Highlights
-
•
Cryptococcus laurentii induces resistance through in concert with key metabolic changes in tomato fruit.
-
•
A total of 59 metabolites were differently abundant in C. laurentii-treated tomato fruit.
-
•
Key metabolites chlorogenic acid, caffeic acid and ferulic acid are involved in phenylpropanoid biosynthesis pathway may play a key role in resistance induction by C. Laurentii in tomato.
Keywords: Tomato (Solanum lycopersicum), Cryptococcus laurentii, Metabolomics, Resistant response, Phenylpropanoid biosynthesis pathway, Biocontrol
Abstract
To investigate the mechanisms underlying inducible resistance in postharvest tomato fruit, non-targeted metabolome analysis was performed to uncover metabolic changes in tomato fruit upon Cryptococcus laurentii treatment. 289 and 149 metabolites were identified in positive and negative ion modes, respectively. A total of 59 metabolites, mainly including phenylpropanoids, flavonoids and phenolic acids, were differently abundant in C. laurentii-treated tomato fruit. Moreover, key metabolites involved in phenylpropanoid biosynthesis pathway, especially chlorogenic acid, caffeic acid and ferulic acid were identified through KEGG enrichment analysis. Enhanced levels of phenolic acids indicated activation of the phenylpropanoid biosynthesis pathway, which is a classic metabolic pathway associated with inducible resistance, suggesting that its activation and consequent metabolic changes contributed to inducible resistence induced by C. laurentii. Our findings would provide new understanding of resistance induction mechanism in tomato fruit from the metabolic perspective, and offer novel insights for new approaches reducing postharvest loss on tomato.
1. Introduction
The cherry tomato (Solanum lycopersicum var. cerasiforme) is a nutritionally and economically important vegetable crop, as well as a model plant for biocontrol research (Tao et al., 2020). As a climacteric fruit, preharvest latent infection and postharvest cross infection are major causes of postharvest rot of tomato (Altuntas & Ozkurt, 2019). The main fungal pathogens responsible for postharvest diseases of tomato are Botrytis cinerea and Alternaria alternata, which would dwell on the surface temporarily or for a long time until the fruit is ripe and senescent, otherwise infect fruit tissues through mechanical wounds (Altuntas & Ozkurt, 2019). Generally, pathogen spores germinate rapidly after entering the fruit wound, then activate related pathogenic mechanisms thereby leading to fruit rot and deterioration(Zang, Jiang, Ma, Li, Yin, & Yuan, 2020). Control of postharvest fungal diseases of cherry tomato currently relies on chemical fungicides, however overuse of these chemical fungicides has enabled increasingly widespread resistance. Moreover, toxic residues of chemical fungicides and their accumulation have posed potential risks to the environment, food safety as well as human health (Cordova, Amiri, & Peres, 2017). Therefore, it is of significance finding new methods to control postharharvest diseases of cherry tomato.
Among non-fungicidal methods reported, application of safe and effective microorganisms, especially biocontrol yeasts with antagonistic effects on pathogens, for biological control of postharharvest diseases has received much research attention during past decade, and is considered as one of the most promising alternatives to chemical fungicides (Droby, Wisniewski, Teixido, Spadaro, & Jijakli, 2016). Cryptococcus laurentii is a postharvest biocontrol microbial strain that has been extensively studied at home and abroad, with mounting evidence that it can not only effectively and directly inhibit a variety of fungal diseases in fruit including apples, pears, peaches (Lai, Renna, Yarema, Ruberti, He, & Brandizzi, 2018), cherry tomatoes (Zang, Jiang, Ma, Li, Yin, & Yuan, 2020), grapesDhanasekaran, citrus (Li, Li, Ji, Chen, Tian, & Qin, 2019), waxberries, cherries and strawberries (Zhang, Sun, Yang, Chen, Li, & Zhang, 2015), but also significantly induce resistant responses to develop and enhance their own abilities of defense against diseases in pear, jujube, peach, grape and cherry tomato fruit (Tang, Zhu, Cao, Zheng, Yu, & Lu, 2019).
We previously investigated effects of C. laurentii inhibiting pathogenic bacteria as well as the mechanism underlying enhanced disease resistance in fruit. First, in tomato induced by C. laurentii in the early stage, it was demonstrated that SlPR5, a resistance gene in tomato, was significantly up-regulated by C. laurentii treatment, and SlPR5 protein is the key resistance protein for inhibition of the infection of Alternaria alternata (Guo, Zhao, Wang, Yu, Miao, & Zheng, 2016). Further studies established that the resistance of tomato induced by C. laurentii was dependent on the concentration and induction time of yeast treatment. For instance, at the concentration of 1 × 108 cells/mL with induction time of 48 h, C. laurentii treatment significantly improved the resistance of tomato fruit against B. cinerea and A. alternata. Mechanisms of C. laurentii-induced resistance in tomato were mainly associated with enhanced activities of resistance-related enzymes (SOD, POD and PAL), activation of signaling pathways of salicylic acid, brassinolide and abscisic acid, and induction of overexpression of disease-resistance genes (Lai, Cao, Yu, Wang, Zhang, Zheng, et al., 2018). In addition, transcriptomics and proteomics results showed that C. laurentii treatment enhanced transcriptional expression of related genes in tomato fruit that were involved in secondary metabolic pathways, disease defense, resistance signaling pathways, and plant hormone signaling pathways, including the ethylene signaling pathway (Tang, Zhu, Cao, Zheng, Yu, & Lu, 2019). Also, previous studies have shown that the cell wall of C. laurentii or chitin extracted from its cell wall could also significantly enhance the resistance of tomato fruit to B. cinerea, and the induction mechanism may be related to increased activities of resistence-related enzymes, callose deposition, reactive oxygen accumulation and activation of salicylic acid signaling pathway (Sun et al., 2018, Sun et al., 2018). Admittedly metabolic changes in tomato fruit during postharvest pathological progression are extremely complex, how C. laurentii treatment induces and enhances disease resistance in tomato fruit from the perspective of metabolic pathways and associated metabolites remains unknown.
