Highlights:
-
•
Plant miRNAs are present in Australian polyfloral and Leptospermum scoparium honey.
-
•
Sequencing shows that honey contains a diverse range of small, non-coding RNAs.
-
•
Honey RNA comes from different phylogenies including invertebrates and prokaryotes.
-
•
Unique small RNA profiles can provide insight into honey production conditions.
Keywords: Honey, Molecular markers, Food authentication, small RNAs, Leptospermum scoparium, Next generation sequencing
Abstract
Honey adulteration is a problem that effects the global honey industry and specifically, has been discovered in the Australian market. Common methods of adulteration include dilution with sugar syrup substitutes and the mislabelling of the floral and geographic origin(s) of honey. Current authentication tools rely on the molecular variability between different honeys, identifying unique chemical profiles and/or DNA signatures characteristic of a particular honey.
Honey is known to contain plant miRNAs derived from its floral source. To explore the composition and variability of honey RNA molecules, this is the first study to catalogue the small RNA content of Australian polyfloral table honey and New Zealand Leptospermum scoparium honey using next generation sequencing. The data shows that in addition to miRNAs, honey contains a variety of small non-coding RNAs including tRNA-derived fragments. Moreover, the honey small RNAs are derived from a range of phylogenetic sources, including from plant, invertebrate, and prokaryotic species. The data indicates that different honeys contain unique small RNA profiles, which suggests a novel avenue in developing molecular-based honey authentication tools.
1. Introduction
Honey is a sweetener produced by honeybees from nectar, which they forage from flowering plants. Rising consumer demand coupled with the limited production of this natural product may explain why commercially available honey is widely adulterated, with one study finding that 27% of honeys tested were diluted with sugar syrup substitutes (Zhou, Taylor, Salouros, & Prasad, 2018). Furthermore, to gain an economic advantage, some honey products are mislabelled with regards to its floral and/or geographical origin. Identifying fraudulent honey is therefore a prevailing challenge.
The molecular composition of honey is highly variable. For example, certain honeys derived from Leptospermum plants native to Australia and New Zealand have high antimicrobial activity due to the presence of specific chemicals methylglyoxal and dihydroxyacetone which are derived from the floral source (Adams et al., 2008, Cokcetin et al., 2016).
This molecular variability allows for the development of authentication tests that can identify unique chemical signatures (Ayton, Prenzler, Raman, Warren-Smith, & Meyer, 2019), to determine the floral source and geographical location of honey production (Chin & Sowndhararajan, 2020). However, these tests can be cheated (Ayton et al., 2019). Melissopalynology (pollen analysis), is an alternative method of honey classification (Bambara, 1991). However, both these tests are unsuitable for Australian honey analysis due to the unique nature of Australian native flora (Kale Sniderman, Matley, Haberle, & Cantrill, 2018). Pollen is however a rich source of plant DNA, and studies have suggested DNA markers as useful tools for honey authentication (McDonald et al., 2018, Prosser and Hebert, 2017).
Gismondi, Di Marco, and Canini (2017) was the first to show that honey also contains RNA molecules. This study identified mRNA and rRNA molecules within monofloral European honeys and determined that they originated from the floral source. Nine plant microRNAs (miRNAs), small (22 nucleotides), conserved, non-coding RNAs that function in post-transcriptional gene regulation (Reinhart, Weinstein, Rhoades, Bartel, & Bartel, 2002), were also isolated from the honeys. Interestingly, the miRNA concentrations varied between honey types, suggesting honeys may contain unique miRNA profiles. Further, as the identified RNAs did not show signs of degradation, Gismondi et al. (2017) concluded honey RNAs are stable and could retain functionality.
To further explore honey RNA, we used small-RNA next generation sequencing (NGS). Using this approach, we determined that the honey samples we tested contain processed tRNA fragments known as tRFs (Keam & Hutvagner, 2015). tRFs have been found in many eukaryotes, including a diverse range of plant species (Gupta, Singh, Zahra, & Kumar, 2018), and in a distinct mechanism from miRNAs, regulate the translation stage of gene expression (Keam & Hutvagner, 2015). In plants, expression of specific tRFs increases in times of stress (Park & Kim, 2018).
Additionally, analyses of the sequencing data revealed that the small RNA molecules within honey originate from a variety of species and is not limited to the floral source. These data suggest that this molecular constituent of honey is more diverse that previously reported.
