Highlights
-
•
Molecular characterization of GM crops with two approaches of WGS-PE and WGS-MP.
-
•
The molecular characterization of GM rice 114-7-2 was deciphered comprehensively.
-
•
The pros and cons of WGS-PE and WGS-MP were compared in molecular characterization analysis.
Keywords: GM rice line 114-7-2, Mate pair, Molecular characterization, Paired-end, Whole-genome sequencing
Abbreviations: GMO, genetically modified organism; WGS, whole-genome sequencing; PE, paired-end; MP, mate-pair; ISAAA, International Service for the Acquisition of Agri-Biotech Applications; NOS, nopaline synthase; FAM, 6-carboxyfluorescein; BHQ, black hole quencher; CTAB, Cetyltrimethyl ammonium bromide; NGS, Next-generation sequencing; WT, Wild type; ddPCR, Droplet digital polymerase chain reaction
Abstract
Basic data for the safety assessment of transgenic line involves the molecular characterization of the integration site of exogenous DNA, flanking sequences, copy number, and unintended plasmid backbone residues. However, performing a full molecular characterization remains challenging, especially for GMOs that possess complex exogenous DNA integrations. We established two whole-genome sequencing strategies: paired-end and mate-pair, to characterize the exogenous DNA integration of a human serum albumin gene into rice line 114-7-2, and evaluated the performance of these two strategies in the molecular characterization of transgenic line. The results showed the existence of two exogenous DNA insertion loci (Chr 01 and Chr 04) and their corresponding flanking sequences, five copies of the exogenous rHSA gene, and the presence of unintended residual plasmid backbone sequences. However, the WGS-MP strategy demonstrated higher efficiency, lower cost, and lower background noise compared with the WGS-PE analysis, especially for identification of the exogenous DNA integration site.
1. Introduction
In the past three decades, recombinant DNA technology has been widely used to generate transgenic plants, and more than 300 genetically modified crops have been approved globally for planting and commercialization (James, 2019). However, all approved GM lines should undergo careful and strict safety assessment, including molecular characterization. Furthermore, environmental safety and food safety should be evaluated, and GM crops should only be cultivated if they are deemed to be safe (Kuiper, Kleter, Noteborn, & Kok, 2010). The molecular characterization of GM crops often includes determining the insertion site, the inserted DNA and associated native flanking sequences, the insert copy number at each insertion site, and the unintended existence of backbone sequences derived from the plasmid vector used for the transgenes, et al. (Alimentarius, 2003) A comprehensive and accurate molecular characterization is a fundamental requirement for each GM transgenesis event as part of the safety assessment (König et al., 2004). In addition, a low copy number and the integration of intact exogenous DNA comprise the most favorable molecular profile for selecting the best events from putative lines that show the stable heritability of inserted DNA (Kovalic et al., 2012). Therefore, a full molecular characterization is necessary not only for safety assessment but also to select transgenic events that confer excellent target traits and to develop event-specific detection methods for GMO monitoring.
The commonly used methods for the molecular characterization of transgenic plants include Southern blot analysis, PCR, and PCR-derived methods combined with Sanger sequencing (Li et al., 2017). Southern blot analysis is one traditional method with which to determine the copy number of inserted DNA fragments, the unintended presence of residual backbone DNA, and generational stability. PCR and its derived methods combined with Sanger sequencing are often used to characterize the insertion site, the native genomic flanking sequences surrounding the insertion site, and the exact sequence of the inserted DNA. These commonly used approaches, such as Southern blotting, TAIL-PCR, Sanger sequencing, real-time PCR, and digital droplet PCR have been adopted in many countries, and have been used to delineate guidelines to generate molecular data for safety assessment (FAOO Nations, 2009). However, these methods are time-consuming and potentially limited by the existence of sequence substitutions, insertions, and deletions, and by the number and/or size of the targets (Zhang et al., 2012). A full molecular characterization of GMOs using these methods remains difficult because current transgenic techniques lead to random DNA integration.
Next-generation sequencing (NGS) allows the complete sequencing of the entire genome, and whole-genome sequencing (WGS), coupled with paired-end (PE) read generation has been used for the molecular characterization of GMOs, with an associated reduction in sequencing costs (Goodwin et al., 2016, van Dijk et al., 2014). Next-generation sequencing platforms have been successfully used to characterize the integration of transgenes (Zastrow‐Hayes et al., 2015). Two transgenic glyphosate-tolerant soybean insertion sites and flanking sequences were identified based on WGS (Guo, Guo, Hong, & Qiu, 2016). The precise insertion loci and copy number of transfer DNA in transgenic rice plant lines SNU-Bt9-5, SNU-Bt9-30, and SNU-Bt9-109 were detected using NGS-based molecular characterization methods, and the data were equivalent to those obtained from Southern blot analysis (Doori, Su-Hyun, Yong, Ban, & Shic, 2017). Furthermore, the corresponding bioinformatics pipelines for the analysis of NGS data have also been developed (Yang et al., 2013, Zhang et al., 2020). NGS technology combined with target enrichment was also used to identify genetic integrations from complex food/feed samples (Debode et al., 2019, Košir et al., 2018). Improvements in sequencing technology and the associated reduction in costs have meant that NGS technology is playing an increasingly important role in GMO analysis, particularly at the level of molecular characterization (Arulandhu et al., 2018). Previous studies have successfully used WGS coupled with PE reads (WGS-PE) to identify transgene insertion sites and their flanking sequences (Guttikonda et al., 2016). However, some problems remain to identify the structure and sequence of entire transgene insertions using the WGS-PE strategy, especially for insertions that have a repetitive and inverse arrangement, of for recipient endogenous DNA, and highly similar DNA sequences.
