Abstract
The colonic microbiome has been implicated in the pathogenesis of colorectal cancer (CRC) and intestinal microbiome alterations are not confined to the tumour. Since data on whether the microbiome normalises or remains altered after resection of CRC are conflicting, we studied the colonic microbiota of patients after resection of CRC. We profiled the microbiota using 16S rRNA gene amplicon sequencing in colonic biopsies from patients after resection of CRC (n = 63) in comparison with controls (n = 52), subjects with newly diagnosed CRC (n = 93) and polyps (i = 28). The colonic microbiota after surgical resection remained significantly different from that of controls in 65% of patients. Genus-level profiling and beta-diversity confirmed two distinct groups of patients after resection of CRC: one with an abnormal microbiota similar to that of patients with newly diagnosed CRC and another similar to non-CRC controls. Consumption levels of several dietary ingredients and cardiovascular drugs co-varied with differences in microbiota composition suggesting lifestyle factors may modulate differential microbiome trajectories after surgical resection. This study supports investigation of the colonic microbiota as a marker of risk for development of CRC.
Graphical Abstract
Graphical Abstract.

Retention after surgery of a colon-cancer related gut microbiome correlates with diet.
INTRODUCTION
Colorectal cancer (CRC) remains one of the major causes of mortality (1). Hereditary cancers account for a minority of cases while environmental and lifestyle factors are the main driver of sporadic CRC (2). The pathogenesis of most cases of sporadic CRC is thought to involve a series of genetic mutations (3), progressing through an adenoma-carcinoma sequence (4),(5). While CRC has been linked to individual pathogens such as (pks+) Escherichia coli (6), Fusobacterium nucleatum (7) and Streptococcus gallolyticus (8), it is probably driven by microbial metabolites and their interaction with the host (9). The microbiota profiles in patients with adenomatous polyps and colorectal tumors differ from those of healthy controls (10–15). It is also noteworthy that changes in the microbiota in CRC occur throughout the colon and are not limited to the tumor itself (13,15). Moreover, similar networks of biofilm-forming bacteria are found on oral and gut mucosa (15) in patients with CRC.
While the presence of colonic adenomatous polyps is a risk factor for developing a new CRC, another high-risk group are those who have previously had CRC. However, there are limited data on which patients are at greatest risk of developing a new CRC after the resection of a primary CRC. In particular, the potential role of the microbiome as a marker of risk of developing a new CRC has received limited attention. We (16) and others (17),(18) have shown persistence of alterations post-resection of CRC in some patients which suggests that the microbiome may have predictive value in this setting, but results have not been uniform (19),(20). However, earlier studies were conflicting, relatively small, with a short interval since surgery and did not identify the relative proportions of patients for whom the microbiome either reverted toward normal or remained abnormal. Therefore, we have assessed the microbiome in patients after surgical clearance of CRC in comparison with that of apparently healthy controls, patients with adenomatous polyps and those with newly diagnosed CRC.
MATERIALS AND METHODS
Patient recruitment and sampling
The study population included 63 patients post-CRC removal who were recruited during surveillance endoscopy (Table 1 and Supplementary File S1). Also recruited were 18 patients with a new diagnosis of CRC. For comparative analyses we included additional data previously published by us (15). These included an additional 75 patients with CRC giving a total of 93 patients newly diagnosed with CRC as a comparator group for patients after resection for CRC (Table 1 and Supplementary File S1). Also included were 28 individuals who had undergone polypectomy (15) (Table 1 and Supplementary File S1). While controls included 58 individuals who had no previous medical condition (non-CRC) (15) (Table 1 and Supplementary File S1). This study was conducted in accordance with the ethical principles set forth in the current version of the Declaration of Helsinki, the International Conference on Harmonization E6 Good Clinical Practice (ICH-GCP). Eligible patients were identified from case lists the day prior to endoscopy. Patients post-CRC removal were eligible for inclusion if they were undergoing surveillance endoscopy after colonic resection for CRC provided that the original diagnosis could be verified by consultation of the patients’ records, radiological data and histology reports. Only patients with sound clinical, radiological or pathological evidence of previous CRC with subsequent surgery were eligible. Patients post-CRC removal were ineligible for inclusion in this study if they suffered from a medical condition, due to which, in the opinion of the investigator, their participation could put them at increased risk of ill health (e.g. blood clotting disorder). Patients were also ineligible if they were receiving an experimental drug or were in the process of partaking in another clinical trial. Ethical approval was granted by The Clinical Research Ethics Committee of the Cork Teaching Hospitals (Cork, Ireland) under study number APC089 and is specifically applicable to the experiments reported in this paper. The study was conducted from July 2017 to January 2019. Overall, we collected data for each patient covering a range of clinical parameters including history of treatment, tumor stage, tumor location, the duration of time since surgery as well as drug treatment. Data pertaining to habitual diet were also collected using a Food Frequency Questionnaire (FFQ).
Table 1.
Overview of study population
| Patients (n) | Samples | Male (%) | Age (mean + SD) | |
|---|---|---|---|---|
| non-CRC | 58 | 120 | 43.1 | 53.1 ± 13.5 |
| Polyp | 28 | 66 | 71.4 | 59.8 ± 14.1 |
| CRC | 93 | 157 | 67.7 | 67.9 ± 11.5 |
| Post-Operative | 63 | 108 | 58.7 | 64.5 ± 12.1 |
For patients undergoing post-operative colonoscopic surveillance, two 1-2 mm biopsies were taken from each patient, one 10 cm proximal and one 10 cm distal to the anastomosis. For some patients who underwent ultra-low anterior resection, or abdominoperineal resection there was insufficient residual tissue to obtain a distal sample. In these cases, a proximal sample alone was taken. If the patient had a stoma with a rectal stump in situ, the proximal sample was taken 10 cm proximal to the stoma entrance and the distal sample was taken in the rectum. In patients with CRC, a biopsy was taken of the tumor with two further samples taken 10 cm proximal and distal to the diseased tissue. Proximal samples were taken first. Biopsy forceps were rinsed with normal saline between each biopsy taken. A disposable sterile needle was used to peel the biopsy off the forceps. Biopsies were placed in empty sterile biobank tubes and stored at −80°C until processing. For patients recruited to the Flemer et al. 2018 study, colonic biopsies were collected as previously described (15) and stored in 3 mL RNAlater, stored at 4°C for 12 h and then stored at −20°C.
DNA Extraction and 16S rRNA amplicon sequencing
Genomic DNA were extracted using the AllPrep DNA kit from Qiagen as described elsewhere (13,15). When preparing each sample, approximately 20 mg tissue was dissected in small fragments from the biopsy and pooled. These pooled fragments were then added to a bead beating tube containing sterile beads and 600 μl of buffer RLT plus was added. Samples were then homogenised for two 15 s at full speed pulses in a MagnaLyzer (Roche, Penzberg, Germany) with rests on ice between pulses. The rest of the DNA extraction was carried out according to the Qiagen AllPrep DNA/RNA extraction kit. Total community DNA underwent 16S rRNA gene PCR. The 16S rRNA gene was amplified using primers for the V3-V4 region; forward (S-D-Bact-0341-b-S-17) and reverse, (S-D-Bact-0785-a-A-21) (21). The PCR thermocycler protocol was as follows: Initiation step of 98°C for 3 min followed by 30 cycles of 98°C for 30 s, 55°C for 60 s, and 72°C for 20 s, and a final extension step of 72°C for 5 min. Oligonucleotide indices were subsequently added to amplicons according to the Illumina 16S Metagenomic Sequencing Protocol (Illumina, CA, USA). Library DNA concentration was quantified using a Qubit fluorometer (Invitrogen) using the High Sensitivity assay and samples were pooled at a standardised concentration. It is important to note that identical DNA extraction and 16S PCR protocols (including all reagents, kits and primers) were used for the Flemer et al. 2018 study data included in our analysis. For both studies the pooled library was sequenced at Eurofins Genomics/GATC Biotech (Konstanz, Germany) on the Illumina MiSeq platform. The current study samples were sequenced utilizing 2×300 bp chemistry while the Flemer et al., 2018 samples were sequenced utilizing 2×250 bp chemistry.
