Highlights
-
•
14 Carotenoids were detected and quantified in three Zucchini fruits by LC-MS/MS.
-
•
Plenty Lutein and less Chlorophyll were identified in yellow and orange fruits.
-
•
A total of 664 DEGs were identified by transcriptome analysis.
-
•
Hub DEGs related to Chlorophyll and Carotenoids were associated by WGCNA.
-
•
A model of gene regulatory network was proposed for Zucchini peel coloration.
Keywords: Zucchini, Peel colouration, Chlorophyll, Carotenoids, Transcriptome, WGCNA
Abstract
To reveal the molecular mechanism underlying peel colouration, carotenoid metabolites and the transcriptome were jointly analysed in zucchini peels with three different colours: light green (Lg), yellow (Y), and orange (O). Our results showed that the carotenoid levels in O (157.075 μg/g) and Y (22.734 μg/g) were both significantly higher than in Lg (7.435 μg/g), while the chlorophyll content was highest in Lg (32.326 μg/g), followed by O (7.294 μg/g) and Y (4.617 μg/g). A total of 14 carotenoids were detected in zucchini peels, primarily lutein (103.167 μg/g in Lg, 509.667 μg/g in Y, and 1543.333 μg/g in O). In particular, significant accumulation of antheraxanthin, zeaxanthin, neoxanthin, and β-cryptoxanthin was first reported in orange zucchini in this study. Furthermore, two modules with hub genes related to carotenoid or chlorophyll content were identified through weighted gene coexpression network analysis. Additionally, the transcription level of some hub genes (PIF4, APRR2, bHLH128, ERF4, PSY1, LCYE2, and RCCR3) was highly correlated with pigment content in the peel, which may be responsible for carotenoid accumulation and chlorophyll degradation in the Y and O varieties. Taken together, the results obtained in this study help to provide a novel mechanism underlying peel colouration in zucchini.
1. Introduction
Carotenoids and chlorophyll are the primary pigments in higher plants. Carotenoids are naturally occurring, light-trapping pigments that protect tissues from photooxidative damage (Tracewell, Vrettos, Bautista, Frank, & Brudvig, 2001). Chlorophylls are also important for photosynthesis in plants. Chlorophylls capture light energy, drive electrons to the reaction centre, and convert and redirect light energy (Fromme, Melkozernov, Jordan, & Krauss, 2003). Importantly, the ratio of carotenoid and chlorophyll content ultimately determines plant colour (Wang et al., 2016).
As the carotenoid synthesis precursor, isopentenyl diphosphate isomerase (IPP) is primarily derived from the methyl-D-erythritol 4-phosphate (MEP) pathway (Kang et al., 2016) and is catalysed by geranylgeranyl diphosphate synthase (GGPPS) to produce geranylgeranyl diphosphate (GGPP) with its isomer dimethylallyl diphosphate (DMAPP). GGPP is a common precursor for the biosynthesis of carotenoids, chlorophyll, gibberellin, and abscisic acid (Beck et al., 2013). GGPP is involved in a series of reactions, including condensation, dehydrogenation, cyclization, hydroxylation, and epoxidation, finally producing such compounds as lutein, violaxanthin, and neoxanthin. Chlorophyll, including chlorophyll a (Chla) and chlorophyll b (Chlb), in higher plants can be produced by two biosynthetic pathways. The first pathway is from L-glutamyl-tRNA to protoporphyrin IX (proto IX) and then to chlorophyll, which requires 15 reactions with 15 enzymes (Nagata, 2005). In the second pathway, GGPP serves as a substrate and synthesizes chlorophyll a by geranylgeranyl reductase (GGR) (Giannino et al., 2004). In rice, the recruiting protein GRP controls GGPP flux towards chlorophyll biosynthesis (Zhou et al., 2017).
Zucchini (also known as American pumpkin) has a variety of peel colours, such as yellow, green, light green, and orange. Yellow-peel zucchini contains markedly higher carotenoid content than the other colours (Paris, Schaffer, Ascarelli, & Burger, 1989). Metabonomic analysis revealed that carotenoids are primarily composed of violaxanthin, lutein, and β-carotene in squash (Nakkanong, Yang, & Zhang, 2012). LCYE and LCYB are branching enzymes that produce lutein or β-carotene, and their ratio determines fruit colour (Wyatt, Strickler, Mueller, & Mazourek, 2015). Furthermore, LCYE regulates both peel and flesh colour (Obrero et al., 2013). Whole genome resequencing analysis of eight zucchini varieties with different peel colours demonstrated that nonsynonymous mutations occurred in the CDSs of Or, CCD, and APRR2 related to fruit pigmentation, which may have an important effect on their function (Xanthopoulou et al., 2019), but the molecular mechanism of carotenoid accumulation in zucchini has not been elucidated. In this study, analyses of the transcriptome and carotenoid metabolites were conducted in zucchini fruits with three peel colours (light green, yellow, and orange), and the related DEGs were further screened using weighted gene coexpression network analysis (WGCNA). The present study attempted to elucidate the molecular mechanism underlying peel colouration in yellow and orange zucchini fruit peels through the expression of the identified hub gene.
2. Materials and methods
2.1. Plant materials and sampling
Zucchini plants were grown from April 2019 to July 2019 in the greenhouse at the Horticultural Experiment Station of Shanxi Agricultural University, Taigu, Shanxi Province, P.R. China. The greenhouse was maintained in a controlled environment at 25/18 °C (day/night) with a 16-hour photoperiod (photosynthetically active radiation = 160 µmol m−2 s−1), and the relative humidity was approximately 70–80%. The homozygous zucchinis included three lines with different peel colours: 19pu07 (light green peel, Lg), 19pu11 (yellow peel, Y), and 19pu14 (orange peel, O), which were sampled 10 days after pollination (dap) (commercial mature stage) (Fig. 1A). Fruit peels were collected and pooled from three individual plants (only one fruit per individual plant was pollinated), and three biological replicates were performed for each zucchini variety. Three colour difference indexes (L*: brightness, black and white, a*: red-green, b*: yellow-blue) of Zucchini peels were measured by a CM-700D colorimeter (Konica Minolta Sensing, Inc., Osaka, Japan), and the CCI value was calculated as 1000 × a*/(L*×b*). The peels of each material were randomly divided into four parts, immediately frozen in liquid nitrogen, and stored at −80 °C for determination of total pigment content, metabolite extraction, transcriptomic sequencing, and real-time quantitative PCR.
