Abstract
Macrophages regulate protective immune responses to infectious microbes, but aberrant macrophage activation frequently drives pathological inflammation. To identify regulators of vigorous macrophage activation, we analyzed RNA-seq data from synovial macrophages and identified SLAMF7 as a receptor associated with a super-activated macrophage state in rheumatoid arthritis. We implicated IFN-γ as a key regulator of SLAMF7 expression. Engaging SLAMF7 drove an exuberant wave of inflammatory cytokine expression. Induction of TNF-α following SLAMF7 engagement amplified inflammation through an autocrine signaling loop. We observed SLAMF7-induced gene programs not only in macrophages from rheumatoid arthritis patients, but in gut macrophages from active Crohn’s disease patients and lung macrophages from severe COVID-19 patients. This suggests a central role for SLAMF7 in macrophage super-activation with broad implications in pathology.
One Sentence Summary:
SLAMF7 is upregulated on macrophages in inflamed tissues and drives super-activation in inflammatory human disease.
Introduction
Macrophages are necessary for protection against infectious microbes (1) but can drive acute inflammation that can become exuberant or chronic and cause significant tissue pathology (2). Dysfunctional macrophage activation is evident in autoimmune diseases including rheumatoid arthritis (RA) (3–5), inflammatory bowel disease (IBD) (6–8) and interstitial lung disease (9, 10). RA is characterized by infiltration of macrophages into the synovium, along with populations of lymphocytes and activated stromal cells (11–15). Macrophage numbers and activation states change and can correlate with response to therapy in RA (16–18). In respiratory infection, macrophages promote inflammation resulting in lung injury (19) and contribute to immune activation in severe acute respiratory disease syndrome (20, 21) associated with coronavirus disease of 2019 (COVID-19) (22, 23).
Macrophage activation states are determined by receptors for an array of environmental signals (24), with cytokines and microbial molecules as the best-known macrophage regulators (25–27). Interferon (IFN)-γ is a key component of classical M1 macrophage activation (27) and potentiates macrophage responses to subsequent stimulation (28–30). Toll-like receptor (TLR) agonists prime macrophages to express inflammasome components that, when activated, result in pyroptotic cell death and release of bioactive interleukin (IL)-1β (31).
Here, we find that an important component of the macrophage response to a primary signal is upregulation of a secondary super-activator receptor that can then transform these primed or potentiated macrophages into a highly activated, potentially pathogenic inflammatory state. As a strategy to find key regulators of super-activated macrophages (SAMs), we evaluated inflammatory human diseases where macrophages are implicated as major drivers of inflammation. Using this approach, we identified SLAMF7 (also known as CD319, CRACC, and CS1) (32–34), and implicate this receptor as having a central role in highly activated macrophage-related inflammatory diseases. We determined that SLAMF7 is selectively expressed by macrophages from sites of inflammation and is strongly regulated. by IFN-γ. Engagement of the receptor drove a strong inflammatory signature, activating NF-κB and MAPK pathways, along with further autocrine amplification by TNF-α. In publicly available single cell RNA-seq data, we identified this SLAMF7 super-activated macrophage (SLAMF7-SAM) population in diverse tissues from patients with rheumatoid arthritis, but also inflammatory bowel disease and COVID-19 pneumonia. This implicates SLAMF7 activation of inflammatory macrophages as a key pathway driving pathology in acute and chronic inflammatory human diseases.
Results
Marked upregulation of SLAMF7 on macrophages from inflamed synovial tissue
We focused on the inflammatory human disease RA to identify signaling receptors that could act as macrophage super-activators. We analyzed bulk RNA-sequencing (RNA-seq) data from phase 1 of the Accelerating Medicines Partnership® (AMP®) Rheumatoid Arthritis / Systemic Lupus Erythematosus Network (15) to examine gene expression on sorted CD14+ synovial tissue macrophages from patients with inflammatory RA (n=11) and relatively non-inflammatory osteoarthritis (OA, n=10). Using DESeq2, we identified 509 differentially expressed genes (log2foldchange (LFC) ≥ 1, Wald adjusted p value (padj) ≤ 0.05) that were upregulated in RA and that we defined as an “Inflamed RA Macrophage Signature” (Data file S1). This signature contained IFN-inducible genes such as GBP1, IFI6, and CD40, as well as inflammatory cytokines and chemokines including TNF, CCL3, and CXCL8, suggesting that both IFN-induced and inflammatory transcriptional programs define the dominant macrophage state in RA. Strikingly, the single most significantly upregulated gene in the “Inflamed RA Macrophage Signature” was SLAMF7 (LFC=3.82, padj=2.23E-21, Fig. 1A), a receptor that regulates leukocyte activation through homotypic interactions with SLAMF7 on other cells (32–34). To confirm this finding, we disaggregated synovial tissue from an independent cohort of individuals with OA (n=8, 87.5% female, mean age 67.1, range 58–85) and RA (n=9, 100% female, mean age 62.1, range 28–86) to quantify SLAMF7 protein expression by flow cytometry (gating in Fig. S1A–D). SLAMF7 was present at very low levels on synovial macrophages from patients with OA but was forty times higher on macrophages from patients with RA (Fig. 1B, Mann-Whitney p = 0.0016). We detected SLAMF7 on up to 55 percent of macrophages (mean 26.4%, 95% confidence interval (CI) 13.3–39.5) from patients with RA compared to less than 6 percent of macrophages (mean 2.4, 95% CI 0.35–4.5) from patients with OA (Fig. 1C). We were also able to obtain discarded synovial fluid from de-identified patients with RA and OA. SLAMF7 levels were twice as high on synovial fluid macrophages from patients with RA compared to OA (Fig. 1D) and on up to 48 percent of macrophages (mean 24.3, 95% CI 17.03–31.56) from patients with RA, compared to less than 25 percent of macrophages (mean 13.3, 95% CI 6.1–20.5) from patients with OA (Fig. 1E). In some patients with RA, only a subset of macrophages expressed SLAMF7, while in others the majority of cells expressed this receptor (Fig. S1E). In contrast, we did not observe major differences in levels of another macrophage-expressed SLAM family member, CD84 (SLAMF5) (35) on macrophages from synovial tissue (Fig. S1F, S2A–B) or synovial fluid (Fig. S2C–D) from patients with RA versus OA. This associates SLAMF7 as a receptor that is selectively expressed by inflammatory macrophages in RA.
Figure 1. Marked upregulation of SLAMF7 on macrophages from inflamed synovial tissue.
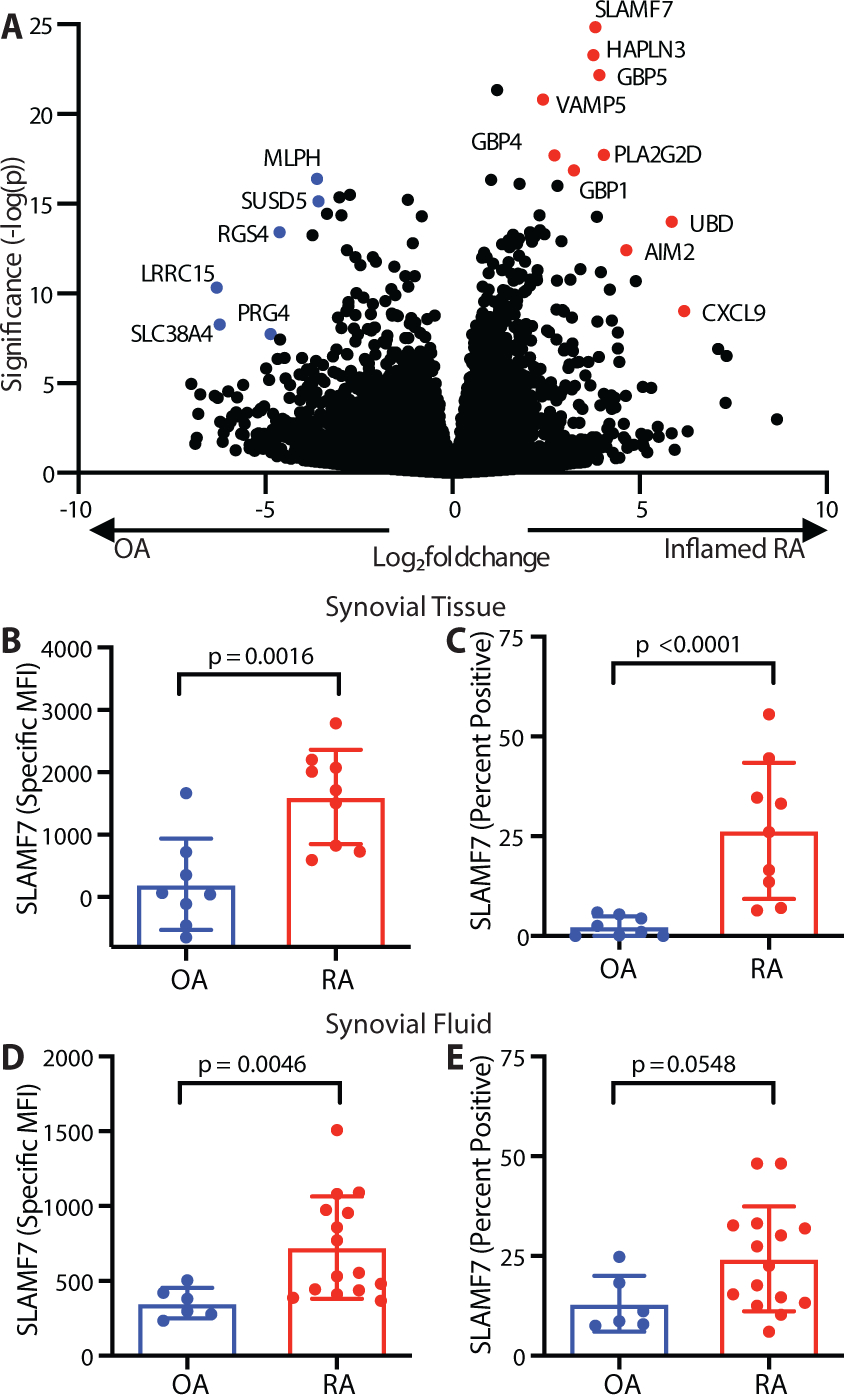
A) Differential gene expression in bulk RNA-seq of synovial tissue macrophages from patients with inflamed RA (n=11) compared to OA (n=10) (15). B) Specific MFI for SLAMF7 and C) percent of macrophages expressing SLAMF7 in synovial tissue from patients with OA (n=8) or RA (n=9). D) Specific MFI for SLAMF7 and E) percent of macrophages expressing SLAMF7 in synovial fluid from patients with OA (n=6) or RA (n=15). Data represent mean ± SD. The Mann-Whitney test was used for statistical comparisons. OA, osteoarthritis; RA, rheumatoid arthritis.
IFN-γ is a dominant driver of macrophage SLAMF7 expression
In the unstimulated state, SLAMF7 is expressed at high levels by plasma cells (36), and can also be expressed on B cells, T cells, and natural killer (NK) cells, but is expressed at low levels on resting macrophages (35, 37). However, elevated SLAMF7 expression has been found on macrophages from atherosclerotic lesions (38) and from patients with myelofibrosis (39). BLIMP-1 regulates SLAMF7 expression in lymphocytes (40), but it is not well understood how this receptor is regulated in macrophages. To address this gap in knowledge, we used RNA-seq to define the transcriptional activation state of macrophages with high levels of SLAMF7 by sorting CD14+ and CD16+ myeloid populations with high and low expression of SLAMF7 (gating in fig. S3) from peripheral blood of healthy controls (table S1: n = 5; 80% female; mean age, 59.6; range, 45 to 74) or patients with RA (n = 7; 57.1% female; mean age, 53.7; range, 37 to 67), as well as synovial fluid from patients with RA (n = 4; 50% female; mean age, 53.5; range, 42 to 67). We then examined genes that were differentially expressed in sorted cells with high versus low expression of SLAMF7 and identified 21 genes that were commonly upregulated (LFC ≥ 1, padj ≤ 0.05) in CD14+ populations of cells with high SLAMF7 expression from both blood and synovial fluid that we defined as a “SLAMF7-High Macrophage Signature” (Data file S2). In addition to SLAMF7, IFN-inducible genes CXCL9, CD40 and IDO1 were upregulated in SLAMF7-high CD14+ cells from blood (Fig. 2A) and synovial fluid (Fig. S4A), with downregulation of genes such as SELL and SERPINB2 (Fig. 2A, S4A). Gene set enrichment analysis revealed that high SLAMF7 expression was significantly associated (padj <0.05) with msigdb Hallmark gene sets for type I and type II IFN response (Fig. 2B), suggesting a prominent in vivo role for IFN in regulation of this receptor on macrophages. Oxidative phosphorylation pathways, which have been associated with anti-inflammatory macrophages rather than the glycolytic M1 state (41), were enriched in cells with high SLAMF7. TNF signaling was decreased in monocytes with high SLAMF7 (Fig. 2B). Indeed, the transcription factor EGR1, which can suppress inflammatory gene expression (42), was expressed at significantly lower levels (LFC −1.53, padj = 0.0017) in synovial fluid cells with high SLAMF7, suggesting that these cells are in a poised or potentiated but not yet fully activated state.
Figure 2. SLAMF7 is a key feature of IFN-γ potentiated macrophages.
