Highlights
-
•
Fermented nozawana pickle with starter culture strains was studied by metabolomics.
-
•
NMR and SPME-GC/MS analyses revealed starter culture-dependent metabolite profiles.
-
•
Various flavor compounds showed significantly different levels among the strains.
-
•
Latilactobacillus curvatus exhibited higher isothiocyanate intensities at a maximum 3.30-fold.
-
•
Levilactobacillus brevis influenced on dimethyl trisulfide and S-methyl thioacetate levels.
Keywords: Fermented food, Fermented pickle, Cruciferous vegetables, Lactic acid bacteria, Metabolomics, NMR, SPME-GC/MS, Isothiocyanates
Abstract
Metabolomic characterization of a lactic-fermented pickle of nozawana (Brassica rapa L. var. hakabura) was conducted to evaluate the effects of different starter culture strains on the chemical profiles. We compared the profiles of water-soluble and volatile compounds obtained by non-targeted nuclear magnetic resonance and solid-phase microextraction gas chromatography/mass spectrometry analyses. Principal component analyses indicated that the fermented samples differed significantly in terms of the levels of various compounds, including taste- and aroma-active components, such as water-soluble residual sugars, organic acids, mannitol, ethanol, dihydroxyacetone, ornithine, γ-aminobutyric acid, choline, volatile isothiocyanates, 3,4-epithiobutyl cyanide, 2,3-butanedione, acetoin, ethyl acetate, dimethyl trisulfide, and S-methyl thioacetate. Fermentation with a Latilactobacillus curvatus culture was associated with a unique metabolite profile characterized by higher levels of isothiocyanates and hexanoic acid and lower levels of lactic acid, acetic acid, acetoin, and 2,3-butanedione. These variations in the chemical profile might be associated with different qualities in fermented nozawana pickle products.
1. Introduction
Nozawana (Brassica rapa L. var. hakabura) is a variety in the cruciferous plant family that is common in foods worldwide and is an economically and culturally important vegetable. Pickled vegetables are popular processed food products in Japan and elsewhere, and pickled nozawana (called nozawana-zuke) enjoys the largest market share in Japan among pickled leafy vegetables. A traditional method for preparing nozawana-zuke is fermentation with lactic acid bacteria (LAB). Currently, however, most nozawana-zuke products are manufactured without fermentation as quick-pickled nozawana-zuke.
Nonetheless, fermented nozawana-zuke remains popular because of its complex flavor profile that develops during lactic acid fermentation and that is unique to each producer. In practical manufacturing, fermented nozawana-zuke is prepared by spontaneous fermentation utilizing the activity of autochthonous LAB rather than a starter culture, likely derived from raw materials. Although conventional spontaneous fermentation is a simple and low-cost alternative to the starter culture method, it involves management of the fermentation process and product quality due to uncontrollable bacterial communities. To stabilize the fermentation process and product quality of fermented nozawana-zuke, it is essential to understand the bacterial community structure and the impact of various LAB on the chemical composition.
Several studies have analyzed the bacterial communities present in spontaneously fermented nozawana-zuke. Recently, a culture-independent analysis of LAB in fermented nozawana-zuke found that Latilactobacillus curvatus (formerly Lactobacillus curvatus) was dominant, followed by Lactiplantibacillus plantarum (formerly Lactobacillus plantarum) and Levilactobacillus brevis (formerly Lactobacillus brevis), and occasionally by Latilactobacillus sakei (formerly Lactobacillus sakei) (Sandagdorj, Hamajima, Kawahara, Watanabe, & Tanaka, 2019). A study employing a culture-dependent method isolated Leuconostoc mesenteroides and Latilactobacillus curvatus as dominant LAB and detected no LAB of the genera Streptococcus, Pediococcus, or Lactococcus (Osawa, Takanami, Kuribayashi, & Kuwahara, 2005). A bacteriocin-producing LAB strain isolated from fermented nozawana-zuke has also been identified as Latilactobacillus curvatus (Kawahara, Iida, Toyama, & Fukuda, 2010). In an earlier study, Leuconostoc mesenteroides and Limosilactobacillus fermentum (formerly Lactobacillus fermentum) were isolated from commercial nozawana-zuke products (Nakagawa et al., 2001). The results of these studies also indicated that the bacterial composition of spontaneously fermented nozawana-zuke changed depending on the duration of fermentation. In the fermentation of sauerkraut, which is one of the most studied fermented pickles of cruciferous vegetable, it is well known that the bacterial composition, in general, shows a successive growth of several different LAB species including Leuconostoc mesenteroides, Levilactobacillus brevis, and Lactiplantibacillus plantarum (Pederson & Albury, 1969). Additionally, the chemical characteristics (total acid production and the ratio of organic acids) are associated with the succession and the changes in composition of LAB species resulting from different fermentation conditions such as temperature and NaCl concentration (Pederson & Albury, 1969).
These microbiological studies on nozawana-zuke suggest that these LAB are important facilitators of nozawana-zuke fermentation and can be potentially used as starter cultures to control chemical composition. However, to the best of our knowledge, the impact of these LAB on the compositional characteristics of nozawana-zuke has not yet been empirically assessed, and the components that exhibit common or different changes among the starter culture strains remain unclear. Considering the biodiversity of LAB (Claesson et al., 2007, Gänzle, 2009), it is hypothesized that LAB play multiple roles in nozawana-zuke fermentation, leading to differences in the metabolite profile. In recent years, comprehensive compositional analysis via the metabolomics approach has been performed in studies on various fermented foods, including fermented pickles (Mozzi et al., 2013, Park et al., 2016, Randazzo et al., 2017). The application of metabolomic analysis to clarify the changes in a wide range of metabolites produced through starter culture fermentation of nozawana-zuke is promising.
Nozawana is a popular, locally cultivated ingredient long since established as a traditional regional vegetable in the Nagano Prefecture of Japan. This study analyzed and compared the compositional profiles of nozawana-zuke samples fermented by four different starter cultures alongside a sample fermented by a non-starter culture to assess the potential impacts of starter culture LAB on the chemical characteristics. Compositional analysis was conducted using non-targeted metabolomics based on a combined application of nuclear magnetic resonance (NMR) spectroscopy and solid-phase microextraction (SPME) gas chromatography/mass spectrometry (GC/MS) to obtain in-depth compositional profiles of water-soluble and volatile components.
2. Materials and methods
2.1. Bacterial strains
The LAB strains used in this study were isolated from homemade and commercially available nozawana-zuke produced in Nagano, Japan, as described previously (Sandagdorj, et al., 2019). The four isolates used for starter cultures were Lactiplantibacillus plantarum K4G4 and K5G3, Latilactobacillus curvatus ♯4G2, and Levilactobacillus brevis K4G1. The taxonomy of the four strains was verified by 16S rRNA gene sequencing and species-specific polymerase chain reaction assays (Berthier and Ehrlich, 1999, Fusco et al., 2016, Torriani et al., 2001). Cultures were grown in Lactobacilli MRS Broth (Difco Laboratories, Detroit, MI, USA). Plate Count Agar with BCP (Nissui Pharmaceutical Co., Tokyo, Japan) was used to measure viable LAB cell counts of the fermented samples.
