPS of 501/888 cases (56.4%) and 296/870 (34.0%) of controls (P < 0.001) regardless of the number of bacteria detected (Table 1). Single detection of HMPV, RSV, PIV, and IV was more frequently detected in case NPS compared with control NPS (P < 0.001 for each), regardless of the number of bacteria detected. Conversely, CoV and BoV were more frequently detected in control NPS compared with case NPS without other viral detection (P = 0.042 and 0.012, respectively).
Table 1.
Single virus detection in nasopharyngeal samples of pneumonia cases and controls with and without bacterial co-detection, the GABRIEL Network pneumonia study, 2010 through 2014
| Cases, nasopharyngeal samples, N = 888 | Controls, nasopharyngeal samples, N = 870 | P value | |
|---|---|---|---|
| Single virus detection without bacterial co-detection, n (%) | |||
| Rhinovirus | 30 (3.4) | 42 (4.8) | 0.18 |
| Respiratory syncytial virus A/B | 29 (3.3) | 5 (0.6) | < 0.001 |
| Bocavirus | 5 (0.6) | 14 (1.6) | 0.034 |
| Parainfluenza virus 1/2/3/4 | 19 (2.1) | 6 (0.7) | 0.010 |
| Coronavirus NL63/229E/OC43/HKU1 | 2 (0.2) | 14 (1.6) | 0.0035 |
| Adenovirus | 4 (0.5) | 4 (0.5) | 0.99 |
| Influenza virus A/A(H1N1)/B | 16 (1.8) | 5 (0.6) | 0.018 |
| Human metapneumovirus A/B | 14 (1.6) | 0 (0.0) | < 0.001 |
| Enterovirus | 2 (0.2) | 7 (0.8) | 0.17 |
| Parechovirus | 0 (0.0) | 1 (0.1) | 0.99 |
| Number of patients with a single virus detection regardless of the number of bacteria detected, n (%) | |||
| Rhinovirus | 140 (15.8) | 116 (13.3) | 0.15 |
| Respiratory syncytial virus A/B | 112 (12.6) | 18 (2.1) | < 0.001 |
| Bocavirus | 22 (2.5) | 41 (4.7) | 0.012 |
| Parainfluenza virus 1/2/3/4 | 59 (6.6) | 16 (1.8) | < 0.001 |
| Coronavirus NL63/229E/OC43/HKU1 | 23 (2.6) | 38 (4.4) | 0.042 |
| Adenovirus | 27 (3.0) | 25 (2.9) | 0.84 |
| Influenza virus A/A(H1N1)/B | 58 (6.5) | 11 (1.3) | < 0.001 |
| Human metapneumovirus A/B | 42 (4.7) | 6 (0.7) | < 0.001 |
| Enterovirus | 17 (1.9) | 20 (2.3) | 0.57 |
| Parechovirus | 1 (0.1) | 5 (0.6) | 0.12 |
GABRIEL = Global Approach for Biological Research, Infectious diseases and Epidemics in Low-Income Countries.
The proportion of single virus detection without bacterial co-detection was not different between pneumonia cases and controls (13.6% versus 11.3%, respectively, P = 0.13) (Table 1). Similarly to single virus detection regardless of the number of bacteria detected, single detection of HMPV, RSV, PIV, and IV without bacteria detection was more frequently detected in case NPS compared with control NPS (P < 0.001, P < 0.001, P = 0.01, P = 0.018, respectively), whereas CoV and BoV were more frequently detected in control NPS compared with case NPS (P = 0.0035 and P = 0.034, respectively). Rhinovirus was similarly detected in cases and control subjects NPS (P = 0.18).
Co-detections of at least two viruses without detection of bacteria are described in Table 2 and site-stratified in Supplemental Figure 2. No statistical difference between the two groups was observed (5.2% of cases [46/888] and 4.0% of controls [35/870], P = 0.25). The most frequent viral co-detections of two viruses were RV + RSV (0.9% of cases and 0.2% of controls), RV + CoV (0.3% of cases and 0.5% of controls), RV + BoV (0.0% of cases and 0.6% of controls), RV + AdV (0.3% of cases and 0.1% of controls), and RV + HMPV (0.3% of cases and 0.1% of controls). No significant difference of detection was noted between cases and controls for each couple studied except for RV + BoV which was more frequently detected in NPS of controls compared with cases (P = 0.03).