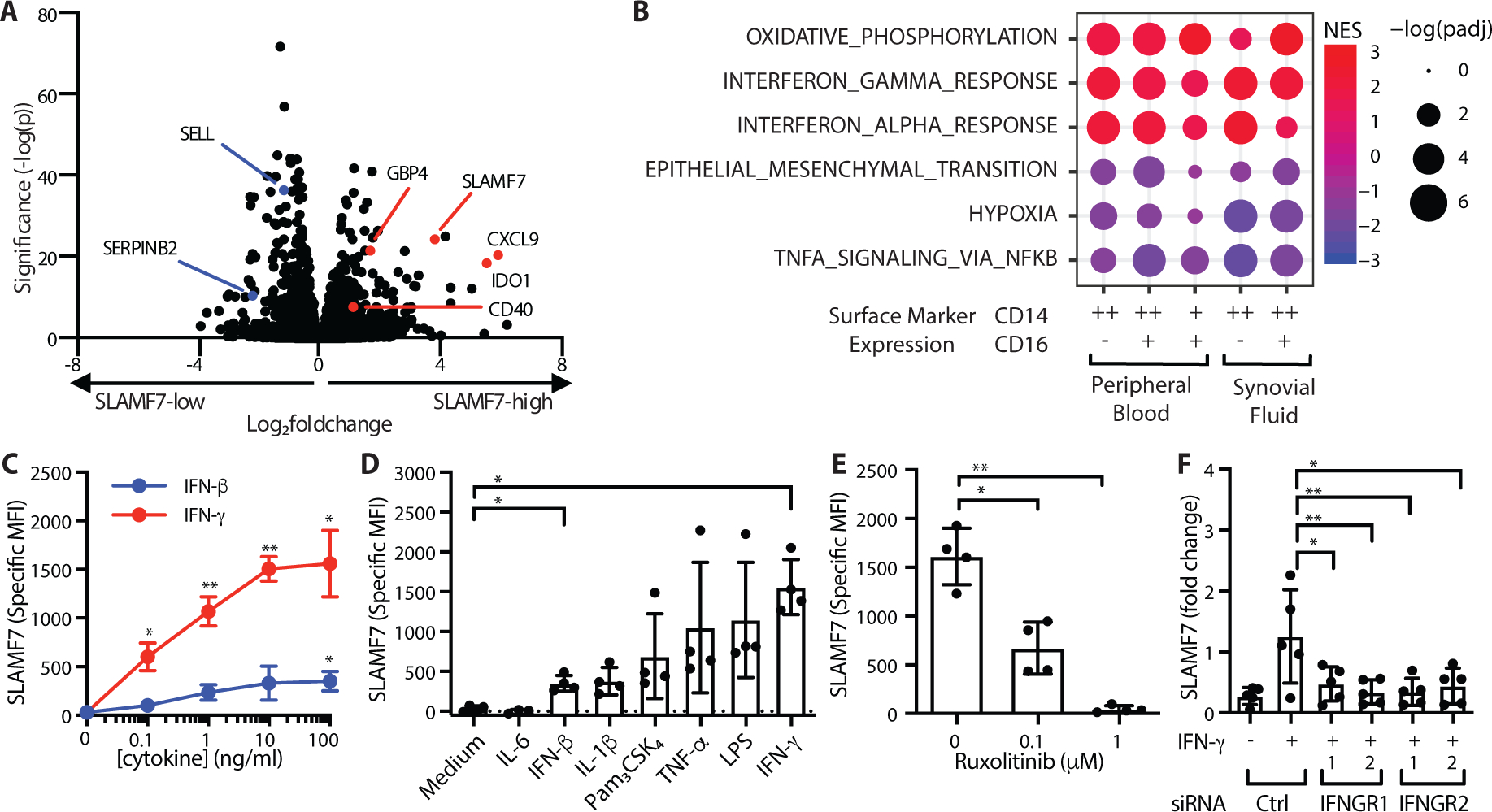
A) Differential gene expression in SLAMF7-high versus SLAMF7-low CD14+CD16- cells from peripheral blood (n=12). B) Gene set enrichment analysis for Hallmark Pathways in SLAMF7-high compared to SLAMF7-low myeloid populations from synovial fluid and peripheral blood. C) Specific MFI for SLAMF7 on macrophages incubated with different doses of IFN-γ or IFN-β. D) Specific MFI for SLAMF7 on macrophages incubated with 100 ng/ml of cytokines or TLR agonists. IFN-β and IFN-γ results are the same as the 100 ng/ml dose in panel C. E) Macrophages were incubated with ruxolitinib or DMSO prior to IFN-γ treatment (10 ng/ml), and the specific MFI for SLAMF7 was measured after 16h. Data in C-E represent mean ± SD of 4 donors. F) Macrophages were treated with a siRNA control or siRNA targeting IFNGR1 or IFNGR2, then potentiated with IFN-γ (5–10 ng/ml) for 24h. SLAMF7 was quantified relative to macrophages treated with control siRNA only. Data represent mean ± SD of 5 donors. Statistics were calculated using the one-way ANOVA with Dunnett’s multiple comparisons test. NES, normalized expression score; Ctrl, control; *, p ≤ 0.05; ** p ≤ 0.01.
We then performed in vitro cytokine stimulation on monocyte-derived macrophages from peripheral blood to determine how SLAMF7 is regulated on macrophages. Treatment of macrophages with IFN-γ resulted in high expression of SLAMF7 (Fig. 2C). Cytokines IFN-β, IL-1β and TNF-α, as well as TLR agonists Pam3CSK4 and lipopolysaccharide (LPS), also increased expression of SLAMF7 on macrophages but to lower levels than IFN-γ, while IL-6 did not affect SLAMF7 expression (Fig. 2D). In contrast, CD84 levels decreased by up to fifty percent after stimulation with IFN-β, IL-1β, TNF-α, Pam3CSK4, LPS and IFN-γ (Fig. S4B–C), while CD45 levels were relatively unchanged (Fig. S4D–E). JAK1 and JAK2 are known to transduce IFN-γ signaling (43). Inhibitors of JAK1 and JAK2 such as ruxolitinib effectively disrupt this pathway and are used clinically to treat patients with myelofibrosis (44) and graft versus host disease (45). We observed profound inhibition of IFN-γ induced SLAMF7 expression in macrophages treated with ruxolitinib in vitro (Fig. 2E), confirming the importance of this pathway and revealing a potential method to disrupt SLAMF7 upregulation. Conversely, there was a doubling in CD84 expression in macrophages treated with ruxolitinib (Fig. S4F), suggesting reciprocal regulation of these two SLAM family members by IFN-γ. CD45 expression was unchanged by ruxolitinib treatment (Fig. S4G). Next, we used siRNA to silence IFN-γ receptors, IFNGR1 (Fig. S4H) and IFNGR2 (Fig. S4I), resulting in decreased SLAMF7 levels (Fig. 2F) and confirming that IFN-γ is a key regulator of SLAMF7 expression in macrophages.
Engagement of SLAMF7 triggers an inflammatory cascade
Next, we asked if the increased expression of SLAMF7 on macrophages from inflamed tissues might play a role in their activation. SLAMF7-high macrophages from blood and synovial fluid expressed higher levels of IFN-induced genes but not the inflammatory genes from the “Inflamed RA Macrophage Signature.” Based on the striking upregulation of SLAMF7 after primary IFN-γ stimulation, we hypothesized that SLAMF7 engagement might provide a special signal to control activation of macrophages primed to upregulate its expression. SLAMF7 has previously been reported to inhibit activation of monocytes (46, 47), and we wondered whether this receptor might serve to downregulate macrophage activation. However, silencing SLAMF7 with siRNA did not result in increased TNF production following IFN-γ treatment (Fig. 3A, S5A), despite a reduction in SLAMF7 expression (Fig. 3B, S5B). Thus, we evaluated the hypothesis that SLAMF7 may instead serve to activate IFN-γ potentiated macrophages. Recombinant SLAMF7 protein (r-SLAMF7) has been reported to drive proliferation of myeloma cells (48), so we tested the response of macrophages using this recombinant protein as a soluble ligand. First, we pretreated macrophages with IFN-γ to cause SLAMF7 upregulation and second, we added r-SLAMF7 protein to engage the receptor. Macrophages exhibited a remarkable eighty-fold increase in TNF RNA (Fig. 3C) and thirty-fold increase in TNF-α protein (Fig. S5C) that was significantly reduced after siRNA silencing of SLAMF7 (Fig. 3C, S5C). We confirmed that endotoxin levels in this protein were less than 1 EU/ml (Fig. S6A) and induced secretion of 10–20 times more TNF-α than other (control) protein preparations from the same company (Fig. S6B), including after endotoxin depletion (Fig.S6C–D). We also evaluated endotoxin levels in the 162.1 SLAMF7 activating monoclonal antibody (mAb) (32), and did not detect endotoxin levels within the range of the assay for this antibody and other antibodies from the same company (<0.1 EU/ml, Fig. S6E). Monocytes treated with this activating antibody produced much higher levels of TNF-α than control antibodies (Fig. S6F), suggesting that engagement of this receptor can drive striking macrophage activation.
Figure 3. Engagement of SLAMF7 triggers an inflammatory program.
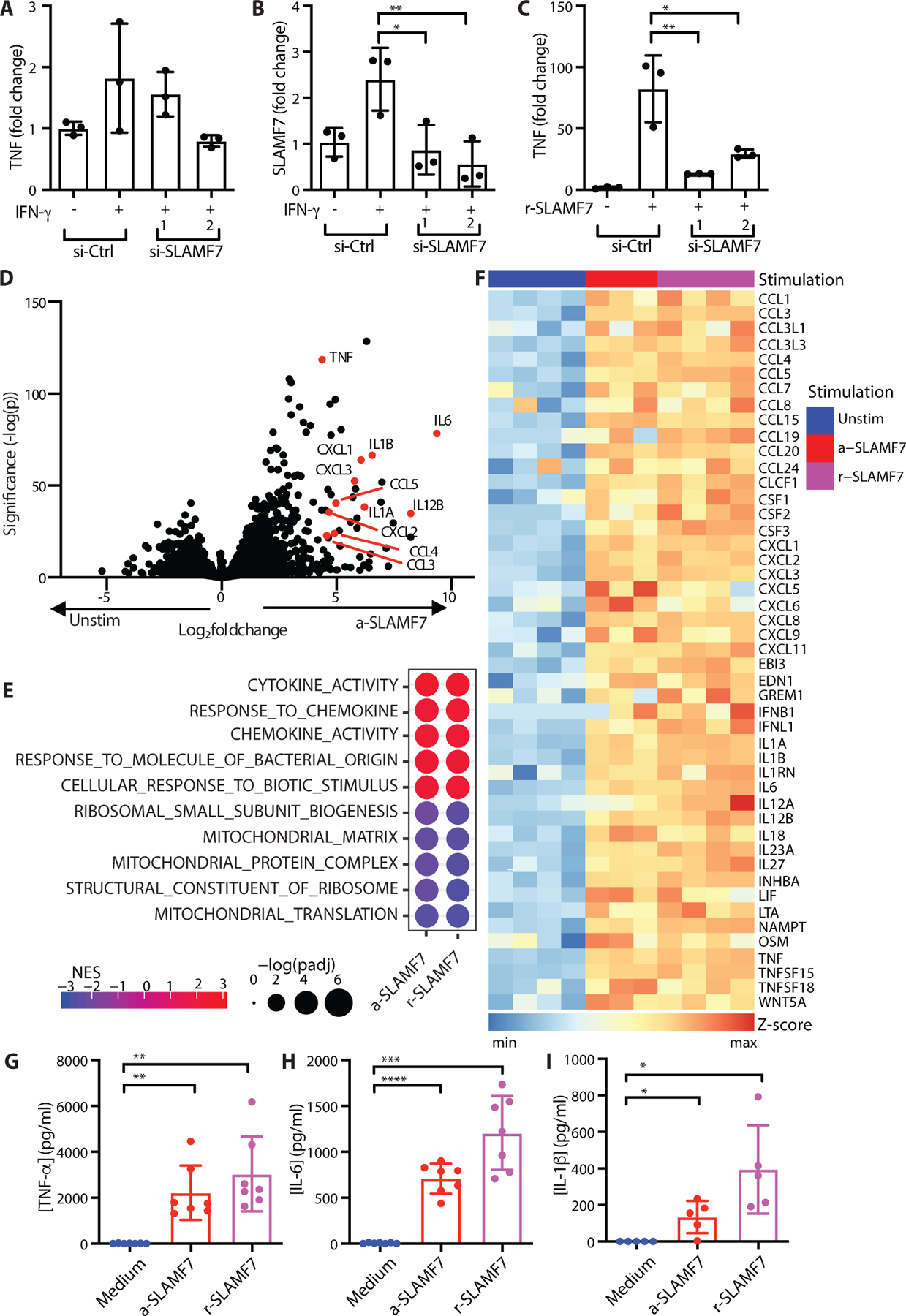
A-C) Macrophages were treated with siRNA control or siRNA targeting SLAMF7. They were then potentiated with IFN-γ (10 ng/ml) for 24 hours. A) TNF or B) SLAMF7 expression was quantified relative to macrophages treated with control siRNA only by RT-PCR. C) After potentiation with IFN-γ, macrophages were stimulated with r-SLAMF7 (100 ng/ml) for 2.5h, and TNF was quantified relative to macrophages treated with control siRNA and IFN-γ only by RT-PCR. Data represent mean ± SD of triplicate samples in an experiment representative of at least 2 independent experiments. D-I) Macrophages were potentiated with IFN-γ (10 ng/ml) for 24 hours prior to treatment with a-SLAMF7 (10 μg/ml) or r-SLAMF7 (1 μg/ml) for 4h. D) Differential gene expression for macrophages incubated with a-SLAMF7 (n=3 donors) compared to macrophages treated with only IFN-γ (n=4 donors). E) Gene set enrichment analysis for macrophages after stimulation with a-SLAMF7 or r-SLAMF7 showing the top and bottom 5 GO categories for macrophages after SLAMF7 engagement. F) Heatmap showing z-scores for gene expression values of differentially expressed genes in the msigdb GO:cytokine activity gene set for IFN-γ pre-treated macrophages without additional stimulation (n=4 donors), stimulated with a-SLAMF7 (n=3 donors), or with r-SLAMF7 (n=4 donors). G) Secreted TNF-α, and H) secreted IL-6 after macrophage incubation with a-SLAMF7 or r-SLAMF7 (n=7 donors). I) Release of IL-1β after macrophage incubation with a-SLAMF7 or r-SLAMF7 for 4h followed by treatment with nigericin (10 μM) for 30m (n=5 donors). Data represent mean ± SD. Statistics were calculated with the one-way ANOVA, using Dunnett’s multiple comparisons test. Unstim, unstimulated; a-SLAMF7, anti-SLAMF7 antibody; r-SLAMF7, recombinant SLAMF7 protein; si-Ctrl, control siRNA; si-SLAMF7, siRNA against SLAMF7; NES, normalized expression score; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
To define transcriptome-wide changes driven by SLAMF7 engagement, we then used the SLAMF7 activating antibody (a-SLAMF7) or r-SLAMF7 protein to engage SLAMF7 in vitro in this two-step process. First, we pretreated macrophages with IFN-γ to induce high SLAMF7 expression and second, we used a-SLAMF7 or r-SLAMF7 to engage cellular SLAMF7. We then performed RNA-seq on these in vitro stimulated macrophages to examine transcriptome-wide changes in gene expression (n=4 donors, 100% female, mean age 34.3, range 22–70). We identified drastic changes in gene expression after SLAMF7 engagement, with 596 upregulated genes driven by both a-SLAMF7 and r-SLAMF7 (LFC ≥ 1, padj ≤ 0.05) that we defined as the “Macrophage SLAMF7 Stimulation Signature” (Data file S3). We observed striking upregulation of inflammatory cytokines TNF, IL1B, IL6, and IL12B as well as chemokines CCL3, CCL4, CXCL1, CXCL2, and CXCL8 after treatment with activating anti-SLAMF7 mAb (Fig. 3D) or r-SLAMF7 protein (Fig. S7A) compared to macrophages treated with IFN-γ alone. SLAMF7 was itself upregulated after stimulation, suggesting a positive feedback loop. In contrast, we detected downregulation of the transcription factor CEBPA, which regulates monocyte development (49) and may inhibit inflammatory pathways in monocytes (50). Gene set enrichment analysis identified strong enrichment of msigdb gene ontology (GO) categories (padj <0.05) including cytokine activity and response to molecule of bacterial origin (Fig. 3E), indicating that triggering SLAMF7 drives a dominant myeloid inflammatory program. Mitochondrial and ribosomal pathways were downregulated (Fig. 3E), suggesting changes in metabolism to support this exuberant inflammatory macrophage state. Indeed, a large number of cytokines were upregulated in macrophages stimulated with either anti-SLAMF7 or r-SLAMF7, as visualized by heatmap (Fig. 3F).
