Abstract
Background
Preventing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2_ infections in healthcare workers (HCWs) is critical for healthcare delivery. We aimed to estimate and characterize the prevalence and incidence of coronavirus disease 2019 (COVID-19) in a US HCW cohort and to identify risk factors associated with infection.
Methods
We conducted a longitudinal cohort study of HCWs at 3 Bay Area medical centers using serial surveys and SARS-CoV-2 viral and orthogonal serological testing, including measurement of neutralizing antibodies. We estimated baseline prevalence and cumulative incidence of COVID-19. We performed multivariable Cox proportional hazards models to estimate associations of baseline factors with incident infections and evaluated the impact of time-varying exposures on time to COVID-19 using marginal structural models.
Results
A total of 2435 HCWs contributed 768 person-years of follow-up time. We identified 21 of 2435 individuals with prevalent infection, resulting in a baseline prevalence of 0.86% (95% confidence interval [CI], .53%–1.32%). We identified 70 of 2414 incident infections (2.9%), yielding a cumulative incidence rate of 9.11 cases per 100 person-years (95% CI, 7.11–11.52). Community contact with a known COVID-19 case was most strongly correlated with increased hazard for infection (hazard ratio, 8.1 [95% CI, 3.8–17.5]). High-risk work-related exposures (ie, breach in protective measures) drove an association between work exposure and infection (hazard ratio, 2.5 [95% CI, 1.3–4.8). More cases were identified in HCWs when community case rates were high.
Conclusions
We observed modest COVID-19 incidence despite consistent exposure at work. Community contact was strongly associated with infections, but contact at work was not unless accompanied by high-risk exposure.
Keywords: COVID-19, SARS-CoV-2, healthcare worker, healthcare personnel
In a longitudinal cohort study of 2435 Bay Area healthcare workers (July 2020–January 2021), coronavirus disease 2019 (COVID-19) incidence was low. COVID-19 was strongly associated with community COVID-19 contacts but not with work contacts, unless accompanied by high-risk exposure.
Many assume that healthcare workers (HCWs) acquire coronavirus disease 2019 (COVID-19) at work [1–3]. While early studies supported work-related risks, more recent studies have shown that other factors, including community-based exposures, race/ethnicity, and residential zip code, are associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) acquisition and may be more consequential than workplace exposures [4–6].
Existing literature and media reports have noted widely varying estimates of prevalence, incidence, and risk factors for infection [7–9]. Few studies have assessed HCW infection and risk longitudinally, despite changes in community prevalence of COVID-19 over time, workplace infection prevention efforts, and dynamic individual adherence to public health measures outside of work. Two European groups reported data from longitudinal HCW screening programs and estimated the prevalence and incidence of infection [2, 10], but, to date, a similar granular approach to describing the prevalence and incidence of COVID-19 in US HCWs has not been reported to our knowledge.
In addition, most seroprevalence studies of HCWs—both cross-sectional and longitudinal—have used a single unconfirmed serological test result without orthogonal confirmation (ie, using a different test) as their main outcome measure. Most studies have also not reported neutralizing antibody titers [11–14]. Orthogonal antibody testing increases specificity, which is critical when testing populations with low disease prevalence [15]. Furthermore, neutralizing antibody titers provide a functional assessment of immune responses.
To address these current gaps in our understanding of SARS-CoV-2, we sought to estimate and characterize the prevalence and incidence of COVID-19 using both reverse-transcription polymerase chain reaction (RT-PCR) and orthogonal antibody testing in a large longitudinal cohort of HCWs during a dynamic phase of the US epidemic and to identify risk factors associated with infection among HCWs.
METHODS
Ethics Statement
The COVID-19 Healthcare Worker Antibody and RT-PCR Tracking (CHART) Study was approved by the Committee on Human Subjects Research at the University of California, San Francisco (UCSF), and the Panel on Human Subjects in Medical Research at Stanford University School of Medicine.
Study Population and Setting
From May to September 2020, we recruited HCWs from Stanford Health Care (SHC), UCSF Health (UCSF), and Zuckerberg San Francisco General Hospital for this longitudinal, prospective cohort study. These 3 medical centers serve large, mostly nonoverlapping catchment populations in the San Francisco Bay Area and implemented similar mitigation policies over time (Supplementary Table 1). Recruitment included medical center-wide email and verbal announcements, targeted email notifications to department leaders, and recruitment flyers.
HCWs completed an electronic screening questionnaire (Supplementary Material). Inclusion criteria were (1) age ≥18 years, (2) employment at any of the 3 medical centers, and (3) not anticipating ending employment or taking leave in the next 6 months. Eligible HCWs provided consent electronically. We collected study data using Research Electronic Data Capture (REDcap) electronic data capture tools hosted at Stanford University [16, 17].
Schedule of Evaluations
The study was conducted from July 2020 to January 2021. Participants completed up to 10 visits: 7 visits at 2-week intervals (±7 days), followed by 3 visits at 4-week intervals (±7 days) up to completion or the end of the study. At all visits, participants completed an electronic survey, and study staff collected nasopharyngeal (NP) swab samples; swab samples were optional for the final 3 visits. Participants underwent phlebotomy monthly for anti–SARS-CoV-2 antibody testing. For individuals who tested positive with either RT-PCR or serology, 4 additional weekly visits were scheduled for serological testing only. Participants received no incentives or compensation for joining the study.
Laboratory RT-PCR and Serology
The UCSF Clinical Laboratories and Chan Zuckerberg Biohub analyzed samples from the UCSF and Zuckerberg San Francisco General Hospital subcohorts. Serology was performed using an assay to detect antinucleocapsid immunoglobulin (Ig) G antibodies (Abbott Architect; Abbott Laboratories) [18]. The Stanford Clinical Virology Laboratory analyzed samples using an assay to detect antispike IgG antibodies (Eurimmune Medizinische Labordiagnostika) [19] as well as a laboratory-derived assay to detect anti–receptor-binding domain IgG [20]. Samples that were positive at one laboratory underwent confirmatory testing at the other laboratory. Serum samples that were positive for antibodies to either spike, nucleocapsid, or both proteins were assayed for the presence of neutralizing antibodies at UCSF or at Vitalant Research Institute (San Francisco, California) by optimizing a lentivirus-based pseudotype neutralization assay [21].