Table 2.
Viral co-detection in nasopharyngeal samples of patients without bacterial co-detection, the GABRIEL Network pneumonia study, 2010 through 2014
| Total N = 81 | Cases N = 46 | Controls N = 35 | P value | |
|---|---|---|---|---|
| Viral co-detection without bacterial co-detection, by number of virus detected, n (%) | ||||
| Two viruses | 65 (80.2) | 37 (80.4) | 28 (80.0) | 0.96 |
| Three viruses | 12 (14.8) | 6 (13.0) | 6 (17.1) | 0.61 |
| Four viruses | 4 (4.9) | 3 (6.5) | 1 (2.9) | 0.83 |
| Viral co-detection without bacterial co-detection, by virus, n (%) | ||||
| Rhinovirus | 42 (51.9) | 24 (52.2) | 18 (51.4) | 0.95 |
| Respiratory syncytial virus A/B | 29 (35.8) | 21 (45.7) | 8 (22.9) | 0.03 |
| Bocavirus | 25 (30.9) | 11 (23.9) | 14 (40.0) | 0.12 |
| Parainfluenza virus 1/2/3/4 | 14 (17.3) | 9 (19.6) | 5 (14.3) | 0.53 |
| Coronavirus NL63/229E/OC43/HKU1 | 23 (28.4) | 8 (17.4) | 15 (42.9) | 0.01 |
| Adenovirus | 13 (16.0) | 7 (15.2) | 6 (17.1) | 0.82 |
| Influenza virus A/A(H1N1)/B | 11 (13.6) | 9 (19.6) | 2 (5.7) | 0.13 |
| Human metapneumovirus A/B | 13 (16.0) | 10 (21.7) | 3 (8.6) | 0.19 |
| Enterovirus | 8 (9.9) | 3 (6.5) | 5 (14.3) | 0.43 |
| Parechovirus | 4 (4.9) | 2 (4.3) | 2 (5.7) | 0.99 |
GABRIEL = Global Approach for Biological Research, Infectious diseases and Epidemics in Low-Income Countries.
Virus–bacteria co-detections.
Virus–bacteria co-detections in NPS were observed in 59.6% of cases (529/888) and in 36.1% of controls (314/870), P < 0.001. Overall and site-stratified numbers of bacteria and viruses detected in cases and controls are shown in Table 3 and Supplemental Figure 2.
Table 3.
Virus/bacteria co-detection in nasopharyngeal samples of pneumonia cases and controls, the GABRIEL Network pneumonia study, 2010 through 2014
| Number/type of bacteria | Number of virus | Cases, n (%) N = 888 | Controls, n (%) N = 870 | P value |
|---|---|---|---|---|
| 0 | 0 | 62 (7.0) | 228 (26.2) | < 0.001 |
| 0 | 1 | 121 (13.6) | 98 (11.3) | 0.13 |
| 0 | 2 | 37 (4.2) | 28 (3.2) | 0.29 |
| 0 | 3 | 6 (0.7) | 6 (0.7) | 0.97 |
| 0 | 4 | 3 (0.3) | 1 (0.1) | 0.64 |
| One S. pneumoniae | 0 | 90 (10.1) | 132 (15.2) | 0.001 |
| One S. pneumoniae | 1 | 296 (33.3) | 130 (14.6) | < 0.001 |
| One S. pneumoniae | 2 | 84 (9.5) | 37 (4.3) | < 0.001 |
| One1 S. pneumoniae | 3 | 21 (2.4) | 8 (0.9) | 0.017 |
| One S. pneumoniae | 4 | 3 (0.3) | 4 (0.5) | 0.98 |
| One S. pneumoniae | 5 | 1 (0.1) | 1 (0.1) | 0.99 |
| One other than S. pneumoniae | 0 | 16 (1.8) | 45 (5.2) | < 0.001 |
| One other than S. pneumoniae | 1 | 24 (2.7) | 37 (4.3) | 0.078 |
| One other than S. pneumoniae | 2 | 10 (1.1) | 10 (1.1) | 0.96 |
| One other than S. pneumoniae | 3 | 2 (0.2) | 0 (0.0) | 0.51 |
| One other than S. pneumoniae | 4 | 1 (0.1) | 0 (0.0) | 0.99 |
| 2 | 0 | 25 (2.8) | 40 (4.6) | 0.047 |
| 2 | 1 | 57 (6.4) | 26 (3.0) | < 0.001 |
| 2 | 2 | 19 (2.1) | 22 (2.5) | 0.59 |
| 2 | 3 | 4 (0.5) | 2 (0.2) | 0.71 |
| 2 | 4 | 0 (0.0) | 4 (0.5) | 0.12 |
| 3 | 0 | 1 (0.1) | 3 (0.3) | 0.61 |
| 3 | 1 | 3 (0.3) | 5 (0.6) | 0.70 |
| 3 | 2 | 2 (0.2) | 3 (0.3) | 0.98 |
GABRIEL = Global Approach for Biological Research, Infectious diseases and Epidemics in Low-Income Countries.
