Highlights
-
•
First report in Bangladesh on species authentication of meat products (n = 64) and cheese (n = 25) using duplex PCR.
-
•
High priced Bangladeshi ‘beef’ product was adulterated (7 out of 22) with low price ‘chicken’ and ‘buffalo’ meat.
-
•
Foreign chicken products were adulterated (66.67%) with buffalo meat.
-
•
Buffalo DNA was detected in 4 out of 12 cheese products labeled as cows’ milk.
-
•
The attention to Bangladesh Food Safety Authority is an inspection of meat and milk products ingredient authenticity.
Keywords: Food authentication, Meat species, Milk species, PCR, Mitochondrial cyt b
Abstract
Food origin authentication is imperative for consumer protection. This study explored meat and milk species identification as a pioneering country-specific report on mislabeling prevalence among processed meat and milk products in Bangladesh. Meat products (64; sausages, burger patty, meatball, kabab) labeled as chicken or beef and Mozzarella-type cheeses (25), made in Bangladesh and of overseas origins, were analyzed for species detection. Two duplex PCR (cattle-buffalo and chicken-pig) were applied with species-specific mitochondrial Cyt b gene primers and determine to be accurate for species identification in meat and milk. Bangladeshi origin beef-labeled products were found to be mixed with buffalo (n = 2) and chicken (n = 5) suggesting up to one third of products might be mislabeled. Such mislabeling would allow these ‘beef’ products to charge a higher price compared with chicken products that command a lower price. Imported meat products were also found mislabeled with buffalo and chicken. Cheese samples, declared as bovine, were found to contain buffalo DNA, and no cattle or buffalo DNA was found in six imported cheese samples. All the meat and cheese products were Halal, as none contained pig DNA. This pilot study shows the majority of products were labelled correctly, but large proportions were not and strict monitoring is recommended to ensure food safety and address consumer preferences, especially religious concerns.
1. Introduction
Food authenticity is a worldwide issue and there is growing awareness and concern about food fraud, adulteration, and authenticity (http://www.fao.org/evaluation/evaluation-digest/evaluations-detail/en/c/1041465/). For more than a decade, species identification for authentication of animal origin foods has received attention from regulatory bodies for ensuring consumer protection. Meat and dairy product adulteration and associated fraud is intentional in most cases, violating consumers’ right and trust and is a matter of great concern for religious groups (choice of Halal for Muslims, and preference for other religious groups), economic, and health aspects (e.g., food allergies). Commonly reported fraudulent practices in the meat and dairy sectors are the use of cheaper or forbidden products to substitute more costly or permitted ones (https://bangladeshpost.net/posts/buffalo-meat-bring-sold-as-beef-in-city-markets-14211).
Authentication of declared species in meat products was highlighted across international markets when horse meat was reported in otherwise labeled meat products by the Food Safety Authority of Ireland and, afterward, pork as an adulterant across Europe (Walker et al., 2013). Horse meat has also been detected in ground meat (mince) sold in the US (Kane & Hellberg, 2016). In Bangladesh, these examples triggered Halal authentication of imported frozen meat products, as an international trade criterion. In the dairy industry, use of cheaper milk is also commonplace, but not always declared. For instance, López-Calleja et al. (2005) reported species adulteration in Mozzarella, which should be made with milk from Italian Mediterranean water buffalo (Bubalus bubalis), identifying the presence of bovine milk (less expensive) as an adulterant/ mislabeling.
Molecular technologies have provided a breakthrough strategy for species detection in food authentication. Use of DNA-based markers and the now universal polymerase chain reaction (PCR) in molecular authentication allow closely related animal species to be identified, and if so desired quantified, in processed food including meat, fish, and cheese (Lo & Shaw, 2018). Although protein-based techniques such as ELISA have been used for species identification, proteins are significantly degraded during processing while DNA is more stable (Asensio et al., 2008). There are some commonly used PCR-based techniques for food forensic analysis, which include restriction fragment length polymorphisms, random amplified polymorphic DNA, and multiplex PCR (Lo & Shaw, 2018). However, multiplex PCR is also well established, where multiple primers are added in a single PCR mixture to amplify several species-specific DNA sequences simultaneously reducing cost and time (Ali et al., 2014).
