Abstract

Glycoproteomics, or the simultaneous characterization of glycans and their attached peptides, is increasingly being employed to generate catalogs of glycopeptides on a large scale. Nevertheless, quantitative glycoproteomics remains challenging even though isobaric tagging reagents such as tandem mass tags (TMT) are routinely used for quantitative proteomics. Here, we present a workflow that combines the enrichment or fractionation of TMT-labeled glycopeptides with size-exclusion chromatography (SEC) for an in-depth and quantitative analysis of the glycoproteome. We applied this workflow to study the cellular glycoproteome of an isogenic mammary epithelial cell system that recapitulated oncogenic mutations in the PIK3CA gene, which codes for the phosphatidylinositol-3-kinase catalytic subunit. As compared to the parental cells, cells with mutations in exon 9 (E545K) or exon 20 (H1047R) of the PIK3CA gene exhibited site-specific glycosylation alterations in 464 of the 1999 glycopeptides quantified. Our strategy led to the discovery of site-specific glycosylation changes in PIK3CA mutant cells in several important receptors, including cell adhesion proteins such as integrin β-6 and CD166. This study demonstrates that the SEC-based enrichment of glycopeptides is a simple and robust method with minimal sample processing that can easily be coupled with TMT-labeling for the global quantitation of glycopeptides.
Introduction
N-Linked glycosylation is the covalent attachment of carbohydrate moieties to a protein at asparagine residues. N-Linked glycan moieties are built on a core structure consisting of two N-acetyl glucosamine residues and three mannose residues. N-Glycans are mainly synthesized by the enzymes in the endoplasmic reticulum where the core structure is formed, and Golgi complex enzymes subsequently expand the core structure to various glycoforms, leading to the potential diversification of functions ranging from protein folding and molecular interactions to signaling.1 The dysregulation of glycosylation has been implicated in several diseases such as cancer,2,3 genetic disorders,4 and autoimmunity,5,6 with several clinically approved biomarkers being glycoproteins.7 Although the role of N-glycosylation as the driver of various disorders has long been recognized, a global and unbiased glycoproteomics analysis has remained challenging owing to the lack of well-developed analytical methods. This is further complicated by the microheterogeneity and macroheterogeneity of the glycoforms at the global level.
Mass spectrometry-based glycoproteomics has been a major analytical tool used for studying both glycosylation patterns and their quantitation across different experimental conditions. Recent improvements in the glycoproteomics workflows, including the enrichment of intact glycopeptides,8,9 and newer fragmentation methods such as stepped collision energy have expanded the coverage of intact glycopeptide analysis. However, the current quantitative workflows, which include the enrichment of intact glycopeptides followed by label-free quantitation, still suffer several issues in the analytical workflows. These include increased analysis time due to the inability to multiplex samples and the limited coverage and depth of the glycoproteomics analysis because fractionation is not commonly employed. The multiplexing of samples using isobaric tags such as iTRAQ or tandem mass tags (TMT) has been extensively applied for global proteomics and phosphoproteomics, thereby increasing the reproducibility and accuracy of the quantitation.10−12 Despite these advantages of TMT-based quantitation, it should be noted that because the samples are multiplexed the MS/MS spectra from individual samples cannot be evaluated separately.
Glycopeptides require enrichment for efficient ionization and subsequent analysis by liquid chromatography–tandem mass spectrometry (LC-MS/MS). Some popular choices for glycopeptide enrichment include lectin-affinity chromatography and hydrophilic-interaction chromatography (HILIC). Lectin-affinity chromatography is widely used for glycopeptide enrichment, followed by LC-MS/MS analysis.13−15 However, lectin-affinity chromatography is only useful when analyzing a glycoproteome with a narrow specificity, with the exact classes of glycopeptides that are enriched depending on the choice of the lectins used. In contrast, HILIC is broader in its glycopeptide specificity, although serine-, threonine-, and tyrosine-containing peptides are retained regardless of glycosylation. Hydrophobic neutral compounds are retained less on HILIC columns. Although TMT reagents are hydrophobic and further increase the net hydrophobicity of the labeled peptides,16 they have been used in conjunction with the HILIC-based enrichment of TMT-labeled glycopeptides.9,17−19 Glycopeptides with attached N-glycans that are generated by trypsin are generally larger than nonglycosylated tryptic peptides, making size-exclusion chromatography (SEC) an effective means of enrichment.20−22 Thus, we used SEC as an alternative method, which could simultaneously enrich and fractionate N-glycopeptides after TMT labeling of the peptides. We utilized this method to enrich and fractionate TMT-labeled N-glycopeptides from whole-cell lysates of mammary epithelial MCF10A cells in a single step. We first ascertained the feasibility and efficiency of enriching labeled glycopeptides by SEC and LC-MS/MS analysis using TMT labeling of whole-cell digests. Subsequently, we applied this novel workflow to study the effects of oncogenic mutations in the catalytic subunit of phosphoinositide 3-kinase (PI3-Kinase) on the glycopeptidome. PI3-Kinase is a family of lipid kinases that comprise of two subunits, an 85 kDa regulatory subunit and a 110 kDa catalytic subunit. Mutations in the regulatory subunit of PI3-Kinase have been implicated in the oncogenesis of several cancers, including cervical, head, neck, and ovarian cancers, among others.23−25
In summary, we have established SEC as a powerful method to simultaneously enrich and fractionate TMT-labeled glycopeptides for deep quantitative glycoproteomics. We identified and quantified 1999 unique N-glycopeptides from an isogenic cell system of MCF10A parental cells and corresponding mutant cells bearing mutations in either exon 9 (E545K) or exon 20 (H1047R) of the PIK3CA gene. We demonstrate that multiplexing the N-glycopeptide analysis using TMT labeling can be applied to study outstanding questions at the interface of cell signaling and protein glycosylation.
Results and discussion
TMT-Labeling of N-Glycopeptides for Quantitation
The major goal of our study was to establish a simplified workflow for the identification and multiplexed quantification of N-glycopeptides to address the role of glycoproteins in biological systems. We first selected a standard glycoprotein (transferrin) and spiked it into cell lysates from MCF10A cells to establish the TMT labeling of the glycopeptides and confirm that quantitation was not affected by background peptides. To test this, human serum transferrin was mixed in a 1:2:1:2:1:2 ratio to constant protein amounts of MCF10A cell lysate aliquots in six separate tubes. These six samples were trypsin-digested individually, labeled with six isotopically labeled versions of TMT 10-plex reagents, and pooled. The N-glycopeptides were enriched using SEC. No cleanup of the total peptides was performed prior to SEC, as we wanted to develop a simple method that could enrich and fractionate glycopeptides in as few steps as possible.
