Highlights
-
•
Sequence-based database Nexplorer describing EU-authorized GMOs was developed.
-
•
Sequences were annotated and presented in structured and extractable formats.
-
•
Workflow for an efficient analysis of NGS data using Nexplorer database was designed.
-
•
Method was successfully tested for various scenarios for routine GMO analysis.
-
•
This paves the way for the use of NGS for routine GMO detection and identification.
Keywords: GMO detection, GMO identification1, NGS, GMO database, Data analysis workflow
Abstract
The increasing number and diversity of genetically modified organisms (GMOs) for the food and feed market calls for the development of advanced methods for their detection and identification. This issue can be addressed by next generation sequencing (NGS). However, the efficiency of NGS-based strategies depends on the availability of bioinformatic methods to find sequences of the transgenic insert and junction regions, which is a challenging topic. To facilitate this task, we have developed Nexplorer, a sequence-based database in which annotated sequences of GM events are stored in a structured, searchable and extractable format. As a proof of concept, we have developed a methodology for the analysis of sequencing data of DNA walking libraries of samples containing GMOs using the database. The efficiency of the method has been tested on datasets representing various scenarios that can be encountered in routine GMO analysis. Database-guided analysis allowed obtaining detailed and reliable information with limited hands-on time. As the database allows for efficient analysis of NGS data, it paves the way for the use of NGS sequencing technology to aid routine detection and identification of GMO.
1. Introduction
1.1. Background
In order to respect freedom-of-choice for consumers, and protect public health and the environment, introduction and the presence of genetically modified organisms (GMOs) in foods and feed chains in the European market are subject to strict regulation (EU Regulations No. 1829/2003 and 1830/2003). Within this regulatory framework, a distinction is made between GMOs authorized for food and feed that must be traceable by labelling when their content is above the EU threshold (i.e. 0.9% relative to mass), and unauthorized GMOs which are subject to zero tolerance. An important component in the enforcement of this regulation is efficient detection, identification and quantification of GMOs in food and feed products.
The current GMO screening and detection approaches are most often based on quantitative polymerase chain reaction (qPCR). The standard workflow in routine GMO detection analysis is to carry out a first-line screening for the presence/absence of taxon-specific sequences and genetic elements that are commonly encountered in the transgenic constructs, such as cauliflower mosaic virus (CaMV) 35S promoter (P-35S) and nos-terminator from Agrobacterium tumefaciens (T-nos) (Fraiture et al., 2015). This screening is designed to ensure a more rapid and cost-efficient detection of samples potentially containing one or several GMOs by covering at least all the authorized GMOs. This approach provides clues on GMO identity, limiting the spectrum of possible transgenic events to identify in the second step. On the basis of the screening results, a list of potential EU-authorized GM events that may be present in the sample is composed and the corresponding event-specific qPCR methods targeting authorized GMO are applied for their subsequent identification and quantification (Fraiture et al., 2015). GMO screening and detection approaches applied in routine are required to show a satisfactory limit of detection: according to the method performance requirements, the sensitivity of a qPCR method must be at least 25 haploid genome equivalents (HGE) (Mazzara et al., 2008).
The described qPCR-based system has a number of flaws, especially concerning the unauthorized GMOs. Even when an unexplained positive signal is observed during the qPCR screening, it does not unambiguously indicate the presence of an unauthorized event because this signal can also be caused by the presence of the naturally occurring organism in the sample (Bak and Emerson, 2019, Broeders et al., 2012, Holst-Jensen et al., 2012). Furthermore, the presence of an unauthorized event can be concealed by the presence of authorized events harboring the same transgenic elements in the mixture. Moreover, due to the increasing number and diversity of GMO (ISAAA, 2019, EU regulation 1830/2003), the number of PCR analyses that need to be carried out for each sample is increasing, rendering the routine qPCR-based screening procedures less time- and labor-efficient and more costly to perform. Finally, for the design of new broad-range qPCR methods used in first line screening accurate sequence information is required. While numerous GMO databases have been created to keep track of the increasing diversity of transgenic events, most of these do not include sequence information. The transgenic elements that are grouped under the same name can, however, demonstrate a high variability in terms of nucleotide sequence. For example, the length of the P-35S promoter encountered in GM constructs can vary between 300 and 1400 base pairs (Podevin & Du Jardin, 2012), which limits the usefulness of the non-sequence-based databases for qPCR method development. These considerations indicate the need for more open and informative approaches for GMO detection (Fraiture et al., 2015).
1.2. Approach
One solution to the previously described methodological issues could be the use of next generation sequencing (NGS) combined with an appropriate database containing not only the name of GM elements but also their sequences. However, at the present time, both NGS and the available databases encounter bottlenecks. It is currently still costly to obtain the NGS sequencing depth necessary for detection of low GM contents, such as encountered in mixtures (Wang et al., 2020, Willems et al., 2016). Therefore, an approach based on initial enrichment of the targets using DNA walking starting from transgenic elements covering a large spectrum of GMO (P-35S, T-nos, the CaMV 35S terminator from the pCAMBIA vector (T-35S pCambia), and cry genes) has been proposed to characterize unknown sequences including the transgenic inserts and junctions (Fraiture et al., 2017, Fraiture et al., 2018, Liang et al., 2014). In the proposed methodology, DNA walking is followed by long-read sequencing such as Pacific Biosciences’ single-molecule real-time sequencing (further referred to as PacBIO) and Oxford Nanopore Technologies’ MinION sequencing (further referred to as MinION). While this methodology showed to be highly efficient for the detection and identification of GMO, the analysis of shotgun sequencing data from a DNA walking library consisting of multiple amplicons appeared challenging. In previous studies, analysis of DNA walking libraries sequencing data has been performed manually (Fraiture et al., 2017), or by a semi-automated strategy (Fraiture et al., 2018) based on the publicly available nucleotide (nt) database from the National Center for Biotechnology Information (NCBI) (Coordinators, 2016). The manual annotation of the data appeared to be very tedious and time-consuming. The semi-automated approach based on a publicly available database was more efficient, as the sequences were sorted based on produced hits with the plant- and non-plant sections of NCBI. The data interpretation remained complex and labor-intensive however because information on each new hit needed to be retrieved from the description of the subject sequences from NCBI. Therefore, we concluded that this type of approach would benefit from the availability of a database containing structured and annotated sequence information of known GMOs (Fraiture et al., 2018).
Although multiple databases containing organized and searchable information on GMOs exist, they are based on transgenic element names instead of nucleotide sequences, limiting their usability for both the development of qPCR-based detection methods as described previously and for the analysis of NGS data (Gerdes et al., 2012, Morisset et al., 2014). To our knowledge, only two GMO databases currently offer sequence information on GMOs namely, the Joint Research Centre (JRC) GMO-Amplicons (Petrillo et al., 2015) and the Euginius (https://euginius.eu) databases. The JRC GMO-Amplicons database contains putative GMO related sequences found by applying existing PCR-based detection methods in silico to screen numerous public nucleotide sequence databases, including patents and available whole plant genomes (Petrillo et al., 2015). The main aim of this database is to allow validation of new qPCR GMO detection methods and verification of existing methods. It includes an Amplicon finder interface designed to list sequences that are detected by various element-, construct- and event-specific methods, and a Blast amplicons interface, where sequences can be blasted against the JRC GMO-Amplicons datasets. Since no description of the amplicons is provided by the database, and no information is readily available about the position of event-specific regions and regions corresponding to transgenic elements, the obtained results cannot be interpreted directly except in the case where a full-length hit is observed. The Euginius database is designed to store accurate information on GMO events, and qPCR detection methods. It allows to perform complex searches, and has powerful functionalities for processing results of various detection methods, including the obtained amplicon sequences. For a subset of transgenic events, a list of associated sequences is provided in the database, e.g. sequences from patents and from NCBI, as well as amplicon sequences generated by various detection methods. However, in most cases the annotation of the sequences is limited, and no link is made with the transgenic element list provided for each event. That the provided information is partial and unharmonized prevents its use in a NGS data analysis pipeline.
Thus, despite that for a substantial number of transgenic events sequencing information can be found in both the corresponding patents and scientific studies, and the described databases, ordered and annotated sequencing information is absent from the public domain for most existing GMOs. Neither is the sequencing data stored in the databases easy to use for identification of GM elements or events using NGS data, because of the lack of standardization and detailed information on the sequences.
Therefore, in the current work, we have collected publicly available sequences of all GMOs authorized in the EU, annotated them by indicating the positions of transgenic elements, and stored these in a created database named Nexplorer. This database was used for the analysis of long-read sequencing data (both PacBio and MinION) of various samples coming from routine GMO analysis, enriched by DNA walking as a case study, demonstrating the benefits of this approach for detection and identification of authorized and unauthorized GMOs using NGS.