At UCSF, RT-PCR testing was performed using (1) the M2000 Abbott RealTime SARS-CoV-2 assay [22], amplifying the RdRP and N genes; (2) the MAGPIX Luminex NxTag CoV Extended Panel assay [23], amplifying the N, the Orf1ab, and E genes; or (3) a Clinical Laboratory Improvement Amendments–validated laboratory-derived test modified from the Centers for Disease Control and Prevention, amplifying the N and E genes [24, 25]. At SHC, RT-PCR testing was performed using an SHC laboratory–derived test amplifying the E gene or the Panther Fusion SARS-CoV-2 assay [26, 27].
Definitions of COVID-19 Exposures, Positive Test Results, and Cases
We defined a low-risk work exposure as providing direct care to or being within 6 feet of a patient with COVID-19, directly interacting with the environment where a patient with COVID-19 received care, or processing laboratory samples from a patient with COVID-19. We defined a high-risk exposure at work as ever interacting with a patient who had COVID-19 without wearing full personal protective equipment (PPE)—the institutionally recommended PPE for care of patients with COVID-19—or while having a breach in PPE (eg, tears or accidental removal).
We defined an RT-PCR result as positive if the result was (1) detected or (2) indeterminate (positive RT-PCR result followed by negative subsequent confirmatory RT-PCR test results(s) obtained according to medical center occupational health protocols).
We defined a confirmed positive serological result as an initial positive serological result (antinucleocapsid or antispike antibody) followed by confirmation with a second positive serological result using a different target (antinucleocapsid, antispike, or neutralizing antibodies). A confirmed positive serological result represented prior COVID-19 infection. We defined an unconfirmed positive serological result as an isolated positive antinucleocapsid or antispike antibody result (ie, a negative result on confirmatory testing) in the absence of RT-PCR positivity.
We defined baseline prevalent cases as those in participants with a positive RT-PCR or confirmed positive serological result at their initial visit. Participants who did not have baseline infection entered an incident cohort. We defined incident cases among this cohort as those in participants with a positive RT-PCR or confirmed positive serological result at any subsequent visit. The date of incident infection was the first date on which either the RT-PCR or the first serological result was positive (if confirmatory testing occurred within 4 weeks).
Statistical Analyses
We estimated the prevalence as the proportion of cases at baseline among the total number of enrolled participants who completed baseline visits. We estimated the cumulative incidence as the number of incident cases divided by the total follow-up time per 100 person-years and assumed a uniform incidence distribution across the 6-month follow-up time. We censored person-time when a participant met the case definition, completed or withdrew from the study, or received a first dose of any COVID-19 vaccine. We calculated the confidence intervals (CIs) using a nonparametric bootstrapping method. We conducted a sensitivity analysis to assess the impact of different case definitions on estimates considering (1) all unconfirmed positive serological results as cases and (2) all individuals with a single positive RT-PCR result, no positive serological result, and ≥1 serological measurement ≥4 weeks after the positive RT-PCR result as potential false-positives and removing them from case counts. We obtained community-wide data on COVID-19 incidence in the 6 Bay Area counties from the California Department of Public Health [28].
We compared characteristics of prevalent and incident cases with those of noncases. For binary time-varying exposures, we used participant self-report at the most recent visit before censoring. For continuous time-varying exposures, we computed median responses across all visits before censoring. We reported symptoms using the most recent reported status at the visit at which infection was identified. We reported standardized mean difference to describe the magnitude of differences in characteristics between incident cases and noncases. The magnitude of effect is considered small, medium, or large with a standardized mean difference of 0.2, 0.5, or 0.8, respectively.
In the incident cohort, we first assessed associations between time to infection and baseline characteristics using multivariable Cox proportional hazards models. We evaluated the impact of prespecified time-varying exposures on time to infection via marginal structural models [29–32]. We implemented a 2-step marginal structural model for each time-varying exposure by first estimating the inverse probability of treatment weights, in which exposure probability was estimated for each participant at each visit, conditioning on fixed and other time-varying exposures up to that time. To stabilize weights, we excluded correlated time-varying variables. Each participant was weighted with the inverse predicted probability of exposure to simulate a counterfactual participant. Second, we applied an extended Cox proportional hazard model with inverse probability of treatment weights and reported hazard ratios (HRs) for the impact of time-varying exposures on time to infection. For all regression analyses, we imputed missing laboratory data using a last observation carried forward method and missing time-invariant or time-varying data using multiple imputation. We controlled family-wise type I error at 0.05 and used the significance level of .05 in hypothesis tests. All analysis were conducted (by Y. W., D. L., and M. D.) using SAS 9.4.3 software (SAS Institute) and R software, version 4.5.3. (R Project for Statistical Computing).
RESULTS
HCW Demographics
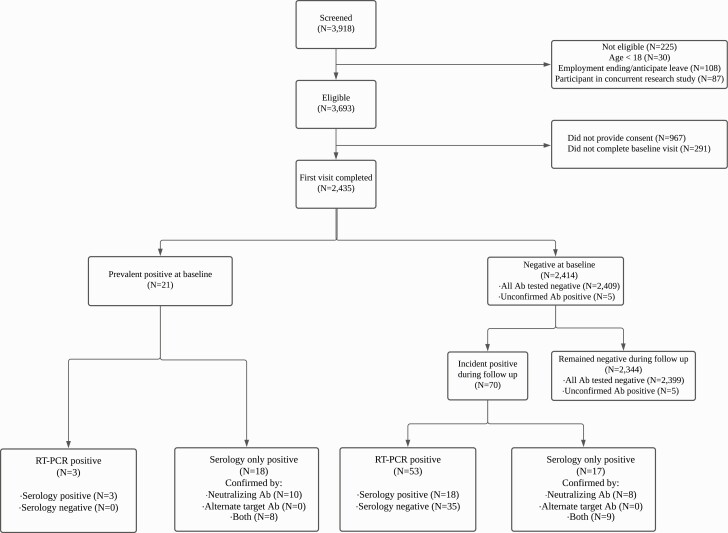
Of 3918 individuals screened, 2435 provided consent and completed the first study visit, contributing 768 total person-years of follow-up time (Figure 1). Baseline demographics are presented in Table 1. Overall, participants’ mean age was 40.4 years (standard deviation, 10.1 years), 1923 of 2435 (79%) were female, and most participants (1921 of 2435 [79%]) reported providing direct patient care, including 701 of 1921 (36%) who performed aerosol-generating procedure . Many participants reported work-related COVID-19 exposure (1477 of 2419 [61%]) with 797 of 1477 (54%) reporting high-risk exposure. Only 176 of 2419 participants (7%) overall reported contact with a COVID-19–positive person outside of work.
Figure 1.
