Abstract

Moracins, a kind of 2-phenyl-benzofuran compound from Moraceae, serve as phytoalexins with antimicrobial, anti-inflammatory, antitumor, and antidiabetes activities and respond to biotic and abiotic stresses, while their biosynthetic pathway and regulatory mechanism remain unclear. Here, we report a de novo transcriptome sequencing for different tissues of seedlings, as well as leaves under different stresses, in M. alba L. A total of 88 282 unigenes were assembled with an average length of 937 bp, and 82.2% of them were annotated. On the basis of the differential expression analysis and enzymatic activity assays in vitro, moracins were traced to the phenylpropanoid pathway, and a putative biosynthetic pathway of moracins was proposed. Unigenes coding key enzymes in the pathway were identified and their expression levels were verified by real-time quantitative reverse transcription PCR (qRT-PCR). Particularly, a p-coumaroyl CoA 2′-hydroxylase was presumed to be involved in the biosynthesis of stilbenes and deoxychalcones in mulberry. Additionally, the transcription factors that might participate in the regulation of moracin biosynthesis were obtained by coexpression analysis. These results shed light on the putative biosynthetic pathway of moracins, providing a basis for further investigation in functional characterization and transcriptional regulation of moracin biosynthesis in mulberry.
1. Introduction
Morus, a genus of flowering plants in the family Moraceae, has been used as traditional herb and silkworm fodder for thousands of years in China, bringing about important economic and medicinal values. Many parts of mulberry, including leaves, fruits, branches, and root barks, have a variety of biological and pharmacological activities that are antioxidant, antimicrobial, antihyperglycemic, glucosidase-inhibiting, hypolipidemic, antiobesity, antiatherosclerosis, and antineoplastic, which are attributed to the bioactive compounds enriched in tissues.1,2 Until now, several kinds of active ingredients, including flavonoids, alkaloids, stilbenoids, coumarins, and phenolic acids, have been identified from mulberry.1 Furthermore, mulberry is also recognized as a rich source of benzofuran derivatives and Diels–Alder adducts.3
Moracins, the derivatives of 2-phenyl-benzofuran, are mainly isolated from Moraceae, especially Morus. Structurally, they all contain a scaffold of 2-phenyl substituted benzo[b]furan-fused-ring. Twenty-six moracins (A–Z) and their derivatives, modified with hydroxylation, methylation, prenylation, cyclization, glycosylation, and Diels–Alder reaction,4 have been identified from Moraceae, and some of them showed valuable biological and pharmacological activities. For example, moracin M was found to inhibit lipopolysaccharide-induced inflammatory responses in nucleus pulposus cells;5 moracin S, with a prenyl moiety, showed the highest BACE1 inhibitory activity compared with the other moracins, which provided valuable information for the design of anti-Alzheimer’s disease drugs;6 Gao et al.7 reported moracin N induced autophagy and apoptosis through ROS generation in lung cancer. In addition, moracins were found to possess antimicrobial, antioxidant, antidiabetes, and phosphodiesterase inhibition properties.4 However, only some of them have been synthesized chemically.8−10 Considering the difficulties in extraction and isolation of moracins from Morus, the development of their medicinal potential is largely restricted. Therefore, there is an emergency need to elucidate their biosynthetic pathway.
Transcriptomics has been an efficient tool for identifying biosynthetic pathways of nature products in the past decade. In contrast to microbes, the biosynthesis genes of plant-derived compounds are usually not enriched in a cluster on chromosomes, making it hard to screen them directly in the genome. Nevertheless, on the metabolic level, metabolites differentially accumulate in some specific tissues and are affected by biotic or abiotic stress, which is thought to result from the differential expression of biosynthesis genes. Besides, biosynthetic pathway genes in plants are often coordinately regulated, so it is possible to identify candidate genes through coexpression analysis with the dedicated pathway gene to be used as a bait. A few studies have been done on the basis of this method, including identification of genes related to biosynthesis of etoposide aglycone,11 seco-iridoid,12 protolimonoid,13 triterpenoid saponin,14 and colchicine.15
To date, there is little information regarding the biosynthetic pathway of moracins. In this study, we performed de novo RNA sequencing toward different tissues of M. alba L. seedlings, as well as leaves, under ultraviolet-B (UV-B) treatment and Botrytis cinerea infection. Differential expression analysis and coexpression analysis were used to identify candidate genes involved in moracin biosynthesis and transcriptional regulation, and a putative biosynthetic pathway was proposed. Our results will provide a foundation for further biological synthetic study.
2. Results and Discussion
2.1. Identification of Compounds in the Moracin Pathway from Different Samples
Several main metabolites in the moracin biosynthetic pathway were identified by high-performance liquid chromatography (HPLC) and found to be differentially distributed in different tissues (Figure 1a) and leaves under different treatments (Figure 1b), and their relative contents were quantified and shown in Table S2. In untreated seedlings, compounds in the moracin pathway were mainly enriched in roots, especially in fibrous roots (FR), while they were undetectable in untreated leaves (CKL) and stems (S). In addition, leaves infected by B. cinerea (BCL) accumulated all interesting compounds in moracin pathway, including moracin M (standard 3), moracin C (standard 4), moracin N (standard 5), as well as oxyresveratrol (standard 2) and chalcomoracin (standard 7), the up- and downstream compound of moracins, respectively. However, in UV-B-treated leaves (UVL), the accumulation of these metabolites was relatively lower, and moracin M was even undetectable. Besides, mulberroside A (standard 1), the diglucoside of oxyresveratrol, was found to be enriched in taproots (TR) and FR significantly, but was rarely present in other groups, which was not inconsistent with the expression of stilbenoid pathway and could be explained by the expression levels of the glycosyltransferases responsible for mulberroside A biosynthesis in different groups.
Figure 1.
HPLC analysis of the metabolites involved in moracin biosynthetic pathway. (a) Metabolites in different tissues of mulberry. FR, fibrous roots; S, stems; CKL, untreated leaves; TR, taproots. (b) Metabolites in different treated mulberry leaves. UVL, UV-B-treated leaves; BCL, B. cinerea-infected leaves. Standards 1–7 represent mulberroside A, oxyresveratrol, moracin M, moracin C, moracin N, morachalcone A, and chalcomoracin, respectively.
As indicated by previous studies, moracins are unique and vital compounds mostly isolated from mulberry barks and root barks, as well as diseased shoots.4 In our study, HPLC analysis showed consistent results that moracins were only detected in the roots of mulberry seedlings but accumulated in leaves under biotic infection and UV-B radiation to different degrees, suggesting their function as phytoalexins.
