Highlights
-
•
A chemical database called ChemTastesDB was created, which consists of 2944 organic and inorganic tastants.
-
•
Tastants have been curated and classified into 5 basic and 4 non-basic taste categories.
-
•
The chemical space of the ChemTastesDB has been analyzed with unsupervised machine learning.
-
•
ChemTastesDB is freely available on line and provides support for decision-making for designing new tastants.
Keywords: ChemTastesDB, Database, Tastes, Chemical space, Foodinformatics
Abstract
The purpose of this work is the creation of a chemical database named ChemTastesDB that includes both organic and inorganic tastants. The creation, curation pipeline and the main features of the database are described in detail. The database includes 2944 verified and curated compounds divided into nine classes, which comprise the five basic tastes (sweet, bitter, umami sour and salty) along with four additional categories: tasteless, non-sweet, multitaste and miscellaneous. ChemTastesDB provides the following information for each tastant: name, PubChem CID, CAS registry number, canonical SMILES, class taste and references to the scientific sources from which data were retrieved. The molecular structure in the HyperChem (.hin) format of each chemical is also made available. In addition, molecular fingerprints were used for characterizing and analyzing the chemical space of tastants by means of unsupervised machine learning. ChemTastesDB constitutes a useful tool to the scientific community to expand the information of taste molecules and to assist in silico studies for the taste prediction of unevaluated and as yet unsynthetized compounds, as well as the analysis of the relationships between molecular structure and taste. The database is freely accessible at https://doi.org/10.5281/zenodo.5747393.
1. Introduction
The sensation of taste plays an important role in the food chemistry field, since it is closely related to the development and selection of food products and food intake. Throughout history, there has been a strong interest in understanding the mechanism by which gustatory sensation is perceived by humans (Damodaran & Parkin, 2017). The extraordinary developments in foodinfomatics (computational food chemistry) and bioinformatics (computational biochemistry) have provided the necessary tools to study the receptor/ligand binding interaction. In order to achieve a particular taste, it is now understood that the structure of the receptors and the specific features of the tastant ligands to interact with receptors must be analyzed (Chandrashekar et al., 2006, Rojas et al., 2016). A molecular tastant is a water-soluble chemical compound (ligand) able to interact with the chemosensory receptors to produce a taste sensation (Di Lorenzo et al., 2009). The taste-receptor cells (TRCs) are located in the gustatory papillae of the tongue and palate epithelium, which react to tastants by means of receptor-ligand interactions along with other mechanisms. These additional mechanisms are associated with the opening of ion channels or through secondary messenger channels associated with nucleotides or phosphorylated inositol (Damodaran and Parkin, 2017, Di Lorenzo et al., 2009, Wong, 2018). Evidence suggests that there are five basic tastes (sweet, bitter, umami, sour and salty), which are also known as “taste modalities” or “receptor-mediated tastes” (Chandrashekar et al., 2006, Morini et al., 2011).
Among the basic tastes, sweetness is probably the most important, since sweeteners evoke a pleasant sensation in several foods and medicines (Chandrashekar et al., 2006, Damodaran and Parkin, 2017). Sucrose is used as a standard to quantify the relative sweetness (RS) of new sweet-tasting molecules (Rojas et al., 2016, Rojas et al., 2016a). The sweet taste chemoreceptor is a G-protein coupled receptor (GPCR) of class C made up of T1R2/T1R3 subunits (Chandrashekar et al., 2006, Morini et al., 2011). In contrast to the pleasant sensation of sweetness, bitterness may be related to the protection of humans from the consumption of toxic compounds (Chandrashekar et al., 2006, Di Lorenzo et al., 2009), although in some foods or products it is perceived as a pleasant taste. Quinine sulfate is used as the bitterness standard (Damodaran and Parkin, 2017, Rojas et al., 2017). Bitterness receptors are comprised of a family of T2Rs proteins, which are located on taste receptor cells. (Chandrashekar et al., 2006, Di Lorenzo et al., 2009). Umaminess is defined as a meaty or savory sensation (Baines and Brown, 2016, Damodaran and Parkin, 2017, Suess et al., 2015, Wong, 2018) and monosodium glutamate (MSG) is used as a standard to quantify the relative intensity of umami tastants (Baines & Brown, 2016). As it is for sweetness, this taste receptor is a GPCR made up of T1R1/T1R3 subunits (Chandrashekar et al., 2006, Morini et al., 2011). The taste sensation of sourness is related to substances that produce hydrogen ions when they are diluted in water, such as citric acid, which is used as the sourness standard (Damodaran and Parkin, 2017, Ley et al., 2012, Wong, 2018). Finally, saltiness is a stimulus produced by soluble salts, particularly salts of low-molecular-weight, such as chlorides (sodium, potassium and calcium). Sodium chloride is the standard for quantifying saltiness (Di Lorenzo et al., 2009, Ley et al., 2012, Wong, 2018).
