Abstract
Background
The epidemiology of nosocomial bloodstream infections (NBSIs) in patients with coronavirus disease 2019 (COVID-19) is poorly understood, due in part to substantial disease heterogeneity resulting from multiple potential pathogens.
Methods
We identified risk factors for NBSIs and examined the association between NBSIs and mortality in a retrospective cohort of patients hospitalized with COVID-19 in 2 New York City hospitals during the height of the pandemic. We adjusted for the potential effects of factors likely to confound that association, including age, race, illness severity upon admission, and underlying health status.
Results
Between January 1 and October 1, 2020, 1403 patients had a positive blood culture, and 79 and 101 met the stringent criteria for NBSI among non-COVID-19 and COVID-19 patients, respectively. NBSIs occurred almost exclusively among patients who were severely ill with COVID-19 at hospital admission. NBSIs were associated with elevated mortality, even after adjusting for baseline differences in COVID-19 illness (55% cases vs 45% controls; P = .13). Mortality was concentrated in patients with early-onset pneumonia caused by S. aureus and gram-negative bacteria. Less virulent Candida (49%) and Enterococcus (12%) species were the predominant cause of NBSI in the latter stages of hospitalization, after antibiotic treatment and COVID-19 treatments that attenuate immune response. Most Enterococcus and Candida infections did not have an identifiable source and were not associated with common risk factors for infection by these organisms.
Conclusions
Pathogen species and mortality exhibited temporal differences. Early recognition of risk factors among COVID-19 patients could potentially decrease NBSI-associated mortality through early COVID-19 and antimicrobial treatment.
Keywords: SARS, CoV2, COVID, 19, nosocomial, bloodstream infection
Since the pandemic onset, coronavirus disease (COVID-19) mortality rates in US hospitals have declined owing to increasing experience with the disease, decreased patient-to-caregiver ratios, new treatments, and vaccination [1–4]. However, vaccine hesitancy, along with the emergence of new viral variants, could undo such progress [5]. A better understanding of the factors associated with poor COVID-19 outcomes could reduce vulnerability during recurrent COVID-19 outbreaks and build resilience against future pandemic pathogens.
Bloodstream infections (BSIs) are among the most serious medical complications in COVID-19 patients [6-10]. Recent work by us and others suggests that gut microbiome dysbiosis is associated with bacterial translocation into the blood during COVID-19 [11, 12]. Consistent with this idea, a high frequency of Candida and Enterococcus species BSI has been described among hospitalized COVID-19 patients [13–15]—a finding otherwise seen primarily in patients with dysbiosis from cancer treatment, in which these microbes translocate from the gut [16–18]. The natural history of BSI is poorly defined, in part because of substantial disease heterogeneity resulting from multiple potential pathogens, sites of infection, and the clinical time course of COVID-19.
In March 2020, New York City emerged as an epicenter of the COVID-19 pandemic [19]. To explore relationships between COVID-19, BSI risk, and mortality, we assembled a retrospective cohort of COVID-19 patients to study BSI epidemiology using both matched and unmatched controls. We excluded patients having a SARS-CoV-2-positive PCR test result but non-COVID illness. We focused on nosocomial BSI (NBSI) because nosocomial infection, rather than coinfection upon hospital admission, is the primary threat to COVID-19 patients [20]. We also excluded skin microbiota from the analysis (eg, coagulase-negative staphylococci, Bacillus), because skin microbiota can represent a high proportion of contaminants in overstretched hospitals managing many COVID-19 patients [21], and these organisms may not contribute to mortality [22]. These stringent criteria enabled us to examine temporal trends between COVID-19 and NBSIs.
METHODS
Setting and Design
We screened for NBSI among all adult COVID-19 patients admitted to 2 New York University Langone Health (NYULH) hospitals—1 that serves an affluent population (Manhattan) and 1 that serves an underserved population (Brooklyn). Floor beds are able to provide intensive care unit (ICU)–level care; consequently, both floors and ICUs managed critically ill patients.
To determine the impact of NBSI on mortality, we compared COVID-19 patients with recorded NBSIs with COVID-19 patients with no record of BSIs. To establish a more comparable control group, we generated a subset of matched COVID-19 patients with no record of NBSI using propensity score matching [23].
Data Collection and Definitions
Patient data were extracted from electronic health records and by manual chart review. We obtained data on baseline clinical characteristics (within 72 hours of admission) and underlying comorbidities. Oxygen use was categorized as follows: none (no supplementary oxygen), low oxygen (low-flow noninvasive supplementary devices), high oxygen (high-flow), noninvasive ventilation (Bi-PAP, CPAP), and invasive ventilation (intubated). Time-varying data (eg, steroid use) were collected from the admission date forward.
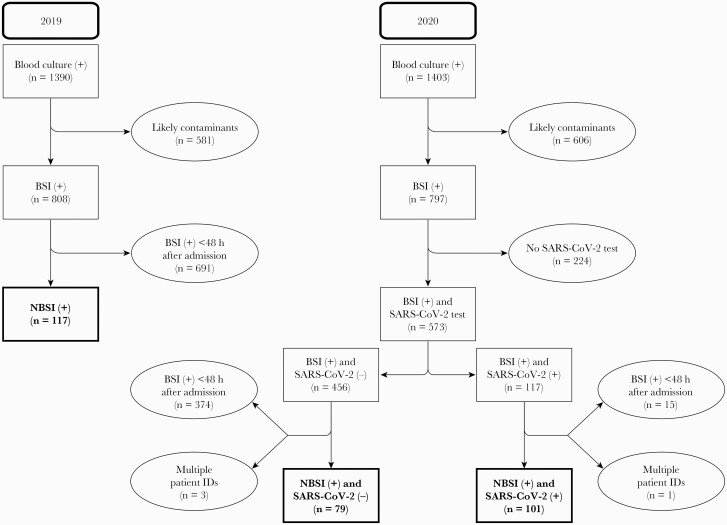
Cases were defined as all patients with a COVID-19 diagnosis at the time of admission, a positive SARS-CoV-2 PCR result, and a positive blood culture obtained >48 hours after admission to the hospital (Figure 1). Infectious disease consultation notes, admission and primary team notes, and discharge summaries were reviewed by 2 infectious diseases physicians (T.D.F. and B.S.) to verify (1) that acute COVID-19 was the primary reason for hospitalization, (2) nosocomial onset of BSI (eg, no signs/symptoms of BSI at the time of hospital admission), and (3) presumed source of NBSI [24]. Patients were categorized as having a central venous catheter if it was present within 48 hours before bacteremia. Mortality was defined as death from any cause occurring after the onset of NBSI until December 31, 2020.
Figure 1.
Flowchart of patient inclusion. Abbreviations: BSI, bloodstream infection; NBSI, nosocomial bloodstream infection; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Microbiology
Microorganisms typically belonging to the skin flora, identified according to Centers for Disease Control and Prevention National Healthcare Safety Network Patient Safety Component Manual criteria (eg, coagulase-negative staphylococci, Micrococcus species, Bacillus species, or diphtheroids [corynebacteria or propionibacteria]) [25], were considered contaminants and were excluded.