2. Materials and methods
2.1. Honey samples
Four honey samples were examined, New Zealand Manuka honey (Leptospermum scoparium, Medihoney Batch #1815; n = 2) kindly provided by Comvita (Paengaroa, New Zealand) and a polyfloral Australian table honey (Batch: PKD Q 03/05/2017 23:54; n = 2) kindly provided by Capilano (Brisbane, Australia).
2.2. Phenol equivalence assay for Antibacterial activity
Antibacterial activity of honey was determined as described in Irish, Blair, and Carter (2011). Using Staphylococcus aureus ATCC 25923 (Oxoid, Hampshire, UK) as the test organism, honey solutions were prepared fresh to a 50% (w/v) solution in sterile water and further diluted to 25% (v/v) in either sterile water to test for total activity, or a 2 mg/ml catalase solution (3187 units/mg; Sigma-Aldrich, St. Louis, USA) for non-peroxide activity. Phenol (Sigma-Aldrich, St. Louis, USA) standards of 2%−7% v/v were prepared fresh in sterile water.
Aliquots (100ul) of each honey sample and phenol solutions were dispensed into the wells of assay plates in duplicate using quasi-Latin squares to randomise sample distribution. Plates were incubated (37 °C, 18–24 h) before the diameter of each zone of inhibition was measured using digital Vernier callipers (Kinechrome, Carole Park, Australia). The mean diameter of the zone of inhibition was calculated and squared, and a standard curve (concentration of phenol versus mean squared diameter) was generated using the phenol standards. The activity (total and non-peroxide) of each honey was calculated using the standard curve, multiplied by 4.69 to account for the dilution and density of honey, and expressed as the equivalent phenol concentration.
2.3. RNA isolation from honey
Honey (10 g) was diluted 1:1 in ultrapure water and shaken in an incubator (37 °C, 180 rpm) until homogenised. Solution was centrifuged (3900 rcf, 15 mins), supernatant was discarded, and pellet resuspended in 1 ml of Trizol (Invitrogen, Carlsbad, USA). Total RNA was purified using Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, USA) following the manufacturer's instructions. Total RNA concentration was measured using Nanodrop Microvolume Spectrophotometer (Thermo Scientific, Waltham, USA) and miRNA levels quantified using the Qubit miRNA Assay (Invitrogen, Carlsbad, USA). RNA quality was assessed with the small RNA kit (Agilent, Santa Clara, USA) on a 2100 Bioanalyser (Agilent, Santa Clara, USA).
2.4. Quantitative PCR
Total RNA (50 ng) from each sample was used as a template to make cDNA using the HiCapacity cDNA Kit (Applied Biosystems, Foster City, USA) following the manufacturer's instructions. qPCR reaction was prepared with TaqManTM Universal PCR Master Mix (Applied Biosystems, Foster City, USA) and run in a QuantStudio Flex 6 (Applied Biosystems, Foster City, USA) with the following Taqman miRNA assays (Applied Biosystems, Foster City, USA): (ath-mir156a-5p; ath-mir162a-3p; ath-miR396a-5p). Individual biological samples were run in technical duplicates, averaged for analysis, and standard error calculated.
2.5. Sequencing of small RNA samples
Total RNA from each sample was extracted and sent to Novogene (Hong Kong) for small RNA sequencing. Small RNA libraries were prepared using the NEBNext® Multiplex Small RNA Library Preparation Kit (Illumina, San Diego, USA) according to the manufacturers protocol and paired-end 150 bp sequencing was performed on a HiSeq 4000 (Illumina, San Diego, USA). Reads were subsequently trimmed to 50 bp single reads.
2.6. Sequencing data analysis
Raw reads were processed so that all adapter sequences were removed and reads smaller than 10 bases were excluded from further analysis. For cross-species annotation, reads were first aligned to all known precursor miRNAs (miRbase v 22) using miraligner with one mismatch allowed. All remaining unmapped reads were then aligned with bowtie (using parameters -v 1 --all --best --strata) to a set of rRNA and tRNAs spanning all three domains of life. rRNAs were downloaded from SILVA (SILVA Ref NR collection v 132) and tRNAs from GtRNAdb (v 17). NCBI’s Taxonomy database (v 2019–07-01) was used for classifying species into divisions.