In addition to molecules that elicit the five basic tastes, there are tastants that evoke other kinds of tastes, such as astringent, chilling, cooling, heating or pungent (Damodaran and Parkin, 2017, Ley et al., 2012, Wong, 2018). Other compounds elicit a complex combination of tastes (multitastes), for instance potassium acid oxalate and protocatechuic acid produce a sour/bitter taste (Wong, 2018), while calcium phenolsulfonate and benzyl acetate exhibit bitter/astringent and bitter/pungent tastes, respectively (Dagan-Wiener et al., 2019). Additionally, tastelessness refers to insipid molecules, that is, chemicals exhibiting the lack of any particular taste. This class of compounds involves non-sweet, non-bitter, non-sour, non-salty or non-umami compounds (Damodaran and Parkin, 2017, Rojas et al., 2017).
The chemical analysis of taste molecules in raw ingredients and in end-products for human consumption play an important role for the assurance of food quality and desirability, as well as to prevent defects (offensive tastes) in consumer food products (Ley et al., 2012). Due to interest in producing new, safe and more potent tastants (particularly sweet and bitter), several freely accessible databases containing information of taste molecules have been reported in the literature in the last decade. These databases include: SuperSweet (http://bioinformatics.charite.de/sweet/) (Ahmed et al., 2011), BitterDB (http://bitterdb.agri.huji.ac.il/) (Dagan-Wiener et al., 2019), TasteDB (by merging portions of the SuperSweet and the BitterDB) (Ruddigkeit & Reymond, 2014), SweetenersDB (http://chemosim.unice.fr/SweetenersDB/) (Bouysset et al., 2020). Additionally, other databases of tastants have been recently developed for in silico modeling, such as BitterX (Huang et al., 2016), expert system (Rojas et al., 2017), BitterPredict (Dagan-Wiener et al., 2017), BitterSweetForest (Banerjee & Preissner, 2018), e-Bitter (Zheng et al., 2018), e-Sweet (Zheng et al., 2019), BitterSweet (Tuwani et al., 2019), structure-based screening (Shoshan-Galeczki & Niv, 2020), BTP640 (Charoenkwan et al., 2020), children’s bitter drug prediction system (CBDPS) (Bai et al., 2021), and multi-layer prediction system (Yang et al., 2022).
Given these advances, we developed an extensive database of molecular structures with associated information on taste. The database is named ChemTastesDB and includes 2944 organic and inorganic tastants. For each tastant, the database includes the following information: PubChem CID, CAS registry number, canonical SMILES, class taste and references to the scientific sources from where data were retrieved, as well as the molecular structure in the HyperChem (.hin) format. The overall aim of the ChemTastesDB is to provide a tool to the scientific community to increase the available information of taste molecules and to support the development of in silico approaches for taste prediction. The database is freely available at the following URL: https://doi.org/10.5281/zenodo.5747393.
2. Materials and methods
2.1. Data collection
Data was collected on 4580 molecular tastants from several scientific sources: 37 papers (including public databases), 3 books and 53 book chapters. Each molecule was associated with an experimental basic taste (sweet, bitter, umami, sour or salty) or other gustatory sensations; for instance, tasteless (neutral taste), non-sweet (lacking sweet), non-bitter, astringent, cooling, scratchy, burning, heating, pungent, and tingling. Initially, we adopted the following criteria for a preliminary screening of the collected data on the specific chemicals:
-
a.
protein tastants were not considered, for instance, miraculin, brazzein, curculin, pentadin, monellin (I and II), thaumatin (I, II, III, a, b and c) and mabinlin (I and II).
-
b.
water molecules were removed from the hydrated compounds, because the sensorial evaluation of the taste is performed by a sip-spit methodology using aqueous (or hydro-alcoholic) solutions (Bassoli et al., 2008, Kelly et al., 2005, Rojas et al., 2016).
-
c.
when dealing with the umami taste, we considered umami compounds, taste-modulating and umami enhancer molecules (Suess et al., 2015, Wong, 2018).
-
d.
the Haworth projection (Damodaran & Parkin, 2017) was used to represent the chemical structure of monosaccharides, such as fructose, glucose, psicose or tagatose.
2.2. Curation and optimization of molecular structures
The 3D molecular structures of 4580 tastants were manually represented using the HyperChem software (Hypercube Inc.). For geometry optimization, the molecular mechanics force field (MM+) and the conjugate gradient algorithm were used. The convergence criteria for geometry optimization was established when the root mean square deviation of the gradient vector was less than 0.01 kcal×(Å×mol)-1. The information of stereocenters was used in order to differentiate stereoisomers (when available). The information of stereochemistry, when not available, was obtained from the PubChem open library (Kim et al., 2019) and other scientific sources. Otherwise, the default structure generated by the model builder of HyperChem was used (no conformational analysis was performed).
Since chemical structures available in scientific papers, books (or chapters) and/or public and commercial databases are not exempt from errors, we performed a further molecular structure curation. The curation process of a query compound constitutes a crucial step during the development of a reliable database to be used in QSAR/QSPR modeling. Identification of errors in molecular structures includes, for example, missing atoms or functional groups, misplaced atoms or rearranged chemical groups. All of these potential errors can negatively influence the calculation of molecular descriptors, which may have deleterious effects on subsequent modelling (Fourches et al., 2010).
