Abstract
Autonomous robotic surgery has the potential to provide efficacy, safety, and consistency independent of individual surgeon’s skill and experience. Autonomous anastomosis is a challenging soft-tissue surgery task because it requires intricate imaging, tissue tracking and surgical planning techniques, as well as a precise execution via highly adaptable control strategies often in unstructured and deformable environments. In the laparoscopic setting, such surgeries are even more challenging due to the need for high maneuverability and repeatability under motion and vision constraints. Here we describe an enhanced autonomous strategy for laparoscopic soft tissue surgery and demonstrate robotic laparoscopic small bowel anastomosis in phantom and in vivo intestinal tissues. This enhanced autonomous strategy allows the operator to select among autonomously generated surgical plans and the robot executes a wide range of tasks independently. We then use our enhanced autonomous strategy to perform in vivo autonomous robotic laparoscopic surgery for intestinal anastomosis on porcine models over a one-week survival period. We compared the anastomosis quality criteria including needle placement corrections, suture spacing, suture bite size, completion time, lumen patency, and leak pressure, between the developed autonomous system, manual laparoscopic surgery, and robot-assisted surgery (RAS). Data from a phantom model indicates that our system outperforms expert surgeons’ manual technique and RAS technique in terms of consistency and accuracy. This was also replicated in the in vivo model. These results demonstrate that surgical robots exhibiting high levels of autonomy have the potential to improve consistency, patient outcomes, and access to a standard surgical technique.
Summary
An autonomous robotic strategy for laparoscopic soft tissue surgery in end-to-end anastomosis of the small bowel.
INTRODUCTION
Autonomous robotic surgery systems have the potential to substantially improve efficiency, safety, and consistency over current tele-operated robotically assisted surgery (RAS) with systems such as the da Vinci robot (Intuitive Surgical Inc., Sunnyvale, CA, USA). Autonomous robotic systems aim to provide access to standard surgical solutions that are independent of individuals’ experience and day-to-day performance changes. Proven examples of autonomous surgical robotic systems include TSolution-One (1) (THINK Surgical, Fremont, CA, USA) for procedures on rigid bony tissues, ARTAS for hair restoration (2) (Restoration Robotics Inc., San Jose, CA, USA), and Veebot (3) (Veebot LLC, Mountain View, CA, USA) for autonomous blood sampling. Currently, the most advanced autonomous capabilities are realized in the CyberKnife robot (4) (Accuracy Inc., Sunnyvale, CA, USA) which performs radiosurgery for brain and spine tumors under human supervision. However, this robot uses a contactless therapy method for tissues that are enclosed in rigid bony structures. Despite such efforts, autonomous soft tissue surgery still poses considerable challenges.
Autonomous soft tissue surgery in unstructured environments requires accurate and reliable imaging systems for detecting and tracking the target tissue, complex task planning strategies that take tissue deformation into consideration, and precise execution of plans via dexterous robotic tools and control algorithms that are adaptable to the dynamic surgical situations (5). In such highly variable environments, preoperative surgical planning such as in rigid tissues is not a viable solution (6). In the case of laparoscopic surgeries, the difficulty further increases due to limited access and visibility of the target tissue and the disturbances from respiratory motion artifacts. Anastomosis is a soft tissue surgery task that involves the approximation and reconstruction of luminal structures and requires high maneuverability and repeatability and hence is a suitable candidate for examining autonomous robotic surgery systems in soft tissue surgery scenarios. Well over a million anastomoses are performed in the USA each year (7–11). Critical factors affecting the anastomotic outcome include: the health of local tissue including perfusion status and contamination; physical parameters in anastomotic techniques such as suture bite size, spacing, and tension; anastomotic materials including suture and staples; and human factors such as the surgeon’s technical decisions and experience (12–17).
The level of autonomy (LoA) of medical robots are categorized in distinct levels ranging from pure teleoperation to full autonomy (5, 18). According to the classification introduced in (5, 19), the levels of autonomy include: LoA 0: no autonomy (e.g., pure teleoperation), LoA 1: robot assistance (continuous control by human with some mechanical guidance or assistance from robot via virtual fixtures or active constraints (20)), LoA 2: task autonomy (robot autonomously performs specific tasks such as running sutures which are initiated by human via a discrete control rather than continuous control), LoA 3: Conditional Autonomy (“A system generates task strategies but relies on the human to select from among different strategies or to approve an autonomously selected strategy” (19)), LoA 4: High autonomy (robot makes medical decisions but it has to be supervised by a qualified doctor), and LoA 5: Full autonomy (a robotic surgeon that performs an entire surgery without the need of a human).
Although high-level classifications for autonomy in medical robots were introduced in (19), it is understood that these categories are broad in scope and lack concrete metrics to properly delineate autonomy between consecutive classifications. In response, (5) expands the definition of each autonomy level to include an empirical evaluation based on the type or quantity of a task performed. Specifically, in LoA 2: Task-level autonomy, the system is trusted to complete a specific task or sub-task in an autonomous manner), in LoA 3: Supervised autonomy, the system autonomously completes the majority of a surgical procedure (such as an anastomosis procedure), and makes low-level decisions, and in LoA 4: High-level autonomy, the robotic system executes complete procedures based on human-approved surgical plans. The most well-known example of LoA 0 autonomy is the da Vinci surgical system in which every motion of the robot during surgery is teleoperated by the surgeon via a master console. Many of the recent works such as SmartArm (21, 22) still include a LoA 0 autonomy and are developed for operation under space limitations such as neonatal chest surgery. LoA 1 is implemented in the form of shared control for reducing the complexity of steering flexible robotic endoscopes (23). The first demonstration of LoA 2 (task autonomy) via in vivo open surgeries (24) was enabled via a robotic suturing tool controlled by a robot arm and a dual channel near infrared (NIR) and plenoptic three-dimensional (3D) camera that allowed the robot to detect the target tissue (stabilized outside of the body) and its landmarks, calculate a linear suture plan on the tissue, and execute the suture placement step by step under human supervision. Recent methods demonstrate LoA 2 laparoscopic in vivo hernia repair for porcine models (25). Machine learning-based techniques mostly automate surgical subtasks such as tumor ablation (26), debridement of viscoelastic tissue phantoms (27), clearing the surgical field (28), autonomous control of magnetic endoscopes for colonoscopy (29), autonomous steering of robotic camera holders (30) (i.e., LoA 2), or imitate surgical procedures via learning by observation on phantom tissues in a LoA 3 (31–34). More examples consistent with similar LoA definitions can be found in (6, 35). Despite these considerable efforts, most works either present low autonomy in complex tasks or high autonomy in simpler tasks using phantom tissues.
