Abstract
The MICs for many oxacillin-resistant (OR) Staphylococcus epidermidis (ORSE) strains are below the Staphylococcus aureus methicillin or oxacillin resistance breakpoint. The difficulty detecting the OR phenotype in S. epidermidis may be due to extreme heterotypy in resistance expression and/or transcriptional repression of mecA, the OR gene, by MecI. To determine the role of these factors in the phenotypic expression of ORSE, 17 geographically diverse mecI+ ORSE isolates representing 14 distinct pulsed-field gel electrophoresis pulse types (>3 band differences) were investigated. Thirteen of the 14 types contained mecI and mecA promoter-operator sequences known to be associated with maximal mecA repression, and in all isolates, mecA transcription was repressed. All 17 were heterotypic in their resistance expression. Oxacillin MICs ranged from 1 to 128 μg/ml and increased for 16 of 17 isolates after β-lactam induction. Allelic replacement inactivation of mecI in three isolates similarly resulted in a four- to sevenfold increase in MIC. In the two of these three isolates producing β-lactamase, mecA transcription was regulated by both mecI and β-lactamase regulatory sequences. Heterotypic expression of resistance in these three isolates was unaffected by either β-lactam induction or mecI inactivation. However, prolonged incubation in concentrations of oxacillin just sufficient to produce a lag in growth (0.5 to 1.0 μg/ml) converted the population resistance expression from heterotypic to homotypic. Homotypic conversion could also be demonstrated in microtiter wells during MIC determinations in one isolate for which the MIC was high. We conclude that the phenotypic expression of S. epidermidis OR in broth can be affected both by mecA transcriptional regulation and by subpopulation resistance expression.
More than 70% of nosocomial Staphylococcus epidermidis isolates are methicillin resistant (MRSE) or oxacillin resistant (ORSE) (39), but resistance is often difficult to detect by conventional susceptibility testing methods. While Staphylococcus aureus clinical isolates tend to be either very susceptible or very resistant to methicillin or oxacillin (MICs of <0.5 or >8 μg/ml, respectively), S. epidermidis strains are not as clearly bimodal in their resistance pattern (21). The oxacillin MICs for many S. epidermidis strains are 0.5 to 2 μg/ml, which is below the breakpoint for oxacillin-resistant S. aureus (ORSA) (4 μg/ml), but these strains are found to contain mecA, the gene that mediates oxacillin resistance. Resistance expression in these S. epidermidis isolates can only be demonstrated when more laborious susceptibility testing techniques are used. When the in vitro expression of oxacillin resistance in S. epidermidis is examined on agar plates containing increasing concentrations of a β-lactam antibiotic, most isolates exhibit a heterotypic phenotype. This type of resistance expression is defined by a small percentage of cells (0.1%) that are able to survive on plates containing 100 μg of oxacillin per ml; the surviving colonies are of different sizes. In contrast, extreme heterotypic expression is relatively uncommon among clinical S. aureus isolates; resistant subpopulations comprise a greater proportion of the overall population than among S. epidermidis strains, and often every member of the population is highly resistant (homotypic or homogeneous resistance) (T. M. Dickinson and G. L. Archer, unpublished data).
As with S. aureus the production of an alternative, β-lactam-resistant penicillin-binding protein, PBP2a, encoded by mecA, confers OR in S. epidermidis (2, 4). In addition, a two-gene operon, mecR1-mecI, that encodes a signal transducer/inducer and repressor, respectively, is divergently transcribed from mecA (14, 35). MecI represses mecA and autorepresses mecR1-mecI transcription by binding to promoter-operator (P-O) sequences (32). The gene products of the mecA regulatory system have amino acid similarity to BlaR1 and BlaI, proteins that regulate transcription of the β-lactamase gene, blaZ. Previous studies have shown that the mec and bla regulatory systems are able to interact and that BlaI can regulate mecA transcription (10, 29, 37). Sequences hybridizing with mecR1-mecI probes are present in one-half to two-thirds of ORSA and ORSE clinical isolates (2), while the β-lactamase gene is found in 90% of clinical isolates (20).
It has been proposed (13) that among early ORSA strains, repression of mecA transcription by mecI yielded a β-lactam-susceptible phenotype, because repression led to little or no production of PBP2a, and mecA transcription was poorly inducible through MecR1. According to this hypothesis, the evolution of clinically resistant ORSA required mutations in either mecI or the mecA P-O. Additional chromosomal mutations were necessary to convert heterotypic resistance expression to homotypic (13). However, a recent analysis of clinical ORSE isolates from one hospital in Japan found that all contained mecI and mecA P-O sequences identical to those associated with maximal mecA repression in ORSA (17).
In the following study, we have attempted to assess the relative contributions of mecA transcriptional regulation and heterotypic resistance expression to the low MICs seen for many clinical ORSE isolates. We have chosen geographically diverse, genetically unrelated ORSE isolates found in a previous study to contain mecI by hybridization with a DNA probe. We have examined the nucleotide sequence of mecI and mecA P-O in these isolates, analyzed mecA transcription, and assessed phenotypic expression in both broth and on agar in the presence and absence of mecA regulation. Finally, we have assessed changes in the size of the OR subpopulation upon oxacillin exposure and the contribution of subpopulation selection to the MIC.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The S. epidermidis strains used in this study are listed in Table 1. Recombinant plasmids were generated and maintained in Escherichia coli TB1 (38) or JM109 (9) cells. The E. coli cloning vector used was pUC19 (38). The E. coli-S. aureus shuttle vectors used were constructed by adding pE194ts (15, 33) to pUC19. The staphylococcal tetracycline and gentamicin resistance genes used for selection of colonies containing recombinant vectors in E. coli or S. aureus or S. epidermidis were tetM (34) and aac/aph (28), respectively.
TABLE 1.