Thus, the accuracy of molecular structures was initially analyzed in PubChem (Kim et al., 2019) or other open libraries. Subsequently, the alvaMolecule software (Alvascience, 2020) was used for molecular curation to identify molecules with multiple structures, unusual valence, covalent/ionic bonds, total charge, isotopes, charged atoms, non-carbon atoms, non-standard atom sets (H, C, N, O, P, S, F, Cl, Br and I), no aromatic ring standardization and radical atoms. These issues were corrected applying some standard criteria as implemented in the software, such as standardization of benzene rings into aromatic form, conversion of unusual covalent bonds to ionic forms, addition of charge to quaternary nitrogen atom, removal or adding excessive or missing hydrogens, standardization of nitro, azide and diazo groups, and NOxide compatibility. Finally, the CAS registry number and the PubChem CID of each tastant was also obtained from the PubChem (when available) along with the search function implemented in alvaMolecule.
For 402 compounds, the name, PubChem CID and CAS registry number were automatically retrieved from the PubChem library by means of alvaMolecule. Additionally, the Marvin Sketch (ChemAxon Ltd., 2021) was used to generate the IUPAC name for 538 molecules, which were not found when applying the alvaMolecule similarity search.
2.3. Database merging and filtering
Data were further filtered to verify replicated compounds. Initially, the canonical SMILES (simplified molecular input line entry system) of the 4580 tastants were generated in alvaMolecule from the HyperChem 3D molecular representation. Subsequently, the chemical name (with the corresponding taste), CAS registry number, PubChem CID, canonical SMILES and scientific reference were merged with an in-house KNIME workflow (Berthold et al., 2008), which included the following filtering steps:
-
a.
molecules labelled as 3,5-dichlorophenyl guanidineacetic acid derivative, 4-cyanophenyl guanidineacetic acid derivative, compound, isovanillyl derivative, perillartine derivative and phenylsulfamate monosubstituted were excluded;
-
b.
molecules exhibiting the exact match of name, CAS number or PubChem CID were merged into a single entry;
-
c.
molecules excluded in step a) were considered together with the molecules processed in step b), and a new curation step was applied to find chemicals with the same molecular structure by comparing canonical SMILES. Stereoisomers (for instance D-glucose and L-glucose) were not considered in this step;
-
d.
molecules exhibiting multiple-valued tastes were assigned to the most frequent taste class with a majority voting approach. When multiple-valued tastes were tied, the tastant was included in the miscellaneous class;
-
e.
molecules with the following tastes were included in the miscellaneous class: astringent, cooling, hot burning, heating, pungent, and tingling. The same criterion was adopted to assign compounds labelled with an ambiguous class (bitter/burning/scratchy, bitter/tasteless, non-bitter, non-bitter/burning, non-sweet/sweet, sweet/bitter, sweet/tasteless).
2.4. Analysis of the chemical space of tastants
Chemical space (Medina-Franco et al., 2021) is a useful concept in diverse areas of computational chemistry including chemoinformatics and foodinformatics. The chemical space is defined by all chemicals represented by a N-dimensional vector of features (for instance MACCS) that captures the most relevant chemical information of compounds. Thus, this multidimensional space aims to conceptualize molecular similarities by identifying regions where molecules are clustered by their features. The most suitable way to visualize and analyze the chemical space is by the projection of similarities/dissimilarities into a low-dimensional space by means of diverse unsupervised machine learning approaches. Previous studies have analyzed the chemical spaces of taste molecules by applying Principal Component Analysis (PCA) (Dagan-Wiener et al., 2017, Di Pizio et al., 2019, Ruddigkeit and Reymond, 2014), Multidimensional Scaling (MDS) (Rojas et al., 2017), and the t-Distributed Stochastic Neighbor Embedding (t-SNE) (Bouysset et al., 2020, Tuwani et al., 2019).
In this study, structural characteristics of molecular tastants were represented by means of the Molecular ACCess System (MACCS) fingerprints (Durant et al., 2002). These are 2D binary fixed size fingerprints associated with a SMART pattern, which is a chemical language able to specify substructures that describe atomic and bond properties by means of well-defined rules based on simple extensions of the SMILES notation. Thus, each bit indicates the presence/absence of a particular molecular feature. These MDL structural keys are suitable fingerprints for substructure searching or molecular similarity. The alvaDesc software (Alvascience, 2021) was used to calculate the binary 166 MACCS fingerprints starting with the molecular SMILES.
The chemical space was defined through molecular similarity/diversity analysis based on the t-Distributed Stochastic Neighbor Embedding (t-SNE) (van der Maaten & Hinton, 2008), which attempts to project tastants fingerprints into a two-dimensional space (ℝN → ℝ2), in such a way as to preserve the local structure. To calculate the pairwise similarities in low-dimensional space, t-SNE uses a symmetrized version of the cost function with simpler gradients to facilitate the optimization process, as well as the heavy-tailed Student-t distribution to overcome the crowding problem. This unsupervised approach is able to match pairwise similarity distributions in both higher-dimensional space and lower-dimensional space to preserve the local structure of data. Consequently, t-SNE efficiently captures the local structure of the high-dimensional space, while eliciting the presence of clusters at several scales (structure of the data). The pairwise similarities were calculated by means of the Jaccard-Tanimoto similarity coefficient (Todeschini et al., 2015). This well-known binary similarity coefficient emphasizes the presence of common features omitting the absence of common features and the simple matching accounting for both presence and absence of common features.
