Abstract
Background
Containing coronavirus disease 2019 (COVID-19) has been difficult, due to both the large number of asymptomatic infected individuals and the long duration of infection. Managing these challenges requires understanding of the differences between asymptomatic vs symptomatic patients and those with a longer duration of infectivity.
Methods
Individuals from Los Angeles were tested for COVID-19, and a group positive for COVID-19 chose to have follow-up testing. Associations between symptoms and demographic factors, viral burden measured by cycle threshold (CT) value, and duration of polymerase chain reaction (PCR) positivity were analyzed.
Results
Eighteen point eight percent of patients were positive for COVID-19. Asymptomatic COVID-19-positive patients were significantly younger than symptomatic patients (2.6 years; P < .001). There were no differences in average CT between asymptomatic and symptomatic patients. The estimated median duration of COVID-19 PCR positivity was 23 days. Being asymptomatic throughout the course of infection was the only factor associated with a shorter course of COVID-19 PCR positivity (21 vs 28 days; P = .002).
Conclusions
We found important differences and similarities between asymptomatic and symptomatic COVID-19-positive patients, the most meaningful being a similar level of virus as measured by PCR, but a shorter duration of PCR positivity for asymptomatic patients. These findings suggest that asymptomatic patients may have more efficient clearance of virus, which may be relevant for management and screening.
Keywords: COVID-19, asymptomatic, viral burden, duration
Since the first case of coronavirus disease 2019 (COVID-19) in late 2019, the novel coronavirus has rapidly spread across the globe. Although frequently manifesting as pneumonia and shortness of breath, with the majority of those who are hospitalized developing fever, it is apparent that patients exhibit a wide range of symptoms as well as severity of illness [1]. As of August 2021, the World Health Organization reported over 215M confirmed cases and 4.48M deaths globally, with over 650 000 deaths in the United States.
A significant issue in mitigating the spread of COVID-19 is the existence of both presymptomatic and asymptomatic COVID-19-positive individuals. Analysis of infection clusters has shown that patients without symptoms can still test positive for the virus [2] and are potentially infectious as they are still shedding virus [3]. The importance of presymptomatic transmission of COVID-19 comes from a study showing that up to 44% of secondary cases of COVID-19 were infected during the index cases’ presymptomatic stage [1, 4]. More recent studies of asymptomatic patients have found that 40%–50% of infected individuals can remain asymptomatic throughout the entire course of their infection [5, 6]. In fact, asymptomatic individuals have been referred to as “the Achilles heel” in the efforts to control COVID-19 [7]. Although initially it was suggested that asymptomatic individuals were “less” infectious, several studies have found that asymptomatic individuals have similar viral levels as those who are symptomatic [8]. Many believe that vaccination is more likely to be associated with asymptomatic COVID-19 infections, even with newer variants such as the Delta variant (lineage B.1.617.2) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [9]. Regardless, better understanding the characteristics of asymptomatic individuals that could lead to their identification and best management is an important step toward a better understanding of and prevention of COVID-19.
In addition, an important and perhaps underappreciated challenge in combatting the spread of COVID-19 is the unusually long time course until COVID-19 viral clearance based on polymerase chain reaction (PCR) testing [10]. Recent reports have found a median of 17 days for viral shedding from the upper respiratory tract, with shedding of up to 83 days [11]. This delay in clearance may be relevant regarding the continued development of new viral variants—or mutants—for example, a long-term infection of an immunocompromised host led to the development of the UK variant [12]. Early evidence found that more severe infections, associated with a longer course of PCR positivity, led to intrahost viral heterogeneity [13]. Severe infections have been shown to predict longer durations of viral shedding in hospitalized patients [14]; however, for outpatients, there are no indicators of duration of COVID-19 PCR positivity.
In this study, we sought to gain a better understanding of the differences between asymptomatic and symptomatic patients, as well as identify any indicators of duration of COVID-19 PCR positivity, through the study of a large cohort of COVID-19-tested individuals.
METHODS
Description of the Full Cohort
The cohort of patients tested was collected through a group of clinics in LA County that offered free testing to individuals who were either symptomatic or were believed to have had a known COVID-19 exposure. The cohort included 26 334 individuals tested for COVID-19 using an Emergency Use Authorization (EUA)–approved PCR assay with demographic and symptom information including age, sex, and ethnicity between April 9 and June 20, 2020. Ethnicity was recorded as either Hispanic or Non-Hispanic. Repeat testing was allowed at these clinics at will, and 433 individuals had ≥2 COVID-19 tests.