Properties of the S. epidermidis strains used in this study
| Strain | Pulse type | Resistancea | mecI | EOP
(10−5) with oxacillin
|
Oxacillin MIC
(μg/ml)b
|
||
|---|---|---|---|---|---|---|---|
| 20 μg/ml | 100 μg/ml | No induction | Inductionc | ||||
| SE12 | I | Tetr | + | 1,300 | 300 | 16 | 64 |
| SE20 | II | Tetr | + | 3 | 2 | 8 | 128 |
| SE20ΔmecI | II | Tetr Minr | − | 8 | 6 | 128 | 512 |
| SE22 | III | Genr | + | 36 | 78 | 64 | 256 |
| SE42 | IV | + | 4 | 2 | 8 | 256 | |
| SE42ΔmecI | IV | Tetr Minr | − | 7 | 5 | 128 | NDd |
| SE43 | V | Tetr | + | 680 | 2 | 1 | 8 |
| SE53 | VIa | Genr Chlr | + | 0.8 | 0.4 | 1 | 128 |
| SE53ΔmecI | VIa | Genr Chlr Tetr Minr | − | 0.4 | 0.4 | 128 | 256 |
| SE54 | VIb | Genr | + | 9 | 0.7 | 64 | 128 |
| SE24 | VIc | Genr Chlr Eryr | + | 1,800 | 65 | 4 | 256 |
| SE34 | VII | Genr Chlr Eryr | + | 2 | 7 | 4 | 32 |
| SE65 | VIII | Tetr Eryr | + | 15 | 3 | 128 | 256 |
| SE1 | IXa | Eryr | Ile66→Asn | 1 | 0.6 | 1 | 32 |
| SE2 | IXb | Tetr Eryr | Ile66→Asn | 2,700 | 600 | 2 | 128 |
| SE13 | X | Chlr Eryr | + | 3,800 | 120 | 64 | 128 |
| SE33 | XI | Genr Eryr | + | 230 | 19 | 64 | 128 |
| SE55 | XII | Tetr Eryr | + | 4,300 | 1,000 | 32 | 128 |
| SE60 | XIII | Tetr Eryr | + | 0.1 | 0.1 | 1 | 1 |
| SE407 | XIV | Tetr Eryr | + | 1,700 | 545 | 4 | 128 |
Gen, gentamicin; Chl, chloramphenicol; Tet, tetracycline; Min, minocycline; Ery, erythromycin.
MIC by broth dilution test.
Bacteria were induced with 5 μg of CBAP per ml overnight before MIC determination.
ND, not determined.
Materials and media.
Mueller-Hinton broth (MHB) and agar (MHA) (BBL Microbiology Systems, Cockeysville, Md.) and brain heart infusion (BHI) broth and agar (Difco Laboratories, Detroit, Mich.), with and without selective additives (Sigma, St. Louis, Mo.; United States Biochemicals, Cleveland, Ohio), were used for the subculture and maintenance of E. coli, S. epidermidis, and S. aureus strains. The antibiotics and concentrations used for E. coli strains for initial selection after transformation were as follows: ampicillin, 50 μg/ml; gentamicin, 5 μg/ml; minocycline, 1 μg/ml; and tetracycline, 5 μg/ml. The antibiotics used for S. aureus and S. epidermidis strains for determining antibiotic resistance and initial selection after electroporation or conjugative mobilization were gentamicin (5 μg/ml), chloramphenicol (10 μg/ml), erythromycin (10 μg/ml), tetracycline (5 μg/ml), minocycline (1 μg/ml), and mupirocin (20 μg/ml). The antibiotics used to select for the recipient S. epidermidis in conjugative mobilization were novobiocin (1 μg/ml) and rifampin (10 μg/ml). Induction experiments were performed with 2-(2′-carboxyphenol) benzoyl-6-amino penicilloic acid (CBAP) (5 μg/ml) and oxacillin (0.1, 0.3, 0.5, 0.6, or 1 μg/ml) in broth. Other selective additives, such as sodium citrate (Sigma) (8 mM, for transductions) or β-d-galactopyranoside (X-Gal) (50 μg/ml; Inalco Spa, Milano, Italy), were added to the media as required.
β-Lactamase production.
β-Lactamase activity was detected by growing single colonies of bacteria for 16 h on BHI agar supplemented with 1% (wt/vol) soluble starch. Five milliliters of a 0.1 N I2 solution in 0.4 M KI containing 20 mg of benzylpenicillin per ml was pipetted over the colonies and allowed to sit for 10 s before the excess was poured away. The presence of a rapidly spreading white halo around the colonies, as the penicilloic acid reacted with the iodine and decolorized the agar around the colony, indicated the presence of β-lactamase in the bacteria.
Quantitative analysis of β-lactamase activity to assess induction by β-lactam antibiotics was accomplished by using colorimetric detection of nitrocefin hydrolysis. Bacteria were grown in BHI at 37°C to an optical density at 600 nm (OD600) of 0.6 with (induced) and without (uninduced) CBAP (5 μg/ml). Bacteria were harvested, pelleted, and resuspended in 2 ml of Z buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4, 0.05 M β-mercaptoethanol, pH 7.0). One milliliter of the cell suspension and 2.5 g of 0.1-mm-diameter Zirconia beads (Biospec Products, Bartlesville, Okla.) were added to a 2-ml screw-cap tube. The samples were bead beaten (Biospec Products) for 5 min at 4°C and centrifuged for 10 min at 26,895 × g at 4°C. One hundred microliters of the supernatant was removed and added to 750 μl of 0.1 M sodium phosphate buffer (pH 7.0) in a visible cuvette. One hundred-fifty microliters of a 0.2-mg/ml concentration of nitrocefin (kindly provided by Bristol-Meyers Squibb) was added to the cuvette, and the sample was mixed and read at an OD490 at 30 min.
Cloning, transformation, and DNA manipulation.
All restriction endonuclease digestions and ligation reactions were performed per the manufacturer's (New England Biolabs, Beverly, Mass.) specifications. Plasmids were electroporated (31; T. B. Luchansky, P. M. Muriana, and T. R. Klaenhammer, Bio-Rad technical bulletin no. 1350, p. 1–3. Bio-Rad Laboratories, Richmond, Calif.) into E. coli in the Bio-Rad Gene Pulser. Shuttle plasmids were moved from E. coli to S. aureus by electroporation into the restriction-deficient S. aureus strain RN4220 (18) as previously described (22). Plasmids were introduced into other S. aureus strains by transduction with the general transduction phage 80α (16, 26). Transductions using phage 80α and the isolation of both E. coli and staphylococcal plasmid and genomic DNA were performed as previously described (16, 25, 26).
Conjugative mobilization.