As our first contribution towards this goal, we achieve the enhanced autonomy necessary to perform robotic laparoscopic anastomosis of the small bowel using the Smart Tissue Autonomous Robot (STAR). This is accomplished by developing several autonomous features including starting/pausing/unpausing the tissue tracking system, detecting the breathing motion of tissue and its deformations and notifying the operator to initiate a replanning step, robot tool failure detection, camera motion control, suture planning in different modes with uniform and non-uniform spacing, pre-filtering the plan for reducing noise and irregularity, predicting the tool collisions with the tissue, and synchronizing the robot tool with breathing motions of the tissue and under a remote center of motion (RCM). The operator selects among the autonomously suggested suture plans or approves a re-planning step and monitors the robot for repeating a stitch as needed. The main objective is to increase the overall accuracy in suture placement and at the same time reduce the operator workload and involvement via the additional autonomous. Although the system does require manual fine adjustment of the robot to correct positioning if a stitch is missed, more than 83% of the suturing task is completed autonomously using this workflow. In our previous work for open surgical intestinal anastomosis (24), the tissue tracking only considered a stationary tissue without the breathing motions, only one linear suture plan option without noise pre-filtering and collision prevention was considered, autonomous replanning suggestions were not included, the operator needed to monitor each sub-step of the suturing procedure, and the tool failure monitoring and autonomous camera motion control were not implemented. Consequently, only 57.8% of the sutures were completed autonomously with no adjustments. Therefore, the developments presented in this work provide notably more autonomy and accuracy during the surgical procedure compared to a step-by-step method. This enhanced autonomy combined with surgeon’s supervision goes hand in hand with several benefits to the patient and the surgeon. Increasing the amount of autonomy in robotic surgical systems has the potential to standardize surgical outcomes that are independent of surgeons’ training, experience, and day-to-day performance changes. This will improve the consistency of performing surgery and reduce the surgeon’s operating workload to taking advantage of their supervisory role to guarantee the safety of the overall surgery. Consistency in anastomotic parameters such as suture bite size, spacing, and tension directly affects quality of a leak-free anastomosis (36) which in turn improves the patient outcomes and reduces the recurrence and complication rates of the surgery (12, 14, 15).
The second contribution of this work is enabling a laparoscopic implementation of the enhanced autonomous strategy for full anastomosis, which imposes various technical and workflow challenges. In the new laparoscopic setting, the tissue is suspended in the peritoneal space of the patient with transabdominal stay sutures. In contrast to open surgical settings, where the motion of target tissue motion is restricted via external fixtures outside the body and the tissue is more accessible, the laparoscopic settings introduce multiple challenges including the effects of breathing motions on the staged tissue location and deformation, kinematic and spatial motion constrains for the robots through the laparoscopic ports, tool deflections and difficulties in force sensing, limited visibility, and a need for miniaturized laparoscopic 3D imaging systems, and other factors such as humidity during the surgery. We develop machine learning, computer vision, and advanced control techniques to track the target tissue movement in response to patient breathing, detect the tissue deformations between different suturing steps, and operate the robot under motion constraints. We demonstrate implementation of such soft tissue robotic surgeries in a series of in vivo survival studies via porcine models for laparoscopic bowel anastomosis.
RESULTS
In this section, we explain the design and workflow of the enhanced autonomous STAR system, then present accuracy testing of the tissue motion tracker, comparison testing between STAR and expert surgeons performing complete end-to-end anastomosis in phantom bowel tissues, a markerless tissue landmark tracking method, and porcine in vivo survival studies using STAR to complete laparoscopic small bowel anastomosis. The in vivo experiment results were compared to a control test condition via manual laparoscopic surgery with standard tools and surgical endoscopes performed by a surgeon as a benchmark (see Section In vivo end-to-end anastomosis for details). Although the in vivo experiments prove the feasibly of the implementation in the pre-clinical settings, we also conducted a thorough set of experiments via tests on phantom tissues that compared the performance of STAR via several metrics that are not measurable after a one-week survival period after the tissue healing process (see Section Phantom end-to-end anastomosis for details). Furthermore, the phantom test conditions enabled controlled disturbance and robustness tests to examine the capabilities of each system in a wider range of conditions. We compared STAR with standards of care including a da Vinci SI based robotic assisted surgery using 3D endoscopic vision as well as manual laparoscopic anastomosis.
System design for enhanced autonomy
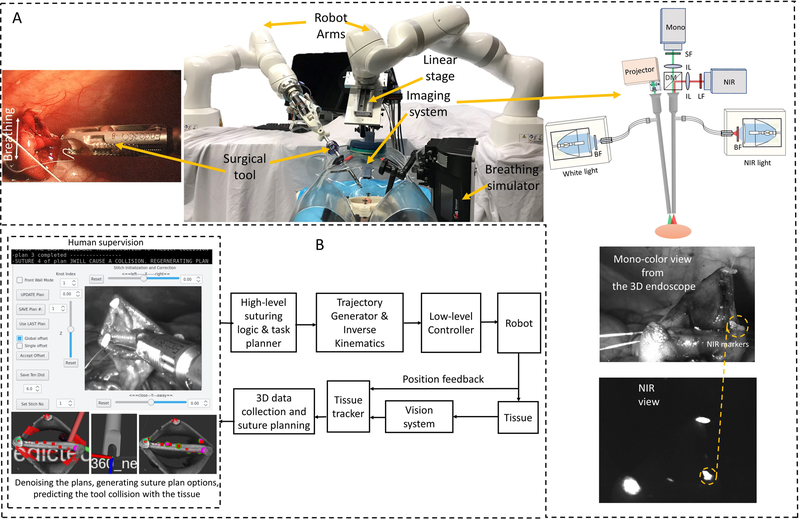
The laparoscopic version of STAR with the proposed autonomous architecture is shown in Fig. 1. The system consists of one KUKA LBR Med robot with a motorized Endo 360 suturing tool (37) for robotic suturing and a second KUKA LBR Med robot that carries an endoscopic dual-camera system consisting of a NIR camera and a 3D mono color endoscope (38). The camera system allows the STAR to autonomously track biocompatible NIR markers (39) on the tissue and reconstruct the 3D surface of the tissue (used for suture planning). NIR markers provided a method for tracking desired landmarks on the tissue (e.g., the start and end points of the suturing process on the target tissue) that is robust to blood and thin tissue occlusions during the surgery (39). The proposed method also allows tracking the breathing motion of a patient regardless of the lighting conditions inside the animal (e.g., the white light used for monitoring the surgery). The two-dimensional (2D) view of the camera was also used as visual feedback for the operator when supervising the autonomous control (Fig. 1A). The view is provided from the 3D mono color endoscope when it is not running in the point cloud collection mode (i.e., projecting fringes to detect the 3D shape of the tissue) which can be confusing to the operator. Since the robot operates with an elevated level of autonomy based on the 3D tissue detection and task planning methods, a 2D view was sufficient for the supervisory tasks using the developed system. Later in this article (in Section “Cascaded U-Net for segmentation assisted landmark detection”) we present robust tissue landmark tracking via this camera that will obviate the need for NIR camera in future works.
Fig. 1. Enhanced autonomous laparoscopic soft tissue surgery.
(A) The components of the Smart Tissue Autonomous Robot (STAR) system including medical robotic arms, actuated surgical tools, and dual-channel NIR and 3D structured light endoscopic imaging system. (B) Control architecture of the enhanced autonomous control strategy for STAR.