The effectiveness of multiplex PCR depends on primer design and intra-species conserved regions, such as mitochondrial DNA (mtDNA), are preferable to nuclear DNA targets (Mohamad et al., 2013) simply because they are more specific. In studying molecular traceability of species origin, mitochondrial genes have been targeted because of their abundance per cell and maternal inheritance, which is advantageous for discrimination of closely related species (Murugaiah et al., 2015, Wang et al., 2006). The widely used mtDNA gene for phylogenic study is cytochrome b (Cyt b), which is highly conserved making it suitable for identifying intra- and inter-species ancestry even in processed foods (Murugaiah et al., 2015). Several studies have been performed using multiplex PCR for mitochondrial genes for verification of meat species (Hou et al., 2015, Zarringhabaie et al., 2011) and identification of milk species (Dalmasso et al., 2011, Csikos et al., 2016, López-Calleja et al., 2005). Others report using ribosomal RNA (Song et al., 2019, Golinelli et al., 2014). Besides species identification, Halal authentication (considering pork) has also been studied in some countries, such as Malaysia, Egypt, and Iran (Murugaiah et al., 2009, Chuah et al., 2016; Yakoobie & Sadek, 2016; Doosti et al., 2014).
In Bangladesh, the choices for ready-to-cook frozen food products of both local and imported brands are increasing especially to working individuals due to the convenience, appearance, taste and availability (Sen et al., 2019). Potential adulterant practices include mixing buffalo or chicken meats in burger patties sold as beef (cattle) or the presence of pork in Halal products. Moreover, bovine milk might be mixed with buffalo milk for cheese production (Bottero et al., 2002).
To the best of our knowledge, this is a pioneering investigation of labeled species using PCR in meat products and cheeses sold in Bangladesh. Rapid and sensitive conventional PCR techniques were used for species identification in both domestic and imported frozen meat products and Mozzarella-type cheeses. Meat products, labeled as beef (Bos Taurus) were chosen because beef is costly (∼7 USD). The approach might also be used to identify product that contain beef and are, therefore, not suitable for of the Hindu religion and are adulterated with cheaper meats, such as chicken (1.7–2.5 USD) and buffalo (Bubalus bubalis) (∼4.0 USD). The presence or absence of pork was also tested to determine Halal authenticity. Most of the Bangladeshi cheeses selected were labeled as made from cows’ milk, although some dairy farms also produce buffalo milk. Thus, there is the potential for adulteration, accidental or deliberate, during production. In addition, some imported cheeses did not declare milk species (labelled as fresh milk). Thus, our study aimed to authenticate both meat and milk origins used in selected frozen meat products and cheeses, respectively, based on amplification of species-specific mitochondrial cyt b gene.
2. Materials and methods
2.1. Meat products and mozzarella cheese samples
A total of 64 meat products (40 chicken + 24 beef as labeled) and 25 cheese samples were purchased (Table 1). Frozen ready-to-cook meat (labeled as beef or chicken; sausages, burger patty, meatball, kabab, kofta, and nugget) and Mozzarella-type cheese (i.e., semi-hard shaped, shredded, cubes, and sliced spreadsheets), both domestic (Bangladeshi) and imported, were purchased from grocery and departmental supermarkets in Dhaka (Bangladesh). Several of each item were purchased and analyzed (Table 1). As positive controls, fresh meat (muscle) samples from cows (Bos taurus), buffalo (Bubalus bubalus), chicken (Gallus gallus), and pig (Sus scrofa domesticus) were collected from slaughterhouse. Also, fresh milks from cows and buffalo were collected from a dairy at the Bangladesh Livestock Research Institute (Savar, Dhaka) to positive controls for milk species authentication. All meat, milk, and cheese samples were kept at −20 °C until analysis.
Table 1.
Samples used in this study.
| Sample | Product type | Origin | Species declared |
|---|---|---|---|
| Meat products | Sausages (n = 11), Kabab (n = 8), Burger patty (n = 10), Kofta (n = 9), Nugget (n = 14), Meatball (n = 12) | Bangladesh | Beef (n = 22); Chicken (n = 28) |
| Foreign | Beef (n = 2); Chicken (n = 12) | ||
| Mozzarella cheese | Shredded (n = 3), Cubes (n = 3), Sliced (n = 6), Semi-hard (n = 13) | Bangladesh | Cow milk (n = 12); Not declared (n = 04) |
| Foreign | Cow milk (n = 0); Not declared (n = 09) |
2.2. PCR analysis of meat products
2.2.1. DNA extraction
Total genomic DNA from the fresh meat (beef, buffalo, pork, and chicken) and frozen meat samples were extracted using a Dneasy Blood and Tissue kit (Qiagen, Manchester, UK). Frozen samples were thawed before starting extraction. Genomic DNA extraction was carried out two times using 20–25 mg of homogenized meat, which was placed in a 2 ml microcentrifuge tube. Extractions were achieved following the Dneasy Blood and Tissue kit manufacturer’s protocol. Concentrations and purities of extracted DNAs (ratio of absorbance at 260 nm and 280 nm) were determined using a NanoDrop UV–Vis spectrophotometer (Thermo Fisher Scientific, NanoDrop 2000c, Waltham, MA, USA). DNA was stored at −20 °C before use in PCR amplification.