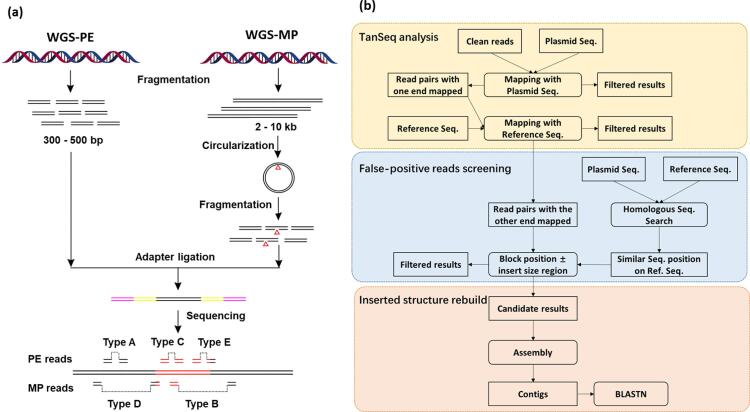
Compared with the WGS-PE strategy, WGS coupled with mate-pair (WGS-MP) is an alternative approach that has been used to detect genome rearrangements and structural variants (Johnson et al., 2018, Srivastava et al., 2014). The difference between WGS-MP and WGS-PE approaches resides in the construction of the sequencing libraries: PE libraries are constructed using short DNA fragments (300–500 bp), whereas MP libraries are constructed using longer DNA fragments (2–10 kb), and the protocol for MP library construction is slightly more complex than that for PE libraries (Illumina, 2009). However, the WGS-MP method provides paired sequences from the ends of comparatively large DNA fragments, which promotes the recovery of reads that span insertion sites and might be helpful for the analysis of complex rearranged exogenous DNA insertions.
The GM rice line 114-7-2 that expresses rHSA was developed to produce recombinant human serum albumin proteins in the seed endosperm at Wuhan University, China. This transgenic line was co-transformed with two expression vectors, pOsPMP114 and pOsPMP02, into recipient line TP309 by Agrobacterium-mediated transformation (He et al., 2011, Ning et al., 2008, Xie et al., 2008). In this study, we have molecularly characterized GM rice line 114-7-2, and provide data on the exogenous DNA insert site, flanking sequence, the copy number of the inserted gene, and the presence of plasmid backbone sequences, using WGS-PE and WGS-MP strategies. We also compare the advantages and disadvantages of these two strategies, which provide a reference for the future selection of an appropriate method for the molecular characterization of transgenic material.
2. Materials and methods
2.1. Plant material and DNA extraction
The GM rice line 114-7-2 was developed by co-transforming pOsPMP114 and pOsPMP02 into rice cultivar TP309 by Agrobacterium-mediated transformation (He et al., 2011, Ning et al., 2008). pOsPMP114 contained the exogenous rHSA gene expression cassette regulated by the rice Gt13a promoter and the NOS terminator, and pOsPMP02 contained the hygromycin phosphortransferase (hpt) gene-expression cassette with a callus-specific promoter from rice, b-cysteine protease (CP), and the NOS terminator (Xie et al., 2008). Mixed fresh leaf samples of five individual plants of GM rice 114-7-2 event and five individual plants of recipient line TP309 were kindly supplied by Wuhan University, China, respectively. Total genomic DNA was extracted from fresh rice leaves using the CTAB method. The quantity and quality of extracted rice genomic DNA were evaluated with a Nanodrop ND-8000 (Thermo Fisher Scientific) and 1% agarose gel electrophoresis, respectively. The extracted DNA of GM rice 114-7-2 and recipient line TP309 were used for WGS-PE and WGS-MP analysis.
2.2. Construction of PE and MP libraries and Illumina sequencing
For PE library construction, approximately 5 µg genomic DNA from GM rice 114-7-2 and WT (TP309 line) were fragmented to a peak size of 500 bp by sonication with a Diagenode Bioruptor UCD-300TM-EX (Denville, NJ, USA). The Illumina TruSeq DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) was used to construct the PE library, with a mean insert size of 500 bp. To construct the MP library, 5 µg DNA from line 114-7-2 was fragmented to a peak size of 5 kb, and the Illumina Mate Pair v2 kit (part number PE-930-1003) was then used for library construction with a mean insert size of 5 kb, according to the manufacturer’s protocol. The procedure of MP library construction was slightly more complex than that for the PE library and included additional steps of circularizing the long fragments, re-fragmenting and isolating the junction fragments. The quality of the constructed PE and MP libraries was evaluated using Pico-Green (Quant-iT; Invitrogen) and an Agilent Bioanalyzer 2100 with DNA 1000 kit and 12,000 kit (Agilent Technologies), respectively. The constructed PE and MP DNA libraries were sequenced using Illumina HiSeq2000 (BGI-Shenzhen, Shenzhen, China). The quality control of the raw data after removing the index sequences was performed by NGS QC Toolkit v2.3 (Patel, Jain, & Liu, 2012). After filtration, the samples with base qualities greater than 30 and a read length over 70 nucleotides were used for further analysis.