Candidemia was identified by both culture and the T2Candida panel, a sensitive molecular assay [26, 27], in patients verified to have infection and to have received treatment by chart review. Polymicrobial NBSI was defined by the isolation of a second microbial species (excluding skin microbiota) within 72 hours of initial positive blood culture. Patients with >1 episode of NBSI during their admission were classified as “multiple” NBSI. S. aureus and Enterococcus were considered antimicrobial-resistant if they were nonsusceptible to methicillin or vancomycin, respectively. Gram-negative bacteria were defined as antimicrobial-resistant if they were nonsusceptible to ceftriaxone, piperacillin-tazobactam, cefepime, ceftazidime, or a carbapenem.
Statistical Analysis
The Pearson χ2 test was used to compare microorganism frequencies. To examine the independent effect of NBSI on mortality, we first created a predictive model of NBSI that included the following set of potentially confounding variables: age, race, illness severity upon admission (eg, supplemental oxygen), and underlying health status (Charlson Comorbidity index) [28–30] using the complete COVID-19 cohort. We then calculated the estimated propensity score for each subject using nearest neighbor propensity score matching (4 controls per case) to identify an NBSI-negative control group [31]. To evaluate the covariate distribution for the unmatched and matched groups, standardized differences were calculated. We also determined the effect of COVID-19 treatments before the positive blood culture (eg, steroids, interleukin [IL]-6 inhibitors) on NBSI risk and mortality.
Kaplan-Meier curves were used to present mortality risk over time. Log-rank tests accounting for matched pairing were used to test the group difference in mortality risk. We ran a Cox proportional hazard model, matching between cases and their respective controls and adjusting for covariates, including type of microorganism (using “none” as a control), age, and sex. We tested the assumption of proportionality using the function “cox.zph()” in R package “survival.” We also performed an accelerated failure time model, which does not require such stringent assumptions, using the Weibull distribution to evaluate survival time [32]. Time ratios (TRs) and 95% CIs were calculated to assess the percent change in survival time (100% * [TR-1]). Logistic regression was used to estimate odds ratios of NBSI onset for different treatments. All statistical analyses were performed in R, version 3.6.3 (R Project for Statistical Computing).
RESULTS
COVID-19 NBSI Study Population
Between January 1 and October 1, 2020, 1403 inpatients had a positive blood culture. We excluded 606 patients with cultures positive for microorganisms likely to be contaminants or low-virulence pathogens (Figure 1). Of the 797 remaining patients, 224 were not tested for SARS-CoV-2, mostly during the period of January to late March 2020, when testing was not widely available. Of the 573 BSI patients with a SARS-CoV-2 PCR test result, 456 tested negative and 117 were positive. Chart review confirmed the clinical diagnosis of COVID-19 and nosocomial acquisition in 101 of 117 cases. Most patients with a negative SARS-CoV-2 PCR test and positive culture results did not meet our >48-hour criteria for nosocomial infection (374/456). Chart review of SARS-CoV-2-negative BSI cases (n = 82) occurring >48 hours after admission identified 79 patients who met inclusion criteria for NBSI controls.
Microbiology of NBSI
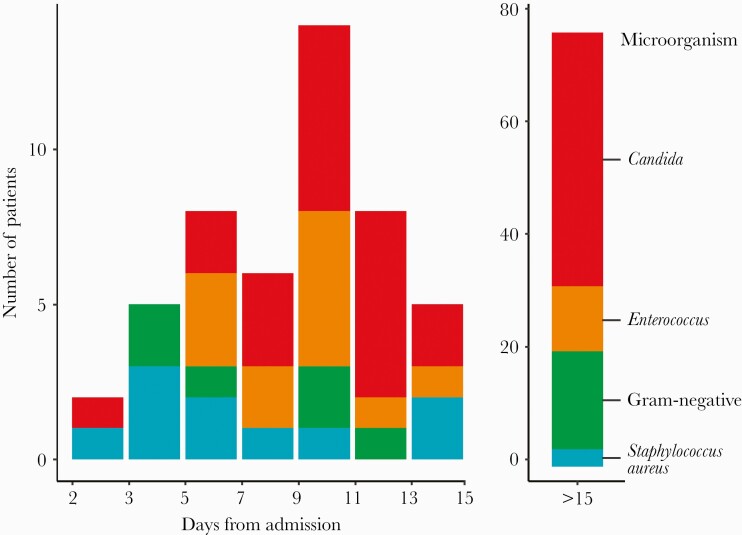
Among the 101 COVID-19 patients with NBSI, Candida (49%) and Enterococcus (12%) were the leading pathogens (Table 1). The proportion of BSIs that were due to Candida (Figure 2) or antimicrobial-resistant bacteria was significantly higher after the first week of hospitalization, post-treatment with empiric antibiotics, steroids, and/or an IL-6 inhibitor, compared with the first week (4/22 [18.2%] vs 51/79 [64.6%]; P = .001). In contrast, BSI cases in the first week of hospitalization were usually caused by antimicrobial-susceptible bacteria. These observations suggest that, over time, COVID-19 patients are at increasing risk of antimicrobial-resistant NBSI pathogens.
Table 1.
Distribution of Pathogens in Bloodstream Infections in Patients With and Without COVID-19, 2019–2020
| Characteristic | 2020 | 2019 | 2019 vs 2020 COVID-19 (+) | 2019 vs 2020 COVID-19 (-) | ||
|---|---|---|---|---|---|---|
| COVID-19 (-) (n = 79), No. (%) | COVID-19 (+) (n = 101), No. (%) | P Value | Pre-COVID-19 (n = 117), No. (%) | P Value | P Value | |
| Organisms | <.001 | <.001 | .102 | |||
| Gram-positive | ||||||
| Overalla | 23 (29.1) | 21 (20.8) | .197 | 36 (30.8) | .095 | .804 |
| Enterococcus spp. | 10 (12.7) | 12 (11.9)c | .874 | 16 (13.7) | .693 | .837 |
| Enterococcus faecalis | 4 (5.1) | 5 (5.0) | 9 (7.7) | |||
| Enterococcus faecium | 6 (7.6) | 7 (6.9) | 7 (6.0) | |||
| Staphylococcus aureus | 13 (16.5) | 9 (8.9)d | 20 (17.1) | |||
| Gram-negative | ||||||
| Overall | 26 (32.9) | 11 (10.9) | <.001 | 35 (29.9) | <.001 | .657 |
| Acinetobacter baumannii | 0 (0.0) | 1 (1.0) | 0 (0.0) | |||
| Enterobacter cloacae | 5 (6.3) | 1 (1.0) | 5 (4.3) | |||
| Escherichia coli | 8 (10.1) | 0 (0.0) | 9 (7.7) | |||
| Klebsiella spp. | 8 (10.1) | 4 (4.0) | 13 (11.2) | |||
| Pseudomonas aeruginosa | 3 (3.8) | 2 (2.0) | 3 (2.6) | |||
| Morganella morganii | 0 (0.0) | 0 (0.0) | 1 (0.9) | |||
| Proteus mirabilis | 2 (2.5) | 0 (0.0) | 1 (0.9) | |||
| Serratia marcescens | 0 (0.0) | 3 (3.0) | 3 (2.6) | |||
| Candida spp. | ||||||
| Overall | 18 (22.8) | 49 (48.5) | <.001 | 18 (22.8) | <.001 | .189 |
| Candida albicans | 3 (3.8) | 11 (10.9) | 9 (7.7) | |||
| Candida glabrata | 2 (2.5) | 1 (1.0) | 5 (4.3) | |||
| Candida parapsilosis | 2 (2.5) | 3 (3.0) | 1 (0.9) | |||
| Other Candida spp. | 2 (2.5) | 1 (1.0) | 3 (2.6) | |||
| T2Candidab | 8 (10.1) | 32 (31.7) | 0 (0.0) | |||
| Candida albicans/Candida tropicalis | 2 (2.5) | 24 (23.8) | ||||
| Candida parasilopsis | 2 (2.5) | 5 (4.95) | ||||
| Candida glabrata/Candida kruseii | 1 (1.3) | 3 (2.97) | ||||
| Anaerobes | ||||||
| Bacteroides spp. | 1 (1.3) | 0 (0.0) | — | 0 (0.0) | ||
| Polymicrobial infectionc | 9 (11.4) | 11 (10.9) | .915 | 17 (14.5) | .423 | .525 |
| Multiple infectiond | 2 (2.5) | 9 (8.9) | .076 | 10 (8.5) | .924 | .085 |
Two-sided Pearson χ2 tests were used to determine P values. Bold formatting indicates statistical significance where the chance of a type I error is 5% or less.