2. Materials and methods
The datasets used in this study (Table 1) were described in Fraiture et al., 2017, Fraiture et al., 2018 in which details on samples, DNA extraction methods and sequencing can be found. The Nexplorer database is publicly available at https://nexplorer.sciensano.be in the form of a web-application, and contains data from public sources on 70 transgenic events authorized on the EU market. First the sequencing datasets will be laid out, after which we discuss the different phases of the data analysis.
Table 1.
Datasets used for the five case studies as possible scenario occurring in routine GMO analysis.
| Sample name | Sample content | Sample type | Study case | NGS Platform |
|---|---|---|---|---|
| Bt rice 100% | Bt rice*,# 100% (200000 HGE) | DNA walking (p35S, tNOS, t35S pCAMBIA) | 1,2 | PacBio |
| Bt rice 1% | Bt rice*,# 1% (2000 HGE) | DNA walking (p35S, tNOS, t35S pCAMBIA) | 3 | PacBio |
| Bt rice 0.1% | Bt rice*,# 0.1% (200 HGE) | DNA walking (p35S, tNOS, t35S pCAMBIA) | 3 | PacBio |
| Bt rice 0.01% | Bt rice*,# 0.01% (20 HGE) | DNA walking (p35S, tNOS, t35S pCAMBIA) | 3 | PacBio |
| Bt noodles 100% | Noodles made from Bt rice*,# 100% | DNA walking (p35S, tNOS, t35S pCAMBIA) | 3 | PacBio |
| Bt noodles 1% | Noodles made from Bt rice*,# 1% | DNA walking (p35S, tNOS, t35S pCAMBIA) | 3 | PacBio |
| Bt rice MinION 100% | Bt rice*,# 100% (200000 HGE) | DNA walking (p35S, tNOS, t35S pCAMBIA) | 3 | MinION |
| Kuwaiti matrix | Food matrix from Kuwaiti market: NK603 and DAS1507 at quantifiable levels, MON810 and Bt11 at trace levels£ | DNA walking (p35S, tNOS, t35S pCAMBIA) | 4 | PacBio |
| Mixture 1 | Bt rice* (2000 HGE) + MON863 (2000 HGE) | DNA walking (p35S, tNOS, t35S pCAMBIA) | 5 | PacBio |
| Mixture 2 | Bt rice* (2000 HGE) + MON863 (2000 HGE) + GTS-40–3-2 (2000 HGE) | DNA walking (p35S, tNOS, t35S pCAMBIA) | 5 | PacBio |
| Mixture 3 | Bt rice* (20 HGE) + MON863 (20 HGE) + GTS-40–3-2 (20 HGE) | DNA walking (p35S, tNOS, t35S pCAMBIA) | 5 | PacBio |
HGE: haploid genome equivalents. Detailed description on the creation of the datasets can be found in (Fraiture et al., 2017). Bt rice may act as a known (#) or unknown (*) GMO depending on the version of the Nexplorer database that is used for the data analysis. All other events represent known GMOs, £according to qPCR analysis described in (Fraiture et al., 2017).
2.1. Sequencing datasets preparation
The dataset sequenced previously using MinION technology (Fraiture et al., 2018) was adapter trimmed with Porechop 0.2.2 (https://github.com/rrwick/Porechop), minimal trim size 4 bp, middle threshold 0.75, minimal read length 100 bp). The PacBio datasets (Fraiture et al., 2017) were adapter-trimmed by the sequencing provider. DNA walking adapters were removed using Cutadapt 2.10 (M. Martin, 2011) (error rate 0.30 for MinION and 0.15 for PacBio, minimal read length 100 bp, allowing the removal of multiple adapter occurrences within a single read).
2.2. Data analysis
2.2.1. Phase I
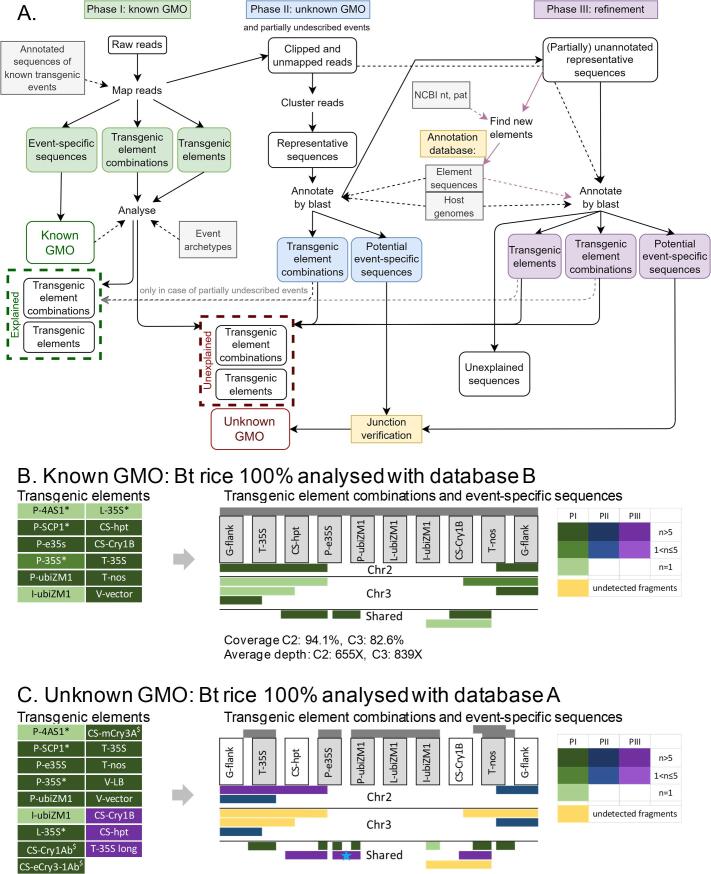
Depending on the data analysis needs, two versions of the Nexplorer database were used, namely version A corresponding to the database as it was available online on October 7th, 2021, and version B corresponding to the same database, upgraded with Bt rice. During phase I of the analysis (Fig. 1A), adapter trimmed reads were mapped to a reference library, consisting of all sequences from the Nexplorer database (127 sequences for version B that included the Bt rice, and 125 sequences for version A that lacked Bt rice) using bwa mem 0.7.17 (-k14 -W 40 -r10 -A1 -B2 –O2 -E 1 -L0 -T 20 -M -Y) (H. Li & Durbin, 2009). The reference library contains annotated sequences of known transgenic events, including full or partial transgenic inserts and in some cases regions of transgenic vectors that were used for transformation. The obtained bam files were processed in R 3.5, using the GenomicAlignments package (Lawrence et al., 2013) to calculate the coverage for each sequence, and ggplot2 (Villanueva & Chen, 2019) to make the coverage plots. The coverage plots were used to record the presence of individual genetic elements, genetic element combinations and event-specific sequences in each dataset. IGV genome viewer (Thorvaldsdóttir, Robinson, & Mesirov, 2013) was used to verify whether the elements effectively occurred on the same reads. The presence of event-specific sequences in the dataset was interpreted as proof for the presence of the corresponding events in the sample. To verify which of the observed elements and element combinations could be explained by the presence of detected events, the event archetypes in the Nexplorer database were consulted. The event archetypes describe the transgenic inserts found in an event by providing the identity and the order of the transgenic elements.
Fig. 1.