Conjugative mobilization was used to introduce plasmid DNA from S. aureus into S. epidermidis and was performed by using a three-plasmid S. aureus donor strain as previously described (36). The recipient S. epidermidis strains were made novobiocin (1 μg/ml) and rifampin (10 μg/ml) resistant by serial passage on selective agar plates. The donor S. aureus strain, RN4220, contained the plasmids pGO626, pC221, and pGO400. To produce the tetracycline- and minocycline-resistant plasmid pGO626 (13.8 kb), a 700-bp nick site from pC221 (27) was added to pGO514. pGO514 is the plasmid containing tetM-inactivated mecI and a temperature-sensitive pE194 origin of replication described previously for allelic replacement of mecI in S. aureus (25). pC221 (4.6 kb) encodes chloramphenicol resistance and provides mobilization genes that act in trans on the nick site of pGO626 (27, 36). pGO400 (33.8 kb), a member of the pGO1 family of conjugative plasmids, encodes resistance to mupirocin and provides the conjugative apparatus (23). Following filter mating (23), colonies were sought that were resistant to novobiocin, rifampin, and minocycline, indicating mobilization of pGO626 into S. epidermidis, and susceptible to chloramphenicol and mupirocin, identifying those colonies that did not receive pC221 and pGO400 by cotransfer. Differentiation of S. epidermidis recipients from S. aureus donors was also achieved by performing matings on plates containing mannitol, resulting in white S. epidermidis and yellow S. aureus colonies.
Plasmid curing and allelic replacement.
S. epidermidis isolates harboring plasmid constructs with the pE194ts replicon (pGO626) were cured of their plasmids in order to detect allelic replacement of chromosomal genes by homologous recombination (25). Briefly, single colonies were inoculated into 10 ml of BHI broth and grown for 16 h at 30°C with antibiotic selection. Following growth of S. epidermidis at the nonpermissive temperature for plasmid replication (43°C), colonies were patched to minocycline, gentamicin, and erythromycin plates. Colonies were sought that were minocycline resistant, indicating chromosomal integration into the replacement locus, and either gentamicin or erythromycin susceptible, indicating secondary recombination to remove plasmid DNA. The inclusion of erythromycin and gentamicin resistance genes in the pGO626 plasmid allowed us to include in this study S. epidermidis strains that were resistant to either erythromycin or gentamicin, but not both. However, all isolates used had to be minocycline susceptible, so that chromosomal insertion of the tetM marker could be identified.
EOP.
Phenotypic expression of OR was determined by the efficiency of plating (EOP) procedure described by Hackbarth et al. (11), except that oxacillin was used instead of methicillin or nafcillin. Staphylococcal strains were inoculated into 5 ml of BHI broth and incubated for 16 h at 37°C with constant shaking. Cultures were then serially diluted and plated on MHA containing 0, 10, 20, 50, 100, 250, 500, and 800 μg of oxacillin per ml. The plates were incubated at 30°C for 96 h, after which CFU were counted and expressed as a ratio of cells growing in plates containing oxacillin to the number of cells on antibiotic-free medium. We defined heterotypic resistance as a population decrease of at least 2 log10 on 20-μg/ml oxacillin plates and of at least 3 log10 on 100-μg/ml oxacillin plates.
Southern blot analysis.
Alkaline capillary transfer, fixation of DNA to positively charged Zeta-Probe nylon membrane (Bio-Rad, Hercules, Calif.), and hybridization were performed per the directions of the manufacturer (Bio-Rad, Hercules, Calif.) and as previously described (1).
Northern blot and PBP analysis.
Cellular RNA isolation, formaldehyde gel separation, 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) capillary transfer, fixation of RNA to neutrally charged nylon membrane (Qiagen, Valencia, Calif.), and hybridization were performed according to established protocols (25). PBPs were analyzed by Michael Pucci at the Bristol-Myers Squibb Pharmaceutical Research Institute by methods previously described (25). Transcript and protein abundance on gels were quantified by scanning densitometry with an AlphaImager 1000 digital imaging system (Alpha Innotech Corp., San Leandro, Calif.).
PCR amplification.
PCR amplification of DNA sequences was performed to generate fragments for cloning into pGO626 and to create the mecA probe for Northern blot analysis. The primer sequences and amplification protocols were performed as previously described (25).
Sequence analysis.
DNA sequence analysis was performed to confirm the sequence of mecI and the mecA-mecR1 or -mecI P-O in the different S. epidermidis isolates. Sequencing was performed by the automated laser fluorescence technique employing fluorescein-labeled oligonucleotides (Applied Biosystems and the Virginia Commonwealth University Nucleic Acid Synthesis and Analysis Core Facility).
PFGE.
Genomic DNA was prepared and cut with the restriction enzyme SmaI for pulsed-field gel electrophoresis (PFGE) by established methods (8). The following parameters (22 h at 14°C) were used to visualize large fragments of DNA: 6 V/cm; initial pulse time, 1 s; final pulse time, 30 s.
Susceptibility testing.
MICs for S. epidermidis isolates were determined in cation-supplemented MHB containing 2% NaCl by using a 105-CFU/ml inoculum, according to National Committee for Clinical Laboratory Standards (NCCLS) guidelines (24). MIC determinations were performed in quadruplicate and read after an 18- to 24-h incubation at 37°C. The only exception to the NCCLS guidelines was incubation of the MIC plates at 37°C; the guidelines stipulate an incubation temperature of 35°C.
RESULTS
Characterization of clinical S. epidermidis.
We chose to study 22 geographically and chronologically diverse clinical ORSE isolates previously shown to contain the mecR1-mecI regulatory region by DNA hybridization. This collection of multiresistant S. epidermidis isolates came from five areas of North America (Richmond, Boston, Toronto, Birmingham, and Portland) collected over 19 years (1970 to 1989). Southern blot analysis of genomic DNA digested with the restriction endonuclease BamHI was probed with a mecR1-mecI DNA probe to confirm the presence of the regulatory region in each of the isolates. Pulsed-field gel (contour-clamped homogeneous electric field [CHEF]) analysis of S. epidermidis genomic DNA digested with SmaI was performed and identified 17 unique banding patterns (at least 2 band differences) representing 14 pulse types (>3 band differences) listed in Table 1. The S. epidermidis isolates were then screened for antibiotic resistance to gentamicin, tetracycline, minocycline, chloramphenicol, erythromycin, and oxacillin. All isolates were resistant to oxacillin, and most of the isolates were resistant to two or three of the additional antibiotics tested (Table 1). All but one isolate (SE42) produced β-lactamase.