Abbreviation: COVID-19, coronavirus disease 2019.
Six Enterococcus were vancomycin-resistant; 1 S. aureus was methicillin-resistant.
T2Candida positivity in 2019 was only observed in polymicrobial or multiple infections.
Defined as isolation of >1 microorganism from blood within 72 hours.
Defined as isolation of >1 microorganism from blood after 72 hours.
Figure 2.
Histogram of time of diagnosis of bloodstream infection by pathogen species among patients with COVID-19. Abbreviation: COVID-19, coronavirus disease 2019.
NBSI Cases Compared With Non-COVID-19 Controls
To determine whether COVID-19 patients are more likely than non-COVID-19 patients to have Candida and Enterococcus BSI, we compared clinical characteristics between patients from 2 time periods: (1) BSI without COVID-19 from February 15 to December 31, 2020, or (2) hospitalized for any reason from January 1 to December 31, 2019 (historical non-COVID-19 cohort) (Table 1). The historical non-COVID-19 cohort consisted of 117 control cases with NBSI from 2019 out of 1390 patients with positive blood cultures. We excluded 581 cases due to potentially contaminating or clinically insignificant organisms and 691 that occurred <48 hours after hospital admission.
The prevalence of Candida was >2 times higher in COVID-19 patients than in patients belonging to either control group (Table 1). We did not find significant differences of Enterococcus BSI frequencies. However, among patients with NBSI due to Candida or Enterococcus (Supplementary Table 1), there were significant differences between COVID-19 cases and non-COVID-19 controls in the 2020 cohort in terms of known risk factors for infection by these organisms, the latter having more gastrointestinal surgeries, active malignancies, chemotherapy, and higher Charlson index score. Gram-negative bacteria were more abundant among non-COVID-19 controls. The abundance of Candida and Enterococcus in the absence of obvious risk factors suggests that Candida and Enterococcus represent distinct complications of COVID-19.
Candida and Enterococcus causing NBSI in COVID-19 patients were recovered from the urine or sputum in only 4 of 62 instances (6%). COVID-19 patients with NBSI were more likely to have a central venous catheter (CVC) than controls (Supplementary Table 1). Less than 5% were associated with femoral catheters, which are highest risk for BSI.
In contrast to NBSI with Candida and Enterococcus, where few patients had these organisms isolated from sites other than blood, COVID-19 patients with S. aureus or gram-negative NBSI had the same organisms recovered in respiratory cultures in 7 of 9 (77%) and 9 of 11 (80%) instances, respectively. NBSI in these cases most often occurred after endotracheal intubation, an important risk factor for bacterial pneumonia [33]. Indeed, all patients with S. aureus or gram-negative NBSI developed clinical signs of pneumonia (eg, fever, purulent secretions, radiology changes) and were treated for nosocomial pneumonia.
Mortality of NBSI Among Patients With and Without COVID-19
During the study period, we identified a control population of 3273 SARS-CoV-2 PCR test-positive patients without BSI who were admitted for COVID-19. Baseline inflammatory markers (eg, C-reactive protein) were elevated in almost all patients (Table 2). COVID-19 patients with NBSI were substantially different from COVID-19 patients without NBSI before matching; patients with NBSI were younger, more likely to be male and non-Hispanic White, required more oxygen at the time of hospitalization, and had different comorbidities.
Table 2.
Patient Demographic and Clinical Characteristics Before and After Propensity Score Matching
| Variablesa | Unmatched | Matched | |||
|---|---|---|---|---|---|
| COVID-19 (+) & BSI (+) 2020 (n = 101) | COVID-19 (+) & BSI (-) 2020 (n = 3273) | SDS | COVID-19 (+) & BSI (-) 2020 (n = 404) | SDS | |
| Demographic characteristic | |||||
| Age, mean (SD), y | 61.0 (15.5) | 64.1 (16.9) | 0.2 | 60.8 (15.4) | 0.01 |
| Sex, No. (%) | 0.36 | 0.03 | |||
| Female | 24 (23.8) | 1327 (40.5) | 91 (22.5) | ||
| Male | 77 (76.2) | 1946 (59.5) | 313 (77.5) | ||
| Race, No. (%) | 0.3 | 0.09 | |||
| Asian | 10 (9.90) | 213 (6.51) | 32 (7.92) | ||
| Black | 7 (6.93) | 497 (15.2) | 30 (7.43) | ||
| Hispanic | 28 (27.7) | 923 (28.2) | 116 (28.7) | ||
| Other/multiracial | 9 (8.91) | 298 (9.10) | 37 (9.16) | ||
| Unknown | 4 (3.96) | 96 (2.93) | 12 (2.97) | ||
| White | 43 (42.6) | 1246 (38.1) | 177 (43.8) | ||
| Clinical characteristic | |||||
| Maximum oxygenb (first 72 h), No. (%) | 1.48 | 0.08 | |||
| None | 2 (1.98) | 426 (13.6) | 11 (2.72) | ||
| Low oxygen | 3 (2.97) | 1315 (42.0) | 11 (2.72) | ||
| High oxygen | 20 (19.8) | 646 (20.6) | 79 (19.6) | ||
| Noninvasive ventilation | 26 (25.7) | 381 (12.2) | 113 (28.0) | ||
| Mechanical ventilation | 50 (49.5) | 365 (11.7) | 190 (47.0) | ||
| Charlson score, mean (SD) | 2.57 (1.84) | 2.13 (2.12) | 0.24 | 2.49 (2.15) | 0.06 |
| BMI, mean (SD), kg/m2 | 29.4 (6.69) | 30.4 (7.70) | 0.15 | 30.2 (7.45) | 0.01 |
| Temperature, mean (SD), °F | 99.9 (1.82) | 99.5 (1.97) | 0.2 | 99.4 (3.57) | 0.18 |