Statistical Analysis on the Full Cohort
The probability of a positive COVID-19 test was examined in relation to a subject’s basic demographic characteristics (age, sex, and ethnicity) and symptomatic profile (cough, fever, gastrointestinal [GI] issues). Univariate associations were assessed using a 2-sample t test for continuous predictors (age) and a z-test for equal proportions for categorical predictors (sex, ethnicity, symptomatic profile). A multivariable analysis for the probability of a positive test vs symptomatic profiles, adjusting for age, sex, and ethnicity, was carried out using a logistic regression model.
The subcohort of patients receiving a positive COVID-19 test was analyzed separately to assess how viral load at detection relates to symptomatic profiles and basic demographic characteristics. A surrogate measure of viral load was defined using cycle threshold (CT) values, considered the standard to measure viral levels [15]. In this study, the smallest value for CT between 2 RT-PCR probes for COVID-19 (N1 and N2) was used. CT values were related to symptomatic profiles, adjusting for demographic characteristics using a linear regression model.
In both analyses, together with a detailed comparison of different symptoms, we also considered a combined indicator of asymptomatic cases by defining a patient to be asymptomatic if they presented with no cough, fever, or GI issues.
Statistical Analysis of the Self-Selected Repeat Testing Cohort Analysis
A subset of the patients who visited the various testing sites chose to have multiple tests conducted over a period of time. We define time to PCR negativity (or CT remission) as the time (in days) between the first positive PCR COVID-19 test and the first negative PCR COVID-19 test. This definition of time to PCR negativity is used as a surrogate end point for duration of COVID-19 infections. Univariate associations between time to PCR negativity, age, sex, ethnicity, and symptomatic profiles were established using a Kaplan-Meier estimator of median survival time. Formal independent tests for univariate association and related P values were obtained using a log-rank test of survival differences. A multivariable analysis of time to PCR negativity as it relates to symptomatic profiles and CT values at first detection, adjusting for demographic characteristics (age, sex, and ethnicity) and total number of repeat tests, was carried out using a Cox proportional hazards model.
Sample Collection, Processing, and Analysis
Sample collection and analysis were performed as detailed in EUA 200380 (MiraDx SARS-CoV-2 RT-PCR assay). In brief, samples were collected through oropharyngeal swabbing using a DNA Genotek RNA collection device, RNA was isolated through magnetic bead-based nucleic acid extraction using the Mag-BIND Viral DNA/RNA Kit from Omega Biotek and the Hamilton Microlab Star MagEx STARlet system, and RT-PCR assays were performed using Centers for Disease Control and Prevention–approved primers from IDT, TaqPath 1-Step RT-qPCR Master Mix, CG (ThermoFisher), and an Applied Biosystems Dx Real-time PCR Instrument. The diagnostic assay targeted the N1 and N2 2019-nCoV markers using RNAseP as the control. A COVID-19 result was defined as positive or DETECTED when either the N1 and/or N2 CT value was <40, indeterminant when the N1 and/or N2 value was ≥40 and <45, and negative or NOT DETECTED when N1 and N2 were ≥45. For this analysis, we classified both indeterminate and not detected values together as NOT DETECTED.
Patient Consent
All patients consented to viral testing for COVID-19, as well as to having their samples and anonymized information used for research through informed consent through MiraDx at the time of testing.
RESULTS
Factors Predicting COVID-19 PCR-Positive Test Results
Of the 26 334 individuals who came for testing, 4940 tests (18.8%) had a positive (detected) COVID-19 result, and 21 394 tests (81.2%) had a negative (not detected or indeterminate) result. The data analyzed in this section contain 28 241 tests, with 26 334 unique patients who took a test, 433 of whom took ≥2 tests. For patients who took >1 test, the test with the lowest CT was used for this first analysis. For patients who had multiple visits, symptoms were considered throughout all their visits to account for presymptomatic patients.