NGS-based target prediction was performed with the miR-PREFeR pipeline with default settings, using processed reads from all honey samples and the Eucalyptus Grandis genome (Egrandis1_0).
3. Results
3.1. Plant miRNAs are present in Australian polyfloral and New Zealand Leptospermum scoparium honey
We isolated total RNA from New Zealand monofloral manuka (Leptospermum scoparium) honey, which has broad spectrum antimicrobial activity (20% phenol equivalence) and is used in honey-based wound care products (Robson, Dodd, & Thomas, 2009). For comparison, we isolated honey from an Australian polyfloral table honey with no measurable antimicrobial activity or known clinical use (<5% phenol equivalence).
We first determined whether the samples yielded sufficient quality RNA for further analysis. Initial investigations (data not shown) indicated the honeys contain only small RNA molecules (<150 nucleotides (nt)). Further analysis revealed an RNA size distribution of approximately 20–100 nt in length (Fig. 1a).
Fig. 1.
RNA isolated from honey. (a) Total RNA isolated from Australian polyfloral table honey (Sample 1 and 2) and New Zealand monofloral manuka (Leptospermum scoparium) honey (samples 3 and 4) was analysed with the small RNA kit (Agilent), showing the size distribution of RNA molecules in the samples. (b) Highly conserved plant miRNAs are present in both honeys. miRNA presence was ascertained using reverse transcription-PCR (Taqman miRNA assays) (n = 2; technical replicates). Nt – nucleotide. Ct – Cycle threshold value.
To verify if the purified small RNAs have maintained their integrity, we investigated if we could detect specific plant miRNAs within the sample. We chose highly conserved plant miRNAs with roles in flower development: miR156a, miR162a, both already detected in distinct honey samples (Gismondi et al., 2017), and miR396a (Shulga et al., 2017). All three miRNAs could be amplified from the samples (Fig. 1b), indicating these miRNAs are present in the honey.
3.2. The small RNA content of honey
While plant miRNAs have been identified within honey ((Gismondi et al., 2017), Fig. 1b), we catalogued the small RNA content of honey using next generation sequencing.
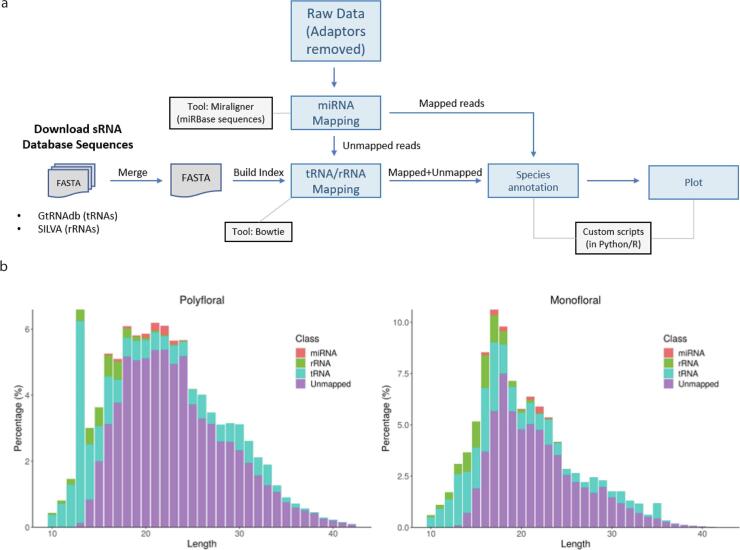
The bioinformatic pipeline used to analyse the small RNA sequencing data is illustrated in Fig. 2a. To identify small RNAs across different species, we aligned our reads to a comprehensive set of miRNAs, ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs). We aimed to i) identify the different types of RNA present and ii) determine the phylogenetic classification of the RNA molecules within the honey.
Fig. 2.
Small RNA sequencing of honey samples. (a) Bioinformatic approach. (b) length distribution (nucleotide) and RNA type (miRNA, ribosomal RNA (rRNA), transfer RNA (tRNA)) of the sequenced reads from polyfloral and monofloral honeys.