Fig. 1.
Peel colour and pigment content of three zucchini materials. A: Peel colour of three materials at 10 dap (day after pollination). 19pu07: Light green peel (Lg); 19pu11: Yellow peel (Y); 19pu14: Orange peel (O). B: The colour difference index (L*, a*, b*) of three zucchini peels. L* represents brightness (black and white), while a* represents red-green and b* represents yellow-blue. C: The colour of anhydrous ethanol extracts of three peels. D: Pigment content of three materials at 10 dap. Chla, Chlb, Chl, and Car represent chlorophyll a, chlorophyll b, total chlorophyll, and total carotenoid content, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. Determination of pigment content in the peel
Peels were cut into small pieces, and 1.0 g samples were placed in 15 mL 96% ethanol in the dark for 24 h at room temperature. After centrifugation at 3000 r/min for 10 min, the supernatants were transferred into a cuvette with a 1-cm optical path and 96% ethanol as a blank control. Absorption values were measured at 665 nm, 649 nm, and 470 nm, with three technical replicates for each treatment. Data were calculated as follows (Wellburn & Lichtenthaler, 1984):
Ca = 13.96D665-6.88D649
Cb = 24.96D649-7.32D665
Cx.c = (1000D470-2.05 Ca-114.8 Cb)/245
Pigment content (μg/g) = 1000C × V/Awhere Ca represents the concentration of chlorophyll a (mg/L), Cb represents chlorophyll b, and Cx.c represents the concentration of lutein and carotenoid (mg/L). C represents the pigment concentration (mg/L), V represents the volume of extracted liquid, and A represents the fresh weight of the sample (g). D665, D649, and D470 are the absorbance values of the extracted liquid at wavelengths 665 nm, 649 nm, and 470 nm, respectively.
2.3. Extraction, identification, and quantification of carotenoids
Carotenoid extraction was performed according to a previous method with some modifications (Ma et al., 2017). Peel samples were freeze-dried in a SCIENTZ-100F freeze dryer (Xinzhi 100F, Ningbo, China) for one week with a minimum temperature of −50 °C and a vacuum of 1 Pa. The lyophilized 100 mg samples were ground to powder using a grinder with 30 Hz for 1 min (MM 400, Retsch, Germany), after which a mixed extraction solution of n-hexane:acetone:ethanol (2:1:1, V/V/V) with 0.01% BHT (g/mL) was added. The sample was vortexed to mix, sonicated for 20 min in an ultrasound apparatus, and later centrifuged while the supernatants were aspirated. The supernatants were collected after repeating the above steps and evaporated to dryness using a nitrogen blowing instrument (Haike DCY-12S, Qingdao, China) at 40 °C. Samples were then dissolved in a solution of acetonitrile:methanol:methyltert-butyl ether, vortexed to mix well, centrifuged for 2 min, filtered through a 0.22-µm PTFE membrane filter, and subsequently stored in a brown bottle until LC-MS/MS analysis.
Ultrahigh-performance liquid chromatography (UHPLC) was performed using a Waters UPLC system with a YMC C30 column and i.d. 2 × 100 mm columns. Mobile phase A was composed of acetonitrile:methanol (3:1, V/V) + 0.01% BHT, mobile phase B was methyl tert-butyl ether +0.01% BHT, and methanol was used as the probe wash. The flow rate was set at 0.8 mL/min at 28 °C. The gradient ran A:B (85:15,V/V) from 0 min, A:B (75:25, V/V) at 2.0 min, A:B (40:60, V/V) at 2.5 min, A:B (5:95, V/V) at 3.0 min, A:B (5:95, V/V) at 4.0 min, A:B (85:15, V/V) at 4.10 min, and A:B (85:15, V/V) to 6.0 min.
MS/MS detection was performed on an Applied Biosystems 6500 Quadrupole Trap (https://sciex.com). The atmospheric pressure chemical ionization (APCI) temperature was 350 °C, and curtain gas (CUR) was set at 25 psi. In a q-Trap 6500, each ion pair was scanned based on the optimized declustering potential (DP) and collision energy (CE).
The plant carotenoid database from a Metware database (MWDB, Wuhan, China) was used to qualitatively analyse the mass spectrometry data. LC-MS/MS data from the carotenoids were analysed using Analyst 1.6.3 software (AB Sciex, Framingham, MA, USA) using the default parameters for automatic identification of the various MRM changes and integrals. The mass spectrum peaks of each carotenoid in different samples were modified according to the carotenoid retention time and ion pair information (Fig. S1). The peak area represents the relative content of the corresponding carotenoids. The mass spectrum peak intensity data of the corresponding quantitative signals of each concentration standard were obtained by preparing different concentrations of carotenoid standard solutions, and the standard curves of different carotenoids were drawn with the standard concentration (μg/mL) as the abscissa and the peak area of the mass spectrum peak (Peak Area) as the ordinate (Table S1). Finally, carotenoid content was calculated using the following formula: B*C/1000/D (μg/g), where B is the concentration value after the carotenoid integrated peak area in the sample was substituted into the standard curve (μg·mL−1), C was the redissolved solution volume (μL), and D was the sample weight (g). The T-test on data was calculated using MetaboAnalyst (https://www.metaboanalyst.ca/).
2.4. RNA-seq analysis
Total RNA from peels was extracted using the RNAprep Pure Polyphenol Plant Total RNA Extraction Kit (TIANGEN, China). A cDNA library was constructed and sequenced on an Illumina HiSeq4000 system. The adapters and low-quality sequences were removed during Fastp with default parameters (Chen, Zhou, Chen, & Gu, 2018), and clean reads were mapped to the reference genome of zucchini V4.1 (http://cucurbitgenomics.org/organism/14) using HISAT2 (Kim, Langmead, & Salzberg, 2015). The numbers of mapped reads and transcript lengths were normalized. FPKM (fragments per kilobase per million) was used as an indicator of transcripts or gene expression (Mortazavi, Williams, McCue, Schaeffer, & Wold, 2008). The Pearson correlation coefficient and principal component analysis (PCA) were used to evaluate the correlation and repetitiveness among samples. Differential expression analysis was performed using DESeq2 (|log2Fold Change| ≥1, and FDR < 0.05) to obtain a set of genes differentially expressed between two biological samples (Varet, Brillet-Gueguen, Coppee, & Dillies, 2016). Heat maps and Venn diagrams were drawn using TBtools software (Chen et al., 2020).
