Graphical abstract
Keywords: Lacticaseibacillus paracasei ZY-1, Exopolysaccharide (EPS), Structural characterization, eps gene cluster, Biosynthesis
Highlights
-
•
L. paracasei ZY-1 produce two exopolysaccharide fractions (EPS1, EPS2).
-
•
EPS1 was an heteropolysaccharide consisting of mannose and glucose with linear repeating units.
-
•
Identification of the EPS1 biosynthesis cluster suggests a Wzy-dependent synthesis.
-
•
The crude and purified EPS were observed with certain antioxidant property.
Abstract
Exopolysaccharide (EPS) was produced by Lacticaseibacillus paracasei ZY-1 isolated from Tibetan kefir grains, and the preliminary structure of two EPS fractions (EPS1 and EPS2) was investigated. NMR analysis revealed that the backbone of higher producing EPS1 was consisted of →6)-α-D-Manp-(1→, →2,6)-α-D-Glcp-(1→, α-D-Manp-(1→, →2)-α-D-Glcp-(1→, →3)-α-D-Manp-(1→, →6)-α-D-Glcp-(1→. Furthermore, an eps gene cluster that encodes the glycosyltransferase and relevant proteins for EPS biosynthesis was identified on the basis of bioinformation analysis of the complete genome. RT-qPCR results indicated that wzd (ZY-1_2260) and wze (ZY-1_2259) might be essential genes involved in EPS production. Meanwhile, the synthetic mechanism of EPS1 in L. paracasei ZY-1 was further proposed. Besides, the crude and purified EPS showed certain scavenging activities against DPPH, hydroxyl and ABTS radicals. Results provided a better understanding of EPS biosynthesis in L. paracasei ZY-1 at the gene level.
1. Introduction
Kefir is a type of slightly carbonated traditional dairy beverage in the Caucasian mountains, Mongolia and Tibet. The physical appearance of kefir grains is usually described as yellowish or white gelatinous irregular masses, varying in size from 3 to 35 mm in diameter (Rosa et al., 2017). Tibetan kefir grains were originally derived from Tibet, China, where they were traditionally used to make popular home-made fermented yoghurt (Zhou, Liu, Jiang, & Dong, 2009). Regarding the chemical composition of kefir grain, polysaccharide is the dominant constituent, followed by protein, ash, fat and minor vitamins (Marshall & Cole, 1985). Throughout the world, kefir has been well known for its myths of healing power and various nutritional claims for keeping the gastrointestinal balance, improving immunity and even resistance to cardiovascular diseases (Chen et al., 2015, Rodrigues et al., 2016). These benefits of kefir are mainly attributable to the endogenous groups of microorganisms, including Lactobacillus, Lactococcus, and yeast, and sometimes in addition, acetic acid bacteria (Rosa et al., 2017). Generally, the complex microbiological composition of kefir grain probably depends on the origin of the grains, storage conditions and processing methods (Wang, Sun, Song, & Guo, 2021). Most research on Tibetan kefir has focused on the isolation and identification of the microflora, the characterization of exopolysaccharides (EPSs) produced during fermentation and its potential probiotic properties (Chen et al., 2015, Rajoka et al., 2019, Wang et al., 2015).
Lactic acid bacteria (LAB) are generally recognized as safe (GRAS) microorganisms due to their wide and safe application in the production of fermented dairy products such as cheese and yogurt (Saadat, Khosroushahi, & Gargari, 2019). EPSs produced by LAB are classified into two groups depending on their chemical composition, both of which show an enormous structural diversity. Homopolysaccharides (HoPSs) are composed of a single type of sugar subunit with different linkages, while heteropolysaccharides (HePSs) consist of repeating units of two or more types of saccharide residue (Angelin and Kavitha, 2020, Rajoka et al., 2020). EPS has attracted a lot of scientific interest due to its numerous health promoting effects and promising applications in food industry. EPSs from different LAB have been proven to participate in LAB adhesion/recognition, and possess immunomodulatory properties, anti-cancer, antioxidant activity, anti-biofilm agents to prevent pathogenic bacteria adhesion, which could be applied widely as health care products and agents in food industry (Saadat et al., 2019). For instance, Lactobacillus helveticus MB2-1 isolated from traditional Sayram ropy fermented milk (SRFM) synthesized a HePS composed of galactose, glucose and mannose and it exhibited strong scavenging activities on three kinds of radicals (Li et al., 2014a). Moreover, several physio-technical characteristics of EPS have been investigated and extensive efforts have been put forth to explore its structural–functional relationships (Korcz and Varga, 2021, Tiwari et al., 2021). Recent developments in the field of EPSs have resulted in a renewed interest in the structure and biosynthesis mechanism of EPSs. The biosynthesis of HePSs is a relatively complex process that contains specific roles for several gene products encoded by specific genes in eps gene clusters, located in chromosomal or plasmid DNA. In general, genes involved in regulation, chain length determination, polymerization, export and assembling of repeating units are present in eps gene clusters. Among Lactobacillus, eps gene clusters have been identified in L. plantarum, L. rhamnosus, L. casei, L. paracasei, L. sakei, L. reuteri, L. delbrueckii and L. helveticus (Cho et al., 2020, Lamothe et al., 2002). Deletion of a putative priming glycosyltransferase gene epsE resulted in a mutant produce only EPS-1 but not EPS-2, confirming that the eps gene cluster is essential for synthesis of EPSs (Dertli et al., 2013). Although many HePS structures have been published so far, these researches are predominantly focused on pure structural characterization or the determination of potential probiotic effects. However, analysis of associated biosynthesis genes is important to comprehensively understand HePS synthesis. Therefore, identification and prediction of gene cluster involved in EPSs biosynthesis may provide valuable information on the production and synthesis mechanism of EPSs from LAB and even assist structural elucidation of EPSs.
In this study, an EPS-producer strain named L. paracasei ZY-1 was isolated from Tibetan kefir grains, we further described the isolation, purification and primary structural characterization of ZY-1-EPS by ultraviolet–visible (UV–vis), fourier-transform infrared spectroscopy (FT-IR), high-performance liquid chromatography (HPLC) and nuclear magnetic resonance spectroscopy (NMR) including one-dimensional- (1D-) and two-dimensional- (2D-) NMR. Meanwhile, the whole genome of ZY-1 was sequenced for elucidation of EPS biosynthesis, and the relationship between several key genes involved in putative eps gene cluster and EPS production was analyzed based on the RT-qPCR results. Finally, the in vitro antioxidant potential of crude and purified ZY-1-EPS was evaluated.
2. Materials and methods
2.1. Materials and reagents
DEAE-cellulose 52 was purchased from Whatman Co., Ltd (Maidstone, Kebt, UK). 3-methyl-1-phenyl-2-pyrazolin-5-one (PMP), mannose (Man), rhamnose (Rha), glucose (Glc), galactose (Gal), arabinose (Ara), 1,1-Diphenyl-2-picryl-hydrazyl (DPPH), vitamin C (Vc), 1,10-phenanthroline and diammonium 2,2′-azino-bis(3-ehtylbenzothiazoline-6-sulfonate) (ABTS) were obtained from Sigma Chemical Co., Ltd. (St. Louis, Mo, USA). All other reagents were of analytical grade.