2.5. Software and code
HyperChem version 8 was used for drawing and displaying chemical structure of molecular tastants, while the chemical structures were checked and curated in the alvaMolecule software. An in-house KNIME workflow was programmed for filtering the database. MACCS fingerprints were calculated in alvaDesc. MATLAB was used to calculate t-SNE models.
3. Results and discussion
3.1. ChemTastesDB description
The curated ChemTastesDB consisted of 2944 compounds grouped into nine classes, which include the five basic tastes (sweet, bitter, umami, sour and salty) and four additional classes (non-sweet, tasteless, multitaste and miscellaneous). Table 1 lists the number of molecules included in each class. The ChemTastesDB is freely available at https://doi.org/10.5281/zenodo.5747393, and includes four files:
-
1.
a pdf file (ChemTastesDB_readme.pdf) containing a complete description of the ChemTastesDB;
-
2.
an excel file (ChemTastesDB_database.xls), where the following data are collected for each tastant: molecular ID, name, PubChem CID, CAS registry number, canonical SMILES string, class taste and reference to the scientific sources from which data were retrieved;
-
3.
an excel file (ChemTastesDB_references.xls), containing a comprehensive list of all scientific references with their extended details;
-
4.
ChemTastesDB_molecules.zip file, which includes the Hyperchem file (.hin) of each compound optimized by the mechanics force field (MM+). Files are named using the molecular IDs of the ChemTastesDB_database excel file.
Table 1.
Number of molecular tastants included in the nine classes of the curated ChemTastesDB.
| Tastant class | Number of molecules |
|---|---|
| Sweetness | 977 |
| Bitterness | 1183 |
| Umaminess | 98 |
| Sourness | 38 |
| Saltiness | 12 |
| Non-sweetness | 233 |
| Tastelessness | 203 |
| Multitaste | 113 |
| Miscellaneous | 87 |
The database will be continuously updated by including new molecular tastants, when available. To the best of our knowledge, ChemTastesDB constitutes the most comprehensive curated database that provides support for decision-making to rationally design new tastants by means of quantitative structure–activity relationships and diverse supervised machine learning approaches.
3.2. Analysis of the chemical space
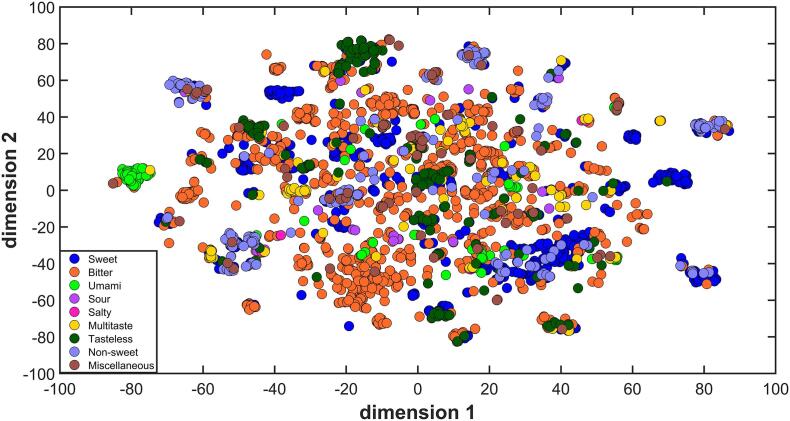
The 2944 molecules included in the ChemTastesDB were used to define the chemical space of tastants based on their structural similarity provided by the 166 MACCS structural keys. The intent of this analysis is a comprehensive characterization of the chemical features of tastants and an evaluation of how these molecules are structurally clustered. Since MACCS is a Boolean vector, molecular similarities/dissimilarities were quantified by means of the Jaccard-Tanimoto distance in the t-Distributed Stochastic Neighbor Embedding (t-SNE). We tested diverse values for the parameters to be set in t-SNE by using the following values for the Exaggeration = [2, 4, 50, 100], Perplexity = [20, 30, 40, 50] and Learning Rate = [100, 500, 900, 1300]. Results were visually inspected and the parameters that generated the t-SNE scatter plot with the best discrimination of the taste classes as well as the formation of consistent clusters inside each class were selected: Exaggeration = 100, Perplexity = 30 and Learning Rate = 100. Fig. 1 presents the chemical space defined by the t-SNE scores of the two coordinates. t-SNE generates interesting low-dimensional clusters of data that represents the distributions in the original multidimensional data space. The chemical space of tastants exhibits a high degree of overlap among the nine classes. However, it is possible to identify some interesting groups, particularly for the basic tastes. In order to thoroughly explore the nature of these clusters, we defined the chemical space in terms of a class/non-class scatter plot for the sweet, bitter, umami and sour classes.
Fig. 1.
Scatter plot of the t-SNE coordinates of tastants included in the ChemTastesDB, as obtained on MACCS structural keys. Molecules are colored based on their taste class.