The NP detection of S. pneumoniae without viral co-detection was most frequent in control subjects compared with cases (18.2% [90/495] versus 43.2% [132/312], P < 0.001). The detection of at least one virus with S. pneumoniae was more frequent in cases compared with controls (81.8% [405/495] versus 57.7% [180/312], respectively, P < 0.001).
In most of the virus–bacteria co-detections, a single virus was detected, without difference between cases and controls (72.1% of pneumonia cases versus 68.5% in controls with virus–bacteria co-detection, P = 0.28). In all, when virus–bacteria co-detection was observed, the median number of viruses detected was not significantly higher in cases compared with controls (median number of viruses detected: 1, IQR [1–1] in cases (mean ± SD: 1.08 ± 0.78) and 1, IQR [0–1] in controls (mean ± SD: 0.80 ± 0.89), P = 0.073).
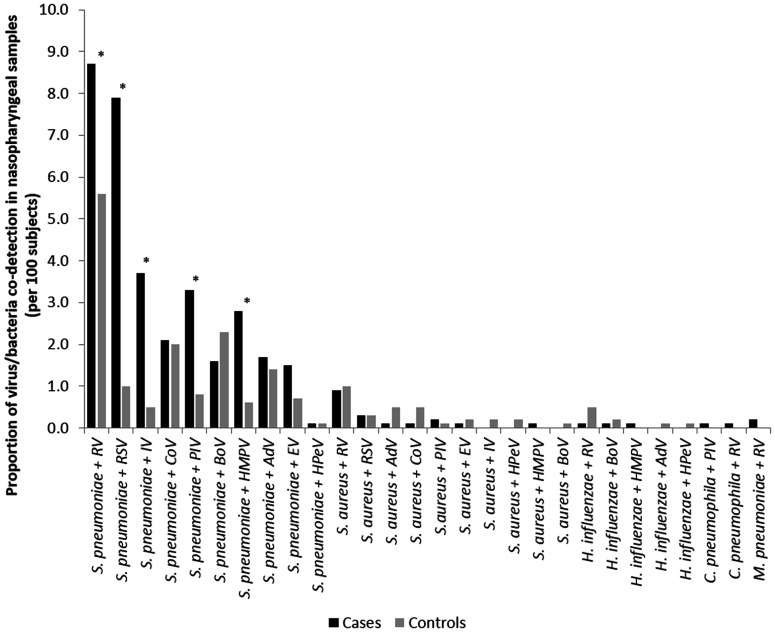
When virus–bacteria co-detection was found, regardless of the number of virus and bacteria detected, three viruses (RSV, IV, and HMPV) were more frequently found in pneumonia cases compared with controls (P < 0.001 for all of the comparisons). Conversely, four viruses (RV, BoV, AdV, and CoV) were more frequently observed in controls compared with pneumonia cases (P = 0.007, P < 0.001, P = 0.018, P = 0.042, respectively) (Supplemental Table 5). The most frequent co-detections of 1 virus and 1 bacterium in NPS are described in Figure 1.
Figure 1.