2.2. Isolation and molecular identification of LAB
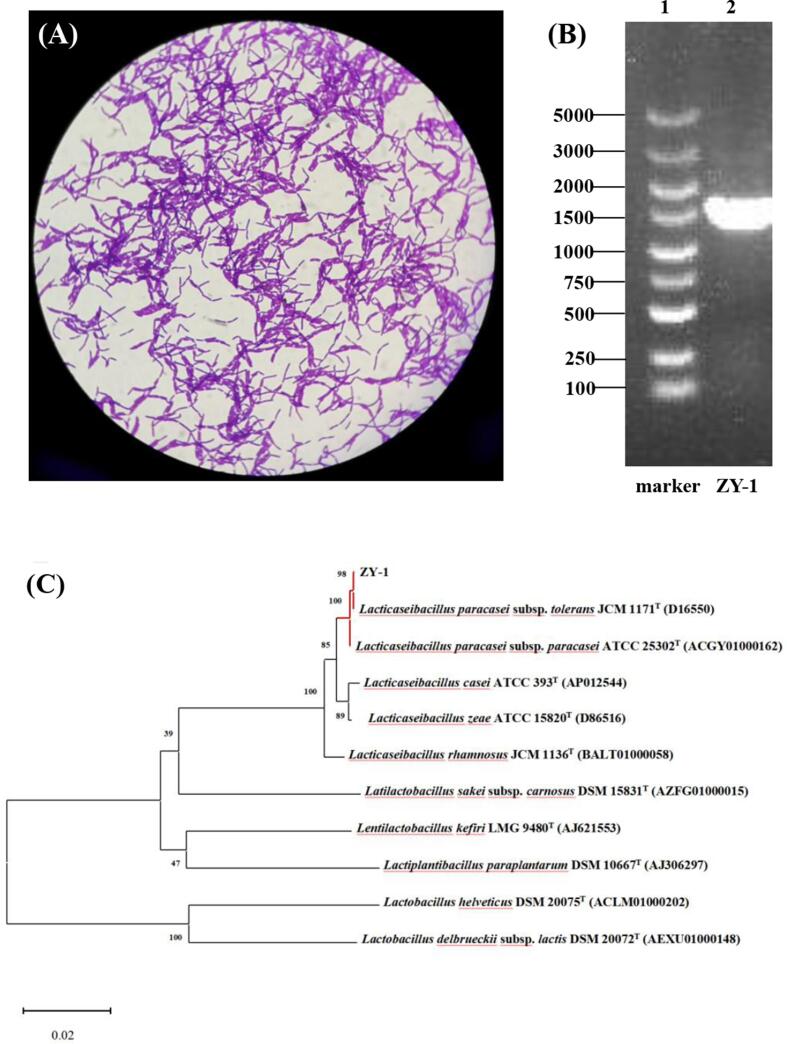
Tibetan kefir grains were obtained from local peasant household in the Nyingchi, Tibet of China. All the samples after collection were immediately placed in ice boxes and transported to laboratory under refrigerated condition within 24 h. The kefir grains were activated in pasteurized milk medium for three times and serially diluted with nine times the volume of sterile physiological saline (0.85 %, w/v). Then, 0.1 mL of serial dilutions (10−3–10−9) were spread in triplicate on De Man, Rogosa and Sharp (MRS, pH 6.2 ± 0.2, Oxoid-CM0361, Unipath, Basingstoke, UK) broth for the isolation of Lactobacillus spp. after incubation at 37 ℃ for 48 h. Gram-positive bacterial isolates were resuspended and maintained in the same medium containing 40% (v/v) glycerol at −80 ℃. The ZY-1 strain was selected and identified based on the 16S rRNA gene sequencing. Briefly, total DNA was isolated using Bacteria Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China) and amplified using the universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGCTACCTTGTTACGACTT-3′). The PCR programs were as follows: 94 ℃ for 5 min; 29 cycles of 94 ℃ for 30 s, 55 ℃ for 30 s and 72 ℃ for 1 min and finally, 72 ℃ for 10 min. The PCR products were electrophoresed at constant voltage (150 V) on 1% (m/v) agarose gel. Sequence similarity was compared using BLAST tool by NCBI and a phylogenetic tree was constructed with Mega 7.0 software based on 16S rRNA gene sequences from 10 Lactobacillus strains according to the neighbor-joining method (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013).
2.3. Analysis of bacterial growth, pH, titratable acidity (TA)
Incubations of L. paracasei ZY-1 for bacterial growth were carried out as batch culture with concentration of 4% (v/v) in MRS broth at 37 °C in 1,000 mL sterile conical flasks. Samples were taken out from the thermostatic incubator every four hours from 0 to 36 h. The viable cell counts were evaluated after incubation of 48 h and calculated by counting colony-forming unit (CFU) on agar plates. A pH Meter (PB-10, Sartorius, Germany) was used to determine the pH values of samples from different time periods and the TA (˚T expressed as Thorner degree) of samples were estimated according to AOAC (Li et al., 2014b).
2.4. Isolation and purification of crude ZY-1-EPS
The EPS isolation was carried out using our previous method (Xiao, Li, et al., 2020b). L. paracasei ZY-1 was typically cultured in MRS broth at 37 ℃ for 28 h. Bacterial cells were removed by centrifugation (12,000 rpm for 15 min) at 4 °C. Afterwards, trichloroacetic acid (TCA) solution was added to a final concentration of 4% (m/v) to remove the precipitated proteins by centrifugation at 12,000 rpm for 15 min. The resulting supernatant was concentrated using rotary evaporator (Heidolph Instruments GmbH & Co, Schwabach, Germany) and then precipitated with three volumes of 99.5% ice cold ethanol and kept at 4 °C for overnight. After centrifugation (12,000 rpm for 15 min at 4 °C), the precipitate was dissolved in deionized water for 72 h and lyophilized for 48 h to afford crude EPS. The freeze-dried crude ZY-1-EPS was fractionated by a DEAE-52 anion exchange column (2.6 × 30 cm) and eluted stepwise with deionized water, 0.1 and 0.3 M NaCl at a flow rate of 1 mL/min. The eluents (10 mL/tube) were then collected automatically and the fractions were assayed for carbohydrate content by the phenol–sulfuric acid method.
2.5. EPS characterization
2.5.1. Chemical composition analysis
Total carbohydrate content was determined by the phenol–sulfuric acid method as mentioned above. Protein content was measured using the Braford method with bovine serum albumin (BSA) as standard. Uronic acid content was evaluated by the carbazole-sulfuric acid method. Sulfate group content was measured as described by our previous method (Tang et al., 2017).
2.5.2. Monosaccharide composition analysis
Monosaccharide composition of the purified EPS was analyzed by HPLC as described previously with minor modification (You et al., 2020). Briefly, 5 mg of EPS sample was hydrolyzed with 2 M trifluoroacetic acid (TFA) at 120 °C for 2 h. The hydrolysate was repeatedly co-concentrated with methanol for three times to dryness and converted to its PMP derivatives. The monosaccharide standards (mannose, rhamnose, glucose, galactose and arabinose) were converted to their derivatives in the same way. After being filtered through 0.22 μm membrane, the hydrolyzate were injected into the Agilent 1200 HPLC system (Palo Alto, CA, USA) with an Eclipse Plus C18 column (4.6 × 250 mm, 5 μm, Agilent) and ultraviolet (UV) detector at 245 nm at 40 °C. Chromatographic separation of PMP derivatives was carried out using phosphate buffer (0.1 M, pH 6.7) and acetonitrile at a ratio of 83 : 17 (v/v) as mobile phase at a flow rate of 0.8 mL/min.
2.5.3. Ultraviolet–visible (UV–vis) and Fourier-transform infrared (FT-IR) analysis
UV–Vis spectroscopy analyses of the purified EPS were recorded using UV-1603 spectrophotometer (Shimadzu Co., Ltd. Kyoto, Japan) in the wavelength range of 190–500 nm. Each sample was dissolved in deionized water at a concentration of 0.5 mg/mL. For determination of the major functional groups in EPS, the infrared spectrum of EPS was performed by a Bruker Tensor-27 FT-IR spectrophotometer (Bruker Co., Ettlingen, Germany), within wave number of 4000–400 cm−1 at a resolution of 4 cm−1 with 64 scans. EPS sample (1 mg) was grounded with dried KBr powder (100 mg) and pressed into 1 mm pellet. Atmospheric compensation, baseline correction and integration were carried out by using the Bruker OPUS 6.5 software package (Bruker Co., Ettlingen, Germany).
2.5.4. Congo red test
The conformational structure of the purified EPS was established by helix-coil transition analysis according to our previous method (Xiao, Li, et al., 2020b). EPS samples were dissolved at 1 mg/mL in dilute NaOH solutions (0.15 M) with a Congo red solution (40 μM). Spectra were recorded on a UV-1603 spectrophotometer (Shimadzu Co., Ltd. Kyoto, Japan) at 190–800 nm.
2.5.5. 1D and 2D NMR spectra analysis
The NMR spectrum of the EPS solution was recorded on a Bruker AVANCE AV-500 spectrometer (Bruker Group, Fällanden, Switzerland) at 500 MHz with deuterium oxide (D2O, 99.9 % D) as the solvent. 1D 1H and 13C NMR experiments were performed at 313 K. 2D 1H–1H correlated spectroscopy (COSY), 1H–13C heteronuclear single quantum coherence (HSQC), 1H–13C nuclear Overhauser effect spectroscopy (NOESY) and 1H–13C heteronuclear multiple quantum coherence (HMBC) measurements were used to assign signals and determine the sequence of sugar residues.
2.6. Whole genome sequencing and annotation of L. paracasei ZY-1
The complete genome sequence of L. paracasei ZY-1 was performed with a combined strategy of long-read Nanopore PromethION48 (Oxford Nanopore Technologies, Oxford, UK) and short-read Illumina NovaSeq 6000 platforms (Illumina, San Diego, CA, USA). The hybrid genome assembly was carried out using Unicycler v0.4.7, which allows for both Nanopore reads and Illumina reads to be used in the conservative mode. The assembled sequence was then corrected using CheckM v1.0.18 to improve the quality of genome assembly. Afterwards, the complete genome sequence of L. paracasei ZY-1 and putative protein-coding sequences (CDS) were annotated using Prokka 1.1.2 (http://www.vicbioinformatics.com/software.prokka.shtml). Functional annotation of CDS was performed by the Clusters of Orthologous Groups of proteins (COG) database (https://www.ncbi.nlm.nih.gov/COG/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg) (Kanehisa et al., 2014, Tatusov et al., 2003). Meanwhile, the secondary metabolism gene clusters were analyzed by the anti-SMASH. Additionally, carbohydrate-active enzymes were predicted by the Carbohydrate-Active Enzymes Database. The eps gene cluster was identified based on BLASTx (https://blast.ncbi.nlm.nih.gov/) and RAST (https://rast.nmpdr.org/) results with the genome data of L. paracasei ZY-1.