Our analyses revealed that the majority of RNA present in the polyfloral (68.9–76.1%) and monofloral (60.6–66.5%) honey samples did not align to either predicted or validated small RNA sequences and therefore their identity is unknown. One reason for this is the lack of fully annotated genomes available for Australian native species, including the plants commonly used as nectar sources for honey production (e.g. Corymbia species). The Leptospermum scoparium genome has been sequenced (Thrimawithana, Jones, Hilario, Grierson, & Ngo, 2019), however due to the cultural importance of this plant to the indigenous Māori of Aotearoa people of New Zealand, the genome sequence has not been published. Furthermore, honey RNA molecules may undergo editing and/or modifications during honey production and processing, which may influence their ability to be accurately identified. Despite these caveats, we confirmed that the plant miRNAs identified by RT-PCR (Table 1) are present, and successfully identified many others (Fig. 2b). We also identified other kinds of small RNA molecules. Interestingly, most were derived from tRNA molecules (22.9% and 27.6% for polyfloral and monofloral respectively), and a significant fraction from rRNAs (3.55% and 7.53% for polyfloral and monofloral respectively; Fig. 2b). miRNAs were the smallest identified fraction at 1.03% for polyfloral and 1.30% for monofloral honey. While it is yet to be discovered if these RNAs have any physiological function within honey, their roles in gene expression regulation and their highly conserved nature (Engels & Hutvagner, 2006), raises further questions about their possible role in the bioactive properties of honey.
Table 1.
Highly conserved miRNAs identified in honey.
| Polyfloral Australian table honey |
Monofloral New Zealand manuka honey |
||
|---|---|---|---|
| miRNA | Total Read Count | miRNA | Total Read Count |
| miR156a | 64.38 | miR156a | 130.34 |
| miR162a | 439.91 | miR162a | 241.40 |
| miR396 | 535.59 | miR396 | 650.88 |
Values represent counts per million (CPM) raw reads normalised to all reads which map to at least one plant sequence.
Finally, to identify plant-specific miRNAs that have not been previously annotated, we employed a miRNA prediction tool. As we are currently unable to access the Leptospermum scoparium genome, we used the phylogenetically related species, Eucalyptus grandis. On average, 58.01% of reads mapped to the E. grandis genome from the polyfloral and 48.17% from the monofloral honey, suggesting a significant portion of the unmapped reads from the cross-species analysis are either unannotated small RNAs or degraded RNAs from plants. This result was expected as eucalypt plants are a common nectar source for Australian honey. In total, this analysis identified 52 predicted plant miRNAs and was generally consistent with the miRNAs identified in our cross-species analysis (Supp Table 1).
3.3. Honeys have unique small RNA expression profiles
The small RNA composition between the two honey samples show clear differences (Fig. 2b). The polyfloral honey has a distinct tRF peak, 13 nucleotides in length. Conversely, the monofloral honey has a distinctive tRF peak, 35 nucleotides in length. A larger data set is required to determine if the RNA profile differences observed here are truly representative of the honeys tested. However, this disparity confirms that RNA composition of honey is not uniform, and therefore has the potential to distinguish between honey types.
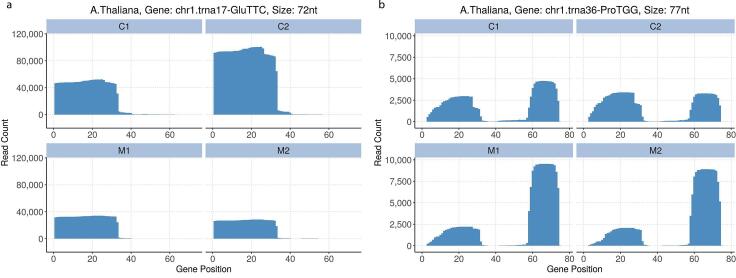
tRFs have been classified into two broad groups according to their biogenesis. Half-tRNAs (30–35 nts) are generated by a single cleavage event in the anticodon loop of a mature tRNA. half-tRNA expression correlates with perceived stress, and function to repress translation in mammalian cells (Yamasaki, Ivanov, Hu, & Anderson, 2009). To determine if the 35-nucleotide tRFs observed in the honey samples are half-tRNAs, we mapped their read distributions across the full length tRNA gene (Fig. 3a). The data indicates that these tRFs are a product of a specific and controlled processing event, producing a stable half-tRNA.
Fig. 3.
tRNA fragments in honey are formed from regulated processing of full length tRNA. (a) Example of a read distribution sequenced read across a full length tRNA gene which indicates a controlled endonucleolytic event forming a stable 35-nt tRNA fragment. (b) Example of a read distribution sequenced read across a full length tRNA gene which indicates controlled endonucleolytic event forming a stable 3ʹ tRF.