2.2. Enzymatic Assays of the Key Step for Biosynthesis of Moracins in Mulberry
Moracins share the structures of 2-phenyl-benzofuran with two C–O bonds at the meta positions of the benzene and another one at the C-6 position of benzofuran. They are mainly isolated from Moraceae plants, and most of their derivatives are modified by hydroxylation, methylation, prenylation, and cyclization. Among them, moracin M is the simplest one in structure and recognized as the precursor for the other moracins (Table S1). Therefore, the biosynthesis of moracin M is important for the biosynthesis of moracins in mulberry. In the chromatographic profile of BCL, a strong peak eluted at 14.8 min was considered as oxyresveratrol because of the same retention time and ultraviolet absorption with the standard 2 (Figure 1). It is worth noting that oxyresveratrol has a similar structure to moracin M, especially for the hydroxy position on benzene ring, which suggests that oxyresveratrol participates in the response to biotic stress along with moracin pathway.
Enzymatic activity assays in vitro showed that oxyresveratrol could be converted into moracin M under the catalyzation of crude enzymes from fibrous roots of mulberry seedlings (Figure S1). Therefore, it was proved that the formation of benzofuran ring of moracins was enzymatic, and the enzyme(s) responsible were named as moracin M synthase(s) (MMS). The MMS activity from other groups was also analyzed. The results showed that the MMS activity of fibrous roots was the highest, followed by taproots and stems; however, no MMS activity was detected in untreated leaves and UV-B-treated leaves (Figure 2). Meanwhile, in B. cinerea-infected leaves, the MMS activity was activated to a detectable level. The combined enzymatic assays and metabolic level (Figure 1) indicated that the biosynthesis of moracins was enriched in roots, not in leaves, and could be significantly activated under B. cinerea infection rather than UV-B radiation.
Figure 2.
Enzymatic activity of moracin M synthase in different groups. FR, fibrous roots; TR, taproots; S, stems; CKL, untreated leaves; UVL, UV-B-treated leaves; BCL, B. cinerea-infected leaves.
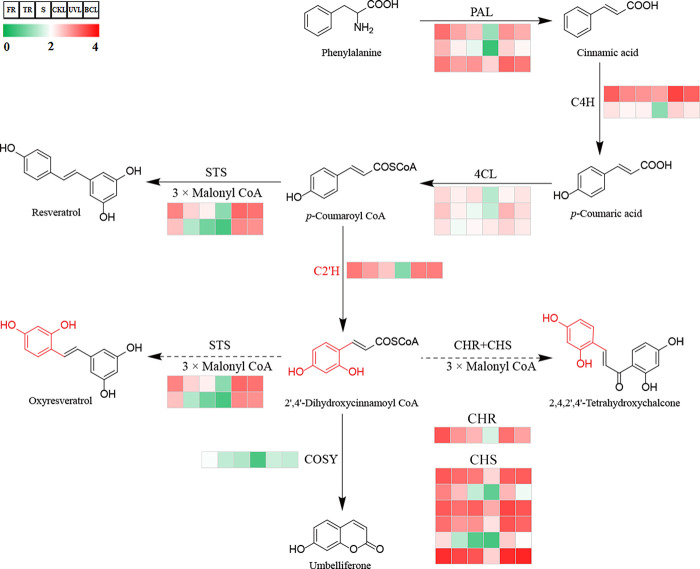
Mulberry contains high levels of stilbenoids, including oxyresveratrol and its diglucoside mulberroside A (Figure 1).16 Stilbenoids are phenylpropanoid phytoalexins produced by plants in response to biotic and abiotic stress.17 Therefore, the MMS relates moracin biosynthesis to stilbenoid and phenylpropanoid pathways (Figure 4).
Figure 4.
Putative phenylpropanoid pathway in M. alba. The expression levels of unigenes coding the enzymes are shown in heatmap. PAL, phenylalanine ammonia-lyase; C4H, trans-cinnamate 4-hydroxylase; 4CL, 4-coumarate-CoA ligase; C2′H, p-coumaroyl CoA 2′-hydroxylase; CHS, chalcone synthase; CHR, chalcone reductase; STS, stilbene synthase; COSY, coumarin synthase. FR, fibrous roots; TR, taproots; S, stems; CKL, untreated leaves; UVL, UV-B-treated leaves; BCL, B. cinerea-infected leaves. The solid and broken lines represent enzymatic reactions demonstrated in other species and putative in M. alba, respectively.
2.3. Transcriptome Sequencing, De Novo Assembly and Functional Annotation
Transcriptome sequencing of 6 groups containing 17 samples of M. alba L. generated 388.3 million raw reads (Table S3). After filtering, 382.3 million clean reads and 115.0 GB clean bases were de novo assembled using Trinity, yielding a total of 192 111 transcripts and 88 282 unigenes. The average length and N50 value of unigenes were 937 bp and 1421 bp, respectively (Table S4). Functional annotation showed that 61 514 (69.7%), 62 154 (70.4%), 21 626 (24.5%), 43 170 (48.9%), 37 643 (42.6%), 37 639 (42.6%), and 11 748 (13.3%) unigenes were annotated in NR (nonredundant protein database), NT (nonredundant nucleotide database), KO (Kyoto Encyclopedia of Genes and Genomes Orthology), SwissProt (annotated protein sequence database), Pfam (protein family), GO (Gene Ontology), and KOG (EuKaryotic Orthologous Groups) databases, respectively (Table S5).
2.4. Differential Expression Analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment
To identify differentially expressed genes (DEGs) in different tissues, as well as leaves under UV-B and B. cinerea treatments, read counts of all unigenes were normalized through DESeq method, and then hypothesis testing was conducted in pairwise groups (FR vs TR, FR vs S, FR vs CKL, UVL vs CKL, and BCL vs CKL). The FR vs TR comparison revealed 8176 DEGs, with 4982 unigenes up-regulated and 3194 unigenes down-regulated. For the FR versus S comparison, 10 447 DEGs were identified, including 5608 up-regulated and 4839 down-regulated unigenes. Furthermore, the most DEGs, 8077 up-regulated and 6893 down-regulated, were identified in the FR versus CKL comparison, suggesting a large biological difference between roots and leaves. In addition, compared to untreated leaves, 13 972 unigenes were differentially expressed in UVL, including 6734 up-regulated unigenes and 7238 down-regulated unigenes, while in BCL the numbers were 8629, 4593, and 4036, respectively.