Participant flow diagram. A confirmed positive serological result was defined as an initial positive result (antinucleocapsid or antispike antibody [Ab] result), followed by confirmation with a second positive serological result using a different target (antinucleocapsid, antispike, or neutralizing Abs). A positive unconfirmed serological result was defined as an isolated positive antinucleocapsid or antispike Ab result (ie, a negative result with confirmatory testing) in the absence of reverse-transcription polymerase chain reaction (RT-PCR) positivity. Prevalent coronavirus disease 2019 (COVID-19) cases were defined as those in participants with a positive RT-PCR or a confirmed positive serological result at baseline, and incident COVID-19 cases as those in participants with a positive RT-PCR or a confirmed positive serological result at any subsequent visit.
Table 1.
Characteristics of Healthcare Workers
| Characteristic | Overall, No. (%)a (N = 2435) | Baseline prevalent COVID-19 cases, No. (%)a (n = 21) | Incident Cohort | ||
|---|---|---|---|---|---|
| Negative, No. (%)a (n = 2344) | Incident COVID-19 cases, No. (%)a (n = 70) | SMDb | |||
| Follow-up time, person-years | 779 | 0 | 750 | 17.5 | 0.60 |
| Age, mean (SD), y | 40.4 (10.1) |
42.7(13.0) | 40.4 (10.05) |
40.6 (11.73) |
0.02 |
| Health system | |||||
| SHC | 891 (36.6) | 5 (23.8) | 855 (36.5) | 31 (44.3) | 0.16 |
| UCSF Health | 826 (33.9) | 7 (33.3) | 798 (34.0) | 21 (30.0) | |
| ZSFGH | 718 (29.5) | 9 (42.9) | 691 (29.5) | 18 (25.7) | |
| Female sex | 1923 (79.0) | 13 (61.9) | 1855 (79.1) | 55 (78.6) | 0.11 |
| Latinx ethnicity | |||||
| No | 2036 (87.8) | 17 (100.0) | 1960 (87.7) | 7 (10.6) | 0.12 |
| Yes | 267 (11.5) | 0 (0.0) | 260 (11.6) | 7 (10.6) | |
| Decline to answer | 15 (0.6) | 0 (0.0) | 15 (0.7) | 0 (0.0) | |
| Race | |||||
| White | 1411 (60.9) | 9 (52.9) | 1364 (61.1) | 38 (57.6) | 0.14 |
| Asian | 530 (22.9) | 3 (17.6) | 511 (22.9) | 16 (24.2) | |
| Black | 37 (1.6) | 1 (5.9) | 34 (1.5) | 2 (3.0) | |
| Multiracial | 137 (5.9) | 3 (17.6) | 131 (5.9) | 3 (4.5) | |
| Other | 143 (6.2) | 0 (0.0) | 138 (6.2) | 5 (7.6) | |
| Decline to answer | 58 (2.5) | 1 (5.9) | 55 (2.5) | 2 (3.0) | |
| Highest educational level | |||||
| Less than college | 33 (1.4) | 0 (0.0) | 33 (1.5) | 0 (0.0) | 0.29 |
| College | 1006 (43.4) | 6 (35.3) | 967 (43.3) | 33 (50.0) | |
| Higher than college | 1264 (54.5) | 11 (64.7) | 1222 (54.7) | 31 (47.0) | |
| Other | 15 (0.6) | 0 (0.0) | 13 (0.6) | 2 (3.0) | |
| Comorbid conditions | |||||
| None reported | 1642 (71.5) | 12 (70.6) | 1582 (71.5) | 48 (72.7) | 0.03 |
| Asthma or COPD | 328 (14.3) | 3 (17.6) | 314 (14.2) | 11 (16.7) | 0.07 |
| Diabetes mellitus, heart disease, high blood pressure, or severe obesity | 266 (11.6) | 2 (11.8) | 258 (11.7) | 6 (9.1) | 0.08 |
| Kidney disease on dialysis, liver disease, cancer, autoimmune disorder, or neurological disease |
161 (7.0) | 1 (5.9) | 154 (7.0) | 6 (9.1) | 0.08 |
| Job role | |||||
| Registered nurse or nurse manager | 1077 (44.2) | 8 (38.1) | 1035 (44.2) | 34 (48.6) | ~ |
| Clinician (MD, MD-equivalent, APP, or trainee) | 804 (33.0) | 8 (38.1) | 780 (33.3) | 16 (22.9) | |
| Research/administrative | 115 (4.7) | 1 (4.8) | 113 (4.8) | 1 (1.4) | |
| Support service | 112 (4.6) | 0 (0.0) | 110 (4.7) | 2 (2.9) | |
| Assistant or phlebotomist | 84 (3.4) | 1 (4.8) | 80 (3.4) | 3 (4.3) | |
| Laboratory or pharmacist | 77 (3.2) | 1 (4.8) | 73 (3.1) | 3 (4.3) | |
| Care coordination | 71 (2.9) | 0 (0.0) | 65 (2.8) | 6 (8.6) | |
| Clinic manager or ward clerk | 45 (1.8) | 2 (9.5) | 42 (1.8) | 1 (1.4) | |
| Respiratory or speech therapist | 33 (1.4) | 0 (0.0) | 31 (1.3) | 2 (2.9) | |
| Environmental/food services | 17 (0.7) | 0 (0.0) | 15 (0.6) | 2 (2.9) | |
| Work duties | |||||
| Direct patient care involved in intubating or suctioning patient airways | 701 (30.2) | 6 (35.3) | 670 (30.0) | 25 (37.9) | 0.30 |
| Direct patient care but not performing any airway procedures | 1220 (52.6) | 10 (58.8) | 1178 (52.7) | 32 (48.5) | |
| Staff with indirect patient contact (eg, reception, environmental services) | 128 (5.5) | 0 (0.0) | 127 (5.7) | 1 (1.5) | |
| Laboratory | 58 (2.5) | 0 (0.0) | 55 (2.5) | 3 (4.5) | |
| Work in healthcare but not with patients or biological samples | 81 (3.5) | 1 (5.9) | 78 (3.5) | 2 (3.0) | |