Consistent with this profoundly inflammatory state identified by RNA-seq, secreted TNF-α levels increased from 12 pg/ml to 2.2 – 3.0 ng/ml after stimulation (Fig. 3G) and IL-6 levels increased from 6 pg/ml to 0.7 – 1.2 ng/ml after stimulation (Fig. 3H). Real-time PCR (RT-PCR) analysis confirmed induction of CCL3 (mean LFC 3.1 – 4.2, Fig. S7B), CXCL1 (mean LFC 4.3 – 5.1, Fig. S7C), and CXCL8 (mean LFC 10.0 – 10.2, Fig. S7D) after SLAMF7 engagement. The inflammatory cytokine IL1B was strongly upregulated after stimulation with SLAMF7. However, macrophages only express high levels of inflammasome components and pro-IL-1β when they are primed by microbial TLR agonists or cytokines like TNF-α, and subsequent inflammasome activation with pyroptotic cell death is required for abrupt release of bioactive IL-1β (31, 51). We detected release of 134 – 395 pg/ml of IL-1β from macrophages stimulated with SLAMF7 when nigericin was added to activate the inflammasome (Fig. 3I), suggesting that SLAMF7 can also prime inflammasomes. We also sought to verify the relevance of this pathway in cells from diseased human tissues by purifying macrophages from synovial fluid of patients with RA. Stimulation of SLAMF7 on synovial fluid macrophages with r-SLAMF7 protein resulted in strong induction of TNF (mean LFC 5.0, Fig. S7E) and IL1B (mean LFC 6.9, Fig. S7F), indicating the potential importance of this receptor in macrophage activation in RA.
We did not observe a strong overlap with the msigdb Immunologic Signatures gene set for classically activated M1 macrophages. The “Classical M1 vs. Alternative M2 Macrophage” upregulated gene set (52) defines activation after 18h combined stimulation with IFN-γ + LPS, but interestingly the array of cytokines driven by SLAMF7 engagement was strikingly absent from that signature. Other M1 activation protocols include initial priming with IFN-γ followed by LPS, which results in production of inflammatory cytokines and gene expression (53) that partially overlaps with those we identified in our conditions. This underscores the fact that this SLAMF7 activation program rests upon and is a separate step after primary stimulation of macrophages by IFN-γ or other M1 differentiation and activation factors. These sequential in vitro conditions include 1) initial macrophage potentiation with IFN-γ to drive high SLAMF7 expression, followed by 2) engagement of SLAMF7 and completion of activation, underscoring this distinct program. We termed this activation state, defined by upregulation of the SLAMF7 receptor followed by SLAMF7 engagement that then triggers profound inflammatory activation, as the SLAMF7 super-activated macrophage (SLAMF7-SAM) state.
SLAMF7 drives an inflammatory signaling cascade
SLAMF7 contains a cytoplasmic immunoreceptor tyrosine switch motif (ITSM) that can either drive inhibitory signaling by phosphatases or can alternatively allow the docking of the adaptor EAT-2 to activate downstream pathways while sterically blocking binding of inhibitory phosphatases (54, 55). SLAMF7 engagement drives activation of NK cells through phospholipase C and Akt (56) and has been reported to activate macrophages independent of EAT-2 by recruiting the immunoreceptor tyrosine activating motif (ITAM) bearing proteins Fc common gamma chain (FcGammaR) and DAP12 (57). To assess which pathways might transduce the profound activation observed in human macrophages, we first assessed the expression of EAT-2 (SH2D1B), FcGammaR (FCER1G) and DAP12 (TYROBP) in the bulk RNA-seq data for synovial tissue macrophages from AMP (15). We detected high levels of FCER1G and TYROBP but not SH2D1B in synovial macrophages (Fig. 4A). There was similar gene expression in the CD14+ cells we sorted from synovial fluid (Fig. S8A) and peripheral blood (Fig. S8B), suggesting that the EAT-2 adaptor is unlikely to contribute to macrophage activation through SLAMF7. We then evaluated whether activating FcGammaR or DAP12 might contribute to activation of macrophages by SLAMF7. Silencing FCER1G (Fig. S8C), but not TYROBP (Fig. S8D), reduced production of TNF-α by macrophages after stimulation with r-SLAMF7 (Fig. 4B), suggesting that FcGammaR plays an important role in macrophage activation by SLAMF7.
Figure 4. SLAMF7 engagement drives an inflammatory signaling cascade.
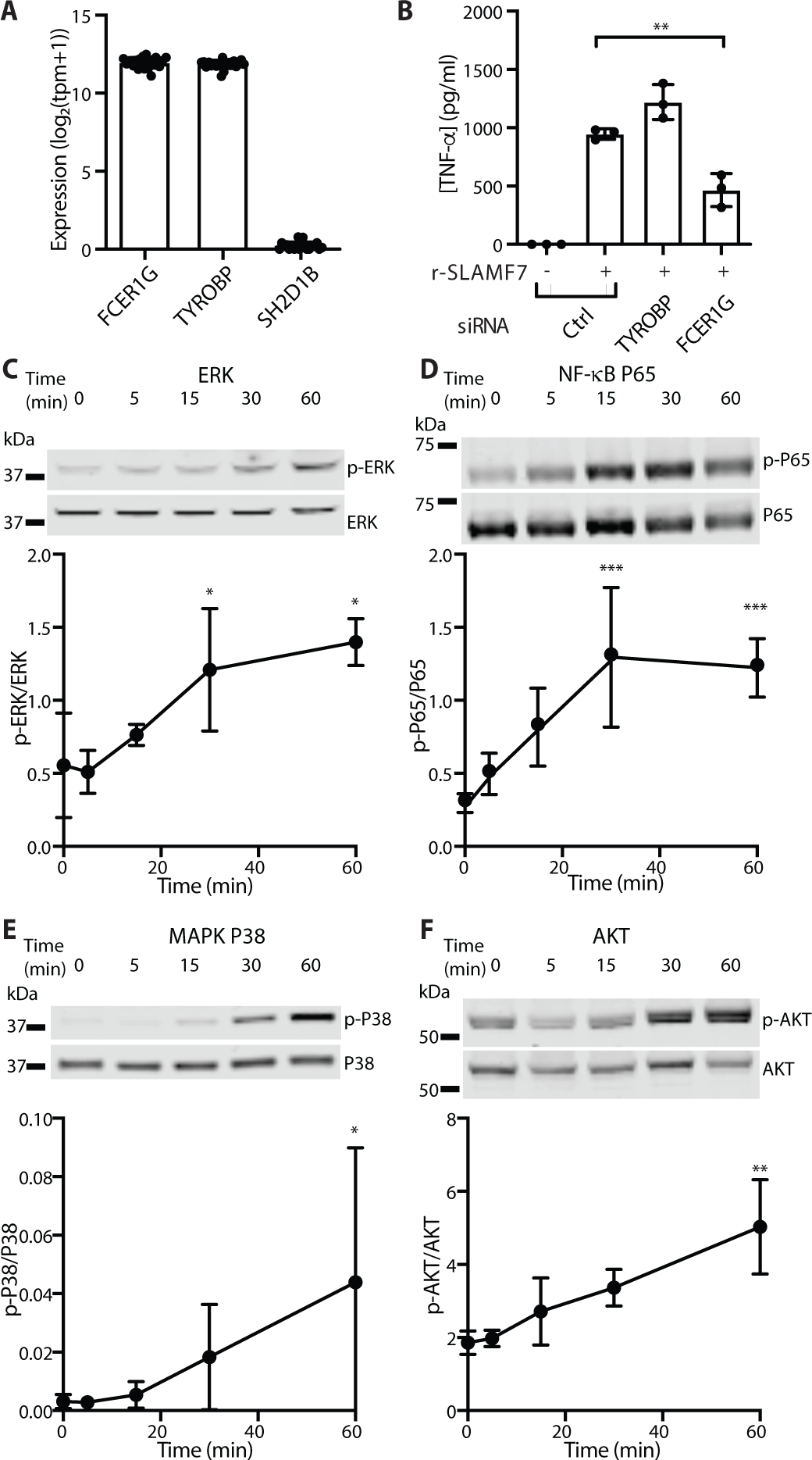
A) Gene expression in bulk RNA-seq of synovial tissue macrophages from patients with arthritis from AMP (n=21 donors) (15). B) Macrophages were treated with siRNA control, or siRNA targeting TYROBP or FCER1G. Cells were potentiated with IFN-γ (5 ng/ml) for 24 hours, then stimulated with r-SLAMF7 (100 ng/ml) for 4 hours. Secreted TNF-α was measured by ELISA. Data represent mean ± SD of triplicate wells from an experiment representative of at least 2 independent experiments. C-F) Macrophages were potentiated with IFN-γ (10 ng/ml) for 24h, then stimulated with r-SLAMF7 (100 ng/ml) for the times indicated. Representative Western blots and densitometry quantification for C) ERK and phospho-ERK, D) P65 and phospho-P65, E) MAPK P38 and phospho-MAPK P38, and F) AKT and phospho-AKT. Data represent mean ± SD of at least 3 donors. Statistics were calculated using the one-way ANOVA with Dunnett’s multiple comparisons test. *, p ≤ 0.05; **, p ≤ 0.01; r-SLAMF7, recombinant SLAMF7 protein; tpm, transcripts per million.
We then explored activation of pathways downstream of FcGammaR, which signals through SYK and SRC pathways (58, 59), resulting in phosphorylation of downstream mediators including RAS/ERK, PI3K/AKT, PLC-γ, NF-κB, and MAPK pathways (60, 61). Engagement of SLAMF7 with r-SLAMF7 resulted in doubling of phosphorylation of ERK (Fig. 4C) and more than four times more phosphorylation of NF-κB P65 (Fig. 4D) at the time points tested. We also detected more than ten times higher phosphorylation of MAPK P38 (Fig. 4E) and an almost three-fold increase in AKT phosphorylation (Fig. 4F). This implicates SLAMF7 as a receptor that activates multiple pathways that reprogram macrophage metabolism to drive inflammatory gene expression.
SLAMF7 amplifies macrophage activation through a TNF-α autocrine loop
The magnitude of macrophage activation by SLAMF7 engagement was so striking that we wondered if it might recruit autocrine amplification pathways. A time course revealed the extremely rapid induction of TNF RNA in 30 minutes, resulting in secretion of TNF-α within 2 hours, with continued accumulation over time (Fig. 5A). Based on the very rapid induction of TNF-α, we hypothesized that this cytokine might provide an autocrine amplification signal as previously noted in other contexts (28, 62). We used siRNA to silence the genes encoding the receptors for TNFR1 (TNFRSF1A, Fig. S9A) and TNFR2 (TNFRSF1B, Fig. S9B), resulting in a marked reduction of TNF-α secretion after stimulation with r-SLAMF7 (Fig. 5B). Antibody blockade of TNF-α decreased expression of TNF (Fig. 5C) and IL1B (Fig. 5D) by fifty percent after SLAMF7 stimulation. Corresponding blockade of the TNFR1 and TNFR2 receptors also reduced levels of TNF (Fig. 5C) and IL1B (Fig. 5D). This implicates TNF-α autocrine signaling as an additional amplification step for inflammatory pathway activation following SLAMF7 engagement in SLAMF7-SAMs.
Figure 5. SLAMF7 amplifies macrophage activation through a TNF-α autocrine loop.