Oxacillin MICs.
Broth microdilution oxacillin MICs were determined for all 17 S. epidermidis isolates (Table 1). The range of MICs obtained was consistent with the findings of others for S. epidermidis clinical isolates containing the OR gene mecA (21). For five isolates (29%), the MIC was ≤2 μg/ml; for three isolates (18%), the MIC was 4 μg/ml (the NCCLS oxacillin resistance breakpoint for S. aureus); and for the remaining nine isolates (53%), the MIC was ≥8 μg/ml.
EOP.
EOPs examined to determine the OR phenotypes of the 17 clinical S. epidermidis isolates revealed that each had a heterotypic phenotype (Table 1). All isolates had at least a 2-log10 drop in colony counts on plates containing ≥20 μg of oxacillin per ml, and all but one isolate (SE55) had at least a 3-log10 drop in colony count on plates containing ≥100 μg of oxacillin per ml. The reduction in colony count on plates containing 10 μg of oxacillin per ml was more variable.
Analysis of mecI and mecR1.
PCR amplification and subsequent nucleotide sequencing of the mecI gene and the mecA-mecR1 P-O revealed that 13 of the 14 S. epidermidis pulse types were identical to the wild-type S. aureus mecI and P-O sequences associated with maximal mecA transcriptional repression (S. aureus strain N315 [14]). The lone mutation observed was a single A-to-T point mutation in the mecI of pulse type IX, resulting in an amino acid change (isoleucine→asparagine) at position 66.
Northern blot analysis of uninduced mecA transcript in the clinical S. epidermidis isolates showed heavily repressed (∼50-fold reduction) transcript in the all isolates containing mecI, including the isolate with the single-amino-acid change, compared to that of the mecA transcript in a mecI mutant unregulated isolate. A PBP profile of one uninduced mecI+ S. epidermidis isolate, SE42, showed little PBP2a production, consistent with the mecA transcript data (Fig. 1). Northern blot analysis also showed that when induced for 16 h with a β-lactam antibiotic, CBAP (5 μg/ml), all but 1 (SE34) of the 17 mecI-positive S. epidermidis isolates produced mecA transcript equivalent to that of an unregulated isolate (∼50-fold increase). mecA transcript in SE34 induced with 5 μg of CBAP per ml increased ∼10-fold over uninduced SE34 mecA transcript. These data indicate that in the S. epidermidis clinical isolates, mecI and mecR1 are largely intact and functional. MecI represses mecA transcription and consequently PBP2a production, while MecR1 provides the signal sensor/transducer necessary to relieve MecI repression following induction with β-lactams.
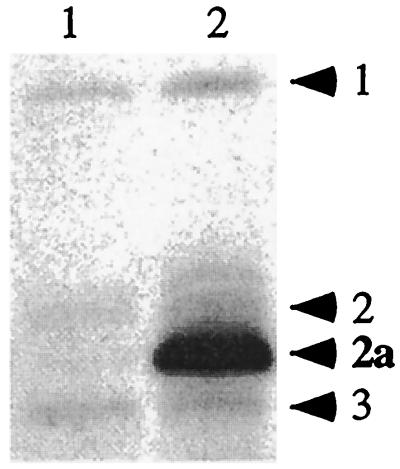
FIG. 1.
PBP profiles. Determination of relative amounts of PBP2a in the cell membranes of S. epidermidis strains SE42 (lane 1) and SE42ΔmecI (lane 2). PBPs are designated by numbers to the right of the panel. This image was scanned from the original film by using an AlphaImager 1000 (Alpha Innotech) and was cropped and labeled by using Canvas 5 graphics software (Deneba, Miami, Fla.). The same systems were used to prepare Fig. 2.
mecI allelic replacement mutagenesis.
mecI allelic replacement mutagenesis was performed with three mecI+ S. epidermidis isolates (SE20, SE42, and SE53) to observe the relationship of mecA transcriptional repression to the OR phenotype. Deletion of mecI was confirmed by Southern blot hybridization by noting the loss of a 0.7-kb BglII fragment from deletion mutants.
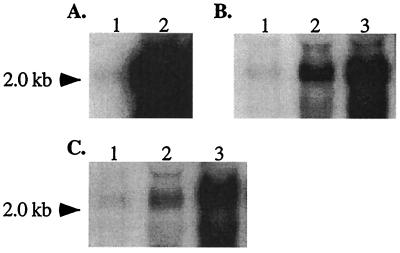
Northern blot analysis of mecA transcript in the mecI mutant S. epidermidis isolates showed an increase in mecA transcript compared to the level of mecA transcript in mecI+ parents. The SE42 mecI knockout resulted in an ∼50-fold increase in mecA transcription, as determined by scanning densitometry (Fig. 2A). The PBP profile of SE42 and SE42ΔmecI also showed a comparable increase in PBP2a after the deletion of mecI (Fig. 1), correlating with the Northern blot data. SE42 produces no β-lactamase and contains no blaZ regulatory sequences. However, in the two S. epidermidis isolates (SE20 and SE53) that produced β-lactamase, there was only a three- to sixfold increase in mecA transcription after mecI inactivation (Fig. 2B and C), presumably due to the presence of the blaZ repressor BlaI, which has also been shown to repress mecA transcription. Both SE20 and SE53 were shown to contain blaI sequences by Southern blot hybridization. Maximal mecA transcription in SE20ΔmecI and SE53ΔmecI was achieved by induction with CBAP (Fig. 2B and C).
FIG. 2.
Northern blot analysis of the 2-kb mecA transcript. The relative abundance of mecA transcript in SE42 (lane 1) and SE42ΔmecI (lane 2) (A); SE20 (lane 1), SE20ΔmecI (lane 2), and SE20ΔmecI grown in 5 μg of CBAP per ml (lane 3) (B); or SE53 (lane 1), SE53ΔmecI (lane 2), and SE53ΔmecI grown in 5 μg of CBAP per ml (lane 3) (C) is shown.