Quantitation of N-Glycopeptides
The enriched TMT-labeled glycopeptides were analyzed by LC-MS/MS on an Orbitrap Fusion Lumos Tribrid mass spectrometer as described under in the Methods section. The raw files were analyzed using the publicly available glycoproteomics software pGlyco (ver. 2.2.2), and the same files were processed by MaxQuant (ver. 1.6.7.0). The glycopeptides identified by pGlyco were matched on a scan-by-scan basis with the reporter ion intensities of the corresponding scans reported by MaxQuant (Figure 1). A total of 3322 glycopeptide peptide spectrum matches (PSMs) were identified and quantified. After removing redundancy, 1951 glycopeptides were quantified by this workflow. Glycopeptides belonging to transferrin were filtered manually, and the ratios of the two groups (1×:2×) were calculated. We identified and quantified 107 glycopeptides (Supplementary Table 1) belonging to transferrin, covering all four known glycosylation sites (Asn432 Asn523, Asn630, and Asn637). Asn432 was the most diverse in terms of glycan heterogeneity (Figure 2A). Asn432 and Asn630 contained consensus glycosylation motifs of NXS and NXT, respectively, where X can be any amino acid except proline. Asn523 and Asn637 belonged to a relatively rare motif of NXC. Ninety-one glycopeptides showed an average significant fold change of 1.85 (t-test p-value <0.05). Plotting the average reporter ion intensities of the 1× group versus those of the 2× group for each glycopeptide returned a R2 value of 0.99, barring one outlier, suggesting an excellent quantitation accuracy (Figure 2B). The identified transferrin glycopeptides were filtered for confidence and the quality of the MS/MS spectra by examining glycan oxonium ions and filtered glycan compositions at Asn432, Asn523, Asn630, and Asn637, which are shown in Figure 2C. This included asialo, monosialo, and disialo glycan compositions, which are used to screen patients suspected of having congenital disorders of glycosylation.26,27 Some advantages of using the SEC-based simultaneous enrichment and fractionation of N-glycopeptides are the reduced number of workflow steps and not needing a C18 cleanup of the peptides prior to fractionation. Using fewer steps reduces the sample-to-analysis time and avoids potential losses incurred in additional steps. These are unavoidable in current protocols, which use a combination of HILIC and basic pH reversed-phase liquid chromatography-based fractionation that involves a C18 cleanup after every step.
Figure 1.

Experimental workflow. The schematic depicts the strategy for the quantitative analysis of N-glycopeptides using TMT labeling. The individual cell lysates are harvested and subjected to in-solution trypsin digestion, followed by TMT labeling as indicated. The labeled peptides were pooled, and all the peptides from the harvested cell lysates were separated by size-exclusion chromatography (SEC). The data were subject to database searches for identification and quantification using pGlyco 2.0 and MaxQuant software, respectively.
Figure 2.
TMT-based quantitation of glycopeptides of transferrin spiked into MCF10A cells in defined ratios. (A) The number of all identified glycopeptides of transferrin at four glycosylation sites are indicated in the bar chart. Four glycosylation sites (Asn432, Asn523, Asn630, and Asn637) are plotted on the x-axis, with the corresponding consensus motif indicated in parentheses. Asn432 and Asn630 had consensus motifs of NXS and NXT, respectively, where X is any amino acid but proline. Asn523 and Asn637 contained the NXC motif, which is relatively rare in mammalian N-glycopeptides. The y-axis shows the number of identified glycopeptides. (B) A scatter plot with the average reporter ion intensity of all glycopeptides from three replicates of spiked-in transferrin from the 1× group (x-axis) and the 2× group (y-axis). A trendline was drawn, and R2 was shown on the plot (0.99). One glycopeptide with discordant values is represented by a red circle. (C) Filtered list of identified glycopeptides of transferrin at the four sites that have unique glycan compositions, where H = hexose, N = N-acetyl hexosamine, F = fucose, and S = N-acetylneuraminic acid.
TMT Labeling-Based Quantitative N-Glycoproteomics Reveals Site-Specific Changes in Glycosylation as an Effect of Oncogenic Mutations in Isogenic Cells
The multiplexed TMT-based analysis workflow for N-glycopeptides was next applied to study the effect of oncogenic mutations in either exon 9 (E545 K) or exon 20 (H1047R) of the PIK3CA gene in MCF10A cells by creating a three-group isogenic cell system.28 The cells were cultured as three independent replicates per group and processed as described in the Methods section in a nine-plex TMT experiment. We identified 3418 glycopeptide PSMs, corresponding to 1999 unique N-glycopeptides with a 1% false discovery rate (FDR) at the peptide, glycan, and glycopeptide levels (Supplementary Table 2). These 1999 glycopeptides belonged to 480 glycosylation sites of 242 proteins (Figure 3A). Of the glycopeptide PSMs, 63 belonged to 36 glycopeptides, with 17 peptide sequences that were shared between proteins. In total, 143 glycan compositions on 480 glycosylation sites were quantified. Significant heterogeneity was found on several glycosylation sites, including Asn365 of CD98 (SLC3A2), which exhibited the greatest diversity. Of all the glycopeptides identified, 68% were complex-type, 21% were high mannose, and 10% were hybrid. Additionally, 1% of the glycopeptides had glycosylation sites that were occupied by truncated glycans (Figure 3A). Sialylated glycans were 75% of the total glycopeptides identified; fucosylated glycans constituted 62% of the total, while 55% were both fucosylated and sialylated (Figure 3B). The median expression value of all complex-type glycopeptides combined as well as hybrid glycopeptides was higher in both mutant cell types than the parental MCF10A cells, whereas high mannose and truncated glycopeptides were lower in abundance. Quantitative values at the site-specific glycoform level were analyzed by separately comparing the parental cells with exon 9- and exon 20-mutated cells using permutation-based FDR-corrected paired t-tests. The log2 fold changes of exon 9/parental and exon 20/parental plotted against each other showed an excellent correlation (R2 = 0.84), with largely similar changes across the two mutated cell lines (Figure 3C). When glycopeptides that were significantly different in both comparisons (p < 0.05) were plotted against each other, the R2 value was found to be 0.93, suggesting that these mutations changed the cellular glycoproteome in a similar direction and with a similar magnitude (Figure 3D).
Figure 3.