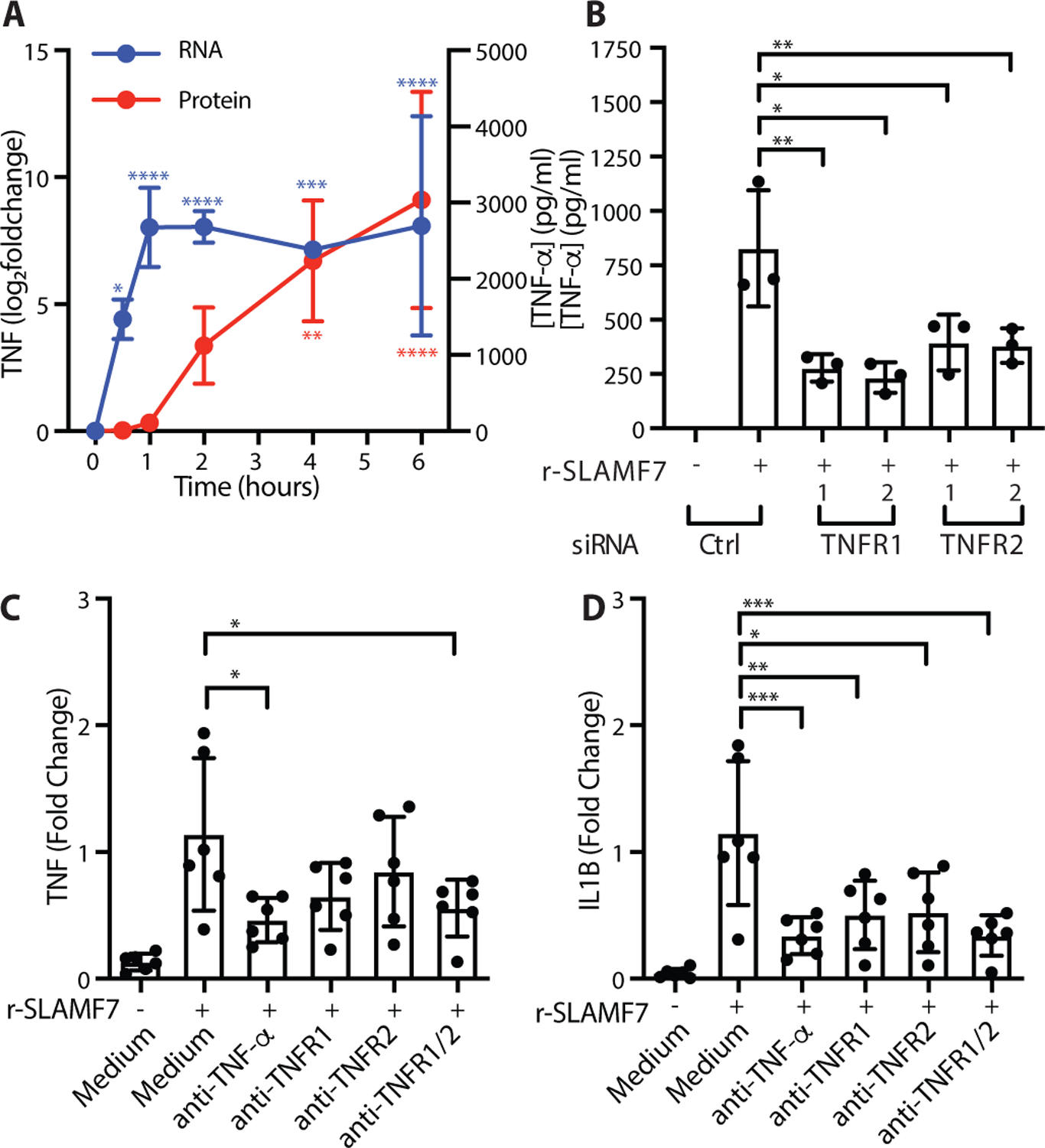
A) Macrophages were potentiated with IFN-γ (10 ng/ml) for 24h, then stimulated with r-SLAMF7 (500 ng/ml). TNF RNA was measured at each time point relative to unstimulated cells (blue) by RT-PCR and secreted TNF-α protein was measured by ELISA (red). Data represent mean ± SD of four donors. B) Macrophages were treated with siRNA control or siRNA targeting TNFR1 (TNFRSF1A) or TNFR2 (TNFRSF1B), potentiated with IFN-γ (5 ng/ml) for 16–18h, and stimulated with r-SLAMF7 (100 ng/ml) for 3h. TNF-α was measured by ELISA. Data represent mean ± SD of triplicate wells from an experiment representative of at least 3 independent experiments. C-D) Macrophages were potentiated with IFN-γ (10 ng/ml) for 24h, treated with antibodies for 30m, and stimulated with r-SLAMF7 (100 ng/ml) for 8h. RT-PCR was used to quantify C) TNF and D) IL1B relative to macrophages without antibody pre-treatment. Data represent mean ± SD of 6 donors. Statistics were calculated using the one-way ANOVA with Dunnett’s multiple comparisons test. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001; r-SLAMF7, recombinant SLAMF7 protein.
SLAMF7 super-activation of macrophages in rheumatoid arthritis
In the data from AMP (15), we had observed much higher SLAMF7 gene expression in synovial tissue macrophages from patients with RA compared to OA (Fig. 1). We then evaluated expression of the 596 genes in the “Macrophage SLAMF7 Stimulation Signature” identified by in vitro stimulation of macrophages (Fig. 3) in the AMP bulk RNA-seq data by calculating the percent of gene expression derived from SLAMF7-induced genes. Expression of these genes as a “SLAMF7 Activation Score” was almost twice as high in RA compared to OA (Fig. 6A), suggesting that SLAMF7-SAM program contributes to this disease. We also analyzed an independent single cell RNA-seq dataset on synovial macrophages (18), focusing on the comparison between healthy controls and untreated patients with rheumatoid arthritis. Macrophages from patients with RA had more than double the levels of SLAMF7 relative to healthy controls (Fig. 6B), along with increased levels of the “SLAMF7 Activation Score” (Fig. 6C). Of note, one untreated individual (SA220) with both low SLAMF7 and the “SLAMF7 Activation Score” (Fig. 6B–C) also had a low disease activity score compared to other untreated RA patients in the study (18), suggesting that SLAMF7 and its activation score are correlated and may relate to disease activity. We then performed clustering of macrophages from healthy controls and untreated RA patients and visualized with a Uniform Manifold Approximation and Projection (UMAP) plot (Fig. 6D). These cells uniformly expressed CD14 and CD68, with one large population expressing MERTK and FOLR2, and another with high levels of CD48 (Fig. S10A–C). We further subdivided these populations into groups defined by expression of TREM2, LYVE1, CLEC10A, S100A12, or SPP1 (Fig. S10D–F). As previously reported (18), the SPP1+ and S100A12+ populations were expanded in most patients with RA (Fig. 6E). We observed prominent expression of the “SLAMF7 Activation Score” in the SPP1+ and S100A12+ clusters that were expanded in RA patients (Fig. 6F). Interestingly, the SPP1+ cluster also had very high expression of SLAMF7, transcription factor NFKB1, and inflammatory cytokine CXCL8 (Fig. 6G, S11A–B, red circle). This population also exhibited high levels of IFN-induced CD40 and GBP1, as well as cytokines such as TNF, CCL3, and CXCL2 (Fig. S11C–E, red circle). This suggests that the high expression of SLAMF7, along with its gene expression signature, represents the super-activated SLAMF7-SAMs that contribute to disease pathology.
Figure 6. SLAMF7 super-activated macrophages drive inflammation in rheumatoid arthritis.
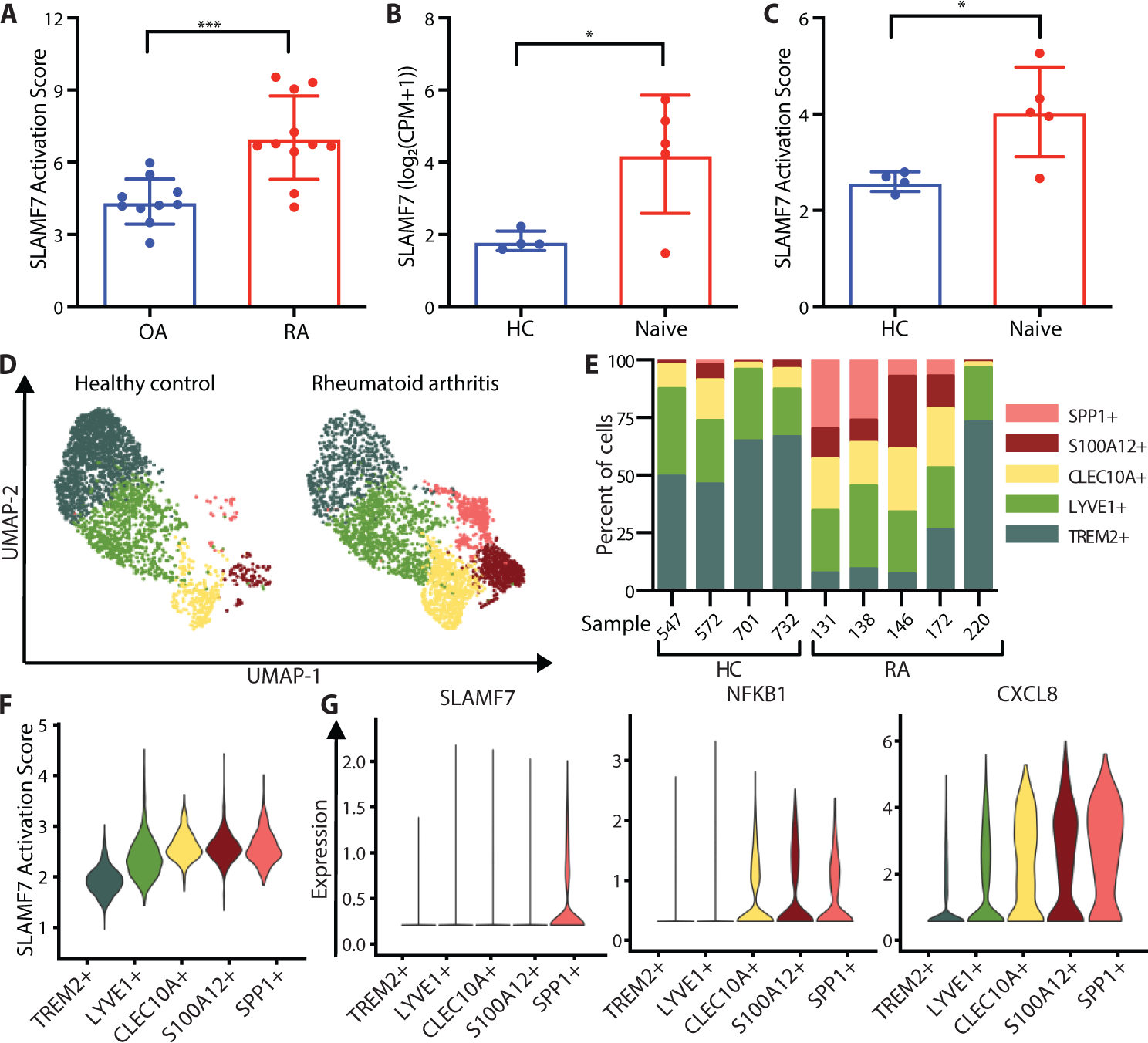
A) SLAMF7 activation score for bulk RNA-seq data on synovial macrophages from patients with OA (n=10) or RA (n=11) (15). B) SLAMF7 expression and C) SLAMF7 activation score for pseudobulk RNA-seq data for synovial macrophages from healthy controls (n=4) or untreated RA patients (n=5) (18). Data in A-C represent mean ± SD. D) UMAP plot of macrophage clusters from synovial tissues of healthy controls or patients with untreated RA. E) Percent of macrophages from each donor assigned to each cluster. F) Violin plot showing the SLAMF7 Activation Score in different macrophage populations. G) Violin plots showing gene expression of synovial macrophage populations. The t-test was used for statistical comparisons. OA, osteoarthritis; RA, rheumatoid arthritis; HC, healthy control; Naive, untreated treatment-naive rheumatoid arthritis; CPM, counts per million; *, p ≤ 0.05; ***, p ≤ 0.001.
SLAMF7 super-activation of macrophages in autoimmune and infectious disease
Based on the striking evidence for the SLAMF7-SAM state in RA, we evaluated public data from other inflammatory diseases. Macrophages contribute to inflammatory pathways in Crohn’s disease (6–8) as part of the GIMATS (IgG plasma cells, inflammatory mononuclear phagocytes, and activated T and stromal cells) module that has been associated with resistance to TNF-blockade in patients with IBD (8), and macrophages also are major contributors to pathological inflammation in individuals with COVID-19 pneumonia (20, 21). We analyzed publicly available single cell RNA-seq data on ileal tissue from patients with Crohn’s disease (8) and bronchoalveolar lavage fluid from individuals infected with COVID-19 (20) to determine whether this SLAMF7 activation program identified in RA contributes to macrophage-driven inflammation in other diseases. First, we used gene set enrichment analysis to compare the gene expression profiles of macrophages from inflamed ileal tissue and infected lungs to genes upregulated in RA that we had defined as the “Inflamed RA Macrophage Signature” (Fig. 1). We observed significant overlap of macrophage gene expression in both IBD and COVID-19 with gene expression in RA (Fig. 7A), suggesting common pathways of macrophage activation across tissues and diseases. We then compared the transcriptomes of macrophages from inflamed ileal tissue and bronchoalveolar lavage cells with the in vivo derived “SLAMF7-High Macrophage Signature” (Fig. 2) and the in vitro derived “Macrophage SLAMF7 Stimulation Signature” (Fig. 3). Importantly, we found a strong correlation between these signatures and gene expression in macrophages from inflamed gut and lung tissues (Fig. 7A). The average levels of SLAMF7 activation were more than twice as high in macrophages from inflamed gut tissue than non-inflamed tissue (Fig. 7B), with the highest levels of SLAMF7 activation in samples categorized as having high (Fig. S12A) rather than low (Fig. S12B) GIMATS module intensity scores. Individuals with mild COVID-19 lung involvement had double the SLAMF7 activation levels compared to healthy controls, and remarkably, the signature was more than four-fold higher in individuals with severe disease (Fig. 7C). This suggests that this SLAMF7-SAM program represents a dominant state in these inflammatory diseases.
Figure 7. SLAMF7 super-activated macrophages drive inflammation in inflammatory bowel disease and COVID-19 infection.