2.2. Fermentation of nozawana-zuke
Nozawana-zuke fermentation was performed based on a two-step, well-established industrial procedure for pre-salting and fermentation. Fresh nozawana raw material was obtained from Takeuchi Nosan Corp. (Nagano, Japan), washed with water, and chopped into 2.5-cm sections. Sections were washed in 100 ppm NaClO (Sankyo Co., Nagano, Japan) for 3 min, thoroughly rinsed with water, and then salted with 10 kg of 6% brine per 20 kg of material for 2 d at 7 °C. The material was again rinsed with water, and the viable cells in the pre-salted material were confirmed to be <1 × 103 CFU/g. Then, portions of 200 g of material and 100 g of brine were placed in heat-sealed nylon bags (BA-1727H; Meiwa Pax Co. Ltd., Osaka, Japan), the salt concentration was adjusted to 1.5%, and starter cultures was added at a population of 1 × 105 CFU/g. Separate bags containing only the material and brine were used as controls. Three bags were prepared for each of the four bacterial strains and non-starter control, and the samples were fermented at 10 °C for 7, 14, or 21 d. Separate bags were used for each fermentation period in single replicate to avoid potential contamination from iterative handling.
After fermentation, the bag components were gently homogenized in a stomacher (Masticator 400S; Gunze Sangyo, Inc., Tokyo, Japan) for 1 min at room temperature, and the liquid was decanted from each bag. The liquid samples were analyzed to measure pH, titratable acidity (TA) calculated as lactic acid, and viable LAB cell counts. The samples were then subjected to metabolomic characterization.
2.3. NMR analysis
Water-soluble compounds were analyzed by NMR spectroscopy as described previously but with minor modifications (Tomita et al., 2015). Briefly, NMR spectra were measured on an Avance III 500 MHz (Bruker, Billerica, MA, USA) equipped with a CryoProbe CPBBO and SampleJet automatic sample changer (Bruker). Analytical sample preparation was performed using a previous method with minor modifications (Tomita, Nakamura, & Okada, 2018). Briefly, 130 µL of sample supernatant centrifuged at 20,500×g for 5 min at 4 °C was mixed with 520 µL of 125 mM potassium phosphate buffer (pH 7.0) in deuterium oxide (99.9% D; Cambridge Isotope Laboratories, Andover MA, USA). After centrifuging again, 600 µL of clear supernatant was transferred to NMR sample tubes (5.0 mm O.D. × 103.5 mm; Norell, Landisville, NJ, USA). Proton (1H) NMR spectra were recorded using a Bruker pulse program zgpr with the following parameters: spectral width, 20 ppm; offset frequency, 4.7 ppm; 1H 90° pulse, 13.5 µs; relaxation delay, 4 s; and number of scans, 128. Metabolite annotation (tentative identification) was performed as described previously (Tomita, et al., 2015). Two-dimensional 1H–13C heteronuclear single quantum coherence spectra were also recorded and used for metabolite annotation as described previously (Tomita et al., 2018, Tomita et al., 2015).
2.4. SPME-GC/MS analysis
Profiles of volatile compounds were obtained using a previously described method with minor modifications (Tomita, et al., 2018). Briefly, 3 mL of sample was transferred to a 20-mL screw cap vial (Shimadzu GLC Ltd., Tokyo, Japan) and stored at 4 °C until use. Then, samples were placed in an agitator unit at 50 °C for 10 min, and volatile components were extracted for 20 min onto a 2-cm long DVB/CAR/PDMS fiber (Sigma-Aldrich, St. Louis, MO, USA). The agitator unit was rotated at 250 rpm during extraction. Volatile compounds were desorbed from the fiber for 3 min at 230 °C in splitless mode and were resolved on an Rtx-WAX capillary column (60 m × 0.25 mm I.D. × 0.25 μm film thickness; Restek, Bellefonte, PA, USA) with helium as the carrier gas. The column temperature program was as follows: 40 °C for 5 min, then increased to 230 °C at 5 °C/min, and maintained at 230 °C for 5 min. Analyses were conducted in duplicate to confirm reproducibility. Detected peaks were annotated based on the mass spectrum similarity and retention index in the NIST 02 MS Library (National Institute of Standards and Technology, Gaithersburg, MD, USA).
2.4.1. Dataset preparation and multivariate analysis
Datasets for multivariate analyses were prepared from 1H NMR spectra and GC/MS chromatograms. For water-soluble compounds, NMR spectra were subdivided into 0.04 ppm integral regions (buckets), and signal intensities in each bucket were calculated using Amix Software (Bruker). Buckets were normalized against the signal intensity of the internal standard DSS-d6 at 0.00 ppm, and buckets with residual solvent signals were replaced with zeroes. Buckets generated from the spectral noise region with a maximum signal intensity of <0.005 were removed to eliminate noise contamination. For volatile compounds, baseline correction and peak alignment of GC/MS chromatograms were performed using MetAlign (Lommen, 2009). Mass peaks derived from a single compound were integrated using AIoutput (Tsugawa, Bamba, Shinohara, Nishiumi, Yoshida, & Fukusaki, 2011) at a height threshold of 5000 and an RSD filter value of 100. After these processes, the NMR and GC/MS datasets comprised 119 buckets and 127 peaks, respectively. Principal component analysis (PCA) was performed using SIMCA software (version 14; Umetrics, Umeå, Sweden) with Pareto scaling.
2.5. Statistical analysis
Integral values from highlighted metabolites were obtained by manually subdividing spectra using the variable-sized buckets option in Amix software (Bruker) by specifying spectral ranges containing an isolated signal of a single compound. For GC/MS analysis, chromatograms were processed using GCMS solution software (Shimadzu Co., Kyoto, Japan), and the peak areas of volatile compounds were recorded for signal intensity values. Statistical significance was analyzed using Tukey’s multiple comparison test with R software (version 3.6.1; R Core Development Team, Vienna, Austria), and a 0.05 level of probability was used as the criterion for significance.
3. Results
3.1. Fermented nozawana-zuke preparation
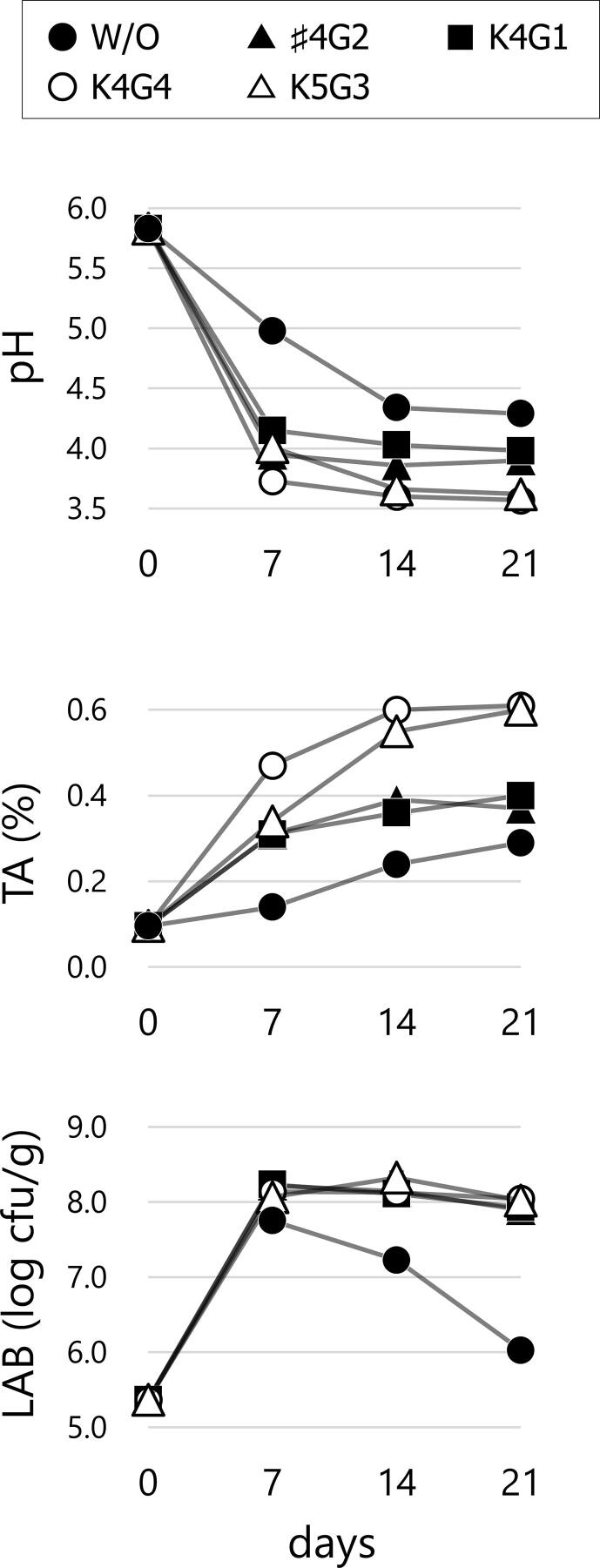
Samples were collected from the pickling liquid of five nozawana-zuke fermentations with one of four starter cultures or without a starter culture (control) at different time points during fermentation. The decreases in pH and increases in TA varied among the samples (Fig. 1). The pH of starter culture samples (S/C) rapidly decreased over the first 7 d to approximately 4.0. The sample without a starter culture (W/O) presented with less prominent TA increases over a greater period of time, and the pH did not decrease to the same level as that in the S/C samples. Viable LAB cell counts were comparable among all samples for the first 7 d. However, LAB cell counts were maintained only in the S/C samples throughout the experimental period, and the W/O sample presented with decreased cell counts after 7 d.