Cellular glycoproteomics of MCF10A cells and isogenic cells with defined mutations in the PIK3CA gene. (A) All identified and quantified glycopeptides were classified according to glycan classes, with the left bar showing the distribution of complex, hybrid, high mannose, and truncated glycans. Glycans were manually drawn for each glycopeptide from their glycan compositions, and broad categories of complex type, hybrid, and high mannose were assigned. Glycan compositions containing either only the N-glycan core or fewer monosaccharides than a core were marked as truncated. To draw the right side of the bar chart, the total number of glycan compositions was considered as a 100% distribution of each composition within each categories. (B) The presence or absence of fucose or N-acetyl neuraminic acid was manually evaluated for each glycopeptide, and their distribution within each glycan category is shown. (C) Fold changes of the identified individual glycopeptides (exon 9/parental and exon 20/parental) were converted to log2 values and plotted against each other in a scatter plot. The R2 value is shown. (D) Only the significantly different glycopeptides were plotted as in panel C. (E) Principal component analysis of all identified glycopeptides was performed using Metaboanalyst 4.0. The three groups of cells are indicated in blue (MCF10A), green (exon 9), or red (exon 20). (F and G) The top 20 glycopeptides that were significantly different between the groups were used to make mirror plots. The left side of the central line shows the exon 9/parental log2 fold change, and the right side of the central line shows the exon 20/parental log2 fold change. The gene symbol, glycosylation site, and manually drawn order of attachment of the glycans are indicated in the plots. Panel F contains the 20 most upregulated glycopeptides in both exon 9/parental and exon 20/parental comparisons, while panel G contains the 20 most downregulated glycopeptides.
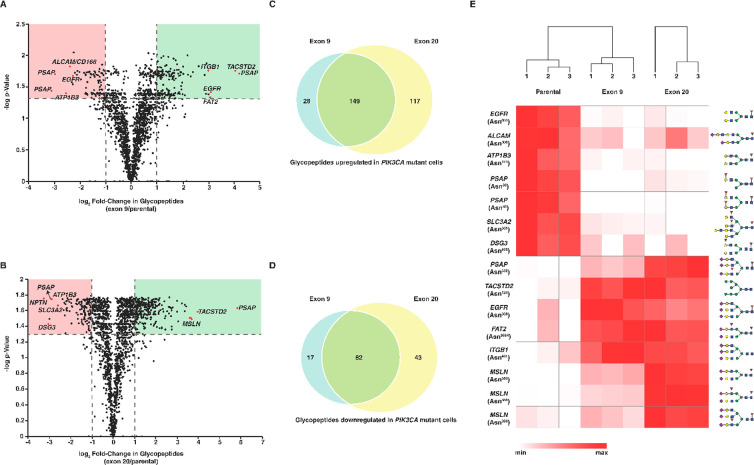
In exon 9/parental and exon 20/parental, 599 and 158 glycopeptides were found to be different, respectively (q < 0.05). Glycopeptides that were different in both comparisons were filtered further with a fold-change cutoff of 1.5 (up ≥1.5 and down ≤0.67). With this cutoff, 300 glycopeptides were found to be upregulated, and 164 were found to be downregulated. Of the upregulated glycopeptides, 300 belonged to 137 sites of 83 glycoproteins, and 164 of the downregulated glycopeptides belonged to 72 sites of 51 glycoproteins. Thirty-one glycosylation sites harbored both up and downregulated glycopeptides. All glycopeptides were analyzed with principal component analysis (PCA), which revealed that all three groups clustered away from each other (Figure 3E). However, parental cells were farther away from both exon 9 and exon 20 mutant cells, which fell in proximity in the variance space of principal components 1 and 2. The variance explained by the first or second component was 56% or 18%, respectively (Figure 3E). PCA separation further lends support to the suggestion that both mutations drive the glycoproteome remodeling in a similar fashion. Glycopeptides common to both the exon 9/parental cells comparison and the exon 20/parental cells comparison were sorted by the magnitude of their change in abundance upon mutations, and mirror plots were generated for both comparisons for both upregulated (Figure 3F) and downregulated (Figure 3G) glycopeptides. Among the most upregulated glycopeptides were the fucosylated and sialylated complex glycan at Asn332 of prosaposin, high-mannose glycans at Asn120 of tumor-associated calcium signal transducer 2 (TROP2), the sialylated complex glycan at Asn481 of integrin β-1, and a core fucosylated biantennary complex sialylated glycan at Asn161 of sortilin, among others (Figure 3F). TROP2 encodes a cell-surface glycoprotein that is highly expressed in carcinomas. This glycoprotein transduces calcium signaling in the cell and contains a phosphatidylinositol binding motif.29 The absence of fucose and sialic acids, which are both calcium-binding sugars,30,31 among the most increased glycopeptides of TROP2 would imply less calcium binding. The high expression of TROP2 is associated with poor prognosis for pancreatic, cervical cancer, and hilar cholangiocarcinoma.32−35 Among the upregulated glycopeptides, 8 of the 10 most-altered ones contained sialic acids, while in the set of downregulated glycoproteins only 3 of 10 had a sialic acid (Figure 3 F and G). This indication is supported by previously published studies, which showed the upregulation of sialic acid metabolism in highly metastatic breast cancer cells and upon the oncogenic transformation of mammary epithelial cells.36,37
A volcano plot was drawn for both exon 9/parental (Figure 4A) and exon 20/parental (Figure 4B) comparisons (Figure 4C and D). The relative abundance levels of the top 15 altered glycopeptides are shown in the heatmap (Figure 4E). The number of asialo complex-type glycans at Asn603 was reduced upon both mutations, while the number of sialylated complex-type glycans increased at Asn568 of epidermal growth factor receptor (EGFR). EGFR is a substrate of a few sialyltransferases, including ST6Gal-I, and its increased sialylation has been observed in breast cancer cells.38 Further, among the upregulated glycopeptides, 15 glycopeptides from four glycosylation sites of CD166 were upregulated in mutant cells. CD166 is a highly conserved glycoprotein from the immunoglobulin superfamily that is expressed in epithelial, immune cells, neuronal cells, and hematopoietic and mesenchymal stem cells. It is implicated in melanoma, prostate, and breast cancers39,40 and is also a candidate prognostic biomarker for cancer of the digestive system41 and pancreatic neuroendocrine cancer.42 CD166 is also overexpressed in prostate cancer cells and is known as a marker that can be used to enrich prostate stem or progenitor cells and cancer-initiating cells.43 Endocytosis of CD166 from the cell surface is driven by endophilin-A, which is independent of clathrin-mediated endocytosis. Instead, it is dependent on CD166 glycosylation and its ensuing interaction with extracellular galectin-8.44
Figure 4.