Sample characteristics are summarized in Supplementary Table 1. All summaries are also reported stratifying by RT-PCR test result (positive/negative). In this descriptive univariate analysis, we found that asymptomatic patients were less likely to have a positive test (4.5% difference; P < .001). Similar associations were found when stratifying by type of symptoms. A positive COVID-19 test result was more likely in patients reporting to have a cough, a fever, or GI symptoms (P < .001), with the largest symptom differences being with cough and fever (9.45% and 8.48% higher in positive vs negative patients). Patients receiving a positive COVID-19 test result were also on average 1 year younger compared with negative COVID-19 test result patients (P < .001). Positivity rates did not change significantly with gender. Finally, there was a higher proportion of Hispanic patients in the positive group vs the negative group (7.8% difference; P < .001).
Results from a multivariable logistic regression analysis focusing on how symptomatic profiles associate with the probability of a positive COVID-19 test are reported in Supplementary Table 2. Among patients of similar age, gender, and ethnicity, lack of symptoms at the time of testing (being asymptomatic) was associated with decreased probability of a positive test (odds ratio [OR], 0.592; P < .001). Stratifying by type of symptom at the time of testing, patients experiencing cough had a meaningfully higher probability of testing positive for COVID-19 (OR, 1.439; P < .001). Similar findings were found in subjects experiencing fever (OR, 2.129; P < .001). GI symptoms were not found to be significantly associated with the odds of a positive test. Considering demographic characteristics as potential risk factors, we found that among patients with a similar symptomatic profile, only ethnicity was meaningfully associated with the probability of a positive test, with non-Hispanic subjects exhibiting a lower probability of positivity (OR, 0.390; P < .001). Age was also statistically associated with PCR positivity (P = .005), but its effect size was relatively small compared with the other predictors (OR, 0.97 for a 10-year age difference; OR, 0.84 for a 60-year age difference).
CT Levels and Symptom Profiles
Within the subcohort of patients with a positive COVID-19 test, we investigated how CT values, as a surrogate measurement of viral load, associated with symptom profiles and demographic characteristics. The sample characteristics of this positive subcohort are summarized in Table 1. Asymptomatic patients were on average 2.6 years younger than symptomatic patients (P < .001). Small but unremarkable differences were found between symptomatic and asymptomatic patients in terms of average CT and ethnic composition.
Table 1.
Sample Characteristics for the Full Patient Cohort; Demographics (Positive Patients)
| Total | Asymptomatic | Symptomatic | P Value | |
|---|---|---|---|---|
| (n = 4940) | (n = 2811) | (n = 2129) | ||
| CT, mean (SD) | 28.4 (7.6) | 28.0 (7.9) | 28.9 (7.1) | <.001 |
| Age, mean (SD), y | 39.2 (17.4) | 38.1 (18) | 40.7 (16.5) | <.001 |
| Hispanic, % | 93.6 | 94.4 | 92.5 | .010 |
| Male, % | 45.0 | 44.8 | 45.3 | .761 |
Abbreviation: CT, cycle threshold.
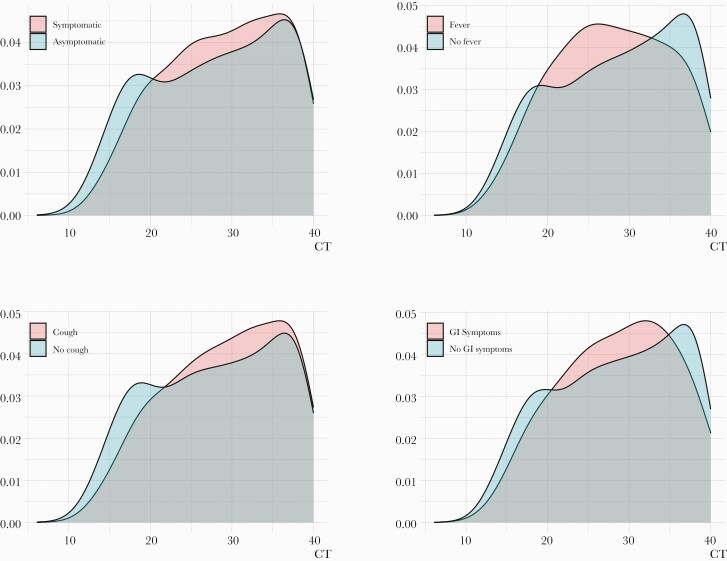
A formal multivariable analysis of CT values vs symptomatic profiles was performed through a linear regression model, adjusting for age, gender, and ethnicity. Detailed results of this analysis are reported in Table 2. The CT values in this analysis were standardized (overall CT mean [SD], 28.39 [7.57]), so the effect of the covariates is in terms of standard deviations of the CT values. Among patients of similar age, gender, and ethnicity, average CT values for asymptomatic patients were slightly lower (indicating more virus) than for symptomatic patients (SD, –0.06; P = .044). A detailed analysis, stratifying by type of symptom, shows that patients who reported having a fever had on average a lower CT (SD, –0.125; P = .004). However, patients who reported having a cough were found to have a slightly higher average CT (less virus; SD, 0.132; P < .001). No statistical differences in average CT values were found to be associated with GI symptoms. Crucially, while small statistically significant differences were found between average CT values between asymptomatic and symptomatic patients, the linear regression models explain <1% (< 0.01) of the variance in the observed CT distribution. In fact, we observe significant overlap in the distribution of observed CT values, independent of symptom profile stratifications (Figure 1).