2.7. Total RNA isolation and cDNA synthesis
The cellular pellet obtained from a 1 mL sample of each culture in different time intervals (every four hours from 0 h to 36 h) was resuspended evenly in 180 μL of Tris-HCL buffer (10 Mm, pH 8.0). The cells were then lysed at 37 °C by addition of 20 μL of lysozyme (100 mg/mL of Tris-HCl) and processed for total RNA extraction using a Universal RNA isolation kit (Accurate Biotechnology Co., Ltd, Hunan, China). The obtained RNA was dissolved in 50 μL of DEPC-treated water and stored at − 80 °C. Purity and quantity of RNA was determined by 260/280 nm ratio and 1% (m/v) agarose gel electrophoresis. An aliquot of 1 μg of RNA was transcribed into cDNA using the Evo M−MLV RT Kit with gDNA Clean for qPCR II (Accurate Biotechnology Co., Ltd, Hunan, China) and random primers.
2.8. Primer design and real-time quantitative PCR analysis
16S rRNA was selected as housekeeping gene for normalizing the expression of several key genes in eps gene cluster. The sequences of primers given in Table S1 were designed for real-time quantitative PCR (RT-qPCR) analysis using Primer Premier 5 based on the genome sequence of L. paracasei ZY-1. RT-qPCR reactions were set up in a QuantStudio 3 (Thermo Fisher, USA) preloaded with relative quantification software (QuantStudio™ Design & Analysis Software) along with SYBR Green qPCR Kit (Sikejie Biotech Co., Shandong, China). Each PCR reaction was performed in a 20 μL reaction mixture containing 10 μL of 2 × SYBR Green qPCR Mix, 4 μL of each primer at 1 μM and 2 μL of properly diluted cDNA (80 ng/μL). The DNA template was replaced with nuclease-free water for each primer set as negative control. The thermal cycling programs were as follows: pre-denaturation at 95 °C for 30 s, 40 cycles each of denaturation at 95 °C for 10 s, annealing and extension (30 s at 60 °C). The measurement of gene expression by RT-qPCR were performed in triplicate, and the mean of all these values for each gene was analyzed and normalized using the 2−△△CT method. Meanwhile, EPS yield in different time intervals (every four hours for 36 h) was measured by the method as mentioned above.
2.9. Antioxidant properties of ZY-1-EPS
2.9.1. DPPH free radical scavenging activity
The DPPH free radical scavenging activity of crude and purified EPS was determined by our previous method (Xiao, Han, et al., 2020a). In brief, 0.2 mL of DPPH radical solution (0.4 mM in ethanol) and 1.8 mL of deionized water were added to 1.0 mL of sample solution with variable concentrations (0.125–4 mg/mL. The mixture was shaken and incubated at 25 °C in the dark for half an hour, and the absorbance was measured at 517 nm against a blank. VC was used as the positive control.
2.9.2. Hydroxyl radical scavenging activity
The scavenging activity of crude and purified EPS on hydroxyl radical was evaluated by the previous method with a minor modification (Hu, Pang, Wang, & Chen, 2019). One milliliter of sample solution (0.125–4 mg/mL) was mixed with 1.0 mL of 1,10-phenanthroline (0.75 mM), 1.0 mL of FeSO4 (0.75 mM), 1.5 mL of sodium phosphate buffer (0.15 M, pH 7.4) and 1.0 mL of H2O2 (0.01%, v/v). The absorbance of the mixture was measured at 536 nm after 30 min of incubation at 37 °C. VC was used as the positive control.
2.9.3. ABTS free radical scavenging activity
The scavenging activity of crude and purified EPS on ABTS free radical was determined (Ma et al., 2020). Briefly, ABTS solution (7.4 mM) was mixed with potassium persulphate solution (2.6 mM) in a volume ratio of 1: 1 at 4 °C in the dark for 12 h. The mixture was diluted with absolute ethanol to an absorbance of 0.7 ± 0.02 at 734 nm. Then, 0.2 mL and 0.8 mL of ABTS working solution were mixed thoroughly, and the absorbance was measured at 734 nm after 6 min of incubation in the dark. VC was used as the positive control.
2.10. Statistical analysis
All experiments were performed in triplicates (n = 3). Results were analyzed by one-way analysis of variance (ANOVA) using SPSS version 23.0 (SPSS Inc., Chicago, IL, USA) and presented as the mean ± standard deviations (SD). Significant differences were determined by Duncan’s multiple-range tests. P < 0.05 was considered statistically significant.
3. Results and discussion
3.1. Isolation, molecular identification of LAB
Using MRS medium, a white and circular colony with colony diameter of 1–2 mm was picked and examined microscopically (Fig. 1A), then it was re-streaked onto MRS agar plated to obtain pure cultures. The purified isolate was confirmed to be Gram-positive and catalase negative. Fig. 1C given the result of phylogenetic analysis of ZY-1 strain using 16S rRNA gene sequence homology, thus ZY-1 strain was identified as L. paracasei because of the high degree of similarity (as high as 99%) between the 16S rRNA gene sequence of ZY-1 and two type strains of Lacticaseibacillus paracasei. It has been reported that kefir grains encompass a complex microbiota, in which LAB occupy the major propotion. Among them, L. plantarum, L. kefir, L. helveticus, L. kefiranofaciens, L. casei and L. paracasei were generally dominant microorganisms of Tibetan kefir grains (Rosa et al., 2017, Zhou et al., 2009).
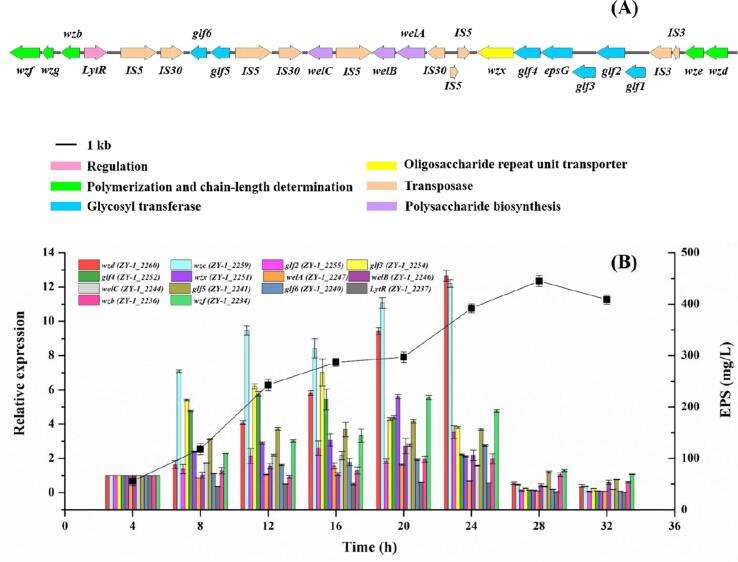
Fig. 1.
Morphology of the ZY-1 strain (10 × 100) (A). PCR product of amplified 16S rDNA gene coding region of ZY-1 strain (B). Phylogenetic tree indicated the relative position of isolate of L. paracasei ZY-1 (C). Phylogenetic tree was carried out using 100 replicates and the number on the nodes represents the support proportion of each branch.
3.2. Time course of bacterial growth by L. paracasei ZY-1
The crude EPS of L. paracasei ZY-1 was obtained after 28 h of incubation, with yield of 527 mg/mL. The crude ZY-1-EPS was subsequently fractionated through DEAE-cellulose 52 anion-exchange chromatography, affording 32.4 % of EPS1 and 25.1 % of EPS2 as detected by phenol–sulfuric acid assay (Fig. S1A). The pH values, TA values and the growth of L. paracasei ZY-1 at different fermentation times were shown in Fig. S1B. The pH values decreased continuously throughout fermentation periods and stabilized at about 3.6 after 24 h. Similarly, the TA values increased with time until of 28 h and the final TA value reached 177.14. In addition, the viable count peaked at 28 h and reached a maximum of 9.59 log10 CFU/mL.
3.3. EPS characterization
3.3.1. Chemical composition and monosaccharide composition analysis
The carbohydrate contents in EPS1 and EPS2 were 85.44% and 77.31%, respectively. The contents of protein and uronic acid were not detected in both of purified EPSs, and the sulfated group content in EPS1 and EPS2 was 0.57% and 0.66%, respectively. The monosaccharides composition of EPS1 and EPS2 was analyzed by HPLC by comparing the retention time with the reference sugar standards. The monosaccharide analysis in Fig S2 showed that EPS1 and EPS2 were composed of glucose, mannose and galactose, but in different molar ratios. The molar ratios of mannose and glucose in EPS1 and EPS2 were 1.17: 1.00 and 5.18: 1.00, respectively. Generally, EPSs from different Lactobacillus comprise repeating units of different monosaccharides, mainly including glucose, galactose, rhamnose and mannose with different molar ratios (Hussain et al., 2017, Min et al., 2019). The differences of monosaccharide compositions might be attributed to different strain and fermentation conditions.