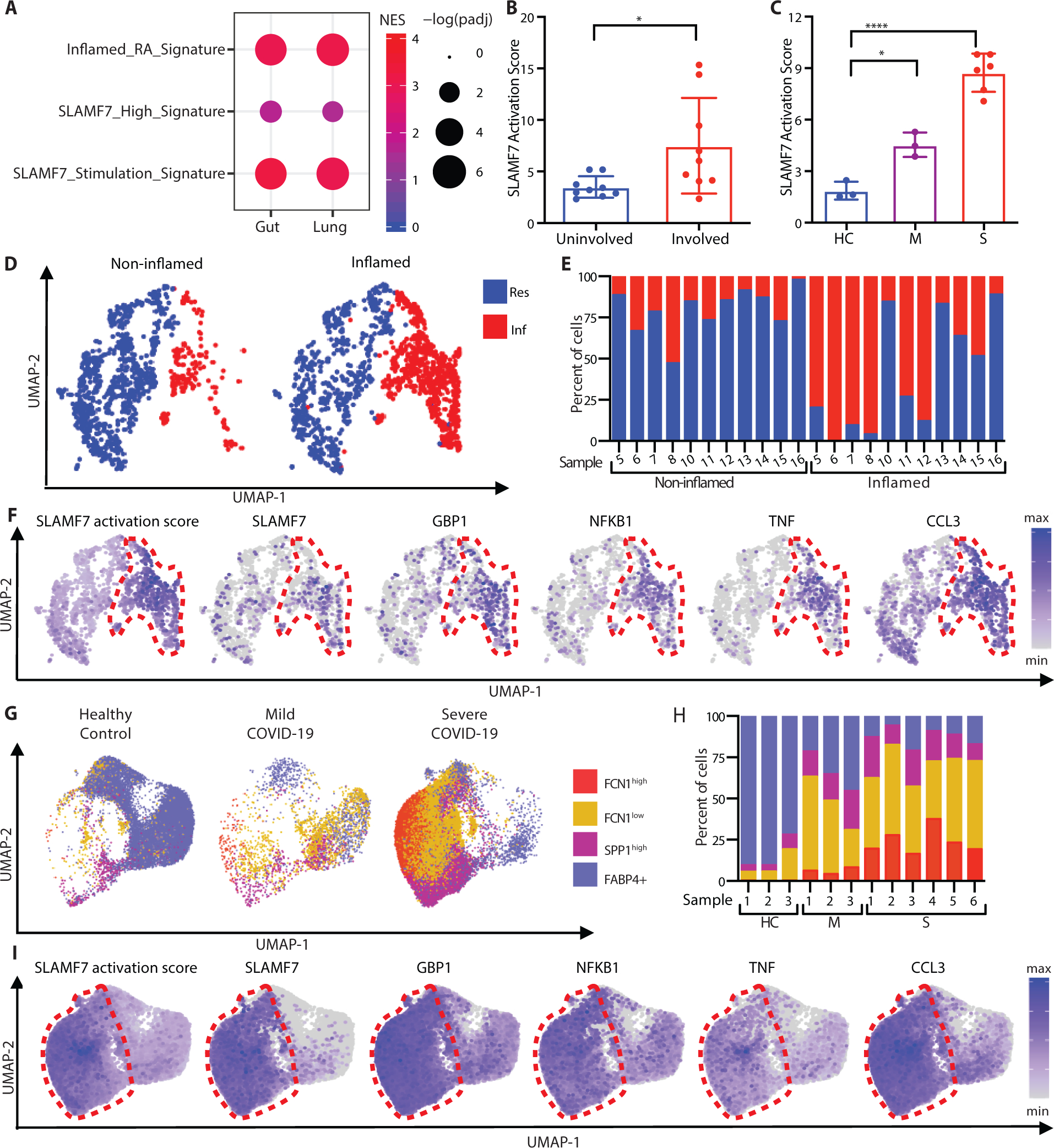
A) Gene set enrichment analysis comparing gene expression from macrophages from inflamed ileal tissues in patients with Crohn’s disease or lungs of patients with severe COVID-19 with the “Inflamed RA Macrophage Signature”, the “SLAMF7-High Macrophage Signature” and the “Macrophage SLAMF7 Stimulation Signature.” B) SLAMF7 activation score for macrophages from non-inflamed (n=9) and inflamed ileal tissues (n=9) (8). C) SLAMF7 activation score for bronchoalveolar lavage macrophages from healthy controls (n=3), or individuals with mild (n=3) or severe COVID-19 (n=6) (20). Data in B-C represent mean ± SD. D) UMAP plot of macrophage clusters from involved and uninvolved ileal tissues. E) Percent of macrophages from each donor assigned to each cluster. F) UMAP plots showing gene expression of ileal macrophage populations. G) UMAP plot of bronchoalveolar lavage macrophage populations. H) Percent of macrophages from each donor assigned to each population. I) UMAP plots showing gene expression for bronchoalveolar lavage macrophage populations. The paired t-test was used to compare inflamed and non-inflamed gut tissues, and the one-way ANOVA with Dunnett’s multiple comparisons test was used to compare mild and severe COVID-19 to healthy controls. NES, normalized expression score; Res, Resident macrophage cluster; Inf, Inflammatory macrophage cluster; HC, healthy control; M, mild COVID-19; S, severe COVID-19; *, p ≤ 0.05; **, p ≤ 0.01; ****, p < 0.0001.
We explored which single cell populations displayed the SLAMF7-SAM state in vivo. First, we visualized macrophage populations from ileal tissue of patients with IBD using UMAP plots. We observed two dominant populations of ileal macrophages with expression of CD14 and CD68, including a population of macrophages with higher expression of MRC1 (Fig. S13) that was present in both inflamed and non-inflamed tissues, consistent with “resident macrophage” populations (Fig. 7D). Another macrophage population with higher cytokine expression was predominantly derived from inflamed tissues, consistent with “inflammatory macrophages” (Fig. 7D,E). Gene expression from the “SLAMF7 Activation Score” was extremely high in the inflammatory macrophage population (Fig. 7F, red circle). This cluster also exhibited higher expression of SLAMF7, as well as GBP1, NFKB1, TNF, CCL3 (Fig. 7F, S14A–B), IFN-induced CD40 and IDO1, and inflammatory chemokines CXCL2 and CXCL8 (Fig. S14C–E).
We then focused on lung macrophages from patients infected with COVID-19, including a monocyte-like FCN1high population, a chemokine-high FCN1low group, fibrosis-associated SPP1high macrophages (10), and FABP4+ alveolar macrophages (Fig. 7G). On the UMAP plot, there was clear separation of the three FCN1+ and SPP1+ macrophage populations from the FABP4+ alveolar macrophages (Fig. S15), with drastic increases in the proportions of FCN1high, FCN1low, and SPP1high macrophage populations in individuals with COVID-19 (Fig. 7H). We observed the highest levels of the “SLAMF7 Activation Score” in cells from the FCN1low and FCN1high populations that were expanded in patients with severe disease, although it was highly expressed in all three FCN1+ and SPP1+ populations compared to FAB4+ alveolar macrophages (Fig. 7I, red circle). There was also a striking increase in SLAMF7 expression in FCN1+ and SPP1+ macrophages from patients with severe COVID-19 compared to extremely low expression in FABP4+ alveolar macrophages from healthy controls (Fig. 7I, S16A–B). The FCN1+ and SPP1+ populations also expressed very high levels of GBP1, NFKB1, TNF and CCL3 (Fig. 7I, S16A–B), as well as IFN-inducible CD40, IDO1, and inflammatory CXCL2 and CXCL8 (Fig. S16C–E). The convergence of higher levels of SLAMF7 and other IFN-induced genes, along with SLAMF7-induced inflammatory genes, implicates the SLAMF7-SAM state as an important contributor to COVID-19 pneumonia. This macrophage activation state, defined by initial potentiation with IFN-γ and upregulation of SLAMF7, followed by subsequent SLAMF7 engagement and amplification through TNF-α, is one that may play a major role in pathological inflammation of RA, IBD and COVID-19 pneumonia.
Discussion
Macrophages orchestrate immune responses to protect against invading microbes, but overly robust immune responses and inflammation can cause autoimmune disease or cytokine storm during infection. Here, we have identified a pathway in which SLAMF7 is a key receptor that defines an activated macrophage, with the highest levels on macrophages exposed to IFN-γ, and lesser but significant induction by other cytokines and TLR agonists. Subsequent triggering of SLAMF7, even in the absence of microbial molecules, drives rapid and explosive production of cytokines and chemokines that we have defined as SLAMF7 superactivated macrophages (SLAMF7-SAMs). While resting macrophages are characterized by selectins and anti-inflammatory genes, IFN-γ reprograms the macrophage state, including high expression of SLAMF7. Subsequent triggering of the SLAMF7 receptor leads to production of inflammatory cytokines with massive autocrine amplification to drive this SLAMF7-SAM state (Fig. S17). A similar inflammatory macrophage population has been reported in several single cell RNA-seq datasets (63, 64), and here we identify a pathway that can drive the profound activation of this macrophage population.
Because macrophages must pass through multiple steps to achieve this complete super-activation state, interventions targeting different parts of this pathway could allow for fine-tuned therapeutic strategies. JAK inhibitors such as ruxolitinib impair macrophage responses to IFN-γ, and would diminish the initial upregulation of SLAMF7, thereby limiting activation of this pathway. TNF-α blockade could impair autocrine amplification of this pathway but would have limited effects on the array of inflammatory effectors driven by direct SLAMF7 stimulation. Indeed, current therapeutics targeting these pathways may partially alleviate different components driving SLAMF7-SAMs, but we did not have access to transcriptional data from treated individuals in sufficient numbers to evaluate this question in this study. The patients with inflamed RA from AMP predominantly represented untreated individuals, but also patients treated with tofacitinib, methotrexate, and etanercept (15), suggesting that this macrophage state is shared across treatment conditions, and is unlikely to be a medication effect. The absence of this receptor on normal resident macrophage populations implicates it as having particular importance as a specific target for inflammatory macrophages.
It is blockade of SLAMF7 itself that would prevent the completion of macrophage super-activation on IFN-γ potentiated macrophages and likely have the most important therapeutic implications. Elotuzumab is an antibody targeting SLAMF7 that is in clinical use for treatment of relapsed multiple myeloma (65), and part of this drug’s efficacy is through activation of NK cells (66) and macrophages (67) indicating it has activating effects. Another antibody against SLAMF7 was used to deplete plasma cells and reduce disease severity in an animal model of arthritis (68), but the importance of this receptor on highly activated macrophages has not previously been appreciated. SLAMF7 and its activation signature are strikingly upregulated in inflammatory RA, IBD, and COVID-19 pneumonia, implicating it as contributing to highly activated macrophage-driven inflammation in autoimmune and infectious diseases. Targeting this receptor and downstream pathways may offer a special opportunity to block severe macrophage-driven inflammation without diminishing more moderate macrophage immune surveillance and helpful homeostatic macrophage functions.
Materials and Methods
Study design
The objective of this study was to identify receptors that regulate macrophage activation in inflammatory human disease. We analyzed publicly available RNA-seq data to identify genes that are selectively expressed in macrophages from inflamed tissues, identifying and validating that SLAMF7 is highly expressed in synovial tissue macrophages from inflamed RA tissues. To define the transcriptional state of macrophages with high SLAMF7, we sorted myeloid cells with high versus low expression of SLAMF7. We used in vitro stimulation to identify IFN-γ as a dominant driver of SLAMF7 expression. We then engaged this receptor on monocyte-derived macrophages in vitro and defined a strong inflammatory gene expression signature driven by SLAMF7. We used siRNA and Western blots to define signaling pathways that drive this signature, along with blocking antibodies to implicate a TNF-α autocrine loop that amplifies this super-activation state. Finally, we analyzed other datasets to identify this SLAMF7-activated macrophage state in single cell RNAseq data not only from synovial tissue macrophages from patients with arthritis, but also gut macrophages from patients with IBD and lung macrophages from individuals with COVID-19 pneumonia.
Human research
Human subjects research was performed with approval from the Institutional Review Board at Brigham and Women’s Hospital. Patients with RA were defined according to American College of Rheumatology 2010 Rheumatoid Arthritis classification criteria (69). Synovial tissue samples were obtained as excess samples from patients undergoing arthroplasty procedures at Brigham and Women’s Hospital, and samples were frozen in Cryostor CS10 preservation medium (Sigma-Aldrich). Synovial fluid samples were obtained sterile prior to arthrotomy as discarded samples from routine clinical care, and peripheral blood samples were collected as discarded samples from routine clinical care. Patient consent for genomic studies was obtained for all samples used for RNA sequencing.
Sample processing
Synovial tissue was disaggregated as previously described (11, 70). Frozen synovial tissue was thawed and minced into small pieces that were digested with 100 μg/ml Liberase TL (Sigma) and 100 μg/ml DNAse I (Sigma) at 37°C for 30 minutes. Samples were passed over a 70 μM cell strainer (Fisher), RBCs were lysed with ACK lysis buffer (Fisher), and cells were stained for flow cytometry. Synovial fluid and peripheral blood were isolated as previously reported (14), with density centrifugation on a Ficoll-Hypaque gradient to isolate mononuclear cells. Cells were cryopreserved for subsequent analysis.
Reagents and antibodies
Recombinant human M-CSF, IFN-β, IFN-γ, IL-1β, IL-6 and TNF-α were from Peprotech. ELISA kits for TNF-α, IL-6, and IL-1β were from R&D Systems. Endotoxin quantification kits and endotoxin removal kits were from Pierce. Pam3CSK4 and LPS were from Invivogen. Ruxolitinib was from Seleckchem. Nigericin was from Invivogen. Fetal bovine serum (FBS) was from Hyclone. Propidium iodide was from Biolegend, and fixable blue dead cell stain kit was from Thermo Fisher. The following 6x-His tagged recombinant proteins, expressed in HEK 293 cells, with at least 95% purity, were from Sigma: SLAMF7 (r-SLAMF7), CD84 (r-CD84), and CD326 (r-CD326). Purified anti-SLAMF7 (162.1), anti-TNF-α (MAb1), anti-CD4 (OKT4), and mouse IgG2b, κ isotype control (MPC-11) were from BioLegend. Anti-TNFR1 (16803) and anti-TNFR2 (22210) were from R&D Systems. The following antibodies were used for flow cytometry and cell sorting: CD16 (CB16, eBioscience), CD45 (HI30, BioLegend), CD14 (61D3, eBioscience), SLAMF7 (162.1, BioLegend), CD84 (CD84.1.21, BioLegend), CD3 (UCHT1, eBioscience), CD19 (HIB19, eBioscience), CD56 (CMSSB, eBioscience), TNF-α (Mab11, BioLegend), mouse IgG2b, κ isotype control (MPC-11, BioLegend). The following antibodies from Cell Signaling were used for Western blot: Akt (40D4), phospho-Akt (Ser473, D9E), ERK1/2 (3A7), phospho-ERK1/2 (Thr 202/Tyr204, D13.14.4E), p38 MAPK (polyclonal, #9212), phospho-p38 MAPK (Thr180/Tyr182, 28B10), NF-κB p65 (L8F6), and phospho-NF-κB p65 (Ser636, 93H1). Fluorescent secondary antibodies anti-rabbit IRDye 680RD and anti-mouse IRDy3 800CW were from Li-Cor.
Cell culture and stimulation
Monocytes were MACs purified from peripheral blood mononuclear cells using CD14 microbeads (Miltenyi). 50,000 cells per well were cultured in flat bottom 96 well plates, or 300,000 cells per well were cultured in 24 well plates in RPMI (Fisher) with 10% FBS, with additives of beta-mercaptoethanol, sodium pyruvate, HEPES, penicillin/streptomycin, and L-glutamine (Fisher) and M-CSF (20 ng/ml). Macrophages were allowed to rest for 24 h. Cytokines were then added for an additional 24 h. For inhibitor experiments, ruxolitinib or an equivalent concentration of DMSO was added 30 minutes prior to IFN-γ. For SLAMF7 stimulation, after 24 h incubation with IFN-γ, macrophages were treated with a-SLAMF7 antibody or r-SLAMF7 protein. For blocking experiments, antibodies were added at 10 μg/ml (TNFR1 and TNFR2) or 20 μg/ml (TNF-α) 30 minutes prior to stimulation with r-SLAMF7.
siRNA
Monocytes were cultured in RPMI with 10% FBS and 20 ng/ml M-CSF. They were transfected with either a control siRNA or siRNA against a gene of interest (Silencer Select, Life Technologies) at 25–30 nM by reverse transfection using RNAiMax (Life Technologies). After 24–72 hours of siRNA treatment, macrophages were treated with IFN-γ for 16–24h prior to analysis or stimulation with r-SLAMF7 protein.