2.3. Bioinformatics pipelines
Sequencing data were preprocessed by removing low-quality reads, the reads lengths were made uniform, and adapters were trimmed, using the Ubuntu X86_64 system. The bioinformatics pipeline was developed by adding the new function for screening false-positive reads to our previous TranSeq pipeline (Fig. 1). Firstly, the qualified reads were aligned with transgene sequences using the Burrows-Wheeler Aligner (BWA) algorithm (version 0.6.2) with default mapping parameters (Li & Durbin, 2009). Secondly, the paired reads with mapped single ends were aligned with the rice reference genome. The reference rice genome (GCF_001433935.1) was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/assembly/GCF_001433935.1). Thirdly, false-positive reads were screened by searching for the position of similar sequences between plasmid and reference using BLASTN. Candidate results were generated after blocking the position ± the insert size of homologous sequences. A python script was generated to filter the reads mapped with the Gt13a and CP promoters (Supplementary File 1). Finally, candidate reads were assembled by SPADES (Bankevich et al., 2012), and the read mapping pattern was depicted by Integrated Genome Viewer (IGV) (Robinson et al., 2011).
Fig. 1.
The bioinformatics pipeline used for molecular characterization.
2.4. Mimicking of sequencing depth, and copy number analysis in silico
To identify the minimal amount of sequencing data required to successfully detect the insertion site and its flanking sequences, a python script (Supplementary File 2) was built to randomly extract the raw PE data, which reduced the paired-end data volume to 5×, 6.19×, 10×, 15× and 20× of the rice genome size. The reduced paired-end data was analyzed using the previously described pipelines to generate insertion site results and assemble the contigs. The copy number of the target gene was calculated using Eq. (1), based on NGS sequencing data. ADT represents the mean sequencing depth of the target gene, D represents the general sequencing depth and R is the relative relationship between mapped reads and the host genome.
| (1) |
2.5. Conventional PCR and Sanger sequencing
Conventional PCR primers were used to confirm the junction DNA sequence between host genome and exogenous DNA insert and were designed using Primer 5, based on the sequences of the assembled contigs from candidate reads across host genome and exogenous DNA sequences. The designed primers were purchased from Invitrogen, Shanghai, China, and are listed in Supplementary Table S1. The PCR reactions were performed using the cycling profile: 5 min at 98 °C, 35 cycles of 10 s denaturation at 98 °C, 30 s annealing at 55 °C, and 1-min extension at 72 °C, followed by a 7 min additional extension step at 72 °C. Following PCR amplification, the expected PCR amplicons were purified by the Qiagen MinElute PCR Purification Kit and subjected to Sanger sequencing by BGI Genomics Co Ltd., Shanghai, China.
2.6. Droplet digital PCR
Two droplet digital PCR (ddPCR) assays that targeted the exogenous rHSA gene and rice endogenous reference gene SPS were developed to evaluate the copy number of rHSA in GM rice line 114-7-2. The copy number of rHSA was calculated based on the ratio of rHSA and SPS (copy/copy). The primers and TaqMan probes for rHSA gene and SPS gene are listed in Supplementary Table S2. The ddPCR reaction was performed in a final volume of 20 µL and consisted of 10 µL of 2× ddPCR Supermix (Bio-Rad, USA), 1 µL 10 µM forward primer, 1 µL 10 µM reverse primer, 0.5 µL 10 µM probes, 1 µL DNA template, and 6.5 µL RNase-free and DNase-free water. The ddPCR was performed on a QX200 Droplet PCR platform (Bio-Rad, USA). The amplification conditions were: 5 min at 95 °C, 40 cycles of 30 s denaturation at 95 °C and 60 s extension at 60 °C, and a 10 min additional step at 72 °C. Droplet fluorescent signals were read on a QX200 Droplet Reader (Bio-Rad, USA). For each sample, the ddPCR reactions were repeated three times with triplicate samples.
3. Results
3.1. Sequencing data from WGS-PE and WGS-MP
For WGS-PE, a total of 21.1 GB and 20.9 GB of sequencing data corresponding to 44,205,804 and 43,894,757 read pairs of 100 bp were generated from GM line 114-7-2 and recipient line TP309 (WT), which represented approximately 23.61× and 23.45× sequence-level coverage, respectively (Table 1). Furthermore, both ends of 42,265,219 and 43,335,205 read pairs were mapped to the rice reference genome. The calibration weight R with values of 0.96 and 0.99 for GM and WT, respectively, was used to modify bias among the mapped sequencing data, which was calculated by the percentage of read pairs that mapped to the reference genome among the number of total trimmed paired reads. For WGS-MP, a total of 5.5G and 4.8G of sequencing data were generated for GM and WT, which corresponded to 11,596,364 and 10,101,309 read pairs, respectively. The sequencing coverage was up to 6.19× and 5.40× for GM and WT, respectively (Table 1). In total, 10,854,467 and 9,732,550 read pairs were mapped to the reference genome at both ends, with calibration weight R values of 0.94 and 0.96 for GM and WT, respectively.