3.3.2. Spectra analysis
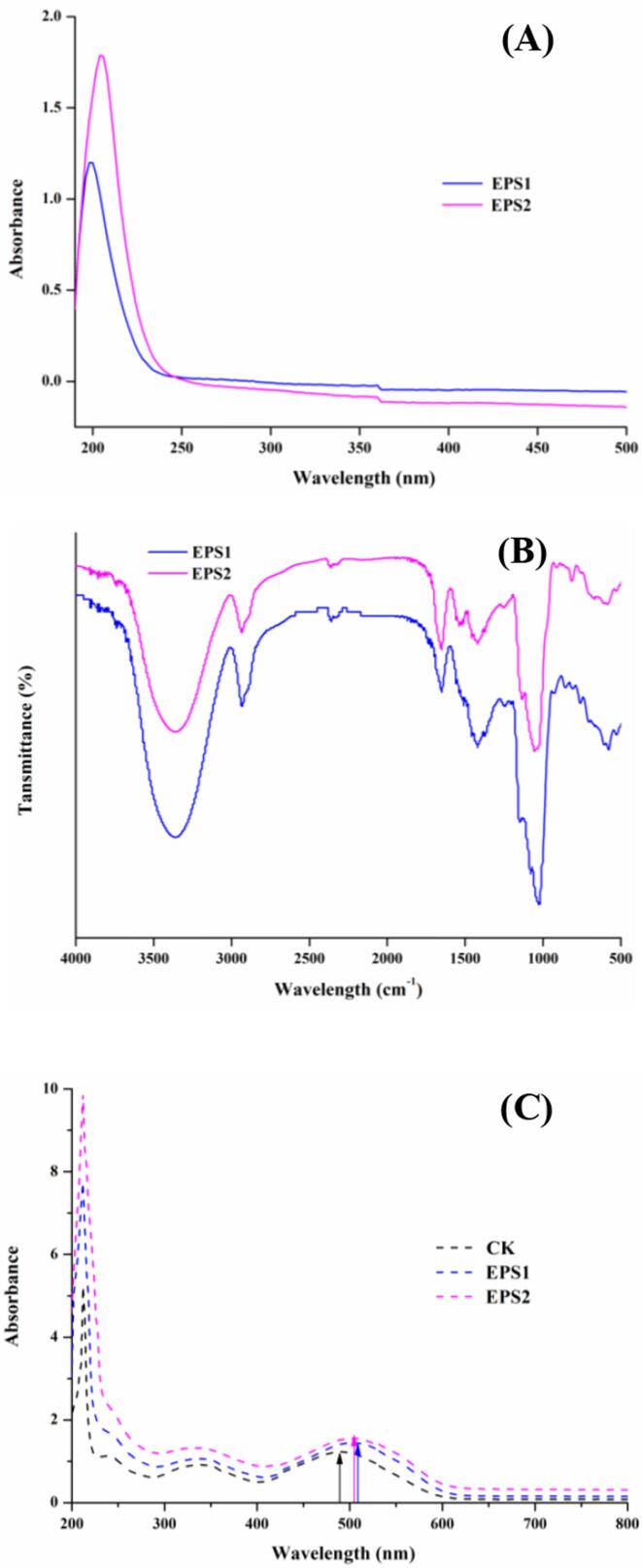
The UV–Vis spectra of EPS1 and EPS2 were shown in Fig. 2A, no obvious absorption at 260 or 280 nm were observed, confirming the absence of nucleic acid and protein. As shown in Fig. 2B, the FT-IR spectra of EPS1 and EPS2 were similar and in good agreement with the typical peaks of polysaccharides. An intense broad band at approximately 3366 cm−1 for EPS1 and EPS2 indicated the presence of (O—H) stretching vibration in hydrogen bonds, and the signal at 2933 cm−1 was attributed to the C—H stretching vibration. Additionally, two asymmetrical stretching band at 1654 and 1419 cm−1 might be characteristic of the absorbance of C O (Zhai et al., 2021). All of the three weak peaks appeared at around 1000–1260 cm−1 on behalf of the presence of the existence of α-pyranose (Wang et al., 2020). Therefore, the FT-IR spectra analysis suggested it was highly likely that the EPS1 and EPS2 belonged to the HePSs with pyranose saccharides.
Fig. 2.
Ultraviolet–Visible (UV–Vis) spectra (A), Fourier-transform infrared (FT-IR) spectra (B) and Congo red test analysis (C) of EPS1 and EPS2 from L. paracasei ZY-1, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3.3. Congo red test
Polysaccharides with a triple-stranded helical conformation can form into a complex with Congo red in a weakly alkaline solution (0.05–1.00 M NaOH solutions), and the complexation is linked with a bathochromic shift in the visible absorption maxima (λmax) of the Congo red spectrum. (Xiao, Li, et al., 2020b). The λmax of Congo red-EPS complexes at 0.15 M NaOH solution were detected by UV–Vis and illustrated in Fig. 2C. Congo red in 0.15 M NaOH without EPS sample was evaluated as the negative control. Compared with blank Congo red solution, EPS1 and EPS2 were exhibited obviously bathochromic shift from 490 nm to 504 nm and 502 nm, respectively, suggesting that both of EPS1 and EPS2 formed a complex with the dye in weak alkaline solution.
3.3.4. NMR spectroscopy
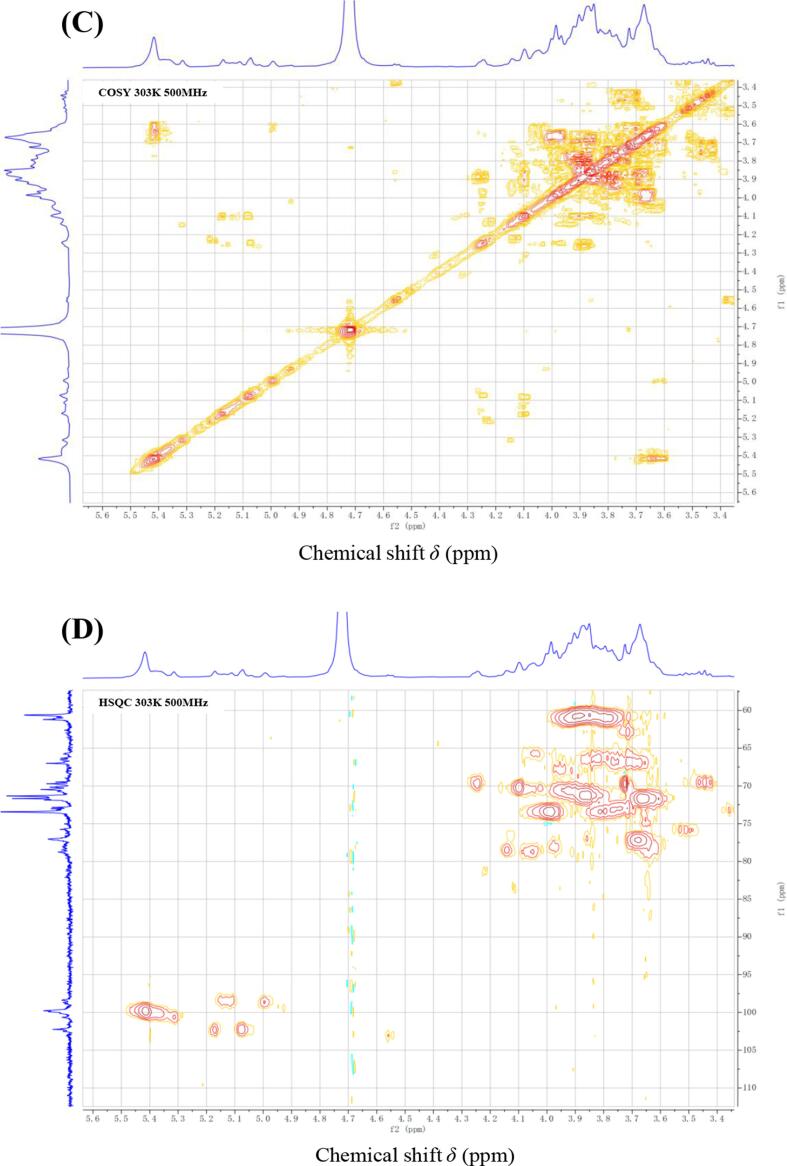
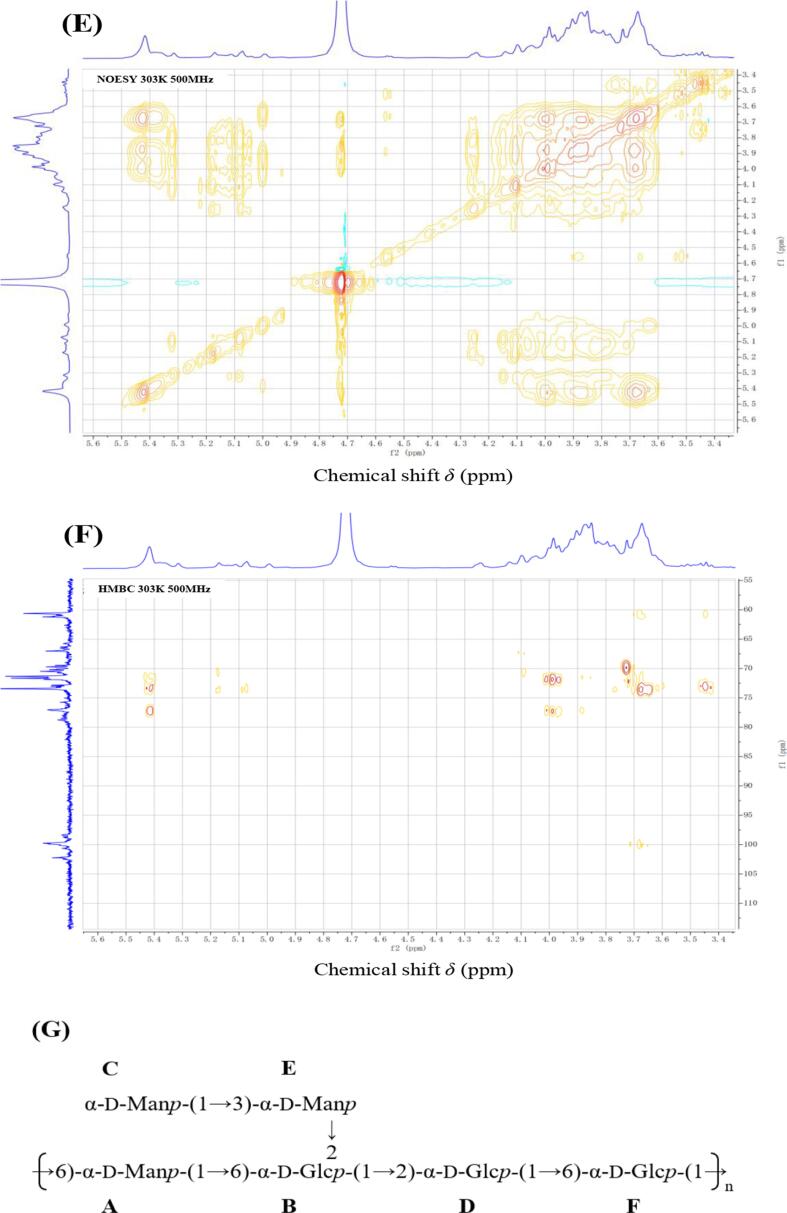
A series of NMR experiments were carried out for EPS1 on account of its relatively higher yield. As shown in the 1D-1H NMR spectrum (Fig. 3A), six clear chemical shift signals of anomeric protons (H-1) were found at δ5.42, 5.31, 5.17, 5.11, 5.07 and 4.99 ppm. They were chosen as A, B, C, D, E and F according to their anomeric proton chemical shifts, respectively. The other signals at δ3.10–4.20 ppm represent the signal peaks of the remaining proton, which were assigned to the H-2–H-6, and the signal at δ4.72 ppm was from the hydrogen of HDO. Based on the chemical shift and coupling constant for the anomeric signals, residues A–F were determined to have α-configurations. Combined with the result of monosaccharide analysis, the A–F residues may reflect six different α-type glycosidic bonds of mannose and glucose residues. In addition, the 1D-13C NMR spectrum (Fig. 3B) showed six signals at δ102.22, 102.10, 100.61, 99.74, 98.65 and 98.35 ppm in the anomeric region. Furthermore, the linkage patterns and complete assignments of 1H and 13C signals for EPS1 were identified on the basis of the 2D-1H–1H COSY, 1H–13C HSQC, 1H–1H NOESY and 1H–13C HMBC experiments. From the HSQC spectrum (Fig. 3D), the anomeric carbon signals at chemical shifts δ102.22, 102.10, 100.61, 99.74, 98.65 and 98.35 ppm cross link to the anomeric proton signals at δ5.17 (C), 5.07 (E), 5.31 (B), 5.42 (A), 4.99 (F) and 5.11 (D), respectively. The chemical shifts of H-2 to H-6 and C-2 to C-6 for A, B, C, D, E and F were further determined from the 1H–1H COSY (Fig. 3C) and 1H–13C spectra according to stepwise magnetization transfers from the anomeric protons. According to the data summarized