| Fever, No. (%) | 92 (91.1) | 2914 (93.3) | 0.08 | 367 (92.0) | 0.03 |
| Demographic characteristic | |||||
| Age group, No. (%) | 0.22 | 0.13 | |||
| 0–39 | 12 (11.9) | 268 (8.57) | 44 (10.9) | ||
| 40–49 | 9 (8.91) | 317 (10.1) | 47 (11.6) | ||
| 50–59 | 15 (14.9) | 572 (18.3) | 70 (17.3) | ||
| 60–69 | 31 (30.7) | 737 (23.6) | 124 (30.7) | ||
| 70+ | 34 (33.7) | 1234 (39.5) | 119 (29.5) | ||
| Comorbidity, No. (%) | |||||
| Myocardial infarction | 23 (22.8) | 458 (14.0) | 0.23 | 67 (16.6) | 0.16 |
| Congestive heart failure | 11 (10.9) | 572 (17.5) | 0.18 | 43 (10.6) | 0.01 |
| PVD | 12 (11.9) | 485 (14.8) | 0.08 | 68 (16.8) | 0.14 |
| Cerebrovascular disease | 31 (30.7) | 424 (13.0) | 0.45 | 98 (24.3) | 0.15 |
| Dementia | 9 (8.91) | 324 (9.90) | 0.03 | 26 (6.44) | 0.02 |
| Pulmonary disease | 0.1 | ||||
| Chronic disease | 21 (20.8) | 824 (25.2) | 0.1 | 88 (21.8) | 0.02 |
| Asthma | 5 (4.95) | 377 (12.03) | 0.52 | 47 (11.6) | 0.51 |
| COPD | 10 (9.90) | 369 (11.8) | 0.07 | 32 (7.9) | 0.11 |
| RA/CVD | 2 (1.98) | 114 (3.48) | 0.09 | 15 (3.71) | 0.1 |
| Liver disease | 9 (8.91) | 57 (1.74) | 0.33 | 15 (3.71) | 0.26 |
| Peptic ulcer disease | 6 (5.94) | 106 (3.24) | 0.14 | 17 (4.21) | 0.08 |
| Diabetes | 49 (48.5) | 1256 (38.4) | 0.2 | 214 (53.0) | 0.09 |
| Hemiplegia/paraplegia | 14 (13.9) | 94 (2.87) | 0.42 | 18 (4.46) | 0.3 |
| Renal disease | 24 (23.8) | 865 (26.4) | 0.06 | 131 (32.4) | 0.18 |
| End-stage renal disease | 3 (2.97) | 198 (6.32) | 0.14 | 40 (9.90) | 0.08 |
| Cancer | 11 (10.9) | 360 (11.0) | 0.00 | 45 (11.1) | 0.03 |
| Metastatic tumor | 2 (1.98) | 105 (3.21) | 0.07 | 13 (3.22) | 0.02 |
| AIDS/HIV | 1 (0.99) | 19 (0.58) | 0.05 | 5 (1.24) | 0.05 |
| Index | 0.42 | ||||
| ≥5 | 31 (30.7) | 836 (25.5) | 133 (32.9) | ||
| 3–4 | 26 (25.7) | 516 (15.8) | 52 (12.9) | 0.34 | |
| 1–2 | 33 (32.7) | 1112 (34.0) | 175 (43.3) | ||
| 0 | 11 (10.9) | 809 (24.7) | 44 (10.9) | ||
| DNR status | 0.17 | ||||
| DNR | 30 (29.7) | 718 (21.9) | 121 (30.0) | ||
| No DNR | 71 (70.3) | 2555 (78.1) | 283 (70.0) | 0.03 | |
| Laboratory values | |||||
| WBC, mean (SD), 1 × 109 cells | 9.39 (5.49) | 8.46 (10.2) | 0.12 | 10.6 (26.0) | |
| Neutrophil, mean (SD), % | 81.8 (9.89) | 76.9 (11.4) | 0.44 | 80.7 (10.5) | |
| Lymphocyte, mean (SD), % | 10.7 (7.97) | 14.1 (9.36) | 0.38 | 11.8 (9.22) | 0.05 |
| Hemoglobin, mean (SD), g/dL | 13.5 (1.90) | 12.9 (2.06) | 0.3 | 12.9 (2.23) | 0.07 |
| Platelets (×1000), mean (SD), µ/L | 230 (108) | 230 (102) | 0.01 | 227 (101) | 0.08 |
| Troponin, mean (SD), ng/mL | 0.19 (0.43) | 0.26 (1.62) | 0.06 | 0.37 (1.08) | 0.27 |
| AST, mean (SD), U/L | — | 97.0 (87.5) | 0.13 | 59.6 (62.9) | 0.03 |
| LDH, mean (SD), U/L | 514 (193) | 566 (305) | 0.18 | 529 (275) | 0.22 |
| PT/INR, mean (SD) | 16.9 (14.4) | 1.39 (1.05) | 0.21 | 14.6 (5.99) | 0.72 |
| Inflammatory markers | 0.4 | ||||
| Procalcitonin, mean (SD), ng/mL | 2.84 (15.9) | 1.22 (5.58) | 0.13 | 2.24 (11.7) | 0.19 |
| CRP, mean (SD), mg/dL | 163 (99.3) | 159 (86.9) | 0.07 | 158 (90.1) | |
| D-dimer, mean (SD), ng/mL | 1783 (2705) | 1322 (3694) | 0.13 | 2078 (5315) | 0.14 |
| Ferritin, mean (SD), ng/mL | 1414 (1638) | 1982 (3184) | 0.21 | 1741 (2846) | 0.44 |
| Outcome, No. (%) | 0.07 | ||||
| Length of stay | 44.0 (31.9) | 10.6 (15.3) | 1.41 | 17.2 (18.4) | 0.09 |
| Alive | 64.6 (32.8) | 9.66 (12.3) | 20.8 (21.4) | ||
| Deceased | 26.8 (17.9) | 11.7 (11.0) | 12.9 (12.7) | 1.07 | |
| In-hospital mortality | 0.67 | ||||
| Alive | 46 (45.5) | 2521 (77.0) | 222 (55.0) | ||
| Deceased | 55 (54.5) | 752 (23.0) | 182 (45.0) | 0.13 | |
Abbreviations: AST, aspartate aminotransferase; BMI, body mass index; BSI, bloodstream infection; CHF, congestive heart failure; COPD, chronic obstructive pulmonary disease; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; CTD, connective tissue disease; DNR, do not resuscitate; INR, international normalized ratio; PT, prothrombin time; PVD, peripheral vascular disease; RA, rheumatoid arthritis; SDS, standard difference score; WBC, white blood cells.
All values obtained at hospital admission.
Low oxygen, <4L; high oxygen, >4L; noninvasive, high-flow nasal canula or bilevel/continuous positive airway pressure.
Propensity score matching resulted in a well-balanced control group of 404 patients (SDS < 0.1 for all matching covariates). The number of patients requiring a ventilator at admission between groups was not significantly different (49.5% among cases vs 47.0% among matched controls; SDS = 0.08). The absolute difference in in-hospital mortality risk between BSI (+) patients and BSI (-) unmatched patients was 31.5% (54.5% vs 23.0%; 95% CI, 21.7%–40.9%). When compared with propensity-scored BSI (-) matched controls, mortality risk among those with NBSI remained higher by 9.5% (54.5% vs 45.0%), but the adjusted observed difference was not statistically significant, as confidence intervals were wide and overlapped (95% CI, 44.2%–64.4% vs 40.1%–50.0%). Thus, while NBSI may be associated with moderately increased risk of mortality in patients with COVID-19, we cannot rule out the possibility that the observed difference was due to chance.