| Other | 131 (5.6) | 0 (0.0) | 128 (5.7) | 3 (4.5) | |
| Performed AGPc | 385 (16.0) | 3 (18.8) | 368 (15.9) | 14 (20.0) | 0.11 |
| COVID-19 exposure at work | |||||
| No exposure | 942 (38.9) | 8 (47.1) | 908 (38.9) | 26 (37.1) | 0.28 |
| Low-risk exposured | 680 (28.1) | 6 (35.3) | 661 (28.3) | 13 (18.6) | |
| High-risk exposured | 797 (32.9) | 3 (17.6) | 763 (32.7) | 31 (44.3) | |
| Time spent in the healthcare workplace | |||||
| 0 h/wk | 28 (1.2) | 0 (0.0) | 28 (1.2) | 0 (0.0) | 0.26 |
| <10 h/wk | 90 (3.7) | 2 (11.8) | 85 (3.6) | 3 (4.3) | |
| 10–20 h/wk | 169 (7.0) | 1 (5.9) | 165 (7.1) | 3 (4.3) | |
| 21–30 h/wk | 296 (12.2) | 0 (0.0) | 287 (12.3) | 9 (12.9) | |
| 31–40 h/wk | 1148 (47.5) | 5 (29.4) | 1104 (47.3) | 39 (55.7) | |
| >40 h/wk | 688 (28.4) | 9 (52.9) | 663 (28.4) | 16 (22.9) | |
| Time spent providing direct patient-facing care | |||||
| 0 h/wk | 234 (9.7) | 1 (5.9) | 226 (9.7) | 7 (10.0) | 0.38 |
| <10 h/wk | 305 (12.6) | 3 (17.6) | 299 (12.8) | 3 (4.3) | |
| 10–20 h/wk | 400 (16.5) | 4 (23.5) | 386 (16.6) | 10 (14.3) | |
| 21–30 h/wk | 397 (16.4) | 0 (0.0) | 381 (16.3) | 16 (22.9) | |
| 31–40 h/wk | 825 (34.1) | 5 (29.4) | 791 (33.9) | 29 (41.4) | |
| >40 h/wk | 258 (10.7) | 4 (23.5) | 249 (10.7) | 5 (7.1) | |
| No. in household, mean (SD) | 2.3 (17.2) | 1.5 (1.1) | 2.3 (1.7) | 2.3 (1.9) | 0.02 |
| Any children aged <18 y in household | 841 (36.5) | 2 (11.8) | 817 (36.8) | 22 (33.8) | 0.06 |
| Any adults aged ≥65 y in household | 256 (11.1) | 1 (5.9) | 245 (11.0) | 10 (15.4) | 0.13 |
| Extent of avoiding contact with people who live outside your home | |||||
| All of the time | 80 (3.3) | 2 (11.8) | 72 (3.1) | 6 (8.6) | 0.34 |
| Most of the time; I only leave my home to buy food or other essentials or to walk/exercise | 1232 (50.9) | 8 (47.1) | 1197 (51.3) | 27 (38.6) | |
| Some of the time; I have reduced the amount of time I am in public spaces, social gatherings, or at work | 1096 (45.3) | 7 (41.2) | 1052 (45.1) | 37 (52.9) | |
| None of the time | 11 (0.5) | 0 (0.0) | 11 (0.5) | 0 (0.0) | |
| Frequency of mask wearing when leaving home | |||||
| All of the time | 1778 (73.5) | 10 (58.8) | 1719 (73.7) | 49 (70.0) | 0.25 |
| Most of the time | 621 (25.7) | 7 (41.2) | 596 (25.6) | 18 (25.7) | |
| Some of the time | 17 (0.7) | 0 (0.0) | 14 (0.6) | 3 (4.3) | |
| Never | 3 (0.1) | 0 (0.0) | 3 (0.1) | 0 (0.0) | |
| Had community exposure with a person who tested positive for COVID-19 | 176 (7.3) | 1 (5.9) | 158 (6.8) | 17 (24.3) | 0.50 |
| Spent time in another country/state | 825 (34.1) | 3 (17.6) | 798 (34.2) | 24 (34.3) | <0.01 |
Abbreviations: AGPs, aerosol-generating procedures; APP, advanced practice providers; COPD, chronic obstructive pulmonary disease; COVID-10, coronavirus disease 2019; MD, medical doctor; SD, standard deviation; SHC, Stanford Health Care; SMD, standardized mean difference; UCSF, University of California, San Francisco; ZSFGH, Zuckerberg San Francisco General Hospital.
Data represent no. (%) of healthcare workers, unless otherwise stated.
The SMD is a comparative measure of effect size between groups. The magnitude of effect is considered small if the SMD is 0.2, medium at 0.5, and large at 0.8.
AGPs include intubation, extubation, chest physiotherapy, noninvasive ventilation, open suction, nebulized medications, manual ventilation, bronchoscopy, pulmonary function tests, high-frequency ventilation, laryngoscopy, autopsy, cardiopulmonary resuscitation, tracheostomy, and high-flow nasal cannula use.
Low-risk exposure at work included interacting with a patient who had COVID-19, with no reported breach in personal protective equipment (PPE) or other safety protocols. High-risk exposure included ever interacting with a patient who had COVID-19, without wearing full PPE or with a breach in any safety protocol.
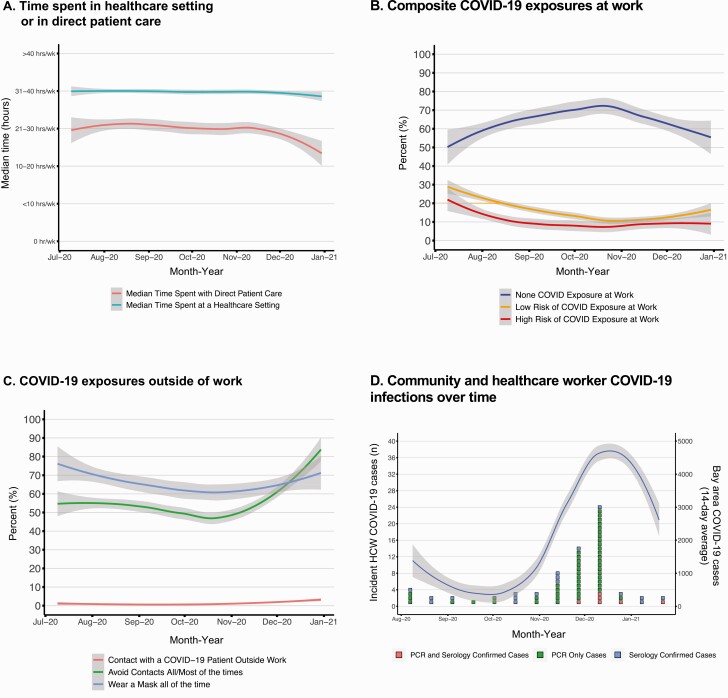
Overall, demographic and behavior characteristics of participants with prevalent and incident COVID-19 reflected overall cohort characteristics (Table 1), including 73 of 91 (80%) providing direct patient care, mostly as nurses (42 of 91 [46%]) or clinicians (MD, MD-equivalent, APP, or trainee) (24 of 91 [26%]). During the course of the study, time spent in the healthcare environment and work-related exposures to COVID-19 were both stable for HCWs (Figure 2A and 2B).