in Table 1 and literatures data (Agrawal, 1992, Chen et al., 2021, Xie et al., 2020, Yu et al., 2009;), it has been considered that residues A, C and E were assigned as α-D-Manp, and residues B, D and F were assigned as α-D-Glcp. Meanwhile, on the basis of the transition of chemical shifts for carbon under the condition of substitution (Rozi et al., 2019, Zhang et al., 2021), it might be concluded that residues A and F was substituted at C-6 position, and residue D was substituted at C-2 position, whereas residue B was substituted at C-2 and C-6 position. It has been reported that the linkages between backbone and branching of each residue could be obtained from the 1H–1H NOESY and 1H–13C HMBC spectra (Doost, Akbari, Stevens, Setiowati, & Meeren, 2019). Inspection of the cross-peaks in 1H–1H NOESY and 1H–13C HMBC spectra (Fig. 3E and Fig. 3F) indicated that the A H-1 to B H-6, C H-1 to E H-3, E H-1 to B H-2, B H-1 to D H-2, D H-1 to F H-6 were in agreement with the A-(1 → 6)-B, C-(1 → 3)-E, E-(1 → 2)-B, B-(1 → 2)-D and D-(1 → 6)-F linkages, respectively. Thus, residues A, B, C, D, E and F could be further assigned to →6)-α-D-Manp-(1→, →2,6)-α-D-Glcp-(1→, α-D-Manp-(1→, →2)-α-D-Glcp-(1→, →3)-α-D-Manp-(1→, →6)-α-D-Glcp-(1→, respectively. In summary, the predicted repeating unit of EPS1 was proposed as shown in Fig. 3G.
Fig. 3.
The 1H NMR (A) and 13C NMR (B), 1H–1H COSY (C), 1H–13C HSQC (D), 1H–1H NOESY (E), 1H–13C HMBC (F) spectra and proposed structure (G) of purified EPS1 from L. paracasei ZY-1.
Table 1.
Chemical shifts (ppm) of 1H and 13C signals for EPS1 from L. paracasei ZY-1, recorded in D2O at 313 K.
| Glycosyl residues | H-1/C-1 | H-2/C-2 | H-3/C-3 | H-4/C-4 | H-5/C-5 | H-6/C-6 |
|---|---|---|---|---|---|---|
| A | 5.42 | 3.63 | 3.97 | 3.65 | 3.87 | 3.81 |
| →6)-α-D-Manp (1→ | 99.81 | 71.67 | 73.41 | 74.63 | 71.33 | 66.30 |
| B | 5.31 | 4.14 | 3.92 | 3.79 | 3.87 | 3.67 |
| →2,6)-α-D-Glcp (1→ | 100.61 | 78.48 | 70.62 | 73.41 | 71.33 | 67.06 |
| C | 5.17 | 4.10 | 3.87 | 4.26 | 3.90 | 3.85 |
| α-D-Manp (1→ | 102.22 | 70.11 | 71.33 | 69.85 | 70.73 | 60.71 |
| D | 5.11 | 4.05 | 3.63 | 3.70 | 3.97 | 3.72 |
| →2)-α-D-Glcp (1→ | 98.35 | 78.74 | 71.67 | 70.03 | 73.41 | 62.74 |
| E | 5.07 | 4.24 | 3.97 | 3.67 | 3.85 | 3.88 |
| →3)-α-D-Manp (1→ | 102.10 | 69.60 | 78.05 | 71.63 | 71.38 | 60.71 |
| F | 4.99 | 3.62 | 3.81 | 3.89 | 3.79 | 3.94 |
| →6)-α-D-Glcp (1→ | 98.65 | 71.63 | 73.28 | 70.85 | 73.41 | 67.82 |
3.4. Genome features of L. paracasei ZY-1
The complete genome sequence of L. paracasei ZY-1 has been deposited at the GenBank under accession number CP065154.1 (chromosome), CP065155.1 (plasmid1), CP065156.1 (plasmid2), CP065157.1 (plasmid3) and CP065158.1 (plasmid4). The complete genome of L. paracasei ZY-1 was composed of a circular chromosome of 3,148,894 bp and four plasmids named as pLPZ1 (47,612 bp), pLPZ2 (46,347 bp), pLPZ3 (6,289 bp) and pLPZ4 (5,087 bp), with GC contents of 46.38%, 44.03%, 49.57%, 40.94%, 39.51%, respectively (Fig. S3A, Table S2). The entire genome of L. paracasei ZY-1 contained 3,242 genes in total, including 3,124 protein-coding genes (CDS), 59 tRNA genes and 15 rRNA operons, which were accomplished using the NCBI Prokaryotic Genomes Annotation Pipeline (PGAP). The gene category analysis of COG and KEGG pathways illustrated that the carbohydrate transport and metabolism related genes accounted for the largest proportion (Fig. S3B and S3C). It is well-known that the presence of specific genes connected with carbon metabolism in LAB may increase carbon transport capacity and promote the growth of LAB (Jia et al., 2017). Besides, two bacteriocin clusters (ZY-1_2529 to ZY-1_2538 and ZY-1_2577 to ZY-1_2604) were identified according to the bioinformatics prediction of anti-SMASH.