Metabolites are metabolic products that directly influence the physiology in plants. Metabolites produced by plants can be roughly divided into two categories: primary and secondary metabolites (Feng, Ding, Li, Wang, & Cui, 2020). Primary metabolites are essential for maintaining plant bioactivity and growth (Mamat, Azizan, Baharum, Noor, & Aizat, 2020), while secondary metabolites are more involved in coping with stress resistance and plant diseases (Carmona-Hernandez, Reyes-Perez, Chiquito-Contreras, Rincon-Enriquez, Cerdan-Cabrera, & Hernandez-Montiel, 2019). Biotic and abiotic stresses in plants usually involves a series of metabolic changes, including start of oxidation protective enzyme system, accumulation of osmotic protective agents, activation of resistance-related pathways and so forth, which will alter numerous associated metabolic pathways in plant tissues and cells, a new metabolic balance would then be rebuilt with alteration of various metabolic products leading to a distinct metabolic profile (Bueno and Lopes, 2020, Feng et al., 2020, Putri et al., 2013). Metabolomics enables comprehensive study of the complex metabolic processes and products, and elucidation of secondary metabolic pathways and networks (Shu et al., 2020). Metabolomics makes qualitative, quantitative and dynamic analysis of low-molecular weight (<1000) metabolites in a given physiological time and environmental condition of organisms, organs, tissues, or cells (Feng, Ding, Li, Wang, & Cui, 2020). In particular, widely-targeted metabolome analysis based on liquid chromatography-tandem mass spectrometry (LC-MS/MS) is a fast and reliable method for the detection of a wide-spectrum of plant metabolites to find the relationship between metabolites and physiological and pathological changes (Osorio et al., 2020).
In order to further elucidate resistance-motivating factors and resistance-inducing mechanisms by which C. laurentii induces disease resistance and associated metabolic responses in fruit, the present study aimed to apply metabolomics approach for identification of key metabolites in C. laurentii-induced internal resistance responses in tomato. The results will provide new evidence on mechanisms of C. laurentii-induced resistance, offering new insights regarding improvement of the biocontrol effectiveness of C. laurentii.
2. Materials and methods
2.1. Plant materials and microorganisms
Tomato (Solanum lycopersicum var. cerasiforme) of the cultivar ‘QianXi’ without infections or injuries were hand-picked during the red-ripening stage from Hainan Lingshui Modern Agriculture Demonstration Base in Lingshui country, Hainan Province, China, and quickly transported to our laboratory in Zhejiang University. C. laurenti (Kufferath) Skinner (CGMCC No. 3590) is a strain stored in our laboratory. Selection of tomato fruit and the cultivation of C. laurentii were conducted as previously described by Tang et al (Tang, Zhu, Cao, Zheng, Yu, & Lu, 2019). Tomato fruits were then dipped in sterile distilled water (control), or 1 × 108 cells mL−1 suspension of C. laurentii for 10 min (treatment). The fruits were stored in enclosed plastic trays with high Relative Humidity (90–95%) at 25 °C. Samples were collected 48 h later.
2.2. Sample preparation and metabolite extraction
The experiment was divided into two groups, each with 100 tomatoes. The treatment of tomato fruit was conducted as previously described by Lai et al (Lai, Cao, Yu, Wang, Zhang, Zheng, et al., 2018). Samples were collected 48 h after treatment. 6 ripe fruit were peeled and combined as a biological sample. 6 biological samples were created per tomato group (total ≥ 36 fruit). Two treatment groups were set up, sterile water control group and C. laurentii treatment group. Obtained samples were stored at −80 °C until metabolite extraction. A sample of 50 mg was weighed and placed in a 1.5 mL Eppendorf tube and added with 800 μL extract (methanol: water = 7:3, V: V, −20 °C precooling) and 20 μL internal standard. Two small steel balls were added in and ground in a tissue grinding machine (50 Hz, 5 min). After ultrasonic treatment at 4 °C for 30 min, the samples were placed in a refrigerator at −20 °C for 1 h. The samples were centrifuged at 4 °C and 14,000 rpm for 15 min. After centrifugation, 600 μL supernatant was taken and filtered through a 0.22 μm membrane. The filtered samples were placed in a bottle for LC-MS analysis. 20 μL of each sample was mixed into QC (Quality control) samples which were used to evaluate the repeatability and stability of LC-MS analysis. QC samples were analysed one in ten samples and the results were shown in Fig. S1.
2.3. Untargeted UPLC-MS/MS analysis
Metabolites in fruit extracts were isolated and detected by the Waters 2D UPLC (Waters, USA) tandem Q Exactive high resolution mass spectrometer (Thermo Fisher Scientific, USA). The chromatographic conditions were as follows. Aliquots (5 μL) were injected on to a Hypersil GOLD aQ column (1.9 µm, 100 mm*2.1 mm, Thermo Fisher Scientific, USA). The mobile phase consisted of aqueous solution (Liquid A) containing 0.1% formic acid and 100% acetonitrile (liquid B) containing 0.1% formic acid.The following steps were used for elution: 0 ∼ 2 min, 5% B solution; 2 ∼ 22 min, 5%∼95% B solution;95% B solution, 22 ∼ 27 min.27.1 ∼ 30 min, 5% B solution.The flow rate was 0.3 mL/min, the column temperature was 40℃, and the injection volume was 5 μL.
Q Exactive mass spectrometer (Thermo Fisher Scientific, USA) was used to collect primary and secondary mass spectrometry data. The range of m/z was 150 ∼ 1500, the first-order resolution was 70,000, the maximum injection time was 100 ms. Top 3 candidates were selected to fracture according to the ionic strength, then the secondary information was collected. The secondary resolution was 35000, AGC for 2e5, maximum injection time (IT) of 50 ms, fracture energy (stepped an nce) was set to: 20,40,60 eV. The parameters of the ion source (ESI) were set as follows: the sheath gas flow rate was 40, aux gas flow rate was 10, spray voltage positive ion mode was 3.80, negative ion mode was 3.20, capillary temperature was 320℃, and aux gas heatertemp was 350 °C.
2.4. MS data and bioinformatics analysis
A summary of the metabolomic studies is shown in Fig. 1.
Fig. 1.
A summary of the metabolomic study.
2.4.1. Data pre-processing
Raw data collected by LC-MS/MS was imported into Compound Discoverer 3.1 (Thermo Fisher Scientific, USA) for data processing. It mainly includes: peak extraction, retention time correction within and between groups, adjoint ion combination, missing value filling, background peak marking and metabolite identification. Finally the molecular weight, retention time, peak area and identification results were obtained. Metabolites were identified in combination with BGI Library and mzCloud database.
The result of Compound Discoverer 3.1 was exported and then imported into metaX for data preprocessing. It mainly included: (a) using the Probabilistic Quotient Normalization (PQN) to normalize data and obtain relative peak area; (b) Using Quality control-based robust LOESS signal correction (QC-RLSC) to correct batch effect; (c) Compounds with a coefficient of variation (CV) of relative peak area greater than 30% in all QC samples were deleted.
2.4.2. Data quality control(QC)
The quality of data was evaluated by QC sample test repeatability. The contents included chromatogram overlap of QC samples, Principle Component Analysis (PCA), peak lift number and peak response intensity difference.
2.4.3. Classification and functional annotation of detected metabolites
Identified metabolites were annotated by classification and functional annotation. The Human Metabolome Database contains chemical, molecular/biochemical, and clinical information of metabolites, supporting metabolic pathway and spectrogram search. Kyoto Encyclopedia of Genes and Genomes(KEGG)PATHWAY database is the core of KEGG database, which can introduce numerous metabolic pathways and their relationships through its powerful graphical function. Pathway functional annotation was carried out through the KEGG PATHWAY database to identify the major biochemical metabolic pathways and signal transduction pathways involved in metabolites.