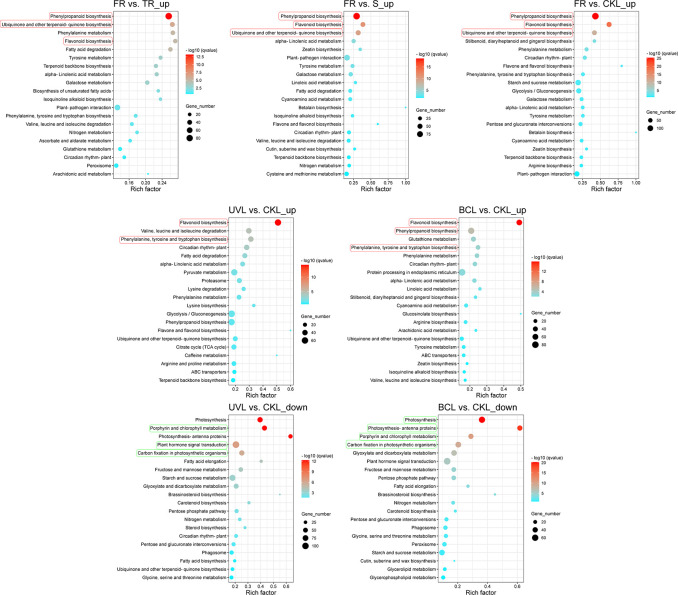
These DEGs were then enriched in KEGG pathways, and the top 20 enrichment pathways in each comparison are shown in Figure 3. Interestingly, we found that up-regulated DEGs in FR compared to the other tissues (TR, S, and CKL) were mostly enriched in “phenylpropanoid biosynthesis,” “ubiquinone and other terpenoid-quinone biosynthesis,” and “flavonoid biosynthesis.” Besides, compared with CKL, the up-regulated DEGs in UVL and BCL were also enriched in “flavonoid biosynthesis;” “phenylalanine, tyrosine and tryptophan biosynthesis;” and “phenylpropanoid biosynthesis” pathways. All of these pathways had a close relation to the biosynthesis of phenylpropanoids. In addition to flavonoids, moracins were also presumed to be derived from phenylpropanoid pathway due to the discovery of MMS. These results were consistent with the enrichment of moracin biosynthesis in FR, UVL, and BCL at the level of transcription. However, the down-regulated DEGs in UVL and BCL were mainly enriched in “photosynthesis,” “porphyrin and chlorophyll metabolism,” “photosynthesis-antenna proteins,” and “carbon fixation in photosynthetic organisms” pathways, indicating the damage of UV-B radiation and B. cinerea infection on photosynthesis in mulberry leaves.
Figure 3.
KEGG enrichment of top 20 pathways of DEGs in different comparisons. FR, fibrous roots; TR, taproots; S, stems; CKL, untreated leaves; UVL, UV-B-treated leaves; BCL, B. cinerea-infected leaves. Rectangles represent the major enriched pathways.
2.5. Identification and Expression Analysis of Known Genes Involved in the Moracin Biosynthetic Pathway
The confirmation of enzymatically converting oxyresveratrol to moracin M enabled us to integrate the biosynthesis of moracins with the downstream of stilbenoids and phenylpropanoid pathways and correlate them legitimately. By analyzing their expressions in different groups, homologous genes involved in putative pathway were filtered and summarized (Table S6). Unigenes coding phenylalanine ammonia-lyase (PAL, EC 4.3.1.24), trans-cinnamate 4-hydroxylase (C4H, EC 1.14.14.91), and 4-coumarate-CoA ligase (4CL, EC 6.2.1.12), common and conserved enzymes of phenylpropanoid pathway, were examined and visualized (Figure 4). Three, two, and three nonredundant unigenes were annotated as PAL, C4H, and 4CL (Table S6), respectively. Chalcone reductase (CHR), combined with chalcone synthase (CHS, EC 2.3.1.70), catalyzed three molecules of malonyl-CoA and one molecule of p-coumaroyl CoA to form deoxychalcone.18 Six and one unigenes were identified to code CHS and CHR, respectively. Another two unigenes were taken as coding stilbene synthase (STS, EC 2.3.1.74), which utilized the same substrates as CHS but produced the resveratrol instead.19−21 Based on the fragments per kilobase million (FPKM) values, most of these unigenes had the highest expression level in FR, followed by TR and S, with relatively low expression in CKL. In addition, most of these unigenes were up-regulated in UVL and BCL compared to CKL (Figure 4 and Table S6).
It was interesting that a unigene annotated as p-coumaroyl CoA 2′-hydroxylase (C2′H) showed a high similar expression pattern to the aforementioned unigenes in phenylpropanoid pathway (Figure 4 and Table S6). C2′H was a 2-oxoglutarate-dependent dioxygenase that catalyzed the ortho-hydroxylation of p-coumaroyl CoA and feruloyl CoA, produced umbelliferone and scopoletin in final via a spontaneous or enzymatic cyclization, respectively, and participated in the biosynthesis of coumarins.22,23 We further determined the transcription levels of genes involved in the biosynthesis of simple coumarins, since no furan- or pyran- coumarins were reported in mulberry.1 Coumarins were also derived from the phenylpropanoid pathway as flavonoids and stilbenoids, and the subsequent efficient trans–cis isomerization and lactonization catalyzed by coumarin synthase (COSY) formed the coumarin skeleton (Figure 4).24 As expected, COSY showed a lower expression level in our transcriptome compared with C2′H (Figure 4 and Table S6), making us reexamine the roles of C2′H in secondary metabolism of mulberry.
In the biosynthesis of moracins, we noticed a pair of similar stilbenes, resveratrol and oxyresveratrol, and the latter had an extra hydroxyl at the C-2’ position. It was similar to the difference between p-coumaroyl CoA and 2′,4′-dihydroxycinnamoyl CoA (Figure 4 and S2). STS catalyzed three molecules of malonyl-CoA and one molecule of p-coumaroyl CoA to produce resveratrol in peanut,19 grape,20 and mulberry.21 STS converted cinnamoyl-CoA and malonyl-CoA to pinosylvin, a stilbene similar to resveratrol and oxyresveratrol, while in scots pine (Figure S2).25 Therefore, oxyresveratrol was reasonably supposed to derive from 2′,4′-dihydroxycinnamoyl CoA and malonyl-CoA under the catalysis of STS. In other words, C2′H might play an important role in the biosynthesis of stilbenoids and moracins in mulberry.
Meanwhile, 2,4,2′,4′-tetrahydroxychalcone was determined in structure by reasoning about the prenylation catalyzed by M. alba isoliquiritigenin 3′-dimethylallyltransferase (MaIDT) in formation of morachalcone A (Figure 5).26 This compound had an extra hydroxyl at the C-2 position compared to isoliquiritigenin, a deoxychalcone formed by malonyl-CoA and p-coumaroyl CoA in the presence of CHS-CHR complexes and involved in the biosynthesis of deoxyflavonoids (Figure S2).18 Similar to oxyresveratrol, 2,4,2′,4′-tetrahydroxychalcone was presumed to be biosynthesized from 2′,4′-dihydroxycinnamoyl CoA and malonyl CoA, which was supported by the identification of a highly expressed unigene coding CHR (Figure 4 and Table S6). In summary, by structure comparison, we assigned C2′H into the biosynthesis of stilbenoids and flavonoids in mulberry, which was supported by our transcriptome data.