Fig. 1.
Changes in pH, titratable acidity, and viable cell counts of nozawana-zuke fermented with or without starter cultures. The result of viable LAB cell counts is depicted as average value of two analytical replicates.
3.2. Compositional analysis of fermented nozawana-zuke
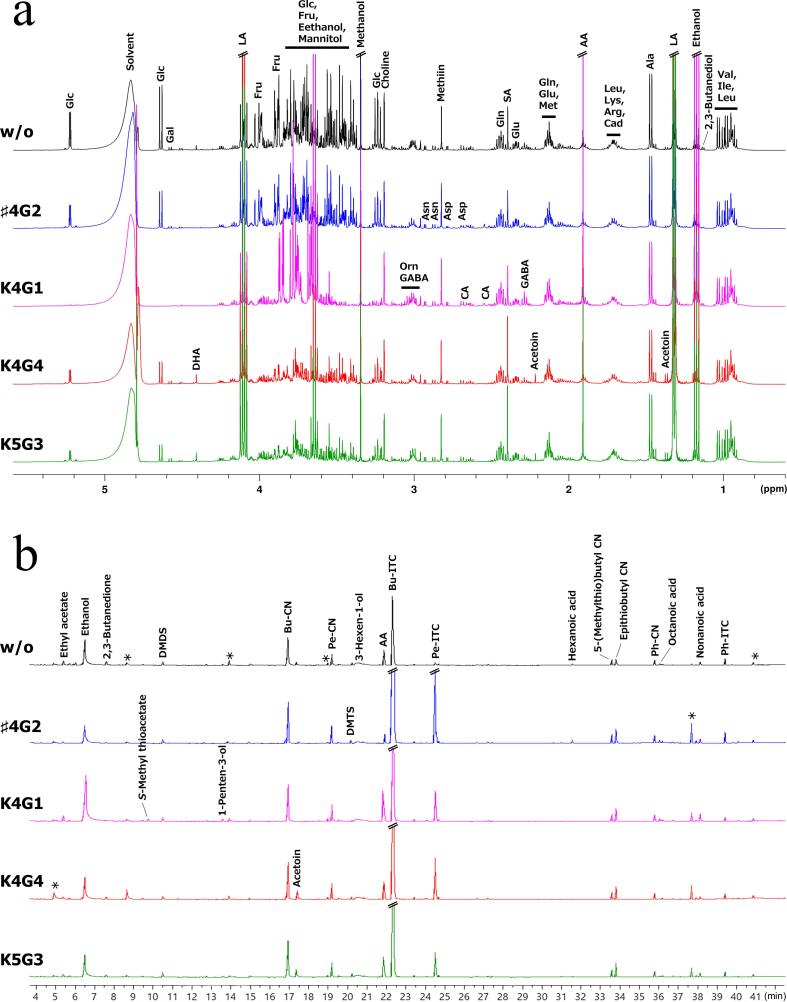
NMR and SPME-GC/MS were used to analyze the chemical compositions of water-soluble and volatile components, respectively. The dominant signals observed in the NMR spectra indicated that the major metabolites were organic acids: lactic acid (LA), acetic acid (AA), and succinic acid (SA); aldoses and alditols: glucose (Glc), fructose (Fru), and mannitol; alcohols: ethanol and methanol; amino acids: alanine (Ala), glutamic acid (Glu), glutamine (Gln), valine (Val), leucine (Leu), and isoleucine (Ile); and choline and methiin (Fig. 2a). Signals with lower intensities included citric acid (CA), formic acid, fumaric acid, γ-aminobutyric acid (GABA), galactose, trehalose, 2,3-butanediol, phenylalanine (Phe), tyrosine (Tyl), aspartic acid (Asp), asparagine (Asn), histidine (His), tryptophan (Trp), acetoin, ornithine (Orn), dihydroxyacetone (DHA), and uracil. In two-dimensional NMR spectra, more metabolites were annotated, including arabinose (Ara), myo-inositol, arginine (Arg), lysine (Lys), methionine (Met), glycine (Gly), serine (Ser), threonine (Thr), proline (Pro), pyroglutamic acid (Glp), and cadaverine (Cad) (Fig. S1).
Fig. 2.
Compositional analyses of water-soluble and volatile compounds of nozawana-zuke pickling juice by NMR and SPME-GC/MS. (a) 1H NMR spectra and metabolite annotations of samples at 21 d. The spectral region ranging from 5.60 to 0.60 ppm is displayed. (b) Total ion chromatograms of SPME-GC/MS analysis and metabolite annotations. The labels U and asterisk (*) represent peaks of unannotated compounds and those observed in blank measurements, respectively. The following bacterial strains were used: ♯4G2, Latilactobacillus curvatus; K4G1, Levilactobacillus brevis; and K4G4 and K5G3, Lactiplantibacillus plantarum.
The GC/MS chromatograms of the samples at 21 d showed that isothiocyanates (ITCs), including 3-butenyl ITC (Bu-ITC) and 4-pentenyl ITC (Pe-ITC), were the common major peaks, followed by AA, ethanol, and cyanides (CNs) such as 3-butenyl CN (Bu-CN) and 4-pentenyl CN (Pe-CN) (Fig. 2b). Lower-intensity peaks were observed for dimethyl disulfide (DMDS), dimethyl trisulfide (DMTS), 3-hexen-1-ol, phenethyl CN (Ph-CN), phenethyl ITC (Ph-ITC), 3,4-epithiobutyl-CN (epithiobutyl-CN), 5-(methylthio)butyl CN, octanoic acid, and nonanoic acid. The levels of ethyl acetate, 2,3-butanedione, S-methyl thioacetate, acetoin, and hexanoic acid varied among samples. The detected GC/MS peaks are listed in Table S1. Minute peaks were also observed, but these peaks did not contribute substantially to the differences among samples in the metabolomic analyses.