Table 1.
Summary of sequencing and bioinformatics data.
| WGS-PE |
WGS-MP |
|||
|---|---|---|---|---|
| 114-7-2 | WT | 114-7-4 | WT | |
| Total trimmed read pairs | 44,205,804 | 43,894,757 | 11,596,364 | 10,101,309 |
| General sequencing depth | 23.61 | 23.45 | 6.19 | 5.39 |
| Q20 (%) | 96.66 | 95.18 | 96.34 | 96.26 |
| GC (%) | 42.96 | 42.29 | 43.92 | 43.73 |
| Type C reads | 33,724 | 5346 | 9238 | 1338 |
| Type B, D & E reads | 11,795 | 2827 | 2036 | 403 |
| False-positive reads from Gt13a and CP promoters | 8951 | 2654 | 1613 | 386 |
| Reads clustered around candidate insertion regions | 41 pairs for chr 1 site, 36 pairs for chr 4 site | / | 71 pairs for chr 1 site, 85 pairs for chr 4 site | / |
3.2. Bioinformatics pipelines used for molecular characterization
The pipeline of the previously developed TranSeq approach for molecular characterization was suitable to reveal the introduction of exogenous DNA in transgenesis events in which all the inserted DNA was derived from other species with low sequence similarity with the recipient genome (Yang et al., 2013). However, increasingly more transgenic elements and genes derived from the recipient genome have been introduced into new GM crops, which has led to difficulties in analyzing the inserted DNA using the previous TranSeq approach. For example, the two native rice promoters Gt13a (Genebank accession No. AP003256) and (CP) (GeneBank accession No. AL732346) were used to generate GM rice line 114-7-2. To reduce the interference from native elements, a single false-positive read screening strategy (Fig. 1B) was developed by searching for the similar position of sequences in the plasmid and reference with BLASTN. The candidate results were generated after blocking this position ± the insert size of homologous sequences. All the trimmed read pairs from WGS-PE and WGS-MP were analyzed according to the pipelines depicted in Fig. 1. For analysis of the WGS-PE data, 9,038 read pairs related to the rice Gt13a and CP promoters were obtained, and 8,951 read pairs were then filtered after screening for false-positive reads. For the analysis of the WGS-MP data, 2,036 read pairs were obtained, including 1,613 false-positive read pairs. The false-positive read ratios for the WGS-PE and WGS-MP data were 99.04% and 79.22%, respectively (Table 1). These results also revealed that more false-positive read pairs were present in the WGS-PE analysis, because the PE reads covered very short DNA fragments and the MP reads extended far beyond the full sequence of the two promoters. These false-positive read pairs closely matched the distribution of homologous sequences of the Gt13a and CP promoters within the rice reference genome (Supplementary Fig. S1). This indicates that the false-positive read-screening pipeline significantly reduced the amount of candidate reads for further analysis.
3.3. The exogenous DNA integration site and its flanking sequence
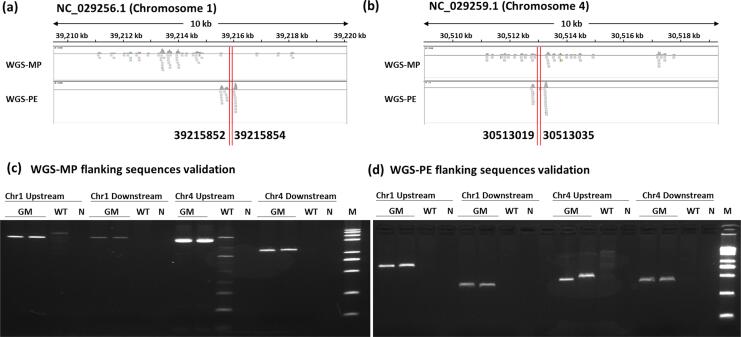
All the trimmed read pairs from WGS-PE and WGS-MP were analyzed according to the pipelines depicted in Fig. 1. For analysis of the WGS-PE data, 77 read pairs were extracted as candidates that might span the integration site of exogenous DNA, including 41 read pairs that mapped to Chr 01, and 36 read pairs that mapped to Chr 04 (Table 1). All candidate read pairs that mapped to Chr 01 and Chr 04 are illustrated by IGV (Fig. 2a), and the clustering pattern showed that exogenous DNA integrated between positions 39,215,496 and 39,216,046 on Chr 01, and the region between 30,512,708 and 30,513,281 on Chr 04. The flanking sequences of the inserted sites were obtained by assembling all 41 and 36 candidate read pairs (Supplementary File 3). For the WGS-MP data, 156 read pairs that targeted the integration site were obtained, among which 71 read pairs clustered on Chr 01 and 85 paired reads clustered on Chr 04. All the target read pairs are depicted in Fig. 2B, and indicate that two integration sites are present in the GM rice line.
Fig. 2.
Comparison of MP resequencing data and PE resequencing data of transgenic rice line 114-7-2. (a) Visualization of the candidate insertion sites I. (b) Visualization of the candidate insertion sites II. (c) PCR results for the insertion site on chromosome 1. Lane GM: GM rice 114-7-2 event; Lane WT: non GM riceTP309 line; Lane N: No template control; Lane M: 1 kb Plus Opti-DNA Marker; (d) PCR results for the insertion site on chromosome 4. Lane GM: GM rice 114-7-2 event; Lane WT: non GM riceTP309 line; Lane N: No template control; Lane M: DL2000 Marker.