Data analysis workflow for detection of known and unknown GMO in DNA walking data. (A) Panel A shows the data analysis workflow. In the first phase, long-read sequencing data is aligned to the annotated sequences of known GMOs from the Nexplorer database, listing all the elements, element combinations and event-specific sequences such as junctions between the transgenic insert and host plant genome that are represented in the sample. Based on the observed event-specific sequences, a list is made of all the events whose presence in a sample is confirmed (Known GMO). The events archetypes (which describe the transgenic elements that are located on an insert in the correct order) are consulted in the database to verify whether all of the observed elements and element combinations can be explained by the presence of detected events. Unexplained elements and element combinations can originate either from known GMOs for which no event-specific sequences were detected in the sample, or from unknown GMOs. During the second phase, which aims at the detection of unknown GMOs, all reads that map partially (clipped reads) and unmapped reads are collected, clustered and annotated by blasting the representative reads of the clusters against the annotation database, consisting of host plant representative genomic sequences (Host genomes), sequences of transgenic elements extracted from the Nexplorer database prior to each analysis and clustered to remove redundant elements (Element sequences) and contaminating sequences such as PacBio internal control sequence and sequences of microorganisms (Contaminants). This phase is performed in case the sample contains clipped or unmapped reads, independent of whether unexplained transgenic elements were observed to avoid potential masking of unknown GMOs by known GMOs with the same elements. Blast results are visualized, allowing to quickly identify the element combinations and potential event-specific sequences that were not observed in phase I, and thus possibly belong to unknown GMOs. The sequences containing potential transgenic junction regions are redirected to the junction verification step. Besides the unknown GMO, phase II allows to reconstruct sequences of known events for which the insert sequence is only partially represented in the database. In the third, optional phase, the obtained annotation results are refined. Representative sequences that were not, or only partially annotated in the previous step are blasted to NCBI nt and pat (patent) databases, and the annotation database is extended with the newly identified transgenic elements and contaminating sequences. The annotation step is repeated with the newly extended annotation database which allows to detect most transgenic elements and element combinations that are not included in the database, and to describe junction regions if the insert portion of the junction does not contain any elements and consists of e.g. transgenic vector sequence. Exceptionally, clipped reads of interest can be extracted and annotated individually, e.g. when all reads containing unexplained transgenic element(s) were discarded during clustering. (B, C) Panels B and C demonstrate the result obtained by applying the described data analysis methodology on rice grain sample containing Bt rice at 100%. The analysis was performed using version B of the database that contains Bt rice sequences (panel B) to represent a scenario where a sample contains a known GMO at 100%, and using version A of the database lacking Bt rice (panel C), representing a scenario where the sample contains an unknown GMO at 100%. Transgenic expression cassettes are shown schematically, elements are not drawn to scale. C2 and C3 stand for chromosomes 2 and 3 respectively. Grey bars above the cassettes indicate which elements and element combinations are present in each version of the database. Elements that are represented in the database that is used for the analysis are displayed in grey boxes. Colored bars below the expression cassette show the longest amplicons that were observed for the first time during phase I, II and III (PI, PII, PIII) of the analysis as described in A. The green/blue/purple colour gradient according to the color legend shows the number of reads (n) representing a given amplicon. Fragments that are known to be present in the sample but that could not be detected in the current analysis are colored yellow. Fragments that were manually reconstructed from reads (instead of clusters) during phase III are marked by a blue star. * Transgenic elements containing homologous regions with P-e35S. $ Transgenic elements containing homologous regions with CS-Cry1B. Transgenic elements are described in Supplementary File 3.
2.2.2. Phase II
The SamJdk module (version 23c0a5c2) from Jvarkit toolkit (Lindenbaum, 2015) was used for filtering the bam files, allowing to retrieve the unmapped reads and reads containing a clipped end of at least 20 bp. The reads were clustered to reduce dataset complexity by applying a two-step binning procedure similar to the one described in (Fraiture et al., 2018). In the first clustering round using CD-HIT 4.6.8 (W. Li & Godzik, 2006), a global alignment was carried out with a minimal sequence similarity of 85% and a minimal length similarity of 98% for PacBio data and with a minimal sequence similarity of 80% and a minimal length similarity of 98% for MinION data. Clusters of at least 5 reads for PacBio datasets and 25 reads for MinION datasets were retained, to exclude underrepresented sequences potentially corresponding to DNA walking library preparation and sequencing artefacts. To further reduce the number of sequences, the output of the first clustering step was subjected to a second clustering round applying a local alignment with a minimal sequence similarity of 85% and a minimal alignment length of the shortest read equaling 98% of its length for PacBio data and a minimal sequence similarity of 70% and a minimal alignment length of 90% for MinION data. Clusters were filtered retaining those containing at least 0.3% of the reads. This threshold was chosen based on the annotation of all clusters in each sample (see further) as the one allowing to discard most clusters from which the representative read is a sequencing or data preparation artifact, while keeping the majority of the informative sequences (Supplementary Table 1, Supplementary File 1).
Each of the obtained clusters, represented by the longest read, was annotated by blasting against a distinct set of blast databases, including a database of reference sequences of plant genomes (Glycine_max: GCF_000004515.5, Oryza sativa: GCF_001433935.1, Zea mays: GCF_000005005.2) and a database of transgenic elements, cumulatively referred to as the annotation database (Fig. 1A). The database of transgenic elements was constructed prior to each analysis using the sequences of the transgenic elements from the Nexplorer database, that were binned with CD-HIT (local alignment with at least 90% identity and 90% of the shortest sequence aligning to the longest sequence) to remove redundant element sequence versions. The nucleotide Blast was performed using blastn 2.7.1+ (Camacho et al., 2009) with a word size of 7, an e-value of 0.1, minimal sequence identity of 85% for PacBio and 70% for MinION, and limiting the number of HSPs to 2 for the host genome database and to 3 for the element sequence database to simplify the output. For the same reason, short hits to regions of T-35S, P-35S (and its derivatives), and T-nos corresponding to the primer sequences were masked. Blast hits were visualized using Alvis (S. Martin & Leggett, 2021), and analyzed for the presence of sequences containing previously undescribed element combinations. Notably, detailed examination of the clusters revealed that not all of them represented novel element combinations: some sequences were classified as clipped because of the fragments of DNA walking primers which failed to be removed during pre-processing of the reads. In case the presence of chimeric sequences was suspected based on occurrence of tandem or inverted repeats, a closer examination of the clusters was performed by mapping of the reads back to the representative sequence. Sequences that produced hits with both the host plant genome, and transgenic elements were extracted and analyzed in more detail to identify new junction regions (see further).
2.2.3. Phase III
The steps carried out during phase III (refinement) were different for each sample, depending on the remaining gaps in the obtained information. For most of the tested samples partially unannotated clusters from phase II were further annotated by blast against the nt database, adding the newly identified transgenic elements to the transgenic elements database from phase II. Some clusters appeared to comprise contaminating sequences, such as the PacBio internal control sequence. These sequences were added to a separate contaminants database, which is part of the annotation database. The annotation described in phase II was redone with the updated databases, and the procedure was repeated until all sequences were fully annotated. Another task that was occasionally performed during phase III was to extract and annotate individual reads. For instance, if all reads containing an unexplained transgenic element were discarded during the clustering procedure of phase II, such reads were in some cases manually extracted and annotated individually during phase III to allow a more complete reconstruction of the transgenic insert.
2.2.4. Verification of the junction regions
To find clusters containing the transgenic junction regions of potentially unknown GMOs, all annotated sequences that produced both hits to transgenic elements, and host plant genomic sequences, were analyzed in more detail. Therefore, the sequences were blasted back to the reference genome of the host plant. The arrangement of the potential junction regions on the chromosomes was visualized in Blast genome viewer, to verify whether the fragments aligned to a genomic region in a way that indicated the presence of a transgene insertion site (i.e. sequences aligning uniformly along two sides of the potential insertion site, with non-aligned overhangs facing towards the insertion site, Supplementary File 1, Bt rice 100% DbA).
3. Results and discussion
3.1. Design and construction of the Nexplorer database
The Nexplorer database is a relational SQL database accessible from a web-application with the same name (https://nexplorer.sciensano.be), containing annotated sequences of transgenic events along with related general information (Supplementary Fig. 1). The database has been filled with data from 70 transgenic events (among which 3 stacked events, i.e. those produced by conventional crossing of two or more GM events) that are authorized on the EU market, which simultaneously contain 89 transgenic inserts (Supplementary File 2). For each insert, we carried out an extensive search for the available sequence information in patent databases such as The Lens (https://lens.org) and Google Patents (https://patents.google.com), NCBI and JRC GMO-Amplicons, resulting in retrieval of 125 sequences of entire inserts or insert parts (sequence sources can be found in Supplementary File 2 as well as in the Nexplorer database). For 46 events a full sequence of all transgenic inserts was found, and for an additional 21 events the sequences of at least one junction region were retrieved along with a portion of the transgenic insert, and often of the transgenic vector used for transformation. The availability of the sequence of at least one transgenic junction for over 95% of the non-stacked events allows for quick and precise identification of a large number of events. The introduced sequences were annotated by indicating the positions of the transgenic elements and the genomic flanking regions. The database contains a total of 157 transgenic elements (e.g. promoters, terminators and coding sequences), whose positions are indicated on one, or several, sequences. This detailed annotation allows to retrieve the sequences of individual functional elements from one, or a selection of transgenic inserts, a feature foreseen to facilitate future qPCR method development and testing. In addition, the database web-application foresees an advanced search module and embeds a Blast (Camacho et al., 2009) functionality that is useful for the identification of amplicons obtained from various existing detection methods.