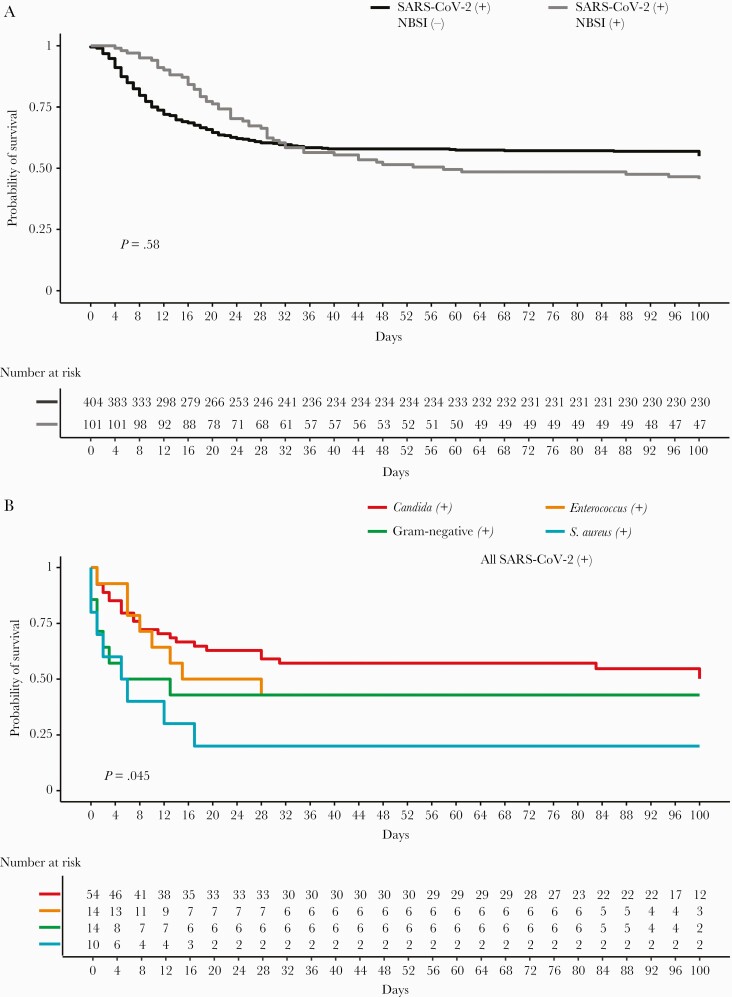
Kaplan-Meier mortality curves for cases and matched controls indicated no overall difference between the 2 curves (Figure 3A). However, curves stratified by microorganism species (Figure 3B) indicated that mortality was significantly concentrated in S. aureus and gram-negative NBSI early after hospitalization. Patients with Candida alone did not have a higher risk of mortality compared with those without NBSI. Thus, although Candida and Enterococcus were the predominant pathogens in NBSI, S. aureus and gram-negative bacteria were possibly the most deadly.
Figure 3.
Kaplan-Meier survival curves. A, Mortality of COVID-19 patients with NBSI vs matched controls without NBSI. B, Mortality of COVID-19 patients with NBSI by microorganism. Log-rank tests were used to determine P values. Abbreviations: COVID-19, coronavirus disease 2019; NBSI, nosocomial bloodstream infection; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
COVID-19-positive patients were administered several types of antibiotics, antifungals, and immunosuppressive agents, namely steroids and the IL-6 inhibitor tocilizumab (the most commonly used IL-6 inhibitor at our institution). Sensitivity analyses using different treatments to stratify mortality revealed that tocilizumab significantly reduced mortality in both the NBSI and non-NBSI COVID-19 patient groups (Supplementary Figure 1).
Adjusted Mortality Models
To define potential independent predictors of mortality, we initially compared COVID-19 NBSI and matched cases using an accelerated failure time (AFT) Weibull model using type of microorganism, age, and sex as covariates, as cases and the matched control group did not meet the assumption of proportionality required for Cox-PH modeling (P < .001). Both the AFT model and the Cox-PH model (shown for comparative purposes) showed that patients with NBSI had a decreased survival time for all organisms except Candida. Additionally, older age and male gender decreased survival (Table 4).
Table 4.
Mortality Survival Time Ratios Using Accelerated Failure Time Model (Adjusted by Sex and Age)
| COEF | Time-Varying HRa | SE | P Value | |
|---|---|---|---|---|
| Microorganisms | ||||
| Candida | 0.286 | 1.33 | 0.35 | .41 |
| Enterococci | –0.125 | 0.88 | 0.59 | .83 |
| Gram-negative | –0.679 | 0.51 | 0.56 | .22 |
| Polymicrobial | –0.829 | 0.44 | 0.75 | .27 |
| S. aureus | –1.899 | 0.15 | 0.61 | .002 |
| Demographics | ||||
| Age | –0.066 | 0.94 | 0.01 | <.001 |
| Sex | ||||
| Female | Ref. | Ref. | ||
| Male | –0.337 | 0.71 | 0.27 | .2 |
| Constant | 9.661 | 15 693.47 | 0.658 | <.001 |
| Scale = 1.64 | ||||
Bold formatting indicates statistical significance where the chance of a type I error is 5% or less.
Abbreviations: COEF, coefficient; HR, hazard ratio.
Time ratios >1 indicate that the effect of a covariate prolongs survival time, while time ratios <1 indicate a shorter survival time.
Gram-negative and S. aureus NBSI time ratios (TRs) were shortened, the latter significantly (TR, 0.51 and 0.15; P = .22 and <.01, respectively). In contrast, the TR for Candida was 1.33 (P = .41). Time ratios were also decreased with age (TR, 0.94; P < .01) and male gender (TR, 0.71; P = .2). In sum, patients with S. aureus and gram-negative NBSI had a shorter survival compared with both patients without NBSI and patients with NBSI due to Candida.
Length of Stay
The average length of stay (SD) was 44 (31.9) days among NBSI cases, compared with 10.6 (15.3) days and 17.2 (18.4) days for unmatched and matched non-NBSI controls, respectively. When stratified by living status, NBSI cases who survived had the longest hospitalizations (64.6 [32.8] days), while those who did not survive averaged 26.8 (17.9) days in the hospital. Likewise, matched survivors had longer lengths of stay compared with those who died (20.8 [21.4] days and 12.9 [12.7] days, respectively). Unmatched controls showed the opposite pattern: Whereas those who died stayed 11.7 (11.0) days, survivors stayed 9.66 (12.3) days. Thus, survival increases with increasing length of stay in NBSI patients and matched controls with severe COVID-19, but not in unmatched controls with mild disease, with implications for prediction of hospital mortality and length of stay from risk factors on admission.