Figure 2.
Work and community-related coronavirus disease 2019 (COVID-19) exposures and incident cases among healthcare workers (HCWs) over time. A–C, Self-reported work and home exposures over time. Each line depicts the 7-day smoothed median responses of each self-reported home or community behavior or exposure. Gray shading represents 95% confidence intervals around the average. D, Incident cases in the context of surrounding community caseload. Boxes represent unique incident cases and are color coded by how they met case definitions, and the line represents the 14-day smoothed average of community-reported cases from the 6 San Francisco Bay Area Counties surrounding the 3 medical centers. Abbreviation: RT-PCR, reverse-transcription polymerase chain reaction.
Prevalence and Incidence of COVID-19
We identified 21 of 2435 individuals with evidence of COVID-19 at baseline and estimated a prevalence of 0.86% (95% CI, .53%–1.32%). We identified 70 of 2414 individuals (2.9%) with incident COVID-19 during follow-up and estimated a cumulative incidence rate of 9.11 cases per 100 person-years (95% CI, 7.11–11.52). The number of incident cases increased with rising prevalence of COVID-19 in the 8-county region in which the study was conducted (Figure 2). Incidence rate estimates did not differ by subgroups of sex, race/ethnicity, or job role (Supplementary Figure 1).
All 21 prevalent COVID-19 cases met the case definition with a positive serological result; only 3 also had a positive RT-PCR result. Most of the 70 incident cases were identified by a positive RT-PCR result (53 of 70 [76%]), with or without a positive serological result. Of the 17 of 70 participants (24%) meeting the case definition by positive serological results alone, only 2 (12%) had a positive RT-PCR result at a later visit (2 or 5 weeks after the positive serological result).
We performed a sensitivity analysis using an alternative incident COVID-19 case definition that included all unconfirmed positive serological results as cases, resulting in 26 prevalent and 71 incident cases. This slightly increased the baseline prevalence to 1.07% (95% CI, .79%–1.56%) and increased the cumulative incidence rate to 9.26 cases per 100 person-years (95% CI, 7.24–11.69).
To examine the impact of potential false-positive RT-PCR results, we performed a second sensitivity analysis using a second alternative case definition that excluded 7 cases meeting this definition. This decreased the cumulative incidence rate to 8.18 cases per 100 person-years (95% CI, 6.29–10.4). Overall, the testing yield of the incident cohort was relatively low: only 30 of 12 007 RT-PCR tests (0.25%) performed in asymptomatic participants had positive results, and 7 of the 30 (23%) met the false-positive case definition.
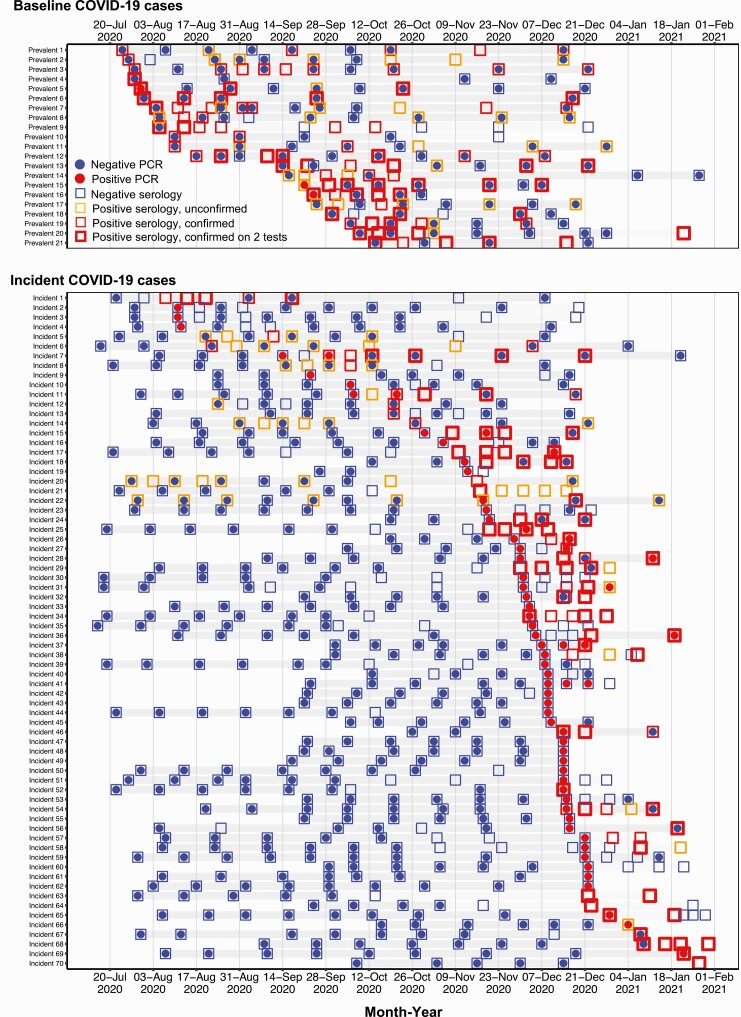
Figure 3 demonstrates the participant-level temporal sequence of testing results for all baseline prevalent cases and all incident cases. We found substantial evolution of antibody responses over time: of the 56 cases initially diagnosed using RT-PCR, 11 had ≥1 positive antibody at diagnosis. By the end of follow-up, this rose to 27 individuals with ≥1 positive antibody test result. Furthermore, 11 individuals who had antibodies detected with one assay subsequently tested positive using another assay.
Figure 3.
Timing and sequence of positive tests among healthcare workers with coronavirus disease 2019 (COVID-19). Each row represents all test results for each prevalent and incident case over the study period. Gray shading indicates each participant’s follow-up time. Dots represent reverse-transcription polymerase chain reaction (RT-PCR) results and boxes represent serological results. Blue coloring represents negative RT-PCR or serological results; red coloring, positive RT-PCR or confirmed positive serological results. Orange boxes represent unconfirmed positive serological results. The thickness of the red boxes is correlated with the number of confirmed positive serological results (eg, 2 or 3 positive antibody test results).
COVID-19 Symptoms
Of the 70 incident cases, 36 (51.4%) were asymptomatic at diagnosis (30 with positive RT-PCR results and 6 with positive serological results only). Of the 36 participants who were asymptomatic at diagnosis, 14 of 22 (64%) who completed a follow-up symptom assessment had remained asymptomatic. Of the 25 participants who were symptomatic at diagnosis, 19 had positive RT-PCR results and 6 had positive serological results only.