For demonstration purposes, this study uses two versions of the database. Version A corresponds to the database as it was available online on October 7th, 2021. Version B corresponds to the same database, upgraded with the sequences of Bt rice, an EU-unauthorized GMO described in (Fraiture et al., 2017). When data analyses are carried out with database A, the Bt rice (Fig. 2) that is present in the analyzed samples represents an unknown GMO, while with database B this event comprises a known GMO. The terms known and unknown GMO thus refer to GM events for which the sequence is respectively present or absent in the Nexplorer database.
Fig. 2.
Bt rice transgenic inserts in chromosomes 2 and 3, drawn to scale. In this event, two copies of the transgenic insert were introduced, at two different places. As a result, the internal portion of the inserts are the same except for a small length difference because of short truncations at the ends, while the junction regions are different. Each copy consists of two transgenic expression cassettes, arranged in a head-to-head orientation: one for the expression of the insect resistance gene CS-Cry1B, directed by the P-ubiZM1 core promoter along with the L-ubiZM1 (leader) and I-ubiZM1 (the first intron) and T-nos terminator, and one for the expression of the hygromycin resistance gene CS-hpt, directed by the P-e35S promoter (the P-35S promoter preceded by a duplicated L-35S enhancer) and the T-35S terminator. Transgenic elements are described in Supplementary File 3.
3.2. Study setup
To illustrate how annotated transgenic insert sequences from a sequence-based database can be used to extract information from long-read sequencing data generated from a DNA walking enrichment approach, we analyzed NGS datasets obtained from different sample types including (1) a known GMO at 100%: Bt rice that harbors two highly similar copies of a transgenic insert into chromosome 2 and 3 (Fig. 2), (2) an unknown GMO at 100%: the same Bt rice absent from the database, (3) a series of samples containing a single known or unknown GMO at different concentrations, (4) a mixture of known GMOs at different concentrations, and (5) a mixture of known GMOs and one unknown GMOs at different concentrations (Table 1). These study cases cover all the scenarios encountered when analyzing GMO samples in food and feed with an increasing degree of complexity. The first two study cases correspond to samples consisting entirely of an authorized GMO (known GMO at 100%) or an unauthorized GMO (unknown GMO at 100%), allowing to test the ability of the workflow to detect and identify these two types of events. The purpose of case study three is to benchmark the sensitivity of the workflow, and evaluate its suitability for the analysis of sequencing data obtained with an alternative long-read sequencing technology. This third case study was carried out with a series of datasets obtained from plant material and processed food samples where a known or unknown GMO is present at different, well-known concentrations. Additionally, it included one dataset generated from a sample containing a single GMO at 100% using the MinION sequencing technology instead of PacBio like all other samples in this study. The fourth and the fifth cases represent the more complex scenarios in which a sample consists of more than one event, either all authorized (i.e. mixture of known GMOs) or authorized and unauthorized (i.e. mixture of known and unknown GMOs). These last two cases are used to verify whether the applied data analysis workflow can detect and identify all events in a sample, and whether presence of authorized events would not impede the detection of an unauthorized GMO.
3.3. Analysis of long-read sequencing data generated by DNA walking enrichment approach
3.3.1. Overview of the data analysis phases
A detailed description of the three-phase data analysis workflow used in this study is provided in Fig. 1A. Briefly, in the first phase of the data analysis, which mainly aims at detection and identification of known transgenic events (Fig. 1A), the reads are mapped to the annotated transgenic insert sequences from the Nexplorer database. Sequence similarity between the reads and regions of transgenic inserts corresponding to genetic elements and transgenic junctions allows to list the transgenic elements, transgenic element combinations and event-specific sequences that are represented in the sample. Because of the high precision of the read mapping procedure, even a single read originating from an insert of a known event will be detected. Annotation of the sequences allows to readily decide whether the observed fragments are specific to a particular event, or could have originated from another GMO, including unknown events.
The second phase of the data analysis aims at identification of unknown transgenic events (Fig. 1A). Reads from unknown transgenic events, at least those which are sufficiently long, will not map fully, or at all against database references. That feature allows to differentiate them from the reads that originate from the known GMOs. During phase II such reads are extracted, clustered and filtered to reduce complexity of the dataset and discard sample preparation and sequencing artefacts. Obtained clusters are annotated using a set of known transgenic element sequences on the one hand, and the genomes of potential host plants for detection of flanks on the other hand. This strategy makes the proposed method suitable for the detection of unknown GMOs which may contain previously unobserved combinations of transgenic elements. In this case the unknown GMOs will not be masked by the known GMOs with similar transgenic elements.
The third phase is performed to complete the information which is missing from the results obtained in the first two phases (Fig. 1A). Sequences that could not be fully annotated during phase II are blasted against the NCBI nt and patent sequences (pat) databases (Coordinators, 2016), allowing to detect transgenic elements that are not represented in the Nexplorer database but can be found in the public databases. This step is important to make the system able to detect any GMO, including those with fully unknown transgenic elements. Additionally, during this phase, reads of interest can be extracted and annotated individually, for example when all reads containing unexplained transgenic element were discarded during clustering.
3.3.2. Scenario 1: sample containing a known GMO at 100%
For the simplest study case, i.e. that of a known GMO that is present in a sample at 100%, the PacBio sequencing data generated from a DNA walking library from a sample containing Bt rice at 100% (i.e. 200,000 haploid genome equivalents, HGE) were used (Table 1). The DNA walking originated from three sites present in the Bt rice insert: P-35S, T-nos, T-35S (pCAMBIA) as described in (Fraiture et al., 2017). To mimic the situation where Bt rice is a known GMO, we have used the version of the database that includes sequences of both transgenic inserts of this event (version B of the Nexplorer database).
Phase I of the data analysis performed with version B of the database revealed a large number of genetic elements that are typical for transgenic constructs (Fig. 1B). The dataset contained left and right flanking regions of both Bt rice inserts (in chromosome 2 and 3). These event-specific sequences confirmed the presence of Bt rice in the sample. As expected, the dataset also contained reads encoding transgenic element combinations from Bt rice (Fig. 1B). Analysis showed that some of the detected transgenic elements were not represented in the confirmed GM event (i.e. Bt rice), more specifically P-35S, P-4AS1, P-SCP1 and L-35S (Fig. 1B, detailed description of transgenic elements provided in Supplementary File 3). All of the additional elements, however, had homologous regions with P-e35S (i.e. the enhanced P-35S promoter that is used in the Bt rice expression cassettes), explaining their appearance in the dataset.
The sample contained 1.5% (3 8 6) unmapped reads, and 4.7% (1234) reads that mapped only partially to the reference sequences, further referred to as clipped reads (Supplementary Table 2). As unmapped and clipped reads can potentially belong to unknown GMOs, phase II of the analysis (detection of unknown GMOs) was performed. The filtering procedure yielded two clusters with a total of 331 reads (Supplementary Table 2). Annotation of the clusters showed that both clusters contained sequences from the internal portion of Bt rice transgenic inserts. Phase III of the analysis was not needed as both clusters from phase II could be fully annotated. Together these findings indicate that no unknown (unauthorized) GMO (i.e. GMO not included in the database) was present in the sample. The data analysis was completed in two hours.
This study case illustrates the efficacy of the proposed data analysis methodology for the detection of sequences from known (and hence present in the database) transgenic events in the sample. While interpretation of the results of phase I has been performed manually, this part of the procedure can potentially be fully automated in the future, on the condition that additional tests are performed to tune the thresholds applied during the automatic detection procedure.
3.3.3. Scenario 2: sample containing an unknown GMO at 100%
The same strategy as in the first scenario described above was subsequently used to analyze a sample in which an unknown GMO was present. To simulate that situation, the Bt rice 100% dataset was reanalyzed using the version of the database lacking the corresponding Bt rice sequences (version A).
Phase I performed with the version A of the database allowed to detect genetic elements that are often used in transgenic constructs, including P-e35S (and elements containing fragments thereof, more specifically P-35S, P-4AS1, P-SCP1 and L-35S), P-ubiZM1, I-ubiZM1, T-35S, T-nos, V-LB (the left border T-DNA repeat), and transgenic vector fragments of various lengths (Fig. 1C). No coding sequences were observed except for short fragments of different cry genes (version A of the database lacks both CS-Cry1B and CS-hpt coding sequences because these elements are not present in any of the other included events). No element combinations were detected besides two consecutive L-35S elements which could originate from P-e35S. However, the sample contained sequences where T-35S and T-nos promoters were joined with short fragments of transgenic vectors, clearly of unnatural origin (Fig. 1C, green). Moreover, the majority of the reads mapped in a clipped fashion (64.7%, 17,076 reads), and the sample contained a high percentage of unmapped reads (34.3%, 9054 reads), further pointing at the possible presence of an unknown GMO in the sample.