Table 2.
CT Values as They Relate to Patient Demographic Characteristics and Symptomatic Profile
| Combined Symptoms | ||||
|---|---|---|---|---|
| Estimate | Std. Error | 95% CI | P Value | |
| (Intercept) | –0.032 | 0.027 | (–0.086 to 0.022) | .243 |
| Asymptomatic | –0.061 | 0.030 | (–0.121 to –0.001) | .044 |
| Sex: male | 0.015 | 0.030 | (–0.044 to 0.074) | .615 |
| Ethnicity: non-Hispanic | 0.314 | 0.061 | (0.194 to 0.434) | <.001 |
| Age (centered) | 0.001 | 0.001 | (0.000 to 0.003) | .121 |
| Stratification by Type of Symptom | ||||
|---|---|---|---|---|
| Estimate | Std. Error | 95% CI | P Value | |
| (Intercept) | –0.096 | 0.023 | (–0.141 to –0.051) | <.001 |
| Cough | 0.132 | 0.033 | (0.067 to 0.197) | <.001 |
| Fever | –0.125 | 0.043 | (–0.210 to –.040) | .004 |
| GI symptoms | 0.002 | 0.064 | (–0.123 to 0.128) | .970 |
| Sex: male | 0.023 | 0.030 | (–0.036 to 0.082) | .439 |
| Ethnicity: non-Hispanic | 0.307 | 0.061 | (0.187 to 0.427) | <.001 |
| Age (centered) | 0.001 | 0.001 | (0.000 to 0.003) | .160 |
The response (CT) was standardized in the linear regression analysis (< 0.01; overall CT mean [SD], 28.39 [7.57]).
Abbreviations: CT, cycle threshold; GI, gastrointestinal.
Figure 1.
CT distribution stratified by patients’ symptomatic profiles. We show that the marginal distributions of CT exhibit significant overlap independently of stratification factor, indicating that CT values (viral burden) are similar in asymptomatic and symptomatic subjects. Abbreviations: CT, cycle threshold; GI, gastrointestinal.
Duration of COVID-19 PCR Positivity
Individuals were allowed to come for multiple tests at these clinics at their request, and 433 patients presented ≥2 times for testing. Using these data, we were able to evaluate the percentage of patients who remained asymptomatic throughout the duration of their illness, vs those who were simply “presymptomatic” at their first test. We found that 51.3% of people in this repeat testing cohort reported remaining asymptomatic throughout the full course of their testing.