Proportion of virus-bacteria co-detection in nasopharyngeal samples of patients, the Global Approach for Biological Research, Infectious diseases and Epidemics in Low-Income Countries (GABRIEL) Network pneumonia study, 2010 through 2014. * P < 0.05, AdV = adenovirus, BoV = bocavirus, CoV = coronavirus NL63/229E/OC43/HKU1, EV = enterovirus, HPeV = parechovirus, HMPV = human metapneumovirus A/B, IV = influenza virus A/A(H1N1)/B, PIV = parainfluenza virus 1/2/3/4RV = rhinovirus, RSV = respiratory syncytial virus A/B.
Association of single virus detection and virus–bacteria co-detection with pneumonia.
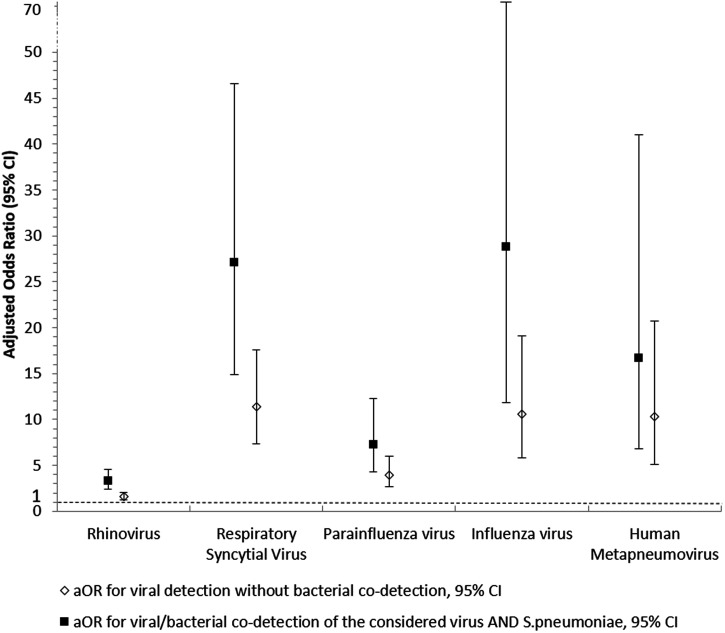
Overall and site-stratified associations between major microorganism detection or co-detection and pneumonia are described in Figure 2 and Supplemental Table 6. For the major viruses (RV, RSV, PIV, IV, and HMPV), adjusted odds ratios (aOR) of pneumonia ranged from 1.57 95% CI [1.26–2.01] for RV to 11.37 95% CI [7.37–17.55] for HMPV. The aOR of pneumonia steadily increased when co-detection between S. pneumoniae and a major virus was found (aOR ranged between 3.31 95% CI [2.38–4.60] for RV and 28.85 95% CI [11.83–70.38] for IV). No interaction effect was found between the microorganisms studied for each co-detection.
Figure 2.
Adjusted odds ratio of the risk of pneumonia according to the microorganisms detected in nasopharyngeal samples, the Global Approach for Biological Research, Infectious diseases and Epidemics in Low-Income Countries (GABRIEL) Network, 2010 through 2014. aOR: adjusted odds ratio. The white diamonds represent adjusted odds ratio of pneumonia for a viral detection, with 95% CI. The black squares represent adjusted odds ratio of pneumonia for a co-detection of Streptococcus pneumoniae + virus, with 95% CI. All aOR are adjusted for gender, age, other microorganisms detected, and time period (year), with study center as the random effect.
Association of microorganism detection and hypoxemic pneumonia.
Only pneumonia cases with oxygen saturation data were included in this analysis; data from the Cambodian site were excluded in this subanalysis since no hypoxemic pneumonia was recorded. In all, of the 712 pneumonia cases, 647 (90.9%) had data for oxygen saturation. Of 647 patients, 88 had hypoxemic pneumonia (13.6%). By study site, the proportion of included patients with hypoxemic pneumonia ranged from 3.7% in Mongolia to 30.5% in Mali. The microorganisms detected in the NPS of patients with hypoxemic pneumonia are described in Table 4 and Supplemental Table 7. Detection of HMPV and co-detection of S. pneumoniae and RSV were significantly more frequent in hypoxemic pneumonia than in nonhypoxemic pneumonia (P = 0.05 and P = 0.014, respectively). However, after adjustment for age, gender, and time period, no virus/bacteria co-detection was found to be associated with hypoxemic pneumonia.