Quantitative Real-Time PCR (RT-PCR)
Primers (Table S1) were from Integrated DNA Technologies. RNA was processed using the RNEasy Mini kit (Qiagen) and Quantitect Reverse Transcription kit (Qiagen). RT-PCR was performed using SybrGreen (Agilent) on the AriaMX (Agilent). Gene expression for each sample was normalized to GAPDH and compared across conditions.
Cell staining
Cells were washed with staining buffer (PBS, 2% FBS, 2mM EDTA) (Fisher), treated with Human Trustain FcX (Biolegend) for 10 minutes on ice, and stained with antibodies for 30 minutes on ice. Propidium iodide (1 μg/ml) was added 15 minutes prior to acquisition. For experiments with stimulated macrophages, cells were stained and then fixed in 4% paraformaldehyde for 10 minutes on ice prior to acquisition. The Tru-Nuclear Transcription Factor Buffer Set (BioLegend) was used for intracellular cytokine staining. Analysis was done on the LSR-Fortessa (BD).
Flow cytometry analysis
Analysis was performed by gating on myeloid cells by forward scatter and side scatter, excluding doublets, and gating on CD45+ leukocytes with exclusion of dead cells. CD14+ cells were selected for analysis (Fig. S1A–D). Specific MFI was determined by subtracting the MFI from an isotype control from the MFI for a specific antibody. Percent positive cells were calculated by setting a gate with ≤ 1% of cells in a sample stained with isotype control as the threshold for positivity.
Cell sorting
Sorting was performed on a FACS-Aria Fusion sorter (BD). Myeloid cells were gated as CD45+ cells with exclusion of live/dead dye, CD3, CD19 and CD56. SLAMF7-low cells were defined as 40% of myeloid cells with lowest expression of SLAMF7. SLAMF7-high cells were defined as those with staining above the level of isotype control. Three populations from blood and two populations from synovial fluid were sorted based on expression of CD14 and CD16 (Fig. S3). Up to 1,000 cells from each population were collected in Eppendorf tubes with 5 μl of TCL buffer and 1% beta-mercaptoethanol. Cell lysates were frozen at −80 °C for further processing.
Western Blot
Monocytes were cultured in M-CSF, treated with IFN-γ for 16–24h and stimulated with recombinant SLAMF7 protein. Cells were lysed in RIPA buffer (Cell Signaling) with 1mM PMSF (Cell Signaling) and protease/phosphatase inhibitor (Cell Signaling) and boiled at 95°C in Laemmli buffer (Biorad). Protein was quantified with BCA (Pierce Technologies) and 25–50 ug protein was loaded in each well for SDS-PAGE separation (Invitrogen). Samples were transferred to nitrocellulose membrane (iBlot2, Invitrogen) for detection with primary antibodies. Fluorescent secondary antibodies were used for detection, and membranes were imaged on a Li-Cor Odyssey Blot Imager (Li-Cor Biotechnology). Blots were processed and analyzed with Li-Cor Image Studio version 4.0.21.
RNA-seq
Cell lysates were collected in TCL buffer with 0.1% beta-mercaptoethanol and frozen at −80°C. Samples were sequenced at the Broad Institute using the Smart-seq2 RNA-seq platform (71–73). This platform uses paired read sequencing and provides around 4 million reads per sample. Samples with detection of at least 10,000 genes were used for subsequent analysis.
Analysis of bulk RNA-seq from synovial tissue macrophages
FASTQ files for bulk RNA-seq data were obtained for sorted CD14+ synovial macrophages from patients with OA (n=10) and inflamed RA (n=11) from AMP (dbGaP phs001457.v1.p1) (15). Reads were quantified using kallisto. To focus on signaling receptors, we selected genes with the “protein_coding” Ensembl biotype with at least 1 count across samples, resulting in 18,304 genes for analysis. DESeq2 was used for differential gene expression analysis comparing macrophages from inflamed RA and OA, including both disease status and processing batch in the model. The 509 genes with LFC ≥ 1, padj ≤ 0.05 for RA compared to OA were considered significantly upregulated genes, defined as the “Inflamed RA Macrophage Signature” (Data file S1).
Analysis of bulk RNA-seq from sorted SLAMF7-high and SLAMF7-low cells
RNA-seq on SLAMF7-high and SLAMF7-low cells sorted from peripheral blood from healthy controls (n=5) or patients with RA (n=7), and synovial fluid from patients with RA (n=4 donors), was performed at the Broad Institute using Smart-seq2 with 25 bp paired reads. Read quantification was performed using the Broad Institute pipeline, with alignment to GRCh38.83 using STAR version 2.4.2a (74) and quantification with RSEM version 1.2.2.1 (75). 37,414 genes were included for analysis. DESeq2 was used to calculate differential gene expression between SLAMF7-high and SLAMF7-low cells for each population, including both SLAMF7 expression and donors in the model. Genes with LFC ≥ 1, padj ≤ 0.05 were considered significantly upregulated, and the “SLAMF7-High Macrophage Signature” was defined as 21 genes that were upregulated in SLAMF7-high compared to SLAMF7-low CD14+CD16- cells from both peripheral blood and synovial fluid (Data file S2). Differentially expressed genes from SLAMF7-high versus SLAMF7-low cells from each population in peripheral blood and synovial fluid were ordered by the Wald statistic calculated by DESeq2. The fgsea package was used for gene set enrichment analysis with 106 permutations for comparison of these ordered genes to msigdb Hallmark gene sets (h.all.v7.0.symbols.gmt).
Analysis of bulk RNA-seq from in vitro SLAMF7 stimulated macrophages
For analysis of macrophages after in vitro SLAMF7 stimulation (n=4 donors), RNA-seq was performed using Smart-seq2 with 50 bp paired reads. Reads were quantified using kallisto. One sample stimulated with a-SLAMF7 was excluded due to low gene counts. 36,513 genes were included for analysis. DESeq2 was used for differential gene expression analysis comparing macrophages potentiated with IFN-γ, followed by stimulation with either anti-SLAMF7 or r-SLAMF7 to macrophages treated with IFN-γ without additional stimulation. Fold-change values were shrunk using the apeglm algorithm (76). Genes with LFC ≥ 1, padj ≤ 0.05 were considered significantly upregulated. The “Macrophage SLAMF7 Stimulation Score” was defined as 596 genes that were commonly upregulated with both a-SLAMF7 and r-SLAMF7 stimulation (Data file S3). Differentially expressed genes from macrophages stimulated with a-SLAMF7 or r-SLAMF7 compared to unstimulated macrophages were ordered by the Wald statistic calculated by DESeq2. The fgsea package was used for gene set enrichment analysis with 106 permutations for msigdb Gene Ontology gene sets (c5.all.v7.0.symbols.gmt), and CollapsePathways was used to combine overlapping GO categories to identify top and bottom pathways.
Analysis of single cell RNA-seq on synovial macrophages from individuals with rheumatoid arthritis
Read count matrices and metadata for single cell RNA-seq for synovial tissue from patients with RA (E-MTAB-8322) (18) were downloaded on July 13, 2020. Harmony reduction was used to correct for differences between donors for clustering and UMAP analysis. Macrophages were selected for analysis based on expression of CD68 and CD14, excluding possible doublets using CD2 and CD3D to identify T cells, COL1A1 and PDPN to identify stromal cells, and VWF and CD34 to identify endothelial cells. Further subclustering was performed only on macrophages from healthy controls and individuals with untreated rheumatoid arthritis, using harmony reduction to correct for differences between donors for clustering and UMAP analysis. There was clear separation of macrophages expressing MERTK and FOLR2 versus CD48. These included subpopulations with MERTK, FOLR2, and TREM2 (TREM2+), MERTK, FOLR2 and LYVE1 (LYVE1+), CD48 and CLEC10A (CLEC10A+), CD48 and S100A12 (S100A12+), and CD48 and SPP1 (SPP1+) expression.
Analysis of single cell RNA-seq on macrophages from ileal samples from individuals with Crohn’s disease
Read count matrices for single cell RNA-seq of ileal biopsies from patients with Inflammatory Bowel Disease (IBD) (GSE134809) (8) were downloaded on December 10, 2019. Cells were filtered to include those with > 500 genes and < 5,000 genes, and < 25% mitochondrial RNA. Harmony reduction was used to correct for differences between samples for clustering and UMAP analysis. Myeloid cells were selected for analysis based on expression of CD68 and CD14. From the myeloid population, we excluded non-macrophage populations of dendritic cells (DCs) and other cells to focus our analysis on macrophages, using CD1C and FCER1A to identify CD1c+ DCs and monocyte-derived DCs, CLEC9A and BATF3 to identify CD141+ DCs, CCR7 and LAMP3 to identify migratory DCs, GZMB and IRF7 to identify plasmacytoid DCs, and CD3D and IGHG1 to exclude possible doublets. The remaining macrophages were positive for CD14, CD68, CSF1R, and MAFB. A cluster of resident macrophages (Res) had higher expression of MRC1, CD163 and MERTK, while a cluster of inflammatory macrophages (Inf) had high expression of inflammatory cytokines and chemokines including TNF and CCL3 (8). As described in the original publication, samples from donor 6 were excluded from additional analysis because of the low number of cells and samples from donor 16 were excluded due to similarity in cells from involved and uninvolved tissues.
Analysis of single cell RNA-seq from bronchoalveolar lavage of individuals with COVID-19 infection
Read count matrices for single cell RNA-seq of bronchoalveolar lavage cells from patients with COVID-19 (GSE145926) (20) were downloaded on May 5, 2020 and metadata provided by the authors was downloaded from github on May 21, 2020. Myeloid cells and populations in the metadata were used as originally described for analysis (20). Cells were filtered to include those with genes > 500 and < 5,000. Harmony reduction was used to correct for differences between donors for UMAP visualization.
Custom gene-set enrichment analysis
For single cell datasets, the sum of counts for each gene from all cells for each sample was combined to generate pseudobulk RNA-seq counts. For ileal macrophages from GSE134809, DESeq2 was used for differential gene expression comparing inflamed tissues (n=9) compared to non-inflamed tissues (n=9), including both disease involvement and donor in the model. For bronchoalveolar lavage macrophages from GSE145926, DESeq2 was used for differential gene expression comparing healthy controls (n=3) to patients with severe COVID-19 infection (n=6), including disease state in the model. Differentially expressed genes for macrophages from inflamed versus non-inflamed tissues from patients with RA or IBD, or healthy versus COVID-19 infected individuals were ordered by the Wald statistic calculated by DESeq2. Genes from the “Inflamed RA Macrophage Signature”, the “SLAMF7-High Macrophage Signature” and “Macrophage SLAMF7 Stimulation Signature” were compiled into a custom derived gmt file. The fgsea package was used with 106 permutations for gene set enrichment analysis of these ordered gene lists against these custom gene sets.
SLAMF7 Activation Score
The “SLAMF7 Activation Score” was calculated using the upregulated genes (LFC ≥ 1, padj ≤ 0.05, n=596 genes) from the “Macrophage SLAMF7 Stimulation Signature.” For bulk RNA-seq data on CD14+ synovial macrophages from AMP, the “SLAMF7 Activation Score” was calculated as the sum of counts for these 596 genes as a percent of total gene counts for each donor. For single cell RNA-seq data on macrophages from patients with RA, IBD or COVID-19, the “SLAMF7 Activation Score” was calculated as the sum of counts for these 596 genes as a percent of total gene counts for each cell, and the median value for all cells from each donor was used for pseudobulk analysis.
Data and statistical analysis
Flow cytometry data were analyzed using Flowjo version 10.4 (Treestar). Graphical and statistical analysis was done in RStudio version 1.1.383 with R version 3.6.0, JMP Pro version 13.0.0 (JMP Inc), and Prism version 6.0.h (GraphPad). Kallisto version 0.46.0 was used for quantification of RNA-seq reads (77) using version GRCh38.97 of the Ensembl transcriptome (downloaded August 13, 2019). Differential gene expression analysis was performed using DESeq2 version 1.24.0 (78). Gene set enrichment analysis was performed with fgsea version 1.10.1 (79) using msigdb gene sets (downloaded December 4, 2019), including Hallmark gene sets (h.all.v7.0.symbols.gmt), Gene Ontology gene sets (c5.all.v7.0.symbols.gmt) and Immunologic Signature gene sets (c7.all.v7.0.symbols.gmt). Heatmaps were generated using pheatmap version 1.0.12. Single cell RNA-seq analysis was performed on a cloud-computing cluster using R version 3.5.3, Seurat version 3.1.4 (80, 81), and harmony version 1.0 (82).
Supplementary Material
List of genes in the “Inflamed Arthritis Signature.”
List of genes in the “SLAMF7-High Signature.”
List of genes in the “SLAMF7 Stimulation Signature.”
Figure S1. Gating for flow cytometric analysis of macrophages.
Figure S2. CD84 expression in synovial macrophages
Figure S3. Gating strategy for cell sorting.
Figure S4. Interferon regulation of surface marker expression on macrophages.
Figure S5. SLAMF7 is necessary for macrophage activation by r-SLAMF7
Figure S6. Validation of reagents used for SLAMF7 activation.
Figure S7. Inflammatory cascade driven by SLAMF7 engagement.
Figure S8. FCER1G and TYROBP are highly expressed by macrophages.
Figure S9. Silencing of TNFRSF1A and TNFRSF1B with siRNA.
Figure S10. Macrophage populations in synovial tissue from healthy controls or patients with untreated rheumatoid arthritis.
Figure S11. Evidence for SLAMF7 activation in macrophages from patients with untreated rheumatoid arthritis.
Figure S12. High SLAMF7 activation in ileal macrophages from samples with high GIMATS module intensity scores.
Figure S13. Macrophage populations in ileal tissue from patients with inflammatory bowel disease.
Figure S14. Evidence for SLAMF7 activation in macrophages from patients with inflammatory bowel disease.
Figure S15. Macrophage populations in bronchoalveolar lavage from patients with COVID-19.
Figure S16. Evidence for SLAMF7 activation in macrophages from patients with COVID-19 infection.
Figure S17. Model for SLAMF7 super-activation of macrophages.