Bioinformatic and biostatistical analysis
Supplementary File S1 provides a complete description of the methodology used for the bioinformatics and biostatistical analysis in this study. Specifically, this includes the methods used for pre-processing of 16S amplicon reads and taxonomic classification (22–26), identification of significant differences in microbiota composition through alpha (α) diversity (Shannon Index) (27), beta (ß) diversity (Bray-Curtis dissimilarity) (28), linear regression, analysis of compositions of microbiome with bias correction (ANCOMBC) (29) and Co-Abundance Group (CAG) analysis (15). Supplementary File S2 also gives a complete description of the methodology used to validate our findings in several previously published datasets as well as associating the microbiome in patients after removal of CRC with diet and drug consumption. All statistical analysis was carried out in R (RStudio version 4.1.2)
RESULTS
The colonic microbiome is altered in some patients after removal of colorectal cancer
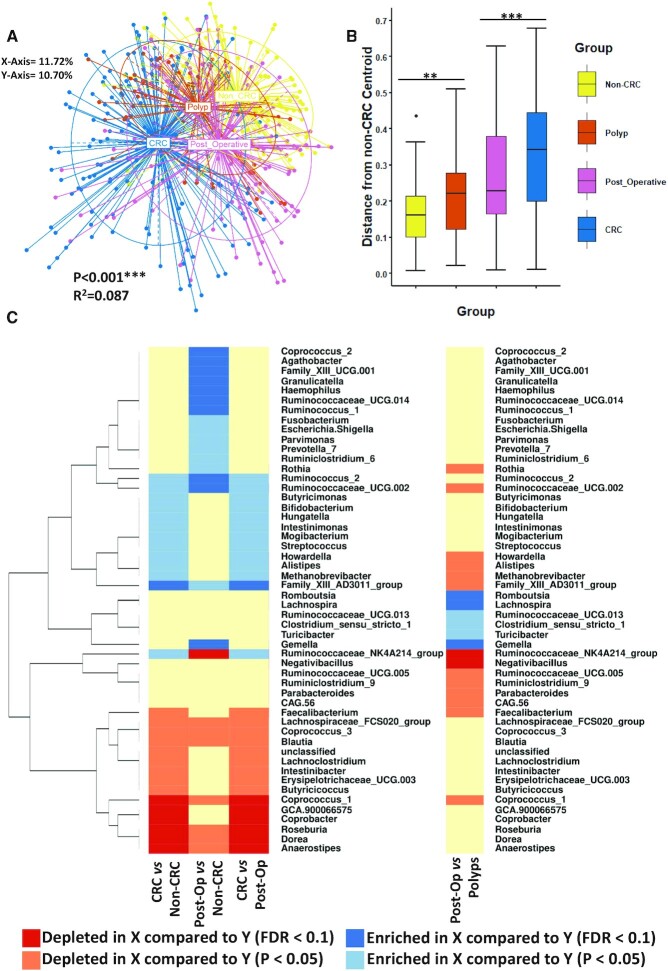
Principal component analysis (PCoA) of ß-diversity (Bray-Curtis dissimilarity) calculated at the genus level revealed statistically significant microbiome separation between the different groups of individuals (non-CRC, patients with newly diagnosed CRC, patients with polyps and patients after resection for CRC) after accounting for the study effect (Figure 1A) (PERMANOVA FDR-corrected P < 0.001: R2 = 0.087). This observation has been previously reported by us and others (10–13). The most extreme differences were observed between patients with newly diagnosed CRC and the non-CRC individuals. In contrast, the microbiomes of patients after surgical resection (as well as patients with polyps), although distinct from the controls, were observed to be closer to the controls than the patients newly diagnosed with CRC. Statistically significant microbiome separation was also observed between the groups at the OTU level (after controlling for the study effect) (PERMANOVA FDR-corrected P > 0.001: R2 = 0.087) (Supplementary Figure S1). Given that the CRC group used in this study contains samples from two different datasets, we compared the microbiome of patients with CRC from both Flemer et al., 2018 study (Flemer_CRC) and the current study (Cronin_CRC) noting that both are significantly different to the non-CRC group at the genus and OTU levels (Supplementary Figure S2A and B). Furthermore, in order to visualize the distances between samples within the respective groups we calculated the median Bray-Curtis distance of each sample from all other samples within in its group (based on Figure 1A) (Supplementary Figure S3). Although all groups were significantly different from one another we observed that samples belonging to those newly diagnosed with CRC had the highest inter-group median Bray-Curtis distances followed by patients after removal of CRC indicating that both groups of patients have higher inter-individual variation in colonic microbiota composition (Supplementary File S3, Supplementary Figure S3). Samples belonging to the non-CRC and polyp groups were observed to have lower inter-group median Bray-Curtis distances indicating these groups of patients have a lower inter-individual variation in microbiota composition compared to the CRC and post-operative groups. To perform a quantitative comparative evaluation of differences in microbiota composition, we next devised a metric that provided a measure of how close a given mucosal microbiota was to that of non-CRC control individuals (based on the Bray-Curtis dissimilarity PCoA coordinates as in Figure 1A). For this purpose, we determined the median PCoA coordinates (median PCoA1 and median PCoA2) corresponding to the microbiomes of all non-CRC individuals and computed the distances of all samples from this point producing a summary statistic for the microbiome distance of all patients from controls (Figure 1B). As expected, the microbiome of patients with newly diagnosed CRC was observed to be the most distinct from those of non-CRC controls (Wilcoxon test FDR corrected P < 0.0001; Figure 1B). The microbiomes of patients who underwent a polypectomy were observed to have the shortest distance from the non-CRC cohort (Wilcoxon test FDR corrected P = 0.006) followed by patients post-resection for CRC (Wilcoxon test FDR corrected P < 0.0001) (Figure 1B; Supplementary File S2). Notably, patients after resection for CRC also had a significantly shorter distance from the non-CRC control group than those with newly diagnosed CRC (Wilcoxon test FDR corrected P < 0.0001; Supplementary File S2). These results indicated that the overall microbiome configurations of post-resection patients were significantly less altered with respect to the non-CRC controls, compared to those newly diagnosed with CRC.
Figure 1.
The colonic microbiome is altered after removal of CRC The microbiota remains altered after removal of CRC. (A) Principal component analysis (PCoA) of Bray-Curtis dissimilarity (ß-diversity) 16S rRNA genus profiles. The PERMAOVA P value (0.001) indicating that there is statistically significant separation between the groups after controlling for the study effect and the patient identifier is shown in addition to the R2 value. The eigenvalues explaining the variation of each axis are expressed as a percentage (X-axis = 11.72%, Y-axis = 10.70%). (B) Based on the PCoA coordinates from (A) the median non-CRC centroid was determined and the distance of all samples from that point was subsequently calculated and shown here as a boxplot. Wilcoxon test (FDR corrected) was used to determine significant differences between the groups for this distance measure. The annotations used for P values are P < 0.05 *; P < 0.01 **; P < 0.001***. (C) Linear regression was used to investigate variations in the prevalence of different genera across the groups after adjusting for study specific variations. Whether a genus was depleted/enriched marginally (P < 0.05) or after FDR correction (<0.1) is annotated by colour while X versus Y is used to explain the directionality of changes in the legend.
Thus, we next investigated these overall differences in microbiome configurations at the increased granularity of the genus level. Given that the samples included in this study were collated from two different data cohorts (the current one and a previous study by Flemer et al., 2018) it was necessary to adjust for the study-specific variations while investigating the genera-specific variations across various individual groups. Consequently, we used linear regression analysis to investigate variations in the abundances of different genera across different groups after adjusting for study-specific variations (Figure 1C). For the analyzed set of samples from patients with newly diagnosed CRC (n = 157 samples) and non-CRC controls (n = 120 samples), a total of 28 genera showed differences in their abundance patterns. Amongst these, 13 were more abundant in patients with newly diagnosed CRC (1 of them significantly with FDR < 0.1 and 12 with P < 0.05). Among the significantly enriched were the typical CRC-enriched markers such as Hungatella and Streptococcus (Figure 1C). Notably, in the microbiome of post-resection patients for CRC the variation with respect to CRC-related genera was reduced. Fusobacterium was more abundant in the post-operative subjects compared to non-CRC controls (P < 0.05). Similarly, several other taxa were significantly elevated in patients after surgical resection compared to non-CRC controls including known pathobionts Parvimonas, Escherichia/Shigella and Fusobacterium (no significant difference was observed with respect to the patients with newly diagnosed CRC). Hungatella and Streptococcus were marginally depleted after removal of CRC with respect to patients newly diagnosed with CRC (while differences with respect to the non-CRC were not significant). Thus, in the microbiome of patients after surgical removal of CRC, there seems to be a gradual (or subtle) reduction in the prevalence of CRC-enriched genera to levels intermediate between patients with newly diagnosed CRC and non-CRC controls. In contrast, the changes with respect to the CRC-depleted group of genera were drastic. Specifically, we observed a sub-group of six CRC-depleted putatively beneficial colonic genera, including Roseburia, Dorea, Coprococcus_1, and Anaerostipes that were significantly enriched in the microbiomes of patients after surgical removal of CRC (all with FDR < 0.1, compared to patients with newly diagnosed CRC) (Figure 1C). These genera have been consistently associated negatively with multiple diseases (30–33). Recognizing that we had applied a non-compositional method (linear regression) to compositional data (relative abundances) for the taxonomic analysis described above, we proceeded to validate our approach by repeating the linear regression analysis using centered-log ratio (clr) transformed genus abundance values and comparing them to the data presented in Figure 1C (using relative abundances) (Supplementary Figures S4,S5 and Supplementary File S4). We found, using clr-transformed genera abundances as an input to the linear regression models, that 46 taxa (compared to 51 for Figure 1C) remained significant in at least one pairwise comparison, constituting a major overlap in the results between methods (Supplementary Figure S4 and Supplementary File S4).