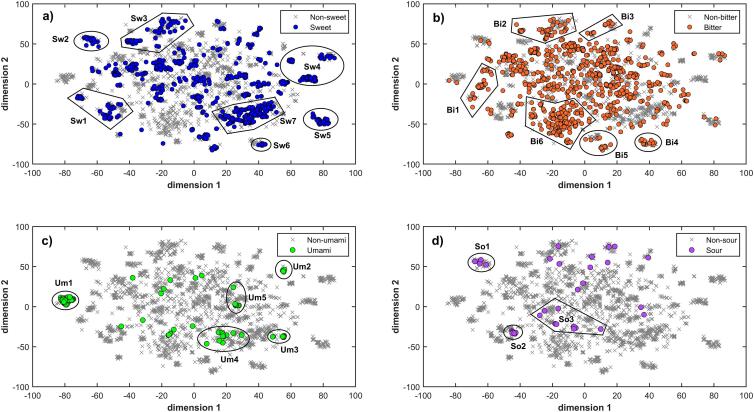
Fig. 2a shows the distribution of compounds from the Sweetness class (Sw) in the chemical space, where some groups with specific structural similarities can be identified. The sucrose standard and some of its derivatives are located in cluster Sw1. Other sweeteners located in this cluster are the D-lactulose, palatinose, raffinose, sedoheptulosan, stachyose, sodium cyclamate, calcium cyclamate chloro-nitroaniline and diverse derivatives of sodium sulfamate. On the other hand, cluster Sw2 includes 22 sodium sulfamate derivatives. The next cluster, Sw3, is formed by the hesperetin DHC, phloroglucinol, resorcinol, trans-anethole, trans-cinnamaldehyde sweeteners, as well as two dihydrochalcone derivatives, some isocoumarin derivatives and diverse guanidineacetic acid derivatives (for instance sucrononic acid). Cluster Sw4 includes three subgroups that comprise other guanidineacetic acid derivatives (for instance bernardame, carrelame and lugduname), acesulfame (and some of its analogues, such as acesulfame K, aspartame-acesulfame or 6-ethyl-acesulfame) and molecules with the phenylsulfonyl fragment in their scaffolds (for instance sulfone, ASA 1, ASA 3 and ASA 5). Another interesting cluster is Sw5, which includes 51 halogenated derivatives (mono-, di-, tri- and tetra- substituted) of both sucrose and galactosucrose, as well as sucralose and three analogues. The saccharin sweetener and ten of its derivatives (including sodium, potassium and calcium salts) are located in cluster Sw6. Cluster Sw7 contains diverse aspartic acid derivatives (for instance aspartame, advantame and neotame), as well as the guanidineacetic acid and two α-amino acids (D-asparagine and D-glutamine).
Fig. 2.
Scatter plot of the t-SNE coordinates. Molecules are colored on the basis of (a) Sweetness, (b) Bitterness, (c) Umaminess and (d) Sourness classes.
The class of Bitterant (Bi) compounds (Fig. 2b) has a great dispersion along the t-SNE scatter plot. However, some consistent clusters of bitterants can be identified. Cluster Bi1 includes diverse type of bitterants, such as butalbital, butethal, hexethal sodium, methyprylon, phenallymal, phenytoin sodium, piperidione, propallylonal. Other compounds located in this group are urea (and 3 derivatives), three sucrose derivatives, butallylonal (and its sodium salt), four thiouracil derivatives and barbital (with 20 analogues). Cluster Bi2 includes essentially the methylergonovine maleate, 13 lupone derivatives (dehydrotricycloadlupone, dehydrotricyclocolupone, dehydrotricyclolupone, hydroperoxytricycloadlupone, hydroxytricycloadlupone, hydroxytricyclocolupone, hydroxytricyclolupone, nortricycloadlupone, nortricyclocolupone, nortricyclolupone, tricycloadlupone, tricyclocolupone and tricyclolupone), as well as the benzaldehyde bitterant and compounds which include the benzaldehyde molecular fragment in their scaffold. Near to this group, cluster Bi3 includes 16 sodium salt sulfamate derivatives. On the contrary side of Bi2 and Bi3, cluster Bi4 comprises the bitterants camphotamide, glimepiride, sulfisoxazole and trimethaphan camsylate, as well as 19 bitter saccharin derivatives (for instance 5-methoxysaccharin, 5-nitrosaccharin, 6-nitrosaccharin, 7-nitrosaccharin and denatonium saccharide). Cluster Bi5 includes 15 bitterants, such as azathioprine, chloramphenicol, chrysamminic acid, m-nitrobenzene, nitrofurazone, picric acid (and ammonium picrate), ranitidine hydrochloride, 1-nitronaphthalene, 2-amino-5-nitrothiazole, 2-nitroaniline, 2-(cyclohexene-4-yl)-1,2-propanediol, 2,4-dinitro-propoxybenzene, 3-(2-(4-nitrophenyl)acetamido)propanoic acid and 3,4-dinitrobenzoic acid. Near to this group, cluster Bi6 includes quinine and its salts; for instance, hydrochloride, dihydrochloride and sulfate (bitterness standard). In this cluster, a large number of molecules was identified including 15 denatonium derivatives, several amino acid sequences (6 linear and 28 cyclic) and other compounds with high molecular similarity among them.