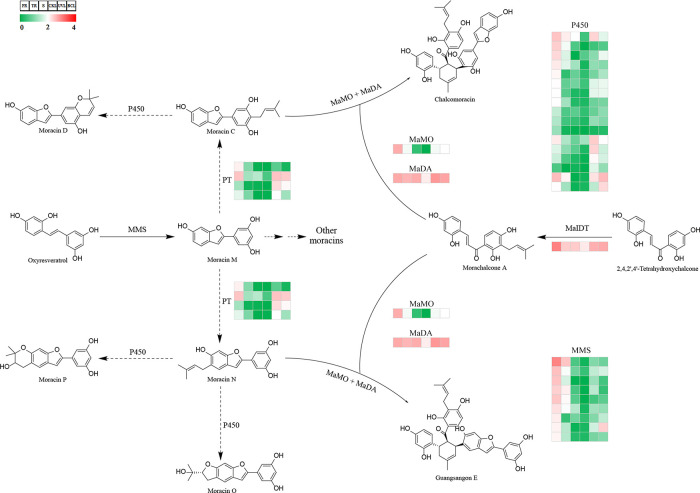
Figure 5.
Putative biosynthetic pathway for moracins and their downstream Diels–Alder adducts in M. alba. The expression levels of unigenes coding the enzymes are shown in heatmap. MMS, moracin M synthase(s); PT, prenyltransferase; P450, cytochrome P450s; MaIDT, M. alba isoliquiritigenin 3′-dimethylallyltransferase; MaMO, M. alba moracin C oxidase; MaDA, M. alba Diels–Alderase. FR, fibrous roots; TR, taproots; S, stems; CKL, untreated leaves; UVL, UV-B-treated leaves; BCL, B. cinerea infected leaves. The solid and broken lines (curves) represent verified and putative enzymatic reactions in M. alba, respectively.
Candidate unigenes coding the other known enzymes in moracin and the downstream pathway were identified and summarized (Figure 5 and Table S6). Three unigenes were annotated as MaIDT, M. alba moracin C oxidase (MaMO), and M. alba Diels–Alderase (MaDA), respectively, and the levels of expression were consistent with that of the former unigenes.
2.6. Identification of Unknown Candidate Genes Involved in the Moracin Biosynthetic Pathway
Transcriptomic analyses also indicated that the transcriptional levels of genes involved in the moracin biosynthetic pathway were highly consistent with the metabolic level, inspiring us to screen the unknown candidate genes according to the following rules. First, the up-regulated DEGs in FR versus TR, FR versus S, FR versus CKL, and BCL versus CKL comparisons were gathered and analyzed, and 717 DEGs were commonly up-regulated in all four comparisons (Figure 6). Most of the inconsistent unigenes were filtered at the first step while the unigenes with high expression levels in FR group and activated by B. cinerea infection were reserved. Second, since moracin biosynthetic pathway was silent in untreated leaves, another 97 DEGs were excluded because their average FPKM values were higher than 10 in the CKL group. The remaining 620 DEGs were used for the further screening of the candidates coding specific enzymes in moracin biosynthetic pathway such as MMS, prenyltransferases (PTs), and cytochrome P450s.
Figure 6.
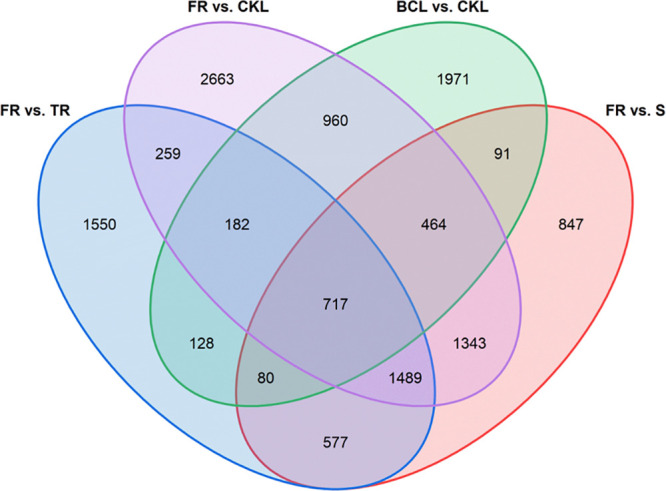
Venn diagram of up-regulated DEGs in different comparisons. FR, fibrous roots; TR, taproots; S, stems; CKL, untreated leaves; UVL, UV-B-treated leaves; BCL, B. cinerea-infected leaves.
Homologous BLAST (basic local alignment search tool) was improper for identification of candidate unigenes for MMS since the reaction type had not been determined. Therefore, a further filter was conducted on the basis of the enzymatic level. DEGs with the average FPKM values ≥10 in UVL group or <10 in FR group were eliminated and 118 unigenes were retained, of which 64 unigenes possessed open reading frames (ORFs) of more than 200 amino acids (Table S7). According to the Pfam annotation and the type of reaction that we could hypothesize, nine candidate unigenes for MMS were proposed, including five cytochrome P450s, as well as one for berberine bridge enzymelike protein, multicopper oxidase (laccase), polyphenol oxidase, and 2-oxoglutarate-dependent oxygenase (Table S7).
Prenylations and oxidative cyclizations were the most common and significant modifications in the process of moracins biosynthesis (Table S1).4 Studies showed that PTs could catalyze the prenylation of various substrates, including flavonoids,27 coumarins,28 stilbenoids,29 and other phenolic compounds.30 Two PT sequences have previously been reported from mulberry. One was MaIDT, which catalyzed the C-3′ and C-6 prenylation on chalcones and flavones, respectively,26 and the other was MaOGT catalyzing the prenylation of oxyresveratrol.31 Most PTs had a strong substrate specificity and only accepted one or several similar prenyl acceptors. However, for moracins, prenylations occurred at different sites on similar substrates. Therefore, there might be several different PTs catalyzing the prenylation of moracins. The identification of PTs involved in moracin pathway was carried out based on the annotation in Pfam database. The Pfam term “UbiA prenyltransferase family” (Pfam ID: PF01040) was used to screen the 620 DEGs, and 4 candidates were obtained (Figure 5 and Table S6).
Similar to furanocoumarins and pyranocoumarins, some moracins also had a furan or pyran ring coupled to the benzenes in structure, such as moracins D, E, K, O, P, and W (Table S1). It has been reported that the furan or pyran ring was formed by the cyclization of isopentenyl and ortho-hydroxyl under the catalysis of cytochrome P450s.32,33 To identify the P450s responsible for the formation of furan and pyran rings in moracins, the same method for PTs screening was used for P450s, and 17 candidate unigenes with a Pfam annotation of “Cytochrome P450” (Pfam ID: PF00067) were acquired (Figure 5 and Table S6).