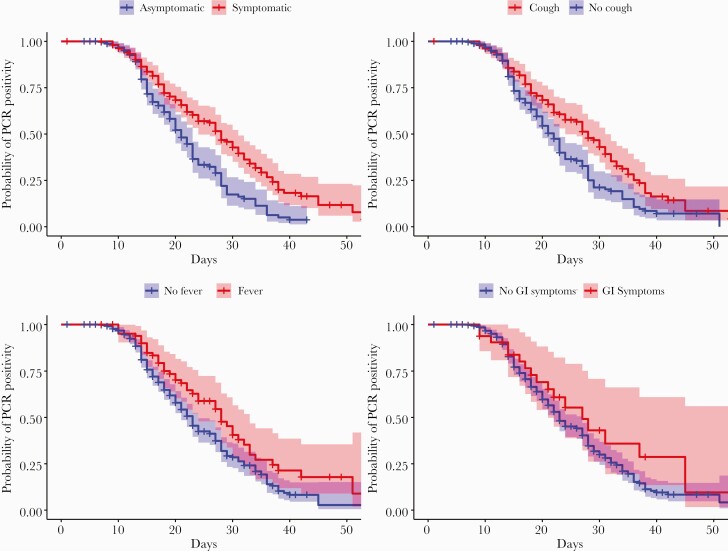
Overall, in this repeat testing cohort, we found that the estimated median length of COVID-19 PCR positivity was 23 days. Table 3 contains descriptive statistics of the repeat testing cohort, along with marginal associations between time to CT remission/PCR negativity and the variables that were used to build the final multivariable model. Patients in our repeat testing cohort who were asymptomatic throughout the entire testing period had an estimated median length of PCR positivity of 21 days, compared with symptomatic patients who had an estimated median length of PCR positivity of 28 days. Similarly, patients who had a cough at any time throughout the testing period had an estimated median length of PCR positivity of 28 days, compared with an estimated median length of PCR positivity of 22 days for patients who did not have a cough. These differences in estimated survival times (PCR negativity) can be visualized in Figure 2, where we show the estimated Kaplan-Meier curves for length until PCR negativity by symptom profile stratification, with pointwise 95% confidence bands.
Table 3.
Sample Characteristics of the Repeat Testing Cohort
| Demographics (Repeat Testing Cohort) | ||||
|---|---|---|---|---|
| Total (n = 433), No. (%) |
Median PCR Positivity, d | 95% CI | P Value | |
| Cough: FALSE | 255 (58.9) | 22 | (20 to 23) | .001 |
| Cough: TRUE | 178 (41.1) | 28 | (24 to 31) | |
| Fever: FALSE | 349 (80.6) | 23 | (21 to 24) | .056 |
| Fever: TRUE | 84 (19.4) | 28 | (24 to 32) | |
| GI: FALSE | 401 (92.6) | 23 | (22 to 27) | .120 |
| GI: TRUE | 32 (7.4) | 27 | (21 to –) | |
| Asymptomatic | 222 (51.3) | 21 | (20 to 23) | <.001 |
| Symptomatic | 211 (48.7) | 28 | (24 to 31) | |
| Age ≤40 y | 224 (51.7) | 23 | (21 to 27) | .351 |
| Age >40 y | 209 (48.3) | 23 | (22 to 28) | |
| Hispanic | 414 (95.6) | 24 | (22 to 27) | .002 |
| Non-Hispanic | 19 (4.4) | 17 | (14 to –) | |
| Female | 256 (59.1) | 23 | (22 to 28) | .343 |
| Male | 177 (40.9) | 23 | (21 to 27) | |
Univariate associations between time to CT remission and baseline covariates were assessed through a log-rank test.
Abbreviations: CT, cycle threshold; GI, gastrointestinal; PCR, polymerase chain reaction.
Figure 2.
Kaplan-Meier Curves for time to CT remission stratified by patients’ symptomatic profiles. Abbreviations: CT, cycle threshold; GI, gastrointestinal; PCR, polymerase chain reaction.
A formal multivariable analysis was performed using a Cox proportional hazards regression model. In this analysis, we estimated the hazard ratio by symptom profile stratifications, controlling for age, ethnicity, gender, initial viral load, and the number of tests an individual took. A detailed summary of this regression analysis is reported in Table 4. Among subjects with similar age, gender, ethnicity, initial CT, and number of tests taken, we found that patients who remained asymptomatic throughout the course of their testing had a significantly shorter time to PCR negativity (symptomatic hazard ratio [HR], 0.645; P = .001). When stratifying by specific symptoms, we found that when controlling for age, ethnicity, sex, other symptoms, initial CT, and the number of tests taken, patients who had a cough were expected to have a significantly longer time to PCR negativity (cough HR, 0.700; P < .001).
Table 4.