3.3. NMR metabolomics of fermented nozawana-zuke
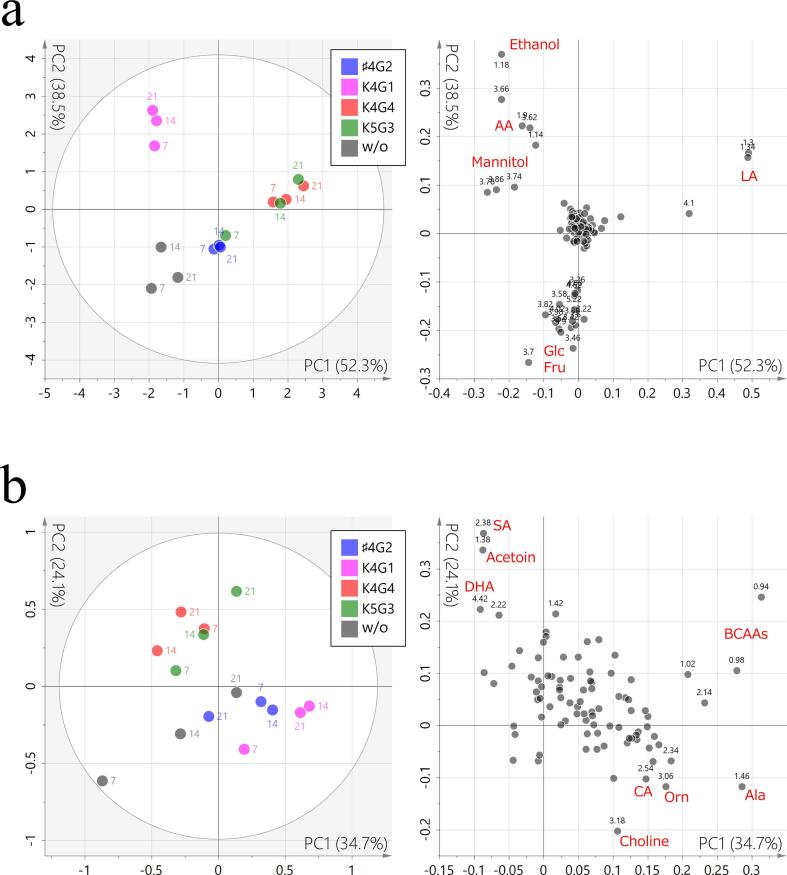
Differences in the chemical composition among the W/O and S/C samples were evaluated by PCA based on the dataset generated from the 1H NMR spectra. In a comparison applying all variables in the dataset (Fig. 3a), the first and second principal component (PC1 and PC2, 52.3% and 38.5% of the total variance, respectively) indicated that the differences depended more on the starter culture than on the fermentation duration. The loading plot explained the different contributions of the major signals observed in the NMR spectra. LA was strongly represented in Lactiplantibacillus plantarum K4G4 and K5G3 starter cultures, followed by that for Latilactobacillus curvatus ♯4G2. Ethanol, AA, and mannitol dominated in Levilactobacillus brevis K4G1 (Fig. 3a), whereas Glc and Fru dominated in W/O (Fig. 3a).
Fig. 3.
PCA of water-soluble compounds in nozawana-zuke samples. PC1–PC2 planes of score (left) and loading (right) plots obtained from (a) PCA and (b) two-step PCA. In the score plots, samples are color-coded as shown in the legend and labeled by fermentation duration in days. In the loading plots, numerical labels in black represent chemical shifts in ppm. The variables contributing to the feature space are labeled with metabolite names in red. The following bacterial strains were used: ♯4G2, Latilactobacillus curvatus; K4G1, Levilactobacillus brevis; and K4G4 and K5G3, Lactiplantibacillus plantarum. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Two-step PCA was then conducted to assess the effects of variables presenting at lower intensities by excluding those derived from major components. The obtained feature space showed a class separation along the PC2 axis between Lactiplantibacillus plantarum K4G4 and K5G3 and Latilactobacillus curvatus ♯4G2, Levilactobacillus brevis K4G1, and W/O (Fig. 3b). SA, acetoin, and DHA contributed to the former class, whereas choline, Orn, GABA, and CA contributed to the latter class. PC1 accounted for differences within strains, such as fermentation duration, as indicated by Ala and branched-chain amino acids, including Val, Leu, and Ile.
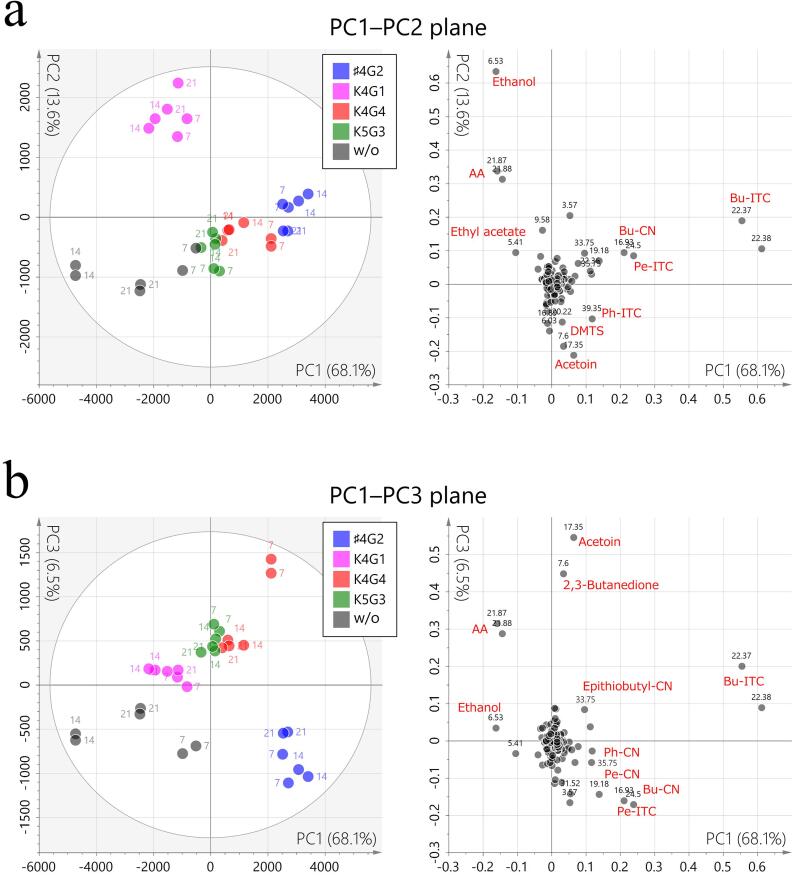
3.4. SPME-GC/MS metabolomics of fermented nozawana-zuke
We explored differences in GC/MS peak profiles among samples to examine the impact of fermentation with starter cultures versus that without starter cultures. In agreement with the PCA results of NMR metabolomics, GC/MS PCA data also revealed more pronounced differences by starter culture type rather than by fermentation duration. The PC1–PC2 plane of the score plot shows a sequential distribution of the strains Lactiplantibacillus plantarum K4G4 and K5G3 and Latilactobacillus curvatus ♯4G2 along PC1 and an isolated class of Levilactobacillus brevis K4G1 along PC2 (Fig. 4a). The loading plot indicated that ITCs and CNs contributed to the former, particularly to Latilactobacillus curvatus ♯4G2. Levilactobacillus brevis K4G1 was characterized by ethanol and AA, consistent with the NMR metabolomics findings. The samples of Levilactobacillus brevis K4G1 and W/O also showed the presence of ethyl acetate. Upon examination of the third principal component (PC3), a clear class separation of Latilactobacillus curvatus ♯4G2 from the others was observed (Fig. 4b). The samples were negatively characterized by low levels of acetoin and 2,3-butanedione, in addition to the presence of Pe-ITC and Bu-CN and Pe-CN.
Fig. 4.