3.5. Sequencing and annotation of the putative eps biosynthetic cluster
The biosynthesis of LAB EPS is a complex process that contains specific roles for unique products that are encoded by eps gene clusters. The specific gene clusters for EPS production in LAB are usually located on chromosomal DNA for thermophilic LAB genera such as Lactobacillus and Streptococcus, but in mesophilic LAB genera like Lactococcus was suggested to be plasmids related (Laws, Gu, & Marshall, 2001). The biosynthesis of EPS in LAB involves several types of functional proteins: regulation; chain-length determination; biosynthesis of repeating units; polymerization and export. Genome analysis of L. paracasei ZY-1 based on NCBI annotation and RAST revealed an eps cluster about 28.6 kb encoded on the chromosomal DNA, which harbor characteristic enzymes of a Wzy-dependent pathway such as glycosyltransferases (GTs) regulatory enzymes and a flippase (Zeidan et al, 2017). Within the eps gene cluster of strain ZY-1, 27 putative open reading frames (QRFs) were identified according to the bioinformatic analysis (Table S3). The predicted gene products are similar to proteins involved in the biosynthesis of various bacteria polysaccharides. Based on these gene sequence similarities, putative encoded function could be attributed to most of the predicted proteins. Within the gene region encoding EPS biosynthesis, ten ORF encodes transposase of the IS3/IS5/IS30 family. The upstream of the eps cluster of L. paracasei ZY-1 contained two genes (wzd and wze) that encode proteins for the chain length determination of EPS in accordance with typical eps clusters in Lacticaseibacillus. rhamnosus GG (74% for wzd; 84% for wze). The epsG (ZY-1_2253) gene was described as a EPSG family protein and displayed 99% identity to the EPSG of L. paracasei (GenBank accession no. WP_196241308). Additionally, six ORFs (glf1-6) in the central region have been predicted to be amino acid sequences that most resemble proteins involved in putative glycosyltransferases genes, which were potentially responsible for the biosynthesis of growing EPS repeating unit. The sequence of wzx (ZY-1_2251) gene showing moderate homology with that of L. rhamnosus GG was assumed as oligosaccharide repeat unit transporter protein (54% with Wzx, GenBank accession no. ACN94850). Particularly, welA (ZY-1_2247), welB (ZY-1_2246) and welC (ZY-1_2244) were predicted as UDP-galactopyranose mutase, UTP-glucose-1-phosphate uridylytransferase and UDP-glucose 4-epimerase, respectively. These genes were involved in the production of sugar nucleotides, which was of great significance for the prophase synthesis of EPS (Laws, Gu, & Marshall, 2001). Moreover, the LytR (ZY-1_2237) gene encoded a protein which had a predicted amino acid sequence with homology to the transcriptional regulator of L. paracasei subsp. paracasei Lpp227 (95% versus LytR (EPC96939)). Finally, the wzb (ZY-1_2236) was identified to encode tyrosine-protein phosphatase, which might also be involved in putative polysaccharide polymerization and chain-length determination (Lebeer et al., 2009). Among them, 14 hypothetical genes in this eps gene cluster were selected for further study because of its different functions in EPS biosynthesis.
3.6. Relationships between expression of key genes and production of EPS
To explore the relationships between the relative expression of hypothetical genes and EPS production during different time points, RNA samples isolated from L. paracasei ZY-1 were evaluated to illustrate the gene expression associated with EPS biosynthesis, and the transcriptional levels for 14 selected EPS genes were investigated by RT-qPCR. Compared with the gene expression at 4 h, all of these genes exhibited prominent differences in relative expression at subsequent time points (Fig. 4B). Notably, the relative expression of wzd (responsible for polysaccharide biosynthesis) was continuously upregulated from 4 h to 24 h, except for a slight dip at 16 h. Meanwhile, both of wzd and wze (responsible for chain length determination) showed significantly higher (>12-fold) transcription at 24 h than that of 4 h, and the production of EPS increased dramatically during the 20–24 h period. This funding suggested that wzd and wze might be essential genes for EPS biosynthesis. At transcriptional level, the expression of glf3 (responsible for glycosyl transferases) was enhanced by more than 7-fold at 16 h compared with control, and the relative expression varied slightly between the genes responsible for different glycosyl transferases in the eps cluster. In addition, results showed that the expression of the all of the 14 genes was significantly reduced from 24 h to 28 h. However, the production of ZY-1-EPS was still accumulating during 24 h to 28 h and reached a maximum concentration of 445.39 mg/L at 28 h, whereas the ZY-1-EPS yield dropped after prolonged incubation, which could be attributed to the action of glycohydrolases in the medium that catalyzed the degradation of EPS.
Fig. 4.
Organization of the eps gene cluster from L. paracasei ZY-1 (CP065154.1) (A). The transcriptional change in the key genes involved in EPS biosynthesis for L. paracasei ZY-1 at different periods of cultivation using RT-qPCR (B). Error bars indicate SD of the mean of triplicates.
3.7. Biosynthesis mechanism of ZY-1-EPS1
According to the bioinformatic analysis of EPS production in the section of 3.5. and structural characterization of EPS1 above, a schematic representation of the putative steps in EPS1 biosynthesis by L. paracasei ZY-1 was proposed in Fig. 5. As we known, EPSs from different LAB strains exhibited unique structures depending on the specific glycosyltransferase enzymes. However, the biosynthesis mechanism of EPS production is relatively conserved among LAB (Zeidan et al., 2017). When glucose was absorbed into the cytoplasm as the main sugar source, glucokinase intermediated the transition from glucose to glucose-6-phosphate, and then glucose-6-phosphate could be converted into glucose-1-phosphate in assistance with the enzyme welB (ZY-1_2246) (Xu et al., 2019). The presence of glucose-1-phosphate uridylyltransferase (welB) and UDP-glucose-4-epimerase (welC) proved the synthesis of remaining activated sugar precursors UDP-glucose. Besides, the eps clusters were speculated to harbour genes responsible for the biosynthesis of a GDP-mannose precursor according to the results of monosaccharide analysis and NMR characterization. Meanwhile, the biosynthesis of lipid carrier on the inner side of the cell membrane was involved in EPS production and assembly of the cell envelope structure (Delcour, Ferain, Deghorain, Palumbo, & Hols, 1999). This step was catalyzed by the priming glycosyltransferase glf2, and a phosphormannosyl residue was transferred from an activated nucleotide sugar to the undecaprenyl phosphate (UndP)-lipid carrier on the cytoplasmic membrane of the cell (Islam & Lam, 2014). This was followed by the sequential addition of the sugar nucleotides by the different unique glycosyltransferases glf3-6 in a sugar and glycosidic linkage-dependent manner. Currently, up to 101 GT families could be retrieved online in the Carbogydrate Active enZymes (CAZy) database (Lombard, Golaconda Ramulu, Drula, Coutinho, & Henrissat, 2014). However, some GTs show promiscuity to different substrates, which makes it difficult to predict their activity based barely on sequence analysis. And a reliable location of the GTs encoded in the eps cluster to the corresponding sugar units of the oligosaccharide is difficult at this stage, further experiments with recombinant enzymes are required. Once an EPS subunit is completed, it was translocated from the inner part of the cytoplasmic membrane to the outer part of the membrane by a flippase (wzx), and then the repeating units were coupled into polysaccharides with the help of specific polymerase (wzb). Finally, several chain length determination proteins detached the polysaccharides complex to stop the polymerization and export process. Tyrosine kinases (wze) and trypsin-like serine protease (wzf) were thought to be involved in the determination of the chain length of the final EPS1, similar pathway was observed in L. rhamnosus GG (Lebeer et al., 2009).
Fig. 5.
Schematic representation of the putative steps in EPS1 biosynthesis by L. paracasei ZY-1 that were encoded within the relevant genes of eps gene cluster.
3.8. Antioxidant properties of ZY-1-EPS
3.8.1. DPPH free radical scavenging activity
The scavenging activity of crude and purified EPS for DPPH free radical was presented in Fig S4A in the concentration range from 0.125 to 4 mg/mL. All the samples exhibited concentration-dependent scavenging activity on DPPH radical, while they showed weaker scavenging ability than that of Vc. Furthermore, the scavenging abilities of the crude EPS was relatively higher than that of its purified fractions (EPS1 and EPS2) at the concentration ranges of 1–4 mg/mL. This might be attributed with the presence of some antioxidant components in the crude EPS, such as protein, sulfate, peptides and microelements, and there may be some synergistic effects and interactions for antioxidant properties among these antioxidant components. At a concentration of 4 mg/mL, the abilities of crude EPS, EPS1 EPS2 and Vc to scavenge DPPH free radicals were 36.03 ± 1.80%, 21.20 ± 1.62%, 25.88 ± 0.84% and 96.78 ± 1.27%, respectively (the difference between groups was significant). Results demonstrated that EPS from L. paracasei ZY-1 had relative scavenging activity on DPPH free radical, suggesting that EPS likely contains substances that are hydrogen donors and react with free radicals to scavenge DPPH (Trabelsi et al., 2017).