2.4.4. Statistical analysis and identification of differentially accumulating metabolites
Multivariate statistical analysis and univariate analysis were used to screen different metabolites between groups. Principal Component Analysis (PCA) and Partial Least Squares Method-Discriminant Analysis (PLS-DA) were used for multivariate statistical Analysis. PCA is an unsupervised pattern recognition method, First, a PCA model was established between the comparative analysis group to observe the distribution and separation trend of the two groups of samples. Before the PCA model was established, log2 transformation was performed on the data, and then Pareto scaling was used to perform scaling on the data. PLS-DA is a supervised statistical method, which calculated Variable Important for Projection (VIP) to measure the influence intensity and explanatory ability of the expression patterns of metabolites on the classification and discrimination of samples of each group, so as to assist the screening of metabolic markers (Westerhuis et al., 2008). The PLS-DA model between the two groups of samples was established after log2 logarithm conversion of the data. The scaling method is Par, and the 7 fold Cross Validation was performed when the model was set up. In order to judge the quality of the model, 200 response permutation tests (RPT) were performed on the PLS-DA model.
Fold- Change (FC) analysis and T test (Student's T test) were performed on the obtained data. FC was obtained through variation multiple analysis, p-value was obtained through T-test, and Q-value was obtained through False Discovery Rate (FDR) correction. The screening criteria for differential metabolites were as follows: (1) The VIP of the first two principal components of the PLS-DA model ≥ 1, (2) fold-change ≥ 1.2 or ≤ 0.83, and(3) P-value < 0.05.
2.4.5. Cluster analysis and enrichment analysis of metabolic pathways of differentially metabolites
Cluster analysis was conducted for differential metabolites, and the log2 conversion and Z-score normalization treatments were used on the data during the analysis. Hierarchical Cluster was used for clustering algorithm and the Euclidian distance was used for distance calculation. The metabolic pathway enrichment analysis of differential metabolites was carried out based on KEGG database, and the metabolic pathway with P-value < 0.05 was a metabolic pathway with significant enrichment of differential metabolites. Moreover, bubble diagrams were drawn for pathways with significant enrichment of differential metabolites.
2.5. Validation and determination of key secondary metabolites in tomato pulp
2.5.1. Isolation of chlorogenic acid, caffeic acid and ferulic acid
Phenolic acids were isolated from extracts according to 2.2 described methods.
2.5.2. HPLC analysis
Ultimate 3000 (Thermo Fisher Scientific, USA) equipped with C18 column (4.6 × 150 mm, 5 μm) and UV detector was adopted for isolation and determination of Phenolic acids. The Water (A liquid) and Acetonitrile (B liquid) were used as mobile phases with under the column temperature of 35 °C. The flow rate was the same as 2.3 described. The detection wavelength and injection volume were 320 nm and 20 μL, respectively. Phenolic acids were identified by the retention time and the UV–vis spectra of standards.
3. Results
3.1. Quality control (QC) of the mass spectrometry data
The base peak chromatogram (BPC) of all QC samples well overlapped in both positive and negative modes, and the retention time and peak response intensity fluctuated little, indicating that the instrument was in a good state during sample detection (Fig. S1). In addition, PCA analysis shows an excellent stability of QC samples (Fig. 2A). RSD ratio refers to the ratio of the number of compounds whose CV of relative peak area is less than or equal to 30% to the number of all detected compounds in QC samples. If the RSD ratio is greater than or equal to 60%, the data quality is qualified. As shown in Fig. 2B, the RSD ratio in positive and negative modes is 94% and 91%, respectively. Therefore, QC results indicated that the experimental instrument was stable during data collection process and the collected data quality is ensured. Under positive ion mode condition, a total of 1821 features were detected (Table S1), 289 of which had identification information, while under the negative ion mode condition, a total of 1092 features were detected, with 149 features being identified (Table S2).