Glycoproteome remodeling in PIK3CA mutants. Volcano plot showing the fold change (log2 values) in the abundance of glycopeptides in (A) exon 9/parental or (B) exon 20/parental against p-values (−log) as indicated. The horizontal dashed line indicates p-values <0.05, and vertical lines indicate a fold-change cutoff of 2 either upregulated (green quadrant) or downregulated (red quadrant) in either comparison. Every red circle indicates a unique glycopeptide that was significantly different between exon 9 and parental cells or exon 20 and parental cells. All red circles are marked with the gene symbol of the glycoprotein from which the glycopeptide was derived. The glycopeptides that were (C) upregulated and (D) downregulated in the exon 9/parental comparison and the exon 20/parental comparison were compared with Venn diagrams. Glycopeptides common to both panel C and panel D were used to extract their abundance for the 15 most-changing glycopeptides, and a heatmap for the triplicate experiment was generated (E) The hierarchical clustering dendrogram is shown at the top. Gene symbols, glycosylation sites, and manually drawn order of attachment of glycans at the sites are depicted. The color gradient from high expression (max, red) to low expression (min, white) is indicated at the bottom of the heatmap.
We previously showed the fractionation of unlabeled glycopeptides using SEC,21 and this technique proved equally suitable for TMT-labeled glycopeptides, as shown in the current study. TMT-Labeled glycopeptides also elute in the earlier fractions of SEC, suggesting that TMT labeling does not affect the fractionation pattern of glycopeptides (Figure 5A). Further, it was apparent that TMT labeling did not affect the characteristic HCD fragmentation pattern of glycopeptides, as shown in annotated spectra for three representative glycopeptides (Figure 5 B–D). Two of the glycopeptides containing an atypical NXC motif with complex-type glycans (Figure 5 C and D) also produced the characteristic glycopeptide fragmentation pattern. As shown in Figure 5C and D, these glycopeptides produced dominant oxonium ions, several b- and y-ions, and glycan-containing Y ions, enabling the ladder sequencing of the glycopeptides. In a comparative glycoproteomic study analyzing glycoproteomic alterations, the differences in glycosylation could be attributed to protein abundance or true glycosylation changes. To establish if changes in the glycopeptides were due to glycosylation alterations or simply protein abundance, total proteomic analysis from an aliquot was also performed. Proteomics data were searched with the Sequest node using Proteome Discoverer 2.4. In total, 7168 proteins were identified and quantified, out of which 2036 were different between both exon 9- and exon 20-mutated cells (p < 0.05) compared to parental MCF10A cells. Exon 9- and exon 20-mutated cells showed coordinate global proteomic changes (R2 = 0.87) in a subset of proteins that were different from MCF10A cells. Another interesting observation was that glutamine-fructose-6-phosphate aminotransferase (GFPT2) was downregulated in both exon 9- and exon 20-mutated cells at the protein level (Supplementary Figure S1). Its upstream regulator, NFkB,45 was also found to be downregulated compared to parental cells, while inhibitor of nuclear factor kappa-B kinase subunit beta was found to be upregulated (Supplementary Figure S1). Glutamine-fructose-6-phosphate aminotransferase is a master regulator of N-glycan branching by virtue of being a first and rate-limiting enzyme of the hexosamine pathway, which controls the synthesis of urine diphosphate N-acetylglucosamine (UDP-GlcNAc).46,47 The branching of N-glycan controls galectin binding and regulates endocytosis and signaling by several cell surface receptors and transporters that affect cell growth and disease states, including cancer.48 Of the 500 glycopeptides described above as changing, 189 displayed significant changes despite the protein level being the same between the mutated cells and the controls. The proteins from which these 189 glycopeptides were derived were analyzed by STRING DB, and the interaction score was set to the highest confidence (0.9). A network comprising several integrins, LAMP1, LAMP2, CD63, and ERBB2, among others, was found to be enriched in these proteins (Supplementary Figure S2). A number of cell adhesion proteins change their glycosylation early in oncogenesis,49 and our results indicate that early events include the modulation of cell adhesion glycoproteins and specific changes in their glycosylation. To illustrate quantitation consistency across a range of proteins, bar plots were drawn for the relative abundance of glycosylation at all detected glycosylation sites of a few representative glycoproteins, including mesothelin, as shown in Supplementary Figures S3 and S4.
Figure 5.
Ultraviolet absorbance chromatogram of size-exclusion chromatography and representative MS/MS spectra of glycopeptides. (A) Chromatogram showing UV absorbance at the 214 nm wavelength. The y-axis represents the absorbance in milli-absorbance units, and the x-axis represents the time in minutes. Dashed lines depict the individual fractions that were collected and separately analyzed by LC-MS/MS. (B) Asn162 of sortilin contains a fucosylated sialylated complex-type glycan, and the fragmentation pattern of this glycopeptide in stepped HCD is shown. Asn158 of NPC intracellular cholesterol transporter 1 contains a sialylated complex-type glycan that was identified as both (C) fucosylated and (D) fucosylated species. The low mass region shows abundant glycan oxonium ions, which are marked according to the glycan units or monosaccharides that produce them; Hex = hexose, HexNAc = N-acetyl hexosamine, Neu5Ac = N-acetylneuraminic acid, and Fuc = fucose. b- and y-ions are fragmented peptide backbone ions, and Y represents ions that contain an intact peptide backbone attached to glycan fragments.
Conclusions
The enrichment of glycopeptides for multiplexed analysis is an important aspect of comprehensive glycoproteomics, which can be used to answer outstanding questions in biology and medicine. We have developed an alternative workflow that combines SEC-based enrichment and fractionation with TMT labeling of glycopeptides. This method is simpler due to having fewer steps from sample to analysis and is suitable for applications ranging from biomarker discovery to mechanistic studies probing the role of glycosylation. Finally, we applied this pipeline to demonstrate site-specific glycosylation changes induced by oncogenic mutations in the PIK3CA gene.