Time to CT Remission
| Combined Symptoms | |||||
|---|---|---|---|---|---|
| Estimate | Std. Error | Exp. (Estimate) | 95% CI (Exp. Estim.) | P Value | |
| Symptomatic | –0.439 | 0.138 | 0.645 | (0.492 to 0.845) | .001 |
| Sex: male | 0.239 | 0.135 | 1.270 | (0.975 to 1.654) | .077 |
| Ethnicity: non-Hispanic | 0.808 | 0.300 | 2.243 | (1.245 to 4.040) | .007 |
| Age (centered) | –0.003 | 0.004 | 0.997 | (0.989 to 1.004) | .385 |
| Initial viral load | 0.014 | 0.009 | 1.014 | (0.997 to 1.031) | .118 |
| No. of tests | –0.637 | 0.127 | 0.529 | (0.412 to 0.679) | <.001 |
| Stratification by Type of Symptom | |||||
|---|---|---|---|---|---|
| Estimate | Std. Error | Exp. (Estimate) | 95% CI (Exp. Estim.) | P Value | |
| Cough | –0.357 | 0.144 | 0.700 | (0.528 to 0.929) | .013 |
| Fever | –0.207 | 0.174 | 0.813 | (0.578 to 1.144) | .235 |
| GI symptoms | –0.007 | 0.253 | 0.993 | (0.604 to 1.632) | .978 |
| Sex: male | 0.240 | 0.137 | 1.271 | (0.972 to 1.662) | .080 |
| Ethnicity: non-Hispanic | 0.737 | 0.300 | 2.089 | (1.161 to 3.760) | .014 |
| Age (centered) | –0.003 | 0.004 | 0.997 | (0.989 to 1.004) | .389 |
| Initial viral load | 0.015 | 0.009 | 1.015 | (0.998 to 1.032) | .093 |
| No. of tests | –0.656 | 0.126 | 0.519 | (0.405 to 0.665) | <.001 |
Cox proportional hazards regression analysis of time to CT remission vs patients’ demographic characteristics and symptomatic profiles.
Abbreviations: CT, cycle threshold; GI, gastrointestinal.
DISCUSSION
In this study of outpatients in Southern California, using a PCR-based viral detection method, we investigated factors predicting a positive COVID-19 PCR test, identified differences between asymptomatic and symptomatic patients, and determined predictors of duration of COVID-19 PCR positivity. We found that being symptomatic with cough or fever and being younger predicted a higher likelihood of having a positive COVID-19 test. Comparing COVID-19-positive individuals who were asymptomatic vs symptomatic, we found that asymptomatic patients were significantly younger; however, importantly, they did not have differences in viral levels based on CT values. Regarding the duration of COVID-19 positivity, we found that the average estimated length of PCR positivity in our cohort was 23 days. However, we found that asymptomatic patients had a significantly shorter duration of COVID-19 PCR positivity.
Our finding of similar viral levels in asymptomatic vs symptomatic patients raises important questions about what the lack of symptoms means in a COVID-19-infected individual. Although it was initially suggested that asymptomatic individuals would be less/noninfectious, it seems unlikely at this point in our understanding of COVID-19 that this is the case. A meta-analysis on the subject found that ≥17% of individuals remain completely asymptomatic throughout their course of COVID-19 infection; it concluded that asymptomatic individuals have similar viral levels, but they cause less “transmission to others” of COVID-19 [16]. Our finding that asymptomatic patients have a significantly shorter duration of PCR positivity could potentially help explain why asymptomatic individuals may have less viral transmission. Regardless, the finding in our study, in agreement with others [8], that there are similar viral levels in asymptomatic and symptomatic individuals as measured by PCR, suggests that these individuals will be as infectious as symptomatic individuals at least at some point in their infection.
One hypothesis for asymptomatic COVID-19 infections is that the host has a different immune response to COVID-19 infection, a hypothesis that is also consistent with our finding of quicker viral clearance. It has been proposed that asymptomatic COVID-19 infections are due to the existence of circulating memory T cells, due to prior infections with other, non-COVID coronaviruses, the presence of which has recently been attributed to vaccinations [17]. This is supported by the finding that CD4+ and CD8+ T-cell responses play important roles in the resolution of SARS-CoV-2 infection and in reducing COVID-19 severity [18], as well as findings that early SARS-CoV-2 T-cell responses are associated with milder COVID-19 and modulating disease severity in humans [19, 20]. Further work to preemptively identify individuals who will have milder or asymptomatic COVID-19 infections looking at baseline T cells would be important but is outside of the scope of this study.