2.5. Functional annotation of genes
Nonredundant transcript sequences identified as genes were further analysed by Gene Ontology (GO) annotation (http://www.geneontology.org/) to identify GO terms among DEGs with significant differences, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) was used to identify significantly enriched pathways.
2.6. WGCNA and visualization of gene networks
Weighted gene coexpression network analysis (WGCNA) (Langfelder & Horvath, 2008) was performed (soft threshold for 6, minimum module size for 30, and combined cutting height for 0.25) in the gene expression profile, including 9 peels (Lg, Y, and O) and 9 corresponding samples of flesh (data not shown). Correlation analysis between the identified modules and the total metabolism of chlorophyll and carotenoids in peel and flesh and the modules with the highest correlation were selected based on the Pearson correlation coefficient (PCC). DEGs with high correlation rates were selected to construct a gene coexpression network map using Cytoscape 3.7.2 (http://www.cytoscape.org/).
2.7. Real-time quantitative PCR
First strand cDNA was synthesized using TAKARA PrimeScript™ RT Master Mix (Perfect Real Time). Real-time quantitative PCR was run on an ABI 7500 instrument using Talent qPCR PreMix (SYBR Green). The zucchini actin gene (GenBank Accession No. MH211008) was used as an internal control. Each reaction was performed in triplicate with a reaction volume of 15 μL. Cycling parameters were 94 °C for 5 min, 40 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s. Quantitative data were calculated as 2−△△CT. Primer sequences are listed in Table S2.
3. Results and discussion
3.1. Determination of pigment content in zucchini peel
As the three materials exhibited three different peel colours, the peel colours were first accurately quantified using a colorimeter, and as illustrated in Fig. 1B, the L* value was largest in Lg (77.49) and smallest in Y (60.08). However, O had the largest a* value (30.62) and b* value (69.7), while Lg had the smallest a* value (-1.89) and b* value (31.15). The colour contribution index (CCI) is also used for colour evaluation; a CCI value > 0 indicates a red-yellow colour, a CCI value < 0 indicates a blue-green colour, and a CCI value = 0 indicates an intermediate mixture of red, yellow, and blue-green. The CCI values for Lg were negative (−0.78), Y for 6.61 and O for 6.93 were both positive, and CCI values of O were slightly higher than Y. In other words, Lg is brighter and greener than the other two types of peels, while O is brighter, redder, and more yellow than Y.
Furthermore, pigment contents, including total chlorophyll and carotenoids, in zucchini for the three different peel colours were measured at 10 dap (Fig. 1C and 1D). Total chlorophyll content was highest (32.326 μg/g) in Lg, approximately 9.67- and 7.84-fold more than Y and O, respectively. Levels of Chla and Chlb were 19.996 μg/g and 12.330 μg/g in Lg, respectively (Chla > Chlb), 2.067 μg/g and 2.550 μg/g in Y, and 2.834 μg/g and 4.460 μg/g in O, respectively (Chlb > Chla). Moreover, total carotenoid content in the peels was also significantly different. Total carotenoid content in the peels of Y (22.734 μg/g) and O (157.075 μg/g) were 3.056- and 21.13-fold higher than in Lg (7.435 μg/g), respectively. As a result, the colour values and pigment contents were in line with the peel colours, suggesting that the higher carotenoid content and lower chlorophyll content in Y and O might result in yellow or orange peel in zucchini.
3.2. Carotenoid identification in three zucchini peels
Fourteen carotenoids were detected from three zucchini materials at 10 dap, and 13 of them were involved in the carotenoid biosynthetic pathway (only apocarotenal was not) (Fig. 2A). Among these carotenoids, 11 of them, including γ-carotene, β-carotene, and lutein, were detected in all three peels, while ε-carotenes were only detected in Y and O, and lycopene and phytoene were only detected in O (Fig. 2B). Moreover, the contents of 14 carotenoids were much higher in O than in Y or Lg (Fig. 2A and Table S3). Lutein was obviously dominant, accounting for 84.7% of the total carotenoid content in O, 88.25% in Y, and 75.02% in Lg. Among the 14 carotenoids, zeaxanthin had the second highest accumulation in O, accounting for 3.94%, while β-carotene had the second highest accumulation in Lg and Y, accounting for 9.99% and 2.68%, respectively.
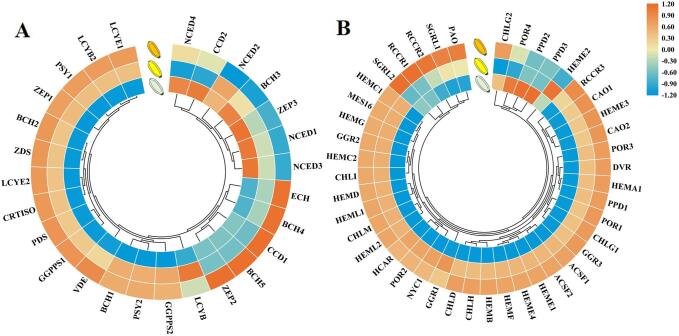
Fig. 2.
Carotenoid content in three zucchini peels. This pathway was constructed based on the KEGG pathway. A: Carotenoid levels of three peel colours at 10 dap. Each bar represents the average of three biological replicates plus the standard deviation. B: Venn diagram of carotenoid types in three zucchini materials. GGPP, geranylgeranyl diphosphates; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; CRTISO, carotenoid isomerase; LCYE, lycopene ε-cyclase; LCYB, lycopene β-cyclase; BCH, β-carotene hydroxylase; ECH, ε-carotene hydroxylase; ZEP, zeaxanthin epoxidase; VDE, violaxanthin de-epoxidase; NXS, neoxanthin synthase; NCED, 9-cis-epoxycarotenoid dioxygenase; CDDs, carotenoid cleavage dioxygenase.