Fig. 2.
Quality control of the data. (A) PCA score graph of QC samples (a) Positive ion mode; (b) Negative ion mode. (B) CV profiles of the compounds (a) Positive ion mode; (b) Negative ion mode. The two lines perpendicular to the X-axis are 20% and 30% CV guides, and the lines parallel to the X-axis are 60% CV guides.
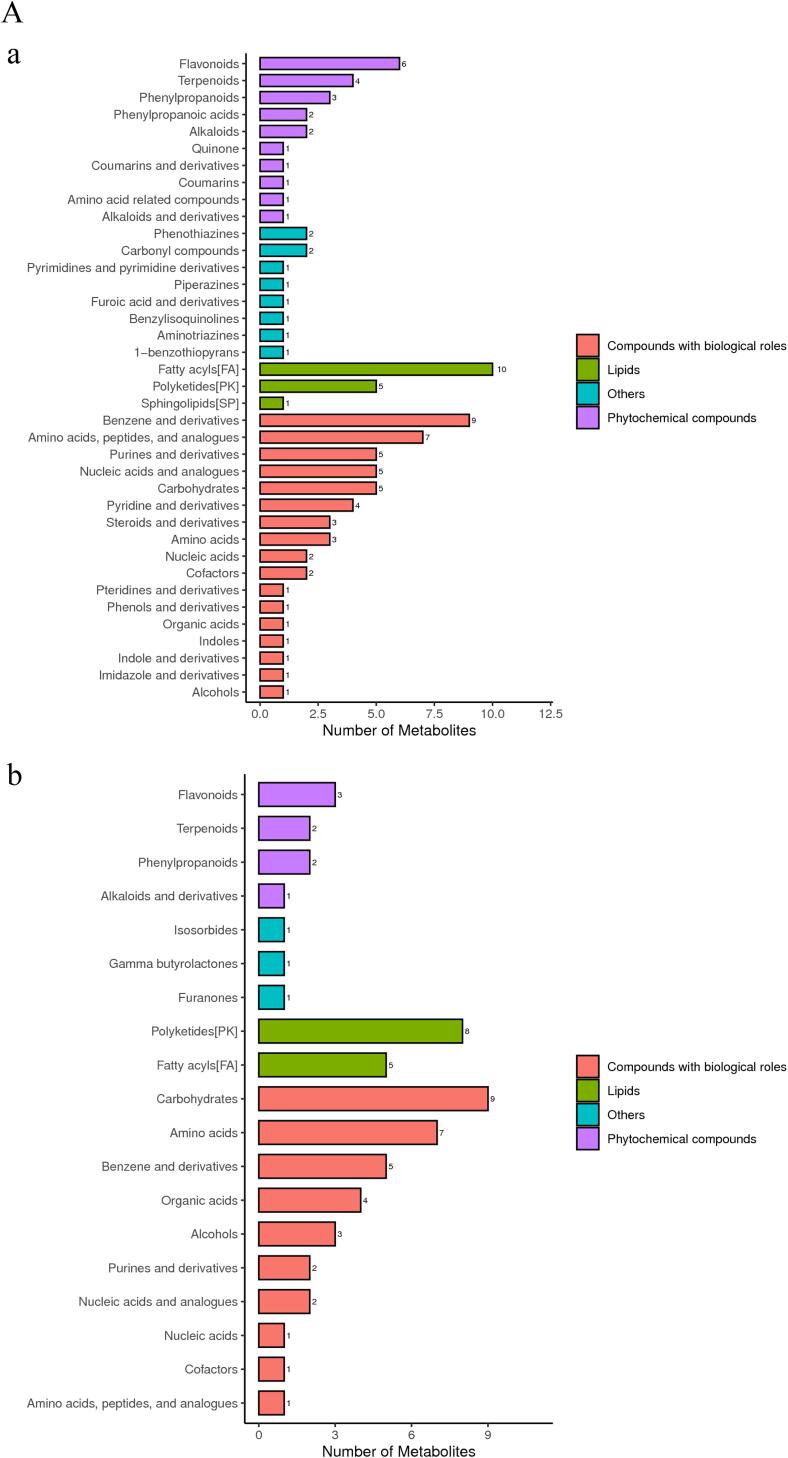
3.2. Classification and functional annotation of identified metabolites
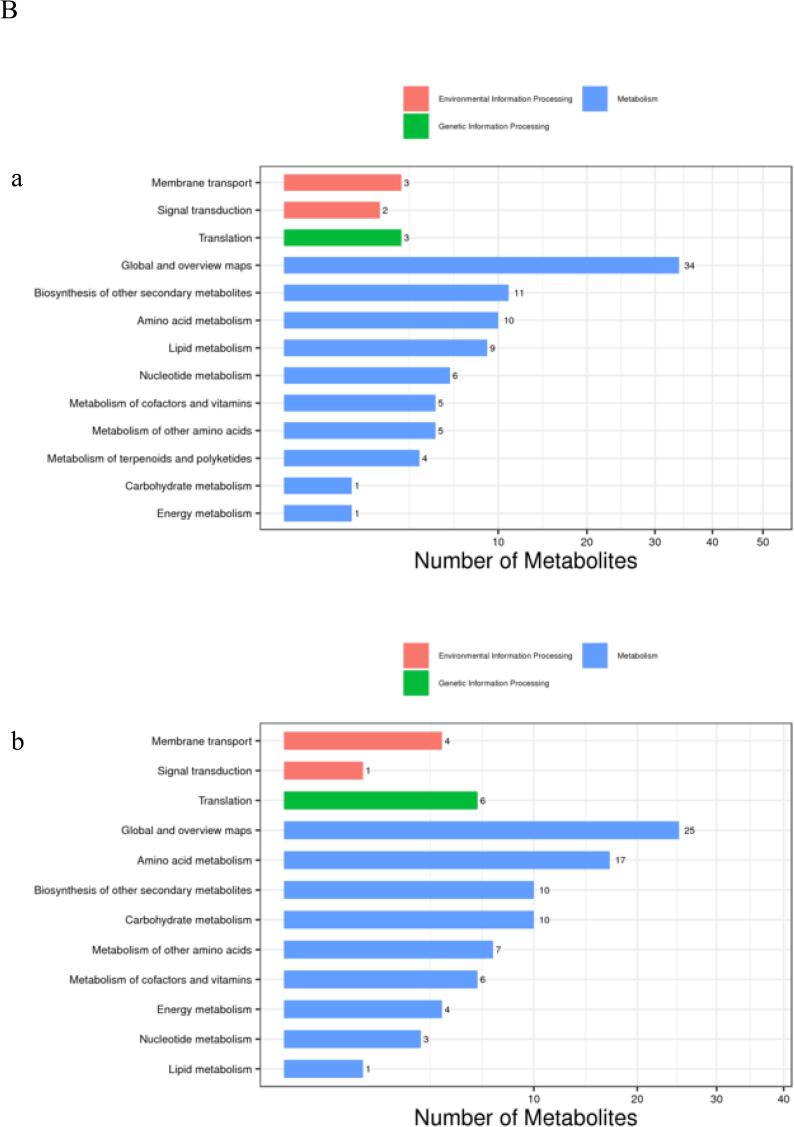
Classification and functional annotation of identified metabolites was conducted. As shown in Fig. 3A, in positive mode, identified metabolites were mainly distributed in fatty acids, compounds and derivatives, amino acids and compounds; while in negative mode, they were mainly centralized in carbohydrates, polyketides and amino acids, which usually are compounds with biological roles. After KEGG pathway annotation (Fig. 3B), identified metabolites were mainly involved in biosynthesis of other secondary metabolites and amino acid metabolism pathways in positive and negative ion modes.
Fig. 3.
Classification and functional annotation of detected metabolites. (A)Bar chart of metabolite classification under the (a) Positive ion mode; (b) Negative ion mode. The X-axis represents the number of metabolite classifications, and the Y-axis represents the metabolite classification entries. (B) Bar chart of KEGG function comment under the (a) Positive ion mode; (b) Negative ion mode. The X-axis represents the number of metabolite annotations, and The Y-axis represents the KEGG athway annotated.
3.3. Multivariate analysis of identified metabolites and screening of the differential metabolites
PLS-DA analysis clearly separated the two cultivars with a significance of 0.01 (p-value) under both positive and negative ion mode conditions (Fig. 4A). Hierarchical cluster analysis also disclosed distinct metabolic patterns associated with control and C. laurentii-treated groups, respectively (Fig. 4B). Together PLS-DA and hierarchical cluster analysis suggested that control and C. laurentii-treated groups had distinct metabolite profiles.
Fig. 4.
Screening and classification of differential metabolites between the control and C. laurentii-treatment groups. (A) core diagram of PLS-DA analysis model under the (a) Positive ion mode; (b) Negative ion mode. The horizontal axis is the first principal component and the vertical axis is the second principal component.The number in brackets is the score of the principal component, indicating the explanatory ability of the principal component to the whole model. (B) Cluster diagram of differential metabolites under the (a) Positive ion mode; (b) Negative ion mode. Each row in the figure represents a differential metabolite, each column represents a sample, the color represents the expression quantity, and the green to red corresponds to the expression quantity from low to high. (C) Volcano diagram of differential metabolites under the (a) Positive ion mode; (b) Negative ion mode. The volcano diagram was used to visually display the selected differential ions. Blue indicated significantly down-regulated differentially expressed ions, and red indicated significantly up-regulated differentially expressed ions. The circle represented the ion with VIP greater than or equal to 1, “×” were the ion with VIP<1, and the insignificant ion is gray. (D) Pie chart depicting the biochemical classification of the differential metabolites identified between the control and C. laurentii-treatment groups.