Next in order to further validate the linear regression analysis we applied another method of differential abundance analysis (ANCOMBC) (29) (Supplementary Figures S6 and S7) (Supplementary File S5). We found using the ANCOMBC approach that 39 taxa remained significant in at least one pairwise comparison when compared to the linear regression analysis in Figure 1C further highlighting an overlap between methods (Supplementary Figure S6). In total 25 genera were found to be significantly differentially abundant between the non-CRC and newly diagnosed CRC groups which included several pathobionts (Parvimonas and Escherichia/Shigella). Similar to the linear regression analysis we found the colonic microbiome of patients after removal of CRC to have significantly lower levels of several CRC-enriched pathobionts (such as Streptococcus and Hungatella) (Supplementary Figures S6 and S7) (Supplementary File S5). Meanwhile, we also found several beneficial taxa to be enriched in the post-resection microbiome (including Dorea, Roseburia and Faecalibacterium) when compared to patients newly diagnosed with CRC (Supplementary Figures S6 and S7) (Supplementary File S5). Thus, the microbiome of patients after surgical resection is characterized by an overall enrichment of multiple putatively beneficial taxa compared to patients with newly diagnosed CRC.
Co-Abundance Group (CAG) signatures are specific to patients after resection for CRC
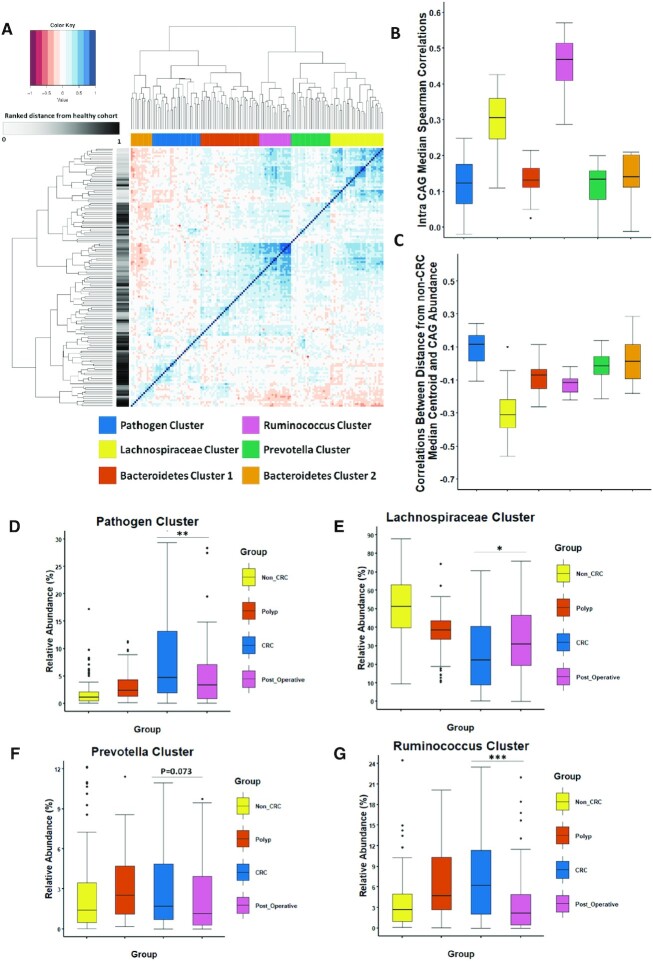
To assess microbiome differences at community level we grouped genera into co-abundance groups (CAGS) using the approach previously reported by us (13, 15) (Supplementary File S2). We also identified six different bacterial CAGs, from all individuals investigated in this study (Figure 2A as reported in Flemer et al. 2017, 2018). These six CAGs (or clusters) were named as the Pathogen cluster, Lachnospiraceae cluster, Bacteroidetes cluster 1, Ruminococcus cluster, Prevotella cluster and Bacteroidetes cluster 2 based on the dominant groups of genera (Figure 2). The genus-level composition of each CAG is shown in Supplementary File S6. Similar to the validation described above for the findings presented in Figure 1C, we repeated the CAG analysis using clr-transformed abundances (Supplementary Figure S8 and Supplementary File S4). Using this approach we identified 6 CAGs each with the same characteristics as the ones described in Figure 2A. We detected a statistically significant similarity between the data derived by the two methods using the adjusted Rand Index (P = 0.001) (as shown in Supplementary Figure S8 and Supplementary File S4).
Figure 2.
Specific Co-Abundance Groups (CAGS) occur after removal of colonic cancer Six different co-abundance groups (CAGs) were identified through hierarchal clustering of Spearman correlations between genera abundances (full methodology described in Supplementary File S2). Patients after surgical resection have a significantly different CAG profile from other groups examined. (A) Heatmap showing the ward.d2 clustering of the spearman correlation coefficients of the relative abundance of genus in the mucosal microbiota of individuals in this study. Each different CAG identified is colour coded by the legend to the left. Row annotation refers to the distance of each sample from the non-CRC group. (B) Boxplot of the Intra Median CAG Spearman Correlations. C) Boxplot of the correlation between the distance from the non-CRC cohort and the CAG abundance. The relative abundance (%) of a number of CAGs was significantly different after surgical resection. Boxplots of four CAGs identified; (D) Pathogen cluster, (E) Lachnospiraceae cluster, (F) Prevotella cluster and (G) Ruminococcus cluster. Kruskal-Wallis followed by Dunns post-hoc test was used to determine significance. Not all significant pairwise comparisons are shown in this Figure. See Supplementary File S6 for complete statistics. The annotations used for P values are P < 0.05 *; P < 0.01 **; P < 0.001***.
The next goal was to examine the robustness and health associations of each CAG. To investigate robustness within each CAG, we computed Spearman correlations between abundances of its various constituent genera. The Ruminococcus cluster followed by the Lachnospiraceae cluster, had the highest observed values of intra-CAG correlations (Figure 2B). Clusters with the highest values have stronger co-occurrence relationships and are generally associated with an un-diseased microbiome. Confirming this hypothesis, both these CAGs have been observed to be negatively associated with CRC in previous studies (Flemer et al. 2017, 2018). To further verify these health associations, for each cluster across the 451 samples, we computed the Spearman correlations between the abundances of the constituent genera with the distance from non-CRC centroid (Figure 1B, Supplementary File S6). The genera belonging to the Lachnospiraceae cluster had the most negative correlations, indicating that the abundances of these genera decreased with increasing distance from non-disease-like microbiome configuration (Figure 2C, Supplementary File S6). Several members of this cluster are also generally associated with functionalities beneficial for the host including fibre fermentation and short-chain fatty acid production. In addition, genera belonging to both the Ruminococcus and (to some extent) Bacteroides cluster 1 also displayed negative correlations indicating their increased association with non-disease microbiome configuration. In contrast, genera belonging to the Pathogen cluster had consistently positive correlations with the distance from the non-CRC centroids (Figure 2C, Supplementary File S6). These results further confirm our previous observations that the Lachnospiraceae cluster and the Ruminococcus cluster are associated with the non-CRC microbiome configuration while the Pathogen cluster is more closely associated with CRC (Figure 2B and C) (Supplementary File S6).
After establishing the health-associations and robustness of the various CAGs, we next compared the abundances of the various CAGs across the four groups of individuals (with a special focus on the post-operative group). We observed several significant differences in the microbiome of patients after removal of CRC (Figure 2D and Supplementary File S6). The Pathogen cluster whose members include CRC-associated taxa such as Fusobacterium and Streptococcus was depleted in patients post-resection compared to patients with newly diagnosed CRC (P = 0.0015) (Figure 2D). Furthermore, the Pathogen CAG had the highest abundance in the newly diagnosed CRC group and the lowest in controls. The putatively-health-associated Lachnospiraceae cluster displayed the highest levels in the non-CRC group and the lowest abundance in patients with newly diagnosed CRC (Figure 2E). The microbiota of patients after removal of CRC displayed significantly higher levels of the Lachnospiraceae cluster compared those with a new diagnosis of CRC (P = 0.019), in line with the observations made in Figure 1C. Contrary to what we expected, the Ruminococcus cluster had the lowest abundance in both non-CRC subjects and patients after resection (Figure 2G). The Ruminococcus abundance values were significantly lower in patients after surgical resection compared to individuals with either newly diagnosed CRC (P = 0) or polyps (P = 0.0001). However, little difference existed between any of the groups for the abundances of Bacteroidetes cluster 1 and 2 (Supplementary Figure S9A,B). The Prevotella cluster which has been previously associated with CRC was detected in lower abundance in post-resection compared with the polyp microbiome (P = 0.013) (Figure 2F) (Supplementary File S6). Thus, CAG analysis reveals significant differences in the microbiome after tumor removal with significant numbers of health-associated microbes (Lachnospiraceae cluster) gained while CRC-associated pathobiont genera are depleted (Pathogen cluster).