To confirm the observed integration sites and their flanking sequences, PCR primers were designed on the basis of target reads that mapped to the upstream and downstream sequences of the potential locus (Table S1). PCR and Sanger sequencing data confirmed that one insert locus was located on Chr 01 at 39,215,852–39,215,854 and another on Chr 04 at 30,513,019–30,513,035, with 2-bp and 17-bp deletions, respectively (Fig. 2C, D). The insertion site on Chr 01 did not localize within currently annotated genes; however, the insertion site on Chr 04 located into the exon region of gene LOC4336906. The results of Sanger sequencing were consistent with those of WGS-PE and WGS-MP.
3.4. The arrangement and sequence of inserted exogenous DNA
Following confirmation of the integrated sites and flanking sequences, the read pairs that mapped to the transformed plasmid DNA sequences were used to reconstruct the arrangement of inserted DNA at the integration site. For example, 33,724 and 9,238 type C read pairs were obtained from WGS-PE and WGS-MP, respectively, for further de novo analysis (Table 1). Assembly of the data revealed that two contigs that mapped to the plasmid sequence were obtained from WGS-PE and WGS-MP data. For WGS-PE, one contig with 3,408 bp in length contained the rHSA gene expression cassette, and another contig of 2,219 bp contained the hpt marker-gene cassette (Supplementary File 4). For the WGS-MP analysis, two contigs contained the rHSA and hpt cassettes and were 4,668 bp and 3,646 bp long, respectively (Supplementary File 4). The contigs from WGS-MP contained additional and partial plasmid sequence information compared with those from WGS-PE. However, no contig that covered the entire integrated region located on Chr 01 and Chr 04 was obtained, indicating that the rearrangement or tandem repeats might exist in the integrated region.
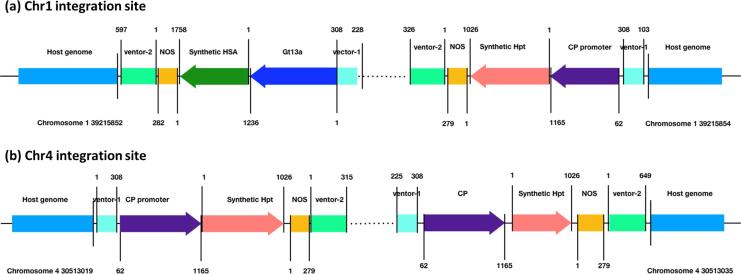
To reveal the internal structure of the integrated region, long-fragment PCR primers were designed according to the obtained flanking sequences and contigs (Supplementary Table S1). Sanger sequencing results of the amplicons revealed the sequence information of the upstream and downstream regions of each integrated site on Chr 01 and Chr 04. The internal structure of the inserted fragments at Chr 01 and Chr 04 are shown in Fig. 3. One rHSA target gene cassette and one hpt marker expression cassette was located at the upstream and downstream of the integration site on Chr 01, respectively. At the Chr 04 integration site, only the hpt marker expression cassette was observed at the upstream and downstream regions. Based on the results of the PCR amplification of long fragments, we believe that the rearrangement or tandem repeats might have occurred at both integration sites. However, we failed to reconstruct the whole inserted exogenous DNA according to the results of WGS-PE, WGS-MP, and long PCR fragments.
Fig. 3.
PCR verification of candidate insertion sites and the structure of exogenous sequences. (a) Exogenous sequence structure of the insertion site on chromosome 1. (b) Exogenous sequence structure of the insertion site on chromosome 4.
3.5. Copy number of exogenous DNA
The rHSA copy number was also evaluated according to formula 1 in Materials and Methods. The copy number of rHSA was calculated to be 5.81 and 5.00 by WGS-PE and WGS-MP, respectively (Supplementary Table S3). Furthermore, the rHSA gene copy number was also analyzed using the digital droplet PCR method using gene-specific primers and probes, and the copy number was calculated as the ratio of rHSA and the rice endogenous reference gene SPS (copy/copy) (Supplementary Fig. S2). The ddPCR results showed that the copy number of rHSA was 4.98 which is closer to the result from WGS-MP (Supplementary Table S3). These results also indicated that tandem repeats were generated by the GM rice transgenesis event.
3.6. Unintended plasmid backbone DNA
To evaluate the presence of residual plasmid backbone DNA, empty pCambia1301 and pUC18 plasmids that were used to construct the transformed plasmids pOsPMP114 and pOsPMP02 were selected, and their sequences were used as a reference against which the reads from WGS-PE and WGS-MP could be aligned. Backbone sequences were identified in WGS-PE and WGS-MP data (Supplementary Fig. S3). These indicated the presence of the KanR gene, the bom sequence from pCambia1301, and the AmpR promoter and AmpR gene from pUC18 in GM rice line 114-7-2.