Furthermore, there were significant differences (−log10(p) > 1, p < 0.05) in the peel contents of α-carotene, zeaxanthin, violaxanthin, γ-carotene, neoxanthin, lutein, ε-carotene and α-cryptoxanthin between Lg and Y, and the contents of α-carotene, violaxanthin, γ-carotene, neoxanthin, lutein, ε-carotene, and α-cryptoxanthin were higher in Y than in Lg. The carotenoids detected in O were present at significantly higher levels than those in Lg, except for β-cryptoxanthin and apocarotenal. Compared to Y, O exhibited significant increases in other types of carotene contents, except for violaxanthin, β-cryptoxanthin and apocarotenal. In addition to violaxanthin, the contents of lutein and β-carotene were consistent with previous reports (Nakkanong et al., 2012, Obrero et al., 2013). In addition, 4 pigments, antheraxanthin, zeaxanthin, neoxanthin, and β-cryptoxanthin, were all reported in O for the first time in the present study. Based on the above results, abundant carotenoids, especially lutein, in O or Y may cause orange or yellow colours in zucchini peels.
3.3. Transcriptomic analysis in three zucchini peels
A total of 9 cDNA libraries from three zucchini peels were constructed for high-throughput sequencing. In total, average clean reads of 71.47 million, 49.84 million, and 49.16 million were obtained after filtering raw data from the peels of Lg, Y, and O, respectively. The average GC contents were approximately 44.53%, 44.44%, 44.44%, and the average Q30 values were 92.85%, 94.52%, 94.64% in Lg, Y and O, respectively. The alignment efficiency of clean reads with the zucchini reference genome (V4.1) varied from 87.12% to 96.07% using HISAT2 (Table S4). Based on the mapped reads, a total of 9513 DEGs were obtained in three comparative components (Fig. S2A, Table S5). Furthermore, 5421 DEGs were identified between Lg and Y, including 3018 upregulated and 2403 downregulated DEGs; 7039 DEGs were identified between Lg and O, containing 4157 upregulated and 2882 downregulated DEGs; and 3470 DEGs were identified between Y and O, 2041 of which were upregulated and 1429 of which were downregulated DEGs. Additionally, a Venn diagram showed that 1004, 2118, and 638 unique DEGs were obtained from Lg vs Y, Lg vs O, and Y vs O, respectively, and a total of 664 DEGs were expressed in the three materials (Fig. S2B). PCA and correlation analysis further confirmed the reproducibility among biological duplications and differences among these materials (Fig. S3). Therefore, these identified DEGs were selected for further analysis with respect to peel colouration.
3.4. Function annotation with GO and KEGG database
The functions of these DEGs were further annotated. Based on the GO database, the DEGs were classified into 56, 55, and 53 subcategories from Lg vs Y, Lg vs O, and Y vs O, respectively. Among these subcategories, cellular components were primarily composed of cells (3529/5421 in Lg vs Y, 4599/7039 in Lg vs O, 2190/3470 in Y vs O), cell parts (3521/5421 in Lg vs Y, 4589/7039 in Lg vs O, 2187/3470 in Y vs O) and organelles (2698/5421 in Lg vs Y, 3543/7039 in Lg vs O 1620/3470 in Y vs O). Molecular function included binding (2547/5421 in Lg vs Y, 3316/7039 in Lg vs O, 1664/3470 in Y vs O), activity (2192/5421 in Lg vs Y, 2845/7039 in Lg vs O, 1364/3470 in Y vs O) and transcription regulator (423/5421 in Lg vs Y, 555/7039 in Lg vs O, 399/3470 in Y vs O). Biological processes consisted of cellular processes (2852/5421 in Lg vs Y, 3717/7039 in Lg vs O, 1797/3470 in Y vs O), metabolic processes (2523/5421 in Lg vs Y, 3299/7039 in Lg vs O, 1578/3470 in Y vs O) and responses to stimuli (1358/5421 in Lg vs Y, 1753/7039 in Lg vs O, 937/3470 in Y vs O) (Table S6). GO enrichment analysis demonstrated that chloroplast thylakoid, plastid thylakoid, thylakoid part, and photosynthetic membrane were significantly enriched in Lg vs Y/O, and photosystem I, photosystem, MCM complex, plastoglobule, and photosystem II were significantly enriched in Y vs O (Fig. S4).
Through enrichment analysis of the KEGG pathway, 2003, 2511 and 1196 DEGs from Lg vs Y, Lg vs O, and Y vs O were annotated on 135, 138, and 134 pathways, respectively, of which the most significantly enriched pathways were photosynthesis, photosynthesis-antenna proteins, biosynthesis of secondary metabolites, porphyrin and chlorophyll metabolism (Table S7). These results all suggest that the light system, chloroplasts, and secondary metabolite synthesis might be critical for pigment synthesis and accumulation in zucchini peels.
3.5. DEGs involved in carotenoid and chlorophyll metabolism
Based on the carotenoid pathway, 27 DEGs were identified among the three peel colours (Fig. 3A and Table S8), of which 5 randomly selected key DEGs were further verified by qRT-PCR (Fig. S5A-E). The average expression levels of 18 upstream synthesis genes, GGPPS1 (Cp4.1LG16g00240), PSY1/2 (Cp4.1LG13g05570/Cp4.1LG01g19670), ZDS (Cp4.1LG04g01620), PDS (Cp4.1LG08g006310), CRTISO (Cp4.1LG14g01900), and LCYE1/2 (Cp4.1LG07g10720/Cp4.1LG11g11790) showed O > Y > Lg. Downstream, the average gene expression levels of CCD2 (Cp4.1LG14g02990) and NCED4 (Cp4.1LG19g05960) exhibited Lg > O > Y, while those of NCED1/3 (Cp4.1LG14g03280/Cp4.1LG10g05900), ECH3 (Cp4.1LG01g01250), and ZEP3 (Cp4.1LG07g02750) were Lg > Y > O. In summary, the upstream genes of carotenoid biosynthesis were upregulated, while the downstream genes, including NCED and CCD, were downregulated in Y and O, resulting in the accumulation of coloured components, such as lutein. In particular, the higher content of carotenoids in O resulted in orange rather than yellow in Y.
Fig. 3.