The majority of patients after surgical resection retain a CRC-like microbiome
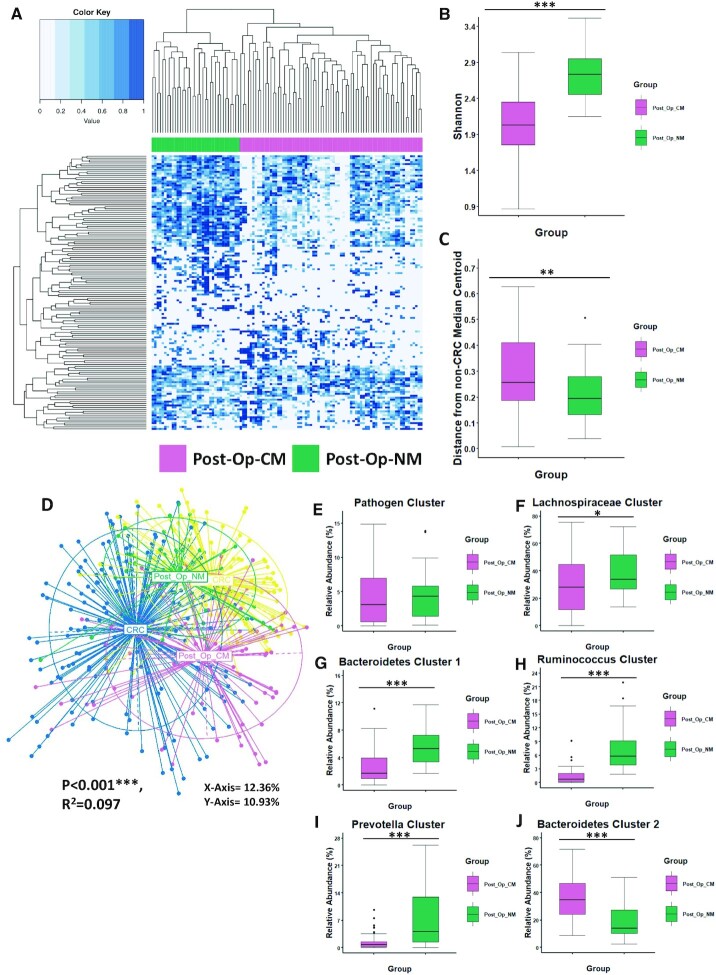
After investigating differences in colonic microbiota composition in patients after removal of CRC (with respect to non-CRC controls, patients with newly diagnosed CRC and patients with polyps), we next focused specifically on the post-operative group. Clustering the biopsy microbiomes of the 63 patients post-resection using the ward.d2 approach at the genus level identified two subject groups with differing genus-level microbiota composition (Figure 3A). The first group of 41 subjects (n = 70 samples) represented 65% of the study cohort while the second group of 22 subjects (n = 38 samples) represented the remaining 35%. We investigated the differential placement of these two sub-groups post-resection with respect to patients with newly diagnosed CRC and the non-CRC controls. PCoA of Bray-Curtis dissimilarity measures revealed significant differences between all groups after controlling for the study effect (PERMANOVA FDR-corrected P < 0.001: R2 = 0.12; Figure 3D). The first group was found to have an abnormal microbiota similar to that of patients with newly diagnosed CRC named Post-Op-CM (Post-Op CRC-like microbiota). The second group had a normal-like microbiota more similar to that of non-CRC controls named Post-Op-NM (Post-Op normal-like microbiota) (Figure 3D). This observation was further evident when comparing the distances of samples belonging to the two sub-groups with the median non-CRC centroid (Figure 3C) as well as by comparing the abundances of the various CAGs (Figure 3E–J). Patients after surgical resection with a microbiota composition most similar to controls (n = 21) were significantly closer to the non-CRC control group than those observed to have an abnormal microbiota (n = 42) (P = 0.008). A visual inspection of the heatmap revealed patients with an abnormal microbiota composition after removal of CRC had lower diversity compared to patients with a normal-like microbiota. This was validated by comparing the genus-level Shannon diversities (Figure 3B). Those with a microbiota composition similar to the control group (Post-Op-NM) had significantly higher Shannon Diversity values than patients with an abnormal microbiota (Post-Op-CM) (P < 0.0001; Figure 3B). For comparison we also wanted to determine the proportion of patients newly diagnosed with CRC who had a microbiota similar to non-CRC controls. Based on the distance from the non-CRC median centroid (as shown in Figure 1B) we calculated the number of patients newly diagnosed with CRC whose samples had a shorter distance than the reported upper quartile for the non-CRC group (0.162172). Using this method, we found that 25% of newly diagnosed CRC patients had a microbiota similar to the non-CRC control group. Thus, these results indicate that a group of patients (65%) after surgical resection have an abnormal microbiota composition similar to that of patients with CRC (Post-Op-CM) while all other patients have a ‘normal’ microbiota composition similar to controls (Post-Op-NM). Thus, despite an overall significant increase in CRC-depleted taxa in patients after removal of CRC compared to patients newly diagnosed with the disease, a majority of patients retained a global microbiota most similar to that of CRC.
Figure 3.
The majority of patients after surgical resection retain a CRC-like microbiome Two groups of patients after surgical resection with differences in microbiota composition were identified. (A) Heatmap showing the genus profiles of all patients after removal of CRC. These groups were identified through ward.d2 clustering as Post-Op-CM (reddish purple) and Post-Op-NM (blueish green) as annotated by legend to the left of the Figure. (B) Boxplot showing significant differences in alpha-diversity (Shannon Index) between the subgroups. (C) Based on the PCoA coordinates from D) the median non-CRC centroid was determined and the distance of all samples from that point subsequently calculated and shown here as a boxplot. (D) Principal component analysis (PCoA) of Bray-Curtis dissimilarity (ß-diversity) 16S rRNA genus profiles. The PERMAOVA P value (0.001) indicates that there is statistically significant separation between the groups after accounting for the student effect and patient identifier. The eigenvalues explaining the variation of each axis are expressed as a percentage (X-axis = 12.36%, Y-axis = 10.93%). (E-J) Comparison of CAG abundance between patients with an abnormal microbiota (Post-Op-CM) and a normal-like microbiota (Post-Op-NM) after removal of CRC. Wilcoxon test (FDR corrected) was used to determine significant differences between the groups. The annotations used for P values are P < 0.05 *; P < 0.01 **; P < 0.001***.
Differential changes in microbiota composition after surgical resection could be replicated in previously published datasets
To validate this observation on a broader scale, we sought to identify a robust CRC-specific microbiome alteration pattern across multiple case-control microbiome datasets investigating CRC. For this purpose, we conducted a meta-analysis of three previously published CRC microbiota datasets obtained from the MicrobiomeHD database (33). These datasets included fecal 16S rRNA sequences for 191 patients with CRC and 202 non-disease controls from France, Canada and the United States (12,34,35). We conducted rank normalisation of genus abundance within each dataset and combined to build a single Random Forest classifier, which in turn was used to identify the top one hundred CRC distinguishing taxa. From this list, we were able to identify seven significant markers which shared the same directionality and whose abundance was highly significantly different between the non-CRC and CRC groups (Table 2). Supplementary File S7 provides a complete description of the methods used to identify these seven microbial markers. Only one of the seven genera was a CRC-depleted taxon namely Anaerostipes (Table 2). The other distinguishing taxa included recognized CRC-associated microbes including Fusobacterium, Peptostreptococcus and Porpyhromonas (Table 2). Interestingly, all seven of these markers had the same directionality between the CRC and non-CRC groups in this study, and also shared the same directionality between patients post-resection with abnormal microbiota (n = 42) and those with a microbiota similar to controls (n = 21) (Supplementary File S7). Thus, this provides further evidence that the majority of patients after removal of CRC have an abnormal microbiota composition similar to patients with CRC (Post-Op-CM) while the remaining patients in this group have a ‘normal’ microbiome similar to non-CRC controls (Post-Op2-NM). Given that many of these taxa like Fusobacterium have been causally implicated with CRC onset, patients after surgical resection with an abnormal microbiota can be hypothesized to be at higher risk of developing a new CRC than those with a microbiota composition similar to the control group.
Table 2.