PCA of volatile compounds in nozawana-zuke samples. (a) PC1–PC2 and (b) PC1–PC3 planes of score (left) and loading (right) plots. In the score plots, the samples are color-coded as shown in the legend and labeled with fermentation duration in days. In the loading plots, numerical labels in black represent retention time in min. The variables contributing to the feature space are labeled with metabolite names in red. The following bacterial strains were used: ♯4G2, Latilactobacillus curvatus; K4G1, Levilactobacillus brevis; and K4G4 and K5G3, Lactiplantibacillus plantarum. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Significance of metabolites and their time-course changes
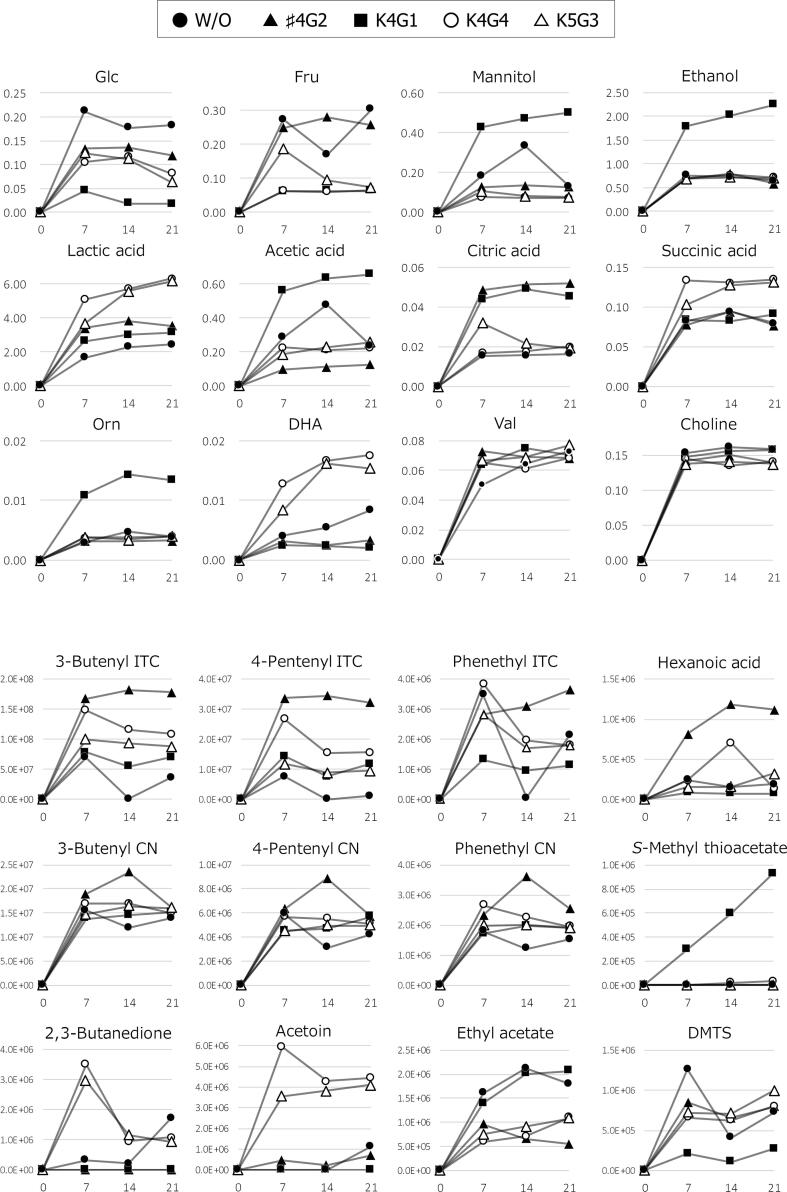
The significance of the differences in the metabolites highlighted in the metabolomic analyses was assessed by subjecting the signal intensity calculated from the peaks of individual compounds to statistical analysis by multiple comparison tests (Table 1). Statistical significance was confirmed for most of the tested metabolites, but patterns of significance among groups were complex. Of these, differences separating the samples into more than three groups were significant for the levels of Glc, Fru, LA, AA, mannitol, choline, Bu-ITC, Pe-ITC, and ethyl acetate. We also found that certain metabolites were characteristic to certain strains, such as Bu-ITC, Pe-ITC, epithiobutyl-CN, and hexanoic acid for Latilactobacillus curvatus ♯4G2; AA, ethanol, mannitol, Orn, S-methyl thioacetate, and DMTS for Levilactobacillus brevis K4G1; and LA, SA, DHA, choline, acetoin, and 2,3-butanedione for Lactiplantibacillus plantarum strains K4G4 and K5G3. Conversely, the other metabolites highlighted by PCA, such as Val, Ile, Leu, Ala, Ph-ITC, Bu-CN, and Pe-CN, did not vary significantly among strains. Time-course data shown in Fig. 5 indicated that the levels of most metabolites in pickling juice, such as LA, AA, SA, ethanol, Orn, Bu-ICT, Bu-CN, hexanoic acid, and ethyl acetate, reached around maximum intensity by 7 d and remained at this intensity throughout the 3-week experimental period (Fig. 5). However, there were exceptions: the 2,3-butanedione intensity decreased sharply from 7 to 14 d in Lactiplantibacillus plantarum K4G4 and K5G3 and the S-methyl thioacetate intensity gradually increased over the 21-d period in Levilactobacillus brevis K4G1.
Table 1.
Results of multiple comparison test for the compounds highlighted in metabolomic characterization.
| Signal intensity ± standard deviation* |
|||||
|---|---|---|---|---|---|
| Compound | W/O starter culture | Latilactobacillus curvatus ♯4G2 | Levilactobacillus brevis K4G1 | Lactiplantibacillus plantarum K4G4 | Lactiplantibacillus plantarum K5G3 |
| NMR peaks | |||||
| Glc | 0.190 ± 0.019 a | 0.130 ± 0.009 b | 0.027 ± 0.016 c | 0.101 ± 0.018 b | 0.100 ± 0.032 b |
| Fru | 0.248 ± 0.069 a | 0.262 ± 0.016 b | 0.062 ± 0.001 c | 0.063 ± 0.003 c | 0.117 ± 0.060 c |
| LA | 2.131 ± 0.406 a | 3.596 ± 0.223 ab | 2.928 ± 0.272 a | 5.706 ± 0.606 c | 5.126 ± 1.311 bc |
| AA | 0.335 ± 0.127 a | 0.113 ± 0.013 bc | 0.617 ± 0.050 d | 0.221 ± 0.007 ac | 0.225 ± 0.036 ac |
| CA | 0.016 ± 0.001 a | 0.051 ± 0.002 b | 0.046 ± 0.003 b | 0.018 ± 0.002 a | 0.024 ± 0.007 a |
| SA | 0.085 ± 0.008 a | 0.083 ± 0.010 a | 0.086 ± 0.005 a | 0.133 ± 0.002 b | 0.121 ± 0.015 b |
| Ethanol | 0.712 ± 0.059 a | 0.689 ± 0.107 a | 2.016 ± 0.230 b | 0.728 ± 0.038 a | 0.699 ± 0.015 a |
| Mannitol | 0.216 ± 0.105 a | 0.129 ± 0.004 ab | 0.468 ± 0.037 c | 0.073 ± 0.002 bd | 0.087 ± 0.015 ad |
| Orn | nd | nd | 0.013 ± 0.002 | nd | nd |
| DHA | 0.006 ± 0.002 a | nd | nd | 0.016 ± 0.003 b | 0.013 ± 0.004 b |
| Ala | 0.286 ± 0.034 ab | 0.326 ± 0.018 a | 0.297 ± 0.009 ab | 0.261 ± 0.016 b | 0.281 ± 0.014 ab |
| Val | 0.062 ± 0.011 | 0.070 ± 0.002 | 0.070 ± 0.006 | 0.065 ± 0.004 | 0.071 ± 0.006 |
| Ile | 0.049 ± 0.009 | 0.056 ± 0.003 | 0.053 ± 0.004 | 0.062 ± 0.005 | 0.062 ± 0.004 |
| Leu | 0.190 ± 0.043 | 0.233 ± 0.010 | 0.233 ± 0.028 | 0.229 ± 0.016 | 0.242 ± 0.023 |
| Choline | 0.157 ± 0.004 a | 0.144 ± 0.006 bc | 0.153 ± 0.006 ab | 0.140 ± 0.005 c | 0.138 ± 0.002 c |
| GC/MS peaks** | |||||
| Bu-ITC | 35659 ± 34193 a | 175589 ± 7433 b | 67603 ± 12035 ac | 124327 ± 21067 bd | 93700 ± 5806 cd |
| Pe-ITC | 2984 ± 4070 a | 33411 ± 1103 b | 11211 ± 3442 ac | 19263 ± 6457 c | 10121 ± 1550 ac |
| Ph-ITC | 2438 ± 927 ab | 3182 ± 409 a | 1140 ± 185 b | 2538 ± 1136 ab | 2103 ± 618 ab |
| Bu-CN | 13742 ± 1819 a | 19531 ± 3678 b | 14464 ± 686 ab | 16284 ± 1124 ab | 15684 ± 912 ab |
| Pe-CN | 4399 ± 1420 | 6963 ± 1668 | 4966 ± 622 | 5432 ± 290 | 4791 ± 288 |
| Ph-CN | 1509 ± 290 a | 2825 ± 685 b | 1859 ± 130 ab | 2289 ± 350 ab | 1960 ± 49 ab |
| Epithiobutyl-CN | 2474 ± 241 a | 5964 ± 394 b | 4176 ± 177 a | 5374 ± 1058 b | 4311 ± 1530 a |
| Acetoin | 376 ± 651 a | 471 ± 240 a | nd | 4902 ± 919 b | 3832 ± 272 b |
| 2,3-Butanedione | 756 ± 843 | nd | nd | 1852 ± 1439 | 1691 ± 1104 |
| Ethyl acetate | 1838 ± 262 a | 723 ± 216 b | 1820 ± 370 a | 804 ± 263 bc | 912 ± 168 bc |
| Hexanoic acid | 195 ± 47 a | 1039 ± 197 b | 79 ± 6 a | 363 ± 304 a | 212 ± 97 a |
| S-Methyl thioacetate | nd | nd | 606 ± 314 a | 20 ± 18 b | nd |
| DMTS | 796 ± 431 a | 765 ± 100 a | 194 ± 80 b | 696 ± 93 a | 807 ± 159 a |
*The average intensities of the three samples from each strain. The superscript letters indicate significant differences (p < 0.05) by Tukey's multiple comparison test.
**The intensity values of GC/MS peaks are displayed in units of 1000. nd = not detected.
Fig. 5.
Changes to compounds in pickling juice during nozawana-zuke fermentation. The x-axis indicates fermentation duration in days, and the y-axis represents signal intensities of displayed metabolites. Symbols represent different bacterial strains, as shown in the legend. The initial addition of brine was considered day 0 and used as such to construct the plot.
4. Discussion