Heatmap of the expression of genes related to carotenoid biosynthesis (A) and chlorophyll metabolism (B) in three zucchini species (Lg, Y, and O). Gene expression level is expressed as the mean value of FPKM of three biological replicates for each material. Orange and blue indicate high expression and low expression. GGPP, geranylgeranyl diphosphates; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; CRTISO, carotenoid isomerase; LCYE, lycopene ε-cyclase; LCYB, lycopene β-cyclase; BCH, β-carotene hydroxylase; ECH, ε-carotene hydroxylase; ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; CDDs, carotenoid cleavage dioxygenase; MES16, glutamyl-tRNA synthetase; HEML, glutamate-1-semialdehyde 2,1-aminomutase; HEMA, glutamyl-tRNA reductase; HEMB, porphobilinogen synthase; HEMC, hydroxymethylbilane synthase; HEMD, uroporphyrinogen-III synthase; HEME, uroporphyrinogen decarboxylase; HEMF, coproporphyrinogen III oxidase; HEMG, menaquinone-dependent protoporphyrinogen oxidase; CHLD, magnesium chelatase subunit D; CHLH, magnesium chelatase subunit H; CHLI, magnesium chelatase subunit I; CHLM, magnesium–protoporphyrin O-methyltransferase; ACSF, magnesium–protoporphyrin IX monomethyl ester (oxidative) cyclase; POR, protochlorophyllide reductase; DVR, divinyl chlorophyllide an 8-vinyl-reductase; GGR, geranylgeranyl reductase; CHIG, chlorophyll/bacteriochlorophyll a synthase; CAO, chlorophyllide an oxygenase; SGRL, magnesium dechelatase; NYC1, chlorophyll(ide) b reductase; HCAR, 7-hydroxymethyl chlorophyll a reductase; PAO, pheophorbide an oxygenase; RCCR, red chlorophyll catabolite reductase; PPD, pheophorbidase. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In previous studies, the PSY and LCYE genes were demonstrated to be key steps of carotenoid metabolism pathways (Stanley & Yuan, 2019). Three PSY genes in zucchini exhibited distinct expression patterns in multiple tissues (Obrero et al., 2015). Heterologous expression of two tomato PSY genes in Arabidopsis and bacterial systems revealed distinct biosynthetic abilities between PSY1 and PSY2, which could result from their specific expression patterns (Cao et al., 2019). In addition, gene expression of LCYE may regulate the ratio of α-carotene and β-carotene (Harjes et al., 2008). The LCYE mutant lut2 led to lutein deficiency and higher beta-carotene accumulation (Pogson, McDonald, Truong, Britton, & DellaPenna, 1996). Furthermore, expression of the LCYE gene was highly correlated with carotenoid biosynthesis in zucchini (Obrero et al., 2013). In summary, most DEGs (18/27) related to carotenoid synthesis, especially LCYE, which is highly expressed in Y and O (Fig. 3A), may directly promote carotenoid accumulation in zucchini.
Additionally, 43 DEGs were identified in the chlorophyll (chl a and b) biosynthesis pathway (Fig. 3B, Table S8), and 38 DEGs exhibited higher expression in Y and O than in Lg. Regardless of whether L-glutamyl-tRNA or GGPP was used as the synthetic precursor, expression levels of the key genes that edited enzymes in the chlorophyll biosynthesis pathway were all higher in Y and O than in Lg. Among these genes, five key genes, CHLH, HEMA1, RCCR3, CAO1, and NYC1, were all confirmed by qRT-PCR (Fig. S5F-J), suggesting that chlorophyll accumulates in Y and O. However, genes related to chlorophyll degradation, such as NYC, PPH, PAO, and RCCR, were also highly expressed in Y and O, which may directly lead to rapid degradation of chlorophyll in Y and O.
In the chlorophyll degradation pathway, pheophorbide a oxygenase (PAO) can use pheophorbide a as a substrate to synthesize red chlorophyll catabolite (RCC), and red chlorophyll catabolite reductase (RCCR) catalyses RCC into primary fluorescent chlorophyll catabolites (pFCCs) (Pruzinska et al., 2007). The PAO and RCCR pathways are key pathways of chlorophyll degradation, resulting in etiolation and even bleaching in plants (Matile, Hortensteiner, & Thomas, 1999). As a result, higher expression of PAO and RCCR genes may be a direct cause of the lower chlorophyll contents in Y and O compared to Lg.
3.6. Weighted gene coexpression network analysis of transcriptomic data from zucchini peel and flesh
WGCNA can identify characteristic genes of a module, key genes in the module, the association between modules, and sample phenotypes (Langfelder et al., 2008). Using WGCNA, a total of 5646 DEGs (FPKM value > 1) in 18 samples (Lg, Y, O with three repeat samples of peel and flesh each) were divided into 12 coexpression modules (Fig. 4A, Table S9), and 2662 coexpressed genes were in the largest blue module and had a significantly positive correlation with total carotenoid content (r = 0.82, P = 0.003), while only 63 coexpressed genes were in the sky blue module and had a negative correlation with chlorophyll a (r = −0.28, P = 0.3), chlorophyll b (r = −0.31, P = 0.2) and total chlorophyll content (r = -0.29, P = 0.2). In the green module, 952 genes were significantly positively correlated with chlorophyll a (r = 0.98, P = 4e-10), chlorophyll b (r = 0.9, P = 5e-05) and total chlorophyll (r = 0.97, P = 2e-08) (Fig. 4B). The gene expression patterns of the blue and green modules were significantly correlated with pigment accumulation in zucchini peels.
Fig. 4.
WGCNA of transcriptomic dates. A: Tree maps of 5646 DEGs were constructed by hierarchical clusters with topological overlap dissimilarity. Each gene was represented by leaves in a branch of the tree map. The coexpression distance between two genes represents the height on the y-axis. The first and second lines below the tree indicate the module members identified by the dynamic tree using the cutting method and the combined dynamic tree identity with a 0.85 merge threshold, respectively. The main branches are made up 27 different coexpression modules. B: The relationship between pigment content in peel and flesh (column) and the module characteristic gene (ME, row). The colour indicates the relevant intensity and direction. The numbers in parentheses are partial Pearson correlations and corresponding P-values.
When PCC was set as ≥0.90 or ≤−0.90, 961 and 741 DEGs were used to construct a coexpression network of blue and green modules, respectively (Fig. 5A-B). The expression levels of 18 genes from the blue module and 7 genes from the green module that were highly correlated with pigment content of the peel may be regarded as highly connected genes (hub genes) and involved in three categories: carotenoid biosynthesis (6), chlorophyll metabolism (2) and transcription factors (16) (Fig. 5C-D, Table S10).
Fig. 5.