Antibiotic and Tocilizumab Use Before NBSI and Risk of NBSI
Ceftriaxone, azithromycin, and hydroxychloroquine were widely prescribed among both cases and controls (Table 3). In contrast, NBSI cases received tocilizumab and steroids, broad-spectrum antimicrobials (vancomycin, meropenem, and piperacillin-tazobactam), and antifungals (micafungin and fluconazole) more frequently. Thus, NBSI cases received more aggressive treatment than non-NBSI controls. We also analyzed the odds of developing NBSI linked to COVID-19 treatments using a univariate logistic model with NBSI as the outcome. Tocilizumab did not predict NBSI (Supplementary Table 2). However, patients treated with piperacillin-tazobactam, vancomycin, meropenem, and hydrocortisone had higher odds of having NBSI compared with those not receiving those treatments.
Table 3.
Treatments in COVID-19 Patients With and Without Bloodstream Infection
| Antimicrobiala | COVID-19 (+) BSI (+) | COVID-19 (+) BSI (-) | |||||
|---|---|---|---|---|---|---|---|
| (Matched Controls) | |||||||
| Days of Treatment, Mean (SD) (n = 101) | No. (%) | Days of Treatment, Mean (SD) (n = 389) | No. (%) | Days, P Value | Patients, P Value | Cases–Controls, % Treated | |
| Gram-positive coverage | |||||||
| Daptomycin | — | 0 (0.00) | 8.25 (6.29) | 4 (1.03) | — | — | –1.03 |
| Linezolid | 3.67 (2.80) | 6 (5.94) | 6.12 (3.83) | 8 (2.06) | .191 | .048 | 3.88 |
| Vancomycin | 6.31 (7.58) | 88 (87.13) | 5.96 (6.33) | 180 (46.27) | .708 | <.001 | 40.86 |
| Gram-negative coverageb | |||||||
| Amoxicillin/clavulanate | — | 0 (0.00) | 3.17 (2.23) | 6 (1.54) | — | — | –1.54 |
| Ampicillin/sulbactam | 2.75 (1.58) | 8 (7.92) | 4.67 (5.82) | 12 (3.08) | .298 | .5 | 4.84 |
| Ampicillin | 6.00 (.) | 1 (0.99) | 6.00 (5.66) | 2 (0.51) | — | — | 0.48 |
| Cefepime | 3.53 (2.61) | 15 (14.85) | 5.20 (3.35) | 50 (12.85) | .053 | .717 | 2.00 |
| Ceftazidime/avibactam | — | 0 (0.00) | 8.00 (2.83) | 2 (0.51) | — | — | –0.51 |
| Ceftolozane/tazobactam | 1.00 (.) | 1 (0.99) | 21.3 (7.77) | 3 (0.77) | — | — | 0.22 |
| Ceftriaxone | 3.89 (2.81) | 65 (64.36) | 3.62 (2.55) | 248 (63.75) | .488 | .9 | 0.60 |
| Levofloxacin | 5.33 (4.03) | 9 (8.91) | 6.50 (6.29) | 16 (4.11) | .578 | .089 | 4.80 |
| Meropenem | 10.2 (15.6) | 50 (49.50) | 10.9 (11.7) | 96 (24.68) | .776 | <.001 | 24.83 |
| Piperacillin/tazobactam | 6.16 (4.05) | 80 (79.21) | 5.97 (4.00) | 144 (37.02) | .735 | <.001 | 42.19 |
| Polymyxin B | 2.00 (.) | 0.99 | 1.00 (.) | 1 (0.26) | — | — | 0.73 |
| Antifungal coverage | |||||||
| Fluconazole | 6.19 (7.32) | 16 (15.84) | 11.3 (12.3) | 25 (6.43) | .103 | .004 | 9.41 |
| Isavuconazonium sulfate | — | 0 (0.00) | 5.00 (.) | 1 (0.26) | — | — | –0.26 |
| Micafungin | 9.93 (14.5) | 30 (29.70) | 8.25 (11.0) | 48 (12.34) | .588 | <.001 | 17.36 |
| Voriconazole | — | 0 (0.00) | 6.00 (5.66) | 2 (0.51) | — | — | –0.51 |
| Atypical coverage | |||||||
| Azithromycin | 4.42 (1.54) | 89 (88.12) | 4.00 (1.69) | 288 (74.04) | .029 | <.001 | 14.08 |
| Doxycycline | 3.38 (3.54) | 8 (7.92) | 3.95 (4.32) | 57 (14.65) | .686 | .107 | –6.73 |
| Minocycline | — | 0 (0.00) | 7.20 (3.49) | 5 (1.29) | — | — | –1.29 |
| Others | |||||||
| Hydroxychloroquine | 5.04 (1.60) | 93 (92.08) | 4.82 (1.42) | 297 (76.35) | .234 | <.001 | 15.73 |
| Trimethoprim/sulfamethoxazole | 12.0 (14.8) | 3 (2.97) | 5.88 (6.76) | 16 (4.11) | .55 | .776 | –1.14 |
Bold formatting indicates statistical significance where the chance of a type I error is 5% or less.
Abbreviations: BSI, bloodstream infection; COVID-19, coronavirus disease 2019; NBSI, nosocomial bloodstream infection.
Antibiotic use was measured before onset of NBSI. For NBSI-negative cases, antibiotic use was measured for the whole period of hospital admission.
Includes gram-positive coverage but not methicillin-resistant S. aureus coverage.
DISCUSSION
Our results suggest that, early on, intubation promotes bacteremic pneumonia by S. aureus and gram-negative bacteria. Later, COVID-19 creates an environment that promotes intestinal acquisition and/or expansion of pathogenic microbes, especially resistant bacteria and organisms intrinsically resistant to antibiotics, such as Candida. Our data also show that NBSI in COVID-19 patients is dominated by Candida and Enterococcus that arise without a defined portal of entry into the host.
Although it is not possible to definitively state to what extent translocation played a role in Candida and Enterococcal NBSIs, at least 4 observations suggest that such infections are primarily translocation-associated. First, use of femoral catheters that are high risk for catheter-associated NBSI was uncommon in our population. Second, COVID-19 patients with Candida or Enterococcus NBSI showed an imbalance in conditions known to facilitate translocation (eg, abdominal surgery, cancer chemotherapy) and alternative routes (eg, urinary tract infection) that predispose patients to infection with these organisms, compared with COVID-19-negative controls. Third, NBSIs due to Candida and antimicrobial-resistant bacteria, especially vancomycin-resistant Enterococcus, arose in the later stages of hospitalization, after treatment with antimicrobials, steroids, or IL-6 inhibitors. SARS-CoV-2 infection and treatments targeting the COVID-19 hyperinflammatory state may fuel gut injury (eg, steroids, IL-6 inhibitors) [34–37], and intestinal overgrowth under antimicrobial pressure is known to precede translocation of Candida and Enterococcus [16–18]. Finally, an analysis of gut microbiota in patients with COVID-19 belonging to our study cohort showed that BSI was associated with gastrointestinal overgrowth by the same organisms [12], as seen in patients undergoing allogeneic hematopoietic stem cell transplantation [17, 18]. Additionally, recent work demonstrates that COVID-19 patients have increased bacterial products [38] and gut microbes [11] in their bloodstream. Collectively, these observations support the hypothesis that gut translocation may contribute to the pathogenesis of NBSI in patients with COVID-19. An alternative but non–mutually exclusive hypothesis is that environmental contamination leads to exogenous entry of the organism into the bloodstream via catheter use [39].