2.2.2. Duplex PCR of species-specific mitochondrial cyt b gene in meat samples
To amplify species-specific Cyt b genes, reference primers for buffalo (127 bp), chicken (227 bp), pork (398 bp), and beef (472 bp) were used. Primer sequences were selected from reference studies (Table 2) and purchased from Integrated DNA Technologies (Singapore). Conditions for the PCR were ascertained by running authentic meat samples (beef, buffalo, chicken, and pork). Two duplex PCR protocols were selected from different multiplex references: cattle-buffalo (Zarringhabaie et al., 2011) and chicken-pork (Yacoub & Sadek, 2016), and were followed to amplify target species-specific sequences in whole DNA extracts from the meat products.
Table 2.
Species-specific reference primer sequences of mitochondrial Cyt b gene used in this study.
| PCR | Primer | Sequences (5′ to 3′) | product size (bp) | Cyclic conditions | Reference |
|---|---|---|---|---|---|
| Duplex 1 | Chicken-R | AAG ATA CAG ATG AAG AAG AAT GAG GCG | 227 | Pre-Dn (93 °C, 2 min); 35 cycles of Dn (93 °C, 30 s), An (64 °C, 30 s), Ext (72 °C, 45 s); and a final extension at 72 °C for 5 min | Yacoub & Sadek, 2016 |
| Pig-R | GCT GAT AGT AGA TTT GTG ATG ACC GTA | 398 | |||
| Common_F | CCT CCC AGC TCC ATC AAA CAT CTC ATC TTG ATG AAA | ||||
| Duplex 2 | Buffalo-F | TCC TCA TTC TCA TGC CCC TG | 127 | Pre-Dn (94 °C, 3 min); 35 cycles of Dn (94 °C, 45 s), An (62 °C, 1 min), Ext (72 °C, 45 s); and a final extension at 72 °C for 10 min | Zarringhabaie et al., 2011 |
| Cattle-F | TCC TTC CAT TTA TCA TCA TAG CAA | 472 | |||
| Common-R | TGT CCT CCA ATT CAT GTG AGT GT | ||||
Dn = Denaturation; An = Annealing; Ext = Extension.
Briefly, PCR mixtures were prepared in 20 μl volumes containing 1.5 mM of a premixed ready-to-use Go Taq master mix (Promega, Madison, WI, USA), 1 µM of each primer, and 30–40 ng of genomic DNA. PCR assays were carried out using a thermocycler (VWR, UN096, Darmstadt, Germany) according to the conditions summarized in Table 2. Besides samples and positive controls, a negative control (master mix without DNA) was also used as well as a 100 bp DNA ladder (Promega, Madison, WI, USA) to determine the sizes of PCR products. PCR products were run in 2% agarose gel (Agarose Standard ROTI®Garose, Roth, Karlsruhe, Germany) electrophoresis stained with SYBR Safe DNA gel stain (Invitrogen, Ref:S33102, Lot:1988947, Thermo Fisher Scientific, Carlsbad, CA, USA) at 90 V for 30 min. Amplified fragments were viewed with a gel documentation system (VILBER, Quantum-CX5, Eberhardzell, Germany).
2.3. Analysis of milk (cheese) products
2.3.1. DNA extraction from positive milk and cheese samples
DNA was extracted from fresh cow and buffalo milks as positive controls for the cheeses. Somatic cells present in milk are typically the source of DNA in milk and milk products. DNA was extracted as described by Volk et al. (2014). Briefly, 1 ml of milk was centrifuged at 5000×g, and the upper fatty layer and supernatant discarded. The residual pellet underwent DNA extraction as described earlier (Spin-Column Protocol). Total DNA from cheese samples was extracted using ∼50 mg of cheese using the same protocol. Extractions were done triplicate of the same cheese to ensure representative sampling. The quality of the total DNA extracts were determined as previously described. Extracts were kept at −20 °C prior to amplification.