3.7. The minimum resequencing data required for the analysis of exogenous DNA insertion loci
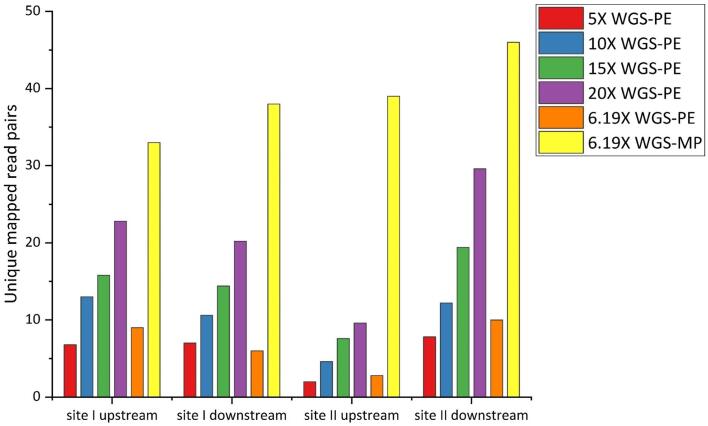
The mean depth of the WGS-PE data was higher than that for WGS-MP. The WGS-PE sequencing depths of 5×, 10×, 15×, 20× and 6.19× (the same for WGS-MP depth) were generated by randomly selecting reads from the WGS-PE data, and logarithmic statistics from the number of paired-end reads that mapped to the junctions of two insertion sites were used to evaluate the efficiency of detecting the insertion site. A total of five repeats were performed for the generation of the random reads from the WGS-PE data and corresponding logarithmic statistics. The mean number of read pairs from the WGS-PE data that mapped to the regions of exogenous DNA integration decreased with a decreasing depth of sequencing (Fig. 4). For example, the candidate reads could still be screened even at a low sequence depth of 5×, and 28 read pairs covered the integration sites from the 6.19× sequencing data. The number of candidate reads in the WGS-PE data was much lower than that in WGS-PE with the same sequencing depth. These results suggest that the minimum sequencing depth should be at least five-fold to effectively characterize the exogenous DNA integration.
Fig. 4.
Inferred number of read pairs across insertion sites under different input data volumes. Candidate read pairs from the PE sequencing data are uniquely mapping reads around two insertion sites under the different sequencing coverages of 5×, 6.19×, 10×, 15×, and 20×. The number of mapped reads is the total of type B, D and E read pairs that span the up- and downstream flanking sequences of two insertion sites; site I: chromosome 1, 39,215,852–39,215,854 and site II: chromosome 4, 30,513,019–30,513,035.
3.8. Comparison of the performance of WGS-PE and WGS-MP in molecular characterization
The above results showed that the WGS-PE and WGS-MP strategies were both successful in effectively characterizing GM rice line 114-7-2 molecularly, although the efficacy of both approaches differed in some respects. The advantages and disadvantages of WGS-PE and WGS-MP are summarized in Table 2. Firstly, the WGS-MP was more efficient than WGS-PE in identifying the DNA integration sites. For example, 77 read pairs that covered the integration sites were identified from the WGS-PE data with a sequencing depth of 23.61×, whereas only 29 read pairs were identified from the WGS-PE data with 6.19× depth. However, 156 read pairs were identified from the WGS-MP data with 6.19× depth, indicating that the WGS-MP was more effective than WGS-PE, although the sequencing depth of the WGS-PE data was much higher than that for the WGS-MP data. Secondly, the WGS-PE data were more effective than those of WGS-MP in assembling and validating the flanking sequence of the integration site, because the PE reads (500 bp) were much shorter than the MP reads (5,000 bp), and therefore, the flanking sequence could be identified directly, based on the isolated read pairs that covered the integration sites. However, the flanking sequence could not be easily obtained by assembling the isolated read pairs that covered the integrated sites, except for the WGS-MP data at a relatively high sequencing depth. For example, the flanking sequences of two integration sites on Chr 01 and Chr 04 were obtained by assembling 77 PE paired reads without further PCR amplification. Moreover, the validation of flanking sequences using PE reads was easier than that using MP reads because the efficiency and specificity of amplifying short DNA fragments by PCR were higher than that for long fragments. Thirdly, the WGS-MP data resulted in fewer false-positive results than the WGS-PE data in distinguishing the transgenes from endogenous DNAs of the recipient. For example, when the native rice Gt13a and CP promoters were transformed into GM rice line 114-7-2, the false-positive ratio was 79.22% according to the WGS-MP analysis, which was much lower than that from the WGS-PE analysis (99.04%). Finally, the WGS-PE approach showed a relatively high overall cost compared with WGS-MP, since the WGS-MP can provide more reads relating to the exogenous DNA integration than WGS-PE at the same sequencing depth. The sequencing costs of WGS-PE and WGS-MP were similar, although the construction of the MP library is relatively complex and time-consuming.
Table 2.