Seven taxa identified as markers of CRC in three different studies
| Taxon | Directionality | P value |
|---|---|---|
| Anaerostipes | CRC-depleted | 0.002 |
| Eisenbergiella | CRC-enriched | 0.0001 |
| Fusobacterium | CRC-enriched | 0 |
| Hungatella | CRC-enriched | 0.0018 |
| Parvimonas | CRC-enriched | 0 |
| Peptostreptococcus | CRC-enriched | 0 |
| Porphyromonas | CRC-enriched | 0 |
To validate the differences between patients with a ‘normal’ microbiome and an abnormal microbiome groups after surgical resection we identified seven taxa as markers of CRC in three different studies representing individuals from France, America and Canada. Wilcoxon test was used to determine the individual P values for each dataset. Significance of taxa was identified using Fisher's exact test of the P values from each Wilcoxon test. See Supplementary File S2 and S7 for a full description of the methods used to identify these markers.
We next investigated whether the identified differences in microbiota composition in the two post-surgical resection sub-groups identified in the current study could be detected in another post-resection microbiome dataset (36). Thus, we analyzed fecal shotgun metagenomic sequencing data for 40 individuals from Japan who had a history of colorectal surgery (36). We conducted Principal coordinate analysis (PCoA) of the gut microbiomes of these individuals based on Spearman distances obtained from the species profiles. Based on the first two Principal coordinates (PCo1 and PCo2), we observed that even in this geographically distinct cohort, individuals who had surgical treatment for CRC could be classified into two distinct groups with differences in microbiota composition (which we refer to as Yashida Post-Op-CM and Yashida Post-Op-NM) (Supplementary Figure S10A,B). Furthermore, we noted that the Yashida Post-Op-CM group had significantly higher abundances of several CRC-enriched taxa (genera identified in Table 2) (P < 0.03) (Supplementary Figure S10C) (similar to our Post-Op-CM group), and lower abundances of several CRC-depleted taxa including Blautia, Faecalibacterium and Coprococcus (genera identified in Figure 1C) (P < 0.05) (Supplementary Figure S10D). It is interesting to find that two distinct post-resection microbiome groups are also evident in another sample type. Previous work by us (13, 15) and others (37) has identified significant differences between the microbiota composition of faeces and colonic biopsies. Specifically in the case of CRC patients, several genera are known to be significantly differentially abundant in biopsy samples compared to fecal samples (including Fusobacterium and Streptococcus). This analysis further validates that two distinct groups of patient exist after removal of CRC with differences in their colonic microbiota composition independently of sample type.
Association of post-resection microbiome groups with diet and medication
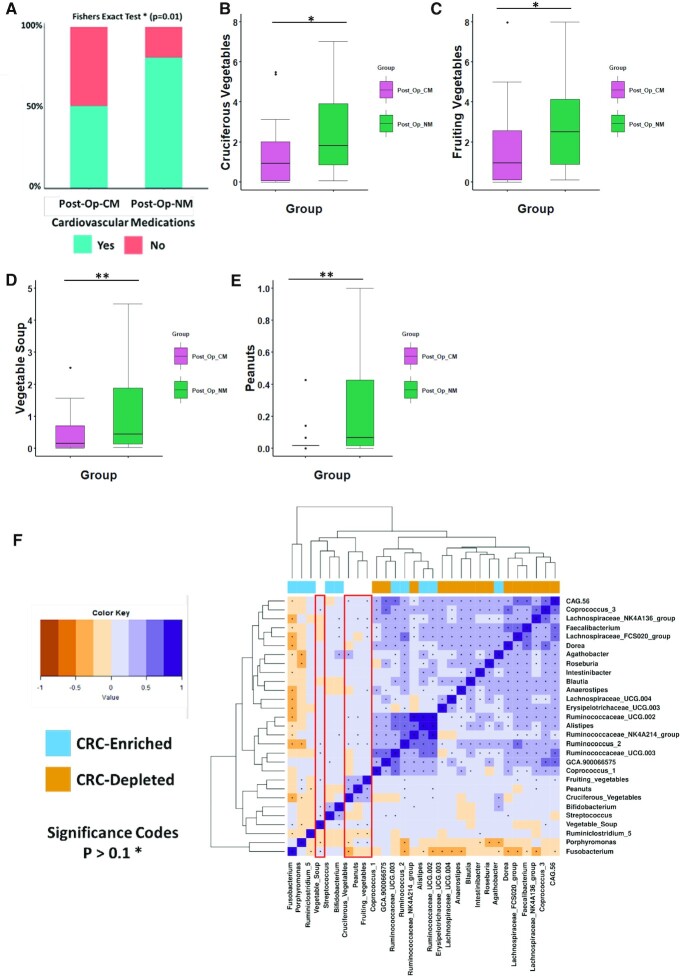
We next sought to identify host-associated or lifestyle factors that covaried with differences in microbiota composition between sub-groups (Post-Op-CM and Post-Op-NM) after removal of CRC. The host-associated metadata available to us for performing these investigations are provided in Supplementary File S8. These include information pertaining to demographics (age and gender), drugs, history of treatment (chemotherapy and radiotherapy), tumour location, tumour stage and habitual diet. We adopted a hierarchical approach, where we first compared the various demographic factors between the two sub-groups post-resection. Gender distribution was significantly different between the two groups, with patients observed to have a ‘normal’ microbiota being predominantly male (P = 0.034) and older in age (P = 0.056) (Supplementary Figure S11A,B). Patients after removal of CRC who had a microbiome similar to controls also had an insignificantly longer duration of time since the surgery. However, this factor had a significant strong positive correlation with age (P = 0.03). Given these observations, we hypothesised that robust associations between the two microbiome-types (high-risk abnormal microbiome versus the ‘normal’ microbiome group) and all the other metadata could only be identified after adjusting for the effects of age, gender and time since surgery. In addition, we also adjusted for the effects of several clinical characteristics of the patient cohort including treatment history (chemotherapy and radiotherapy), tumour stage and tumour location, as these factors have previously been shown to interact with gut microbiota composition (36,38,39). First, we wanted to examine the association of drug treatment with the two colonic microbiome sub-types in patients after removal of CRC. In order to examine a set of ‘informative’ factors we removed drugs which were infrequently consumed (consumed in less than one third of the post-operative group) among patients post-CRC removal. Through logistic regression accounting for age, the duration of time since surgery gender, history of treatment, tumour location and tumour stage as confounding factors we identified a group of cardiovascular drugs as a factor covarying with differential taxa in the microbiome after removal of CRC (Supplementary File S9). This group of drugs specifically includes those used to treat hypertension such as angiotensin II receptor antagonist, beta-blockers and angiotensin-converting enzyme (ACE) inhibitors. We then compared the consumption levels of cardiovascular drugs between the two sub-groups of patients after removal of CRC and found that consumption levels were significantly higher in patients with a normal-like microbiota (Fishers exact test P = 0.01) (Figure 4A). Our logistic regression models also highlighted a negative association with cancer stage, meaning that individuals in Post-Op-CM group tended to have a more advanced stage of cancer (Supplementary File S9). We also identified a weak positive association with the location of cancer. Specifically, we found that patients in the Post-Op-NM group tended to have had a tumour in the descending colon (Supplementary File S9).
Figure 4.
Association of post-resection microbiome groups with diet and medication Three different dietary ingredients and cardiovascular drugs are shown. B)-E) Boxplots of each of the three dietary groups including: B) cruciferous vegetables, C) fruiting vegetables, D) vegetable soup and E) peanuts. Wilcoxon test (FDR corrected) was used to determine significant differences between the groups for the dietary groups. (A) Bar plot showing consumption level of cardiovascular drugs between individuals in the group. Fisher's exact test was to test for significance. The annotations used for P values are P < 0.05 *; P < 0.01 **; P < 0.001***. F) Heatmap showing correlations between specific taxa and four dietary ingredients (cruciferous vegetables, vegetable soup, peanuts and fruiting vegetables). The taxa used in this correlation analysis were identified as the top twenty-five distinguishing taxa between CRC non-CRC groups from a random forest classifier. CRC-enriched genera (sky blue) and CRC-depleted genera (orange) are annotated by the legend to the left. Correlations of interest are outlined in red. Significance was assumed at <0.1*.