2.3.2. Duplex PCR of species-specific mitochondrial Cyt b gene in cheese samples
Since Mozzarella-type cheeses are usually produced from cattle or buffalo milks, the protocol for duplex 2 (Table 2) was used to amplify species-specific Cyt b genes for buffalo (127 bp) and cattle (472 bp). First, two separate PCRs were run with known milk samples (cow and buffalo milks). Then, duplex procedure 2 (Table 2) was applied for a mixture of cow and buffalo milks to establish the protocol for authentication of cheeses in this study. PCR mixtures were prepared as described previously for meat samples, and the conditions as described for duplex 2 PCR (cattle-buffalo). All imported cheese products were also tested with the pig primer for Halal authentication. Amplified fragments were identified as described previously.
3. Results and discussion
3.1. Setting of duplex PCR for meat and milk species
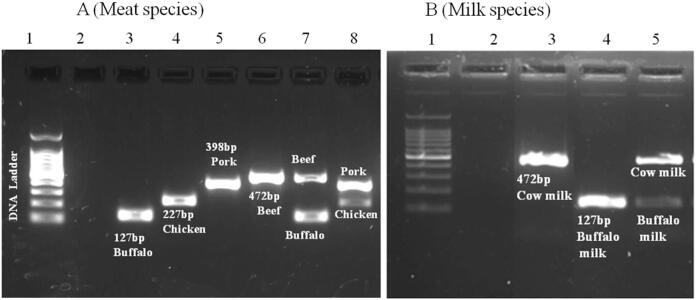
Amplifications of species-specific mitochondrial Cyt b genes of the four known meat samples (beef, buffalo, chicken, and pork) were clearly demarcated (Fig. 1; lane 3–6), indicating that primers and protocols were well suited for their identification in unknown origin samples. The duplex PCR of known meat samples produced clear bands proving the amplification was successful (Fig. 1; lane 7, 8). Likewise, the use of primers for beef and buffalo confirmed correctly the sources of know milk samples (Fig. 1B; lane 3–5). Thus, the selected mitochondrial Cyt b gene primers can be used as reference primers for samples of unknown origins in the authentication of cheese or other milk products.
Fig. 1.
Electrophoresis gel bands for simplex and duplex PCR of meat species (A) and milk species (B) as positive control. Lane indications for A-1: Ladder 100 bp, 2: Negative control, 3: Buffalo(+), 4: Chicken(+), 5: Pork(+), 6: Beef(+), 7: Duplex beef + buffalo, 8: Duplex pork + chicken. Lane indications for B-1: Ladder 100 bp, 2: Negative control, 3: Cow milk(+), 4:Buffalo milk(+), 5: Duplex cow milk + buffalo milk.
3.2. Authentication of ready-to-cook frozen meat products by species identification
Authenticity of the meat products was tested for all four species (beef, buffalo, chicken, and pork) to determine the accuracy of labels. The results are summarized in Table 3, which shows the predominant (as labeled) and adulterant species with percent. Bangladeshi branded chicken products (n = 28) contained only chicken, as labeled. No beef or buffalo DNA was found, which was as expected since chicken is cheaper than beef or buffalo in Bangladesh. However, some imported chicken products were mixed with buffalo meats (n = 8, Table 3). These mislabeled chicken products were made in Malaysia, as stated on the packaging, and this result supports previous studies on mislabeling of poultry products sold in Malaysia, which were also mixed with buffalo meat (Chuah et al., 2016). This type of adulteration is illegal in respect to international trade and, therefore, is a regulatory concern for both national authorities and importers.
Table 3.
Results of identified meat species in studied meat products (n = 64).
| Product origin | Species labeled | Detection object |
% of mislabeling | |||
|---|---|---|---|---|---|---|
| Beef | Buffalo | Chicken | Pork | |||
| Bangladesh | Beef (n = 22) | 22** | 2* | 5* | 0 | 31.82 |
| Chicken (n = 28) | 0 | 0 | 28** | 0 | 0 | |
| Foreign (Imported) | Beef (n = 2) | 2* | 2** | 0 | 0 | 100 |
| Chicken (n = 12) | 0 | 8* | 12** | 0 | 66.67 | |
Principal species (mostly present).