Overall, the results indicated that starter cultures were associated with better fermentation progress, stable cell counts, and compositional profile variation in nozawana-zuke samples. The findings strongly suggested that the application of starter culture LAB improved the quality of fermented nozawana-zuke. To the best of our knowledge, this is the first study to assess Japanese salted pickle made from Brassica rapa L., including nozawana-zuke, via a metabolomics approach. The variation in the compositional profile could also be associated with differences in sensory quality. The overview of the comprehensive profiles of water-soluble and volatile compounds in this study may help in understanding the potential starter culture-dependent effects on the chemical characteristics and resulting quality of fermented pickle products.
In contrast to the S/C nozawana-zuke samples, the W/O samples exhibited delayed fermentation, higher terminal pH, more residual sugars, and lower overall acid production. The W/O samples also did not maintain viable LAB counts after the first 7 d of the 3-week fermentation period. These findings corroborate those observed in the starter culture fermentation of Chinese sauerkraut (Xiong, Li, Guan, Peng, & Xie, 2014), suggesting that autochthonous LAB with acid susceptibility, such as heterolactic-fermentative Leuconostoc mesenteroides, only grew transiently. The results of metabolomics analyses indicated that the chemical profile in W/O samples clearly differed to that in S/C samples. In terms of volatile compounds, W/O samples were characterized by substantially lower levels of Bu-ITC, Pe-ITC, and epithiobutyl-CN. Relative to those in S/C samples, the ratios of Bu-ITC, Pe-ITC, and epithiobutyl-CN levels in W/O samples were only 0.20–0.53-fold, 0.09–0.29-fold, and 0.41–0.59-fold, respectively. ITCs and CNs are major aroma-active volatiles in cruciferous vegetable pickles and are degraded from glucosinolates by the catalytic action of myrosinase (β-thioglucosidase glucohydrolase; EC 3.2.3.1) (Latte, Appel, & Lampen, 2011). The intensity of ethyl acetate, a well-known aroma-active component in fermented pickles (Yang et al., 2020, Zhao et al., 2016), was 2.02–2.54-fold higher in the W/O sample than in the S/C samples, except for Levilactobacillus brevis K4G1. This distinct profile with these major aroma-active volatiles may result in odor differences between the S/C and W/O samples. In fact, after the fermentation of W/O samples, the samples were found to emit an unpleasant smell accompanied by a strong fermentation odor. As a potential off-flavor component in nozawana-zuke, epithiobutyl-CN has been identified to negatively contribute to the flavor via its heavy, stale odor (Uda, Suzuki, & Maeda, 1992). However, the level of this volatile was lower in W/O samples than in S/C samples in this study. This might suggest the possibility that the fermentation odor in W/O samples is influenced by other volatile compound(s) derived from starter culture fermentation.
Fermentation with Latilactobacillus curvatus ♯4G2, a facultative heterofermentative species, resulted in moderate titration acidity (Fig. 1), which may contribute to a mild sour taste. Fermentation with this strain was associated with higher levels of residual sugars (particularly Fru) that persisted through the 21-d fermentation period. Because Latilactobacillus curvatus can produce acid from Fru but is relatively sensitive to low pH (Klein, Dicks, Pack, Hack, Zimmermann, Dellaglio, & Reuter, 1996), the moderate LA production from Glc may have inhibited the cells and its Fru metabolization during fermentation, resulting in lower acid accumulation than that in Lactiplantibacillus plantarum strains K4G4 and K5G3. In addition to the lower acidity, Latilactobacillus curvatus ♯4G2 exhibited a characteristic volatile profile. The level of AA, a key flavor compound that confers a sour pickle-like smell (Zhao, et al., 2016), was the lowest in the ♯4G2 sample, potentially because of the homolactic fermentation pathway of this species. In Latilactobacillus curvatus ♯4G2, higher levels of ITCs and lower intensities of acetoin and 2,3-butanedione also contributed to the characteristic volatile profile. The levels of Bu-ITC, Pe-ITC, and Ph-ITC in the ♯4G2 sample were higher than those in the other S/C samples by 1.41–2.60-fold, 1.73–3.30-fold, and 1.25–2.79-fold, respectively. Tolonen et al. reported similar observations for sauerkraut fermentation, obtaining maximal levels of ITCs upon fermentation with Latilactobacillus sakei (Tolonen et al., 2004), a species closely related to Latilactobacillus curvatus (Klein, et al., 1996). The degradation products of glucosinolate strongly depend on its side chain properties and reaction conditions, including pH value. ITC primarily forms at pH 6–7, whereas CN is favored at pH 2–5 (Latte et al., 2011). The levels of ITCs detected in this study, however, did not directly correlate with the pH values, implying that ITC levels were influenced depending on the starter culture. Although ITCs are known to be degradation products of glucosinolate derived from the enzymic reaction in raw material, it is intriguing that certain bacterial species can possibly influence ITC levels in fermented pickles. In addition, Latilactobacillus curvatus ♯4G2 might engage a bacterial myrosinase in nozawana-zuke fermentation as glucosinolate degradation has been reported in several Lactobacillus spp. (Nugon-Baudon et al., 1990, Palop et al., 1995), although it has not been reported for Latilactobacillus curvatus. The persistent CA content during fermentation with Latilactobacillus curvatus ♯4G2 indicates that this strain lacks CA assimilation ability in nozawana-zuke fermentation. This result may be associated with the fact that the levels of acetoin and 2,3-butanedione were low because these compounds are conversion products from CA (Gänzle, 2015).