A colorimetric β-lactamase assay was performed to determine if the β-lactamase repressor BlaI was functional in isolates SE20ΔmecI and SE53ΔmecI. The mecI mutant isolates known to contain a β-lactamase plasmid were induced for 3 h with and without 5 μg of CBAP per ml. β-Lactamase was barely detectable in the negative control (SE42ΔmecI, OD600 of 0.042) and the β-lactamase-positive isolates (SE20ΔmecI, OD600 of 0.085; SE53ΔmecI, OD600 of 0.08) in the absence of inducer. In the presence of inducer, the β-lactamase-positive isolates (SE20ΔmecI, OD600 of 0.65; SE53ΔmecI, OD600 of 0.86) produced 8- and 11-fold more β-lactamase, respectively, while the negative control (SE42ΔmecI, OD600 of 0.043) showed no increase in activity. Thus, the β-lactamase repressor BlaI was functional in these isolates until inducer was added to the media.
Induction of mecA transcription.
SE42 mecA transcription was induced by growing the isolate in various concentrations of oxacillin (0.1, 0.3, and 0.5 μg/ml) to an OD600 of 0.6. A progressive increase in mecA transcription, as determined by scanning densitometry measurement of transcripts on Northern blots, was seen as the amount of oxacillin in the medium increased (Table 2). SE42 mecA transcript was repressed in the absence of inducer and was 86% of the maximum seen in SE42ΔmecI when grown with 0.5 μg of oxacillin per ml, while 0.1 and 0.3 μg of oxacillin per ml provided relatively less induction (17 and 65% of maximal, respectively). Those strains inducibly producing β-lactamase (SE20 and SE53) displayed a stepwise pattern of mecA induction similar to that of SE42 at the same concentrations of oxacillin and CBAP. However, in SE20ΔmecI and SE53ΔmecI with only partially repressed mecA transcription, presumably due to the presence of the β-lactamase regulators, maximal induction was achieved with only 0.1 μg of oxacillin per ml. The data suggest that transcription of mecA is induced approximately fivefold more easily through bla than through mec regulatory sequences.
TABLE 2.
Northern blot analysis of mecA induction with oxacillin
| Strain | % of maximal mecA induction at
oxacillin concna:
|
||
|---|---|---|---|
| 0.1 μg/ml | 0.3 μg/ml | 0.5 μg/ml | |
| SE42 | 17 | 65 | 86 |
| SE42ΔmecI | 100 | NDb | ND |
| SE20 | 37 | 87 | 97 |
| SE20ΔmecI | 95 | ND | ND |
| SE53 | 52 | 81 | 94 |
| SE53ΔmecI | 94 | ND | ND |
Maximal mecA transcript normalized to SE42ΔmecI.
ND, not determined.
MICs and EOPs of induced isolates.
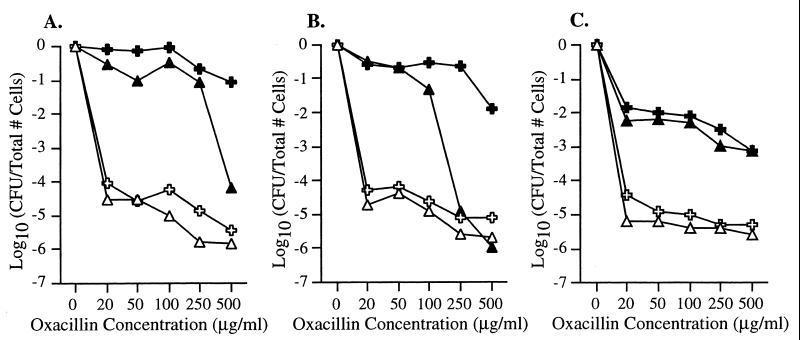
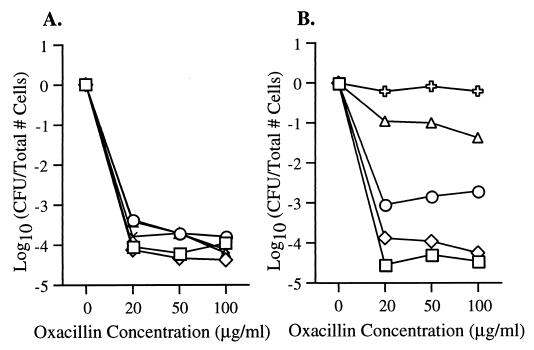
Oxacillin MICs (Table 1) for the mecI mutant strains increased in each of the isolates from either 8 μg/ml (SE20 and SE42) or 1 μg/ml (SE53) to 128 μg/ml (SE20ΔmecI, SE42ΔmecI, and SE53ΔmecI). However, all three of the mecI knockout strains still had heterotypic EOP phenotypes that were within 1 log10 of the parent at all oxacillin concentrations (Fig. 3).
FIG. 3.
EOP curves. Shown on the y axis are the numbers of S. epidermidis cells (in log10 CFU per milliliter on oxacillin/CFU per milliliter on MHA) remaining on the plates containing various concentrations of oxacillin (micrograms per milliliter [shown on the x axis]). Open triangles and crosses represent bacteria grown first in BHI broth without antibiotic, while solid triangles and crosses represent bacteria grown in BHI broth containing concentrations of oxacillin that caused a lag in growth (0.5 μg/ml for mecI+ isolates and 1.0 μg/ml for mecI mutant isolates). Broth-grown bacteria were then diluted and plated on agar containing increasing concentrations of oxacillin. (A) EOP results for SE20 (▵) and SE20ΔmecI ( ) with no oxacillin, SE20 (▴) grown with oxacillin at 0.5 μg/ml, and SE20ΔmecI (✚) grown with oxacillin at 1.0 μg/ml. (B) Results for SE42 (▵) and SE42ΔmecI ( ) with no oxacillin, SE42 (▴) grown with oxacillin at 0.5 μg/ml, and SE42ΔmecI (✚) grown with oxacillin at 1.0 μg/ml. (C) Results for SE53 (▵) and SE53ΔmecI ( ) with no oxacillin, SE53 (▴) grown with oxacillin at 0.5 μg/ml, and SE53ΔmecI (✚) grown with oxacillin at 1.0 μg/ml.