The following describes the general workflow of the enhanced autonomous STAR system used in this study (for further details, please refer to the Materials and Methods section). First, the operator initiated a planning sequence with STAR using the graphical user interface (GUI) shown in Fig. 1B. The tissue tracking algorithm (detailed and tested in “Tissue motion tracking” Section) detected when the patient was temporarily not breathing (i.e., target tissue has reached a stop position due to a pause in the breathing cycle). When the tissue was stationary, STAR generated two initial suture plans to connect adjacent biocompatible NIR markers placed on the corners of the tissue. The suture plans were then filtered for noise and STAR predicted tool-tissue collisions. Unusable suture plans were discarded, and a new set of plans were generated for filtering and collision predictions. Once the set of plans was usable, the operator selected a suture plan that creates either a uniform suture spacing across the entire sample, or a uniform suture spacing with additional stitches in the corners of the tissue to elicit leak free closure in difficult geometry. Once the operator selected and approved one of the plans, STAR executed the suture placement routine by moving the tool to the location of the first planned suture, applied a suture to the tissue, waited for the assistant to clear loose suture from the field, and tensioned the suture. After completion of the suture routine, STAR reimaged the surgical field to determine the amount of tissue deformation. If STAR detected a change in tissue position greater than 3mm compared to the current surgical plan, it notified the operator to initiate a new suture planning and approval step. Otherwise, STAR suggested reusing the initial surgical plan and continued to the next suture routine. This process was repeated for every suture in the surgical plan. In the instance when a planned stitch was not placed correctly or if the suture did not extend across two tissue layers, the operator repeated the last stitch with a slight position adjustment via the GUI before the STAR continued to execute the suture plan. It is worth noting that even though direct intervention from the operator was used to correct missed stitches, the majority of the workflow was completed in an autonomous manner. This strategy can be expanded in future works whereby the STAR system autonomously detects, adjusts, and repeats a missed stich to result in a completely autonomous anastomosis procedure. A more detailed technical block diagram of the overall control strategy is shown in Fig. S1 of the methods section.
Tissue motion tracking
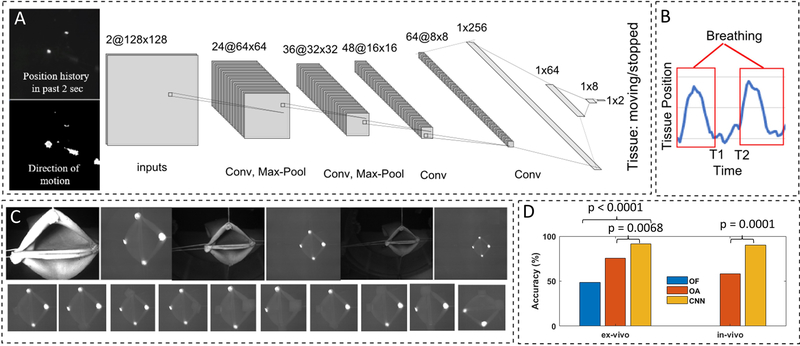
To track the motion of the tissue due to breathing and other tissue motion during laparoscopic surgery, we developed a machine learning algorithm based on the convolutional neural networks (CNN) (40) and the feedback from the NIR camera. We labeled a total of 9294 examples of different motion profiles (4317 for the breathing examples and 4977 for the stopped breathing examples) via data collected from laparoscopic porcine small bowel anastomosis procedures. The labeled data from the live animal surgeries was collected to train the system in a more complex motion tracking setting. Our hypothesis was that if the trained system performs well using in vivo data where reflections and obstruction can cause artifacts in the training set, then the results would perform as well or better in the controlled phantom test conditions (examined later in this section). The inputs to the CNN included the position history of the NIR markers in the past 2 seconds, and the direction of motion of the markers between now and 2 seconds ago which included 2 channels of 128 by 128 down sampled frames (see examples in Fig. 2A). The CNN included 4 convolutional, 3 dense layers, and two outputs (for moving/stopped tissue labeling). The activation function for the convolutional layers and the first three dense layers was ReLU and the activation function for the final dense layer was SoftMax (for labeling the moving and not moving states). Same padding and a 25% dropout were used in the convolution layers. A cross-entropy loss function was used for training the network. This network predicted the motion profiles with an accuracy of 93.56% during the training phase. The result was used to synchronize the motion of robot with tissue and trigger data collection and planning the right times (e.g., on the bottom of breathing cycles) to improve the control algorithm accuracy.
Fig. 2. Tissue motion tracking.
(A) The CNN-based breathing motion tracker. (B) Examples of the vertical motion of NIR marker during in vivo tests. (C) Robustness test configurations for the phantom conditions. (D) The accuracy test results for the breathing motion tracker via optical flow with fixed threshold (OF), optical flow with adjustable threshold (OA), and the CNN-based method (CNN).
The results of the accuracy tests and comparison with an optical flow method are shown in Fig. 2D. The optical flow methods tracked the differences of the last two frames in the NIR image, applied a low pass filter, and used a threshold to detect when the breathing motion ends (i.e., the image flow drops below a certain threshold). Since this method was sensitive to light intensity and distance, two variations were tested. A variation with a fixed threshold for a mid-range distance and light intensity called optical flow with fixed threshold (OF), as well as a variation with adjustable threshold for each measurement distance called optical flow with adjustable threshold (OA). For the OF method with fixed threshold, the target tissue was placed at the mid-range location (here 6.5cm) and the decision threshold was fine-tuned experimentally for the given test conditions to achieve the maximum number of correct detections in 5 breathing cycles in a row. The mid-range detection threshold was then used for targets at close (here 3.5 cm distance) far (here 10cm distance) locations. For the OA method with adjustable thresholds, a similar process as for the mid-range OF threshold adjustment was used separately for each test location/distance to record the best possible motion detection results for this method. For each test, we evaluated whether transitions between the breathing/moving state of the tissue and non-breathing state (i.e., at T1 and T2 in Fig. 2B) were detected correctly in a 14 breaths per minute breathing cycle. If the tested methods labeled any of the transitions incorrectly, a mistake was counted. Combinations of distance to the target tissue (3 cm, 6.5 cm and 10 cm) for 11 marker orientations (Fig. 2C) were recorded for five breathing cycles for each combination using synthetic colon samples (N = 165 per condition). The results shown in Fig. 2D illustrate that the CNN-based method was more robust to tests in different conditions owing to the generalization capability of the neural networks. The CNN method resulted in 91.52% test accuracy which was significantly higher than OF with 75.76% (P = 0.0068) and OA with 48.48% accuracies (P < 0.0001). Accuracy of the CNN remained unchanged when images obtained during in vivo anastomosis (N = 60 representative samples) in four porcine models were analyzed (91.52% for phantom vs 90.00% for in vivo), whereas the OA method’s accuracy dropped to 58.33% from the 75.76% in the phantom tests due to a lack of robustness to variations in the test conditions.