Among the 1170 participants who reported symptoms at any visit, 58 (5%) were confirmed as having prevalent or incident cases. Among 1252 participants who never reported symptoms, 32 (3%) were confirmed as having prevalent or incident cases.
While participants with incident cases more commonly reported ever having symptoms (48 of 70 [69%]), many categorized as noncases (1112 of 2344 [48%]) reported symptoms at least once (Supplementary Table 2). The most common symptoms reported by those categorized as noncases were fatigue (326 [14%]), headache (46 [20%]), nasal congestion (325 [14%]), and rhinorrhea (412 [17%]); those categorized as noncases infrequently reported fever, chills, or decreased sense of taste or smell, while case patients reported them more commonly.
Predictors of COVID-19 Infection
In a multivariable Cox proportional hazards model, we did not find an association of incident COVID-19 with fixed variables, including baseline age, sex, race, ethnicity, household size, role, and work category (Supplementary Table 3). In marginal structural models of self-reported time-varying variables, community contact with a known COVID-19 case was strongly correlated with increased hazard for COVID-19 (HR, 8.1 [95% CI, 3.8–17.5]; Table 2). Self-reported exposure to a patient with COVID-19 at work was associated with infection (P = .01), but this appeared to be primarily driven by high-risk exposures (ie, a PPE failure or breach or an exposure to patient biological material; HR, 2.5 [95% CI, 1.3–4.8]). Increasing community COVID-19 case rate showed a trend toward elevated adjusted hazard of HCW infection, but this finding did not reach statistical significance (HR, 1.3 [95% CI, .97–1.8]). Time spent in the healthcare workplace, time spent providing direct patient-facing care, and adherence to community mitigation strategies were not associated with COVID-19 infection.
Table 2.
Marginal Structural Model of Variables Associated With Incident Coronavirus Disease 2019a
| Self-Reported Time-Varying Variable | Adjusted HR (95% CI) | P Value |
|---|---|---|
| COVID-19 exposure at work | ||
| No exposure | Reference | .01 |
| Low riskb | 0.8 (.4–1.7) | |
| High riskb | 2.5 (1.3–4.8) | |
| Time spent in healthcare workplace | ||
| <10 h/wk | Reference | .68 |
| 10–20 h/wk | 1.9 (.5–7.9) | |
| 21–30 h/wk | 2.5 (.7–8.9) | |
| 31–40 h/wk | 2.2 (.7–7.0) | |
| >40 h/wk | 2.0 (.6–6.6) | |
| Time spent providing direct patient-facing care | ||
| <10 h/wk | 0.5 (.2–.9) | .21 |
| 10–20 h/wk | 0.6 (.3–1.3) | |
| 21–30 h/wk | 0.9 (.5–1.7) | |
| 31–40 h/wk | Reference | |
| >40 h/wk | 0.9 (.4–2.0) | |
| Direct contact outside the workplace with someone who tested positive for COVID-19 (yes vs no) | 8.1 (3.8–17.5) | <.001 |
| Extent of avoidance of people who live outside your home when not at work (all/most of the time vs some/none) | 1.0 (.6–1.6) | .91 |
| Mask adherence when not at work (all of the time vs most/some/never) | 0.8 (.5–1.6) | .59 |
| Average daily no. of new COVID-19 cases in 6 Bay Area counties in 14-d period before visit (1-unit increase per 10 000 cases) | 1.3 (.97–1.8) | .08 |
Abbreviations: CI, confidence interval; COVID-19, coronavirus disease 2019; HR, hazard ratio.
For each marginal structural model, we estimated the inverse probability of treatment weights in which exposure probability was estimated for each participant conditioning on fixed variables (age, sex, race, ethnicity, household size, role, and work category) and the time-varying variables shown in the table.
Low-risk exposure at work included interacting with a patient who had COVID-19, with no reported breach in personal protective equipment (PPE) or other safety protocols. High-risk exposure at work was defined as ever interacting with a patient with COVID-19 without wearing full personal protective equipment (PPE) or while having a breach in PPE (eg, tears or accidental removal).
DISCUSSION
In this large observational cohort of HCWs, we observed modest COVID-19 infection rates despite consistent COVID-19 exposure at work. Changes in COVID-19 incidence tracked most closely with community infection rates and self-reported community contact with known COVID-19 cases rather than work-related factors, except when breaches in standard safety protocols or PPE occurred [5]. Our data provide evidence of the overall safety of standard healthcare work environment protocols and PPE guidelines and are concordant with emerging literature showing that the main COVID-19 related risks to HCWs are those coming from home- and community-based factors [33].
By combining longitudinal and orthogonal RT-PCR and serological testing, our study allowed for a robust granular estimation of the true incidence of COVID-19 infection among HCWs. Unlike many studies based on a single serological test, we used confirmatory serology and also measured neutralizing antibody responses [34]. As our data show, the serological response to infection is multifaceted and evolves over time; measuring a single antibody response to one target may result in inaccurate estimates of true infection rates [35]. By testing serially and confirming antibody responses, we captured some COVID-19 cases that would have likely been missed with a single test in time and excluded others that were likely false-positives.
In our sensitivity analysis accounting for potential false-positive COVID-19 cases, our incidence estimates were 10% lower. This may have been an underestimate because many cases were diagnosed at the end of the study and lacked follow-up time to differentiate true-positives from false-positives. Misclassifying false-positive test results as true cases can affect the ability of a healthcare system to operate by limiting critical staffing and can also have adverse implications for household contacts of HCWs. COVID-19 screening programs for HCWs must balance the value of prompt diagnosis with the downside of potential false-positive results.
Our study is subject to several limitations. We enrolled volunteer participants and had a high fraction of MD, MD-equivalent, and RN practitioners. This cohort composition did not comprehensively reflect the occupational diversity within our medical centers. Thirty-eight percent of those screened did not enroll in the study; because we did not assess reasons for nonparticipation, it is unclear to what degree this may have introduced any bias in our study population. We relied on self-reporting of COVID-19–related risks both at work and home, which may have resulted in overreporting of adherence to protective measures. In addition, our institutional PPE recommendations changed over time; as such, not all breaches in PPE are considered equivalent. However, unlike many studies that have used information from employee health and safety offices, our study was independent of the medical centers in order to foster confidential no-fault reporting.