2.7. Identification of Transcription Factors (TFs) Related to Moracin Biosynthetic Pathway
TFs are a group of specific DNA-binding proteins that can regulate gene expression. In our transcriptome, a total of 2564 unigenes were identified as putative TFs, and most of them were annotated to MYB (216, 8%), AP2/ERF (171, 7%), C2H2 (132, 5%), bHLH (119, 5%), WRKY (107, 4%), GRAS (101, 4%), and C2C2 (100, 4%) families (Table 1). To identify TFs regulating moracin biosynthesis, the unigene “Cluster_14943.34115,” annotated as STS with an intact ORF of 391 amino acids and serving as the specific structural gene in moracin pathway, was used as the query for coexpression analysis of putative TF-coding unigenes. There were 69 TF-coding unigenes that showed high coexpression (Pearson’s r > 0.80 or < −0.80) with Cluster_14943.34115 (Table S8), including 66 positive ones and 3 negative ones. Furthermore, these coexpressed TFs mostly consisted of AP2/ERF family (16, 23%), WRKY family (11, 16%), and NAC family (9, 13%) (Table 1). Although no study about TFs regulating the biosynthesis of moracins or stilbenoids in mulberry was reported, several TFs involved in stilbenoids biosynthesis have been cloned and characterized in Vitis vinifera. These TFs belong to MYB, AP2/ERF, WRKY, and bZIP families and regulate the expression of STS directly or indirectly.34−37 Therefore, the regulation of stilbenoids and moracins in mulberry might be complicated and interlaced, and much work remains to be done.
Table 1. Type and Number of Transcription Factor (TF) Families.
| coexpressed
TFs |
|||
|---|---|---|---|
| TF family | total TFs | positive | negative |
| MYB superfamily | 216 | 4 | 0 |
| AP2/ERF | 171 | 16 | 0 |
| C2H2 | 132 | 4 | 0 |
| bHLH | 119 | 4 | 0 |
| WRKY | 107 | 11 | 0 |
| GRAS | 101 | 3 | 0 |
| C2C2 | 100 | 1 | 0 |
| HB | 90 | 1 | 0 |
| C3H | 89 | 2 | 0 |
| NAC | 88 | 9 | 0 |
| bZIP | 82 | 1 | 0 |
| B3 superfamily | 78 | 1 | 2 |
| AUX/IAA | 54 | 0 | 1 |
| mTERF | 51 | 0 | 0 |
| FAR1 | 50 | 0 | 0 |
| Trihelix | 49 | 0 | 0 |
| GARP | 45 | 2 | 0 |
| NF-Y | 42 | 0 | 0 |
| LOB | 39 | 0 | 0 |
| others | 861 | 7 | 0 |
| total | 2564 | 66 | 3 |
2.8. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) Validation of DEGs Involved in the Moracin Biosynthetic Pathway
The expression levels of nine DEGs involved in the proposed moracin biosynthetic pathway were verified by qRT-PCR analysis. Most of unigenes showed a high expression trend in FR, low in CKL, and up-regulated in UVL and BCL groups (Figure 7). The results were consistent with the transcriptomic data.
Figure 7.
Validation for DEGs by qRT-PCR. The relative expression of qRT-PCR was indicated on the left y-axis and the FPKM normalized expression level (log2FPKM) of RNA-Seq was indicated on the right y-axis. FR, fibrous roots; TR, taproots; S, stems; CKL, untreated leaves; UVL, UV-B-treated leaves; BCL, B. cinerea-infected leaves.
3. Conclusions
On the basis of differential expression analysis and enzymatic assay in vitro, a putative biosynthesis pathway of moracins was proposed in M. alba L. In addition, a total of 51 structural unigenes and 69 TFs were putatively identified to participate in moracin biosynthesis and could be further confirmed by functional validation and DNA-binding assays.38 We provided evidence that moracins are derived from stilbene and phenylpropanoid pathway, and there could be two novel pathways for stilbene and deoxychalcone biosynthesis in mulberry. The presented work provides a basis for further investigation in functional characterization and transcriptional regulation of moracin biosynthesis in mulberry.
4. Methods
4.1. Plant Materials and Treatments
Mature seeds of M. alba L. were collected from the experimental plot in Zijingang Campus of Zhejiang University (Hangzhou, China), washed with running water, and sown directly in soil for germination. Mulberry seedlings were grown in a green house at 25 ± 1 °C with a 16 h light/8 h dark cycle for 5 to 6 weeks. The TR, FR, and S tissues were harvested. At the same time, the middle 3∼4 pieces of leaves were detached, followed by wrapping petioles with wet cotton balls, and used for either of the following treatments: (a) the detached leaves were treated with UV-B (290–320 nm) radiation according to our previous method,39 transferred to covered Petri dishes to retain moisture, and incubated at 25 °C in darkness for 36 h; (b) each mulberry leaf was inoculated with 44 mm diameter mycelium disks taken from 10-day-old B. cinerea grown on potato dextrose agar medium, placed in a covered Petri dish, and incubated at 25 °C with a 16 h light/8 h dark cycle for 48 h. The CKL, UVL, and BCL samples were harvested at the given points in time. All samples were immediately snap-frozen in liquid nitrogen and stored at −80 °C for future use. Tissues from each of the four plants were pooled into one biological replicate, and the same samples were used for metabolite analysis, RNA sequencing (RNA-seq), and qRT-PCR.
4.2. Metabolite Extraction and HPLC Analysis
Mulberry samples were grinded into powder in liquid nitrogen and lyophilized overnight. A portion (0.5 mL) of absolute methanol was added to a 1.5 mL EP tube containing 10 mg of dried sample, followed by sonication for 60 min and centrifugation at 12 000g for 20 min to precipitate plant debris. The supernatants were then filtered through a 0.22 μm nylon filter and used for HPLC analysis. The standards of mulberroside A, oxyresveratrol, and moracin M were purchased from Yuanye Bio-Technology (Shanghai, China), and the others (moracin C, moracin N, morachalcone A, and chalcomoracin) were isolated and identified by our lab.40
A 10 μL aliquot of extracted samples and the mixed standard solution (10 μM for each compound in methanol) was used for HPLC analysis on an Agilent Series 1260 liquid chromatograph (Agilent) with a C18 column (250 mm × 4.6 mm, inner dimension: 5 mm, Agilent). Acetonitrile (solvent A) and 0.1% formic acid in water (solvent B) were used as the mobile phase with a flow rate of 1.0 mL min–1 at 40 °C. The gradient conditions were optimized as follows: 0–25 min, 10∼50% A; 25–40 min, 50∼95% A; 40–45 min, 95% A; 45–47 min, 95∼100% A; 47–52 min, 100% A. Spectra were measured at a wavelength of 320 nm.