Raw data file
Acknowledgments:
We thank Fan Zhang, Ilya Korsunsky and Soumya Raychaudhuri for advice on bioinformatic analysis, Mike Gurish and Greg Keras for assistance recruiting patients and processing samples, Bill Apruzzese for helpful discussions, Jonida Toska for help with Western blots, and patients for their participation. Sorting was performed in the BWH Human Immunology Center.
We thank the members of the AMP RA/SLE Phase 1 Network for their contributions:
Jen Anolik, William Apruzzese, Joan M. Bathon, Ami Ben-Artzi, David L. Boyle, Brendan Boyce, S. Louis Bridges, Vivian Bykerk, Kevin Deane, Edward DiCarlo, Laura Donlin, Tom Eisenhaure, Andrew Filer, Gary S. Firestein, Lindsy Forbess, Susan Goodman, Ellen Gravallese, Peter K. Gregersen, Joel Guthridge, V. Michael Holers, Diane Horowitz, Laura Hughes, Judith James, James Lederer, Arthur Mandelin, Mandy McGeachy, Larry Moreland, Nir Hacohen, Harris Perlman, Costantino Pitzalis, Soumya Raychaudhuri, Christopher Ritchlin, Bill Robinson, Jennifer Seifert, PJ Utz, Kevin Wei, Fan Zhang
Funding:
This work was supported by NIH NIAID P01AI148102, NIH NIAMS P30 AR070253, and the Accelerating Medicines Partnership (AMP) in Rheumatoid Arthritis and Lupus Network. AMP is a public-private partnership (AbbVie Inc., Arthritis Foundation, Bristol-Myers Squibb Company, Foundation for the National Institutes of Health, GlaxoSmithKline, Janssen Research and Development, LLC, Lupus Foundation of America, Lupus Research Alliance, Merck Sharp & Dohme Corp., National Institute of Allergy and Infectious Diseases, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Pfizer Inc., Rheumatology Research Foundation, Sanofi and Takeda Pharmaceuticals International, Inc.) created to develop new ways of identifying and validating promising biological targets for diagnostics and drug development Funding for AMP was provided through grants from the National Institutes of Health (UH2-AR067676, UH2-AR067677, UH2-AR067679, UH2-AR067681, UH2-AR067685, UH2- AR067688, UH2-AR067689, UH2-AR067690, UH2-AR067691, UH2-AR067694, and UM2- AR067678). D. Simmons was supported by Institutional Training Grant NIH NHLBI T32 HL007627 and NIH NIAMS K08 AR075850.
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability:
RNA-seq of sorted monocyte populations with high and low expression of SLAMF7 are available at NCBI GEO GSE185508. RNA-seq from in vitro stimulated monocyte-derived macrophages is available at NCBI GEO GSE185509.
References and Notes:
- 1.Shi C, Pamer EG, Monocyte recruitment during infection and inflammation. Nature reviews. Immunology. 11, 762–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathan C, Ding A, Nonresolving Inflammation. Cell. 140 (2010), pp. 871–882. [DOI] [PubMed] [Google Scholar]
- 3.Mulherin D, Fitzgerald O, Bresnihan B, Synovial tissue macrophage populations and articular damage in rheumatoid arthritis. Arthritis and rheumatism. 39, 115–24 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Misharin A. v, Cuda CM, Saber R, Turner JD, Gierut AK, Haines GK, Berdnikovs S, Filer A, Clark AR, Buckley CD, Mutlu GM, Budinger GRS, Perlman H, Nonclassical Ly6C(−) monocytes drive the development of inflammatory arthritis in mice. Cell reports. 9, 591–604 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smiljanovic B, Grützkau A, Sörensen T, Grün JR, Vogl T, Bonin M, Schendel P, Stuhlmüller B, Claussnitzer A, Hermann S, Ohrndorf S, Aupperle K, Backhaus M, Radbruch A, Burmester GR, Häupl T, Synovial tissue transcriptomes of long-standing rheumatoid arthritis are dominated by activated macrophages that reflect microbial stimulation. Scientific reports. 10, 7907 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West NR, Hegazy AN, Owens BMJ, Bullers SJ, Linggi B, Buonocore S, Coccia M, Görtz D, This S, Stockenhuber K, Pott J, Friedrich M, Ryzhakov G, Baribaud F, Brodmerkel C, Cieluch C, Rahman N, Müller-Newen G, Owens RJ, Kühl AA, Maloy KJ, Plevy SE, S. Oxford IBD Cohort Investigators, Keshav S, Travis SPL, Powrie F, Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nature medicine. 23, 579–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, Sud M, Andrews E, Velonias G, Haber AL, Jagadeesh K, Vickovic S, Yao J, Stevens C, Dionne D, Nguyen LT, Villani A-C, Hofree M, Creasey EA, Huang H, Rozenblatt-Rosen O, Garber JJ, Khalili H, Desch AN, Daly MJ, Ananthakrishnan AN, Shalek AK, Xavier RJ, Regev A, Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell. 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin JC, Chang C, Boschetti G, Ungaro R, Giri M, Grout JA, Gettler K, Chuang L-S, Nayar S, Greenstein AJ, Dubinsky M, Walker L, Leader A, Fine JS, Whitehurst CE, Mbow ML, Kugathasan S, Denson LA, Hyams JS, Friedman JR, Desai PT, Ko HM, Laface I, Akturk G, Schadt EE, Salmon H, Gnjatic S, Rahman AH, Merad M, Cho JH, Kenigsberg E, Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell. 178, 1493–1508.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, Fernandez R, Akbarpour M, Chen C-I, Ren Z, Verma R, Abdala-Valencia H, Nam K, Chi M, Han S, Gonzalez-Gonzalez FJ, Soberanes S, Watanabe S, Williams KJN, Flozak AS, Nicholson TT, Morgan VK, Winter DR, Hinchcliff M, Hrusch CL, Guzy RD, Bonham CA, Sperling AI, Bag R, Hamanaka RB, Mutlu GM, v Yeldandi A, Marshall SA, Shilatifard A, Amaral LAN, Perlman H, Sznajder JI, Argento AC, Gillespie CT, Dematte J, Jain M, Singer BD, Ridge KM, Lam AP, Bharat A, Bhorade SM, Gottardi CJ, Budinger GRS, v Misharin A, Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 199, 1517–1536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morse C, Tabib T, Sembrat J, Buschur KL, Bittar HT, Valenzi E, Jiang Y, Kass DJ, Gibson K, Chen W, Mora A, v Benos P, Rojas M, Lafyatis R, Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. The European respiratory journal. 54, 1802441 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizoguchi F, Slowikowski K, Wei K, Marshall JL, Rao DA, Chang SK, Nguyen HN, Noss EH, Turner JD, Earp BE, Blazar PE, Wright J, Simmons BP, Donlin LT, Kalliolias GD, Goodman SM, Bykerk VP, Ivashkiv LB, Lederer JA, Hacohen N, Nigrovic PA, Filer A, Buckley CD, Raychaudhuri S, Brenner MB, Functionally Distinct Disease-Associated Fibroblast Subsets in Rheumatoid Arthritis. Nature Communications. 9, 789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croft AP, Campos J, Jansen K, Turner JD, Marshall J, Attar M, Savary L, Wehmeyer C, Naylor AJ, Kemble S, Begum J, Dürholz K, Perlman H, Barone F, McGettrick HM, Fearon DT, Wei K, Raychaudhuri S, Korsunsky I, Brenner MB, Coles M, Sansom SN, Filer A, Buckley CD, Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 570, 246–251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo D, Ding J, Cohn IS, Zhang F, Wei K, Rao DA, Rozo C, Sokhi UK, Shanaj S, Oliver DJ, Echeverria AP, DiCarlo EF, Brenner MB, Bykerk VP, Goodman SM, Raychaudhuri S, Rätsch G, Ivashkiv LB, Donlin LT, HBEGF+ macrophages in rheumatoid arthritis induce fibroblast invasiveness. Science translational medicine. 11, eaau8587 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao DA, Gurish MF, Marshall JL, Slowikowski K, Fonseka CY, Liu Y, Donlin LT, Henderson LA, Wei K, Mizoguchi F, Teslovich NC, Weinblatt ME, Massarotti EM, Coblyn JS, Helfgott SM, Lee YC, Todd DJ, Bykerk VP, Goodman SM, Pernis AB, Ivashkiv LB, Karlson EW, Nigrovic PA, Filer A, Buckley CD, Lederer JA, Raychaudhuri S, Brenner MB, Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 542, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, Goodman SM, Tabechian D, Hughes LB, Salomon-Escoto K, Watts GFM, Jonsson AH, Rangel-Moreno J, Meednu N, Rozo C, Apruzzese W, Eisenhaure TM, Lieb DJ, Boyle DL, Mandelin AM, Boyce BF, DiCarlo E, Gravallese EM, Gregersen PK, Moreland L, Firestein GS, Hacohen N, Nusbaum C, Lederer JA, Perlman H, Pitzalis C, Filer A, Holers VM, Bykerk VP, Donlin LT, Anolik JH, Brenner MB, Raychaudhuri S, Raychaudhuri S, B. F. Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium, Boyce BF, DiCarlo E, Gravallese EM, Gregersen PK, Moreland L, Firestein GS, Hacohen N, Nusbaum C, Lederer JA, Perlman H, Pitzalis C, Filer A, Holers VM, Bykerk VP, Donlin LT, Anolik JH, Brenner MB, Raychaudhuri S, Defining Inflammatory Cell States in Rheumatoid Arthritis Joint Synovial Tissues by Integrating Single-Cell Transcriptomics and Mass Cytometry. Nature immunology. 20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haringman JJ, Gerlag DM, Zwinderman AH, Smeets TJM, Kraan MC, Baeten D, McInnes IB, Bresnihan B, Tak PP, Synovial tissue macrophages: a sensitive biomarker for response to treatment in patients with rheumatoid arthritis. Annals of the rheumatic diseases. 64, 834–8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandelin AM, Homan PJ, Shaffer AM, Cuda CM, Dominguez ST, Bacalao E, Carns M, Hinchcliff M, Lee J, Aren K, Thakrar A, Montgomery AB, Louis Bridges S, Bathon JM, Atkinson JP, Fox DA, Matteson EL, Buckley CD, Pitzalis C, Parks D, Hughes LB, Geraldino-Pardilla L, Ike R, Phillips K, Wright K, Filer A, Kelly S, Ruderman EM, Morgan V, Abdala-Valencia H, Misharin A. v, Budinger GS, Bartom ET, Pope RM, Perlman H, Winter DR, Bridges SL, Bathon JM, Atkinson JP, Fox DA, Matteson EL, Buckley CD, Pitzalis C, Parks D, Hughes LB, Geraldino-Pardilla L, Ike R, Phillips K, Wright K, Filer A, Kelly S, Ruderman EM, Morgan V, Abdala-Valencia H, v Misharin A, Budinger GS, Bartom ET, Pope RM, Perlman H, Winter DR, Transcriptional Profiling of Synovial Macrophages using Minimally Invasive Ultrasound-Guided Synovial Biopsies in Rheumatoid Arthritis. Arthritis & rheumatology (Hoboken, N.J.). 70, 841–854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alivernini S, MacDonald L, Elmesmari A, Finlay S, Tolusso B, Gigante MR, Petricca L, di Mario C, Bui L, Perniola S, Attar M, Gessi M, Fedele AL, Chilaka S, Somma D, Sansom SN, Filer A, McSharry C, Millar NL, Kirschner K, Nerviani A, Lewis MJ, Pitzalis C, Clark AR, Ferraccioli G, Udalova I, Buckley CD, Gremese E, McInnes IB, Otto TD, Kurowska-Stolarska M, Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis. Nature Medicine (2020), doi: 10.1038/s41591-020-0939-8. [DOI] [PubMed] [Google Scholar]
- 19.Coates BM, Staricha KL, Koch CM, Cheng Y, Shumaker DK, Budinger GRS, Perlman H, Misharin A. v., Ridge KM, Inflammatory monocytes drive influenza A virus-mediated lung injury in juvenile mice. Journal of immunology (Baltimore, Md. : 1950). 200, 2391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z, Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nature medicine (2020), doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 21.Merad M, Martin JC, Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nature reviews. Immunology. 20, 355–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, Xing F, Liu J, Yip CC-Y, Poon RW-S, Tsoi H-W, Lo SK-F, Chan K-H, Poon VK-M, Chan W-M, Ip JD, Cai J-P, Cheng VC-C, Chen H, Hui CK-M, Yuen K-Y, A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet (London, England). 395, 514–523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team, A Novel Coronavirus from Patients with Pneumonia in China, 2019. The New England journal of medicine. 382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue J, v Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, de Nardo D, Gohel TDD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MRR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TCC, Ulas T, Schultze JLL, de Nardo D, Gohel TDD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MRR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TCC, Ulas T, Schultze JLL, Transcriptome-based Network Analysis Reveals a Spectrum Model of Human Macrophage Activation. Immunity. 40, 274–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim EY, Ner-Gaon H, Varon J, Cullen AM, Guo J, Choi J, Barragan-Bradford D, Higuera A, Pinilla-Vera M, Short SA, Arciniegas-Rubio A, Tamura T, Leaf DE, Baron RM, Shay T, Brenner MB, Post-sepsis immunosuppression depends on NKT cell regulation of mTOR/IFN-γ in NK cells. The Journal of clinical investigation. 130, 3238–3252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrt S, Schnappinger D, Bekiranov S, Drenkow J, Shi S, Gingeras TR, Gaasterland T, Schoolnik G, Nathan C, Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. The Journal of experimental medicine. 194, 1123–40 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosser DM, Edwards JP, Exploring the Full Spectrum of Macrophage Activation. Nature Reviews Immunology. 8, 958–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Held TK, Weihua X, Yuan L, v Kalvakolanu D, Cross AS, Gamma interferon augments macrophage activation by lipopolysaccharide by two distinct mechanisms, at the signal transduction level and via an autocrine mechanism involving tumor necrosis factor alpha and interleukin-1. Infection and immunity. 67, 206–12 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C, Xue Y, Wang P, Lin L, Liu Q, Li N, Xu J, Cao X, IFN-γ primes macrophage activation by increasing phosphatase and tensin homolog via downregulation of miR-3473b. Journal of immunology (Baltimore, Md. : 1950). 193, 3036–44 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Su X, Yu Y, Zhong Y, Giannopoulou EG, Hu X, Liu H, Cross JR, Rätsch G, Rice CM, Ivashkiv LB, Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nature immunology. 16, 838–849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latz E, Xiao TS, Stutz A, Activation and regulation of the inflammasomes. Nature reviews. Immunology. 13, 397–411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouchon A, Cella M, Grierson HL, Cohen JI, Colonna M, Activation of NK cell-mediated cytotoxicity by a SAP-independent receptor of the CD2 family. Journal of immunology (Baltimore, Md. : 1950). 167, 5517–21 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Kumaresan PR, Lai WC, Chuang SS, Bennett M, Mathew PA, CS1, a novel member of the CD2 family, is homophilic and regulates NK cell function. Molecular immunology. 