Because sequencing of virus was beyond the scope of the study, the association between self-reported breach in PPE and incident COVID-19 cases remains solely an association and not proof that the breach itself led to the incident infection. In addition, we did not perform orthogonal SARS-CoV-2 antibody testing on samples that were initially antibody negative, and we thus could have missed certain incident cases. We also did not include confirmatory testing of RT-PCR results so could have inadvertently included false-positive RT-PCR results in incidence rate estimates. We addressed this with a sensitivity analysis and found that incidence rates were minimally affected. Finally, our study was conducted before more recent variants of concern with increased transmissibility and immune escape emerged. One key strength of our study was our use of marginal structural modeling, using detailed longitudinal data to better estimate risks.
Within a large group of frontline HCWs, our data indicate that healthcare workplaces pursuing comprehensive mitigation strategies can operate safely despite facing sequential waves of COVID-19 cases. However, HCWs do face community-based risks for acquiring COVID-19. Medical center infection control practices, vaccination programs, and community mitigation approaches should be sustained and maximized to protect HCWs and health systems during periods of future risk related to rising caseloads and emerging SARS-CoV-2 variants.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank all participants for generously volunteering their time and effort to this study. The authors thank all study staff and our participating health systems, including occupational health and infection control and prevention teams, medical center staff, and university and medical center leadership. The authors also acknowledge Salu Ribiero and the Bay Area Phlebotomy Group; UCSF colleagues Tim Moriarty, Chantal Giunta, Erik Stewart, Robert Cusack, Alexander Lee, Miguel Ortiz, Peter Suslow, Don Rose, Pilar Collins, Eunice Stephens, Tatiana Urisman, Dan Henroid, Pamela Hudson, Jennifer Dearman, Leigh Ann Ambrose, Tim Judson, and Sabrina Mann; Zuckerberg San Francisco General Hospital colleagues Geary Wong, Jeff Whitman, Barbara Haller, Kara Lynch, Jeff Schmidt, Lisa Winston, Elaine Dekker, and Mark Jacobson; Stanford Health Care colleagues Tim Morrison and Karen Baines; and the Chan Zuckerberg Initiative for their support.
REDCap platform. The Stanford Research Electronic Data Capture (REDCap) platform (http://redcap.stanford.edu) is developed and operated by the Stanford Medicine Research IT team, and the platform’s services at Stanford are subsidized by the Stanford School of Medicine Research Office and the National Center for Research Resources and National Center for Advancing Translational Sciences, National Institutes of Health (grant UL1 TR001085). REDCap is a secure, web-based software platform designed to support data capture for research studies, providing an intuitive interface for validated data capture; audit trails for tracking data manipulation and export procedures; automated export procedures for seamless data downloads to common statistical packages; and procedures for data integration and interoperability with external sources.
Financial support. This work was supported by the Chan Zuckerberg Initiative Foundation (grant CZIF2020-003870).
CHART Study Consortium collaborators. University of California, San Francisco (UCSF): Parul Bhargava, Markus Bohn, Jessica Chao, Charles S. Craik, Sarah B. Doernberg, Jacob Ghahremani, David Glidden, Ralph Gonzales, Beatrice Huang, Sravya Jaladanki, Aida Julien, Daniel Lowenstein, Steve Miller, Audrey Mustoe, Marcus Paoletti, George W. Rutherford, Hannah Sample, Rodolfo Villa, Emerald Wan, and Aimee Williams. San Francisco General Hospital, UCSF: Lillian Brown, Jessica Chuang, Vivek Jain, Carina Marquez, Guntas Padda, Luis Rubio, and Daisy Valdivieso. Stanford Medicine/Stanford Health Care: Rosebay Abad, Anthony Bet, Jenna Bollyky, Manisha Desai, Jeffrey Fung, Anna Graber, Cole Holderman, Marisa Holubar, Hannah Kelley, Amanda Kempema, Christina Kong, Christopher Leung, Joseph Lohmann, Di Lu, Yvonne Maldonado, Lloyd Minor, Lorena Orozco, Benjamin A. Pinsky, Jamie Saxeena, Matthew Sklar, Hilary Tang, Jasmine Wiese, and Yingjie Weng. Chan Zuckerberg Biohub: Emily Crawford, Joe DeRisi, and all members of the CLIAHUB Consortium.
Contributor Information
Sarah B Doernberg, Division of Infectious Diseases, University of California, San Francisco, San Francisco, California, USA.
Marisa Holubar, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Vivek Jain, Division of HIV, Infectious Diseases & Global Medicine, San Francisco General Hospital, University of California, San Francisco, San Francisco, California, USA.
Yingjie Weng, Quantitative Sciences Unit, Stanford University School of Medicine, California, USA.
Di Lu, Quantitative Sciences Unit, Stanford University School of Medicine, California, USA.
Jenna B Bollyky, Division of Pediatric Infectious Diseases, Stanford University School of Medicine, California, USA.
Hannah Sample, Department of Biochemistry and Biophysics, University of California, San Francisco, San Francisco, California, USA.
Beatrice Huang, Department of Family and Community Medicine, San Francisco General Hospital, University of California, San Francisco, San Francisco, California, USA.
Charles S Craik, Department of Pharmaceutical Chemistry, University of California, San Francisco, San Francisco, California, USA.
Manisha Desai, Quantitative Sciences Unit, Stanford University School of Medicine, California, USA.
George W Rutherford, Division of Infectious Disease and Global Epidemiology, Department of Epidemiology and Biostatistics, University of California, San Francisco, San Francisco, California, USA.
Yvonne Maldonado, Division of Pediatric Infectious Diseases, Stanford University School of Medicine, California, USA.