Our data also suggest that NBSI increases mortality; COVID-19 patients who developed NBSI had an increased risk of death compared with propensity-matched controls. However, while mortality between the COVID-19 patients with NBSI and matched controls without NBSI was not significant, the slope declined most precipitously in patients without NBSI. This finding could reflect survivor bias (patients must survive long enough to be classified as acquiring NBSI). However, patients without NBSI who died early were older and more frail than those with NBSI, which suggests a potential interaction between COVID-19 treatment and subsequent NBSI. In this scenario, treatment improves survival of otherwise healthy but critically ill COVID-19 patients but might also have negative consequences for immune and microbiota-based clearance mechanisms that control infection, thereby predisposing patients to NBSI. Consistent with this idea, length of stay and mortality were inversely correlated in COVID-19 patients with NBSI. This finding may explain why mortality does not increase with candidemia, which primarily occurred after a prolonged length of hospital stay.
NBSI mortality was found to be significantly concentrated in patients with S. aureus and gram-negative nosocomial pneumonia (NP). S. aureus NP is a frequent complication in mechanically ventilated COVID-19 patients [40–42], and bacteremia is an independent risk factor for mortality in NP [43]. Evidently, COVID-19/bacteremic NP is a combination that kills patients at high frequency.
The present work has several limitations. First, our observational cohort data were obtained from 2 hospitals during the pandemic surge; whether NBSI patterns predict outcome in different scenarios, such as in a partially vaccinated population, will need to be tested. Second, correlations between NBSI and outcome may reflect factors we did not account for, such as managing high numbers of critically unwell patients and digression from infection prevention procedures. Third, use of clinical criteria and the T2Candida panel for the diagnosis of pneumonia and candidemia, respectively, may lead to overdiagnosis, potentially increasing the rate of overall disease detected and the proportion of cases that may be linked to NBSI and mortality.
In conclusion, NBSI is associated with mortality in hospitalized patients with severe COVID-19, depending on the species. Our findings suggest that early recognition of risk factors and treatment of NBSI are indicated. However, without accurate predictors of NBSI to guide clinicians, increased use of active agents could further shift species from a preponderance of susceptible organisms to more frequent isolation of antibiotic-resistant and Candida species. Accordingly, improved prediction models, potentially using immunological and microbiota markers, are needed. Improved prediction of NBSI could decrease morbidity and mortality by increasing early antimicrobial treatment for COVID-19 patients at risk for BSI and by reducing unnecessary antimicrobials for patients at low risk of infection.
Supplementary Material
Acknowledgments
We thank Karl Drlica, Robert Ulrich, and Jeffrey Weiser for critical comments on the manuscript.
Financial support. This work was supported in part by National Institutes of Health grants AI137336 and AI140754 (B.S. and V.J.T.); grants CDC U01CK000590 (B.S.); U48 DP006396-01 (L.E.T.); DP2AI164318 (J.S.), and AI149350 (V.J.T.); NYU Grossman School of Medicine COVID-19 seed research funds (V.J.T.); and funds from the NYU Langone Health Antimicrobial-Resistant Pathogens Program (V.J.T.).
Potential conflicts of interest. B.S. has consulted for Regeneron and MicroGenDx. J.S. is cofounder of Postbiotics Plus Reseach LLC. K.C. has consulted for or received honoraria from Puretech Health, Genentech, and AbbVie. K.C. has received research support from Pfizer, Takeda, Pacific Biosciences, Genentech, and AbbVie. K.C. holds US patent 10 722 600 and provisional patents 62/935 035 and 63/157 225. V.J.T. is an inventor on patents and patent applications filed by New York University, which are currently under commercial license to Janssen Biotech Inc. Janssen Biotech Inc. provides research funding and other payments associated with a licensing agreement. All other authors certify no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. This study does not include factors necessitating patient consent.
Contributor Information
Juan Gago, Vilcek Institute of Graduate Biomedical Sciences, New York University Grossman School of Medicine, New York, New York, USA; Division of Epidemiology, Department of Population Health, New York University Grossman School of Medicine, New York, New York, USA.
Thomas D Filardo, Division of Infectious Diseases and Immunology, Department of Medicine, New York University Grossman School of Medicine, New York, New York, USA.
Sarah Conderino, Division of Epidemiology, Department of Population Health, New York University Grossman School of Medicine, New York, New York, USA.
Samuel J Magaziner, Division of Infectious Diseases and Immunology, Department of Medicine, New York University Grossman School of Medicine, New York, New York, USA.
Yanina Dubrovskaya, Division of Infectious Diseases and Immunology, Department of Medicine, New York University Grossman School of Medicine, New York, New York, USA; Tisch Hospital Department of Pharmacy, NYU Langone Health, New York, New York, USA.
Kenneth Inglima, Department of Pathology, New York University Grossman School of Medicine, New York, New York, USA.
Eduardo Iturrate, Department of Medicine, New York University Grossman School of Medicine, New York, New York, USA.
Alejandro Pironti, Department of Microbiology, New York University Grossman School of Medicine, New York, New York, USA.
Jonas Schluter, Department of Microbiology, New York University Grossman School of Medicine, New York, New York, USA; Institute for Computational Medicine, NYU Langone Health, New York, New York, USA.
Ken Cadwell, Department of Microbiology, New York University Grossman School of Medicine, New York, New York, USA; Kimmel Center for Biology and Medicine at the Skirball Institute, New York University Grossman School of Medicine, New York, New York, USA; Division of Gastroenterology and Hepatology, Department of Medicine, New York University Grossman School of Medicine, New York, New York, USA.
Sarah Hochman, Division of Infectious Diseases and Immunology, Department of Medicine, New York University Grossman School of Medicine, New York, New York, USA; Department of Infection Prevention and Control, NYU Langone Health, New York, New York, USA; Antimicrobial-Resistant Pathogens Program, NYU Langone Health, New York, New York, USA.
Huilin Li, Division of Biostatistics, Department of Population Health, New York University Grossman School of Medicine, New York, New York, USA.
Victor J Torres, Department of Microbiology, New York University Grossman School of Medicine, New York, New York, USA; Antimicrobial-Resistant Pathogens Program, NYU Langone Health, New York, New York, USA.
Lorna E Thorpe, Division of Epidemiology, Department of Population Health, New York University Grossman School of Medicine, New York, New York, USA.
Bo Shopsin, Division of Infectious Diseases and Immunology, Department of Medicine, New York University Grossman School of Medicine, New York, New York, USA; Department of Microbiology, New York University Grossman School of Medicine, New York, New York, USA; Antimicrobial-Resistant Pathogens Program, NYU Langone Health, New York, New York, USA.