Mixed component (minor present).
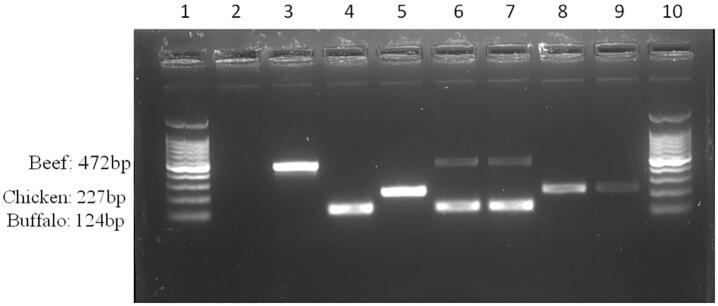
Adulteration or mislabeling was found in nine (out of 24) beef-labeled products. Chicken and buffalo DNA was detected in beef products of Bangladesh origin (Table 3). It is assumed this might be because buffalo and chicken are cheaper than beef. Moreover, chicken is widely available and easy to process. Among the meat samples from Bangladesh (n = 22), five beef ‘burger patty’ were adulterated with chicken (Fig. 2, Lane 8, 9), and two beef ‘kebab’ were mixed with buffalo meat. Only two foreign beef products (burger patty) were mixed with buffalo meat, as determined by duplex beef-buffalo PCR (Fig. 2, Lane 6, 7). The bands (Fig. 2, Lane 6, 7) that are parallel with buffalo positive indicate that buffalo meat was the principal component while beef was only a minor ingredient in the imported burger patty. Incidences of adulteration in beef products have been found in other reports. In Malaysia, Chuah et al. (2016) identified buffalo and chicken DNA in beef products, such as sausages, cold cuts, meatballs, and ground meats while Naaum et al. (2018) detected undeclared pork species in beef sausage products in Canada. Duck is the most common undeclared species in ham sausages produced in China with ca. quarter mislabeled (Song et al., 2019).
Fig. 2.
Electrophoretic gel band of mislabeled meat species found in the present study. The lane indications are – 1: Ladder 100 bp; 2: Negative control; 3: Beef(+); 4: Buffalo(+); 5: Chicken(+); 6,7:Imported beef burger patty; 8,9:Bangladeshi brand beef burger patty; 10: Ladder 100 bp.
Occurrences of undeclared species might be intentional for profit, by mixing the cheaper species with more expensive products, such as chicken (low cost) with beef (high cost), or unintentional cross-contamination during processing. In both beef- and chicken-labeled imported products, buffalo DNA was detected, which might be intentional, due to lower prices in these countries. Other potential sources of these adulterants include mixing of ‘waste’ parts from several species, exploited for more expensive products, rendering them unrecognizable as one species or another, which are used for cheaper products. Whatever the reason, labeling should be correct and followed domestic and international food law to ensure food safety and quality.
3.3. Authentication of milk species in mozzarella cheese
Dairy products are important sources of proteins and fat for human nutrition but allergic reactions, especially among children, are relatively common (Odedra, 2015, Flom and Sicherer, 2019). Therefore, substitution or adulteration of milk or milk products with undeclared species raises the potential for food safety concerns. The present study aimed to detect possible adulteration or mislabeling of commercially available cheeses of both Bangladesh and foreign origins. The optimized duplex PCR assay worked well for discriminating cow or buffalo milks in Mozzarella-type cheeses. Results for the 16 cheese products of Bangladeshi origin (Table 4) showed the presence of cattle DNA (cows’ milk) in 12 samples, two contained buffalo milk and both cattle and buffalo DNAs were found in two cheese samples. These results indicate mislabeling practices in cheese products from Bangladesh. Thus it is a food preference concern because buffalo milk is not likely consumed in this subcontinent region. Moreover, anaphylactic or allergic reaction (angioedema) been reported after consuming buffalo mozzarella (Herz and Kopp, 2020, Broekaert et al., 2008, Baha et al., 2017). According to the Food and Agriculture Organization of United Nations (FAOSTAT, 2013–2015, Handford et al. 2016) India, Pakistan, and China are recognized as three of the main producers of cows’ and buffalo milks in Asia and the purity of Asian cows’ milk is in doubt, as there is evidence it is often mixed with buffalo milk. Cottenet et al. (2011) detected cow and buffalo species in milks from China, India, and Pakistan using multiplex real-time PCR and reported that approximately 20% (mainly from India) of cows’ milk contain contained buffalo milk, and vice versa.