During phase II, 18,988 clipped and unmapped reads (71.94%) were retained in 23 clusters (Supplementary Table 2). Annotation of the clusters revealed several types of junction-like sequences, i.e. simultaneously containing regions of rice chromosomes 2 or 3 and functional genetic elements (Fig. 1C, blue). Several different types of sequences were observed containing long unannotated regions adjacent to T-nos and P-35S. Besides, several sequences showing homology to rice chromosomes 2 and 3 contained shorter unannotated regions. Additionally, one of the transgenic elements observed during phase I at a considerable copy number, P-ubiZM1, was not found on any of the sequences resulting from clustering. Consequently, the missing information was completed by performing phase III.
Annotation of representatives of each type of these partially annotated sequences during phase II indicated that the longer unannotated regions showed homology to two transgenic elements: CS-Cry1B and CS-hpt, revealing several additional combinations of two or more transgenic elements in the dataset (Fig. 1C, purple). The short unannotated regions adjacent to rice sequences corresponded to a tail of an unusual, slightly longer version of T-35S which was not present in Nexplorer (added to the annotation database under the name “T-35S long”). Furthermore, to retrieve and describe the sequence fragments containing P-ubiZM1 that were discarded during the clustering in phase II, reads mapping to P-ubiZM1 were manually extracted from the dataset, and annotated. This procedure revealed that they were part of amplicons consisting of fragments of P-ubiZM1 and P-e35S (Fig. 1C, indicated by a blue star).
Finally, verification of potential transgenic junctions showed that all junction-like sequences, belonging to four different types of amplicons, aligned to chromosomes 2 or 3 in a pattern indicating presence of a transgene insertion site on each chromosome. The applied approach thus allowed to detect an unauthorized GMO in the sample and describe it. The data analysis was completed in two and a half hours, and allowed to detect almost all types of sequence fragments observed in the previous study case (four types of sequence fragments were missed, Fig. 1C, yellow), including the transgenic junction regions and a considerable portion of the two inserts, which was sufficient for univocal identification of the unauthorized GMO. All of the undetected sequences were present at a low copy number (<5 reads).
The second study case illustrates that the proposed method is suitable for the analysis of data from unknown (unauthorized) GMOs, including those with unknown transgenic elements. The detection threshold (at least five similar reads of nearly the same length for PacBio, and at least 0.3% of the reads of various length being assigned to the cluster, see 2.2.2. Phase II section) is, however, slightly higher compared to the known GMOs which can be detected during phase I based even on the presence of a single read. Similar to phase I, at least some of the steps performed during phase III can be automated, including for example selection and blasting of partially annotated amplicons against NCBI nt. and ordering of annotated amplicons according to the database hit in the report. This will further decrease the hands-on time required for the analysis. As phase III is the most labor-intensive part of the analysis, the efficiency of sample processing depends on the diversity of the transgenic events and transgenic elements that are included in the database, and will increase as new events are added.
3.3.4. Scenario 3: benchmarking using samples containing a single GMO at varying concentrations, processed food samples, and a different type of sequencing platform
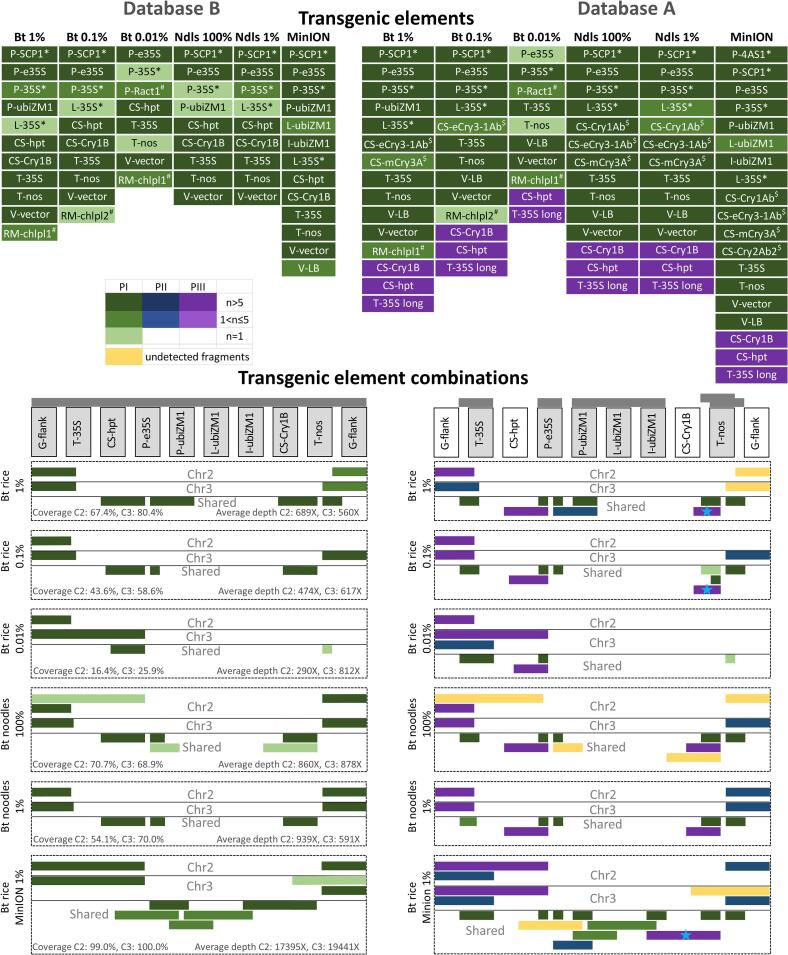
In routine GMO analysis, it is important to be able to detect and identify GMOs that are present at trace level and in processed food (e.g. noodles). After having evaluated the performance of the method using the two relatively simple cases described in the previous two scenarios, and prior to the more complex cases involving mixtures of different GM events at sub-optimal levels, we evaluated the sensitivity of the proposed method with a series of benchmarking datasets from (Fraiture et al., 2017). These datasets consisted of DNA walking data generated using P-35S, T-nos and T-35S (pCAMBIA) primers from wild type rice grain samples containing a low percentage of Bt rice grain material: 2000 HGE (Bt rice 1%), 200 HGE (Bt rice 0.1%), and 20 HGE (Bt rice 0.01%), and Bt noodles 100% and 1% prepared from rice grains with high (100%) or low (1%) percentage of GMOs, respectively (Table 1), all sequenced with the PacBio sequencing technology. Additionally, the method was tested using one dataset sequenced with MinION sequencing technology, Bt rice MinION 100%, obtained from a sample consisting entirely of Bt rice (200000 HGE) (Fraiture et al., 2018). As the data analysis workflow performed differently with respect to the known and unknown GMOs in the previous two scenarios, all samples were analyzed using the versions of the database with (B), and without (A) the transgenic inserts of Bt rice.
During phase I of the analysis carried out with version B of the database, each sample showed to contain at least one transgenic junction region of Bt rice at a very high sequencing depth (>1000X, Supplementary Table 3), implying that a strong and informative signal for the presence of GMO was observed even at the lowest concentration of the transgene. The average sequencing depth of the inserts in the different samples was between 290X and 939X, and correlated poorly with the concentration of Bt rice in the sample (Fig. 3, left panel, Supplementary Table 3). The coverage (i.e. the fraction of the transgenic insert covered by reads), on the other hand, increased with the increasing concentration of Bt rice, from 16.4% for 0.01% Bt rice to 80.4% for 1% Bt rice and 100% Bt noodles. Therefore, more and in some cases longer fragments of the two transgenic inserts were detected for samples with a higher concentration of the GMO (Fig. 3, left panel).
Fig. 3.