Umami compounds (Um) are highlighted in Fig. 2c. It is possible to identify five consistent clusters. The first one (Um1) is comprised of the majority of umami tastants (49 molecules), with salts (disodium, dipotassium and calcium) of guanylate, inosine, adenylate, adenosine, riboside and xanthosine. Cluster Um2 includes five umami compounds with the presence of amide groups in their scaffolds as a common characteristic. The umami standard, monosodium L-glutamate (MSG), is grouped in cluster Um3 together with three other glutamates (monopotassium glutamate, monoammonium glutamate and monosodium D,L-threo-β-hydroxy glutamate), two diglutamates (calcium diglutamate and magnesium diglutamate), two amino acid sequences (Thr-Glu and Glu-Asp-Glu), as well as the monosodium L-aspartate and the monosodium L-α-amino adipate. Near to this group, cluster Um4 includes 13 umami taste molecules: L-ibotenic acid, L-theanin, L-tricholomic acid (erythro-form), Asp-Glu-Ser, γ-L-glutamyl-L-(S-methyl) methionine, γ-L-glutamyl-L-cysteinyl-glycine, ethyl 4-((2-isopropyl-5-methylcyclohexyloxy)carbonyl)butanoate, N-(3-methoxy-4-hydroxy-benzyl)-5-hydroxypentanamide, N-2,4-dimethoxybenzyl-N-(2-pyridyl)ethyl oxalamide, N-phenethyl-4-hydroxypentanamide, as well as three N-(4-hydroxyphenethyl) derivatives (of the erythronamide, gluconamide and succinamide). On the other hand, cluster Um5 contains the 2-mercaptoinosine 5'-monophosphate and seven inosinate derivatives, which were divided into sodium salts (disodium 2-methoxy-5'-inosinate, disodium 2-methyl-5'-inosinate, disodium N1-methyl-5'-inosinate and disodium Ν1-methyl-2-methylthio-5'-inosinate) and calcium salts (calcium inosinate and calcium 2-allyloxy-5'-inosinate).
Fig. 2d shows the distribution of sourness tastants (So) in the chemical space. Cluster So1 consists of four sulfamate sodium salts, while cluster So2 contains 8 imidodisulfuric acid disodium salt derivatives. On the other hand, sour tastants found in foods (for instance, acetic acid, citric acid, lactic acid, malic acid, propionic acid, tartaric acid), as well as carbonic acid, formic acid, phosphoric acid and two sodium salts (sodium 3-(sulfonatoamino)benzene-1-sulfonate and sodium N-[4-(butan-2-yl)phenyl]sulfamate) are located in cluster So3.
The remaining sweet, bitter, umami and sour tastants are more scattered along the t-SNE chemical space and overlap with molecules of other classes. Sensory data is subject to a high degree of variation due to wide differences in human perception as measured by sensory panelists. Diverse factors can affect taste perceptions; for instance, presence of taste modifiers, differences in psychology, anatomy or receptor functionality, as well as the reception, transduction and neural processing of electrical impulse information. In fact, many compounds imprint a complex sensation of diverse tastes (basic and non-basic) (Damodaran and Parkin, 2017, Rojas et al., 2016b, Wong, 2018). From a chemical point of view, during the synthesis of new tastants, small variations in the scaffold could result in the loss of a specific taste. For instance, the sweetener saccharin became bitter when modified with a chloride or a methyl fragment in the meta position (overlapped by bitter tastants), and became tasteless when replacing the imino fragment by a methyl, ethyl, or bromoethyl radical (nearest to tasteless compounds) (Rojas et al., 2016b, Rojas et al., 2017).
To the best of our knowledge, only two published studies exist regarding the definition of the chemical space of tastants based on the t-SNE unsupervised learning approach. One was published in 2019 when defining the BitterSweet classifier (Tuwani et al., 2019). The chemical space developed for the curated molecules and random bioactive compounds (ChEBI) reveals the molecular diversity of bitter, sweet and tasteless molecules in comparison to random bioactive compounds. The chemical space also captures clusters in the general chemical domain by reflecting the molecular distribution of taste molecules taken from several bibliographic sources. The second case is based on 316 sweeteners from the SweetenersDB, 4796 molecules from the Super-Natural II and PhytoLab, and three experimentally tested compounds (namely arctiin, ginsenoside Rd and jujuboside A) (Bouysset et al., 2020). The 2D chemical space was developed in Python using the default parameters (Perplexity = 30, Exaggeration = 12, Learning Rate = 200 and 1000 iterations). This chemical domain reflects a negligible superposition of the natural compounds with sweet-tasting molecules, which suggest that a great portion of the natural chemical space remains for further explorations. In addition, it had been stated that the lignan chemical family constitutes a new chemical space for eliciting new sweet tastants through machine learning approaches.
4. Conclusions
In this work the authors present the ChemTastesDB, an open-access database of 2944 molecular tastants, which are grouped in nine classes, including the five basic tastes and four other categories. Curation of molecules and data filtering allowed the collection of information to cover a more complete chemical domain with respect to existing databases. This database constitutes a novel tool to increase the information of taste molecules and to assist in silico studies for the taste prediction of new compounds. The database is freely accessible at https://doi.org/10.5281/zenodo.5747393.