Phantom end-to-end anastomosis
Experiments for end-to-end anastomosis were conducted using phantom bowel tissues (by 3D Med, Franklin, OH, USA). The study groups included, enhanced autonomous robotic anastomosis via STAR (n = 5), a manual laparoscopy (LAP, n = 4) method, and a da Vinci SI based robotic assisted teleoperation method (RAS, n = 4) (see Fig. S2 for the testbeds). For STAR tests, tissue orientation was set between −20 and 20 degrees with 10-degree increments for each sample, whereas LAP and RAS tissue orientations were randomly selected between −20 and 20 degrees. For all study groups, the test tissues were attached to a linear stage that was programmed to move with a 14 breath/minute frequency and 3mm amplitude to replicate the in vivo breathing cycles. For the STAR study group, a NIR marker tracking algorithm was used to trigger robot motion to the planned suture point, and to enforce that suturing occurs during the rest phase of the breathing cycle. A target suture spacing of 3 mm was selected based on our previous results in (24), which considers the tissue thickness T, bite depth H, and suture spacing S for a leak-free anastomosis (see Fig. 1G in this reference). For the LAP and RAS study groups, surgeons were instructed to suture with the same spacing and consistency that they would perform in a human patient. To simulate tissue movement that occurs during in vivo surgeries, random deformation of the tissue was induced once during back wall suturing, and twice during front wall suturing by loosening and tensioning the stay sutures in a different direction for all study groups. LAP and RAS data was collected from four different surgeons from either George Washington University Hospital (Washington, DC, USA) or Children’s National Hospital (Washington, DC, USA.) The order of surgeons was randomized with half of the participants starting with LAP followed by RAS tests and vice versa.
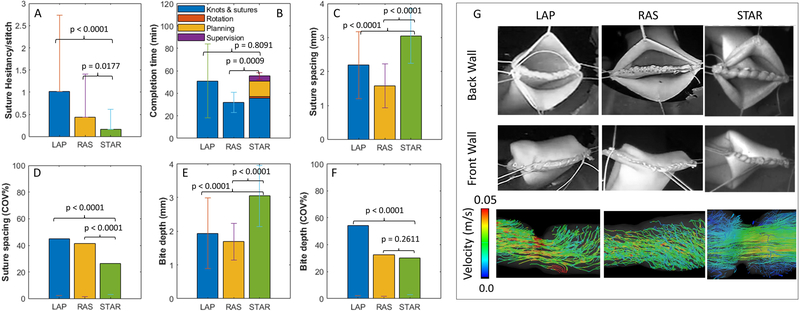
The performance metrics for evaluating the outcomes of test conditions for phantom tissues included: task completion time, number of suture hesitancy events per stitch, suture spacing, and suture bit depth. Individual STAR, LAP, and RAS tests were recorded and analyzed to measure the total time to complete knots (STAR, LAP, RAS), complete sutures (STAR, LAP, RAS), planning (STAR), and supervision (STAR). Suture planning was defined as moving the camera to measure distance, calculating, and saving suture plans, and returning to the safe suture distance. Supervision was defined as slight position adjustments for the robot after hesitancy events. The number of suture hesitancy events per stitch was defined as needle placement corrections for STAR or when surgeons inserted and immediately retracted a needle from tissue for LAP and RAS experiments. This metric measured the collateral tissue damage during surgery. Furthermore, suture spacing, defined as the shortest distance between any two consecutive sutures, and bite depth, defined as the perpendicular distance from a suture to the tissue edge, were collected for each sample (see Fig. S3A). We report the coefficient of variance for suture spacing and bite depth as a measure of consistency for each metric since both are known to directly affect the quality of a leak-free anastomosis (36). All results from phantom testing are summarized in Fig. 3.
Fig. 3. The results of phantom end-to-end anastomosis via LAP (n = 4), RAS (n = 4), and STAR (n = 5).
(A) Suture hesitancy events per stitch (additional suturing attempts per stitch). (B) Task completion time. (C) Suture spacing. (D) consistency of suture spacing via the coefficient of variance (COV). (E) Suture bite depth. (F) Consistency of bite depth via the coefficient of variance (COV). (G) Representative examples of the phantom end-to-end anastomosis test via LAP, RAS, and STAR including three-dimensional flow fields within each sample.
STAR completed a total of 118 stitches (10 knots, 108 running stitches, N=5) with an average suture hesitancy per stitch of 0.17 ± 0.44. The total number of suture hesitancy events among all STAR samples was 20 indicating that STAR correctly placed stitches under the enhanced autonomous mode at the first attempt 83.05% of the time. Twelve of the hesitancy events occurred during a corner stich, whereas the remaining eight hesitancy events occurred on either the back or front wall of the tissue. Observed hesitancy events were a result of small planning errors, tool positioning errors, difficult to reach corner stitches, and shallow bite depth. In each of these cases, the operator would interrupt the suture plan to correct the positioning of the suture tool, and then restart the enhanced autonomous mode. The average three-dimensional norm of offset adjustments was 3.1 ± 1.39 mm with a maximum value of 6.0 mm. For the LAP method, a total of 111 stitches (18 knots, 93 running stitches, N=4) were completed with an average hesitancy per stitch of 1.03 ± 1.71 whereas RAS samples had a total of 139 stitches (14 knots, 125 running stitches, N=4) with an average hesitancy per stitch of 0.44 ± 0.97. As summarized in Fig. 3A, the hesitancy per stitch for STAR was significantly less than that of LAP (P < 0.0001) and RAS (P = 0.0177).
The average total time to complete the end-to-end anastomosis task is shown in Fig. 3B. It was observed that RAS was the fastest method for completing the task (31.96 ± 9.01 minutes), followed by LAP (51.08 ± 32.72 minutes), and STAR (55.41 ± 2.94 minutes). Although STAR had similar total task time to LAP (P = 0.8091), STAR was significantly slower than RAS (P=0.0009). Differences in total completion time were primarily due to additional re-planning due to random tissue deformations and motions, and overall supervision time (approving plans and sporadic slight position adjustments) in the STAR system that were not needed in the RAS technique which account for an additional 19.66 minutes per sample. Additionally, STAR was operated under conservative velocity limits to minimize the force interactions between the surgical tool and the laparoscopic ports to guarantee the smoothness of motion profiles and the accuracy of the suture positioning. The smooth motion profiles had the added benefit of reducing the chance that small planning errors in tight corners would cause the suture tool to damage the stay sutures or native tissues.