39, 1–8 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Murphy JJ, Hobby P, Vilarino-Varela J, Bishop B, Iordanidou P, Sutton BJ, Norton JD, A novel immunoglobulin superfamily receptor (19A) related to CD2 is expressed on activated lymphocytes and promotes homotypic B-cell adhesion. The Biochemical journal. 361, 431–6 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karampetsou MP, Comte D, Kis-Toth K, Kyttaris VC, Tsokos GC, Expression patterns of signaling lymphocytic activation molecule family members in peripheral blood mononuclear cell subsets in patients with systemic lupus erythematosus. PloS one. 12, e0186073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Salort J, Sintes J, Llinàs L, Matesanz-Isabel J, Engel P, Expression of SLAM (CD150) cell-surface receptors on human B-cell subsets: from pro-B to plasma cells. Immunology letters. 134, 129–36 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Veillette A, SLAM-family receptors: immune regulators with or without SAP-family adaptors. Cold Spring Harbor perspectives in biology. 2, a002469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Z, Gu M, Jia X, Wang X, Wu C, Guo J, Zhang L, Du Y, Wang J, Integrated DNA methylation and gene expression analysis identifies SLAMF7 as a key regulator of atherosclerosis. Aging. 10, 1324–1337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maekawa T, Kato S, Kawamura T, Takada K, Sone T, Ogata H, Saito K, Izumi T, Nagao S, Takano K, Okada Y, Tachi N, Teramoto M, Horiuchi T, Hikota-Saga R, Endo-Umeda K, Uno S, Osawa Y, Kobayashi A, Kobayashi S, Sato K, Hashimoto M, Suzu S, Usuki K, Morishita S, Araki M, Makishima M, Komatsu N, Kimura F, Increased SLAMF7high monocytes in myelofibrosis patients harboring JAK2V617F provide a therapeutic target of elotuzumab. Blood. 134, 814–825 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Kim JR, Mathew SO, Mathew PA, Blimp-1/PRDM1 regulates the transcription of human CS1 (SLAMF7) gene in NK and B cells. Immunobiology. 221, 31–9 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Viola A, Munari F, Sánchez-Rodríguez R, Scolaro T, Castegna A, The Metabolic Signature of Macrophage Responses. Frontiers in immunology. 10, 1462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trizzino M, Zucco A, Deliard S, Wang F, Barbieri E, Veglia F, Gabrilovich D, Gardini A, EGR1 is a gatekeeper of inflammatory enhancers in human macrophages. Science advances. 7 (2021), doi: 10.1126/SCIADV.AAZ8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhat MY, Solanki HS, Advani J, Khan AA, Keshava Prasad TS, Gowda H, Thiyagarajan S, Chatterjee A, Comprehensive network map of interferon gamma signaling. Journal of cell communication and signaling. 12, 745–751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plosker GL, Ruxolitinib: a review of its use in patients with myelofibrosis. Drugs. 75, 297–308 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Hui L, Qi L, Guoyu H, Xuliang S, Meiao T, Ruxolitinib for treatment of steroid-refractory graft-versus-host disease in adults: a systematic review and meta-analysis. Expert review of hematology. 13, 565–575 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Kim JR, Horton NC, Mathew SO, Mathew PA, CS1 (SLAMF7) inhibits production of proinflammatory cytokines by activated monocytes. Inflammation research : official journal of the European Histamine Research Society... [et al. ]. 62, 765–72 (2013). [DOI] [PubMed] [Google Scholar]
- 47.O’Connell P, Pepelyayeva Y, Blake MK, Hyslop S, Crawford RB, Rizzo MD, Pereira-Hicks C, Godbehere S, Dale L, Gulick P, Kaminski NE, Amalfitano A, Aldhamen YA, O’Connell P, Pepelyayeva Y, Blake MK, Hyslop S, Crawford RB, Rizzo MD, Pereira-Hicks C, Godbehere S, Dale L, Gulick P, Kaminski NE, Amalfitano A, Aldhamen YA, SLAMF7 Is a Critical Negative Regulator of IFN-α-Mediated CXCL10 Production in Chronic HIV Infection. Journal of immunology (Baltimore, Md. : 1950). 202, 228–238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kikuchi J, Hori M, Iha H, Toyama-Sorimachi N, Hagiwara S, Kuroda Y, Koyama D, Izumi T, Yasui H, Suzuki A, Furukawa Y, Soluble SLAMF7 promotes the growth of myeloma cells via homophilic interaction with surface SLAMF7. Leukemia. 34, 180–195 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Friedman A, C/EBPalpha induces PU.1 and interacts with AP-1 and NF-kappaB to regulate myeloid development. Blood cells, molecules & diseases. 39, 340–343 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J, Li H, Xia X, Herrera A, Pollock N, Reebye V, Sodergren MH, Dorman S, Littman BH, Doogan D, Huang KW, Habib R, Blakey D, Habib NA, Rossi JJ, Anti-inflammatory Activity of MTL-CEBPA, a Small Activating RNA Drug, in LPS-Stimulated Monocytes and Humanized Mice. Molecular therapy : the journal of the American Society of Gene Therapy. 27, 999–1016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donado CA, Cao AB, Simmons DP, Croker BA, Brennan PJ, Brenner MB, A Two-Cell Model for IL-1β Release Mediated by Death-Receptor Signaling. Cell reports. 31, 107466 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez FO, Gordon S, Locati M, Mantovani A, Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. Journal of immunology (Baltimore, Md. : 1950). 177, 7303–11 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Kang K, Bachu M, Park SH, Kang K, Bae S, Park-Min K-H, Ivashkiv LB, IFN-γ selectively suppresses a subset of TLR4-activated genes and enhancers to potentiate macrophage activation. Nature communications. 10, 3320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu N, Veillette A, SLAM family receptors in normal immunity and immune pathologies. Current Opinion in Immunology. 38, 45–51 (2016). [DOI] [PubMed] [Google Scholar]
- 55.van Driel BJ, Liao G, Engel P, Terhorst C, Responses to Microbial Challenges by SLAMF Receptors. Frontiers in immunology. 7, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tassi I, Colonna M, The cytotoxicity receptor CRACC (CS-1) recruits EAT-2 and activates the PI3K and phospholipase Cgamma signaling pathways in human NK cells. Journal of immunology (Baltimore, Md. : 1950). 175, 7996–8002 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Chen J, Zhong M-C, Guo H, Davidson D, Mishel S, Lu Y, Rhee I, Pérez-Quintero L-A, Zhang S, Cruz-Munoz M-E, Wu N, Vinh DC, Sinha M, Calderon V, Lowell CA, Danska JS, Veillette A, SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature. 544, 493–497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joshi T, Butchar JP, Tridandapani S, Fcgamma receptor signaling in phagocytes. International journal of hematology. 84, 210–216 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Fodor S, Jakus X, Mócsai A, ITAM-based signaling beyond the adaptive immune response. Immunology letters. 104, 29–37 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Mócsai A, Ruland J, Tybulewicz VLJ, The SYK tyrosine kinase: a crucial player in diverse biological functions. Nature reviews. Immunology. 10, 387–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yi YS, Son YJ, Ryou C, Sung GH, Kim JH, Cho JY, Functional roles of Syk in macrophage-mediated inflammatory responses. Mediators of inflammation. 2014 (2014), doi: 10.1155/2014/270302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gane JM, Stockley RA, Sapey E, TNF-α Autocrine Feedback Loops in Human Monocytes: The Pro- and Anti-Inflammatory Roles of the TNF-α Receptors Support the Concept of Selective TNFR1 Blockade In Vivo. Journal of immunology research. 2016, 1079851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacDonald L, Alivernini S, Tolusso B, Elmesmari A, Somma D, Perniola S, Paglionico A, Petricca L, Bosello SL, Carfì A, Sali M, Stigliano E, Cingolani A, Murri R, Arena V, Fantoni M, Antonelli M, Landi F, Franceschi F, Sanguinetti M, McInnes IB, McSharry C, Gasbarrini A, Otto TD, Kurowska-Stolarska M, Gremese E, COVID-19 and RA share an SPP1 myeloid pathway that drives PD-L1+ neutrophils and CD14+ monocytes. JCI insight. 6 (2021), doi: 10.1172/JCI.INSIGHT.147413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang F, Mears JR, Shakib L, Beynor JI, Shanaj S, Korsunsky I, Nathan A, Donlin LT, Raychaudhuri S, IFN-γ and TNF-α drive a CXCL10+ CCL2+ macrophage phenotype expanded in severe COVID-19 lungs and inflammatory diseases with tissue inflammation. Genome medicine. 13, 64 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malaer JD, Mathew PA, CS1 (SLAMF7, CD319) is an effective immunotherapeutic target for multiple myeloma. American journal of cancer research. 7, 1637–1641 (2017). [PMC free article] [PubMed] [Google Scholar]
- 66.Pazina T, James AM, MacFarlane AW, Bezman NA, Henning KA, Bee C, Graziano RF, Robbins MD, Cohen AD, Campbell KS, The anti-SLAMF7 antibody elotuzumab mediates NK cell activation through both CD16-dependent and -independent mechanisms. Oncoimmunology. 6, e1339853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurdi AT, Glavey S. v., >Bezman NA, Jhatakia A, Guerriero JL, Manier S, Moschetta M, Mishima Y, Roccaro A, Detappe A, Liu C-J, Sacco A, Huynh D, Tai Y-T, Robbins MD, Azzi J, Ghobrial IM, Antibody-Dependent Cellular Phagocytosis by Macrophages is a Novel Mechanism of Action of Elotuzumab. Molecular Cancer Therapeutics. 17(7), 1454–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woo J, Vierboom MP, Kwon H, Chao D, Ye S, Li J, Lin K, Tang I, Belmar NA, Hartman T, Breedveld E, Vexler V, ‘t Hart BA, Law DA, Starling GC, PDL241, a novel humanized monoclonal antibody, reveals CD319 as a therapeutic target for rheumatoid arthritis. Arthritis Research & Therapy. 15, R207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, Birnbaum NS, Burmester GR, Bykerk VP, Cohen MD, Combe B, Costenbader KH, Dougados M, Emery P, Ferraccioli G, Hazes JMW, Hobbs K, Huizinga TWJ, Kavanaugh A, Kay J, Kvien TK, Laing T, Mease P, Ménard HA, Moreland LW, Naden RL, Pincus T, Smolen JS, Stanislawska-Biernat E, Symmons D, Tak PP, Upchurch KS, Vencovský J, Wolfe F, Hawker G, 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis and rheumatism. 62, 2569–81 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Donlin LT, Rao DA, Wei K, Slowikowski K, McGeachy MJ, Turner JD, Meednu N, Mizoguchi F, Gutierrez-Arcelus M, Lieb DJ, Keegan J, Muskat K, Hillman J, Rozo C, Ricker E, Eisenhaure TM, Li S, Browne EP, Chicoine A, Sutherby D, Noma A, Nusbaum C, Kelly S, Pernis AB, Ivashkiv LB, Goodman SM, Robinson WH, Utz PJ, Lederer JA, Gravallese EM, Boyce BF, Hacohen N, Pitzalis C, Gregersen PK, Firestein GS, Raychaudhuri S, Moreland LW, Holers VM, Bykerk VP, Filer A, Boyle DL, Brenner MB, Anolik JH, Methods for high-dimensional analysis of cells dissociated from cryopreserved synovial tissue. Arthritis Research & Therapy. 20, 139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.S P, ÅK B, OR F, S S, G W, R S, Smart-seq2 for Sensitive Full-Length Transcriptome Profiling in Single Cells. Nature methods. 10 (2013), doi: 10.1038/NMETH.2639. [DOI] [PubMed] [Google Scholar]
- 72.Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R, Full-length RNA-seq from single cells using Smart-seq2. Nature protocols. 9, 171–81 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, Regev A. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Current protocols in molecular biology. 107:4.22.1–17 (2014). doi: 10.1002/0471142727.mb0422s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England). 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li B, Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu A, Ibrahim JG, Love MI, Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics (Oxford, England). 35, 2084–2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification. Nature biotechnology. 34, 525–7 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Korotkevich G, Sukhov V, Sergushichev A, Fast gene set enrichment analysis. bioRxiv, 060012 (2019). [Google Scholar]
- 80.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell. 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, Raychaudhuri S, Fast, sensitive and accurate integration of single-cell data with Harmony. Nature methods. 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes in the “Inflamed Arthritis Signature.”
List of genes in the “SLAMF7-High Signature.”
List of genes in the “SLAMF7 Stimulation Signature.”
Figure S1. Gating for flow cytometric analysis of macrophages.
Figure S2. CD84 expression in synovial macrophages
Figure S3. Gating strategy for cell sorting.
Figure S4. Interferon regulation of surface marker expression on macrophages.
Figure S5. SLAMF7 is necessary for macrophage activation by r-SLAMF7
Figure S6. Validation of reagents used for SLAMF7 activation.
Figure S7. Inflammatory cascade driven by SLAMF7 engagement.
Figure S8. FCER1G and TYROBP are highly expressed by macrophages.
Figure S9. Silencing of TNFRSF1A and TNFRSF1B with siRNA.
Figure S10. Macrophage populations in synovial tissue from healthy controls or patients with untreated rheumatoid arthritis.
Figure S11. Evidence for SLAMF7 activation in macrophages from patients with untreated rheumatoid arthritis.
Figure S12. High SLAMF7 activation in ileal macrophages from samples with high GIMATS module intensity scores.
Figure S13. Macrophage populations in ileal tissue from patients with inflammatory bowel disease.
Figure S14. Evidence for SLAMF7 activation in macrophages from patients with inflammatory bowel disease.
Figure S15. Macrophage populations in bronchoalveolar lavage from patients with COVID-19.
Figure S16. Evidence for SLAMF7 activation in macrophages from patients with COVID-19 infection.
Figure S17. Model for SLAMF7 super-activation of macrophages.
Raw data file
Data Availability Statement
RNA-seq of sorted monocyte populations with high and low expression of SLAMF7 are available at NCBI GEO GSE185508. RNA-seq from in vitro stimulated monocyte-derived macrophages is available at NCBI GEO GSE185509.