The chemical space of the molecules included in the database was explored and characterized by means of MACCS keys molecular fingerprints analyzed with unsupervised machine learning based on t-SNE. The analysis enabled the comprehensive characterization of the tastants chemical space by looking at similarities among chemicals and their derived clusters. This analysis constitutes a useful approach to visualize the similarities/dissimilarities of tastants in multidimensional space and allows a better understanding of the relationships between molecular structure and taste.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Wayne R. Hanson for his valuable revision of the manuscript and for providing some useful comments for improving the technical quality.
References
- Ahmed J., Preissner S., Dunkel M., Worth C.L., Eckert A., Preissner R. SuperSweet-a resource on natural and artificial sweetening agents. Nucleic Acids Research. 2011;39(Database):D377–D382. doi: 10.1093/nar/gkq917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvascience. (2020). alvaMolecule (software to view and prepare chemical datasets) (Version 1.0.4). https://www.alvascience.com.
- Alvascience. (2021). alvaDesc (software for molecular descriptors calculation) (Version 2.0.6). https://www.alvascience.com.
- Bai, G., Wu, T., Zhao, L., Wang, X., Li, S., & Ni, X. (2021). CBDPS 1.0: A Python GUI application for machine learning models to predict bitter-tasting children’s oral medicines. Chemical and Pharmaceutical Bulletin, 69, 989-994. 10.1248/cpb.c20-00866. [DOI] [PubMed]
- Baines D., Brown M. In: Encyclopedia of food and health. Caballero B., Finglas P.M., Toldrá F., editors. Academic Press; 2016. Flavor enhancers: Characteristics and uses; pp. 716–723. [Google Scholar]
- Banerjee P., Preissner R. BitterSweetForest: A random forest based binary classifier to predict bitterness and sweetness of chemical compounds. Frontiers in Chemistry. 2018;6 doi: 10.3389/fchem.2018.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassoli A., Laureati M., Borgonovo G., Morini G., Servant G., Pagliarini E. Isovanillic sweeteners: Sensory evaluation and in vitro assays with human sweet taste receptor. Chemosensory Perception. 2008;1(3):174–183. doi: 10.1007/s12078-008-9027-z. [DOI] [Google Scholar]
- Berthold M.R., Cebron N., Dill F., Gabriel T.R., Kötter T., Meinl T.…Wiswedel B. In: Data analysis, machine learning and applications. Preisach C., Burkhardt H., Schmidt-Thieme L., Decker R., editors. Springer; Berlin Heidelberg: 2008. KNIME: The konstanz information miner; pp. 319–326. [Google Scholar]
- Bouysset C., Belloir C., Antonczak S., Briand L., Fiorucci Sébastien. Novel scaffold of natural compound eliciting sweet taste revealed by machine learning. Food Chemistry. 2020;324:126864. doi: 10.1016/j.foodchem.2020.126864. [DOI] [PubMed] [Google Scholar]
- Chandrashekar, J., Hoon, M. A., Ryba, N. J. P., & Zuker, C. S. (2006). The receptors and cells for mammalian taste. Nature, 444, 288-294. 10.1038/nature05401. [DOI] [PubMed]
- Charoenkwan P., Yana J., Schaduangrat N., Nantasenamat C., Hasan M.M., Shoombuatong W. iBitter-SCM: Identification and characterization of bitter peptides using a scoring card method with propensity scores of dipeptides. Genomics. 2020;112(4):2813–2822. doi: 10.1016/j.ygeno.2020.03.019. [DOI] [PubMed] [Google Scholar]
- ChemAxon Ltd. (2021). MarvinSketch (Version 21.17.0). http://www.chemaxon.com.
- Dagan-Wiener, A., Nissim, I., Abu, N. B., Borgonovo, G., Bassoli, A., & Niv, M. Y. (2017). Bitter or not? BitterPredict, a tool for predicting taste from chemical structure. Scientific Reports, 7, Article 12074. 10.1038/s41598-017-12359-7. [DOI] [PMC free article] [PubMed]
- Dagan-Wiener A., Di Pizio A., Nissim I., Bahia M.S., Dubovski N., Margulis E., Niv M.Y. BitterDB: Taste ligands and receptors database in 2019. Nucleic Acids Research. 2019;47(D1):D1179–D1185. doi: 10.1093/nar/gky974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damodaran S., Parkin K.L. 5th ed. CRC Press; 2017. Fennema's food chemistry. [Google Scholar]
- Di Lorenzo P.M., Chen J.-Y., Rosen A.M., Roussin A.T. In: Encyclopedia of neuroscience. Binder M.D., Hirokawa N., Windhorst U., editors. Springer; 2009. Tastant; pp. 4014–4019. [Google Scholar]
- Di Pizio A., Shoshan-Galeczki Y.B., Hayes J.E., Niv M.Y. Bitter and sweet tasting molecules: It's complicated. Neuroscience Letters. 2019;700:56–63. doi: 10.1016/j.neulet.2018.04.027. [DOI] [PubMed] [Google Scholar]
- Durant J.L., Leland B.A., Henry D.R., Nourse J.G. Reoptimization of MDL keys for use in drug discovery. Journal of Chemical Information and Computer Science. 2002;42(6):1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- Fourches D., Muratov E., Tropsha A. Trust, but verify: On the importance of chemical structure curation in cheminformatics and QSAR modeling research. Journal of Chemical Information and Modeling. 2010;50(7):1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Shen, Q., Su, X., Ji, M., Liu, X., Chen, Y., Lu, S., Zhuang, H., & Zhang, J. (2016). BitterX: A tool for understanding bitter taste in humans. Scientific Reports, 6, Article 23450. 10.1038/srep23450. [DOI] [PMC free article] [PubMed]
- Hypercube Inc. HyperChem Professional (Version 8). http://www.hyper.com.