Precursor and mature tRNAs are processed via several mechanisms to form distinct tRF molecules. 5ʹ tRFs are formed by the endonucleolytic cleavage of the TψC-arm of a tRNA, while 3ʹ CCA tRFs are generated by cleaving the D-arm. These fragments typically range from 13 to 22 nucleotides long, and some function to regulate gene expression in human cells (Sobala & Hutvagner, 2013). From our read distribution data, the tRFs present in the polyfloral honey appear to be processed from the 3ʹ end of a full length tRNA (Fig. 3b).
3.4. RNA in honey comes from a variety of sources
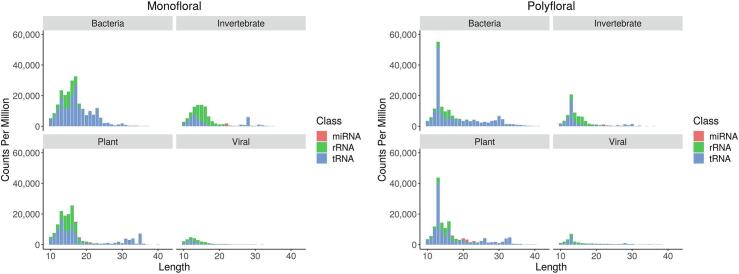
The only documented origin of RNA found in honey is from the floral source (Gismondi et al., 2017). Our data confirms this as 21.1% and 17.0% of annotated polyfloral and monofloral small RNAs mapped exclusively to plants. However, when we performed species annotation of the sequenced reads, we determined that plants are not the only source of honey RNA (Fig. 4). We identified that many reads mapped predominately to species external to the plant kingdom, including invertebrate, bacterial and viral species. This includes miRNA sequences that map to invertebrate genomes such as mir-14, a miRNA conserved across invertebrates which has an important role in insect development (Mead & Tu, 2008) and mir-276, a critical regulator of the reproductive cycle (Lampe, Jentzsch, Kierszniowska, & Levashina, 2019). A miRNA specific to the genomes of eusocial Hymenoptera insects, which includes Apis mellifera (the European honeybee), is also present in the honey. European honeybees underpin the Australian and New Zealand honey industry, therefore identifying these reads gives credence to our method of species assignation of the RNA molecules within the honey.
Fig. 4.
RNA in honey derives from a wide variety of species. Species annotation of the mapped and unmapped reads from the sequencing data shows that the RNA aligns with a wide variety of species from different phylogenetic domains. Counts were normalised and averaged across biological replicates.
Due to the conservation of sequences across species, this analysis cannot conclusively assign an RNA molecule to a particular species. However, this data does suggest the small RNA in honey originates from a range of species that span across different evolutionary kingdoms, which may provide further insight into honey composition and comprise a future avenue for honey authentication.
4. Discussion
The comprehensive approach of next generation sequencing revealed that honey contains several RNA types which derive from a range of different species. The RNA preserved within the New Zealand Leptospermum scoparium honey and the polyfloral Australian honey used in this study were small molecules of less than 150 nts in length. The absence of larger RNA molecules differs from Gismondi et al. (2017), who isolated an rRNA molecule that was 844 nts long from European honey. This discrepancy may be due to the distinct floral, geographic, and environmental conditions of honey production. Varied beekeeping practices, honey processing procedures and the conditions and duration of honey storage prior to RNA isolation may also affect the honey components. Furthermore, the differing chemical profiles of honey may influence its suitability in preserving RNA molecules of different lengths.
We identified small regulatory RNAs, including miRNAs and tRFs, within honey. Further research is required to determine if these molecules retain any physiological function or if they contribute to the nutritional properties of honey. It is possible that they have a role in attracting and manipulating mutualistic species and protecting the plant from species which would exploit the nectar resource. These qualities are an inherent aspect of nectar in plant-insect relationships (Nepi, Grasso, & Mancuso, 2018).
The successful amplification of plant miRNAs (Fig. 1b) indicates that the floral source(s) of the honey likely contributes a proportion of the RNA content. However due to the highly conserved nature of the RNAs identified, species-specific markers are required to confirm the plant source(s). Honey small RNAs derive from several phylogenetic groups including invertebrates and prokaryotes. The discovery of miRNAs which map to the honeybee genome (and other invertebrates) is a novel finding. These miRNAs may be suitable markers capable of validating unadulterated honey.