To identify key differential metabolites induced by C. laurentii treatment, differential metabolites were screened with a fold change ≥ 1.2 (upregulated) or ≤ 0.83 (downregulated) in C. laurentii-treated group compared to the control group. These metabolites were further screened using a variable importance in projection (VIP) with a VIP value ≥ 1 from PLS-DA model in addition to a p-value < 0.05. A total of 59 differential metabolites were identified (Table S3). Of these, 32 metabolites were upregulated and 7 metabolites were downregulated in positive ion mode, while 16 metabolites were upregulated and 4 metabolites were downregulated in negative ion mode, which were visually displayed by volcano plots (Fig. 4C). These 59 key differential metabolites can be categorized into 14 classes, mainly including phenylpropanoids, flavonoids and phenolic acids (Fig. 4D).
3.4. Classification and KEGG enrichment analysis of differential metabolites
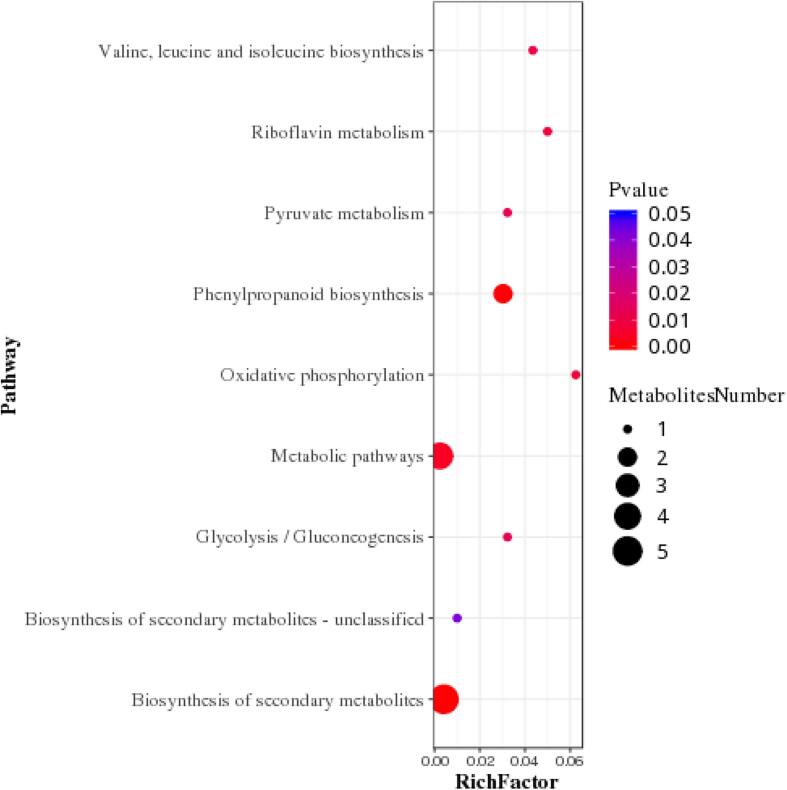
We next mapped these 59 metabolites into the KEGG database. Majority of these metabolites were mapped to metabolic pathways and metabolites biosynthesis, which was as expected (Fig. S3). Subsequently, we performed KEGG pathway enrichment analysis, in order to identify significantly-altered metabolic pathways between these two groups. Enrichment analysis distinguished 8 specialized plant pathways as significantly different (p < 0.05) (Fig. 5). Key metabolites involved in phenylpropanoid biosynthesis pathway, especially chlorogenic acid, caffeic acid and ferulic acid were identified through KEGG enrichment analysis. Enhanced levels of phenolic acids indicated activation of the phenylpropanoid biosynthesis pathway, which is a classic metabolic pathway associated with inducible resistance (Oliva, Guy, Galili, Dor, Schweitzer, Amir, et al., 2021).
Fig. 5.
Bubble diagram of the metabolic pathway enrichment analysis. The X-axis enrichment factor is the number of different metabolites annotated to this pathway divided by all identified metabolites annotated to this pathway. The higher the value, the higher the ratio of different metabolites annotated to this pathway. Dot size represents the number of differentially expressed metabolites annotated to this Pathway.
3.5. Key metabolites and metabolic pathways following C. laurentii application
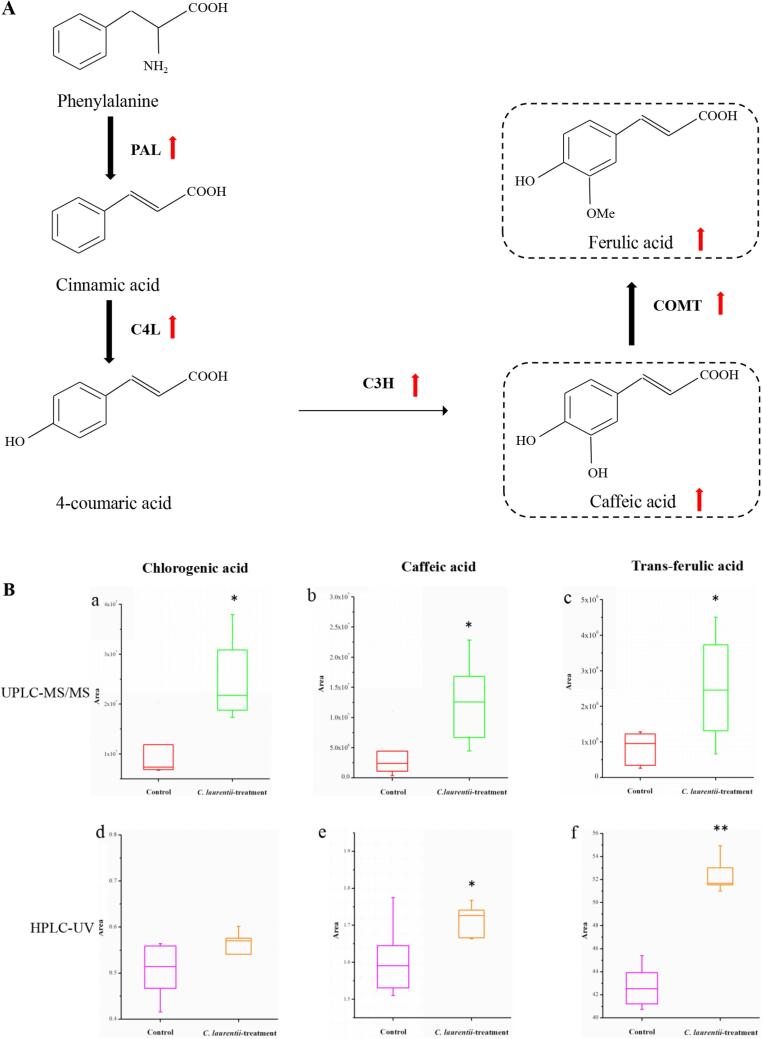
We focused on key metabolites and metabolic pathways that are potential contributors to disease resistance in tomato induced by C. laurentii. Phenylpropanoid biosynthesis pathway is paramount in resistance induction of tomato fruit against biotic and abiotic stresses(reference). 8 phenylpropanoids were identified as differentially abundant (Table 1), suggesting activation of phenylpropanoid biosynthesis pathway in tomato fruit following C. laurentii application. Levels of caffeic acid, chlorogenic acid and ferulic acid (key metabolites of phenylpropanoid biosynthesis pathway) were significantly higher in tomato upon C. laurentii treatment compared to control according to results of mass spectrometry-bassed metabolite profiling (Fig. 6B-a, b&c). In order to verify the metabolomiscs results, levels of caffeic acid, chlorogenic acid and ferulic acid in tomato pulp were qualitatively and quantitatively determined by HPLC (Fig. S2). Trends of HPLC determination results were consistent with that of UPLC-MS/MS determination results (Fig. 6B-d, e&f). Taken together, C. laurentii treatment significantly induced production of chlorogenic acid, caffeic acid and ferulic acid in tomato, indicating activation of phenylpropanoid biosynthesis pathway.