The average suture spacing for LAP, RAS and STAR was 2.28 ± 1.04 mm, 1.58 ± 0.65 mm, and 3.05 ± 0.8 mm, respectively (see Fig. 3C). Suture spacing resulting from STAR was significantly larger than LAP (P < 0.0001) and RAS (P < 0.0001) but was statistically similar to the 3.0 mm spacing suggested by the uniform suture plan (P = 0.4163). Because smaller suture spacing can lead to smaller variance in the data, suture consistency was compared by the coefficient of variance for each study group (i.e., the ratio of standard deviation to mean converted to percentage). Based on the results in Fig. 3D, the coefficient of variance for LAP, RAS, and STAR was 45.37%, 41.42%, and 26.36% respectively, which indicated higher consistency of STAR compared to LAP (P<0.0001) and RAS (P<0.0001). The average bite depth for LAP, RAS, and STAR was 2.02 ± 1.10 mm, 1.69 ± 0.55 mm, and 3.05 ± 0.91 (see Fig. 3E). Although the average bite depth for STAR was significantly larger than LAP (P < 0.0001), and RAS (P < 0.0001), it was similar to the target bite depth of 3 mm programmed by the uniform suture plan (P = 0.4414). As with suture spacing, the consistency of the bite depth can be reported as the coefficient of variation by normalizing the standard deviation of each study group by its mean. As illustrated in Fig. 3F, the coefficient of variation for LAP, RAS, and STAR was 54.41%, 32.57%, and 29.99% respectively, indicating that STAR is more consistent in bite depth than LAP (P < 0.0001) and similar to RAS (P = 0.2611) (see Table S1 for further details).
A representative anastomosis for each study group is shown in Fig. 3G. Back wall suturing is illustrated in the top row, front wall suturing is illustrated in the middle row, and three-dimensional flow fields of fluid passing through the resulting anastomosis are illustrated in the third row. To obtain images of the resulting flow fields, samples from each study group were connected to a MR conditional pump and circulated with a 60/40 glycerin to water mixture at a flow rate of 10 ml/s. Flow fields were acquired using a Siemens Aera 1.5T MRI scanner programmed to a four-dimensional (4D) flow sequence with 1.25 mm x 1.25 mm x 1.3 mm spatial resolution, with the samples placed at the isocenter of the magnet. Post processing, including segmentation and display of flow field lines was performed with iTFlow version 1.9.40 (Cardio Flow Design Inc, Tokyo, Japan). A single phase of the 4D flow was collected for all models. Resulting flow lines are most laminar for STAR and most turbulent for LAP, indicating that STAR reapproximates tissue better than either surgical technique.
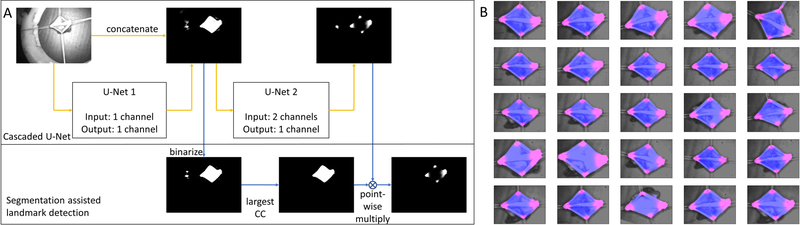
Cascaded U-Net for segmentation assisted landmark detection
To minimize the complexity of the camera system and to eliminate the need for markers which are difficult to get approved for long-term implantation in clinical translation, we further aimed to replace the fluorescence marker-based imaging with a CNN-based landmark detection algorithm. The overall data processing contained a CNN architecture and a post-processing step as shown in Fig. 4A. The CNN architecture was adopted from U-Net, since U-Net has demonstrated superior performances in the segmentation task due to its capability of the multi-resolution analysis (40), and it requires much fewer training data compared to a fully connected network due to its fully convolutional properties (41). Two U-Nets were cascaded for segmentation assisted landmark detection. The first U-Net took each individual grayscale intestine image as the independent input and output a segmentation result of the intestine tissue. Its main architecture was mostly the same as the one in (40), except that we used the same padding in all pooling layers to avoid image cropping, and that we used sigmoid activation instead of the soft-max activation for the final segmentation results. The second U-Net took both the intestine image and the segmentation result from the first U-Net as the input and it output a landmark heatmap, since the landmark heatmap regression has demonstrated better performances than regressing the landmark coordinates directly (42). The final landmark coordinates were obtained as the maximum responses of the predicted heatmaps. The second U-Net architecture was mostly the same as the first U-Net, except that its input contains two channels. Since the cascaded U-Net had two outputs, the total loss function was the summation of the binary cross entropy of both the segmentation and the landmark heatmap with the same weight. In the post-processing step, we first binarized the segmentation results by applying a threshold. Then, we found the largest connected component (CC) from the binarized segmentation. In the end, we multiplied the largest CC with the landmark heatmap using a point-wise approach to eliminate any random landmarks detected in the background.
Fig. 4. Segmentation assisted landmark detection.
(A) Overall data processing scheme based on cascaded U-nets, (B) Final results on the whole testing set. Blue shows the intestine segmentation results, and pink shows the landmark heatmap results.
To evaluate the performance of the algorithm, we recorded a 45-second video of a phantom intestine tissue with various positions and orientations using our camera system. During the recording, we moved the tissue fast enough such that a wide range of positions and orientations were captured to avoid overfitting. The video had a frame rate of 68 Hz and contained 3037 frames in total. We took 50 image frames out of the video every 0.9 second. The 25 image frames with an odd serial number were used as the training set, and the remaining 25 frames with an even serial number were used as the testing set. During the robotic anastomosis in this work, once the tissue was staged (for example for the back wall or the front wall suturing steps), it does not undergo drastic changes in position and orientation until the corresponding step of the anastomosis is completely done. Moreover, 3D tissue and landmark tracking tasks needed to be performed intermittently (i.e., before/after a suture was autonomously placed when the tool was not blocking the tissue and when target tissue had reached a stop position due to a pause in the breathing cycle). Therefore, major temporal information changes did not appear in the current setting and hence the developed CNN was suitable for this task.
We manually segmented the intestine tissue using a binarized hard mask as the segmentation ground truth. Similarly, we manually segmented four corners of the intestine tissue using binarized hard masks, and we used the center of each corner segment as the ground truth for the corner landmark coordinate (i.e., for replacing the NIR markers used for path planning). Moreover, we fit each corner hard mask with an elliptical gaussian probability as the heatmap ground truth. After training, the trained weights were applied to both the training set and the testing set followed by the post-processing step. The predicted landmark coordinates of the four corners were obtained as the four local maximum heatmap responses. Compared with the ground truth, it was found that the landmark detection accuracy to the training set was 2.3±1.0 pixels, and the landmark detection accuracy to the testing set was 2.6±2.1 pixels. Due to the different shapes and sizes of the four intestine corners, we defined the maximum allowable error as the statistical average of the effective radius of all the corners, which was found to be 11.3±4.8 pixels. Furthermore, out of 25 testing frames, there were 3 frames where apart from the correct four corners, extra landmarks in the background were mistakenly detected. With the segmentation assisted landmark detection post-processing, these extra landmarks were effectively removed. The final segmentation and heatmap results were cropped and combined with the intestine image to a color image as shown in Fig. 4B. From these results, we concluded that the cascaded U-Net correctly segmented the intestine tissue and generated the intestine landmark heatmap. Moreover, with the segmentation assisted landmark detection post-processing, the noise from the background was effectively removed.