Next, we investigated the association between habitual diet and the two microbiome sub-types within the cohort of patients after surgical resection after adjusting for the covariates identified above. To investigate a set of ‘informative’ dietary components, we first removed dietary ingredients that had a small effect size (of difference) in consumption patterns across the two groups of patient post-removal of CRC. For this purpose, we computed the cohen's d on the consumption frequencies of all the individual food items between the groups removing those with an absolute value of less than 0.5 (Supplementary File S9). We built logistic regression models on the remaining set of informative dietary components accounting for the previously mentioned confounders as well as total energy consumption (Table 3). We identified four dietary ingredients as being statistically significant using this approach (Table 3). These included cruciferous vegetables (P = 0.02), fruiting vegetables (P = 0.03), peanuts (P = 0.001) and vegetable soup (P = 0.01) the consumption of which was significantly higher in patients with a normal-like microbiota after removal of CRC (Table 3; Figure 4B–E). Interestingly, it was found that diet as a whole was not significantly different between the groups (PERMANOVA P = 0.195, R2 = 0.1051; Supplementary Figure S12). This indicates that consumption levels of specific dietary ingredients rather than major differences in the overall habitual diet provide the major correlational differences between microbiota profiles after surgical resection.
Table 3.
Specific dietary ingredients co-vary with differences in the microbiota of patients after removal of CRC
| Cruciferous vegetables | Peanuts | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Std | Std | |||||||||
| Estimate | error | Z value | Pr(>Z) | Pr(>Chi) | Estimate | error | Z value | Pr(>Z) | Pr(>Chi) | |
| Intercept | -1.28 | 1.74 | -0.73 | 0.46 | -10.82 | 3.40 | -3.18 | 0.00 | ||
| Patient | 0.04 | 0.02 | 2.18 | 0.03* | 0.14 | 0.03 | 0.02 | 1.66 | 0.10 | 0.14 |
| Sample Location | 0.04 | 0.50 | 0.09 | 0.93 | 0.92 | 0.10 | 0.56 | 0.17 | 0.86 | 0.91 |
| Cancer Stage | -0.78 | 0.45 | -1.74 | 0.08 | 0.04* | -0.86 | 0.48 | -1.80 | 0.07 | 0.04* |
| Cancer Location | 0.93 | 0.57 | 1.61 | 0.11 | 0.02* | 0.11 | 0.65 | 0.18 | 0.86 | 0.02* |
| Total energy consumption | 0.30 | 0.88 | 0.34 | 0.74 | 0.154 | -2.75 | 1.31 | -2.10 | 0.04* | 0.15 |
| Gender | 0.31 | 0.55 | 0.57 | 0.57 | 0.25 | 0.60 | 0.62 | 0.97 | 0.33 | 0.25 |
| Duration since surgery | 0.00 | 0.00 | -0.71 | 0.48 | 0.78 | 0.00 | 0.01 | -0.43 | 0.67 | 0.78 |
| Age | -0.01 | 0.02 | -0.41 | 0.68 | 0.65 | 0.15 | 0.05 | 3.10 | 0.00** | 0.65 |
| Chemotherapy | 0.55 | 0.87 | 0.63 | 0.53 | 0.22 | 1.35 | 0.91 | 1.50 | 0.13 | 0.22 |
| Radiotherapy | -0.36 | 0.68 | -0.53 | 0.60 | 0.44 | -0.58 | 0.83 | -0.70 | 0.49 | 0.44 |
| Dietary group | 0.49 | 0.17 | 2.82 | 0.00** | 0.00** | 10.85 | 2.95 | 3.68 | 0.00** | 0.00*** |
| Q Value | 0.03* | 0.00** | ||||||||
| Vegetable soup | Fruiting vegetables | |||||||||
| Std | Std | |||||||||
| Estimate | error | Z value | Pr(>Z) | Pr(>Chi) | Estimate | error | Z value | Pr(>Z) | Pr(>Chi) | |
| Intercept | -1.05 | 1.85 | -0.57 | 0.57 | -2.36 | 1.73 | -1.36 | 0.17 | ||
| Patient | 0.01 | 0.02 | 0.88 | 0.38 | 0.14 | 0.03 | 0.02 | 1.70 | 0.09 | 0.14 |
| Sample Location | -0.09 | 0.51 | -0.18 | 0.86 | 0.92 | -0.14 | 0.50 | -0.29 | 0.77 | 0.92 |
| Cancer Stage | -0.92 | 0.41 | -2.23 | 0.03* | 0.04* | -0.92 | 0.45 | -2.06 | 0.04* | 0.04* |
| Cancer Location | -1.25 | 0.48 | -2.61 | 0.01** | 0.02* | 1.19 | 1.40 | 2.09 | 0.04* | 0.02* |
| Total energy consumption | 0.00 | 0.00 | 1.09 | 0.28 | 0.15 | -0.89 | 1.00 | -0.89 | 0.37 | 0.15 |
| Gender | 0.45 | 0.56 | 0.81 | 0.42 | 0.25 | 0.57 | 0.54 | 1.05 | 0.29 | 0.25 |
| Duration since surgery | 0.00 | 0.01 | 0.35 | 0.73 | 0.78 | 0.00 | 0.00 | 0.35 | 0.73 | 0.78 |
| Age | 0.01 | 0.02 | 0.23 | 0.82 | 0.65 | 0.02 | 0.02 | 0.71 | 0.48 | 0.65 |
| Chemotherapy | 1.35 | 0.83 | 1.63 | 0.10 | 0.22 | 0.94 | 0.82 | 1.15 | 0.25 | 0.22 |
| Radiotherapy | -0.49 | 0.66 | -0.74 | 0.46 | 0.44 | -0.42 | 0.69 | -0.61 | 0.54 | 0.44 |
| Dietary group | 1.01 | 0.40 | 2.52 | 0.01* | 0.01* | 0.33 | 0.12 | 2.66 | 0.01* | 0.00** |
| Q Value | 0.01* | 0.04* | ||||||||
Logistic regression was conducted on dietary ingredients which had a cohens d value of >0.2. Only the four dietary ingredients determined to be statistically significant are shown. We controlled for several confounding factors including tumour stage, tumour location, treatment history, age, time since surgery, gender total energy consumption. Given some patients had more than one sample available, this was also accounted for as a confounding factor. Refer to Supplementary File S9 to see results for all dietary ingredients analysed. Q value refers to the adjusted Pr(>Z) value obtained for each dietary group. The annotations used for P values are P < 0.05 *; P < 0.01 **; P < 0.001***.
We also wanted to establish what relationship these four food items have with the most important taxa in distinguishing between the newly diagnosed CRC and control microbiomes. In order to do so we conducted Spearman correlations between diet and the top twenty five microbial features from the RF classifier (Figure 4F) (Supplementary File S10) (Supplementary Figure S13). The identification of the top 25 microbial features was also validated by the analysis of clr-transformed genus abundances (Supplementary Figure S14 and Supplementary File S4). We found a number of significant associations between specific dietary ingredients and genera. For example, fruiting vegetable consumption was positively associated with several CRC-depleted genera (Lachnospiraceae_FCS020_group, Lachnospiraceae_NK4A136_group, Faecalibacterium and Anaerostipes) (Figure 4F). Meanwhile vegetable soup consumption was found to negatively associate with CRC-enriched genera (Porphyromonas) while positively associating with the CRC-depleted Coprococcus_3 (Figure 4F). Furthermore, cruciferous vegetable consumption was negatively correlated with the CRC-enriched Fusobacterium and Porphyromonas (Figure 4F). Thus, we observed that higher consumption of cruciferous vegetables, fruiting vegetables, peanuts and vegetable soup in patients with a normal-like microbiome after removal of CRC is positively associated with a primary core group of genera, which are linked to control group microbiota composition. These dietary ingredients were also negatively linked to a small group of taxa which are enriched in CRC.
DISCUSSION
The results confirm that the colonic microbiome remains abnormal in most, but not all, patients after removal of a colorectal cancer which may be a risk factor for development of a new CRC. However, in about one third of patients, the microbiome seems to revert toward normal. Unsurprisingly, consumption of specific dietary ingredients and cardiovascular drugs co-varied with the differences in microbiota composition. Thus, the microbiome may be a biomarker for identifying those patients at greatest risk of developing a new CRC for whom surveillance and interventions should be prioritized.
Differences in the microbiomes of newly diagnosed patients with CRC and those post-resection of CRC are to be anticipated and likely due to several factors including the surgical procedure itself (40,41), pre-operative antibiotics, chemotherapy and radiotherapy (39,42). We found that the microbiome of patients after removal of CRC had higher levels of putatively beneficial colonic genera (including Faecalibacterium, Roseburia, Dorea, Coprococcus, Blautia, Butyricoccus, Lachnoclostridium, and Anaerostipes) alongside lower levels of CRC-associated genera (Hungatella and Streptococcus) when compared to the microbiota composition in CRC patients. This was further confirmed by differences in the prevalence of the fiber-fermenting Lachnospiraceae cluster and the CRC-associated Pathogen cluster. This suggests that changes in both diet and lifestyle may also drive microbiome alterations after surgical resection and that the microbiome is significantly altered in the direction of the non-CRC controls, for a sub-set of the survivors. In previous small-scale studies comparing the microbiota of patients with newly diagnosed CRC with that of patients after surgical removal of the cancer, results seemed conflicting (16–20) but the current study clarifies that while the microbiome remains altered in the majority, it reverts toward normal in about one-third of patients after resection of the cancer.