- Kelly D.P., Spillane W.J., Newell J. Development of structure-taste relationships for monosubstituted phenylsulfamate sweeteners using classification and regression tree (CART) analysis. Journal of Agriculture and Food Chemistry. 2005;53(17):6750–6758. doi: 10.1021/jf0507137. [DOI] [PubMed] [Google Scholar]
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., Shoemaker, B. A., Thiessen, P. A., & Yu, B. (2019). PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res., 47(D1), D1102-D1109. 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed]
- Ley, J., Reichelt, K., Obst, K., Krammer, G., & Engel, K.-H. (2012). Important tastants and new developments. In H. Jeleń (Ed.), Food Flavors. Chemical, sensory and technological properties (pp. 19-33). CRC Press.
- Medina-Franco J.L., Sánchez-Cruz N., López-López E., Díaz-Eufracio B.I. Progress on open chemoinformatic tools for expanding and exploring the chemical space. Journal of Computer-Aided Molecular Design. 2021 doi: 10.1007/s10822-021-00399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morini G., Bassoli A., Borgonovo G. Molecular modelling and models in the study of sweet and umami taste receptors. A review. Flavour and Fragrance Journal. 2011;26(4):254–259. doi: 10.1002/ffj.v26.410.1002/ffj.2054. [DOI] [Google Scholar]
- Rojas C., Duchowicz P.R., Pis Diez R., Tripaldi P. In: Chemometrics applications and research: QSAR in medicinal chemistry. Mercader A.G., Duchowicz P.R., Sivakumar P.M., editors. Apple Academic Press; 2016. Applications of quantitative structure-relative sweetness relationships in food chemistry; pp. 317–339. [Google Scholar]
- Rojas, C., Tripaldi, P., & Duchowicz, P. R. (2016b). A new QSPR study on relative sweetness. International Journal of Quantitative Structure-Property Relationships, 1(1), 78-92. 10.4018/IJQSPR.2016010104.
- Rojas, C., Ballabio, D., Consonni, V., Tripaldi, P., Mauri, A., & Todeschini, R. (2016c). Quantitative structure-activity relationships to predict sweet and non-sweet tastes. Theoretical Chemistry Accounts, 135, Article 66. 10.1007/s00214-016-1812-1.
- Rojas C., Todeschini R., Ballabio D., Mauri A., Consonni V., Tripaldi P., Grisoni F. A QSTR-based expert system to predict sweetness of molecules. Frontiers in Chemistry. 2017;5:Article 53. doi: 10.3389/fchem.2017.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddigkeit L., Reymond J.-L. In: Foodinformatics: Applications of chemical information to food chemistry. Martinez-Mayorga K., Medina-Franco J.L., editors. Springer; 2014. The chemical space of flavours; pp. 83–96. [Google Scholar]
- Ben Shoshan-Galeczki Y., Niv M.Y. Structure-based screening for discovery of sweet compounds. Food Chemistry. 2020;315:126286. doi: 10.1016/j.foodchem.2020.126286. [DOI] [PubMed] [Google Scholar]
- Suess B., Festring D., Hofmann T. In: Flavour development, analysis and perception in food and beverages. Parker J.K., Elmore J.S., Methven L., editors. Woodhead Publishing; 2015. Umami compounds and taste enhancers; pp. 331–351. [Google Scholar]
- Todeschini R., Ballabio D., Consonni V. In: Encyclopedia of analytical chemistry: Applications, theory and instrumentation. Meyers R.A., editor. JohnWiley & Sons Ltd; 2015. Distances and other dissimilarity measures in chemometrics; pp. 1–34. [Google Scholar]
- Tuwani, R., Wadhwa, S., & Bagler, G. (2019). BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules. Scientific Reports, 9, Article 7155. 10.1038/s41598-019-43664-y. [DOI] [PMC free article] [PubMed]
- van der Maaten L., Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- Wong D.W. 2nd ed. Springer; 2018. Mechanism and theory in food chemistry. [Google Scholar]
- Yang Z.-F., Xiao R., Xiong G.-L., Lin Q.-L., Liang Y., Zeng W.-B.…Cao D.-s. A novel multi-layer prediction approach for sweetness evaluation based on systematic machine learning modeling. Food Chemistry. 2022;372:131249. doi: 10.1016/j.foodchem.2021.131249. [DOI] [PubMed] [Google Scholar]
- Zheng S., Jiang M., Zhao C., Zhu R., Hu Z., Xu Y., Lin F. e-Bitter: Bitterant prediction by the consensus voting from the machine-learning methods. Frontiers in Chemistry. 2018;6:Article 82. doi: 10.3389/fchem.2018.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Chang W., Xu W., Xu Y., Lin F. e-Sweet: A machine-learning based platform for the prediction of sweetener and its relative sweetness. Frontiers in Chemistry. 2019;7:Article 35. doi: 10.3389/fchem.2019.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]