Benchmarking of the method using samples containing Bt rice at different concentrations. The data analysis workflow described in Fig. 1A was applied on PacBio sequencing data of DNA walking amplicons obtained from rice grain material containing Bt rice at different concentrations (1%, 0.1% and 0.01%), and noodles prepared from rice grains with 100% and 1% of Bt rice (Table 1) and MinION sequencing data of DNA walking amplicons obtained from rice grain material of Bt rice. The left panel shows analysis results obtained with database version B, containing both Bt rice insert sequences, whereby Bt rice represents as a known GMO. The right panel shows results obtained with database version A, lacking the Bt rice insert sequences, whereby Bt rice plays the role of an unknown GMO. Transgenic expression cassettes are shown schematically, elements are not drawn to scale. The same cassette is used to represent both transgenic inserts (Chr2 and Chr3, corresponding to inserts in chromosome 2 and 3 from the rice genome, respectively). Grey bars above the cassettes indicate schematically which elements and element combinations are present in the database. Elements that are represented in the database are indicated in grey colored boxes. Colored bars below the expression cassette show the longest amplicons that were observed for the first time during phase I, II and III (PI, PII, PIII) of the analysis as described in Fig. 1A. The green/blue/purple color gradient according to the color legend shows the number of reads (n) representing a given amplicon. Fragments that were manually reconstructed from reads (instead of clusters) during phase III are marked by a blue star. Fragments that are known to be present in the sample but that could not be detected in the current analysis are colored yellow. * Transgenic elements containing homologous regions with P-e35S. $ Transgenic elements containing homologous regions with CS-Cry1B. # Transgenic elements likely originating from the host plant genome. Transgenic elements are described in Supplementary File 3.
Clipped and unmapped reads were present in all datasets. The fraction of unmapped reads was higher for samples with lower concentration of Bt rice, reaching up to 25% for Bt rice 0.01% (Supplementary Table 2). To identify the nature of these sequences, phases II and III were performed. Similarly to Bt rice 100%, all samples contained sequences from the Bt rice transgenic inserts which were classified as clipped because of short non-mapping overhangs (Supplementary File 1). Additionally, Bt rice 1%, Bt rice 0.1% and Bt noodles 100% contained one or two clusters corresponding to sequencing or PCR artefacts, such as a PCR chimera consisting of three consecutive P-35S copies formed in a process whereby a PCR product from one cycle functions as a primer in a following cycle. A more detailed examination of the reads within each cluster showed that while the clusters contained sufficient reads to pass the threshold of 0.3%, the majority of the reads within the clusters did not align along the full length to the chimeric reads. The abundance of the actual chimeric reads in the sample was very low. Finally, all samples, except for 100% Bt Noodles, showed presence of clusters containing rice genomic sequences, which were more numerous in 0.01% Bt rice (Supplementary Table 1, Supplementary File 1).
Subsequently, we verified whether the same results could be obtained in case Bt rice represents an unknown GMO by repeating the analysis with the database version A. During phase I, datasets typically contained several of the following elements: commonly used transgenic promoter and terminator regions, fragments of different cry genes, V-LB, and L-35S dimers that could originate from the P-e35S element (Fig. 3, right panel). In this case, the sequences of T-nos and T-35S terminators were often joined with short stretches of vector sequences, confirming their transgenic origin. All samples contained high numbers of unmapped (>40%) and clipped (>50%) reads, which along with the detected transgenic elements served as a clear indication for the presence of an unauthorized GMO. The reads were extracted, clustered, and annotated. All amplicons that were observed in the datasets with the version B of the database were also detected with the version A of the database, except for six types of sequence fragments over all tested samples (Fig. 3, right panel, yellow). While fragments consisting of T-nos and CS-Cry1B were lost from the 1% Bt rice and 0.1% Bt rice samples during filtering compared to the data analysis results obtained with database B, their presence could be predicted based on the presence of portions of other cry genes, allowing to retrieve these fragments from unclustered read data (Fig. 3, right panel, indicated by a blue star).
Finally, we tested the method using Bt rice MinION 100% dataset. This dataset contained 20–30 times more reads than the previously tested PacBio datasets (Supplementary Table 2). As a consequence, analysis with database B showed that 99% of both Bt insert sequences were covered by at least one read, with an average sequencing depth of 17–19 thousands of reads. Similar output was obtained with database A, where all but two sequence fragments (represented by 1 and 2 reads respectively, Fig. 3, right panel, yellow) could be retrieved.
The intended use of the developed methodology is to process data obtained from routine samples. The benchmarking tests have shown that the integrated approach is sensitive enough to detect Bt rice present slightly below the detection limit (25 HGE) imposed by competent authorities. Moreover, the signal strength appeared to be equally high for samples containing high and low amounts of Bt rice, as the copy number of the transgene only affected the diversity of the amplicons, but not the copy number of the junction regions. The integrated approach appeared to be equally suitable for processed food samples as for grain material, and sensitive enough to detect GMO present at the labelling threshold (1%) in a processed sample. Moreover, the proposed methodology showed to be suitable for the analysis of PacBio and MinION sequencing data.
3.3.5. Scenario 4: Sample containing a mixture of known GMOs
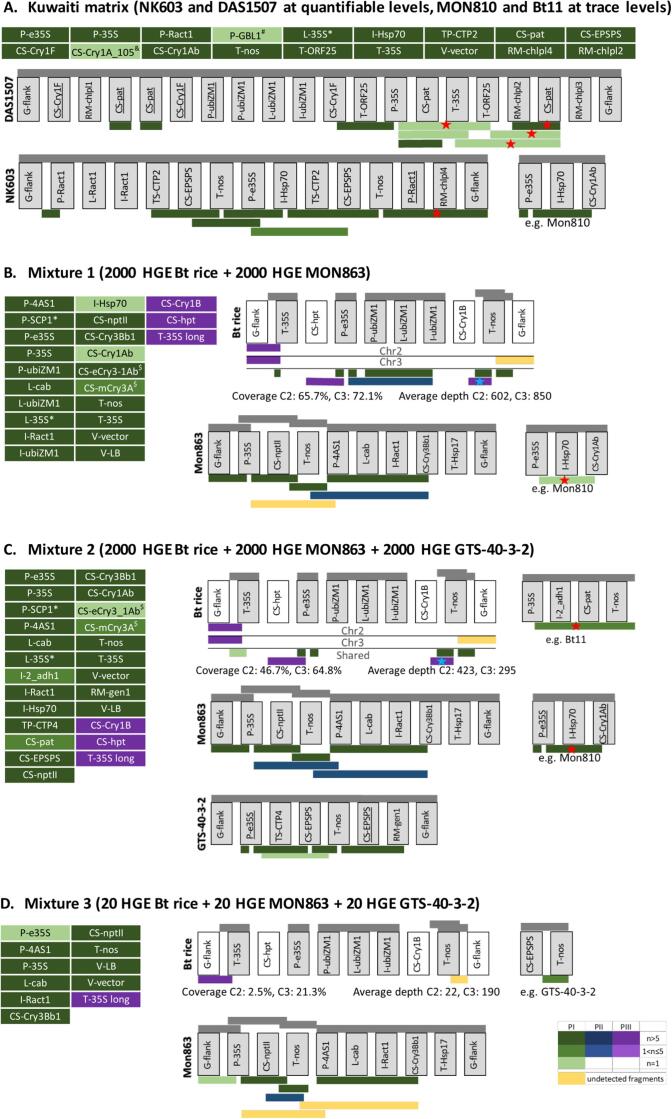
The fourth study case involved a real-life sample from a Kuwaiti food market. This sample has previously been shown using qPCR to contain the NK603 and DAS1507 events at quantifiable levels and MON810 and Bt11 at trace levels, representing a case of a mixture of known GMOs (Fraiture et al., 2017). The corresponding dataset has been generated by carrying out DNA walking with P-35S, T-nos and T-35S (pCAMBIA) primers, followed by PacBio sequencing.
Phase I of the analysis allowed to detect event-specific sequences of NK603 (i.e. transgenic junction region) and DAS1507 (i.e. event-specific rearrangement of the insert and host DNA, consisting of RM-chlpl2 which is a fragment of the maize chloroplast genome, followed by a truncated copy of CS-pat) (Fig. 4A). A small discrepancy was observed for DAS1507, for which all reads mapping to the event-specific region at the 3′ end of the insert were clipped. The clipped portion corresponded to a region of the DAS1507 insert containing a fragment of P-e35S (starting from the primer annealing region), followed by CS-pat. This suggests that because of the homology of the CS-pat regions, a fragment of the internal portion of the DAS1507 insert served as a primer for the amplification of the transgenic junction region, resulting in the creation of a chimeric sequence. For both events, multiple amplicons originating from the internal portion of the transgenic insert were found. Some of the observed sequences were not detected during previous analyses (Fig. 4A, indicated by a red star) (Fraiture et al., 2017), which confirmed the efficiency of the proposed data analysis methodology. Finally, one last observed element combination could not be explained by the presence of the two confirmed events, showing homology to an internal portion of MON810 instead. While MON810 was the only candidate event in the database from which the element combination could have been derived, in the absence of the junction region, the presence of MON810 could not be univocally confirmed. Although the sample showed to contain traces of Bt11 event by qPCR [8], no DNA walking amplicons from Bt11 were observed in this analysis and in the work of Fraiture et al. [8]. Phases II and III, performed with the clipped and unmapped reads which were present in the sample, revealed no new sequences. Despite the high complexity of the sample, the data analysis was completed in 1.5 h. These observations suggest that the proposed strategy is highly efficient for the identification of known GMOs in a mixture.