Methods
Cell Culture
MCF10A cells were purchased from the American Type Culture Collection. The PIK3CA mutant knockin cell lines (E545K and H1047R) were generated by gene targeting methods that were described previously.28 The cells were grown in DMEM/F12 (1:1) supplemented with 5% horse serum with growth factors in 5% CO2 at 37 °C as previously described.50
Sample Processing and Trypsin Digestion
Cell lysates were harvested by scraping the cells in modified radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1 mm EDTA; 1% Nonidet P-40; and 0.25% sodium deoxycholate). Protein concentration was determined by a bicinchoninic (BCA) assay. Two parallel experiments were done. In one, MCF10A cell lysates were spiked by human serotransferrin protein in a 1:2:1:2:1:2 ratio, and MCF10A cell lysate protein amounts were kept constant in all six aliquots. In the other experiment, MCF10A cell lysates and exon 9- and exon 20-mutated knockin cells lysates were used in equal amounts from three independent replicates, generating nine samples. Equal amounts of the total protein were precipitated by ice-cold acetone. The samples were then incubated at −20 °C for 2 h and centrifuged at 14 000 × g for 20 min. The protein pellet was dissolved in 100 μL of 8 M urea in 50 mM triethylammonium bicarbonate (TEAB), pH 8.5. To the sample was added 10 mM dithiothreitol (final concentration, Sigma), and the samples were incubated for 45 min at 37 °C. After cooling the samples to room temperature, 40 mM iodoacetamide (Sigma) was added (final concentration), and samples were incubated for 15 min in the dark. The samples were diluted with triethylammonium bicarbonate (TEAB) buffer, pH 8.5, to reduce the urea concentration to less than 1 M. To the samples was added sequencing-grade trypsin at a final ratio of 1:50 (trypsin/total protein). The digested peptides were labeled with TMT reagents as per the manufacturer’s instructions. In brief, peptides resuspended in 50 mM TEAB (pH 8.0) were mixed with TMT reagents, which were previously dissolved in anhydrous acetonitrile. After incubating for 1 h, the reaction was quenched by adding 1 M tris buffer, pH 8.5. All labeled samples were pooled and subjected to either SEC or basic pH reversed-phase fractionation.
Size-Exclusion Chromatography
Dried peptides from both experiments were resuspended in 100 μL of 0.1% formic acid by water bath sonication without cleanup. A Superdex Peptide 10/300 column (GE Healthcare) was equilibrated with 0.1% formic acid. Two separate isocratic runs were performed at a flow rate of 0.2 mL/min. Early fractions were collected starting at 10 min after injection (total run time of 130 min) and were analyzed by LC-MS/MS as described in the following sections.
Basic pH Reversed-Phase Liquid Chromatography
An aliquot of the total peptides was fractionated by basic pH reversed-phase liquid chromatography (bRPLC) on a reversed-phase C18 column (4.6 × 100 mm column) using an Ultimate 3000 UHPLC System. Solvents A and B were comprised of 5 mM ammonium formate, pH 9, and 5 mM ammonium formate, pH 9, in 90% acetonitrile, respectively. The 96 fractions collected for a total run time of 120 min were concatenated into 12 fractions. These concatenated 12 fractions were dried down in a Speed Vacuum system and resuspended in 0.1% formic acid for LC-MS/MS analysis.
Liquid Chromatography–Tandem Mass Spectrometry
Standard LC-MS/MS parameters were published previously51 and have been specifically modified for the current study as described below. Based on the UV profile (214 and 220 nm) of the earliest-eluting peptides, 21 early fractions from SEC were selected, and 12 fractions from bRPLC were individually analyzed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Liquid chromatography for peptide separation was performed on an Ultimate 3000 system (Thermo Fisher Scientific). An EASY-Spray column (75 μm × 50 cm, PepMap RSCL C18, Thermo Fisher Scientific) packed with 2 μm C18 particles was used as a separating device at 50 °C. Solvents A and B were comprised of 0.1% formic acid in water and 0.1% formic acid in acetonitrile, respectively. Injected peptides were trapped on a trap column (100 mm × 2 cm, Acclaim PepMap100 Nano-Trap, Thermo Fisher Scientific) at a flow rate of 20 mL/min. Every run was 130 min long, and the flow rate used was 300 nL/min. The gradient used for separation was as follows: equilibration at 3% solvent B from 0 to 4 min, 3–10% solvent B from 4 to 10 min, 10–35% solvent B from 10.1 to 125 min, and 35–80% solvent B from 125 to 145 min, followed by equilibration for next run at 5% solvent B for 5 min. Ionization of the eluting peptides was performed using an EASY-Spray source kept at an electric potential of 2 kV. All experiments were done in the data-dependent acquisition mode, with the top 15 ions isolated at a window of 0.7 m/z and a default charge state of +2. Only precursors with charge states ranging from +2 to +7 were considered for MS/MS events. A stepped collision energy was applied to the fragment precursors at normalized collision energies of 15%, 25%, and 40%. The MS precursor mass range was set from 375 to 2000 m/z, and that for MS/MS was set from 100 to 2000 m/z. Automatic gain control (AGC) for MS and MS/MS was at 106 and 1 × 105, and the injection time to reach AGC were 50 and 250 ms, respectively. The exclude isotopes feature was set to “ON”, and 60 s of dynamic exclusion was applied. Data acquisition was performed with the lock mass option (445.1200025 m/z) for all data.
Database Searching and Analysis
Database searching was performed using the publicly available software pGlyco ver. 2.2.0.52,53 An in-built glycan database containing 8092 entries, available with the software, was used, and Uniprot Human Reviewed protein sequences (20 432 entries) were used as a protein sequence file. Cleavage specificity was set to fully tryptic with three missed cleavages. Precursor and fragment tolerances were set to 5 and 20 ppm, respectively. Cysteine carbamidomethylation was set as a fixed modification, and the oxidation of methionine and protein N-terminal acetylation were set as variable modifications. The results were considered at 1% FDR at the peptide, glycan, and glycopeptide levels. Reporter ion quantification was performed in MaxQuant, and identities were matched with quantitation on a scan-to-scan basis (MS/MS). Briefly, the desired TMT channels (TMT 10-plex) were specified as isobaric labels with a reporter mass tolerance of 0.0003 Da. Manufacturer-supplied correction factors were specified. The msmscans table output by MaxQuant was used to extract scan-by-scan impurity-corrected reporter ion intensity values. These intensity values were matched to the scan numbers of identifications from pGlyco to match the identification and quantitation. Glycopeptide PSMs were combined to reflect the unique glycopeptides per search, and reporter ion intensities were summed up. Individual spectra were manually verified for glycan oxonium ions and quality. Additionally, all sialic acid containing glycopeptide spectra were verified for the presence of sialic acid and specific glycan oxonium ions (274.09, 292.1, and 657.23). The spectra of core-fucosylated glycopeptides were checked for at least one peptide+HexNAc+Fuc ion. The proteomics data set was searched using Sequest in Proteome Discoverer 2.4.
Acknowledgments
This work was supported by a DBT/Wellcome Trust India Alliance Margdarshi Fellowship Grant (IA/M/15/1/502023) awarded to A.P.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c06970.