4.3. Enzymatic Assay of Moracin M Synthase In Vitro
To extract crude enzymes from mulberry samples, a portion (0.1 g) of fresh tissue was ground with 1 mL of 100 mM sodium phosphate buffer (pH = 7.5) on ice, added with 1% PVPP. The mixture was then centrifuged at 12 000 g at 4 °C for 20 min, and the supernatant was collected as crude enzymes and tested for protein concentration using the Bradford method.41 The enzymatic reaction was conducted in the mixture (100 μL) that contained 100 mM sodium phosphate buffer (pH = 7.5), 500 μM oxyresveratrol as substrate, and 50 μg crude enzymes. The reactions were incubated at 30 °C for 3 h, then terminated by the addition of 100 μL of methanol and centrifuged at 15 000g for 20 min. The crude enzymes without substrate and boiled crude enzymes were used as controls. Supernatants were analyzed by HPLC using the same method as metabolite analysis, and the peak area of moracin M was calculated to evaluate the activity of MMS.
4.4. RNA Extraction and cDNA Library Preparation
Total RNA was extracted using the RNAprep Pure Plant Plus Kit (TIANGEN BIOTECH, Beijing, China) from FR, S, CKL, UVL, BCL (3 biological replicates), and TR (2 biological replicates). RNA integrity was assessed by 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Strand-specific cDNA libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) according to the manufacturer’s instructions. The quality and insert size of cDNAs were determined by 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA).
4.5. Transcriptome Sequencing, De Novo Assembly, and Annotation
Libraries were sequenced (PE150) on an illumina Novaseq 6000 sequencer. Raw reads of FASTQ files were trimmed using fastp,42 and clean reads were obtained by removing reads containing adapter, reads containing ploy-N, and reads of low quality. Clean reads from all libraries were then combined and assembled into de novo transcriptomes using Trinity (v2.4.0) in strand-specific mode,43,44 and unigenes were finally obtained. ORFs were identified by BLAST in NR and SwissProt protein database or predicted by Estscan (v3.0.3) for those unmapped or mapped with no predicted sequences.45 Unigenes were annotated in NR, NT, SwissProt, KEGG, KOG, Pfam, and GO databases. The KEGG pathway identification and enrichment were conducted by KOBAS (v2.0.12).46 The ORFs of unigenes were mapped to the plant TF database (PlantTFDB) by Hmmsearch (v3.0) using iTAK software (v1.2)47 to identify the potential TFs.
4.6. Analysis of DEGs
Data sets of samples from same tissue or treatment were considered as a group and differential expression analysis of two groups was performed using the DESeq2 R package (v1.6.3).48 In this study, the resulting P values were adjusted by the Benjamini–Hochberg approach to control the false discovery rate. Finally, an adjusted p-value <0.05 and |log2(FoldChange)| > 1 were used as the threshold to determine DEGs between two groups.
4.7. Expression Levels and Coexpression Analysis
Clean reads were mapped to the Trinity unigenes using RSEM (v1.2.15) with the bowtie2 parameter set at 0 mismatch.49 The number of mapped clean reads for each unigene was then counted and normalized into number of FPKM to estimate the expression level of the unigenes.50 Pearson correlation analysis was carried out to identify genes coexpressed with the gene of interest using the stats package available in R (v1.3.1093) based on FPKM values.
4.8. Quantitative Real-Time PCR Validation
The expression levels of nine unigenes related to moracin biosynthesis were validated by qRT-PCR. Total RNA (1.0 μg) was utilized for cDNA synthesis using 5X All-In-One RT MasterMix Kit (Applied Biological Materials Inc., Richmond, British Columbia, Canada) following the manufacturer’s instructions. The specific qRT-PCR primers were designed using NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome; Table S9). The qRT-PCR analysis was performed using BlasTaq 2X qPCR MasterMix (Applied Biological Materials Inc., Richmond, British Columbia, Canada) on the BIO-RAD CFX Connect Real-Time system (Bio-Rad, Hercules, CA, USA). Each 20 μL reaction contained 100 μg cDNA, 10 μL BlasTaq 2X qPCR MM, and 0.25 μM primers. The reaction conditions were as follows: 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. The experiment was conducted in three technical replicates each, using beta actin gene (XM_010112836.2) as an internal reference control. The relative gene expression and fold change were calculated with the 2–ΔΔCt method.51
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No.81872973) and Zhejiang Provincial Public Welfare Technology Research Project (No.LGJ22H280001).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c00409.
Chemical structures of moracins A–Z and their derivatives; peak area of seven interested metabolites in different groups based on HPLC analysis; summary of transcriptome sequencing data; assembly result of clean reads; annotation of unigenes against public databases; identification and expression analysis of unigenes involved in moracin biosynthesis; candidate unigenes coding moracin M synthase(s); identification and expression analysis of transcription factors involved in moracin biosynthesis; primers used for qRT-PCR; HPLC analysis for the enzymatic activity of crude enzymes to convert oxyresveratrol to moracin M; biosynthesis of different stilbenes and droxychalcones (PDF)
Author Contributions
# S.L. and Z.Z. contributed equally to this paper.
The authors declare no competing financial interest.
Notes
The raw data of RNA-Seq have been submitted to the NCBI SRA (PRJNA775672).