Fig. 4.
Characterization of samples containing mixtures of known and unknown GM events. The data analysis workflow described in Fig. 1A was applied on long-read sequencing data of DNA walking amplicons obtained from rice grain material containing a mixture of (A) known, or (B, C, D) known, and unknown events. Transgenic expression cassettes are shown schematically, and the same cassette is used to represent both transgenic inserts of Bt rice (C2 and C3, corresponding to inserts in chromosome 2 and 3 from rice genome respectively). Grey bars above the cassettes indicate schematically which elements and element combinations are present in the database. Elements that are represented in the database are indicated in grey colored boxes. Truncated transgenic elements are underlined. Colored bars below the expression cassette show the longest amplicons that were observed for the first time during phase I, II and III (PI, PII, PIII) of the analysis as described in Fig. 1A. The green/blue/purple colour gradient according to the color legend shows the number of reads (n) representing a given amplicon. Fragments that are known to be present in the sample but that could not be detected in the current analysis are colored yellow. Fragments that were manually reconstructed from reads (instead of clusters) during phase III are marked by a blue star. Fragments that were not found previously by Fraiture et al (Fraiture et al., 2017) are marked by a red star. *Transgenic elements containing homologous regions with P-35S, P-e35S and P-4AS1. $Transgenic elements containing homologous regions with CS-Cry1B. &Transgenic elements containing homologous regions with CS-Cry1Ab. #Transgenic elements likely originating from the host plant genome. Transgenic elements are described in Supplementary File 3.
Notably, while the threshold of 0.3% reads in a cluster allowed highly efficient filtering of all sequencing or sample preparation artefacts from the samples containing 100% Bt rice, in some of the samples from scenario 3 and in the Kuwaiti matrix some clusters with a chimeric representative sequence were present upon filtering. These sequencing artifacts can be distinguished from the other reads by the specific arrangement of the elements (e.g. overlapping tandem or inverted repeats), and confirmed based on their frequency in the dataset. However, despite the very low frequency of the occurrence of chimeric sequences, each newly detected sequence is preferably verified by PCR, possibly in combination with Sanger sequencing of the resulting amplicon. In the future, analysis of a larger number of datasets will permit to identify optimal values of the filtering thresholds for the different types of input, which is a prerequisite for the automation of the current workflow.
3.3.6. Scenario 5: Sample containing a mixture of known and unknown GMOs
The fifth study case, corresponding to the most complex scenario of a mixture of known and unknown GMOs, was performed using Mixture 1 (2000 HGE Bt rice + 2000 HGE MON863), Mixture 2 (2000 HGE Bt rice + 2000 HGE MON863 + 2000 HGE GTS-40-3-2), and Mixture 3 (20 HGE Bt rice + 20 HGE MON863 + 20 HGE GTS-40–3-2) (Table 1). The samples underwent DNA walking with the same three primers as used for the other cases, and were sequenced using the PacBio methodology. All datasets were tested with database version A so that Bt rice functioned as an unknown GMO. Additionally, the database lacked the full insert sequence of MON863. Instead, the insert of MON863 was represented by two sequences, the first covering the left transgenic junction and the second covering the right transgenic junction, with each of the two sequences additionally containing the adjacent regions of the host plant genome and transgenic insert (Fig. 4B, C, D).
During phase I (performed for each mixture separately), all three samples showed to harbor one of the two transgenic junction regions of MON863, and two fragments of the transgenic insert of that event (Fig. 4B, C, D green). In Mixture 3, containing the lowest levels of GMOs, the transgenic junction region was represented by one read only. Mixture 2 also harbored five amplicons derived from the GTS-40–3-2 insert, including event-specific sequences consisting of rearranged insert and host DNA (a fragment of T-nos, followed by a truncated copy of CS-EPSPS and RM-gen1, which is a fragment of maize genome introduced as a result of event-specific rearrangement). Mixture 3 contained only two reads mapping uniquely to GTS-40-3-2 insert, namely to the T-nos region with a small portion of the transgenic vector sequence, but no event-specific sequences of the event. Mixtures 1 and 2 also contained reads homologous to an internal region of MON810 insert at different concentrations, and Mixture 1 contained an amplicon potentially originating from an internal portion of Bt11 although these events have not been intentionally included in the mixtures.
All three samples contained additional transgenic elements and transgenic element combinations whose presence was not explained by the confirmed transgenic events (Fig. 4B, C, D). Mixture 1 contained a combination of P-ubiZM1, L-ubiZM1 and L-ubiZM1 at a copy number higher than would be expected from a contamination with host DNA (>100X), a range of very short fragments of various cry genes, T-35S, V-LB and L-35S dimers that could originate from P-e35S. For Mixtures 2 and 3, this list was limited to V-LB and T-35S, as well as T-35S fused with a short transgenic vector sequence represented by one read for Mixture 2. All three samples contained several different fragments of T-nos surrounded at one or both sides by a short fragment of the transgenic vector, at least some of which could, however, be derived from the portion of MON863 insert that is absent from the database.
Annotation of clustered clipped and unmapped reads (phases II and III) revealed additional one (for Mixture 1 and 3) or two (for Mixture 2) amplicons from MON863 insert, that were not initially detected because the database lacked the full insert sequence (Fig. 4B, C, D blue). Mixtures 1 and 2 also contained an amplicon where P-e35S was joined with CS-hpt, which was found during annotation refinement (Fig. 4B, C purple). Additionally, fragments consisting of T-nos and CS-Cry1B could be retrieved from reads mapping to short portions of various cry genes in Mixtures 1 and 2. All three samples also showed presence of one (Mixture 3) or two (Mixture 1 and 2) junction-like amplicons, that aligned to regions of rice chromosomes 2 and 3 in an arrangement that could be expected from a transgene insertion site (Fig. 4B, C, D, purple). Mixtures 1 and 2 also contained respectively 3 and 1 clusters from which the representative sequence resembled a sequencing artifact, namely a P-35S trimer. A more detailed analysis of the cluster showed that only a small number of reads had this arrangement of elements, allowing to confirm the chimeric origin of the reads. The analysis of each dataset lasted between 1.5 and 3 h. The results of the analysis of Mixtures 1–3 (i.e. rapid identification of transgenic junctions and internal portions of transgenic inserts of known and unknown transgenic events) suggest that the method allows to efficiently deconvolute the contents of mixtures when one of the components is an unknown GMO.
The two last case studies further confirm that the proposed methodology permits an efficient and thorough analysis of long-read sequencing data of DNA walking libraries obtained from samples containing a mixture of events. This is illustrated by the detection of sequence fragments from known GMOs that have not been observed in the previous analyses without the Nexplorer database, and detection of event-specific sequences of known GMOs even when these are represented by a single read. Moreover, the efficiency of detection of unknown GMOs is demonstrated by the fact that accurate results are obtained for complex mixtures with one unknown, and one partially described GMO. The detection limit for unknown GMOs is slightly higher than for the known ones, necessitating at least 5 reads for PacBio, and at 25 reads for MinION datasets due to the filtering at the beginning of phase 2 to remove chimeric reads and other artifacts.
4. Conclusion and future perspectives
Increasing complexity and diversity of GMO events on the market calls for the development of novel tools to help the enforcement laboratories to continue executing their tasks according to the standards set by current regulations. NGS is one of the most promising approaches, offering the enforcement laboratories control over the increasing complexity and diversity of GMOs. Another one is a database that organizes information available on GM events and associated detection methods. An important type of data whose availability in the public domain is increasing, is sequencing information of transgenic inserts. The existing databases are not sequence-based, or when available, the sequences are in most cases not annotated and poorly interlinked with the other database instances, such as the inserts and elements associated with each event. This impedes the use of these databases to analyze NGS data. Therefore, we have developed Nexplorer, a sequence-based database, in which annotated sequences of GM events are presented in a structured, searchable and extractable format. The availability of pre-organized and annotated sequencing information allows to streamline bulk analysis of different types of NGS data, including third-generation sequencing data obtained from targeted sequencing GM detection methods.