Our findings were further supported by the identification of the seven CRC markers which validated that the differences between patients with a normal and abnormal microbiome after removal of CRC are analogous to that of the CRC and non-CRC microbiome. In addition, we were able to show that two distinct microbiome subgroups (Yachida Post-Op-CM and Yachida Post-Op-NM) also exist in a cohort of Japanese patients who underwent surgical treatment for CRC. The Yachida Post-Op-CM group had significantly higher abundance of CRC-enriched genera as well as lower levels of CRC-depleted genera when compared to the Yachida Post-Op-NM group providing further evidence that the colonic microbiota remains abnormal in most, but not all, patients after removal of CRC.
Of note, consumption levels of several dietary ingredients and cardiovascular drugs co-varied with differences in the microbiota post-resection independently of several important clinical confounders (including history of treatment, tumour stage, duration of time since the surgery, age and gender). Knowledge of how specific dietary ingredients influence the colonic microbiome is limited and studies of associations with the microbiome have been inconsistent due to differences in trial design and methodological limitations. Two studies have shown that cruciferous vegetables can significantly alter the microbiota (43,44). Most cruciferous vegetables are a source of vitamins A and C and contain phytonutrients, which are plant-based compounds known to reduce cancer risk through inflammation attenuation (45). This food group is also known to significantly reduce risk of CRC through the action of its secondary metabolites including glucoinolates (46,47). Little evidence is available regarding the effects of peanuts on the microbiome but the legume family has been associated with reduced CRC risk (48). In our study, these dietary ingredients have numerous associations with the top taxa that distinguish between newly diagnosed CRC and non-CRC groups. Various cardiovascular drugs which are commonly prescribed for patients in the age range linked with CRC alter the colonic microbiome, including ACE inhibitors (49), beta-blockers (50), angiotensin II receptor antagonists (51) and aspirin (52). Interestingly, we found that cruciferous vegetables and cardiovascular drugs, specifically aspirin, co-varied with differences after removal of CRC given that both are known to exhibit chemoprotective properties.
Our previous work showed no significant difference between paired samples from the tumour and nearby non-cancerous tissue (13,53). Here we also showed that no major microbiome differences between paired samples were detectable. However, patients with a normal-like microbiome after removal of CRC displayed significantly more variation between their paired samples than those observed to have an abnormal microbiome (Supplementary Figure S15). This is consistent with changes to the spatial gradient of microbiota in the colonic mucosa in a subset of patients observed to have a normal microbiome after resection. Emerging evidence suggests that spatial reprogramming of the colonic microbiota may occur in chronic diseases (54) potentially explaining the low level of variation observed in patients with an abnormal microbiome after removal of CRC.
There are limitations to our study with regard to size and duration. Future studies should be longitudinal for greater periods of follow up and preferably with a controlled intervention such as structured dietary advice aimed at restoring the post-operative microbiota.
In conclusion, following removal of CRC the microbiomes of patients are heterogeneous with most retaining alterations similar to those of newly diagnosed CRC but a substantial minority reverting toward normal and similar to that of controls. This adds support to the pursuit of the colonic microbiota as a marker of risk for development of CRC and not simply as a diagnostic marker. In particular, the microbiome may enable resources to be focused on a subset of patients at greatest risk of developing a new CRC after surgical removal of the primary CRC.
DATA AVAILABILITY
Data for the 16S rRNA gene amplicon sequencing have been uploaded to the European Nucleotide Archive under accession number PRJEB47197. All other data used in this study is available in the supplementary files accompanying the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients who consented to participate in this study, and the support staff of relevant departments in Cork University Hospital and the Mercy University Hospital Cork.
Specific author contributions: PC: data analysis and manuscript writing, CLM: sample handling, carrying out of experiments and sample acquisition, MB: acquisition and analysis of data, TSG: data analysis and manuscript writing, PP: sample handling, EMOC: conceiving the study and manuscript writing; SAZ: sample acquisition, SK: sample acquisition, EA: sample acquisition, MM: sample acquisition, MGOR: study design and sample acquisition, FS: study design and manuscript writing, PWOT: conceiving the study, study supervision, study design and manuscript writing. All authors have read and agreed to the published version of the manuscript.
Contributor Information
Peter Cronin, Department of Biological Science, University of Limerick, Limerick, V94 T9PX, Ireland; APC Microbiome Ireland, University College Cork, Cork, T12 YT20, Ireland.
Clodagh L Murphy, APC Microbiome Ireland, University College Cork, Cork, T12 YT20, Ireland; School of Microbiology, University College Cork, Cork, T12 K8AF, Ireland; Cork University Hospital, Cork, T12 DC4A, Ireland.
Maurice Barrett, APC Microbiome Ireland, University College Cork, Cork, T12 YT20, Ireland; School of Microbiology, University College Cork, Cork, T12 K8AF, Ireland.
Tarini Shankar Ghosh, APC Microbiome Ireland, University College Cork, Cork, T12 YT20, Ireland; School of Microbiology, University College Cork, Cork, T12 K8AF, Ireland.
Paola Pellanda, APC Microbiome Ireland, University College Cork, Cork, T12 YT20, Ireland; School of Microbiology, University College Cork, Cork, T12 K8AF, Ireland.
Eibhlis M O’Connor, Department of Biological Science, University of Limerick, Limerick, V94 T9PX, Ireland; APC Microbiome Ireland, University College Cork, Cork, T12 YT20, Ireland; Health Research Institute, University of Limerick, Limerick, V94 T9PX, Ireland.
Syed Akbar Zulquernain, APC Microbiome Ireland, University College Cork, Cork, T12 YT20, Ireland; Cork University Hospital, Cork, T12 DC4A, Ireland.
Shane Kileen, Cork University Hospital, Cork, T12 DC4A, Ireland; Department of Surgery, Mercy University Hospital, Cork, T12 WE28, Ireland.
Morgan McCourt, Cork University Hospital, Cork, T12 DC4A, Ireland.
Emmet Andrews, Cork University Hospital, Cork, T12 DC4A, Ireland.
Micheal G O’Riordain, Department of Surgery, Mercy University Hospital, Cork, T12 WE28, Ireland.
Fergus Shanahan, APC Microbiome Ireland, University College Cork, Cork, T12 YT20, Ireland; School of Medicine, University College Cork, Cork, T12 AK54, Ireland.
Paul W O’Toole, APC Microbiome Ireland, University College Cork, Cork, T12 YT20, Ireland; School of Microbiology, University College Cork, Cork, T12 K8AF, Ireland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Cancer Online.