In contrast to samples fermented with Latilactobacillus curvatus ♯4G2, samples fermented with Lactiplantibacillus plantarum strains K4G4 and K5G3 showed CA catabolism, resulting in high levels of the conversion products, acetoin and 2,3-butanedione (also known as diacetyl). Acetoin and 2,3-butanedione are strong aroma-active volatiles and are associated with yogurt and butter-like odors (Gänzle, 2015). These are desirable in fermented dairy foods and wine but not in beer and Japanese sake. Although these volatiles are commonly found in fermented vegetable pickles, their influence on the flavor quality of nozawana-zuke remains unclear. The vigorous fermentation of Glc and Fru and the strong resistance to acids led to an abundance of LA and a particularly low terminal pH value. Excess lactic fermentation is generally not preferred in fermented pickles. Their strong pH-lowering capacity, however, could be useful in nozawana-zuke production to increase preservability during storage and aid fermentation in difficult conditions, such as conditions with limited amounts of fermentable sugars and nutrients. Samples fermented with both Lactiplantibacillus plantarum strains were also characterized by increased production of DHA, as reported in a study of fermented mixed vegetables (Kim, Choi, Park, & Kim, 2019). DHA is associated with a sweet, cooling taste and ether-type odor and is known to affect the sensory quality of wine (Du Toit & Pretorius, 2000); however, its effect on fermented pickle taste remains unclear.
The sample fermented with Levilactobacillus brevis K4G1 presented with a metabolite profile that was different from that of samples fermented with the other strains. Levilactobacillus brevis is an obligatory heterofermentative species that ferments Glc and produces LA together with AA, ethanol, and CO2. In the nozawana-zuke fermentation in this study, heterolactic fermentation may have caused moderate pH reduction and acidity, contrary to the highest consumption of Glc and Fru among the four strains. Fru was likely not directly utilized for lactic fermentation but instead for mannitol interconversion to maintain the bacterial redox balance (Gänzle, 2015). The intensities of ITCs were the lowest among the S/C samples (Fig. 5, Table 1), whereas that of ethyl acetate, a fruity aroma metabolite, was substantially higher than that in the samples fermented with other strains (at 2.06–2.59-fold). With respect to other compounds, it is noteworthy that fermentation with Levilactobacillus brevis K4G1 influences the levels of DMTS and S-methyl thioacetate. S-methyl thioacetate has been detected in Chinese fermented pickle (paocai) and cheese (Peres et al., 2001, Zhao et al., 2016). The present study, however, is the first to report that fermentation with Levilactobacillus brevis specifically resulted in S-methyl thioacetate accumulation in pickles. Sulfur compounds are associated with unpleasant odors at a very low sensory threshold (Zhao, et al., 2016). The control of sulfur compound composition by starter culture LAB could be useful for flavor modification of fermented pickles. In cabbage sauerkraut, sulfur-containing volatiles such as DMDS and DMTS are derived from S-methyl-l-cysteine sulfoxide in raw material through mechanical damage and heating without bacterial reaction (Chin & Lindsay, 1994). Further investigation is necessary to assess the starter culture-dependent influence on the changes in sulfur-containing volatile composition in nozawana-zuke fermentation.
In conclusion, this study assessed the impact of various starter culture LAB on water-soluble and volatile compound profiles of fermented nozawana-zuke, compared these profiles with those obtained after fermentation without a starter culture, and further evaluated these results using a metabolomics approach. Our findings demonstrate that the levels of a wide range of metabolites in nozawana-zuke could be controlled using a specific starter culture strain. Many of the metabolites highlighted herein are taste- and/or aroma-active compounds, which suggests the possibility that the selection of starter culture can strongly influence the sensory profile of fermented nozawana-zuke products. Specifically, our results suggest that starter-culture-dependent changes in glucosinolate degradation products should be assessed in greater depth to examine their contributions to taste and aroma. In addition, we observed that different starter cultures produce different levels of beneficial components, such as Orn and GABA, indicating the starter culture-dependent influence on health benefits of fermented nozawana-zuke in addition to immunomodulatory effects reported in previous studies (Kawahara and Otani, 2006, Sandagdorj et al., 2019). Further research is needed to substantiate the preliminary findings obtained in this study regarding the impact on flavor and health benefits. A wide range of LAB are used in the fermentation of vegetable pickles, and their fermentation characteristics differ even among strains of the same species. High-throughput screening by metabolomic analysis could facilitate a broader choice of starter culture candidates and contribute to improving the quality of fermented pickles.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The NMR analyses were performed with technical support from the Advanced Analysis Centre, NARO (Tsukuba, Japan), NARO. This work was supported by a grant from the commissioned project study on “Research Development for Discovering of Regional Agricultural Products and Foods Having a Beneficial Impact on Health,” Ministry of Agriculture, Forestry and Fisheries, Japan (JPJ005336). We thank Enago (https://www.enago.jp/) for the language editing of a draft of this manuscript.
Conflict of interest statement
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochms.2021.100019.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
1H–13C HSQC spectrum and metabolite annotation of nozawana-zuke pickling juice. Data represent the results for the sample fermented without a starter culture (W/O) over 21 d.
Annotated volatile compounds in nozawana-zuke pickling juice.