The limitations of our study include our measure of positivity using a very sensitive oropharyngeal collection and PCR-based assay. Virus was not cultured in this work as collection buffers used lysed viable virus. That said, viral shedding as measured by PCR-based technology is considered a proxy for infectiousness [21], with higher titer (lower CT) being an indicator of a higher risk of infectivity [22]. While the exact cutoff for “infectiousness” of COVID-19 between humans will likely never be determined, our findings of average CTs in the high 20s for both asymptomatic and symptomatic individuals is unarguably in an infectious range. In addition, our high proportion of individuals classified as “asymptomatic” in this study may be due to a limited number of symptoms recorded, which was due to the timing of when the data set was collected, early in the pandemic. However, our findings agree with several others and are not outside of the range of asymptomatic cases that they also reported [5]. Finally, our repeat testing cohort was a “convenience sample” and self-selected, vs planned, which could have led to some bias.
Our findings here show that while asymptomatic individuals on average will clear virus more quickly, they will be as infectious as symptomatic individuals, due to similar viral levels, at some point in the course of their infection. The impact of these findings in the setting of vaccination, which likely increases the proportion of asymptomatic individuals who are infected yet still infectious but likely with a shortened period of infectiousness, should be further evaluated in continued efforts to combat COVID-19.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Financial support. This work was supported by an R01 from the National Cancer Institution to Dr. Weidhaas (CA238998). Testing was performed at MiraDx.
Potential conflicts of interest. The authors do not have any reportable conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Guan W-J, Ni Z-Y, Hu Y, et al. Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med. 2020; 382:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ye F, Xu S, Rong Z, et al. Delivery of infection from asymptomatic carriers of COVID-19 in a familial cluster. Int J Infect Dis. 2020; 94:133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thi Quynh Mai L, Taichiro T, Meng Ling M, et al. Severe acute respiratory syndrome coronavirus 2 shedding by travelers, Vietnam, 2020. Emerg Infect Dis J 2020; 26:1624–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arons MM, Hatfield KM, Reddy SC, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med 2020; 382:2081–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Letizia AG, Ramos I, Obla A, et al. SARS-CoV-2 Transmission among marine recruits during quarantine. N Engl J Med 2020; 383:2407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Denny T, Andrews L, Bonsignori M, et al. Implementation of a pooled surveillance testing program for asymptomatic SARS-CoV-2 infections on a college campus — Duke University, Durham, North Carolina, August 2–October 11, 2020. MMWR Morb Mortal Wkly Rep 2020; 69:1743–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gandhi M, Yokoe DS, Havlir DV.. Asymptomatic transmission, the Achilles’ heel of current strategies to control Covid-19. N Engl J Med 2020; 382:2158–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee S, Kim T, Lee E, et al. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern Med 2020; 180:1447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown C, Vostok J, Johnson H, et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings — Barnstable County, Massachusetts. MMWR Morb Mortal Wkly Rep 2021; 70:1059–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ling Y, Xu SB, Lin YX, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl). In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A.. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2021; 2:e13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Comin-Anduix B, Gualberto A, Glaspy JA, et al. Definition of an immunologic response using the major histocompatibility complex tetramer and enzyme-linked immunospot assays. Clin Cancer Res 2006; 12:107–16. [DOI] [PubMed] [Google Scholar]
- 13. Al Khatib HA, Benslimane FM, Elbashir IE, et al. Within-host diversity of SARS-CoV-2 in COVID-19 patients with variable disease severities. Front Cell Infect Microbiol 2020; 10:575613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu X, Xing Y, Jia J, et al. Factors associated with negative conversion of viral RNA in patients hospitalized with COVID-19. Sci Total Environ 2020; 728:138812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microb Infect Dis 2020; 39:1059–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Byambasuren O, Cardona M, Bell K, et al. Estimating the extent of asymptomatic COVID-19 and its potential for community transmission: systematic review and meta-analysis. JAMMI. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tarke A, Sidney J, Methot N, et al. Impact of SARS-CoV-2 variants on the total CD4 + and CD8 + T cell reactivity in infected or vaccinated individuals. Cell Rep Med 2021; 2:100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sette A, Crotty S.. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021; 184:861–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rydyznski Moderbacher C, Ramirez SI, Dan JM, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 2020; 183:996–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tan AT, Linster M, Tan CW, et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep 2021; 34:108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lau LLH, Ip DKM, Nishiura H, et al. Heterogeneity in viral shedding among individuals with medically attended influenza A virus infection. J Infect Dis 2013; 207:1281–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inagaki K, Song MS, Crumpton JC, et al. Correlation between the interval of influenza virus infectivity and results of diagnostic assays in a ferret model. J Infect Dis 2016; 213:407–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.