Coexpression and regulatory networks of DEGs involved in carotenoid biosynthesis and chlorophyll metabolism. A: Gene coexpression network of the blue module. B: Gene coexpression network of the green module. C: Subnetwork of putative transcription factors (TFs) and structural genes related to carotenoid biosynthesis and chlorophyll degradation from the blue module. D: Subnetwork of putative TFs or other genes from the green module. Green circles denote target genes, and red circles denote TFs. The lines between the two dots indicate their interactions. The colours of lines represent the strength of correlations between the target genes and TFs; the bluer the stronger and the yellower the weaker the interactions between TFs and target genes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
According to the expression patterns of DEGs in carotenoid and chlorophyll pathways, some transcription factors (TFs) may be involved in the regulation of carotenoid and chlorophyll pathways in fruit peels and result in various colours, such as light green, yellow, and orange. Previous studies have demonstrated that bHLH, bZIP, WRKY, MADS, ERF, ARF, and other genes play important roles in regulating pigment accumulation in plants (Stanley et al., 2019). Based on key enzymatic genes of carotenoids and chlorophyll synthesis, 17 differentially expressed transcription factor genes (DETFs) were identified in the gene coexpression network. Expression of six randomly selected genes, PIF4, HY5, APRR2, APRR9, OR, and mTERF4, was verified by qRT-PCR (Fig. S5K-P).
Nine genes of 17 DETFs, APPR2 (Cp4.1LG05g02070), APRR9 (Cp4.1LG07g07580), OR (Cp4.1LG10g03920), WRKY2 (Cp4.1LG08g08130), PIF4 (Cp4.1LG17g09790), bHLH66 (Cp4.1LG04g09000) and mTERF4/5/9 (Cp4.1LG07g08650/Cp4.1LG10g02900/ Cp4.1LG06g08460) are co-expression up-regulated with the synthesis of carotenoids or the degradation of chlorophyll in Y and O (Fig. 5C), while the other 8 genes, including MADS23 (Cp4.1LG01g02990), AP2-like (Cp4.1LG05g11500), ERF4 (Cp4.1LG04g02810), ERF9 (Cp4.1LG12g07240), IAA3 (Cp4.1LG08g00060), bHLH128 (Cp4.1LG03g12350), HY5 (Cp4.1LG11g03290) and chlorophyll A-B binding protein (Cp4.1LG10g03050) are co-expression up-regulated in Lg (Fig. 5D).
The bHLH transcription factor PIF5 positively regulates expression of key enzymatic genes in the MEP pathway and improves the accumulation of chlorophyll and carotenoids in cultured cells (Mannen et al., 2014). Transcription levels of the PIF3 and PIF4 genes were positively correlated with carotenoid content in apricot peel (Zhang, Zhang, Li, Zhang, & Xi, 2019). On the other hand, the transcription factor HY5 in tomato and Arabidopsis also positively activates the synthesis genes of chlorophyll biosynthesis and chlorophyll accumulation (Liu et al., 2004, Zhu et al., 2015), although HY5 competes with PIF1 by combining the cis-element G-box of the PSY promoter to regulate carotenoid accumulation (Toledo-Ortiz et al., 2014). In this study, expression of PIF4 (Cp4.1LG17g09790) in Y and O was significantly higher than in Lg, while expression of HY5 (Cp4.1LG11g03290) was lower in Y and O (Table S10), but both were highly correlated with carotenoid biosynthesis and chlorophyll degradation. These results all indicate the vital roles of the PIF4 and HY5 genes in regulating peel colouration in Y and O.
APRR2 and Or genes also positively regulate the expression of genes involved in chlorophyll and carotenoid synthesis by increasing the storage capacity of pigments. APPR2 modulates chlorophyll accumulation in cucumber (Liu et al., 2016) and chlorophyll and carotenoid accumulation in melons (Oren et al., 2019), while the Or gene promotes carotenoid accumulation in cauliflower (Lu et al., 2006) and melon (Tzuri et al., 2015). In our study, the upregulated genes CpAPPR2 (Cp4.1LG05g02070), CpAPRR9 (Cp4.1LG07g07580), and CpOR (Cp4.1LG10g03920) may be associated with the high expression of key structural genes (e.g., PSY and LCYE) of carotenoid biosynthesis pathways, leading to carotenoid accumulation in Y and O.
In addition, Arabidopsis mitochondrial transcription termination factors (mTERFs) activate leaf colour development by affecting expression of both chloroplast and nuclear genes (Quesada et al., 2011). The colours of cotyledons, leaves, stems, and sepals are lighter in the mTERF5 mutant (Robles, Micol, & Quesada, 2012), and the early development of chloroplasts was blocked in the mTERF6 mutant, which led to bleaching and seedling death (Pfalz, Liere, Kandlbinder, Dietz, & Oelmuller, 2006). In this study, mTERF4/5/9 (Cp4.1LG07g08650/Cp4.1LG10g02900/ Cp4.1LG06g08460) were all upregulated in Y and O and were also considered to be involved in carotenoid accumulation or chlorophyll degradation in peels.
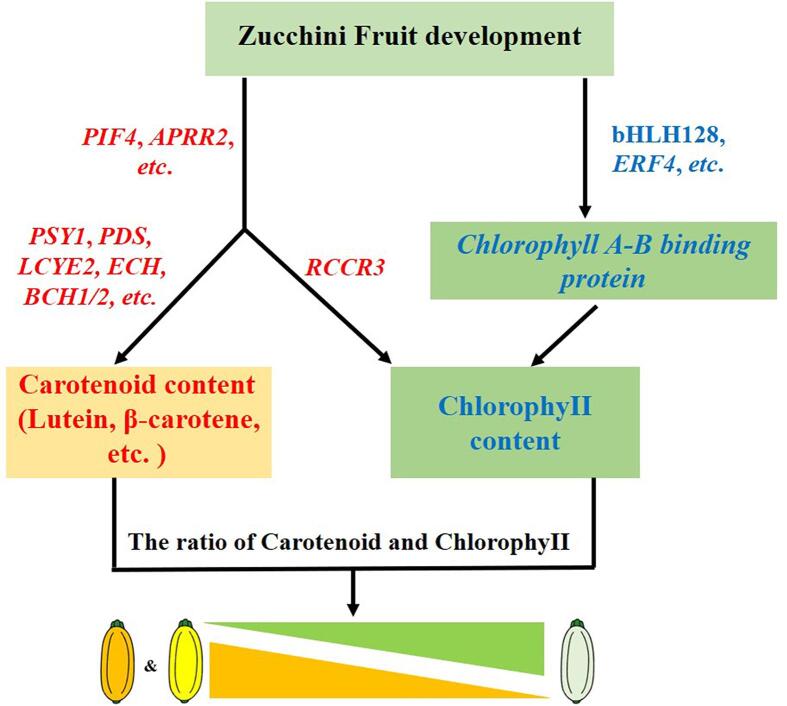
In summary, pigment accumulation in zucchini peel is primarily determined by the proportion of chlorophyll and carotenoid contents, especially lutein (Wang et al., 2016, Wyatt et al., 2015). If the carotenoid content, especially lutein, was higher than chlorophyll, the peel would be yellow or even orange; otherwise, it would be green or light green. During pigment accumulation, expression of some transcription factors (e.g., PIF4, APRR2, bHLH128, and ERF4) and some structural genes (e.g., PSY1, LCYE2, and RCCR3) was highly related to pigment content in the peel, which may lead to the accumulation of carotenoids (lutein, β-carotene) and the degradation of chlorophyll in Y and O (Fig. 6). The proposed model is constructed based on the hub genes screened by gene coexpression network analysis, but its specific regulatory mechanisms remain to be explored in depth.