Table 4.
Microorganisms detected in nasopharyngeal samples of patients with hypoxemic pneumonia, the GABRIEL Network, 2010 through 2014
| Cases with hypoxemic pneumonia (N = 88) | Cases with nonhypoxemic pneumonia (N = 559) | P value | |
|---|---|---|---|
| Detection of at least one bacterium | 62 (70.5) | 400 (71.6) | 0.832 |
| Detection of at least one virus | 67 (76.1) | 433 (77.5) | 0.783 |
| Detection of at least the following microorganisms | |||
| S. pneumoniae | 56 (63.6) | 368 (65.8) | 0.687 |
| S. aureus | 13 (14.8) | 72 (12.9) | 0.625 |
| H. influenzae | 6 (6.8) | 30 (5.4) | 0.581 |
| M. pneumoniae | 0 (0.0) | 7 (1.3) | 0.602 |
| C. pneumophila | 1 (1.1) | 2 (0.4) | 0.356 |
| Rhinovirus | 23 (26.1) | 136 (24.3) | 0.714 |
| Respiratory syncytial virus A/B | 22 (25.0) | 101 (18.1) | 0.123 |
| Bocavirus | 5 (5.7) | 42 (7.5) | 0.662 |
| Parainfluenza virus 1/2/3/4 | 10 (11.4) | 71 (12.7) | 0.725 |
| Coronavirus NL63/229E/OC43/HKU1 | 6 (6.8) | 44 (7.9) | 0.99 |
| Adenovirus | 4 (4.6) | 44 (7.9) | 0.380 |
| Influenza virus A/A(H1N1)/B | 7 (8.0) | 63 (11.3) | 0.352 |
| Human metapneumovirus A/B | 11 (12.5) | 37 (6.6) | 0.05 |
| Enterovirus | 5 (5.7) | 29 (5.2) | 0.798 |
| Parechovirus | 1 (1.1) | 16 (2.9) | 0.492 |
| Virus/bacteria co-detection | |||
| S. pneumoniae + Rhinovirus | 10 (11.4) | 44 (7.9) | 0.27 |
| S. pneumoniae + Respiratory syncytial virus A/B | 10 (11.4) | 27 (4.8) | 0.014 |
| S. pneumoniae + Parainfluenza virus 1/2/3/4 | 3 (3.4) | 19 (3.4) | 0.99 |
| S. pneumoniae + Influenza virus A/A(H1N1)/B | 2 (2.3) | 25 (4.5) | 0.56 |
| S. pneumoniae + Human metapneumovirus A/B | 1 (1.1) | 12 (2.2) | 0.99 |
GABRIEL = Global Approach for Biological Research, Infectious diseases and Epidemics in Low-Income Countries.
DISCUSSION
This large, multicenter study in eight low- and middle-income countries provides important information about the microorganisms detected in the NPS of children under 5 with pneumonia and in control subjects. Overall, S. pneumoniae, RSV, PIV, HMPV, and IV were more frequently detected in the NPS of pneumonia cases compared with controls. Microbial colonization of the respiratory tract with S. pneumoniae, H. influenzae, and M. catarrhalis has been shown elsewhere to be associated with severe pulmonary infections. 12, 20 The recent results of the PERCH study also showed that viruses, especially RSV, PIV, HMPV, and IV were predominant causes of pneumonia requiring hospitalization in children under 5 in low- and middle-income countries. 2
At least one bacterium was detected in 60.3% and one virus in 50.1% of the control subjects in our study; and different microorganisms (S. aureus, BoV, and CoV) were more frequently detected in control subjects compared with cases. It is well known that numerous microorganisms naturally colonize the respiratory tract. 8 This large number explains why the etiology of pneumonia is challenging to determine. Collecting cultures of the lower respiratory tract of the young child is difficult, and neither clinical signs, nor radiologic features, nor biological markers make the differentiation between a viral or bacterial etiology of pneumonia possible. 21
The results highlighted that NP colonization with S. pneumoniae as a single microorganism was more frequent in control subjects compared with cases (15.2% versus 10.1%, P = 0.001). It showed also a high frequency of co-detections of S. pneumoniae and RV, RSV, PIV, IV, or HMPV in NPS of pneumonia cases and an association of the co-detection with pneumonia, compared with the microorganisms without co-detection. Today, improvement in our knowledge of interactions between commensal and pathogenic microorganisms in the lung microbiome has revealed that viruses and bacteria are probably not mutually exclusive in the pathogenesis of pneumonia. 22 Although microbial coinfections were once considered as rare, studies have shown that they are frequent in children with pneumonia. 23 For instance, a study carried out in 1999 showed viral and bacterial coinfection of microorganisms in 23% of 154 children with pneumonia. 