In plants, miRNAs are highly mobile molecules, capable of moving the length of the plant as they can enter and travel through the phloem (De Felippes, Ott, & Weigel, 2011). As plant miRNAs can survive and function in a variety of tissues it is perhaps not unsurprising that they are present in sugar-rich nectar and consequently in honey.
miRNAs are signalling molecules which form a highly intricate regulatory network essential to the development, growth and survival of the plant, and function to coordinate plant-wide responses (Skopelitis et al., 2018). Most of the plant-derived miRNAs that we identified from honey are highly conserved with understood functions. We investigated the known roles of the 10 most abundant plant-derived miRNAs identified in the honey samples. While the known functions of these miRNAs have not been confirmed in Leptospermum or Australian native species, reported regulatory functions of these miRNAs in other plant species can be broadly categorised into three main groups: tissue development; immunity, disease resistance and defence mechanisms; and stress responses, including drought (Table 2). Therefore, these miRNAs may have potential as markers which indicate the developmental stage, specific environmental stresses or health status of the plant at the time nectar was harvested for honey production. This insight into the health status of native Australian flora used as a nectar source may provide valuable information which could help to maximise Australian beekeeping practises and the maintenance of native Australian flora. Further research is required to determine if these factors influence the quality of honey produced.
Table 2.
Functions of the 10 most abundant, identified, plant-derived miRNAs found in honey.
| Function | miRNA(s) | References |
|---|---|---|
| Regulates plant tissue development | miR-166; miR-156; miR-482; miR-894; miR-396 | Bai et al., 2017, Li et al., 2010, Liu et al., 2018, Shulga et al., 2017 |
| Regulates plant immunity/defence mechanisms | miR-396; miR-482; miR-2089 | Soto-Suárez et al., 2017, Zhai et al., 2011 |
| Regulates plant stress response | miR-172c; miR-1511; miR-6300; miR-8175 | Anna et al., 2019, Ferdous et al., 2015, Htwe et al., 2015;Liang, Liu, Hao, Li, & Luo, 2019 |
miRNAs are critical regulators of gene expression in both plants and animals. However, there are significant differences in the biogenesis and function of miRNAs between the two evolutionary kingdoms (Axtell, Westholm, & Lai, 2011). Plant miRNAs are methylated at their 3ʹ ends, whereas most animal miRNAs do not have this modification (Yu et al., 2005). It has been suggested that miRNAs have bioactivity potential i.e. being absorbed into blood from consumed food, entering the miRNA regulatory pathway and regulating gene expression. However, subsequent studies failed to confirm ingested miRNAs have regulatory function (Dickinson et al., 2013), therefore, there is no evidence that honey miRNAs contribute to its bioactive properties once consumed.
While many of the RNA molecules within the honey samples remain unmapped, our data indicates that honeys contain distinct RNA signatures, composed of a variety of RNA types and derive from a range of species. Expression of regulatory small RNAs is dynamic and changes in response to specific environments and stresses. Therefore, a comprehensive analysis of honey RNA signatures may provide further insight into the manufacture of honey, for example the geographical location or climate conditions of the floral source(s) when the nectar was collected.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Comvita New Zealand and Capilano Honey Ltd. provided materials in the form of honey samples for the work described in the manuscript. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
The authors would like to thank the Blue Sky Research Scheme and the Cross-Faculty Collaborative Scheme run by the Faculty of Engineering and IT, University of Technology Sydney for funding this project. The authors also thank Comvita New Zealand and Capilano Honey Ltd. for supplying the honey samples used in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochms.2021.100014.
Contributor Information
Christopher Smith, Email: Christopher.M.Smith@student.uts.edu.au.
Nural Cokcetin, Email: Nural.Cokcetin@uts.edu.au.
Thuyen Truong, Email: quynhthuyen@gmail.com.
Elizabeth Harry, Email: Elizabeth.Harry@uts.edu.au.
Gyorgy Hutvagner, Email: Gyorgy.Hutvagner@uts.edu.au.