Fig. 6.
Proposed model of carotenoids and chlorophyll metabolic pathways in zucchini peel. The red and blue genes represent upregulation and downregulation in yellow peel or orange peel, respectively. The yellow triangle and the green triangle indicate the contents of carotenoids, especially lutein and chlorophyll, from high to low, respectively, and the ratio of chlorophyll and carotenoid (lutein, β-carotene) contents finally determined the peel colour of zucchini. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Conclusions
In this study, carotenoid metabolites and transcriptomes were compared in three zucchini peel types. Fourteen carotene metabolites were detected in zucchini peels, and lutein was the primary component in Y and O (509.667 μg/g, accounting for 88.25% in Y, and 1543.333 μg/g, accounting for 84.7% in O). Moreover, in the present study, significant accumulation of antheraxanthin, zeaxanthin, neoxanthin, and β-cryptoxanthin was detected for the first time in O. In total, 27 enzymatic genes in chlorophyll biosynthesis and degradation and carotenoid synthesis were upregulated in Y and O, resulting in higher carotenoid content (157.075 ± 7.078 μg/g in O, 22.734 ± 8.72 μg/g in Y) and lower chlorophyll content (7.01 ± 3.152 μg/g in O, 4.901 ± 1.399 μg/g in Y). WGCNA revealed that some hub genes (e.g., PIF4, APRR2, bHLH128, ERF4, PSY1, LCYE2, and RCCR3) may be involved in pigment accumulation in zucchini peels, which may affect the formation of peel colour.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We would like to thank associate professor Jiangli Shi from Henan Agricultural University for English language editing.
This research was supported by the Shanxi Province Key Research and Development Program Key Projects (201703D211001-04-01) and the Biological Breeding Project from Shanxi Academy of Agricultural Sciences (17yzgc008).
Author contribution statement
XX and XL designed the experiment and drafted the manuscript. ZT and XZ helped in plant management and data analysis. FL provided the materials and helped design the experiments. PH and ML supervised the research and revised the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochms.2021.100021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Beck G., Coman D., Herren E., Ruiz-Sola M.A., Rodriguez-Concepcion M., Gruissem W., Vranova E. Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant Molecular Biology. 2013;82(4–5):393–416. doi: 10.1007/s11103-013-0070-z. [DOI] [PubMed] [Google Scholar]
- Cao H., Luo H., Yuan H., Eissa M.A., Thannhauser T.W., Welsch R., Hao Y.-J., Cheng L., Li L. A Neighboring aromatic-aromatic amino acid combination governs activity divergence between tomato phytoene synthases. Plant Physiology. 2019;180(4):1988–2003. doi: 10.1104/pp.19.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y.i., Thomas H.R., Frank M.H., He Y., Xia R. TBtools - an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme P., Melkozernov A., Jordan P., Krauss N. Structure and function of photosystem I: Interaction with its soluble electron carriers and external antenna systems. FEBS Letters. 2003;555(1):40–44. doi: 10.1016/s0014-5793(03)01124-4. [DOI] [PubMed] [Google Scholar]
- Giannino D., Condello E., Bruno L., Testone G., Tartarini A., Cozza R., Mariotti D. The gene geranylgeranyl reductase of peach (Prunus persica [L.] Batsch) is regulated during leaf development and responds differentially to distinct stress factors. Journal of Experimental Botany. 2004;55(405):2063–2073. doi: 10.1093/jxb/erh217. [DOI] [PubMed] [Google Scholar]
- Harjes C.E., Rocheford T.R., Bai L., Brutnell T.P., Kandianis C.B., Sowinski S.G., Stapleton A.E., Vallabhaneni R., Williams M., Wurtzel E.T., Yan J., Buckler E.S. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science. 2008;319(5861):330–333. doi: 10.1126/science:1150255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang A., George K.W., Wang G., Baidoo E., Keasling J.D., Lee T.S. Isopentenyl diphosphate (IPP)-bypass mevalonate pathways for isopentenol production. Metabolic Engineering. 2016;34:25–35. doi: 10.1016/j.ymben.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nature Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Jiao J., Liang X., Liu J., Meng H., Chen S.…Cheng Z. Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus L.) Theoretical and Applied Genetics. 2016;129(7):1247–1256. doi: 10.1007/s00122-016-2700-8. [DOI] [PubMed] [Google Scholar]
- Liu Y., Roof S., Ye Z., Barry C., van Tuinen A., Vrebalov J.…Giovannoni J. Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(26):9897–9902. doi: 10.1073/pnas.0400935101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Van Eck J., Zhou X., Lopez A.B., O'Halloran D.M., Cosman K.M.…Li L. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. The Plant Cell. 2006;18(12):3594–3605. doi: 10.1105/tpc.106.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Zhang L., Iida K., Madono Y., Yungyuen W., Yahata M.…Kato M. Identification and quantitative analysis of beta-cryptoxanthin and beta-citraurin esters in Satsuma mandarin fruit during the ripening process. Food Chemistry. 2017;234:356–364. doi: 10.1016/j.foodchem.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Mannen K., Matsumoto T., Takahashi S., Yamaguchi Y., Tsukagoshi M., Sano R.…Nakayama T. Coordinated transcriptional regulation of isopentenyl diphosphate biosynthetic pathway enzymes in plastids by phytochrome-interacting factor 5. Biochemical and Biophysical Research Communications. 2014;443(2):768–774. doi: 10.1016/j.bbrc.2013.12.040. [DOI] [PubMed] [Google Scholar]