7 An association between the severity of H1N1 influenza and S. pneumoniae detection has been described in the literature. 24, 25 Also, an influenza infection could trigger secondary pneumococcal pneumonia in a subject previously colonized by S. pneumoniae. 26 Some studies have also reported that the coinfection of RSV and bacteria in lower tract respiratory infections appears to increase the severity and mortality compared with RSV without coinfection. 27, 28 Our results tend to confirm the assumption since the co-detection of S. pneumoniae + RSV was associated with hypoxemic pneumonia, even if the trends were not significant after adjustment of covariables. The literature also shows an increase of nasopharyngeal pneumococcal density by viral coinfection; however, these data were not studied in our work. 11, 29, 30 Our results also highlighted that RV was frequently detected in control subjects, particularly, when a single virus was detected. However, the role of RV as a true etiology of pneumonia remains unclear. It suggests that RV contributes more frequently to pneumonia in patients with a NP co-detection of S. pneumoniae. 31
An increased severity of pneumonia has not been demonstrated in viral coinfections compared with single viral infections. 32 Moreover, AdV, BoV, CoV, EV, and HPeV were mostly co-detected with other viruses and/or bacteria in pneumonia cases. They were also frequently detected in controls. This confirms data from other studies and suggests an uncertain pathogenic role of these viruses in the disease. 33 – 36 However, viral loads were not measured, and it is known that for viruses such as BoV, high viral loads are potentially associated with respiratory symptoms and low viral loads indicate asymptomatic shedding. 37 This raises the question of the threshold to use to ensure relevance of the test for clinical interpretation. 38
This study has several limitations. First, it remains unclear if a detection of specific pathogens in the nasopharynx is a useful proxy indicator of the same pathogens in the lower respiratory tree (i.e., a cause of pneumonia). Moreover, no classical analytic method can take sensitivity, specificity for pathogen detection and the detection of multiple pathogens into account. The development of relevant modeling techniques provides better understanding of the results. For example, to more accurately determine pneumonia etiologies, the PERCH Study Group created a partial latent class analysis with a Bayesian approach. 39 Nevertheless, assumptions of the model do not make it possible to conclude on the etiology of pneumonia when more than one pathogen is detected. 40 Secondly, the study was adjusted to major covariables, namely gender, age group, study site, and other microorganisms. However, other possible confounding factors such as previous antibiotic intake were not introduced in the models owing to a great deal of missing data, or a low frequency of the variable in the control group (i.e., antibiotic intake). The clinical data on severity were of uncertain quality owing in large part to subjective clinical signs (i.e., chest indrawing, cyanosis). In addition, severity was assessed using an objective variable: oxygen saturation.
The strengths of the study are its large sample size and its multicenter design. The inclusion of pneumonia cases and control subjects enabled the adjustment of positivity rates and odds ratios calculation.
Our findings report extensive data on the viral and bacterial co-detection in pneumonia in children under 5 years of age from low- and middle-income countries. The results suggest that coinfection of S. pneumoniae and certain viruses (RV, RSV, PIV, HMPV, and IV) may play a role in the pathophysiology of pneumonia in children. It suggests also a minor role of other viruses such as AdV, BoV, CoV, EV, and HPeV, even in the case of a coinfection with a bacterium. Nevertheless, further studies focused on the major coinfections of microorganisms are required to improve the understanding of their role in the pathogenesis of pneumonia. This could lead to new putative targets for therapeutic interventions.