Heatmap of experssion proteins in the NFkB pathway, protein–protein interaction network of altered glycoproteins, bar plots of the relative abundance of three glycopeptides derived from mesothelin, and relative abundance of three different glycoproteins at one glycosylation site each (PDF)
Lists of all quantified transferrin (spiked-in standard into MCF10A cell lysate) glycopeptides and all quantified glycopeptides from the comparative analysis of MCF10A, exon9, and exon20 PIK3CA mutant cells (XLSX)
Author Contributions
M.S.: conceptualization, investigation, methodology, formal analysis, writing the original draft, review, editing, and supervision. K.K.M.: investigation, methodology, review, and editing. K.G.: investigation, methodology, review, editing, and visualization. A.P.: conceptualization, formal analysis, funding acquisition, project administration, resources, supervision, review, and editing.
The authors declare no competing financial interest.
Notes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE54 partner repository with the data set identifier PXD025901 under project name “TMT-based multiplexed quantitation of N-glycopeptides reveals glycoproteome remodeling induced by oncogenic mutations”.
Supplementary Material
References
- Helenius A.; Aebi M. Intracellular functions of N-linked glycans. Science 2001, 291 (5512), 2364–9. 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Peixoto A.; Relvas-Santos M.; Azevedo R.; Santos L. L.; Ferreira J. A. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front Oncol 2019, 9, 380. 10.3389/fonc.2019.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S.; Catarino T. A.; Dias A. M.; Kato M.; Almeida A.; Hessling B.; Figueiredo J.; Gartner F.; Sanches J. M.; Ruppert T.; Miyoshi E.; Pierce M.; Carneiro F.; Kolarich D.; Seruca R.; Yamaguchi Y.; Taniguchi N.; Reis C. A.; Pinho S. S. Preventing E-cadherin aberrant N-glycosylation at Asn-554 improves its critical function in gastric cancer. Oncogene 2016, 35 (13), 1619–31. 10.1038/onc.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng B. G.; Freeze H. H. Perspectives on Glycosylation and Its Congenital Disorders. Trends Genet 2018, 34 (6), 466–476. 10.1016/j.tig.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui D.; Sellakumar G.; Green R.; Sutton-Smith M.; McQuistan T.; Marek K.; Morris H.; Dell A.; Marth J. Genetic remodeling of protein glycosylation in vivo induces autoimmune disease. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (3), 1142–7. 10.1073/pnas.98.3.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmi Y.; Ise W.; Harazono A.; Takakura D.; Fukuyama H.; Baba Y.; Narazaki M.; Shoda H.; Takahashi N.; Ohkawa Y.; Ji S.; Sugiyama F.; Fujio K.; Kumanogoh A.; Yamamoto K.; Kawasaki N.; Kurosaki T.; Takahashi Y.; Furukawa K. Sialylation converts arthritogenic IgG into inhibitors of collagen-induced arthritis. Nat. Commun. 2016, 7, 11205. 10.1038/ncomms11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan A.; Utratna M.; O’Dwyer M. E.; Joshi L.; Kilcoyne M. Glycosylation-Based Serum Biomarkers for Cancer Diagnostics and Prognostics. Biomed Res. Int. 2015, 2015, 490531. 10.1155/2015/490531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagglund P.; Bunkenborg J.; Elortza F.; Jensen O. N.; Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 2004, 3 (3), 556–66. 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- Sun S.; Shah P.; Eshghi S. T.; Yang W.; Trikannad N.; Yang S.; Chen L.; Aiyetan P.; Hoti N.; Zhang Z.; Chan D. W.; Zhang H. Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides. Nat. Biotechnol. 2016, 34 (1), 84–8. 10.1038/nbt.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe G.; Mangalaparthi K. K.; Jain A.; Darrow J.; Troncoso J.; Albert M.; Moghekar A.; Pandey A. Multiplexed Phosphoproteomic Study of Brain in Patients with Alzheimer’s Disease and Age-Matched Cognitively Healthy Controls. OMICS 2020, 24 (4), 216–227. 10.1089/omi.2019.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe G.; Na C. H.; Renuse S.; Madugundu A. K.; Albert M.; Moghekar A.; Pandey A. Quantitative Proteomic Profiling of Cerebrospinal Fluid to Identify Candidate Biomarkers for Alzheimer’s Disease. Proteomics Clin Appl. 2019, 13 (4), 1800105 10.1002/prca.201800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P.; Tang L. C.; Krug K.; Clark D. J.; Gritsenko M. A.; Chen L.; Clauser K. R.; Clauss T. R.; Shah P.; Gillette M. A.; Petyuk V. A.; Thomas S. N.; Mani D. R.; Mundt F.; Moore R. J.; Hu Y.; Zhao R.; Schnaubelt M.; Keshishian H.; Monroe M. E.; Zhang Z.; Udeshi N. D.; Mani D.; Davies S. R.; Townsend R. R.; Chan D. W.; Smith R. D.; Zhang H.; Liu T.; Carr S. A. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc 2018, 13 (7), 1632–1661. 10.1038/s41596-018-0006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E.; Zhu R.; Hammoud Z. T.; Mechref Y. LC-MS/MS quantitation of esophagus disease blood serum glycoproteins by enrichment with hydrazide chemistry and lectin affinity chromatography. J. Proteome Res. 2014, 13 (11), 4808–20. 10.1021/pr500570m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley N. M.; Hebert A. S.; Westphall M. S.; Coon J. J. Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nat. Commun. 2019, 10 (1), 1311. 10.1038/s41467-019-09222-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakhrushev S. Y.; Steentoft C.; Vester-Christensen M. B.; Bennett E. P.; Clausen H.; Levery S. B. Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol. Cell Proteomics 2013, 12 (4), 932–44. 10.1074/mcp.O112.021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogrebe A.; von Stechow L.; Bekker-Jensen D. B.; Weinert B. T.; Kelstrup C. D.; Olsen J. V. Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat. Commun. 2018, 9 (1), 1045. 10.1038/s41467-018-03309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.; Shah P.; Hu Y.; Toghi Eshghi S.; Sun S.; Liu Y.; Zhang H. Comparison of Enrichment Methods for Intact N- and O-Linked Glycopeptides Using Strong Anion Exchange and Hydrophilic Interaction Liquid Chromatography. Anal. Chem. 2017, 89 (21), 11193–11197. 10.1021/acs.analchem.7b03641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlmann J.; Taubenschmid J.; Wenzel D.; Gattinger A.; Durnberger G.; Dusberger F.; Elling U.; Mach L.; Mechtler K.; Penninger J. M. Comparative glycoproteomics of stem cells identifies new players in ricin toxicity. Nature 2017, 549 (7673), 538–542. 10.1038/nature24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P.; Ji Y.; Silbern I.; Doebele C.; Ninov M.; Lenz C.; Oellerich T.; Pan K. T.; Urlaub H. A streamlined pipeline for multiplexed quantitative site-specific N-glycoproteomics. Nat. Commun. 2020, 11 (1), 5268. 10.1038/s41467-020-19052-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Manilla G.; Atwood J. 3rd; Guo Y.; Warren N. L.; Orlando R.; Pierce M. Tools for glycoproteomic analysis: size exclusion chromatography facilitates identification of tryptic glycopeptides with N-linked glycosylation sites. J. Proteome Res. 2006, 5 (3), 701–8. 10.1021/pr050275j. [DOI] [PubMed] [Google Scholar]
- Saraswat M.; Garapati K.; Mun D. G.; Pandey A. Extensive heterogeneity of glycopeptides in plasma revealed by deep glycoproteomic analysis using size-exclusion chromatography. Mol. Omics 2021, 17, 939. 10.1039/D1MO00132A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswat M.; Joenvaara S.; Musante L.; Peltoniemi H.; Holthofer H.; Renkonen R. N-linked (N-) glycoproteomics of urinary exosomes. [Corrected]. Mol. Cell Proteomics 2015, 14 (2), 263–76. 10.1074/mcp.M114.040345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.; Bader A. G.; Zhao L.; Vogt P. K. Mutated PI 3-kinases: Cancer targets on a silver platter. Cell Cycle 2005, 4 (4), 571–574. 10.4161/cc.4.4.1593. [DOI] [PubMed] [Google Scholar]
- Zhao L.; Vogt P. K. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (7), 2652–7. 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Roberts T. M. Human tumor mutants in the p110alpha subunit of PI3K. Cell Cycle 2006, 5 (7), 675–7. 10.4161/cc.5.7.2605. [DOI] [PubMed] [Google Scholar]
- Jaeken J.; van Eijk H. G.; van der Heul C.; Corbeel L.; Eeckels R.; Eggermont E. Sialic acid-deficient serum and cerebrospinal fluid transferrin in a newly recognized genetic syndrome. Clin. Chim. Acta 1984, 144 (2–3), 245–7. 10.1016/0009-8981(84)90059-7. [DOI] [PubMed] [Google Scholar]
- Lefeber D. J.; Morava E.; Jaeken J. How to find and diagnose a CDG due to defective N-glycosylation. J. Inherit Metab Dis 2011, 34 (4), 849–52. 10.1007/s10545-011-9370-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin J. P.; Karakas B.; Weiss M. B.; Abukhdeir A. M.; Lauring J.; Garay J. P.; Cosgrove D.; Tamaki A.; Konishi H.; Konishi Y.; Mohseni M.; Wang G.; Rosen D. M.; Denmeade S. R.; Higgins M. J.; Vitolo M. I.; Bachman K. E.; Park B. H. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (8), 2835–40. 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapani E.; Sacchetti A.; Corda D.; Alberti S. Human Trop-2 is a tumor-associated calcium signal transducer. Int. J. Cancer 1998, 76 (5), 671–676. . [DOI] [PubMed] [Google Scholar]
- Jaques L. W.; Brown E. B.; Barrett J. M.; Brey W. S. Jr.; Weltner W. Jr. Sialic acid. A calcium-binding carbohydrate. J. Biol. Chem. 1977, 252 (13), 4533–4538. 10.1016/S0021-9258(17)40194-3. [DOI] [PubMed] [Google Scholar]
- Cook W. J.; Bugg C. E. Calcium-carbohydrate bridges composed of uncharged sugars. Structure of a hydrated calcium bromide complex of alpha-fucose. Biochim. Biophys. Acta 1975, 389 (3), 428–35. 10.1016/0005-2736(75)90153-4. [DOI] [PubMed] [Google Scholar]
- Fong D.; Moser P.; Krammel C.; Gostner J. M.; Margreiter R.; Mitterer M.; Gastl G.; Spizzo G. High expression of TROP2 correlates with poor prognosis in pancreatic cancer. Br. J. Cancer 2008, 99 (8), 1290–5. 10.1038/sj.bjc.6604677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning S.; Guo S.; Xie J.; Xu Y.; Lu X.; Chen Y. TROP2 correlates with microvessel density and poor prognosis in hilar cholangiocarcinoma. J. Gastrointest Surg 2013, 17 (2), 360–8. 10.1007/s11605-012-2105-1. [DOI] [PubMed] [Google Scholar]
- Liu T.; Liu Y.; Bao X.; Tian J.; Liu Y.; Yang X. Overexpression of TROP2 predicts poor prognosis of patients with cervical cancer and promotes the proliferation and invasion of cervical cancer cells by regulating ERK signaling pathway. PLoS One 2013, 8 (9), e75864 10.1371/journal.pone.0075864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong D.; Spizzo G.; Gostner J. M.; Gastl G.; Moser P.; Krammel C.; Gerhard S.; Rasse M.; Laimer K. TROP2: a novel prognostic marker in squamous cell carcinoma of the oral cavity. Mod. Pathol 2008, 21 (2), 186–91. 10.1038/modpathol.3801001. [DOI] [PubMed] [Google Scholar]