Sarah Bajan, Email: SBajan@usc.edu.au.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adams C.J., Boult C.H., Deadman B.J., Farr J.M., Grainger M.N.C., Manley-Harris M., Snow M.J. Isolation by HPLC and characterisation of the bioactive fraction of New Zealand manuka (Leptospermum scoparium) honey. Carbohydrate Research. 2008;343(4):651–659. doi: 10.1016/j.carres.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Anna B.B., Grzegorz B., Marek K., Piotr G., Marcin F. Exposure to high-intensity light systemically induces micro-transcriptomic changes in arabidopsis thaliana roots. International Journal of Molecular Sciences. 2019;20(20):1–14. doi: 10.3390/ijms20205131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Westholm J.O., Lai E.C. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biology. 2011;12(4):1–13. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton, J., Prenzler, P., Raman, H., Warren-Smith, A., & Meyer, R. (2019). Review of chemistry associated with honey (Issue June). www.agrifutures.com.au.
- Bai B., Shi B., Hou N., Cao Y., Meng Y., Bian H., Zhu M., Han N. microRNAs participate in gene expression regulation and phytohormone cross-talk in barley embryo during seed development and germination. BMC Plant Biology. 2017;17(1):1–16. doi: 10.1186/s12870-017-1095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambara S.B. Using pollen to identify honey. American Bee Journal. 1991;131(4):242–243. http://europepmc.org/abstract/AGR/IND91020412 [Google Scholar]
- Chin, N. L., & Sowndhararajan, K. (2020). A Review on Analytical Methods for Honey Classification, Identification and Authentication. In Honey Analysis - New Advantages and Challenges (pp. 1–33). https://doi.org/10.5772/intechopen.9023.
- Cokcetin N.N., Pappalardo M., Campbell L.T., Brooks P., Carter D.A., Blair S.E., Harry E.J. The antibacterial activity of Australian Leptospermum honey correlates with methylglyoxal levels. PLoS ONE. 2016;11(12):1–13. doi: 10.1371/journal.pone.0167780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B., Zhang Y., Petrick J.S., Heck G., Ivashuta S., Marshall W.S. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nature Biotechnology. 2013;31(11):965–967. doi: 10.1038/nbt.2737. [DOI] [PubMed] [Google Scholar]
- Engels B.M., Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25(46):6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- De Felippes F.F., Ott F., Weigel D. Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Research. 2011;39(7):2880–2889. doi: 10.1093/nar/gkq1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous J., Hussain S.S., Shi B.J. Role of microRNAs in plant drought tolerance. Plant Biotechnology Journal. 2015;13(3):293–305. doi: 10.1111/pbi.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gismondi A., Di Marco G., Canini A. Detection of plant microRNAs in honey. PLoS ONE. 2017;12(2):1–12. doi: 10.1371/journal.pone.0172981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Singh A., Zahra S., Kumar S. PtRFdb: A database for plant transfer RNA-derived fragments. Database. 2018;2018(2018):1–8. doi: 10.1093/database/bay063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htwe N.M.P.S., Luo Z.-Q., Jin L.-G., Nadon B., Wang K.-J., Qiu L.-J. Functional marker development of miR1511-InDel and allelic diversity within the genus Glycine. BMC Genomics. 2015;16(1):1–11. doi: 10.1186/s12864-015-1665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish J., Blair S., Carter D.A. The antibacterial activity of honey derived from Australian flora. PLoS ONE. 2011;6(3) doi: 10.1371/journal.pone.0018229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale Sniderman J.M., Matley K.A., Haberle S.G., Cantrill D.J. Pollen analysis of Australian honey. PLoS ONE. 2018;13(5):1–24. doi: 10.1371/journal.pone.0197545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam S.P., Hutvagner G. Trna-derived fragments (Trfs): Emerging new roles for an ancient RNA in the regulation of gene expression. Life. 2015;5(4):1638–1651. doi: 10.3390/life5041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe L., Jentzsch M., Kierszniowska S., Levashina E.A. Metabolic balancing by miR-276 shapes the mosquito reproductive cycle and Plasmodium falciparum development. Nature. Communications. 2019;10(1) doi: 10.1038/s41467-019-13627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Deng Y., Wu T., Subramanian S., Yu O. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiology. 2010;153(4):1759–1770. doi: 10.1104/pp.110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Liu H., Hao J., Li J., Luo L. Expression profiling and regulatory network of cucumber microRNAs and their putative target genes in response to cucumber green mottle mosaic virus infection. Archives of Virology. 2019;164(4):1121–1134. doi: 10.1007/s00705-019-04152-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu H., Tang G., Huang T. Small but powerful: Function of microRNAs in plant development. Plant Cell Reports. 2018;37(3):515–528. doi: 10.1007/s00299-017-2246-5. [DOI] [PubMed] [Google Scholar]