Table 4.
Summery results of milk species identification in Mozzarella cheese samples (n = 25).
| Product origin | Species labeled | Detection object |
||||
|---|---|---|---|---|---|---|
| Cattle | Buffalo | Cattle + Buffalo | Pork | No DNA | ||
| Bangladesh | Cow milk (n = 12) | 8** | 2** | 2* | 0 | 0 |
| Not declared (n = 04) | 4 | 0 | 0 | 0 | 0 | |
| Foreign (Imported) | Not declared (n = 09) | 2** | 0 | 1* | 0 | 6 |
Principal species (mostly present).
Mixed component (minor present).
Regarding the imported cheeses (9), there were no declared species (labeled as ‘fresh milk’). The duplex PCR assay detected the presence of cows’ milk in two samples, and one sample was identified as containing both cow and buffalo DNA (Table 4). No cow or buffalo DNA was found in the remaining six imported cheese samples, which suggests the presence of other milk species (e.g., sheep or goats, Table 4). Several researchers have identified undeclared milk species in cheeses of different countries, such as Brazil (Golinelli et al., 2014), Italy (Di Pinto et al., 2004), and Iran (Zarei et al., 2016). Simultaneous detection of milk species using multiplex PCR is, thus, feasible in commercial cheeses and for confirming authenticity of milk products.
3.4. Halal authentication of meat products and Mozzarella cheese
The major religion in Bangladesh is Islam (88.23%) and, for Muslims, Halal authentication important. In this study, meat and cheese samples were investigated for the presence of pig DNA, because gelatin is used to create and maintain a creamy texture in some products and about 80% of edible gelatin is produced from pig skin. Demirhan et al. (2012) determined porcine DNA in gelatin-containing processed food products previously, so the authors also investigated whether there was pig DNA in the imported cheese samples. PCR results showed that pig DNA was absent in the meat and cheese samples, thus confirming the Halal authenticity of all Mozzarella-type cheeses and ready-to-cook meat products available in Bangladesh markets currently.
This is a significant finding from the religious point-of-view as well as reassuring for international Halal markets. Bangladesh has a huge scope for meat export around the world and a share of the global Halal meat market, and emerging meat industries are now exporting meat on a small-scale (around 12–25 tons a month) to Middle East countries and the Maldives (https://www.theindependentbd.com/post/212545). Although the samples sizes were limited, this study could serve as a preliminary report on the Halal sanctity of meat and cheese products produced in Bangladesh. Moreover, the methods that have been established in this study can be adopted to assure product quality.
4. Conclusion
Authentication of animal species origins of foods is vital, not only for the prevention of fraud but also human health and religious concerns. The present study showed that duplex PCR with mitochondrial Cyt b gene is feasible for meat and milk species authentication in processed meat and milk products. Approximately 32% mislabeling was found in beef-labeled Bangladeshi products, perhaps because beef is a more economically valuable. Foreign origin meat products were adulterated/ mislabeled with buffalo meat. Cows’ cheese was also found to be adulterated with buffalo milk. Reassuringly, no pig DNA was found in any meat and cheese samples.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The research work was supported by International Atomic Energy Agency through the implementation of the Technical Cooperation Project (BGD 5032).