- Matile P., Hörtensteiner S., Thomas H. Chlorophyll degradation. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50(1):67–95. doi: 10.1146/annurev.arplant.50.1.67. [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nagata N. Identification of a vinyl reductase gene for chlorophyll synthesis in arabidopsis thaliana and implications for the evolution of prochlorococcus species. The Plant Cell. 2005;17(1):233–240. doi: 10.1105/tpc.104.027276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakkanong K., Yang J.H., Zhang M.F. Carotenoid accumulation and carotenogenic gene expression during fruit development in novel interspecific inbred squash lines and their parents. Journal of Agricultural & Food Chemistry. 2012;60(23):5936–5944. doi: 10.1021/jf3007135. [DOI] [PubMed] [Google Scholar]
- Obrero Á., González-Verdejo C.I., Die J.V., Gómez P., Del Río-Celestino M., Román B. Carotenogenic gene expression and carotenoid accumulation in three varieties of Cucurbita pepo during fruit development. Journal of Agricultural & Food Chemistry. 2013;61(26):6393–6403. doi: 10.1021/jf4004576. [DOI] [PubMed] [Google Scholar]
- Obrero A., Gonzalez-Verdejo C.I., Roman B., Gomez P., Die J.V., Ampomah-Dwamena C. Identification, cloning, and expression analysis of three phytoene synthase genes from Cucurbita pepo. Biologia Plantarum. 2015;59(2):201–210. doi: 10.1007/s10535-015-0504-3. [DOI] [Google Scholar]
- Oren, E., Tzuri, G., Vexler, L., Dafna, A., Meir, A., Faigenboim, A., … Gur, A. (2019). The multi-allelic APRR2 gene is associated with fruit pigment accumulation in melon and watermelon. Journal of Experimental Botany, 70(15), 3781-3794. https://doi.org/10.1093/jxb/erz182. [DOI] [PMC free article] [PubMed]
- Paris H., Schaffer A., Ascarelli I.M., Burger Y. Heterozygosity of gene B and the carotenoid content of Cucurbita pepo. Crop Research. 1989;29:11–18. [Google Scholar]
- Pfalz J., Liere K., Kandlbinder A., Dietz K.J., Oelmuller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. The Plant Cell. 2006;18(1):176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B., McDonald K.A., Truong M., Britton G., DellaPenna D. Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. The Plant Cell. 1996;8(9):1627–1639. doi: 10.1105/tpc.8.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinska, A., Anders, I., Aubry, S., Schenk, N., Tapernoux-Luthi, E., Muller, T., … Hortensteiner, S. (2007). In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell, 19(1), 369-387. https://doi.org/10.1105/tpc.106.044404. [DOI] [PMC free article] [PubMed]
- Quesada, V., Sarmiento-Manus, R., Gonzalez-Bayon, R., Hricova, A., Perez-Marcos, R., Gracia-Martinez, E., … Micol, J. L. (2011). Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J, 68(4), 738-753. https://doi.org/10.1111/j.1365-313X.2011.04726.x. [DOI] [PubMed]
- Robles P., Micol J.L., Quesada V., Blazquez M.A. Arabidopsis MDA1, a nuclear-encoded protein, functions in chloroplast development and abiotic stress responses. PLoS ONE. 2012;7(8):e42924. doi: 10.1371/journal.pone.0042924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley L., Yuan Y.W. Transcriptional regulation of carotenoid biosynthesis in plants: So many regulators, so little consensus. Frontiers in Plant Science. 2019;10:1017. doi: 10.3389/fpls.2019.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Johansson H., Lee K.P., Bou-Torrent J., Stewart K., Steel G.…Qu L.-J. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genetics. 2014;10(6):e1004416. doi: 10.1371/journal.pgen.1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracewell C.A., Vrettos J.S., Bautista J.A., Frank H.A., Brudvig G.W. Carotenoid photooxidation in photosystem II. Archives of Biochemistry and Biophysics. 2001;385(1):61–69. doi: 10.1006/abbi.2000.2150. [DOI] [PubMed] [Google Scholar]
- Tzuri G., Zhou X., Chayut N., Yuan H., Portnoy V., Meir A.…Tadmor Y. A 'golden' SNP in CmOr governs the fruit flesh color of melon (Cucumis melo) The Plant Journal. 2015;82(2):267–279. doi: 10.1111/tpj.2015.82.issue-210.1111/tpj.12814. [DOI] [PubMed] [Google Scholar]
- Varet H., Brillet-Guéguen L., Coppée J.-Y., Dillies M.-A., Mills K. SARTools: A DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE. 2016;11(6):e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.X., Yu Y.F., Chen L., Qin H.Y., Liu Y.X., Ai J.…Fan S.T. Advances in leaf pigment composition, structure and photosynthetic characteristics of colored-leaf plants. Plant Physiology Journal. 2016;52:1–7. doi: 10.13592/j.cnki.ppj.2015.0490. [DOI] [Google Scholar]
- Wellburn, A., & Lichtenthaler, H. (1984). Formulae and Program to Determine Total Carotenoids and Chlorophylls A and B of Leaf Extracts in Different Solvents. In (pp. 9–12).
- Wyatt L.E., Strickler S.R., Mueller L.A., Mazourek M. An acorn squash (Cucurbita pepo ssp. ovifera) fruit and seed transcriptome as a resource for the study of fruit traits in Cucurbita. Horticulture Research. 2015;2:14070. doi: 10.1038/hortres.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthopoulou A., Montero-Pau J., Mellidou I., Kissoudis C., Blanca J., Picó B.…Ganopoulos I. Whole-genome resequencing of Cucurbita pepo morphotypes to discover genomic variants associated with morphology and horticulturally valuable traits. Horticulture Research. 2019;6(1) doi: 10.1038/s41438-019-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang Q., Li W., Zhang S., Xi W. Identification of key genes and regulators associated with carotenoid metabolism in apricot (Prunus armeniaca) fruit using weighted gene coexpression network analysis. BMC Genomics. 2019;20(1):876. doi: 10.1186/s12864-019-6261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Wang C.Y., Gutensohn M., Jiang L., Zhang P., Zhang D.…Lu S. A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(26):6866–6871. doi: 10.1073/pnas.1705689114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Bu Q., Xu X., Paik I., Huang X., Hoecker U.…Huq E. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nature Communications. 2015;6(1) doi: 10.1038/ncomms8245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.