References
- Berthier F., Ehrlich S.D. Genetic diversity within Lactobacillus sakei and Lactobacillus curvatus and design of PCR primers for its detection using randomly amplified polymorphic DNA. International Journal of Systematic Bacteriology. 1999;49:997–1007. doi: 10.1099/00207713-49-3-997. [DOI] [PubMed] [Google Scholar]
- Chin H.-W., Lindsay R.C. Mechanisms of formation of volatile sulfur-compounds following the action of cysteine sulfoxide lyases. Journal of Agricultural and Food Chemistry. 1994;42(7):1529–1536. [Google Scholar]
- Claesson M.J., van Sinderen D., O'Toole P.W. The genus Lactobacillus–a genomic basis for understanding its diversity. FEMS Microbiology Letters. 2007;269(1):22–28. doi: 10.1111/j.1574-6968.2006.00596.x. [DOI] [PubMed] [Google Scholar]
- Du Toit M., Pretorius I.S. Microbial spoilage and preservation of wine: Using weapons for nature's own arsenal. South African Journal of Enology and Viticulture. 2000;21(1):74–96. [Google Scholar]
- Fusco V., Quero G.M., Chieffi D., Franz C.M.A.P. Identification of Lactobacillus brevis using a species-specific AFLP-derived marker. International Journal of Food Microbiology. 2016;232:90–94. doi: 10.1016/j.ijfoodmicro.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Gänzle M.G. From gene to function: Metabolic traits of starter cultures for improved quality of cereal foods. International Journal of Food Microbiology. 2009;134(1-2):29–36. doi: 10.1016/j.ijfoodmicro.2009.05.018. [DOI] [PubMed] [Google Scholar]
- Gänzle M.G. Lactic metabolism revisited: Metabolism of lactic acid bacteria in food fermentations and food spoilage. Current Opinion in Food Science. 2015;2:106–117. [Google Scholar]
- Kawahara T., Iida A., Toyama Y., Fukuda K. Characterization of the bacteriocinogenic lactic acid bacteria Lactobacillus curvatus strain Y108 Isolated from nozawana-zuke pickles. Food Science and Technology Research. 2010;16(3):253–262. [Google Scholar]
- Kawahara T., Otani H. Stimulatory effect of lactic acid bacteria from commercially available Nozawana-zuke pickle on cytokine expression by mouse spleen cells. Bioscience Biotechnology and Biochemistry. 2006;70(2):411–417. doi: 10.1271/bbb.70.411. [DOI] [PubMed] [Google Scholar]
- Kim J., Choi K.B., Park J.H., Kim K.H. Metabolite profile changes and increased antioxidative and antiinflammatory activities of mixed vegetables after fermentation by Lactobacillus plantarum. PLoS One. 2019;14(5) doi: 10.1371/journal.pone.0217180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G., Dicks L.M.T., Pack A., Hack B., Zimmermann K., Dellaglio F., Reuter G. Emended descriptions of Lactobacillus sake (Katagiri, Kitahara, and Fukami) and Lactobacillus curvatus (Abo-Elnaga and Kandler): Numerical classification revealed by protein fingerprinting and identification based on biochemical patterns and DNA-DNA hybridizations. International Journal of Systematic Bacteriology. 1996;46(2):367–376. [Google Scholar]
- Latté K.P., Appel K.-E., Lampen A. Health benefits and possible risks of broccoli - An overview. Food and Chemical Toxicology. 2011;49(12):3287–3309. doi: 10.1016/j.fct.2011.08.019. [DOI] [PubMed] [Google Scholar]
- Lommen A. MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Analytical Chemistry. 2009;81(8):3079–3086. doi: 10.1021/ac900036d. [DOI] [PubMed] [Google Scholar]
- Mozzi F., Ortiz M.E., Bleckwedel J., De Vuyst L., Pescuma M. Metabolomics as a tool for the comprehensive understanding of fermented and functional foods with lactic acid bacteria. Food Research International. 2013;54(1):1152–1161. [Google Scholar]
- Nakagawa H., Mizuno T., Shimizu T., Kaneko J., Kadono M., Itoh T.…Terada A. Lactic acid bacteria flora isolated from salted vegetables. Japanese Journal of Food Microbiology. 2001;18(2):61–66. [Google Scholar]
- Nugon-Baudon L., Rabot S., Wal J.M., Szylit O. Interactions of the intestinal microflora with glucosinolates in rapeseed meal toxicity - 1st Evidence of an Intestinal Lactobacillus possessing a myrosinase-like activity in vivo. Journal of the Science of Food and Agriculture. 1990;52(4):547–559. [Google Scholar]
- Osawa, K., Takanami, S., Kuribayashi, T., & Kuwahara, H. (2005). Research reports of Nagano Prefecture General Industrial Technology Center Food Technology Department, 33, 9-13.
- Palop M.L., Smiths J.P., Tenbrink B. Degradation of sinigrin by Lactobacillus agilis strain R16. International Journal of Food Microbiology. 1995;26(2):219–229. doi: 10.1016/0168-1605(95)00123-2. [DOI] [PubMed] [Google Scholar]
- Park S.E., Yoo S.A., Seo S.H., Lee K.I., Na C.S., Son H.S. GC-MS based metabolomics approach of Kimchi for the understanding of Lactobacillus plantarum fermentation characteristics. Lwt-Food Science and Technology. 2016;68:313–321. [Google Scholar]
- Pederson C.S., Albury M.N. The sauerkraut fermentation. New York State Agricultural Experiment Station (Geneva) Bulletin. 1969;824:1–84. [Google Scholar]
- Peres C., Viallon C., Berdague J.L. Solid-phase microextraction spectrometry: A new approach to the rapid characterization of cheeses. Analytical Chemistry. 2001;73(5):1030–1036. doi: 10.1021/ac001146j. [DOI] [PubMed] [Google Scholar]
- Randazzo C.L., Todaro A., Pino A., Pitino I., Corona O., Caggia C. Microbiota and metabolome during controlled and spontaneous fermentation of Nocellara Etnea table olives. Food Microbiology. 2017;65:136–148. doi: 10.1016/j.fm.2017.01.022. [DOI] [PubMed] [Google Scholar]
- Sandagdorj B., Hamajima C., Kawahara T., Watanabe J., Tanaka S. Characterization of microbiota that influence immunomodulatory effects of fermented Brassica rapa L. Microbes and Environments. 2019;34(2):206–214. doi: 10.1264/jsme2.ME19003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen M., Rajaniemi S., Pihlava J.-M., Johansson T., Saris P.E.J., Ryhänen E.-L. Formation of nisin, plant-derived biomolecules and antimicrobial activity in starter culture fermentations of sauerkraut. Food Microbiology. 2004;21(2):167–179. [Google Scholar]
- Tomita S., Nakamura T., Okada S. NMR- and GC/MS-based metabolomic characterization of sunki, an unsalted fermented pickle of turnip leaves. Food Chemistry. 2018;258:25–34. doi: 10.1016/j.foodchem.2018.03.038. [DOI] [PubMed] [Google Scholar]
- Tomita S., Nemoto T., Matsuo Y., Shoji T., Tanaka F., Nakagawa H.…Sekiyama Y. A NMR-based, non-targeted multistep metabolic profiling revealed L-rhamnitol as a metabolite that characterised apples from different geographic origins. Food Chemistry. 2015;174:163–172. doi: 10.1016/j.foodchem.2014.11.028. [DOI] [PubMed] [Google Scholar]
- Torriani S., Felis G.E., Dellaglio F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Applied and Environmental Microbiology. 2001;67(8):3450–3454. doi: 10.1128/AEM.67.8.3450-3454.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa H., Bamba T., Shinohara M., Nishiumi S., Yoshida M., Fukusaki E. Practical non-targeted gas chromatography/mass spectrometry-based metabolomics platform for metabolic phenotype analysis. Journal of Bioscience and Bioengineering. 2011;112(3):292–298. doi: 10.1016/j.jbiosc.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Uda Y., Suzuki K., Maeda Y. Off-flavor constituents generated in pickled nozawana (Brassica campestris L var. rapifera) leaves during early pickling process. Journal of the Japanese Society for Food Science and Technology-Nippon Shokuhin Kagaku Kogaku Kaishi. 1992;39(2):200–202. [Google Scholar]
- Xiong T., Li X., Guan Q., Peng F., Xie M. Starter culture fermentation of Chinese sauerkraut: Growth, acidification and metabolic analyses. Food Control. 2014;41:122–127. [Google Scholar]
- Yang X., Hu W., Xiu Z., Jiang A., Yang X., Sarengaowa…Feng K. Microbial dynamics and volatilome profiles during the fermentation of Chinese northeast sauerkraut by Leuconostoc mesenteroides ORC 2 and Lactobacillus plantarum HBUAS 51041 under different salt concentrations. Food Research International. 2020;130:108926. doi: 10.1016/j.foodres.2019.108926. [DOI] [PubMed] [Google Scholar]
- Zhao N., Zhang C., Yang Q., Yang B.o., Lu W., Li D.…Chen W. Multiple roles of lactic acid bacteria microflora in the formation of marker flavour compounds in traditional chinese paocai. Rsc Advances. 2016;6(92):89671–89678. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1H–13C HSQC spectrum and metabolite annotation of nozawana-zuke pickling juice. Data represent the results for the sample fermented without a starter culture (W/O) over 21 d.
Annotated volatile compounds in nozawana-zuke pickling juice.