In vivo end-to-end anastomosis
Finally, laparoscopic autonomous surgery was performed in vivo on porcine small bowel using STAR (n = 4 independent animals). After surgery, the animals were monitored for a one-week survival period, and subject to limited necropsy to compare the healed anastomosis to a laparoscopic control (n = 1) (Table 1). For these tests, STAR utilized the same suture algorithm described in the phantom tests. To guarantee the safety of the procedure and comply with animal care protocols, we increased the safety measures by adding to the number of tasks that the operator was required to monitor and intervene (the phantom tissue tests already allowed higher risk tolerance for testing the system features). This included an additional number of human interventions for initial robot position fine-tuning (before the first suture) and approval of firing the needle. We also reduced the number of plan options and replanning steps and implemented more conservative velocity limits to further prevent collateral tissue damage.
Table 1.
In vivo end-to-end anastomosis. Bowel anastomoses were carried out in pigs using STAR (n = 4) and manual laparoscopic control (n = 1).
| Pig No | Weight at surgery (kg) | Weight at sacrifice (kg) | Leak pressure (psi) | Lumen patency (%) | Completion Time (min) | No of sutures | Suture Hesitancy |
|---|---|---|---|---|---|---|---|
| STAR 1 | 32.7 | 36.4 | 0.23 | 85 | 59.71 | 24 | 4 |
| STAR 2 | 35.4 | 37.3 | 0.12 | 85 | 55.64 | 17 | 7 |
| STAR 3 | 35.3 | 33.5 | 1.2 | 90 | 65.73 | 24 | 11 |
| STAR 4 | 35.5 | 33.8 | 1.2 | 95 | 67.03 | 21 | 7 |
| 5 (Control) | 30 | 32 | 1.2 | 90 | 25.6 | 21 | 9 |
The performance metrics for evaluating the outcomes of test conditions for in vivo experiments included: task completion time, number of suture hesitancy events per stitch, lumen patency, leak pressure, and quality of wound healing and inflammatory changes. Upon completion of the one-week survival period following surgery, limited necropsy was performed. Gross examination of the anastomosis identified scar tissue and adhesions consistent with the control animal. Additionally, residual NIR markers used during the surgery had to be encapsulated and remained outside of the anastomosis. A portion of the small bowel was resected and subject to leak and lumen patency testing. Lumen patency was defined as the largest diameter gauge pin that could be passed through the anastomosis. Leak pressure was defined as the maximum internal pressure achieved by the anastomosis before rupture. Leak pressure testing was limited to a maximum of 1.2 psi to preserve the sample for histology testing (see Fig. 5).
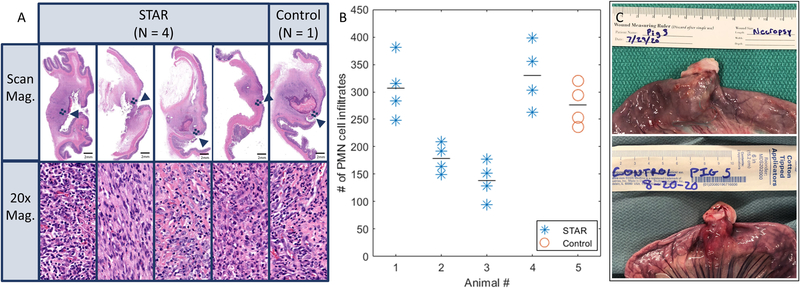
Fig. 5. The results of in vivo experiments.
(A) Representative histology examples from each anastomosis tissue operated on with STAR (n = 4), and manual laparoscopic control test (n = 1). The approximate location of each anastomosis is indicated with an arrow. Dashed boxes near each anastomosis represent the location of the magnified images. (B) PMN cell as a surrogate measure of inflammation for each sample. (C) Representative examples of the anastomosis collected at necropsy for STAR and control tests.
In this study, STAR achieved a leak pressure of 0.69 ± 0.59 psi and a lumen patency of 88.75 ± 4.79 percent which are both consistent with our previous results in open surgeries (24). The average task completion time for STAR was 62.03 ± 5.32 minutes compared to 25.6 minutes for the control. Similar to the phantom tests, STAR was slower to complete the task primarily due to the additional planning (8.5 minutes on average for 3 plans), safe tool rotations (1.13 minutes), and task supervisions (10.2 minutes). Moreover, STAR was operated under conservative speed limits to minimize the interaction forces at the laparoscopic ports and reduce the chance of collisions between the suture tool, stay sutures, and non-targeted tissue due to breathing motions which further increased the total time. Histology carried out on day 7 (Fig. 5A) illustrated that there was no observable difference in wound healing and inflammatory changes as observed in continuity of mucosal edges, degree of tissue injury such as hematoma, and similar counts of polymorphic nucleated cell (PMN) infiltrates (P = 0.4543) (Fig. 5B) between STAR and the control.
In all four in vivo tests, STAR completed a total of 86 stitches (8 knots, 78 running stitches). The total number of sutures requiring manual adjustments were 29 (66.28% correctly placed stitches in the first attempt) which corresponded to an average of 0.34 suture hesitancy per stitch. These attempts included 14 corner stitches and 15 stitches along the back and front walls. Compared to the phantom tests, a higher average number of attempts were required to adjust the offsets manually via the GUI (0.34 additional attempts for in vivo vs 0.17 for phantom). Additional sources of extra attempts observed during the in vivo experiments included animal size variations that affected the optimal laparoscopic port placement and tissue reachability, extra tissue motion induced by the stay sutures on the flexible skin of animal, sporadic insufflation leaks due to the motion of robot tool as well as imperfect sealing of the gel port, edema causing the tissue not to fit in the jaw, harder tissue staging depending on the animal, and tool flex under forces from the trocar. The average norm of offset adjustments in three dimensions was 3.85 ± 1.88 mm with a maximum value of 8.15 mm. For the control test condition, a total of 21 stitches were completed (4 knots and 17 running stitches). The total number of needle repositioning was 9 (57.14% correctly placed stitches in the first attempt), which corresponds to an average of 0.43 suture hesitancy per stitch. These needle repositioning adjustments included 4 corner stitches and 5 stitches on the front and back walls.
DISCUSSION
In this article, we demonstrated laparoscopic autonomous soft tissue bowel surgery in unstructured and deformable environments under motion and visual constraints. Advanced imaging systems, machine vision and machine learning techniques, and real-time control strategies were developed and tested to track tissue position and deformation, perform complex surgical planning, interact with the human user, and adaptively execute the surgical plans. In the robotic laparoscopic anastomosis experiments, the developed system outperformed surgeons using LAP and RAS surgical techniques in metrics including the consistency of suture spacing and bite depth as well as the number of suture hesitancy events which directly affect the quality of a leak-free end-to-end anastomosis. The STAR system encapsulates the autonomous control functionality and reduces the involvement of the human operator, with the robot reacting to tissue deformation and various test condition changes. Although the role of human supervision cannot be eliminated in the complex and unpredictable surgical scenarios, advancing through the levels of autonomy can reduce the individual-based experience and performance variations and result in higher safety, access to a standard surgical technique, and consistency of the surgical outcomes. We also demonstrated in vivo robotic laparoscopic anastomosis which involved surmounting several challenges including soft tissue tracking, surgical planning, and execution in a highly dynamic and variable environment with restricted access and visibility. The survival study results indicated that the developed robotic system could match the performance of expert surgeons in metrics including leak-free anastomosis and lumen patency and at the same time exhibit an elevated level of consistency.