Supplementary Material
References
- Chan E. W.; Lye P. Y.; Wong S. K. Phytochemistry, pharmacology, and clinical trials of Morus alba. Chin J. Nat. Med. 2016, 14 (1), 17–30. 10.3724/sp.J.1009.2016.00017. [DOI] [PubMed] [Google Scholar]
- Yuan Q.; Zhao L. The mulberry (Morus alba L.) fruit-a review of characteristic components and health benefits. J. Agric. Food Chem. 2017, 65 (48), 10383–10394. 10.1021/acs.jafc.7b03614. [DOI] [PubMed] [Google Scholar]
- Dat N. T.; Jin X.; Lee K.; Hong Y. S.; Kim Y. H.; Lee J. J. Hypoxia-inducible factor-1 inhibitory benzofurans and chalcone-derived diels-alder adducts from Morus species. J. Nat. Prod. 2009, 72 (1), 39–43. 10.1021/np800491u. [DOI] [PubMed] [Google Scholar]
- Naik R.; Harmalkar D. S.; Xu X.; Jang K.; Lee K. Bioactive benzofuran derivatives: moracins A-Z in medicinal chemistry. Eur. J. Med. Chem. 2015, 90, 379–393. 10.1016/j.ejmech.2014.11.047. [DOI] [PubMed] [Google Scholar]
- Guo F.; Zou Y.; Zheng Y. Moracin M inhibits lipopolysaccharide-induced inflammatory responses in nucleus pulposus cells via regulating PI3K/Akt/mTOR phosphorylation. Int. Immunopharmacol. 2018, 58, 80–86. 10.1016/j.intimp.2018.03.015. [DOI] [PubMed] [Google Scholar]
- Seong S. H.; Ha M. T.; Min B. S.; Jung H. A.; Choi J. S. Moracin derivatives from Morus Radix as dual BACE1 and cholinesterase inhibitors with antioxidant and anti-glycation capacities. Life Sci. 2018, 210, 20–28. 10.1016/j.lfs.2018.08.060. [DOI] [PubMed] [Google Scholar]
- Gao C.; Sun X.; Wu Z.; Yuan H.; Han H.; Huang H.; Shu Y.; Xu M.; Gao R.; Li S.; et al. A novel benzofuran derivative moracin N induces autophagy and apoptosis through ROS generation in lung cancer. Front. Pharmacol. 2020, 11, 391. 10.3389/fphar.2020.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann I. S.; Widdowson D. A.; Clough J. M. Transition metal mediated synthesis of some prenylated phytoalexins of Morus alba Linn. Tetrahedron 1991, 47 (37), 7991–8000. 10.1016/S0040-4020(01)81952-9. [DOI] [Google Scholar]
- Matsuyama S.; Kuwahara Y.; Nakamura S.; Suzuki T. Oviposition Stimulants for the Lesser Mulberry Pyralid, Glyphodes pyloalis (WALKER), in Mulberry Leaves; Rediscovery of Phytoalexin Components as Insect Kairomones. Agric. Biol. Chem. 1991, 55 (5), 1333–1341. 10.1080/00021369.1991.10870748. [DOI] [Google Scholar]
- Kaur N.; Xia Y.; Jin Y. L.; Dat N. T.; Gajulapati K.; Choi Y.; Hong Y. S.; Lee J. J.; Lee K. The first total synthesis of moracin O and moracin P, and establishment of the absolute configuration of moracin O. Chem. Commun. 2009, (14), 1879–1881. 10.1039/b823340c. [DOI] [PubMed] [Google Scholar]
- Lau W.; Sattely E. S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 2015, 349 (6253), 1224–1228. 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen K.; Dong L. M.; Navrot N.; Schneider T.; Burlat V.; Pollier J.; Woittiez L.; van der Krol S.; Lugan R.; Ilc T.; et al. The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 2014, 5, 3606. 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson H.; De La Pena R.; Stephenson M. J.; Thimmappa R.; Vincent J. L.; Sattely E. S.; Osbourn A. Identification of key enzymes responsible for protolimonoid biosynthesis in plants: Opening the door to azadirachtin production. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (34), 17096–17104. 10.1073/pnas.1906083116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozwiak A.; Sonawane P. D.; Panda S.; Garagounis C.; Papadopoulou K. K.; Abebie B.; Massalha H.; Almekias-Siegl E.; Scherf T.; Aharoni A. Plant terpenoid metabolism co-opts a component of the cell wall biosynthesis machinery. Nat. Chem. Biol. 2020, 16 (7), 740–748. 10.1038/s41589-020-0541-x. [DOI] [PubMed] [Google Scholar]
- Nett R. S.; Lau W.; Sattely E. S. Discovery and engineering of colchicine alkaloid biosynthesis. Nature 2020, 584 (7819), 148–153. 10.1038/s41586-020-2546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Li S. X.; Wang W.; Guo X. Y.; Lu X. Y.; Yan X. P.; Huang D.; Wei B. Y.; Cao L. Variations in the levels of mulberroside A, oxyresveratrol, and resveratrol in mulberries in different seasons and during growth. Scientific World Journal 2013, 2013, 380692. 10.1155/2013/380692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovina A. S.; Kiselev K. V. Regulation of stilbene biosynthesis in plants. Planta 2017, 246 (4), 597–623. 10.1007/s00425-017-2730-8. [DOI] [PubMed] [Google Scholar]
- Welle R.; Schroder G.; Schiltz E.; Grisebach H.; Schroder J. Induced plant responses to pathogen attack. Analysis and heterologous expression of the key enzyme in the biosynthesis of phytoalexins in soybean (Glycine max L. Merr. cv. Harosoy 63). Eur. J. Biochem. 1991, 196 (2), 423–430. 10.1111/j.1432-1033.1991.tb15833.x. [DOI] [PubMed] [Google Scholar]
- Schöppner A.; Kindl H. Purification and properties of a stilbene synthase from induced cell suspension cultures of peanut. J. Biol. Chem. 1984, 259 (11), 6806–6811. 10.1016/S0021-9258(17)39799-5. [DOI] [PubMed] [Google Scholar]
- Sparvoli F.; Martin C.; Scienza A.; Gavazzi G.; Tonelli C. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mol. Biol. 1994, 24 (5), 743–755. 10.1007/BF00029856. [DOI] [PubMed] [Google Scholar]
- Wang C.; Zhi S.; Liu C.; Xu F.; Zhao A.; Wang X.; Ren Y.; Li Z.; Yu M. Characterization of stilbene synthase genes in mulberry (Morus atropurpurea) and metabolic engineering for the production of resveratrol in Escherichia coli. J. Agric. Food Chem. 2017, 65 (8), 1659–1668. 10.1021/acs.jafc.6b05212. [DOI] [PubMed] [Google Scholar]
- Kai K.; Mizutani M.; Kawamura N.; Yamamoto R.; Tamai M.; Yamaguchi H.; Sakata K.; Shimizu B. I. Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J. 2008, 55 (6), 989–999. 10.1111/j.1365-313X.2008.03568.x. [DOI] [PubMed] [Google Scholar]
- Vialart G.; Hehn A.; Olry A.; Ito K.; Krieger C.; Larbat R.; Paris C.; Shimizu B.-I.; Sugimoto Y.; Mizutani M.; et al. A 2-oxoglutarate-dependent dioxygenase from Ruta graveolens L. exhibits p-coumaroyl CoA 2’-hydroxylase activity (C2’H): a missing step in the synthesis of umbelliferone in plants. Plant J. 2012, 70 (3), 460–470. 10.1111/j.1365-313X.2011.04879.x. [DOI] [PubMed] [Google Scholar]
- Vanholme R.; Sundin L.; Seetso K. C.; Kim H.; Liu X.; Li J.; De Meester B.; Hoengenaert L.; Goeminne G.; Morreel K.; et al. COSY catalyses trans-cis isomerization and lactonization in the biosynthesis of coumarins. Nat. Plants 2019, 5 (10), 1066–1075. 10.1038/s41477-019-0510-0. [DOI] [PubMed] [Google Scholar]