Table 1.
Differentially abundant phenylpropanoids induced by C. laurentii treatment in tomato fruit.
| Ion mode | Name | Fold change | p.value | VIP | RT [min] | Molecular Weight | Formula | Compound Class | |
|---|---|---|---|---|---|---|---|---|---|
| Pos | Skimmin | 2.6 | 0.014 | 2.3 | 4.20 | 324.08439 | C15 H16 O8 | Phenylpropanoids | Up |
| Pos | Chlorogenic acid | 2.4 | 0.004 | 2.0 | 4.64 | 354.09496 | C16 H18 O9 | Phenylpropanoids | Up |
| Pos | 7-Methoxycoumarin | 1.9 | 0.033 | 1.3 | 4.82 | 176.04733 | C10 H8 O3 | Phenylpropanoids | Up |
| Neg | N-feruloyloctopamine | 5.3 | 0.025 | 2.8 | 7.25 | 329.12636 | C18 H19 N O5 | Phenylpropanoids | Up |
| Neg | Caffeic acid | 3.5 | 0.018 | 2.3 | 4.57 | 180.04221 | C9 H8 O4 | Phenylpropanoids | Up |
| Neg | Ferulic acid | 3.0 | 0.026 | 1.9 | 6.15 | 194.0578 | C10 H10 O4 | Phenylpropanoids | Up |
| Neg | Isochlorogenic acid B | 2.8 | 0.001 | 2.0 | 6.66 | 516.12683 | C25 H24 O12 | Phenylpropanoids | Up |
| Neg | Cryptochlorogenic acid | 2.3 | 0.001 | 2.0 | 4.42 | 354.09519 | C16 H18 O9 | Phenylpropanoids | Up |
Differentially abundant compounds were identified using thresholds of VIP (variable importance in projection) ≥ 1.0 and fold change ≥ 1.6 (up-regulated) in C. laurentii-treated tomato fruit compared to control.
4. Discussion
Tomato in postharvest storage would be seriously affected by pathogen infections, which leads to tomato rot and tremendous economic losses (Chaouachi et al., 2021). Induced resistance to pathogens in postharvest tomato by biotic elicitors such as C. laurentii has become a promising alternative to synthetic fungicides (Gu et al., 2021, Tang et al., 2019). Postharvest pathological changes in fruit involve a number of key metabolites, meanwhile, induction of disease resistance in plants is intertwined with regulations of multi-gene interactions and multi-metabolic processes (Li, Cao, Wang, Lei, Ji, Xu, et al., 2021). Metabolite profiling enables comprehensive study of the complex metabolic processes as well as metabolic products in fruit, thereby identifying key functional metabolites and elucidating the secondary metabolic network. Non-targeted metabolomics has been successfully used for large-scale metabolite profiling and comparative metabolomics of many important plant species (Zou et al., 2020). However, mechanisms underlying enhanced disease resistance of tomato fruit after C. laurentii-induced treatment has not been investigated thoroughly, in particular, changes in metabolites and metabolic pathways have not been studied. In this study, we used UPLC-MS/MS-based non-targeted metabolomics to profile differential metabolites by C. laurentii-induced treatment. We identified 438 metabolites (289 under the positive ion mode and 149 under the negative ion mode), 59 (39 under the positive ion mode and 20 under the negative ion mode) of which were differentially accumulated in C. laurentii-induced treatment group compared to control. This study offers evidence of resistance-induced mechanism by C. laurentii through the lens of metabolic changes.
Plants use complex defense systems to fight pests and diseases, and production of a large number of low-molecular secondary metabolites with antibacterial activity to enhance the structural defense barrier is one of them (Lu, Wang, Zhu, Lu, Zheng, & Yu, 2015). Secondary metabolites in plants mainly comprise phenylpropanoids, terpenoids, steroids, terpenoids and flavonoids (Magalhaes, Borges, Laumann, Caulfield, Birkett, & Blassioli-Moraes, 2020). It is previously reported that phenylpropanoids, including trans-cinnamic acid, para-coumaric acid, caffeic acid, ferulic acid and benzoid acid derivatives, play an pivotal role in plant defense against the invasion of pathogens and herbivorous insects (Oliva, et al., 2021). Moreover, phenylpropanoids are also involved in regulation of plant growth (Isah, 2019). Among phenylpropanoids responding to C. laurentii treatment, our results show that levels of chlorogenic acid, caffeic acid and ferulic acid were significantly higher in C. laurentii-treated groups, which may be associated with enhanced disease resistance in tomato induced by C. laurentii (Fig. 6). Chlorogenic acid has certain inhibitory effect on both bacteria and fungi resulting from its effects of destroying the cell wall and membrane structure of bacteria. Meanwhile, when the hydroxyl group on the quinine group of chlorogenic acid combine with the free amino group, its inhibitory effect on fungi would become stronger (Tajner-Czopek, Gertchen, Rytel, Kita, Kucharska, & Sokol-Letowska, 2020). In addition to possessing strong antioxidant capability, ferulic acid also has outstanding antibacterial activity. Lattanzio et al. detected 12 kinds of phenolic acids against 5 fungi (Sclerotinia sclerotiorum, Fusarium oxysporum, Altenaria sp., Botrytis cinerea, Penicillium digitatum), and it was found that ferulic acid had the strongest bacteriostatic activity (Ou, 2002). In conclusion, C. laurentii treatment might enhance the disease resistance of tomato via increased contents of chlorogenic acid, caffeic acid and ferulic acid in fruit.
Fig. 6.
Effect of Cryptococcus laurentii treatment on the phenylpropanoid biosynthesis pathway (A) and the level of three phenolic acids (B), including chlorogenic acid, caffeic acid and ferulic acid in tomato fruit. Asterisks indicates significant differences (P ≤ 0.05) according to Student́s t-test.