FUNDING
Science Foundation Ireland through a Centre award [APC/SFI/12/RC/2273_P2 to APC Microbiome Ireland]; Health Research Board of Ireland [ILP-POR-2017-034]; FIRM award scheme of the (Govt. Ireland) Department of Agriculture, Food and Marine [17F251].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D.. Global cancer statistics. CA. Cancer J. Clin. 2011; 61:69–90. [DOI] [PubMed] [Google Scholar]
- 2. Haggar F.A., Boushey R.P.. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 2009; 22:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fearon E.R. Molecular Genetics of Colorectal Cancer. Annu. Rev. Pathol. Mech. Dis. 2011; 6:479–507. [DOI] [PubMed] [Google Scholar]
- 4. Leslie A., Carey F.A., Pratt N.R., Steele R.J.C.. The colorectal adenoma-carcinoma sequence. Br. J. Surg. 2002; 89:845–860. [DOI] [PubMed] [Google Scholar]
- 5. Hiraoka S., Kato J., Fujiki S., Kaji E., Morikawa T., Murakami T., Nawa T., Kuriyama M., Uraoka T., Ohara N.et al.. The presence of large serrated polyps increases risk for colorectal cancer. Gastroenterology. 2010; 139:1503–1510. [DOI] [PubMed] [Google Scholar]
- 6. Pleguezuelos-Manzano C., Puschhof J., Rosendahl Huber A., van Hoeck A., Wood H.M., Nomburg J., Gurjao C., Manders F., Dalmasso G., Stege P.B.et al.. Mutational signature in colorectal cancer caused by genotoxic pks + E. coli. Nature. 2020; 580:269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bullman S., Pedamallu C.S., Sicinska E., Clancy T.E., Zhang X., Cai D., Neuberg D., Huang K., Guevara F., Nelson T.et al.. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science (80-.). 2017; 358:1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boleij A., van Gelder M.M.H.J., Swinkels D.W., Tjalsma H.. Clinical Importance of Streptococcus gallolyticus Infection Among Colorectal Cancer Patients: Systematic Review and Meta-analysis. Clin. Infect. Dis. 2011; 53:870–878. [DOI] [PubMed] [Google Scholar]
- 9. Louis P., Hold G.L., Flint H.J.. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014; 12:661–672. [DOI] [PubMed] [Google Scholar]
- 10. Castellarin M., Warren R.L., Freeman J.D., Dreolini L., Krzywinski M., Strauss J., Barnes R., Watson P., Allen-Vercoe E., Moore R.A.et al.. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012; 22:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M., Clancy T.E., Chung D.C., Lochhead P., Hold G.L.et al.. Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe. 2013; 14:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zeller G., Tap J., Voigt A.Y., Sunagawa S., Kultima J.R., Costea P.I., Amiot A., Böhm J., Brunetti F., Habermann N.et al.. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014; 10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flemer B., Lynch D.B., Brown J.M.R., Jeffery I.B., Ryan F.J., Claesson M.J., O’Riordain M., Shanahan F., O’Toole P.W.. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017; 66:633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hibberd A.A., Lyra A., Ouwehand A.C., Rolny P., Lindegren H., Cedgård L., Wettergren Y.. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017; 4:e000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flemer B., Warren R.D., Barrett M.P., Cisek K., Das A., Jeffery I.B., Hurley E., O’Riordain M., Shanahan F., O’Toole P.W.. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018; 67:1454–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flemer B., Herlihy M., O’Riordain M., Shanahan F., O’Toole P.W.. Tumour-associated and non-tumour-associated microbiota: Addendum. Gut Microbes. 2018; 9:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin Y., Liu Y., Zhao L., Zhao F., Feng J., Li S., Chen H., Sun J., Zhu B., Geng R., Wei Y.. Gut microbiota in patients after surgical treatment for colorectal cancer. Environ. Microbiol. 2019; 21:772–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Y., Geng R., Liu L., Jin X., Yan W., Zhao F., Wang S., Guo X., Ghimire G., Wei Y.. Gut microbiota-based algorithms in the prediction of metachronous adenoma in colorectal cancer patients following surgery. Front. Microbiol. 2020; 11:1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sze M.A., Baxter N.T., Ruffin M.T., Rogers M.A.M., Schloss P.D.. Normalization of the microbiota in patients after treatment for colonic lesions. Microbiome. 2017; 5:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shiroma H., Shiba S., Erawijantari P.P., Yamada M., Kanemitsu Y., Shibata T., Fukuda S., Yachida S., Yamada T.. Surgical Treatment for Colorectal Cancer Partially Restores Gut Microbiome and Metabolome Traits. 2021; Res. Square. doi:15 April 2021,preprint: not peer reviewed 10.21203/rs.3.rs-400629/v1. [DOI] [PMC free article] [PubMed]
- 21. Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O.. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013; 41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P.. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016; 13:581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Magoc T., Salzberg S.L.. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011; 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pẽa A.G., Goodrich J.K., Gordon J.I.et al.. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010; 7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010; 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 26. Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O.. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013; 41:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shannon C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 27:623–656. [Google Scholar]
- 28. Bray J.R., Curtis J.T.. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957; 27:325–349. [Google Scholar]
- 29. Lin H., Peddada S.D.. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020; 11:3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shanahan F., Ghosh T.S., O’Toole P.W.. The healthy Microbiome—What is the definition of a healthy Gut microbiome. Gastroenterology. 2021; 160:483–494. [DOI] [PubMed] [Google Scholar]
- 31. Ghosh T.S., Das M., Jeffery I.B., O’Toole P.W.. Adjusting for age improves identification of gut microbiome alterations in multiple diseases. Elife. 2020; 9:e50240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pasolli E., Truong D.T., Malik F., Waldron L., Segata N.. Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLOS Comput. Biol. 2016; 12:e1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A., Alm E.J.. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017; 8:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zackular J.P., Rogers M.A.M., Ruffin M.T., Schloss P.D.. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014; 7:1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baxter N.T., Ruffin M.T., Rogers M.A.M., Schloss P.D.. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016; 8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yachida S., Mizutani S., Shiroma H., Shiba S., Nakajima T., Sakamoto T., Watanabe H., Masuda K., Nishimoto Y., Kubo M.et al.. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019 256. 2019; 25:968–976. [DOI] [PubMed] [Google Scholar]
- 37. Shah M.S., DeSantis T., Yamal J.M., Weir T., Ryan E.P., Cope J.L., Hollister E.B.. Re-purposing 16S rRNA gene sequence data from within case paired tumor biopsy and tumor-adjacent biopsy or fecal samples to identify microbial markers for colorectal cancer. PLoS One. 2018; 13:e0207002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim K., Castro E.J.T., Shim H., Advincula J.V.G., Kim Y.-W.. Differences Regarding the Molecular Features and Gut Microbiota Between Right and Left Colon Cancer. Ann. Coloproctol. 2018; 34:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murphy C.L., O’Toole P.W., Shanahan F.. The Gut Microbiota in Causation, Detection, and Treatment of Cancer. Am. J. Gastroenterol. 2019; 114:1036–1042. [DOI] [PubMed] [Google Scholar]
- 40. Lederer A.K., Pisarski P., Kousoulas L., Fichtner-Feigl S., Hess C., Huber R.. Postoperative changes of the microbiome: Are surgical complications related to the gut flora? A systematic review. BMC Surg. 2017; 17:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fang X., Vázquez-Baeza Y., Elijah E., Vargas F., Ackermann G., Humphrey G., Lau R., Weldon K.C., Sanders J.G., Panitchpakdi M.et al.. Gastrointestinal surgery for inflammatory bowel disease persistently lowers microbiome and metabolome diversity. Inflamm. Bowel Dis. 2021; 27:603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alexander J.L., Wilson I.D., Teare J., Marchesi J.R., Nicholson J.K., Kinross J.M.. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017; 14:356–365. [DOI] [PubMed] [Google Scholar]
- 43. Li F., Hullar M.A.J., Schwarz Y., Lampe J.W.. Human Gut Bacterial Communities Are Altered by Addition of Cruciferous Vegetables to a Controlled Fruit- and Vegetable-Free Diet. J. Nutr. 2009; 139:1685–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kaczmarek J.L., Liu X., Charron C.S., Novotny J.A., Jeffery E.H., Seifried H.E., Ross S.A., Miller M.J., Swanson K.S., Holscher H.D.. Broccoli consumption affects the human gastrointestinal microbiota. J. Nutr. Biochem. 2019; 63:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aune D., Giovannucci E., Boffetta P., Fadnes L.T., Keum N.N., Norat T., Greenwood D.C., Riboli E., Vatten L.J., Tonstad S.. Fruit and vegetable intake and the risk of cardiovascular disease, total cancer and all-cause mortality-A systematic review and dose-response meta-analysis of prospective studies. Int. J. Epidemiol. 2017; 46:1029–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pan J.H., Abernathy B., Kim Y.J., Lee J.H., Kim J.H., Shin E.C., Kim J.K.. Cruciferous vegetables and colorectal cancer prevention through microRNA regulation: A review. Crit. Rev. Food Sci. Nutr. 2018; 58:2026–2038. [DOI] [PubMed] [Google Scholar]
- 47. Higdon J.V., Delage B., Williams D.E., Dashwood R.H.. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol. Res. 2007; 55:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aranda-Olmedo I., Rubio L.A.. Dietary legumes, intestinal microbiota, inflammation and colorectal cancer. J. Funct. Foods. 2020; 64:103707. [Google Scholar]
- 49. Vich Vila A., Collij V., Sanna S., Sinha T., Imhann F., Bourgonje A.R., Mujagic Z., Jonkers D.M.A.E., Masclee A.A.M., Fu J.et al.. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 2020; 11:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weersma R.K., Zhernakova A., Fu J.. Interaction between drugs and the gut microbiome. Gut. 2020; 69:1510–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhernakova A., Kurilshikov A., Bonder M.J., Tigchelaar E.F., Schirmer M., Vatanen T., Mujagic Z., Vila A.V., Falony G., Vieira-Silva S.et al.. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science (80-.). 2016; 352:565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Prizment A.E., Staley C., Onyeaghala G.C., Vivek S., Thyagarajan B., Straka R.J., Demmer R.T., Knights D., Meyer K.A., Shaukat A.et al.. Randomised clinical study: oral aspirin 325 mg daily vs placebo alters gut microbial composition and bacterial taxa associated with colorectal cancer risk. Aliment. Pharmacol. Ther. 2020; 52:976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murphy C.L., Barrett M., Pellanda P., Killeen S., McCourt M., Andrews E., O’ Riordain M., Shanahan F., O’Toole P.. Mapping the colorectal tumor microbiota. Gut Microbes. 2021; 13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tropini C., Earle K.A., Huang K.C., Sonnenburg J.L.. The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host Microbe. 2017; 21:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for the 16S rRNA gene amplicon sequencing have been uploaded to the European Nucleotide Archive under accession number PRJEB47197. All other data used in this study is available in the supplementary files accompanying the manuscript.