Supplemental Material
ACKNOWLEDGMENTS
We acknowledge the Fondation Mérieux for its support and Thomas Bénet, Florence Ader, René Écochard, Étienne Javouhey and Gérard Lina for their precious advice. We also thank Peter Tucker and Michelle Grange for editing the manuscript.
Note: Supplemental tables and figures appear at www.ajtmh.org.
REFERENCES
- 1. GBD 2019 under-5 Mortality Collaborators , 2021. Global, regional, and national progress towards Sustainable Development Goal 3.2 for neonatal and child health: all-cause and cause-specific mortality findings from the Global Burden of Disease Study 2019. Lancet 398: 870–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pneumonia Etiology Research for Child Health (PERCH) Study Group , 2019. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet 394: 757–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Principi N Esposito S , 2011. Management of severe community-acquired pneumonia of children in developing and developed countries. Thorax 66: 815–822. [DOI] [PubMed] [Google Scholar]
- 4. Ruuskanen O Lahti E Jennings LC Murdoch DR , 2011. Viral pneumonia. Lancet 377: 1264–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bosch AATM Biesbroek G Trzcinski K Sanders EAM Bogaert D , 2013. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog 9: e1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. King PT Buttery J , 2018. Emerging role of viral and bacterial co-infection in early childhood. Respirology 23: 128–129. [DOI] [PubMed] [Google Scholar]
- 7. Michelow IC Olsen K Lozano J Rollins NK Duffy LB Ziegler T Kauppila J Leinonen M McCracken GH Jr , 2004. Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children. Pediatrics 113: 701–707. [DOI] [PubMed] [Google Scholar]
- 8. Kwambana BA Barer MR Bottomley C Adegbola RA Antonio M , 2011. Early acquisition and high nasopharyngeal co-colonisation by Streptococcus pneumoniae and three respiratory pathogens amongst Gambian new-borns and infants. BMC Infect Dis 11: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bakaletz LO , 2017. Viral-bacterial co-infections in the respiratory tract. Curr Opin Microbiol 35: 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCullers JA , 2014. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol 12: 252–262. [DOI] [PubMed] [Google Scholar]
- 11. Wolter N et al. 2014. High nasopharyngeal pneumococcal density, increased by viral coinfection, is associated with invasive pneumococcal pneumonia. J Infect Dis 210: 1649–1657. [DOI] [PubMed] [Google Scholar]
- 12. Vissing NH Chawes BLK Bisgaard H , 2013. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am J Respir Crit Care Med 188: 1246–1252. [DOI] [PubMed] [Google Scholar]
- 13. Madhi SA Klugman KP , Vaccine Trialist Group , 2004. A role for Streptococcus pneumoniae in virus-associated pneumonia. Nat Med 10: 811–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Komurian-Pradel F et al. 2013. Enhancing research capacities in infectious diseases: the GABRIEL network, a joint approach to major local health issues in developing countries. Clin Epidemiol Glob Health 1: 40–43. [Google Scholar]
- 15. Picot VS et al. 2014. Multicenter case-control study protocol of pneumonia etiology in children: Global Approach to Biological Research, Infectious diseases and Epidemics in Low-income countries (GABRIEL network). BMC Infect Dis 14: 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bénet T et al. 2017. Microorganisms associated with pneumonia in children <5 years of age in developing and emerging countries: the GABRIEL pneumonia multicenter, prospective, case-control study. Clin Infect Dis 65: 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. World Health Organization , 2013. Pocket Book of Hospital Care for Children: Guidelines for the Management of Common Illnesses with Limited Resources, 2nd edition. Geneva, Switzerland: WHO. Available at: https://apps.who.int/iris/bitstream/handle/10665/81170/9789241548373_eng.pdf?sequence=1&isAllowed=y. Accessed August 31, 2021.