References
- 1. Anesi GL, Jablonski J, Harhay MO, et al. Characteristics, outcomes, and trends of patients with COVID-19-related critical illness at a learning health system in the United States. Ann Intern Med. 2021; 174:613–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Christie A, Henley SJ, Mattocks L, et al. Decreases in COVID-19 cases, emergency department visits, hospital admissions, and deaths among older adults following the introduction of COVID-19 vaccine - United States, September 6, 2020-May 1, 2021. MMWR Morb Mortal Wkly Rep 2021; 70:858–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Horwitz LI, Jones SA, Cerfolio RJ, et al. Trends in COVID-19 risk-adjusted mortality rates. J Hosp Med 2021; 16:90–2. [DOI] [PubMed] [Google Scholar]
- 4. Nguyen NT, Chinn J, Nahmias J, et al. Outcomes and mortality among adults hospitalized with COVID-19 at US medical centers. JAMA Netw Open 2021; 4:e210417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med 2021; 385:585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhatt PJ, Shiau S, Brunetti L, et al. Risk factors and outcomes of hospitalized patients with severe coronavirus disease 2019 (COVID-19) and secondary bloodstream infections: a multicenter case-control study. Clin Infect Dis 2021; 72:e995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Engsbro AL, Israelsen SB, Pedersen M, et al. Predominance of hospital-acquired bloodstream infection in patients with COVID-19 pneumonia. Infect Dis (Lond) 2020; 52:919–22. [DOI] [PubMed] [Google Scholar]
- 8. Giacobbe DR, Battaglini D, Ball L, et al. Bloodstream infections in critically ill patients with COVID-19. Eur J Clin Invest 2020; 50:e13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khatri A, Malhotra P, Izard S, et al. Hospital-acquired bloodstream infections in patients hospitalized with severe acute respiratory syndrome coronavirus 2 infection (coronavirus disease 2019): association with immunosuppressive therapies. Open Forum Infect Dis 2021; 8: ofab339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nori P, Cowman K, Chen V, et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect Control Hosp Epidemiol 2021; 42:84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prasad R, Patton MJ, Floyd JL, et al. Plasma microbiome in COVID-19 subjects: an indicator of gut barrier defects and dysbiosis. bioRxiv 2021.04.06.438634 [Preprint]. 6 April 2021. Available at: 10.1101/2021.04.06.438634. Accessed 2 May 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Venzon M, Bernard L, Klein J, et al. Gut microbiome dysbiosis during COVID-19 is associated with increased risk for bacteremia and microbial translocation. bioRxiv 2021.07.15.452246 [Preprint]. 27 July 2021. Available at: https://www.researchsquare.com/article/rs-726620/v1. Accessed 8 August 2021. [Google Scholar]
- 13. Bonazzetti C, Morena V, Giacomelli A, et al. Unexpectedly high frequency of enterococcal bloodstream infections in coronavirus disease 2019 patients admitted to an Italian ICU: an observational study. Crit Care Med 2021; 49:e31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seagle EE, Jackson BR, Lockhart SR, et al. The landscape of Candidemia during the coronavirus disease 2019 (COVID-19) pandemic. Clin Infect Dis. 2022; 74:802–11. [DOI] [PubMed] [Google Scholar]
- 15. White PL, Dhillon R, Cordey A, et al. A national strategy to diagnose COVID-19 associated invasive fungal disease in the ICU. Clin Infect Dis. 2021; 73:e1634–e1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ubeda C, Taur Y, Jenq RR, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120:4332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhai B, Ola M, Rolling T, et al. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat Med 2020; 26:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez-Reiche AS, Hernandez MM, Sullivan MJ, et al. Introductions and early spread of SARS-CoV-2 in the New York City area. Science 2020; 369:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Westblade LF, Simon MS, Satlin MJ.. Bacterial coinfections in coronavirus disease 2019. Trends Microbiol. 2021; 29:930–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sepulveda J, Westblade LF, Whittier S, et al. Bacteremia and blood culture utilization during COVID-19 surge in New York City. J Clin Microbiol 2020; 58:e00875-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rupp ME, Archer GL.. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin Infect Dis 1994; 19:231–43; quiz 244. [DOI] [PubMed] [Google Scholar]
- 23. Austin PC. The use of propensity score methods with survival or time-to-event outcomes: reporting measures of effect similar to those used in randomized experiments. Stat Med 2014; 33:1242–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horan TC, Andrus M, Dudeck MA.. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 2008; 36:309–32. [DOI] [PubMed] [Google Scholar]
- 25. Centers for Disease Control and Prevention. National Healthcare Safety Network (NHSN) Patient Safety Component Manual. 2021. Available at: https://www.cdc.gov/nhsn/pdfs/pscmanual/pcsmanual_current.pdf. Accessed 10 January 2022.
- 26. Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis 2015; 60:892–9. [DOI] [PubMed] [Google Scholar]
- 27. Neely LA, Audeh M, Phung NA, et al. T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci Transl Med 2013; 5:182ra–54. [DOI] [PubMed] [Google Scholar]
- 28. Navaratnam AV, Gray WK, Day J, Wendon J, Briggs TWR.. Patient factors and temporal trends associated with COVID-19 in-hospital mortality in England: an observational study using administrative data. Lancet Respir Med 2021; 9:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Price-Haywood EG, Burton J, Fort D, Seoane L.. Hospitalization and mortality among Black patients and White patients with COVID-19. N Engl J Med 2020; 382:2534–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raschke RA, Agarwal S, Rangan P, Heise CW, Curry SC.. Discriminant accuracy of the SOFA score for determining the probable mortality of patients with COVID-19 pneumonia requiring mechanical ventilation. JAMA 2021; 325:1469–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rassen JA, Shelat AA, Myers J, Glynn RJ, Rothman KJ, Schneeweiss S.. One-to-many propensity score matching in cohort studies. Pharmacoepidemiol Drug Saf 2012; 21:69–80. [DOI] [PubMed] [Google Scholar]
- 32. Patel K, Kay R, Rowell L.. Comparing proportional hazards and accelerated failure time models: an application in influenza. Pharm Stat 2006; 5:213–24. [DOI] [PubMed] [Google Scholar]
- 33. Levine SA, Niederman MS.. The impact of tracheal intubation on host defenses and risks for nosocomial pneumonia. Clin Chest Med 1991; 12:523–43. [PubMed] [Google Scholar]
- 34. Bruce-Hickman D, Sajeed SM, Pang YH, et al. Bowel ulceration following tocilizumab administration in a COVID-19 patient. BMJ Open Gastroenterol 2020; 7:e000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. RECOVERY Collaborative Group. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet 2021; 397:1637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Group RC, Horby P, Lim WS, et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med 2021; 384:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lamers MM, Beumer J, van der Vaart J, et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020; 369:50–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arunachalam PS, Wimmers F, Mok CKP, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020; 369:1210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Climo MW, Sepkowitz KA, Zuccotti G, et al. The effect of daily bathing with chlorhexidine on the acquisition of methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus, and healthcare-associated bloodstream infections: results of a quasi-experimental multicenter trial. Crit Care Med 2009; 37:1858–65. [DOI] [PubMed] [Google Scholar]
- 40. Elabbadi A, et al.. Bacterial coinfection in critically ill COVID-19 patients with severe pneumonia. Infection 2021; 49:559–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kreitmann L, Monard C, Dauwalder O, Simon M, Argaud L.. Early bacterial co-infection in ARDS related to COVID-19. Intensive Care Med 2020; 46:1787–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Somers EC, Eschenauer GA, Troost JP, et al. Tocilizumab for treatment of mechanically ventilated patients with COVID-19. Clin Infect Dis 2021; 73:e445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Agbaht K, Diaz E, Munoz E, et al. Bacteremia in patients with ventilator-associated pneumonia is associated with increased mortality: a study comparing bacteremic vs. nonbacteremic ventilator-associated pneumonia. Crit Care Med 2007; 35:2064–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.