The abundance of flavonoids is a key indicator of antioxidant activity (Zhang, Yang, Tong, Hu, Zhang, Tian, et al., 2021). Our metabolomics analysis identified 14 flavonoids (Table S1 and Table S2), and 2 of them (Mulberrin and Morin) were differentially accumulated in C. laurentii-treated group (Table S3). Differences in levels of flavonoids suggested that the enhanced resistance of tomato induced by C. laurentii could originated from increased antioxidant activity. In addition, the levels of salicin and isobutyl 4-hydroxybenzoate were remarkably increased, suggesting that these phenolic acids may also contribute to C. laurentii-induced resistance in tomato fruit.
KEGG enrichment analysis revealed that metabolites involved in phenylpropanoid biosynthesis pathway were significantly altered in C. laurentii-treated group compared to control, which was consistent with our previous experimental results of high-throughput transcriptome sequencing (Tang, Zheng, et al., 2019), providing evidence that phenylpropanoid biosynthesis pathway plays an important role in C. laurentii-induced postharvest resistance of tomato from perspectives of both gene and metabolite levels (Fig. 6A). PAL, the key branch point enzyme, catalyses the first step of the phenylpropanoid pathway, leading to production of phenolic compounds, and is believed to be activated by JA/ET signaling in plant defense responses (Lu, Wang, Zhu, Lu, Zheng, & Yu, 2015). In our previous research, expression levels of the key genes involved in phenylpropanoid biosynthesis pathway (e.g., PAL1, C4H, 4CL3, ATOMT, and cinnamoyl-CoA reductase) were up-regulated. In the present study, metabolite profiling results confirmed that contents of caffeic acid and ferulic acid involved in phenylpropanoid biosynthesis pathway were significantly increased (Fig. 6B). Strong relevance was observed between increased mRNA expression levels of genes related to phenolic acid synthesis (Tang, Zheng, et al., 2019) and their increased levels (Fig. 6B) in C. laurentii-treated tomato fruit. Given the importance of phenylpropanoid biosynthesis pathway and associated phenolics in the defensive response against postharvest pathogenic attack (Bennett & Wallsgrove, 1994), combined with increased levels of key enzymes (PAL, C4L, C3H, COMT) and metabolites (caffeic acid and ferulic acid) in tomato following C. laurentii treatment, it is possible that activaiton of phenylpropanoid biosynthesis pathway is one major contributor to the resistance induction mechanisms for C. laurentii. Admittedly future studies are warranted for further validation, nevertheless, our results provided evidence supporting the role of phenylpropanoid biosynthesis, which is of significance for the community of biocontrol research.
5. Conclusions
Taken together, in the present study, LC-MS/MS-based metabolomics analysis was applied to systematically study enhanced resistance of tomato induced by C. laurentii. These results provide comprehensive information on metabolite compositions, abundances and pathways in tomato upon C. laurentii treatment. In conclusion, metabolic alterations including differential levels of phenylpropanoids, flavonoids and phenolic acids in addition to activaiton of phenylpropanoid biosynthesis pathway might be one of the underlying mechanisms of resistance enhancement in tomato induced by C. laurentii.
Funding
This research was supported by the National Key Technology R&D Program of China (No. 2016YFD0401201), National Natural Science Foundation of China (31571897).
Declaration of Competing Interest
The authors declare that they have no conflicts of interest or personal relationships that could have appeared to influence the work reported in the paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochms.2021.100066.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Altuntas O., Ozkurt H. The assessment of tomato fruit quality parameters under different sound waves. Journal Of Food Science And Technology-Mysore. 2019;56(4):2186–2194. doi: 10.1007/s13197-019-03701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.N., Wallsgrove R.M. Secondary metabolites in plant defense-mechanisms. New Phytologist. 1994;127(4):617–633. doi: 10.1111/j.1469-8137.1994.tb02968.x. [DOI] [PubMed] [Google Scholar]
- Bueno P.C.P., Lopes N.P. Metabolomics to characterize adaptive and signaling responses in legume crops under abiotic stresses. ACS Omega. 2020;5(4):1752–1763. doi: 10.1021/acsomega.9b03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Hernandez S., Reyes-Perez J.J., Chiquito-Contreras R.G., Rincon-Enriquez G., Cerdan-Cabrera C.R., Hernandez-Montiel L.G. Biocontrol of postharvest fruit fungal diseases by bacterial antagonists: A review. Agronomy-Basel. 2019;9(3) [Google Scholar]
- Chaouachi M., Marzouk T., Jallouli S., Elkahoui S., Gentzbittel L., Ben C., Djebali N. Activity assessment of tomato endophytic bacteria bioactive compounds for the postharvest biocontrol of Botrytis cinerea. Postharvest Biology and Technology. 2021;172 [Google Scholar]
- Cordova L.G., Amiri A., Peres N.A. Effectiveness of fungicide treatments following the Strawberry Advisory System for control of Botrytis fruit rot in Florida. Crop Protection. 2017;100:163–167. [Google Scholar]
- Droby S., Wisniewski M., Teixido N., Spadaro D., Jijakli M.H. The science, development, and commercialization of postharvest biocontrol products. Postharvest Biology and Technology. 2016;122:22–29. [Google Scholar]
- Feng Z., Ding C.Q., Li W.H., Wang D.C., Cui D. Applications of metabolomics in the research of soybean plant under abiotic stress. Food Chemistry. 2020;310 doi: 10.1016/j.foodchem.2019.125914. [DOI] [PubMed] [Google Scholar]
- Gu N., Zhang X.Y., Gu X.Y., Zhao L.N., Godana E.A., Xu M.Q., Zhang H.Y. Transcriptomic and proteomic analysis of the mechanisms involved in enhanced disease resistance of strawberries induced by Rhodotorula mucilaginosa cultured with chitosan. Postharvest Biology and Technology. 2021;172 [Google Scholar]
- Guo J., Zhao X., Wang H.L., Yu T., Miao Y., Zheng X.D. Expression of the LePR5 gene from cherry tomato fruit induced by Cryptococcus laurentii and the analysis of LePR5 protein antifungal activity. Postharvest Biology and Technology. 2016;111:337–344. [Google Scholar]