Different properties presented in mate-pair and paired-end strategies.
| Different properties | WGS-MP | WGS-PE |
|---|---|---|
| Recommended sequencing depth | About 5× genome size | Not less than 10× genome size |
| Flanking sequence information | Many and valuable | Little |
| Insertion sites and flanking sequence validation | Easy | Very easy |
| Number of false positives | Few | Extremely high |
| True positives in total results | High | Intermediate |
| Evaluation of copy number | Easy | Easy |
| Plasmid backbone residue analysis | Easy | Easy |
4. Discussion
The molecular characterization of GM rice line 114-7-2, including identification of the insertion site, flanking sequence, copy number of the exogenous target gene, and the presence of unintended plasmid backbone residues, was effective using both WGS-PE and WGS-MP strategies. There were two independent insertion sites in the GM rice line analyzed: one integration was located on Chr 01, and the other was on Chr 04. The Chr 01 insertion consisted of the insertion of the rHSA gene cassette and hpt gene expression cassette at Chr 01 position 39,215,852. The Chr 04 insertion consisted of integration of the hpt gene expression cassette at Chr 04 position 30,513,019. The flanking sequences of these two insertion sites were identified and are shown in Supplementary File 3, and the sequences could be used to develop event-specific detection methods for routine GMO analysis. The copy number of rHSA was determined to be five by WGS-PE, WGS-MP, and ddPCR analysis. The presence of plasmid backbone residues was also confirmed, including a partial KanR gene, bom sequence, the AmpR promoter, and the AmpR gene.
Although the GM rice line 114-7-2 was well characterized using the methodologies here, it was difficult to obtain the entire sequence and identify the internal DNA arrangement for each integration. The de novo analysis of isolated read pairs that mapped to transformed plasmids revealed only two contigs that were related to the exogenous DNA; one mapped to the rHSA gene expression cassette, and the other to the hpt cassette. Together with the copy number and flanking sequence, we presume that the tandem repeats of rHSA and hpt gene cassettes were present in the integration region. Although MP reads with a length of ∼5,000 bp were effective in analyzing complex genome structures (Srivastava et al., 2014), the WGS-MP approach could not effectively resolve multiple DNA repeats. Available techniques for sequencing long DNA fragments without additional PCR amplification, such as SMAT-Seq and nanopore sequencing, might reveal the internal arrangement of exogenous DNA insertions that contain a high number of repeats.
The native transgenic elements from the recipient genome in transformation plasmids increase the difficulty of bioinformatics analysis to molecularly characterize GM crops, and the analysis of cisgenesis and RNAi transgene crop lines remains challenging. In GM rice 114-7-2, the native rice Gt13a and CP promoters were used in the rHSA gene and hpt gene cassettes, which generated many false-positive read pairs and introduced much background noise into the isolation of the reads that covered the junction of the insertion sites. Among the isolated reads, 99.04% read pairs were falsely positive and derived from the Gt13a and CP promoters in the WGS-PE data, and this value was 79.22% for the WGS-MP data. The high false-positive rate made it difficult to identify the expected integration site. Therefore, a specific pipeline to block the false-positive reads resulting from endogenous DNA in the recipient genome will improve molecular characterization analyses. We developed a false-positive read screening pipeline to exclude the related reads from Gt13a and CP promoters, in the analysis of both WGS-PE and WGS-MP. In total, 8,951 read pairs were excluded from 9,038 isolated read pairs from the WGS-PE data, and 1,613 read pairs were excluded from the WGS-MP data, indicating that the screening pipeline for false-positive reads dramatically improved the efficiency and accuracy of detection. These results suggest that the developed screening pipeline might be useful to analyze GM lines transformed with endogenous recipient DNA, even for cisgenesis and RNAi transgenic lines.
In summary, the WGS-PE and WGS-MP strategies could successfully molecularly characterize the GM rice 114-7-2. The evaluation of the performance of both strategies indicated that the WGS-MP has a much higher cost performance than WGS-PE. Therefore, we believe that the WGS-MP strategy is more effective and suitable for the molecular characterization of GM crops.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Transgenic Plant Special Fund (2016ZX08012-002 and 2016ZX08012-003), and the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochms.2021.100061.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alimentarius C. Guideline for the conduct of food safety assessment of foods derived from recombinant-DNA plants. CAC/GL. 2003;45:1–18. [Google Scholar]
- Arulandhu A.J., van Dijk J., Staats M., Hagelaar R., Voorhuijzen M., Molenaar B.…Kok E. NGS-based amplicon sequencing approach; towards a new era in GMO screening and detection. Food Control. 2018;93:201–210. doi: 10.1016/j.foodcont.2018.06.014. [DOI] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S.…Pevzner P.A. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debode F., Hulin J., Charloteaux B., Coppieters W., Hanikenne M., Karim L., Berben G. Detection and identification of transgenic events by next generation sequencing combined with enrichment technologies. Scientific Reports. 2019;9(1) doi: 10.1038/s41598-019-51668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doori P., Su-Hyun P., Yong W., Ban Y., Shic K. A bioinformatics approach for identifying transgene insertion sites using whole genome sequencing data. BMC Biotechnology. 2017 doi: 10.1186/s12896-017-0386-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOO Nations Codex alimentarius: Food hygiene, basic texts. Codex Alimentarius Food Hygiene Basic Texts. 2009 [Google Scholar]
- Goodwin S., McPherson J.D., McCombie W.R. Coming of age: Ten years of next-generation sequencing technologies. Nature Review Genetics. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Guo Y., Hong H., Qiu L.J. Identification of Genomic Insertion and Flanking Sequence of G2-EPSPS and GAT Transgenes in Soybean Using Whole Genome Sequencing Method. Frontiers in Plant Science. 2016;7:1009. doi: 10.3389/fpls.2016.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttikonda S.K., Marri P., Mammadov J., Ye L., Soe K., Richey K.…Jain M. Molecular Characterization of Transgenic Events Using Next Generation Sequencing Approach. PLoS ONE. 2016;11(2):e0149515. doi: 10.1371/journal.pone.0149515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Ning T., Xie T., Qiu Q., Zhang L., Sun Y.…Yang D. Large-scale production of functional human serum albumin from transgenic rice seeds. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(47):19078–19083. doi: 10.1073/pnas.1109736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina (2009) Mate Pair Library v2 Sample Preparation Guide For 2–5 kb Libraries. https://support.illumina.com.cn/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_legacy/MatePair_v2_2-5kb_SamplePrep_Guide_15008135_A.pdf.