To determine if the increase in MICs seen in the mecI mutant isolates could be reproduced by induction of mecA transcription in the mecI+ strains, all 17 mecI+ S. epidermidis isolates were grown for 16 h with and without the inducer CBAP (5 μg/ml), and the MICs were measured the following day (Table 1). The MICs for 5 of the 17 isolates either did not increase at all following CBAP induction (1 isolate) or only increased 1 dilution (4 isolates). The MICs for the remaining 12 isolates increased by 2 dilutions (3 isolates) or >2 dilutions (9 isolates).
In contrast to the increase in MICs seen after induction, EOPs of the 17 S. epidermidis isolates grown with 5 μg of CBAP per ml overnight showed phenotypes that were within 1 log10 of the isolates grown without CBAP, with one exception. SE33 induced with CBAP overnight showed a 2-log10 increase in EOP at oxacillin concentrations of 20, 50, 100, and 500 μg/ml over SE33 grown without CBAP.
Oxacillin growth curves.
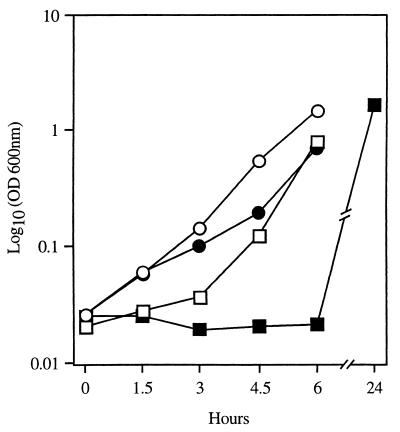
The effect of mecI-mediated mecA repression on the growth of S. epidermidis exposed to β-lactam antibiotics in broth was next sought. SE42 and SE42ΔmecI were both grown for 6 h at 37°C in the presence and absence of 0.6 μg of oxacillin per ml in BHI broth. Growth of the bacteria was monitored at OD600. In the presence of oxacillin, SE42 mecI+ displayed a considerable lag in early growth, with an OD600 at 6 h of 0.026 (Fig. 4). In contrast, the OD600 of SE42ΔmecI at 6 h in the presence and absence of oxacillin was similar to that seen for SE42 in the absence of antibiotics. Results similar to those described above for SE42 were seen when SE20 and SE53 and their mecI-deleted derivatives were grown in the presence of oxacillin.
FIG. 4.
Growth curve of SE42 and SE42ΔmecI with and without oxacillin (0.6 μg/ml). Cell density was measured by OD600 (y axis). □ and ○, SE42 (□) and SE42ΔmecI (○) grown in BHI without oxacillin; ■ and ●, SE42 (■) and SE42ΔmecI (●) grown in BHI with 0.6 μg of oxacillin per ml.
Heterotypic-to-homotypic conversion in the presence of oxacillin.
When sufficient oxacillin was added to S. epidermidis to cause a lag in growth, the EOP phenotype increased between 3 and 5 log10, changing the isolates from a heterotypic to a more homotypic resistance expression (Fig. 3). Incubation of S. epidermidis in concentrations of oxacillin or CBAP that caused no growth lag produced no change in the EOP phenotype despite maximal induction of mecA transcription.
The presence or absence of mecI regulation of mecA transcription influenced the concentration of oxacillin required to produce a growth lag. For all three mecI+ strains, growth inhibition and conversion from heterotypic to homotypic EOP expression could be achieved by incubation in 0.5 μg of oxacillin per ml. However, for each mecI mutant derivative, growth inhibition was only achieved by incubation in 1.0 μg of the antibiotic per ml.
Relationship between MIC and the size of the resistant population.
Oxacillin MICs were determined for isolates SE42, SE20, and SE53 before and after converting expression from heterotypic to homotypic growth. In all cases, the MICs for isolates exhibiting homotypic resistance expression were 128 or 256 μg/ml. In addition, two isolates, SE33 and SE65, that exhibited extremely heterotypic resistance expression, yet for which the MICs were high (64 and 128 μg/ml, respectively [Table 1]), were examined following oxacillin MIC determination. Bacteria were removed from microtiter wells containing 1, 4, 16, and 32 (SE33) or 64 (SE65) μg of oxacillin per ml following overnight incubation, and EOP experiments were performed. SE33 from the MIC microtiter wells became progressively more homotypic in resistance expression with increasing concentrations of oxacillin. In contrast, colonies from wells of SE65 showed no change in the heterotypic subpopulation profile, even at oxacillin concentrations as high as 64 μg/ml (Fig. 5). Colonies taken from 1-μg/ml-oxacillin wells of two isolates for which MICs were low (SE53 and SE60) exhibited no change in EOP.
FIG. 5.
EOP curves of bacteria taken from the microtiter wells of an oxacillin MIC. Shown on the y axis are the numbers of S. epidermidis cells (in log10 CFU per milliliter on oxacillin/CFU per milliliter on MHA) remaining on the plates containing various concentrations of oxacillin (in micrograms per milliliter [shown on the x axis]). (A) EOP results for SE65 taken from the 0-μg/ml oxacillin microtiter well (□), the 1-μg/ml oxacillin well (◊), the 4-μg/ml oxacillin well (○), the 16-μg/ml oxacillin well (▵), and the 64-μg/ml oxacillin well (×). (B) EOP results for SE33 taken from the 0-μg/ml oxacillin microtiter well (□), the 1-μg/ml oxacillin well (◊), the 4-μg/ml oxacillin well (○), the 16-μg/ml oxacillin well (▵), and the 32-μg/ml oxacillin well ( ).
DISCUSSION
The ORSE isolates chosen for this study were previously shown to contain DNA sequences that hybridized with a probe containing the mecA transcriptional repressor, mecI (2). In that study, 48% of the clinical isolates chosen hybridized with the mecI probe; in the other isolates, mecI was deleted and replaced by an IS element (IS1272). In the present study, we have shown that all 17 unique ORSE pulse types with mecI-hybridizing sequences had intact and functional mecA regulators: mecI and mecA P-O DNA sequences in 16 of 17 were the same as wild-type, functional ORSA sequences; baseline, uninduced mecA transcription was heavily repressed; transcription could be induced by β-lactam compounds; and inactivation of mecI led to an increase in both mecA transcription and MICs. The presence of intact mecA regulators in ORSE, in contrast to mutation of these regulators in ORSA, has been previously reported for a group of isolates from a single Japanese hospital (17). The role of these regulators in determining the phenotypic expression of β-lactam resistance in ORSE had not, however, been examined.