- Isah T. Biological Research; 2019. Stress and defense responses in plant secondary metabolites production; p. 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., Cao X., Yu T., Wang Q., Zhang Y.W., Zheng X.D., Lu H.P. Effect of Cryptococcus laurentii on inducing disease resistance in cherry tomato fruit with focus on the expression of defense-related genes. Food Chemistry. 2018;254:208–216. doi: 10.1016/j.foodchem.2018.01.100. [DOI] [PubMed] [Google Scholar]
- Lai Y.S., Renna L., Yarema J., Ruberti C., He S.Y., Brandizzi F. Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proceedings Of the National Academy Of Sciences Of the United States Of America. 2018;115(22):E5203–E5212. doi: 10.1073/pnas.1802254115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. H., Cao, S. F., Wang, K. T., Lei, C. Y., Ji, N. N., Xu, F., Jiang, Y. B., Qiu, L. L., & Zheng, Y. H. (2021). Heat Shock Protein HSP24 Is Involved in the BABA-Induced Resistance to Fungal Pathogen in Postharvest Grapes Underlying an NPR1-Dependent Manner. Frontiers In Plant Science, 12. [DOI] [PMC free article] [PubMed]
- Li J.K., Li H., Ji S.F., Chen T., Tian S.P., Qin G.Z. Enhancement of biocontrol efficacy of Cryptococcus laurentii by cinnamic acid against Penicillium italicum in citrus fruit. Postharvest Biology and Technology. 2019;149:42–49. [Google Scholar]
- Lu L.F., Wang J.X., Zhu R.Y., Lu H.P., Zheng X.D., Yu T. Transcript profiling analysis of Rhodosporidium paludigenum-mediated signalling pathways and defense responses in mandarin orange. Food Chemistry. 2015;172:603–612. doi: 10.1016/j.foodchem.2014.09.097. [DOI] [PubMed] [Google Scholar]
- Magalhaes D.M., Borges M., Laumann R.A., Caulfield J.C., Birkett M.A., Blassioli-Moraes M.C. Inefficient weapon-the role of plant secondary metabolites in cotton defence against the boll weevil. Planta. 2020;252(5) doi: 10.1007/s00425-020-03497-w. [DOI] [PubMed] [Google Scholar]
- Mamat S.F., Azizan K.A., Baharum S.N., Noor N.M., Aizat W.M. GC-MS and LC-MS analyses reveal the distribution of primary and secondary metabolites in mangosteen (Garcinia mangostana Linn.) fruit during ripening. Scientia Horticulturae. 2020;262 [Google Scholar]
- Oliva M., Guy A., Galili G., Dor E., Schweitzer R., Amir R., Hacham Y. Enhanced Production of Aromatic Amino Acids in Tobacco Plants Leads to Increased Phenylpropanoid Metabolites and Tolerance to Stresses. Frontiers In Plant Science. 2021;11 doi: 10.3389/fpls.2020.604349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S., Carneiro R.T., Lytovchenko A., McQuinn R., Sorensen I., Vallarino J.E.G.…Rose J.K.C. Genetic and metabolic effects of ripening mutations and vine detachment on tomato fruit quality. Plant Biotechnology Journal. 2020;18(1):106–118. doi: 10.1111/pbi.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putri S.P., Yamamoto S., Tsugawa H., Fukusaki E. Current metabolomics: Technological advances. Journal Of Bioscience And Bioengineering. 2013;116(1):9–16. doi: 10.1016/j.jbiosc.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Shu H.Z., Zhang W.M., Yun Y.H., Chen W.J., Zhong Q.P., Hu Y.Y.…Chen W.X. Metabolomics study on revealing the inhibition and metabolic dysregulation in Pseudomonas fluorescens induced by 3-carene. Food Chemistry. 2020;329 doi: 10.1016/j.foodchem.2020.127220. [DOI] [PubMed] [Google Scholar]
- Sun C., Fu D., Jin L.F., Chen M.Y., Zheng X.D., Yu T. Chitin isolated from yeast cell wall induces the resistance of tomato fruit to Botrytis cinerea. Carbohydrate Polymers. 2018;199:341–352. doi: 10.1016/j.carbpol.2018.07.045. [DOI] [PubMed] [Google Scholar]
- Sun C., Lin M., Fu D., Yang J., Huang Y.N., Zheng X.D., Yu T. Yeast cell wall induces disease resistance against Penicillium expansum in pear fruit and the possible mechanisms involved. Food Chemistry. 2018;241:301–307. doi: 10.1016/j.foodchem.2017.08.092. [DOI] [PubMed] [Google Scholar]
- Tajner-Czopek A., Gertchen M., Rytel E., Kita A., Kucharska A.Z., Sokol-Letowska A. Study of antioxidant activity of some medicinal plants having high content of caffeic acid derivatives. Antioxidants. 2020;9(5) doi: 10.3390/antiox9050412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Zhu F.H., Cao X., Zheng X.D., Yu T., Lu L.F. Cryptococcus laurentii controls gray mold of cherry tomato fruit via modulation of ethylene-associated immune responses. Food Chemistry. 2019;278:240–247. doi: 10.1016/j.foodchem.2018.11.051. [DOI] [PubMed] [Google Scholar]
- Tao X.Y., Wu Q., Aalim H., Li L., Mao L.C., Luo Z.S., Ying T.J. Effects of Exogenous Abscisic Acid on Bioactive Components and Antioxidant Capacity of Postharvest Tomato during Ripening. Molecules. 2020;25(6) doi: 10.3390/molecules25061346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhuis J.A., Hoefsloot H.C.J., Smit S., Vis D.J., Smilde A.K., van Velzen E.J.J.…van Dorsten F.A. Assessment of PLSDA cross validation. Metabolomics. 2008;4(1):81–89. [Google Scholar]
- Zang H., Jiang K., Ma J., Li M., Yin X.B., Yuan L.X. Selenium research for environment and human health: Perspectives. Technologies And Advancements; 2020. Effect of melatonin and selenium on control of postharvest gray mold of tomato fruit; pp. 125–126. [Google Scholar]
- Zhang Q.H., Yang F.B., Tong H., Hu Y., Zhang X.Y., Tian T.…Su Q. Plant flavonoids enhance the tolerance to thiamethoxam and flupyradifurone in whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) Pesticide Biochemistry And Physiology. 2021;171 doi: 10.1016/j.pestbp.2020.104744. [DOI] [PubMed] [Google Scholar]
- Zhang X.Y., Sun Y., Yang Q.Y., Chen L.L., Li W.H., Zhang H.Y. Control of postharvest black rot caused by Altemaria altemata in strawberries by the combination of Cryptococcus laurentii and Benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester. Biological Control. 2015;90:96–101. [Google Scholar]
- Zou S.C., Wu J.C., Shahid M.Q., He Y.H., Lin S.Q., Liu Z.H., Yang X.H. Identification of key taste components in loquat using widely targeted metabolomics. Food Chemistry. 2020;323 doi: 10.1016/j.foodchem.2020.126822. [DOI] [PubMed] [Google Scholar]
- Tang Q., Zheng X.D. Transcriptome analysis of cherry tomato fruit treated with Cryptoccocus laurentii using high-throughput next-generation sequencing technology. Food Science. 2019;40(18):55–62. [Google Scholar]
- Ou S.Y. Function and application of ferulic acid. Guangzhou Food Science and Technology. 2002;18(4):50–53. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.