Despite these achievements, some limitations exist in the current methods and the results. Currently, a successful implementation of the robot control algorithms depends on the reachability and correct staging of the tissue in a certain working region. This does not allow any arbitrary tissue staging given the limitations of the camera system and robot’s kinematic architecture. Furthermore, once additional staging features are enabled, the tissue landmark detection methods discussed in this article will need to include architectures such as long short-term memory (LSTM) (43) to handle drastic changes in the temporal information caused by rapid position and orientation changes in the tissue. Another limitation of this study was that the comparison between the STAR robot, manual laparoscopic surgery, and tele-operated da Vinci was performed on phantom tissue. There was no possibility of using a da Vinci based test arm for the in-vivo study, since this specific system is only allowed for use in human patients at Children’s National Hospital (Washington, DC, USA). Additionally, due to ethical reasons to limit the number of in vivo experiments and due to dynamic factors, that uniquely occur during each surgery (such as variations in size, port placement, tissue reachability, sporadic insufflation leaks, tissue motion, and edema), we limited the in vivo comparisons to one manual laparoscopic surgery and instead performed an extensive, more controlled comparison study in phantom tissue. However, considering all these factors, we were able to confirm the overall feasibility of implementing the enhanced autonomous strategy of the STAR in the in vivo experiments.
In the current version of the system, the laparoscopic dual-camera system has a 3 cm footprint which requires the use of a gel port for camera insertions. This will also increase the chance of gel port blocking the robotic suture tool or inducing extra pressure and slight positioning errors at the tool control point (TCP). In our future works, we will integrate and test the marker-less tissue tracking techniques to reduce the camera system to an endoscope that will provide quantitative three-dimensional visualization with a smaller footprint than used in this study. Furthermore, we will enable a larger distance for accurate point cloud collection so that the camera system does not require back and forth motion between the imaging and suture positions to reduce the suturing time. It is also worth mentioning that due to additional forces and torques introduced at the laparoscopic port on the robot tool, the force measurements at the tool tip are not observable. More specifically, the 6-axis force/torque sensor of the robot is mounted on the robot flange which measures a combination of tool gravity forces (3 estimated forces based on the tool geometry and mass), interactions at the laparoscopic port (2 unknown forces and 2 unknown torques assuming negligible axial force and torque along the port), and the tension and contact forces at the tool-tip during the suturing process on tissue (3 unknown forces, i.e. a total of at least 7 unknowns with 6 measurements). In future works, we are planning to include a tactile or proximity sensor near the tool tip to decouple the interaction forces and measure the tissue contact and suture tensioning forces locally. Furthermore, we are planning to add a sensor at the tool tip to detect if the two layers of the target tissue are inside the robot tool jaw before firing the needle. This will help eliminate one monitoring task from the operators and guarantee that robot does not miss any stitches on the tissue which will be a key step in enabling a fully autonomous system. We expect that the synergy of these changes will allow a substantially faster task completion time via higher (but safe) robot velocity limits, reduced measurement and (re)planning time, and reduced number of suture hesitancy events.
MATERIALS AND METHODS
Imaging System
A customized laparoscopic imaging system is built for in vivo animal studies via STAR (shown in Fig. 1A). The projector and the monochrome camera are used to reconstruct the 3D point cloud of the sample. The NIR light source and the NIR camera are used for fluorescence marker imaging. Both cameras are enabled simultaneously through the nonoverlapped imaging spectral window. Moreover, a white light source is added to monitor the environment inside the animal body when necessary. NIR and 3D cameras enable the robot to reconstruct the 3D model of the tissue and plan for robotic suture planning. The detailed specifications for the imaging system are presented in the supplementary methods.
Robotic Platform
The platform consists of a KUKA LBR Med lightweight robot that is equipped with a motorized commercial Endo 360 suture tool with pitch control. The camera system allows measurement of the tissue geometry 5–8cm distance from the tissue. The system is capable of dynamic position control of the camera system via a linear stage, which is used to prevent collisions between the camera system and suturing tool when STAR is executing a suture plan. In practice, the camera is positioned 5–8cm from the target tissue and used to collect 3D point clouds, whereas the suture tool is retracted from the field of view to prevent occlusion and collision with the surgical field (imaging configuration), or the camera the camera is positioned at a 4cm distance behind the measurement position so the suture tool can be advanced into the surgical field and execute the surgical plan without collision with the imaging system (suturing configuration).
High-level Control Strategy
A detailed view of the overall control strategy is shown in Fig. S1 which highlights the interaction of the low-level robot motion control with the higher-level control blocks (such as suture planner, operator, and suturing logic). The overall control strategy is based on a feedback control loop with components that include internal feedback mechanisms. In this figure, the first and the second time-derivatives of the variables are shown with the notation and vär respectively. In this control loop, real-time video frames from the 3D endoscope and NIR cameras are collected and processed via a raytracing technique that were developed in (39) to track the 3D motion of the tissue (denoted by xx ∈ R6 for translations and orientations). A tissue motion tracking algorithm (detailed and tested in the Results Section) tracks the position of target tissue via the NIR fluorescent markers in the NIR view of the camera, and autonomously detects when the tissue is stationary. At this point, the motion tracker triggers the imaging system to reconstruct the surgical field based on the image of the stationary scene to produce tissue position estimations (denoted by . A path planning algorithm, developed in our previous works (44, 45), generates multiple suture plan options on the 3D point cloud of the tissue which can be selected by the user from a GUI. This is denoted by xp1 ∈ R3n and xp2 ∈ R3m, which represents the 3D positions of the suture points in plan 1 (with n points) and 2 (with m points), respectively. This method also projects the robot tool on each planned suture point and predicts the chance of tool collision with the tissue, and autonomously generates a new suture plan if the original image is noisy. Once STAR generates a usable suture plan that is selected by the operator (i.e., the approved plan xpa from the two available options), reference waypoints and real-time robot positions (x ∈ R6) are used in a high-level suture logic and task planner via the methods developed in (46). This planner generates the strategy for knots, running stitch indices, and produces motion primitives (such as target position and orientation for approaching the tissue, biting/firing the suture, waiting for the assistant, and tensioning briefly denoted as xs ∈ R6). These stepwise target positions are then sent to the low-level control block to guarantee that the desired position and orientation is followed according to the following details.
Low-level Control
The motion primitives xs are sent to a trajectory generator to obtain smooth time-based desired trajectories in the task-space (shown as xd ∈ R6 here) via Reflexxes Motion Libraries (47). Kinematics and Dynamics Library (KDL) of Open Robot Control Software (OROCOS) convert the task-space trajectories of the robot into to the joint-space trajectories (48) (denoted as qd ∈ R9 since it includes 7 DoF of the KUKA robot in addition to 1 DoF for the tool pitch angle and 1 DoF for the linear stage that carries the camera system). A remote center of motion (RCM) constraint is also implemented in the kinematic solver (i.e., forward and inverse kinematics functions f and f−1 respectively) at the site of the laparoscopic tool port. Finally, closed-loop controllers enabled via KUKA robot controllers in IIWA stack (49), OROCOS Real-time toolkit (RTT) (50), and Fast Research Interface (FRI) (51) (here shown as ‘Robot Driver’) guarantee the joint-space trajectories control of the robots and the mounted suture tool and linear stage. These drivers follow the standard closed-loop control strategies such as in (52), based on the robot dynamics (i.e., inertia H, Coriolis and viscous damping effects c and external disturbances fext to produce the joint torque inputs τ ∈ R9 (implemented via robot motors) to follow the desired trajectories.