CHART Study Consortium:
Parul Bhargava, Markus Bohn, Jessica Chao, Charles S Craik, Sarah B Doernberg, Jacob Ghahremani, David Glidden, Ralph Gonzales, Beatrice Huang, Sravya Jaladanki, Aida Julien, Daniel Lowenstein, Steve Miller, Audrey Mustoe, Marcus Paoletti, George W Rutherford, Hannah Sample, Rodolfo Villa, Emerald Wan, Aimee Williams, Lillian Brown, Jessica Chuang, Vivek Jain, Carina Marquez, Guntas Padda, Luis Rubio, Daisy Valdivieso, Rosebay Abad, Anthony Bet, Jenna Bollyky, Manisha Desai, Jeffrey Fung, Anna Graber, Cole Holderman, Marisa Holubar, Hannah Kelley, Amanda Kempema, Christina Kong, Christopher Leung, Joseph Lohmann, Di Lu, Yvonne Maldonado, Lloyd Minor, Lorena Orozco, Benjamin A Pinsky, Jamie Saxeena, Matthew Sklar, Hilary Tang, Jasmine Wiese, Yingjie Weng, Emily Crawford, and Joe DeRisi
References
- 1. Self WH, Tenforde MW, Stubblefield WB, et al. Decline in SARS-CoV-2 antibodies after mild infection among frontline health care personnel in a multistate hospital network—12 states, April–August 2020. MMWR Morb Mortal Wkly Rep 2020; 69:1762–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weinberger T, Steffen J, Osterman A, et al. Prospective longitudinal serosurvey of health care workers in the first wave of the SARS-CoV-2 pandemic in a quaternary care hospital in Munich, Germany. Clin Infect Dis 2021; 73:e3055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moncunill G, Mayor A, Santano R, et al. SARS-CoV-2 seroprevalence and antibody kinetics among health care workers in a Spanish hospital after 3 months of follow-up. J Infect Dis 2021; 223:62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baker JM, Nelson KN, Overton E, et al. Quantification of occupational and community risk factors for SARS-CoV-2 seropositivity among health care workers in a large U.S. health care system. Ann Intern Med 2021; 174:649–54. doi: 10.7326/M20-7145. Published 29 January 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fell A, Beaudoin A, D'Heilly P, et al. SARS-CoV-2 Exposure and infection among health care personnel—Minnesota, March 6–July 11, 2020. MMWR Morb Mortal Wkly Rep 2020; 69:1605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jacob JT, Baker JM, Fridkin SK, et al. Risk factors associated with SARS-CoV-2 seropositivity among US health care personnel. JAMA Netw Open 2021; 4:e211283–e211283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chou R, Dana T, Buckley DI, Selph S, Fu R, Totten AM.. Update alert 7: epidemiology of and risk factors for coronavirus infection in health care workers. Ann Intern Med doi: 10.7326/L21-0034. Published 9 February 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Our key findings about US healthcare worker deaths in the pandemic’s first year. 8 April 2021. The Guardian. Available from: https://www.theguardian.com/us-news/ng-interactive/2020/dec/22/lost-on-the-frontline-our-findings-to-date. Accessed 1 July 2021.
- 9. Centers for Disease Control and Prevention. COVID data tracker. Available from: https://covid.cdc.gov/covid-data-tracker. Accessed 1 July 2021.
- 10. Lumley SF, O’Donnell D, Stoesser NE, et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med 2021; 384:533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herzberg J, Vollmer T, Fischer B, et al. Prospective sero-epidemiological evaluation of SARS-CoV-2 among health care workers in a German secondary care hospital. Int J Infect Dis 2021; 102:136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jespersen S, Mikkelsen S, Greve T, et al. SARS-CoV-2 seroprevalence survey among 17,971 healthcare and administrative personnel at hospitals, pre-hospital services, and specialist practitioners in the Central Denmark Region. Clin Infect Dis 2020; 73:e2853–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Patel MM, Thornburg NJ, Stubblefield WB, et al. Change in antibodies to SARS-CoV-2 over 60 days among health care personnel in Nashville, Tennessee. JAMA 2020; 324:1781–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin C, Montesinos I, Dauby N, et al. Dynamics of SARS-CoV-2 RT-PCR positivity and seroprevalence among high-risk healthcare workers and hospital staff. J Hosp Infect 2020; 106:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu G, Emanuel AJ, Nadig S, et al. Evaluation of orthogonal testing algorithm for detection of SARS-CoV-2 IgG antibodies. Clin Chem 2020; 66:1531–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG.. Research Electronic Data Capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform 2019; 95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abbott. SARS-CoV-2 IgG assay—for use with Architect. Available from: https://www.fda.gov/media/137383/download. Accessed 1 September 2021.
- 19. Food and Drug Administration. Fact sheet for healthcare providers—EUROIMMUN anti-SARS-CoV-2 ELISA (IgG). Available from: https://www.fda.gov/media/137607/download. Accessed 1 September 2021.
- 20. Röltgen K, Powell AE, Wirz OF, et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol 2020; 5:eabe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ng DL, Goldgof GM, Shy BR, et al. SARS-CoV-2 seroprevalence and neutralizing activity in donor and patient blood. Nat Commun 2020; 11:4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abbott. Abbott RealTime SARS-CoV-2. Emergency use authorization (EUA). Available from: https://www.molecular.abbott/sal/9N77-095_SARS-CoV-2_US_EUA_Amp_PI.pdf. Accessed 1 September 2021.
- 23. Luminex. NxTAG® CoV Extended Panel Assay package insert. Available from: https://www.fda.gov/media/136500/download. Accessed 1 September 2021.
- 24. Centers for Disease Control and Prevention Division of Viral Diseases. CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. Available from: https://www.fda.gov/media/134922/download. Accessed 1 September 2021.
- 25. Crawford ED, Acosta I, Ahyong V, et al. Rapid deployment of SARS-CoV-2 testing: the CLIAHUB. PLoS Pathog 2020; 16:e1008966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stanford Health Care Clinical Virology Laboratory. Stanford Health Care Clinical Virology Laboratory SARS-CoV-2 test EUA summary. Available from: https://www.fda.gov/media/136818/download. Accessed 1 September 2021.
- 27. Hologic. SARS-CoV-2 assay (Panther Fusion System). Available from: https://www.hologic.com/sites/default/files/2020-03/AW-21159-001_002_01.pdf. Accessed 1 September 2021.
- 28. Rosser JI, Röltgen K, Dymock M, et al. Severe acute respiratory coronavirus virus 2 (SARS-CoV-2) seroprevalence in healthcare personnel in northern California early in the coronavirus disease 2019 (COVID-19) pandemic. Infect Control Hosp Epidemiol 2021; 42:1053–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robins JM, Hernán MA, Brumback B.. Marginal structural models and causal inference in epidemiology. Epidemiology 2000; 11:550–60. [DOI] [PubMed] [Google Scholar]
- 30. Hernán MA. A definition of causal effect for epidemiological research. J Epidemiol Community Health 2004; 58:265–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van der Wal WM, Geskus RB.. ipw: an R Package for inverse probability weighting. J Stat Softw 2011; 43:1–23. [Google Scholar]
- 32. Cole SR, Hernán MA.. Constructing inverse probability weights for marginal structural models. Am J Epidemiol 2008; 168:656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nguyen LH, Drew DA, Graham MS, et al. Risk of COVID-19 among front-line health-care workers and the general community: a prospective cohort study. Lancet Public Health 2020; 5:e475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 2020; 383:1724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kamat D, Kamat A, Mathur A.. Immune responses in patients with COVID-19: an overview. Pediatr Ann 2021; 50:e222–e226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.