3.8.2. Hydroxyl radical scavenging activity
The scavenging activity of EPS and Vc on hydroxyl radicals were in a concentration-dependent way as illustrated in Fig S4B. All the three EPS samples exerted a very similar scavenging effects at a low concentration of 0.125 mg/mL and 0.25 mg/mL (no significant differences, p < 0.05). A slight increase in scavenging ability with the concentration of EPS was observed at higher than 2.0 mg/mL, which might be due to the decreased solubility of EPS. At 4 mg/mL, the scavenging abilities of all EPS samples increased in the order of EPS1 (12.44 ± 0.77%), crude EPS (14.41 ± 1.15%) and EPS2 (20.89 ± 1.57%). The scavenging effect of EPS2 was stronger than that of crude EPS and EPS1 in the same concentration, which might be associated with its relatively higher content of sulfate (Zhang et al., 2013). Moreover, the antioxidant activity of EPS can be affected by the combined effects of monosaccharide composition, sulfate and protein content, as well as other factors.
3.8.3. ABTS free radical scavenging activity
As shown in Fig S4C, the scavenging activities of crude and purified EPSs for ABTS free radical followed a dose-dependent manner at 0.125–4 mg/mL. The crude EPS possessed significantly strong scavenging ability, followed by EPS2, and then EPS1. At a concentration of 4 mg/mL, the scavenging activities on ABTS free radical for crude EPS (55.99 ± 2.73%), EPS2 (52.46 ± 0.92%) and EPS1 (52.25 ± 1.21%) and Vc were similar (no significant differences, p < 0.05). As compared with DPPH radical scavenging assay, all of the crude and purified EPSs displayed stronger ability to scavenge ABTS radical. This was probably due to the difference properties of the radical model. Furthermore, the relative higher content of protein may have some impact on the ABTS free radical scavenging activity as reported earlier (Raza et al., 2012). Results suggested that ZY-1-EPS could be an effective scavenger for ABTS free radicals, it further proved the necessity to conduct diverse assays of scavenging of various free radical models to estimate the antioxidant activity of exopolysaccharides.
4. Conclusion
In the present study, L. paracasei ZY-1 was isolated and identified from Tibetan kefir grains, and ZY-1-EPS from this novel strain was derived, purified and characterized by HPLC, UV–vis, FT-IR and Congo red test. EPS1 possessed complex structures consisting of mannose and glucose with linear repeating units according to analysis of 1D- and 2D-NMR experiments. To understand the relationship between the genotype and phenotype of L. paracasei ZY-1 in terms of EPS biosynthesis, the putative eps gene cluster of strain ZY-1 was sequenced and the gene related to EPS biosynthesis was investigated through bioinformatic analysis. The transcriptional change in 14 key genes involved in EPS biosynthesis were determined and RT-qPCR. Besides, the biosynthetic model of EPS1 was speculated based on the 1D- and 2D-NMR. Further study on engineering the key gene in EPS gene cluster could be an efficient strategy for increasing EPS production and elucidating the influence of EPS on probiotic properties of L. paracasei ZY-1.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was co-financed by National Natural Science Foundation of China (No. U1903108, 31871771 and 31571818), Natural Science Foundation of Jiangsu Province (No. BK20201320), Jiangsu Agriculture Science and Technology Innovation Fund (No. CX(20)3043), Key R & D plan of Jiangsu Province (BE2020305 and XZ-SZ202032), Qing Lan Project of Jiangsu Province and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochms.2021.100054.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Agrawal P.K. NMR spectroscopy in the structural elucidation of oligosaccharides and glycosides. Phytochemistry. 1992;31:3307–3330. doi: 10.1016/0031-9422(92)83678-R. [DOI] [PubMed] [Google Scholar]
- Angelin J., Kavitha M. Exopolysaccharides from probiotic bacteria and their health potential. International Journal of Biological Macromolecules. 2020;162:853–865. doi: 10.1016/j.ijbiomac.2020.06.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang T., Zhang X., Zhang F., Linhardt R.J. Structural and immunological studies on the polysaccharide from spores of a medicinal entomogenous fungus Paecilomyces cicadae. Carbohydrate Polymers. 2021;254:117462. doi: 10.1016/j.carbpol.2020.117462. [DOI] [PubMed] [Google Scholar]
- Chen Z., Shi J., Yang X., Nan B., Liu Y., Wang Z. Chemical and physical characteristics and antioxidant activities of the exopolysaccharide produced by Tibetan kefir grains during milk fermentation. International Dairy Journal. 2015;43:15–21. doi: 10.1016/j.idairyj.2014.10.004. [DOI] [Google Scholar]
- Cho S.W., Yim J., Seo S.W. Engineering Tools for the Development of Recombinant Lactic Acid Bacteria. Biotechnology Journal. 2020;15:1900344. doi: 10.1002/biot.201900344. [DOI] [PubMed] [Google Scholar]
- Delcour J., Ferain T., Deghorain M., Palumbo E., Hols P. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Lactic Acid Bacteria: Genetics, Metabolism and Applications. 1999;159–184 doi: 10.1007/978-94-017-2027-4_7. [DOI] [PubMed] [Google Scholar]
- Dertli E., Colquhoun I.J., Gunning A.P., Bongaerts R.J., Gall G.L., Bonev B.B.…Narbad A. Structure and biosynthesis of two exopolysaccharides produced by Lactobacillus johnsonii FI9785. Journal of Biological Chemistry. 2013;288:31938–31951. doi: 10.1074/jbc.M113.507418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doost A.S., Akbari M., Stevens C.V., Setiowati A.D., Meeren P.V. A review on nuclear overhauser enhancement (NOE) and rotating-frame overhauser effect (ROE) NMR techniques in food science: Basic principles and applications. Trends in Food Science & Technology. 2019;86:16–24. doi: 10.1016/j.tifs.2019.02.001. [DOI] [Google Scholar]
- Hu X., Pang X., Wang P.G., Chen M. Isolation and characterization of an antioxidant exopolysaccharide produced by Bacillus sp. S-1 from Sichuan Pickles. Carbohydrate Polymers. 2019;204:9–16. doi: 10.1016/j.carbpol.2018.09.069. [DOI] [PubMed] [Google Scholar]
- Hussain A., Zia K.M., Tabasum S., Noreen A., Ali M., Iqbal R., Zuber M. Blends and composites of exopolysaccharides; properties and applications: A review. International Journal of Biological Macromolecules. 2017;94:10–27. doi: 10.1016/j.ijbiomac.2016.09.104. [DOI] [PubMed] [Google Scholar]
- Islam S.T., Lam J.S. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Canadian Journal of Microbiology. 2014;60:697–716. doi: 10.1139/cjm-2014-0595. [DOI] [PubMed] [Google Scholar]
- Jia F.F., Zhang L.J., Pang X.H., Gu X.X., Abdelazez A., Liang Y.…Meng X.C. Complete genome sequence of bacteriocin-producing Lactobacillus plantarum KLDS1. 0391, a probiotic strain with gastrointestinal tract resistance and adhesion to the intestinal epithelial cells. Genomics. 2017;109:432–437. doi: 10.1016/j.ygeno.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Research. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korcz E., Varga L. Exopolysaccharides from lactic acid bacteria: Techno-functional application in the food industry. Trends in Food Science & Technology. 2021;110:375–384. doi: 10.1016/j.tifs.2021.02.014. [DOI] [Google Scholar]
- Lamothe G., Jolly L., Mollet B., Stingele F. Genetic and biochemical characterization of exopolysaccharide biosynthesis by Lactobacillus delbrueckii subsp. bulgaricus. Archives of Microbiology. 2002;178:218–228. doi: 10.1007/s00203-002-0447-x. [DOI] [PubMed] [Google Scholar]
- Laws A., Gu Y., Marshall V. Biosynthesis, characterisation, and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnology Advances. 2001;19:597–625. doi: 10.1016/S0734-9750(01)00084-2. [DOI] [PubMed] [Google Scholar]
- Lebeer S., Verhoeven T.L.A., Francius G., Schoofs G., Lambrichts I., Dufrêne Y.…De Keersmaecker S.C.J. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Applied and Environmental Microbiology. 2009;75:3554–3563. doi: 10.1128/AEM.02919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Ji J., Chen X., Jiang M., Rui X., Dong M. Structural elucidation and antioxidant activities of exopolysaccharides from Lactobacillus helveticus MB2-1. Carbohydrate Polymers. 2014;102:351–359. doi: 10.1016/j.carbpol.2013.11.053. [DOI] [PubMed] [Google Scholar]