References
- Asensio L., González I., Pavón M.A., García T., Martín R. An indirect ELISA and a PCR technique for the detection of Grouper (Epinephelus marginatus) mislabeling. Food Additives & Contaminants: Part A. 2008;25(6):677–683. doi: 10.1080/02652030701765731. [DOI] [PubMed] [Google Scholar]
- Ali M.E., Razzak M.A., Hamid S.B.A. Multiplex PCR in species authentication: probability and prospects - A review. Food Analytical Methods. 2014;7(10):1933–1949. doi: 10.1007/s12161-014-9844-4. [DOI] [Google Scholar]
- Bottero M.T., Civera T., Anastasio A., Turi R.M., Rosati S. Identification of cow's milk in “buffalo” cheese by duplex polymerase chain reaction. Journal of Food Protection. 2002;65(2):362–366. doi: 10.4315/0362-028x-65.2.362. [DOI] [PubMed] [Google Scholar]
- Broekaert S.M., Darsow U., Ollert M., Ring J., Krause I., Schulmeister U., Spitzauer S., Valenta R. Anaphylactic shock caused by buffalo's mozzarella cheese. Annals of Allergy, Asthma & Immunology. 2008;101(1):105–107. doi: 10.1016/S1081-1206(10)60844-7. [DOI] [PubMed] [Google Scholar]
- Baha S., Bansal R., Bansal A. Second reported case of buffalo milk protein allergy without reactivity to cow’s milk. Journal of Allergy and Immunology. 2017;1:005. [Google Scholar]
- Cottenet G., Blancpain C., Golay P.-A. Simultaneous detection of cow and buffalo species in milk from China, India, and Pakistan using multiplex real-time PCR. Journal of Dairy Science. 2011;94(8):3787–3793. doi: 10.3168/jds.2011-4195. [DOI] [PubMed] [Google Scholar]
- Chuah L., He X.B., Effarizah M.E., Syahariza Z.A., Shamila-Syuhada A.K., Rusul G. Mislabelling of beef and poultry products sold in Malaysia. Food Control. 2016;62:157–164. doi: 10.1016/j.foodcont.2015.10.030. [DOI] [Google Scholar]
- Csikos A., Hodzic A., Pasic-Juhas E., Javor A., Hrković-Porobija A., Goletic T., Gulyas G., Czegledi L. Applicability and sensitivity of PCR-SSCP method for milk species identification in cheese. Acta Alimentaria. 2016;45(1):69–76. doi: 10.1556/066.2016.45.1.9. [DOI] [Google Scholar]
- Doosti A., Ghasemi D.P., Rahimi E. Molecular assay to fraud identification of meat products. Journal of Food Science and Technology. 2014;51(1):148–152. doi: 10.1007/s13197-011-0456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pinto A., Conversano M.C., Forte V.T., Novello L., Tantillo G.M. Detection of cow milk in buffalo “mozzarella” by polymerase chain reaction (PCR) assay. Journal of Food Quality. 2004;27:428–435. doi: 10.1111/j.1745-4557.2004.00662.x. [DOI] [Google Scholar]
- Dalmasso, A., Civera, T., La Neve, F., & Bottero, M. T. (2011). Simultaneous detection of cow and buffalo milk in mozzarella cheese by Real-Time PCR assay. Food Chemistry, 124(1), 362-366. http://doi.org/10.1016/j.foodchem.2010.06.017.
- Demirhan Y., Ulca P., Senyuva H.Z. Detection of porcine DNA in gelatine and gelatine-containing processed food products-Halal/Kosher authentication. Meat Science. 2012;90(3):686–689. doi: 10.1016/j.meatsci.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Flom J.D., Sicherer S.H. Epidemiology of cow's milk allergy. Nutrients. 2019;11(5):1051. doi: 10.3390/nu11051051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinelli L.P., Carvalho A.C., Casaes R.S., Lopes C.S., Deliza R., Paschoalin V.M., Silva J.T. Sensory analysis and species-specific PCR detect bovine milk adulteration of frescal (fresh) goat cheese. Journal of Dairy Science. 2014;97(11):6693–6699. doi: 10.3168/jds.2014-7990. [DOI] [PubMed] [Google Scholar]
- Handford C.E., Campbell K., Elliott C.T. Impacts of Milk Fraud on Food Safety and Nutrition with Special Emphasis on Developing Countries. Comprehensive Reviews in Food Science and Food Safety. 2016;15(1):130–142. doi: 10.1111/1541-4337.12181. [DOI] [PubMed] [Google Scholar]
- Hou B., Meng X., Zhang L., Guo J., Li S., Jin H. Development of a sensitive and specific multiplex PCR method for the simultaneous detection of chicken, duck and goose DNA in meat products. Meat Science. 2015;101:90–94. doi: 10.1016/j.meatsci.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Herz A., Kopp M.V. Anaphylactic reaction at a pizzeria in a 13-year-old female patient. Allergo Journal International. 2020;29(5):165–167. [Google Scholar]