The relevance of assessing the relationship of mecA transcriptional repression to phenotype in ORSE is based on several differences between β-lactam resistance in ORSE and that in ORSA. First, as noted above, those ORSE isolates that have mecA regulators contain intact and functional genes in contrast to the usual presence of mutations and insertions that inactivate these genes in ORSA. Second, the oxacillin MICs for as many as 30% of clinical, mecA-positive S. epidermidis isolates, as determined by broth microdilution, are ≤2 μg/ml, below the NCCLS breakpoint that identifies more than 95% of mecA-positive S. aureus strains (21). Finally, the majority of ORSE strains exhibit extreme heterotypy when examined by EOP on oxacillin-containing agar; this expression class is less common among clinical ORSA isolates (T. M. Dickinson and G. L. Archer, unpublished data).
This study documented a direct relationship between resistance to oxacillin and mecA transcription when S. epidermidis was grown in broth. This relationship was best shown when low concentrations of the antibiotic (0.5 to 1.0 μg/ml) were used during rapid growth in broth over 6 h, but was also seen at higher antibiotic concentrations during low inoculum overnight broth MIC determinations. These data imply that, under conditions of rapid bacterial growth in broth, the presence of increasing amounts of PBP2a in membranes provides increasing resistance to β-lactam antibiotics. However, our data also showed that exposure of bacteria in broth to concentrations of oxacillin that suppressed growth selected a highly oxacillin-resistant subpopulation. This selection of a more highly β-lactam-resistant population upon β-lactam exposure has also been noted for mecA-positive S. aureus (6, 12) and has been attributed to changes in the bacterial cell wall that improve the efficiency of PBP2a in cell wall construction. The nature of the conversion of heterotypic to homotypic resistance expression among staphylococci has eluded molecular definition for some time. Data remain, therefore, largely descriptive. However, preliminary studies suggest that growth in β-lactam antibiotics selects a mutant population rather than inducing a regulatory pathway (J. E. Finan, T. M. Dickinson, A. E. Rosato, and G. L. Archer, unpublished data).
The selection of the resistant subpopulation was related to mecA transcription only in that strains with repressed transcription required lower concentrations of oxacillin to select high-level resistance than did strains with unregulated mecA transcription. The size of the highly resistant subpopulation (heterotypic expression), as determined by EOP, was unrelated to mecA transcription and thus to PBP2a quantity. The lack of correlation between heterotypic resistance expression and mecA transcription or PBP2a quantity has been noted by other investigators studying S. aureus (5, 25).
These observations suggest that there are at least two mechanisms that determine the survival of mecA-positive S. epidermidis strains following their exposure to penicillinase-resistant penicillins. Upon initial exposure to a β-lactam antibiotic, the growth rate of the planktonic cells that constitute the majority of the bacterial population may be directly related to their expression of PBP2a. The expression of PBP2a, regulated at the level of mecA transcription, is affected by both the MecR1-MecI and BlaR1-BlaI sensor-transducers and repressors. In contrast, selection of the more highly resistant minority subpopulation occurs more slowly, is likely to require the participation of additional chromosomal factors (3, 7, 30), and may be favored by growth on solid surfaces or in biofilms.
The contribution of β-lactamase regulators to mecA transcriptional regulation was also observed in this study. When mecI was inactivated in isolates SE20 and SE53, blaI regulation persisted, affording partial transcriptional repression. However, blaI repression was easily and rapidly removed following induction with concentrations of β-lactam antibiotics that were one-fifth those required for induction of mecA transcription through mecR1 and mecI. The difference in β-lactam induction of mecA through mecR1 (slow and partial) versus that through blaR1 (rapid and complete) has been noted previously (19, 29) in S. epidermidis and S. aureus. The inefficiency of mecA induction through mecR1 by oxacillin may further explain why isolates containing these regulators have persistent phenotypic repression following relatively short incubation times in broth. In contrast, in ORSE isolates without mecI, most of which will contain β-lactamase regulatory sequences, mecA transcription is rapidly induced following β-lactam exposure, and, therefore, β-lactamase regulators contribute very little to mecA-related variations in phenotypic expression.
Our data showed that the ORSE oxacillin MIC, as determined in broth with the low standard inoculum, could be independently affected both by mecA transcriptional regulation and by a change in the size of the resistant subpopulation. Inactivation of mecI and induction of mecA transcription by CBAP were able to raise the MIC without increasing the size of the highly resistant subpopulation, as determined by EOP. However, since the MICs for only 8 of the 17 unique mecI-positive ORSE pulse types that we examined were ≤4 μg/ml and the MICs for some isolates with naturally occurring mecI deletions are low (T. M. Dickinson and G. L. Archer, unpublished data), transcriptional repression of mecA is not a complete explanation for low broth dilution MICs.
We also showed that even though there was no correlation between the initial size of the resistant subpopulation (baseline EOPs) and MICs, when isolates for which MICs were low were converted from heterotypic to homotypic resistance expression, MICs increased markedly. In addition, colonies removed from microtiter wells of one ORSE isolate for which the MIC was high and with a very heterotypic baseline EOP became progressively more homotypic in resistance expression as the oxacillin concentrations in the wells increased. This suggests that, with the low standard inoculum used for MIC testing, the ability of some isolates to increase the size of their highly resistant subpopulations upon β-lactam exposure can determine the MICs for them. It further suggests that the differences in MICs for some ORSE isolates may be due to genetically determined variations in their capacity to convert from heterotypic to homotypic resistance expression upon β-lactam exposure. However, the failure of SE65 to exhibit homotypic conversion in microtiter MIC wells containing high concentrations of oxacillin suggests that there are mechanisms that determine ORSE broth microtiter MICs other than those examined in this study. Furthermore, since the MIC measures rapid growth in broth and the EOP determines subpopulation distribution on agar, the two assays may be measuring different aspects of the response of some S. epidermidis isolates to β-lactam antibiotics.