- McDonald C.M., Keeling S.E., Brewer M.J., Hathaway S.C. Using chemical and DNA marker analysis to authenticate a high-value food, manuka honey. Npj Science of Food. 2018;2(1) doi: 10.1038/s41538-018-0016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead E.A., Tu Z. Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito, Anopheles stephensi. BMC Genomics. 2008;9:1–13. doi: 10.1186/1471-2164-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepi M., Grasso D.A., Mancuso S. Nectar in plant–insect mutualistic relationships: From food reward to partner manipulation. Frontiers in Plant Science. 2018;9(July):1–14. doi: 10.3389/fpls.2018.01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, E. J., & Kim, T. H. (2018). Fine-tuning of gene expression by trna-derived fragments during abiotic stress signal transduction. International Journal of Molecular Sciences, 19(2). https://doi.org/10.3390/ijms19020518. [DOI] [PMC free article] [PubMed]
- Prosser S.W.J., Hebert P.D.N. Rapid identification of the botanical and entomological sources of honey using DNA metabarcoding. Food Chemistry. 2017;214:183–191. doi: 10.1016/j.foodchem.2016.07.077. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. MicroRNAs in plants. Trends in Plant Science. 2002;7(11):1616–1626. doi: 10.1101/gad.1004402.of. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson V., Dodd S., Thomas S. Standardized antibacterial honey (MedihoneyTM) with standard therapy in wound care: Randomized clinical trial. Journal of Advanced Nursing. 2009;65(3):565–575. doi: 10.1111/j.1365-2648.2008.04923.x. [DOI] [PubMed] [Google Scholar]
- Shulga O.A., Nedoluzhko A.V., Shchennikova A.V., Gruzdeva N.M., Shelenkov A.A., Sharko F.S., Sokolov A.S., Pantiukh E.S., Rastorguev S.M., Prokhortchouk E.B., Skryabin K.G. Profiling of microRNAs in wild type and early flowering transgenic Chrysanthemum morifolium by deep sequencing. Plant Cell, Tissue and Organ Culture. 2017;128(2):283–301. doi: 10.1007/s11240-016-1109-z. [DOI] [Google Scholar]
- Skopelitis D.S., Hill K., Klesen S., Marco C.F., von Born P., Chitwood D.H., Timmermans M.C.P. Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nature. Communications. 2018;9(1) doi: 10.1038/s41467-018-05571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobala A., Hutvagner G. Small RNAs derived from the 5′ end of tRNAs can inhibit protein translation in human cells. RNA Biology. 2013;10(4):553–563. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Suárez, M., Baldrich, P., Weigel, D., Rubio-Somoza, I., & San Segundo, B. (2017). The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Scientific Reports, 7(November 2016), 1–14. https://doi.org/10.1038/srep44898. [DOI] [PMC free article] [PubMed]
- A.H. Thrimawithana D. Jones E. Hilario E. Grierson H.M. Ngo I. Liachko … … K.E. Schwinn A whole genome assembly of Leptospermum scoparium (Myrtaceae) for mānuka research New Zealand Journal of Crop and Horticultural Science 47 4 2019 233 260 10.1080/01140671.2019.1657911.
- Yamasaki S., Ivanov P., Hu G.F., Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. Journal of Cell Biology. 2009;185(1):35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X. Methylation as a Crucial Step in Plant microRNA Biogenesis. Science. 2005;307(5711):932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Jeong D.H., de Paoli E., Park S., Rosen B.D., Li Y., González A.J., Yan Z., Kitto S.L., Grusak M.A., Jackson S.A., Stacey G., Cook D.R., Green P.J., Sherrier D.J., Meyers B.C. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes and Development. 2011;25(23):2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Taylor M.P., Salouros H., Prasad S. Authenticity and geographic origin of global honeys determined using carbon isotope ratios and trace elements. Scientific Reports. 2018 doi: 10.1038/s41598-018-32764-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.