- Kane D.E., Hellberg R.S. Identification of species in ground meat products sold on the U.S. commercial market using DNA-based methods. Food Control. 2016;59:158–163. doi: 10.1016/j.foodcont.2015.05.020. [DOI] [Google Scholar]
- López-Calleja I., Alonso I.G., Fajardo V., Rodríguez M., Hernández P., García T., Martín R. PCR detection of cows’ milk in water buffalo milk and mozzarella cheese. International Dairy Journal. 2005;15(11):1122–1129. [Google Scholar]
- Lo Y.T., Shaw P.C. DNA-based techniques for authentication of processed food and food supplements. Food Chemistry. 2018;240:767–774. doi: 10.1016/j.foodchem.2017.08.022. [DOI] [PubMed] [Google Scholar]
- Murugaiah C., Noor Z.M., Mastakim M., Bilung L.M., Selamat J., Radu S. Meat species identification and Halal authentication analysis using mitochondrial DNA. Meat Science. 2009;83:57–61. doi: 10.1016/j.meatsci.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Murugaiah C., Al-Talib H., Radu S. Forensics: Food authentication using MtDNA. Journal of Nutritional Health & Food Science. 2015;3(4):1–10. [Google Scholar]
- Mohamad N.A., El Sheikha A.F., Mustafa S., Mokhtar N.F.K. Comparison of gene nature used in real-time PCR for porcine identification and quantification: A review. Food Research International. 2013;50(1):330–338. doi: 10.1016/j.foodres.2012.10.047. [DOI] [Google Scholar]
- Naaum A.M., Shehata H.R., Chen S., Li J., Tabujara N., Awmack D.…Hanner R. Complementary molecular methods detect undeclared species in sausage products at retail markets in Canada. Food Control. 2018;84:339–344. doi: 10.1016/j.foodcont.2017.07.040. [DOI] [Google Scholar]
- Odedra K. M. (2015). Milk allergy in adults and children. Nursing Standard (Royal College of Nursing (Great Britain): 1987), 29(44), 43-48. https://doi.org/10.7748/ns.29.44.43.e9729. [DOI] [PubMed]
- Sen S., Antara N., Sen S. Factors influencing consumers’ to Take Ready-made Frozen Food. Current Psychology. 2019 doi: 10.1007/s12144-019-00201-4. [DOI] [Google Scholar]
- Song Q., Chen Y., Zhao L., Ouyang H., Song J. Monitoring of sausage products sold in Sichuan Province, China: A first comprehensive report on meat species’ authenticity determination. Scientific Reports. 2019;9:19074. doi: 10.1038/s41598-019-55612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk H., Piskernik S., Kurinčič M., Klančnik A., Toplak N., Jeršek B. Evaluation of different methods for DNA extraction from milk. Journal of Food and Nutrition Research. 2014;53(2):97–104. [Google Scholar]
- Walker M.J., Burns M., Burns D.T. Horse meat in beef products-species substitution 2013. Journal of the Association of Public Analysts. 2013;1:67–106. [Google Scholar]
- Wang X., Ma Y.H., Chen H. Analysis of the genetic diversity and the phylogenetic evolution of Chinese sheep based on Cyt b gene sequences. Yi Chuan Xue Bao. 2006;33(12):1081–1086. doi: 10.1016/S0379-4172(06)60145-5. [DOI] [PubMed] [Google Scholar]
- Yacoub H.A., Sadek M.A. Identification of fraud (with pig stuffs) in chicken-processed meat through information of mitochondrial cytochrome b. Mitochondrial DNA Part A. 2016;28(6):855–859. doi: 10.1080/24701394.2016.1197220. [DOI] [PubMed] [Google Scholar]
- Zarringhabaie G.E., Pirany N., Javanmard A. Molecular traceability of the species origin of meats using multiplex PCR. African Journal of Biotechnology. 2011;10(73):16461–16465. doi: 10.5897/AJB11.1250. [DOI] [Google Scholar]
- Zarei M., Maktabi S., Yousefvand A., Tajbakhsh S. Fraud Identification of Undeclared Milk Species in Composition of Sheep Yogurt and Cheese Using Multiplex PCR Assay. Journal of Food Quality and Hazards Control. 2016;3(1):15–19. http://jfqhc.ssu.ac.ir/article-1-225-en.html [Google Scholar]
- Bangladesh Food Safety Cluster Evaluation. (2017). http://www.fao.org/evaluation/evaluation-digest/evaluations-detail/en/c/1041465. Accessed March 07, 2021.
- Buffalo meat bring sold as beef in city markets. (2019). https://bangladeshpost.net/posts/buffalo-meat-bring-sold-as-beef-in-city-markets-14211. Accessed March 07, 2021.
- Bangladesh now plans large-scale meat export. (2019). https://www.theindependentbd.com/post/212545. Accessed March 07, 2021.