- Fliegmann J.; Schröder G.; Schanz S.; Britsch L.; Schröder J. Molecular analysis of chalcone and dihydropinosylvin synthase from Scots pine (Pinus sylvestris), and differential regulation of these and related enzyme activities in stressed plants. Plant Mol. Biol. 1992, 18 (3), 489–503. 10.1007/BF00040665. [DOI] [PubMed] [Google Scholar]
- Wang R.; Chen R.; Li J.; Liu X.; Xie K.; Chen D.; Yin Y.; Tao X.; Xie D.; Zou J.; et al. Molecular characterization and phylogenetic analysis of two novel regio-specific flavonoid prenyltransferases from Morus alba and Cudrania tricuspidata. J. Biol. Chem. 2014, 289 (52), 35815–35825. 10.1074/jbc.M114.608265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K.; Mito K.; Ohara K.; Yamamoto H.; Yazaki K. Cloning and characterization of naringenin 8-prenyltransferase, a flavonoid-specific prenyltransferase of Sophora flavescens. Plant Physiol. 2008, 146 (3), 1075–1084. 10.1104/pp.107.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata R.; Kitajima S.; Nuttens A.; Tatsumi K.; Takemura T.; Ichino T.; Galati G.; Vautrin S.; Bergès H.; Grosjean J.; et al. Convergent evolution of the UbiA prenyltransferase family underlies the independent acquisition of furanocoumarins in plants. New Phytol. 2020, 225 (5), 2166–2182. 10.1111/nph.16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.; Fang L.; Sanders S.; Jayanthi S.; Rajan G.; Podicheti R.; Thallapuranam S. K.; Mockaitis K.; Medina-Bolivar F. Stilbenoid prenyltransferases define key steps in the diversification of peanut phytoalexins. J. Biol. Chem. 2018, 293 (1), 28–46. 10.1074/jbc.RA117.000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H.; Hara R.; Takahashi H.; Iijima M.; Munakata R.; Kenmoku H.; Fuku K.; Sekihara A.; Yasuno Y.; Shinada T.; et al. An Aromatic Farnesyltransferase Functions in Biosynthesis of the Anti-HIV Meroterpenoid Daurichromenic Acid. Plant Physiol. 2018, 178 (2), 535–551. 10.1104/pp.18.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z.; Zhu W.; Liu S.; Guan Q.; Chen X.; Huang W.; Wang T.; Yang B.; Tian J. Molecular characterization of a geranyl diphosphate-specific prenyltransferase catalyzing stilbenoid prenylation from Morus alba. Plant Cell Physiol 2018, 59 (11), 2214–2227. 10.1093/pcp/pcy138. [DOI] [PubMed] [Google Scholar]
- Hamerski D.; Matern U. Elicitor-induced biosynthesis of psoralens in Ammi majus L. suspension cultures. Microsomal conversion of demethylsuberosin into (+)marmesin and psoralen. Eur. J. Biochem. 1988, 171 (1–2), 369–375. 10.1111/j.1432-1033.1988.tb13800.x. [DOI] [PubMed] [Google Scholar]
- Villard C.; Munakata R.; Kitajima S.; van Velzen R.; Schranz M. E.; Larbat R.; Hehn A. A new P450 involved in the furanocoumarin pathway underlies a recent case of convergent evolution. New Phytol. 2021, 231 (5), 1923–1939. 10.1111/nph.17458. [DOI] [PubMed] [Google Scholar]
- Höll J.; Vannozzi A.; Czemmel S.; D’Onofrio C.; Walker A. R.; Rausch T.; Lucchin M.; Boss P. K.; Dry I. B.; Bogs J. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 2013, 25 (10), 4135–4149. 10.1105/tpc.113.117127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Wang Y. Transcription factor VqERF114 regulates stilbene synthesis in Chinese wild Vitis quinquangularis by interacting with VqMYB35. Plant Cell Rep. 2019, 38 (10), 1347–1360. 10.1007/s00299-019-02456-4. [DOI] [PubMed] [Google Scholar]
- Wang D.; Jiang C.; Li R.; Wang Y. VqbZIP1 isolated from Chinese wild Vitis quinquangularis is involved in the ABA signaling pathway and regulates stilbene synthesis. Plant Sci. 2019, 287, 110202. 10.1016/j.plantsci.2019.110202. [DOI] [PubMed] [Google Scholar]
- Jiang J.; Xi H.; Dai Z.; Lecourieux F.; Yuan L.; Liu X.; Patra B.; Wei Y.; Li S.; Wang L. VvWRKY8 represses stilbene synthase genes through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J. Exp. Bot. 2019, 70 (2), 715–729. 10.1093/jxb/ery401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cano F.; Chu Y. H.; Cruz-Gomez M.; Abdullah H. M.; Lee Y. S.; Schnell D. J.; Grotewold E.. Exploring Camelina sativa lipid metabolism regulation by combining gene co-expression and DNA affinity purification analyses. Plant J. 2022. 10.1111/tpj.15682. [DOI] [PubMed] [Google Scholar]
- Guan Q. J.; Yu J. J.; Zhu W.; Yang B. X.; Li Y. H.; Zhang L.; Tian J. K. RNA-Seq transcriptomic analysis of the Morus alba L. leaves exposed to high-level UVB with or without dark treatment. Gene 2018, 645, 60–68. 10.1016/j.gene.2017.12.045. [DOI] [PubMed] [Google Scholar]
- Gu X. D.; Sun M. Y.; Zhang L.; Fu H. W.; Cui L.; Chen R. Z.; Zhang D. W.; Tian J. K. UV-B induced changes in the secondary metabolites of Morus alba L. leaves. Molecules 2010, 15 (5), 2980–2993. 10.3390/molecules15052980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chen S. F.; Zhou Y. Q.; Chen Y. R.; Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34 (17), 884–890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G.; Haas B. J.; Yassour M.; Levin J. Z.; Thompson D. A.; Amit I.; Adiconis X.; Fan L.; Raychowdhury R.; Zeng Q. D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29 (7), 644–652. 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J.; Papanicolaou A.; Yassour M.; Grabherr M.; Blood P. D.; Bowden J.; Couger M. B.; Eccles D.; Li B.; Lieber M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc 2013, 8 (8), 1494–1512. 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli C.; Jongeneel C. V.; Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999, 138–148. [PubMed] [Google Scholar]
- Mao X. Z.; Cai T.; Olyarchuk J. G.; Wei L. P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21 (19), 3787–3793. 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- Zheng Y.; Jiao C.; Sun H.; Rosli H. G.; Pombo M. A.; Zhang P.; Banf M.; Dai X.; Martin G. B.; Giovannoni J. J.; et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant. 2016, 9 (12), 1667–1670. 10.1016/j.molp.2016.09.014. [DOI] [PubMed] [Google Scholar]
- Love M. I.; Huber W.; Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15 (12), 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C.; Williams B. A.; Pertea G.; Mortazavi A.; Kwan G.; van Baren M. J.; Salzberg S. L.; Wold B. J.; Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28 (5), 511–515. 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D.; Livak K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3 (6), 1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.