As long as it remains cheaper and faster than whole genome/shotgun sequencing, DNA walking enrichment coupled with long-read sequencing represents a rational option to detect and characterize GMOs, in particular unauthorized ones, resolving the uninformative signals from routine qPCR screenings. However, the analysis of a large number of sequencing amplicons can be a complex task, which requires considerable time and skill (Fraiture et al., 2018). In the current work, we proposed a data analysis methodology based on the GMO sequence database which provides an efficient solution to deal with this complex analytical task, allowing to obtain detailed and reliable information with a limited hands-on time. This is demonstrated by the analysis of datasets representing real-life scenarios that can be encountered in routine GMO analysis, i.e. samples containing authorized and/or unauthorized GM events at varying concentrations.
It should be noted that while the proposed methodology allows detection and identification of known events, and detection and an at least partial characterization of unknown events in a sample, presence of the transgenic lines needs to be confirmed by conventional methods such as a combination of PCR targeting the transgenic junction followed by Sanger sequencing until NGS becomes a validated method. Besides, an important component of GMO control can be quantification of the detected events. To this end, qPCR- or digital PCR-based methods must be used. An advantage offered by the presented methodology is that it allows to retrieve sequences of the transgenic insert and junction regions of unknown events. This type of information is not available thought PCR-based screening and can be used for PCR/qPCR primer design.
In the future, the database can potentially be used to develop strategies for the analysis of NGS data obtained from shotgun sequencing approaches for detection or characterization of GMO. Importantly, for a final integration of NGS-based methodologies into routine, there is a need to establish a validation approach of the data analysis pipelines designed for the different types of data. Availability of proficiency testing material such as in silico datasets or samples with a well-described composition provided in a centralized way (e.g. at European level) is one of the conditions that would promote the shift from qPCR-based methods to NGS.
Currently, the Nexplorer database contains sequences of the events that are authorized in the EU retrieved from public sources. An initiative that would simplify acquisition of high-quality sequence data for the database is to allow sequencing of certified reference material that is provided officially in the context of the GMO control. In the future, the database could be further extended to include sequences of events authorized in other parts of the world, and for which a description is thus available in the public domain, as well as the ones that are not authorized anywhere. Unfortunately, currently nearly no information is being generated on the worldwide unauthorized events because of the absence of a suitable cost-effective methodology to do so. The use of the enrichment methods like the DNA walking methodology in combination with high throughput sequencing will allow to obtain sequences of a larger number of unauthorized events at a limited cost and effort. Furthermore, while initially not designed to be used for events created using CRISPR-Cas9 and similar genome editing methods, the database can be used to store the sequences of the modifications achieved using those genome editing methods and a dedicated data analysis methodology for the detection of this type of events can be developed. It should be noted that development and maintenance of a sequence database requires a considerable effort that may be too demanding to be accomplished by each single institution. A centralized European database containing the right type of information for an efficient NGS data analysis that could be used by different laboratories is a more easily achievable and therefore a highly attractive concept. While development of a centralized European database is the ultimate goal, the Nexplorer database provides one of the first examples of this type of resource, and demonstrates its added value for the analysis of NGS data.
CRediT authorship contribution statement
Assia Saltykova: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. Julien Van Braekel: Methodology, Software, Visualization, Writing – review & editing. Nina Papazova: Conceptualization, Data curation, Methodology, Software, Validation, Writing – review & editing. Marie-Alice Fraiture: Data curation, Writing – review & editing. Dieter Deforce: Supervision, Writing – review & editing. Kevin Vanneste: Conceptualization, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – review & editing. Sigrid C.J. De Keersmaecker: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. Nancy H. Roosens: Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Véronique Wuyts for her contribution to the development of the initial database concept.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochms.2022.100096.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Bak A., Emerson J.B. Multiplex quantitative PCR for single-reaction genetically modified (GM) plant detection and identification of false-positive GM plants linked to Cauliflower mosaic virus (CaMV) infection. BMC Biotechnology. 2019;19(1):73. doi: 10.1186/s12896-019-0571-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeders S.R.M., De Keersmaecker S.C.J., Roosens N.H.C. How to Deal with the Upcoming Challenges in GMO Detection in Food and Feed. Journal of Biomedicine and Biotechnology. 2012;2012 doi: 10.1155/2012/402418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coordinators N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2016;44(D1):D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraiture M.-A., Herman P., Papazova N., De Loose M., Deforce D., Ruttink T., Roosens N.H. An integrated strategy combining DNA walking and NGS to detect GMOs. Food Chemistry. 2017;232:351–358. doi: 10.1016/j.foodchem.2017.03.067. [DOI] [PubMed] [Google Scholar]
- Fraiture M.-A., Herman P., Taverniers I., De Loose M., Deforce D., Roosens N.H. Current and New Approaches in GMO Detection: Challenges and Solutions. BioMed Research International. 2015;2015 doi: 10.1155/2015/392872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraiture M.-A., Saltykova A., Hoffman S., Winand R., Deforce D., Vanneste K.…Roosens N.H.C. Nanopore sequencing technology: A new route for the fast detection of unauthorized GMO. Scientific Reports. 2018;8(1) doi: 10.1038/s41598-018-26259-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes L., Busch U., Pecoraro S. GMO finder—A GMO Screening Database. Food Analytical Methods. 2012;5(6):1368–1376. [Google Scholar]
- Holst-Jensen A., Bertheau Y., de Loose M., Grohmann L., Hamels S., Hougs L.…Wulff D. Detecting un-authorized genetically modified organisms (GMOs) and derived materials. Biotechnology Advances. 2012;30(6):1318–1335. doi: 10.1016/j.biotechadv.2012.01.024. [DOI] [PubMed] [Google Scholar]
- ISAAA. (2019). Global Status of Commercialized Biotech/GM Crops in 2019: Biotech Crops Drive Socio-Economic Development and Sustainable Environment in the New Frontier. ISAAA Brief No. 55. Cornell University Ithaca, NY, USA.
- Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R.…Carey V.J. Software for Computing and Annotating Genomic Ranges. PLOS Computational Biology. 2013;9(8) doi: 10.1371/journal.pcbi.1003118. Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liang C., van Dijk J.P., Scholtens I.M.J., Staats M., Prins T.W., Voorhuijzen M.M.…Kok E.J. Detecting authorized and unauthorized genetically modified organisms containing vip3A by real-time PCR and next-generation sequencing. Analytical and Bioanalytical Chemistry. 2014;406(11):2603–2611. doi: 10.1007/s00216-014-7667-1. [DOI] [PubMed] [Google Scholar]
- Lindenbaum, P. (2015). Jvarkit: java utilities for bioinformatics.
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. Journal. 2011;17(1):10–12. [Google Scholar]
- Martin S., Leggett R.M. Alvis: A tool for contig and read ALignment VISualisation and chimera detection. BMC Bioinformatics. 2021;22(1):124. doi: 10.1186/s12859-021-04056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzara M., Savini C., Delobel C., Broll H., Damant A., Paoletti C., Van den Eede G. Joint Research Centre; Ispra: 2008. Definition of minimum performance requirements for analytical methods of GMO testing. [Google Scholar]
- Morisset D., Novak P.K., Zupanič D., Gruden K., Lavrač N., Žel J. GMOseek: A user friendly tool for optimized GMO testing. BMC Bioinformatics. 2014;15(1):258. doi: 10.1186/1471-2105-15-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo M., Angers-Loustau A., Henriksson P., Bonfini L., Patak A., Kreysa J. JRC GMO-Amplicons: A collection of nucleic acid sequences related to genetically modified organisms. Database. 2015;2015 doi: 10.1093/database/bav101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podevin N., Du Jardin P. Possible consequences of the overlap between the CaMV 35S promoter regions in plant transformation vectors used and the viral gene VI in transgenic plants. GM Crops & Food. 2012;3(4):296–300. doi: 10.4161/gmcr.21406. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Briefings in Bioinformatics. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva R.A.M., Chen Z.J. Taylor & Francis; 2019. ggplot2: Elegant graphics for data analysis. [Google Scholar]
- Wang, X., Jiao, Y., Ma, S., Yang, J., & Wang, Z. (2020). Whole-Genome Sequencing: An Effective Strategy for Insertion Information Analysis of Foreign Genes in Transgenic Plants. Frontiers in Plant Science. Retrieved from https://www.frontiersin.org/article/10.3389/fpls.2020.573871. [DOI] [PMC free article] [PubMed]
- Willems S., Fraiture M.-A., Deforce D., De Keersmaecker S.C.J., De Loose M., Ruttink T.…Roosens N. Statistical framework for detection of genetically modified organisms based on Next Generation Sequencing. Food Chemistry. 2016;192:788–798. doi: 10.1016/j.foodchem.2015.07.074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.