ACKNOWLEDGMENT
This work was supported in part by USPHS grant R37 AI35705 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Archer G L, Thanassi J A, Niemeyer D M, Pucci M J. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1996;40:924–929. doi: 10.1128/aac.40.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger-Bachi B, Tschierske M. Role of Fem factors in methicillin resistance. Drug Resistance Updates. 1998;1:325–335. doi: 10.1016/s1368-7646(98)80048-4. [DOI] [PubMed] [Google Scholar]
- 4.Chambers H F. Coagulase-negative staphylococci resistant to β-lactam antibiotics in vivo produced penicillin-binding protein 2a. Antimicrob Agents Chemother. 1987;31:1919–1924. doi: 10.1128/aac.31.12.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers H F. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers H F, Hackbarth C J. Effect of NaCl and nafcillin on penicillin-binding protein 2a and heterogeneous expression of methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1987;31:1982–1988. doi: 10.1128/aac.31.12.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lencastre H, Tomasz A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2590–2598. doi: 10.1128/aac.38.11.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fey P D, Climo M W, Archer G L. Determination of the chromosomal relationship between mecA and gyrAin methicillin-resistant coagulase-negative staphylococci. Antimicrob Agents Chemother. 1998;42:306–312. doi: 10.1128/aac.42.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grough J, Murray N. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983;166:1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- 10.Hackbarth C J, Chambers H F. blaI and blaR1 regulate β-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1144–1149. doi: 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hackbarth C J, Miick C, Chambers H F. Altered production of penicillin-binding protein 2a can affect phenotypic expression of methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2568–2571. doi: 10.1128/aac.38.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartman B J, Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1986;29:85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiramatsu K. Molecular evolution of MRSA. Microbiol Immunol. 1995;39:531–543. doi: 10.1111/j.1348-0421.1995.tb02239.x. [DOI] [PubMed] [Google Scholar]
- 14.Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus(MRSA) FEBS Lett. 1992;298:133–136. doi: 10.1016/0014-5793(92)80039-j. [DOI] [PubMed] [Google Scholar]
- 15.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J Bacteriol. 1982;150:804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasatiya S S, Baldwin J. Nature and the determination of tetracycline resistance in Staphylococcus aureus. Can J Microbiol. 1967;13:1079–1086. doi: 10.1139/m67-144. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi N, Taniguchi K, Urasawa S. Analysis of diversity of mutations in the mecI gene and mecA promoter/operator region of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1998;42:717–720. doi: 10.1128/aac.42.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreiswirth B N, Lofdahl A, Betley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock exotoxin structural gene is not detectably transmitted by a phage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 19.Kuwahara-Arai K, Kondo N, Hori S, Tateda-Suzuki E, Hiramatsu K. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP2′ production. Antimicrob Agents Chemother. 1996;40:2680–2685. doi: 10.1128/aac.40.12.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maranan M C, Moreira B, Boyle-Vavra S, Daum R S. Antimicrobial resistance in staphylococci. Infect Dis Clin N Am. 1997;11:813–849. doi: 10.1016/s0891-5520(05)70392-5. [DOI] [PubMed] [Google Scholar]
- 21.McDonald C L, Maher W E, Fass R J. Revised interpretation of oxacillin MICs for Staphylococcus epidermidis based on mecAdetection. Antimicrob Agents Chemother. 1995;39:982–984. doi: 10.1128/aac.39.4.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morton T M, Eaton D M, Johnson J L, Archer G L. DNA sequences and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureusplasmid pGO1. J Bacteriol. 1993;175:4436–4447. doi: 10.1128/jb.175.14.4436-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morton T M, Johnson J L, Patterson J, Archer G L. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob Agents Chemother. 1995;39:1272–1280. doi: 10.1128/aac.39.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. Approved standard M7-A3. Dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 25.Niemeyer D M, Pucci M J, Thanassi J A, Sharma V K, Archer G L. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J Bacteriol. 1996;178:5464–5471. doi: 10.1128/jb.178.18.5464-5471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick R. Properties of high frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 27.Projan S J, Archer G L. Mobilization of the relaxable Staphylococcus aureusplasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol. 1989;171:1841–1845. doi: 10.1128/jb.171.4.1841-1845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouch D A, Byrne M E, Kong Y C, Skurry R A. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn-4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J Gen Microbiol. 1987;133:3039–3052. doi: 10.1099/00221287-133-11-3039. [DOI] [PubMed] [Google Scholar]
- 29.Ryffel C, Kayser F H, Berger-Bächi B. Correlation between regulation of mecAtranscription and expression of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 1992;36:25–31. doi: 10.1128/aac.36.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryffel C, Strässle A, Kayser F, Bächi B-B. Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:724–728. doi: 10.1128/aac.38.4.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenk S, Laddaga R A. Improved method for electroporation of Staphylococcus aureus. FEBS Microbiol Lett. 1992;94:1333–1338. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 32.Sharma V K, Hackbarth C J, Dickinson T M, Archer G L. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. J Bacteriol. 1998;180:2160–2166. doi: 10.1128/jb.180.8.2160-2166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma V K, Johnston J L, Morton T M, Archer G L. Transcriptional regulation by TrsN of conjugative transfer genes on staphylococcal plasmid pGO1. J Bacteriol. 1994;176:3445–3454. doi: 10.1128/jb.176.12.3445-3454.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speer B S, Shoemaker N B, Salyers A A. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin Microbiol Rev. 1992;5:387–399. doi: 10.1128/cmr.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tesch W, Ryffel C, Strässle A, Kayser F H, Berger-Bächi B. Evidence of a novel staphylococcal mec-encoded element (mecR) controlling expression of penicillin-binding protein 2′. Antimicrob Agents Chemother. 1990;34:1703–1706. doi: 10.1128/aac.34.9.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas W D, Archer G L. Mobilization of recombinant plasmids from Staphylococcus aureus into coagulase negative Staphylococcusspecies. Plasmid. 1992;27:164–168. doi: 10.1016/0147-619x(92)90017-5. [DOI] [PubMed] [Google Scholar]
- 37.Ubukata K, Nonoguchi R, Matsuhashi M, Konno M. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol. 1989;171:2882–2885. doi: 10.1128/jb.171.5.2882-2885.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 39.York M K, Gibbs L, Chehab F, Brooks G F. Comparison of PCR detection of mecAwith standard susceptibility testing methods to determine methicillin resistance in coagulase-negative staphylococci. J Clin Microbiol. 1996;34:249–253. doi: 10.1128/jcm.34.2.249-253.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]