- 18. Anderson TP Werno AM Barratt K Mahagamasekera P Murdoch DR Jennings LC , 2013. Comparison of four multiplex PCR assays for the detection of viral pathogens in respiratory specimens. J Virol Methods 191: 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dananché C et al. 2020. Serotypes of Streptococcus pneumoniae in children aged <5 years hospitalized with or without pneumonia in developing and emerging countries: a descriptive, multicenter study. Clin Infect Dis 70: 875–883. [DOI] [PubMed] [Google Scholar]
- 20. Ege M von Mutius E , 2013. Microbial airway colonization: a cause of asthma and pneumonia? Am J Respir Crit Care Med 188: 1188–1189. [DOI] [PubMed] [Google Scholar]
- 21. Haq IJ Battersby AC Eastham K McKean M , 2017. Community acquired pneumonia in children. BMJ 356: j686. [DOI] [PubMed] [Google Scholar]
- 22. Almand EA Moore MD Jaykus LA , 2017. Virus-bacteria interactions: an emerging topic in human infection. Viruses 9: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cilla G Oñate E Perez-Yarza EG Montes M Vicente D Perez-Trallero E , 2008. Viruses in community-acquired pneumonia in children aged less than 3 years old: high rate of viral coinfection. J Med Virol 80: 1843–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Palacios G et al. 2009. Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PLOS ONE 4: e8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Centers for Disease Control and Prevention , 2009. Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1)-United States, May–August 2009. MMWR Morb Mortal Wkly Rep 58: 1071–1074. [PubMed] [Google Scholar]
- 26. Sender V Hentrich K Henriques-Normark B , 2021. Virus-induced changes of the respiratory tract environment promote secondary infections with Streptococcus pneumoniae. Front Cell Infect Microbiol 11: 643326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brealey JC Chappell KJ Galbraith S Fantino E Gaydon J Tozer S Young PR Holt PG Sly PD , 2018. Streptococcus pneumoniae colonization of the nasopharynx is associated with increased severity during respiratory syncytial virus infection in young children. Respirology 23: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leung TF et al. 2014. Epidemiology and risk factors for severe respiratory syncytial virus infections requiring pediatric intensive care admission in Hong Kong children. Infection 42: 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan RR et al. 2016. Nasopharyngeal pneumococcal density and evolution of acute respiratory illnesses in young children, Peru, 2009– 2011. Emerg Infect Dis 22: 1996–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vu HT Yoshida LM Suzuki M Nguyen HA Nguyen CD Nguyen AT Oishi K Yamamoto T Watanabe K Vu TD , 2011. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J 30: 11–18. [DOI] [PubMed] [Google Scholar]
- 31. Hartiala M Lahti E Forsström V Vuorinen T Ruuskanen O Peltola V , 2019. Characteristics of hospitalized rhinovirus-associated community-acquired pneumonia in children, Finland, 2003–2014. Front Med (Lausanne) 6: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scotta MC et al. 2016. Respiratory viral coinfection and disease severity in children: a systematic review and meta-analysis. J Clin Virol 80: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Crom SCM Rossen JWA van Furth AM Obihara CC , 2016. Enterovirus and parechovirus infection in children: a brief overview. Eur J Pediatr 175: 1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chiang GPK et al. 2017. Clinical features and seasonality of parechovirus infection in an Asian subtropical city, Hong Kong. PLOS ONE 12: e0184533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Calvo C García-García ML Pozo F Carballo D Martínez-Monteserín E Casas I , 2016. Infections and coinfections by respiratory human bocavirus during eight seasons in hospitalized children. J Med Virol 88: 2052–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nascimento-Carvalho AC Vilas-Boas AL Fontoura MSH Vuorinen T Nascimento-Carvalho CM , PNEUMOPAC-Efficacy Study Group , 2018. Respiratory viruses among children with non-severe community-acquired pneumonia: a prospective cohort study. J Clin Virol 105: 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Allander T et al. 2007. Human bocavirus and acute wheezing in children. Clin Infect Dis 44: 904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jansen RR Wieringa J Koekkoek SM Visser CE Pajkrt D Molenkamp R de Jong MD Schinkel J , 2011. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol 49: 2631–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hammitt LL Feikin DR Scott JAG Zeger SL Murdoch DR O’Brien KL Deloria Knoll M , 2017. Addressing the analytic challenges of cross-sectional pediatric pneumonia etiology data. Clin Infect Dis 64 (Suppl 3): S197–S204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deloria Knoll M et al. 2017. Bayesian estimation of pneumonia etiology: epidemiologic considerations and applications to the pneumonia etiology research for child health study. Clin Infect Dis 64 (Suppl 3): S213–S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.