- Li W., Ji J., Rui X., Yu J., Tang W., Chen X.…Dong M. Production of exopolysaccharides by Lactobacillus helveticus MB2-1 and its functional characteristics in vitro. LWT-Food Science and Technology. 2014;59:732–739. doi: 10.1016/j.lwt.2014.06.063. [DOI] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.S., Liu H., Han C.R., Zeng S.J., Xu X.J., Lu D.J., He H.J. Extraction, characterization and antioxidant activity of polysaccharide from Pouteria campechiana seed. Carbohydrate Polymers. 2020;229:115409. doi: 10.1016/j.carbpol.2019.115409. [DOI] [PubMed] [Google Scholar]
- Marshall V.M., Cole W.M. Methods for making kefir and fermented milks based on kefir. Journal of Dairy Research. 1985;52:451–456. doi: 10.1017/S0022029900024353. [DOI] [Google Scholar]
- Min W.H., Fang X.B., Wu T., Fang L., Liu C.L., Wang J. Characterization and antioxidant activity of an acidic exopolysaccharide from Lactobacillus plantarum JLAU103. Journal of Bioscience and Bioengineering. 2019;127:758–766. doi: 10.1016/j.jbiosc.2018.12.004. [DOI] [PubMed] [Google Scholar]
- Rajoka M.S.R., Mehwish H.M., Fang H., Padhiar A.A., Zeng X., Khurshid M.…Zhao L. Characterization and anti-tumor activity of exopolysaccharide produced by Lactobacillus kefiri isolated from Chinese kefir grains. Journal of Functional Foods. 2019;63:103588. doi: 10.1016/j.jff.2019.103588. [DOI] [Google Scholar]
- Rajoka M.S.R., Wu Y., Mehwish H.M., Bansal M., Zhao L. Lactobacillus exopolysaccharides: New perspectives on engineering strategies, physiochemical functions, and immunomodulatory effects on host health. Trends in Food Science & Technology. 2020;103:36–48. doi: 10.1016/j.tifs.2020.06.003. [DOI] [Google Scholar]
- Raza W., Yang W., Jun Y., Shakoor F., Huang Q., Shen Q. Optimization and characterization of a polysaccharide produced by Pseudomonas fluorescens WR-1 and its antioxidant activity. Carbohydrate Polymers. 2012;90:921–929. doi: 10.1016/j.carbpol.2012.06.021. [DOI] [PubMed] [Google Scholar]
- Rodrigues K.L., Araújo T.H., Schneedorf J.M., Ferreira C.S., Moraes G.O., Coimbra R.S., Rodrigues M.R. A novel beer fermented by kefir enhances anti-inflammatory and anti-ulcerogenic activities found isolated in its constituents. Journal of Functional Foods. 2016;21:58–69. doi: 10.1016/j.jff.2015.11.035. [DOI] [Google Scholar]
- Rosa D.D., Dias M.M.S., Grześkowiak Ł.M., Reis S.A., Conceicao L.L., Peluzio M.C.G. Milk kefir: Nutritional, microbiological and health benefits. Nutrition Research Reviews. 2017;30:82–96. doi: 10.1017/S0954422416000275. [DOI] [PubMed] [Google Scholar]
- Rozi P., Abuduwaili A., Mutailifu P., Gao Y., Rakhmanberdieva R., Aisa H.A., Yili A. Sequential extraction, characterization and antioxidant activity of polysaccharides from Fritillaria pallidiflora Schrenk. International Journal of Biological Macromolecules. 2019;131:97–106. doi: 10.1016/j.ijbiomac.2019.03.029. [DOI] [PubMed] [Google Scholar]
- Saadat Y.R., Khosroushahi A.Y., Gargari B.P. A comprehensive review of anticancer, immunomodulatory and health beneficial effects of the lactic acid bacteria exopolysaccharides. Carbohydrate Polymers. 2019;217:79–89. doi: 10.1016/j.carbpol.2019.04.025. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Dong M., Wang W., Han S., Rui X., Chen X.…Li W. Structural characterization and antioxidant property of released exopolysaccharides from Lactobacillus delbrueckii ssp. bulgaricus SRFM-1. Carbohydrate Polymers. 2017;173:654–664. doi: 10.1016/j.carbpol.2017.06.039. [DOI] [PubMed] [Google Scholar]
- Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V.…Natale D.A. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:1–14. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S., Kavitake D., Devi P.B., Shetty P.H. Bacterial exopolysaccharides for improvement of technological, functional and rheological properties of yoghurt. International Journal of Biological Macromolecules. 2021;183:1585–1595. doi: 10.1016/j.ijbiomac.2021.05.140. [DOI] [PubMed] [Google Scholar]
- Trabelsi I., Ktari N., Slima S., Triki M., Bardaa S., Mnif H., Salah R.B. Evaluation of dermal wound healing activity and in vitro antibacterial and antioxidant activities of a new exopolysaccharide produced by Lactobacillus sp. Ca6. International Journal of Biological Macromolecules. 2017;103:194–201. doi: 10.1016/j.ijbiomac.2017.05.017. [DOI] [PubMed] [Google Scholar]
- Wang H., Sun X., Song X., Guo M. Effects of kefir grains from different origins on proteolysis and volatile profile of goat milk kefir. Food Chemistry. 2021;339:128099. doi: 10.1016/j.foodchem.2020.128099. [DOI] [PubMed] [Google Scholar]
- Wang J., Zhao X., Tian Z., Yang Y., Yang Z. Characterization of an exopolysaccharide produced by Lactobacillus plantarum YW11 isolated from Tibet Kefir. Carbohydrate Polymers. 2015;125:16–25. doi: 10.1016/j.carbpol.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu N., Xue X., Li Q., Sun D., Zhao Z. Purification, structural characterization and in vivo immunoregulatory activity of a novel polysaccharide from Polygonatum sibiricum. International Journal of Biological Macromolecules. 2020;160:688–694. doi: 10.1016/j.ijbiomac.2020.05.245. [DOI] [PubMed] [Google Scholar]
- Xiao L., Han S., Zhou J., Xu Q., Dong M., Fan X.…Li W. Preparation, characterization and antioxidant activities of derivatives of exopolysaccharide from Lactobacillus helveticus MB2-1. International Journal of Biological Macromolecules. 2020;145:1008–1017. doi: 10.1016/j.ijbiomac.2019.09.192. [DOI] [PubMed] [Google Scholar]
- Xiao L., Li Y., Tian J., Zhou J., Xu Q., Feng L.…Li W. Influences of drying methods on the structural, physicochemical and antioxidant properties of exopolysaccharide from Lactobacillus helveticus MB2-1. International Journal of Biological Macromolecules. 2020;157:220–231. doi: 10.1016/j.ijbiomac.2020.04.196. [DOI] [PubMed] [Google Scholar]
- Xie X., Shen W., Zhou Y., Ma L., Xu D.J., He L., Shen B., Zhou C. Characterization of a polysaccharide from Eupolyphaga sinensis walker and its effective antitumor activity via lymphocyte activation. International Journal of Biological Macromolecules. 2020;162:31–42. doi: 10.1016/j.ijbiomac.2020.06.120. [DOI] [PubMed] [Google Scholar]
- Xu Y., Cui Y., Yue F., Liu L., Shan Y., Liu B.…Lü X. Exopolysaccharides produced by lactic acid bacteria and Bifidobacteria: Structures, physiochemical functions and applications in the food industry. Food Hydrocolloids. 2019;94:475–499. doi: 10.1016/j.foodhyd.2019.03.032. [DOI] [Google Scholar]
- You X., Yang L., Zhao X., Ma K., Chen X., Zhang C.…Li W. Isolation, purification, characterization and immunostimulatory activity of an exopolysaccharide produced by Lactobacillus pentosus LZ-R-17 isolated from Tibetan kefir. International Journal of Biological Macromolecules. 2020;158:408–419. doi: 10.1016/j.ijbiomac.2020.05.027. [DOI] [PubMed] [Google Scholar]
- Yu R., Yin Y., Yang W., Ma W., Yang L., Chen X.…Song L. Structural elucidation and biological activity of a novel polysaccharide by alkaline extraction from cultured Cordyceps militaris. Carbohydrate Polymers. 2009;75:166–171. doi: 10.1016/j.carbpol.2008.07.023. [DOI] [Google Scholar]
- Zeidan A.A., Poulsen V.K., Janzen T., Buldo P., Derkx P.M.F., Oregaard G., Neves A.R. Polysaccharide production by lactic acid bacteria: From genes to industrial applications. FEMS Microbiology Reviews. 2017;41:S168–S200. doi: 10.1093/femsre/fux017. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang X., Gu S., Pan L., Sun H., Gong E.…Elkhateeb W.A. Structure analysis and antioxidant activity of polysaccharide-iron (III) from Cordyceps militaris mycelia. International Journal of Biological Macromolecules. 2021;178:170–179. doi: 10.1016/j.ijbiomac.2021.02.163. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Lv G., He W., Shi L., Pan H., Fan L. Effects of extraction methods on the antioxidant activities of polysaccharides obtained from Flammulina velutipes. Carbohydrate Polymers. 2013;98:1524–1531. doi: 10.1016/j.carbpol.2013.07.052. [DOI] [PubMed] [Google Scholar]
- Zhai Z., Chen A., Zhou H., Zhang D., Du X., Liu Q.…Su P. Structural characterization and functional activity of an exopolysaccharide secreted by Rhodopseudomonas palustris GJ-22. International Journal of Biological Macromolecules. 2021;167:160–168. doi: 10.1016/j.ijbiomac.2020.11.165. [DOI] [PubMed] [Google Scholar]
- Zhou J., Liu X., Jiang H., Dong M. Analysis of the microflora in Tibetan kefir grains using denaturing gradient gel electrophoresis. Food Microbiology. 2009;26:770–775. doi: 10.1016/j.fm.2009.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.