- Teoh S. T.; Ogrodzinski M. P.; Ross C.; Hunter K. W.; Lunt S. Y. Sialic Acid Metabolism: A Key Player in Breast Cancer Metastasis Revealed by Metabolomics. Front Oncol 2018, 8, 174. 10.3389/fonc.2018.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnz R. A.; Roberts L. S.; DeTomaso D.; Bideyan L.; Yan P.; Bandyopadhyay S.; Goga A.; Yosef N.; Nomura D. K. Protein Sialylation Regulates a Gene Expression Signature that Promotes Breast Cancer Cell Pathogenicity. ACS Chem. Biol. 2016, 11 (8), 2131–9. 10.1021/acschembio.6b00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbar C.; Mascaux C.; Merrouche Y.; Bensussan A. Triple-negative and HER2-overexpressing breast cancer cell sialylation impacts tumor microenvironment T-lymphocyte subset recruitment: a possible mechanism of tumor escape. Cancer Manag Res. 2018, 10, 1051–1059. 10.2147/CMAR.S162932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihnen M.; Kilic E.; Kohler N.; Loning T.; Witzel I.; Hagel C.; Holler S.; Kersten J. F.; Muller V.; Janicke F.; Milde-Langosch K. Protein expression analysis of ALCAM and CEACAM6 in breast cancer metastases reveals significantly increased ALCAM expression in metastases of the skin. J. Clin Pathol 2011, 64 (2), 146–52. 10.1136/jcp.2010.082602. [DOI] [PubMed] [Google Scholar]
- Hein S.; Muller V.; Kohler N.; Wikman H.; Krenkel S.; Streichert T.; Schweizer M.; Riethdorf S.; Assmann V.; Ihnen M.; Beck K.; Issa R.; Janicke F.; Pantel K.; Milde-Langosch K. Biologic role of activated leukocyte cell adhesion molecule overexpression in breast cancer cell lines and clinical tumor tissue. Breast Cancer Res. Treat 2011, 129 (2), 347–60. 10.1007/s10549-010-1219-y. [DOI] [PubMed] [Google Scholar]
- Ni C.; Zhang Z.; Zhu X.; Liu Y.; Qu D.; Wu P.; Huang J.; Xu A. X. Prognostic value of CD166 expression in cancers of the digestive system: a systematic review and meta-analysis. PLoS One 2013, 8 (8), e70958 10.1371/journal.pone.0070958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachezy M.; Zander H.; Marx A. H.; Gebauer F.; Rawnaq T.; Kaifi J. T.; Sauter G.; Izbicki J. R.; Bockhorn M. ALCAM (CD166) expression as novel prognostic biomarker for pancreatic neuroendocrine tumor patients. J. Surg Res. 2011, 170 (2), 226–32. 10.1016/j.jss.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Jiao J.; Hindoyan A.; Wang S.; Tran L. M.; Goldstein A. S.; Lawson D.; Chen D.; Li Y.; Guo C.; Zhang B.; Fazli L.; Gleave M.; Witte O. N.; Garraway I. P.; Wu H. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One 2012, 7 (8), e42564 10.1371/journal.pone.0042564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard H. F.; Tyckaert F.; Lo Giudice C.; Hirsch T.; Valades-Cruz C. A.; Lemaigre C.; Shafaq-Zadah M.; Wunder C.; Wattiez R.; Johannes L.; van der Bruggen P.; Alsteens D.; Morsomme P. Endophilin-A3 and Galectin-8 control the clathrin-independent endocytosis of CD166. Nat. Commun. 2020, 11 (1), 1457. 10.1038/s41467-020-15303-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymura S. J.; Zaemes J. P.; Allison D. F.; Clift S. H.; D’Innocenzi J. M.; Gray L. G.; McKenna B. D.; Morris B. B.; Bekiranov S.; LeGallo R. D.; Jones D. R.; Mayo M. W. NF-κB upregulates glutamine-fructose-6-phosphate transaminase 2 to promote migration in non-small cell lung cancer. Cell Commun. Signal 2019, 17 (1), 24. 10.1186/s12964-019-0335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mkhikian H.; Mortales C. L.; Zhou R. W.; Khachikyan K.; Wu G.; Haslam S. M.; Kavarian P.; Dell A.; Demetriou M. Golgi self-correction generates bioequivalent glycans to preserve cellular homeostasis. eLife 2016, 5, e14814. 10.7554/eLife.14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo L.; Khim P.; Mkhikian H.; Mortales C. L.; Demetriou M. Glycolysis and glutaminolysis cooperatively control T cell function by limiting metabolite supply to N-glycosylation. Elife 2017, 6, e21330. 10.7554/eLife.21330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granovsky M.; Fata J.; Pawling J.; Muller W. J.; Khokha R.; Dennis J. W. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat. Med. 2000, 6 (3), 306–12. 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- Li D. M.; Feng Y. M. Signaling mechanism of cell adhesion molecules in breast cancer metastasis: potential therapeutic targets. Breast Cancer Res. Treat 2011, 128 (1), 7–21. 10.1007/s10549-011-1499-x. [DOI] [PubMed] [Google Scholar]
- Wu X.; Renuse S.; Sahasrabuddhe N. A.; Zahari M. S.; Chaerkady R.; Kim M. S.; Nirujogi R. S.; Mohseni M.; Kumar P.; Raju R.; Zhong J.; Yang J.; Neiswinger J.; Jeong J. S.; Newman R.; Powers M. A.; Somani B. L.; Gabrielson E.; Sukumar S.; Stearns V.; Qian J.; Zhu H.; Vogelstein B.; Park B. H.; Pandey A. Activation of diverse signalling pathways by oncogenic PIK3CA mutations. Nat. Commun. 2014, 5, 4961. 10.1038/ncomms5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun D. G.; Renuse S.; Saraswat M.; Madugundu A.; Udainiya S.; Kim H.; Park S. R.; Zhao H.; Nirujogi R. S.; Na C. H.; Kannan N.; Yates J. R. 3rd; Lee S. W.; Pandey A. PASS-DIA: A Data-Independent Acquisition Approach for Discovery Studies. Anal. Chem. 2020, 92 (21), 14466–14475. 10.1021/acs.analchem.0c02513. [DOI] [PubMed] [Google Scholar]
- Zeng W. F.; Liu M. Q.; Zhang Y.; Wu J. Q.; Fang P.; Peng C.; Nie A.; Yan G.; Cao W.; Liu C.; Chi H.; Sun R. X.; Wong C. C.; He S. M.; Yang P. pGlyco: A pipeline for the identification of intact N-glycopeptides by using HCD- and CID-MS/MS and MS3. Sci. Rep 2016, 6, 25102. 10.1038/srep25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. Q.; Zeng W. F.; Fang P.; Cao W. Q.; Liu C.; Yan G. Q.; Zhang Y.; Peng C.; Wu J. Q.; Zhang X. J.; Tu H. J.; Chi H.; Sun R. X.; Cao Y.; Dong M. Q.; Jiang B. Y.; Huang J. M.; Shen H. L.; Wong C. C. L.; He S. M.; Yang P. Y. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 2017, 8 (1), 438. 10.1038/s41467-017-00535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y.; Csordas A.; Bai J.; Bernal-Llinares M.; Hewapathirana S.; Kundu D. J.; Inuganti A.; Griss J.; Mayer G.; Eisenacher M.; Perez E.; Uszkoreit J.; Pfeuffer J.; Sachsenberg T.; Yilmaz S.; Tiwary S.; Cox J.; Audain E.; Walzer M.; Jarnuczak A. F.; Ternent T.; Brazma A.; Vizcaino J. A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47 (D1), D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.