- James C. (2019). Global Status of Commercialized Biotech/GM Crops:2019. ISAAA Brief No. 55, International Service for the Acquisition of Agri-biotech Applications, Ithaca, NY.
- Johnson S.H., Smadbeck J.B., Smoley S.A., Gaitatzes A., Murphy S.J., Harris F.R.…Vasmatzis G. SVAtools for junction detection of genome-wide chromosomal rearrangements by mate-pair sequencing (MPseq) Cancer Genet. 2018;221:1–18. doi: 10.1016/j.cancergen.2017.11.009. [DOI] [PubMed] [Google Scholar]
- König A., Cockburn A., Crevel R.W.R., Debruyne E., Grafstroem R., Hammerling U.…Wal J.M. Assessment of the safety of foods derived from genetically modified (gm) crops. Food & Chemical Toxicology. 2004;42(7):1047–1088. doi: 10.1016/j.fct.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Košir A.B., Arulandhu A.J., Voorhuijzen M.M., Xiao H., Hagelaar R., Staats M.…Dijk J.V. ALF: A strategy for identification of unauthorized GMOs in complex mixtures by a GW-NGS method and dedicated bioinformatics analysis. Scientific Reports. 2018;8(1):17645. doi: 10.1038/s41598-018-35950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalic D., Garnaat C., Guo L., Yan Y., Groat J., Silvanovich A.…Bannon G. The Use of Next Generation Sequencing and Junction Sequence Analysis Bioinformatics to Achieve Molecular Characterization of Crops Improved Through Modern Biotechnology. The Plant Genome Journal. 2012;5(3) doi: 10.3835/plantgenome2012.10.0026. [DOI] [Google Scholar]
- Kuiper H.A., Kleter G.A., Noteborn H.P., Kok E.J. Assessment of the food safety issues related to genetically modified foods. Plant Journal. 2010;27(6):503–528. doi: 10.1046/j.1365-313X.2001.01119.x. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Quan S., Yan X., Biswas S., Zhang D., Shi J. Molecular characterization of genetically-modified crops: Challenges and strategies. Biotechnology Advances. 2017;35(2):302–309. doi: 10.1016/j.biotechadv.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Ning T., Xie T., Qiu Q., Yang W., Zhou S., Zhou L.…Yang D. Oral administration of recombinant human granulocyte-macrophage colony stimulating factor expressed in rice endosperm can increase leukocytes in mice. Biotechnology Letters. 2008;30(9):1679–1686. doi: 10.1007/s10529-008-9717-2. [DOI] [PubMed] [Google Scholar]
- Patel R.K., Jain M., Liu Z. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7(2):e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nature Biotechnology. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Philip V.M., Greenstein I., Rowe L.B., Barter M., Lutz C., Reinholdt L.G. Discovery of transgene insertion sites by high throughput sequencing of mate pair libraries. BMC Genomics. 2014;15(1):367. doi: 10.1186/1471-2164-15-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends in Genetics. 2014;30(9):418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Xie T., Qiu Q., Zhang W., Ning T., Yang W., Zheng C.…Yang D. A biologically active rhIGF-1 fusion accumulated in transgenic rice seeds can reduce blood glucose in diabetic mice via oral delivery. Peptides. 2008;29(11):1862–1870. doi: 10.1016/j.peptides.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Yang L., Wang C., Holst-Jensen A., Morisset D., Lin Y., Zhang D. Characterization of GM events by insert knowledge adapted re-sequencing approaches. Scientific Reports. 2013;3:2839. doi: 10.1038/srep02839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zastrow‐Hayes G.M., Lin H., Sigmund A.L., Hoffman J.L., Alarcon C.M., Hayes K.R.…Beatty M.K. Southern-by-sequencing: A robust screening approach for molecular characterization of genetically modified crops. The Plant Genome. 2015;8(1) doi: 10.3835/plantgenome2014.08.0037. [DOI] [PubMed] [Google Scholar]
- Zhang R., Yin Y., Zhang Y., Li K., Zhu H., Gong Q.…Zhao S. Molecular characterization of transgene integration by next-generation sequencing in transgenic cattle. PLoS ONE. 2012;7(11):e50348. doi: 10.1371/journal.pone.0050348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang H., Qu Z., Zhang X., Cui J., Wang C., Yang L. Comprehensive analysis of the molecular characterization of gm rice g6h1 using a paired-end sequencing approach. Food Chemistry. 2020;309:125760. doi: 10.1016/j.foodchem.2019.125760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.