Suture planning
The suture planning and tissue deformation tracking logic is shown in Fig. S4. This method allows interactions with the operator for approving a suture plan or initiating a (re)planning step. The workflow is according to the following. The breathing motion tracker tracks the motion of tissue xt at the measure mode/distance. When a plan update command is issued by the operator, the motion tracker detects the end of the breathing motion via a sequence of NIR image frames and triggers a 3D point cloud collection from the target tissue . Collecting the point cloud when the tissue is not moving is essential to remove blurry and noisy data that is caused by the motion of the tissue during the fringe projection process. The projected NIR marker positions on the point cloud (via raytracing) are used for determining the start and end points of the suturing path. For the backwall suturing, the NIR marker order is detected automatically via blob tracking techniques such that the top marker is identified first, followed by the left marker, and then the right marker. For the front wall, the order is left marker followed by right marker. The suture path planning method then plans suture points in between the NIR markers in the sequence mentioned above with 3 mm spacings and an optional extra corner stitch for preventing leaks at the corners. For each of the planned suture points (xp1 and xp2), the system autonomously detects potential collisions of the suture tool with the target tissue by projecting the geometry of suture tool with a fixed bounding box onto the point cloud data from the camera. If the ratio between the number of points predicted to collide with the tool and the number of points projected to correctly fit inside the tool jaw exceeds an 80% threshold, the system warns the operator that suture misplacement or tool collision is possible, and the operator can select to generate a new suture plan.
Tissue Deformation and Motion Tracking
Once a suture plan is selected by the operator xpa, the robot saves the plan and autonomously switches to the suture mode. At the end of the next breathing cycle, a first snapshot of the NIR view is recorded (i.e., ) and the suturing process starts. Once a suture has been placed by STAR, the system waits until the end of the next breathing cycle and the camera system captures a new image via the NIR camera (i.e., if the kth suture from the planned path is complete). The new image is then compared to the initial image, and if the norm distance of any marker exceeds 3 mm (i.e., , where τh is the 3 mm decision threshold here), STAR suggests to the operator that a new suture plan should be generated. A new suture plan is recommended in this instance since a tissue deformation greater than 3 mm exceeds more than half of the tool jaw resulting in suture positioning errors if the previous plan is used. If the detected motion between images is less than 3mm, a message is shown to the operator indicating that the previous plan is still usable, and the operator can continue to use the existing plan (i.e., xpak + 1 = xpa. Finally, the motion of the robot is synchronized with the breathing motion such that the suture tool reaches the target tissue at the stationary point of the breathing cycle. The synchronization is achieved by calculating a trigger based on the current time t, distance to target d, the average robot velocity v, and the breathing cycle duration T. For the robot to reach the target at the nth breathing cycle, the trigger time can be calculated according to , where n is . At this point in time, the robot tool starts moving and reaches the target point after seconds. When the robot reaches the target suture point at the end of breathing cycle, the target tissue is enclosed within the tool jaw and a needle fire command is issued at the end of the next breathing cycle. The tissue stays stationary for 1.143 seconds at the end of the breathing cycle. The needle firing mechanism takes 0.5 seconds to insert the needle through the tissue which is considerably less than the 1.143 seconds long stationary period.
Surgical methodology
All animal testing adhered to the NIH Guide for the Use and Care of Animals and was performed under Institutional Animal Care and Use Committee Approval (Protocol 30759) at Children’s National Hospital (CNH) in Washington, DC, USA. The anesthetized animal was positioned supine, and the abdominal/surgical site was prepped with alcohol, followed by chlorhexidine scrub and sterile drapes. On the abdomen, four trocars (Ethicon, Somerville, NJ) were placed laterally, and a gel port (Applied Medical, Rancho Santa Margarita, CA) was inserted medially (e.g., as shown in Fig. S3B). The trocars provided sterile access to the peritoneal space for STAR’s suture tool and the assistant’s grasper, whereas the gel port provided sterile entry for the laparoscopic camera. STAR’s suture tool and imaging system were disinfected using MetriCide 28 (Metrex, Orange, CA). Using insufflation and laparoscopic technique, a surgeon identified and transected a loop of small bowel. The two open ends of the intestine were reapproximated and suspended with transabdominal stay sutures (e.g., as shown in Fig. S3C). Near Infrared (NIR) markers (indocyanine green and Permabond) that can be visualized by STAR’s laparoscopic camera were placed on the corners of the tissue, and robotic suturing was performed. After the anastomosis was completed, the abdominal wall was closed in a multi-layer fashion. The total procedure time was approximately 4 hours. See supplementary methods for the preoperative setup and the postoperative care.
Statistical analysis
GraphPad Prism 7.04 statistical software was used for all analysis in this study. Suture spacing and bite depth for each modality were subjected to the D’Agostino & Pearson test for normality and then compared using the non-parametric Mann-Whitney comparison test. Mann-Whitney tests are also used to compare the hesitancy per stitch, and time per stitch between STAR and the surgical modalities. Levene’s test for variance was performed on total suture times, followed by Welch’s t-test assuming unequal variance to compare STAR and LAP total suture times, and the unpaired t-test assuming equal variance to compare STAR and RAS total suture times. Single comparisons are chosen for all comparison tests because our hypothesis only considers relationships between STAR and each of the surgical techniques individually. Variation in suture spacing and bite depth for each test modality is normalized using the sample mean and reported as the coefficient of variation, with statistical differences calculated as in (53). A Kruskal-Wallis test for analysis of variance followed by Dunnett’s tests for multiple comparisons was performed for phantom motion tracking accuracy, and Mann-Whitney comparison tests were performed for in vivo tracking accuracy. Unpaired t-tests are used for comparing PMN counts. P-values are reported for all comparison tests, with P < 0.05 considered to be statistically significant.
Supplementary Material
Movie S1. Enhanced autonomous intestinal anastomosis.
Materials and Methods
Fig. S1. Detailed block diagram of the overall control strategy.
Fig. S2. Ex-vivo experiment testbeds.
Fig. S3. Examples of suture metrics and animal procedures.
Fig. S4. Suture (re)planning workflow and deformation tracking.
Table S1. Quantitative geometry quality of ex vivo end-to-end anastomosis.
Acknowledgments:
The authors would like to thank Mr. Jiawei Ge and Vincent Cleveland for their help with material preparation and analyzing the flow data in iTFlow software. We would also like to thank Dr. Yun Chen, Mr. Matthew Pittman, and Junjie Chen for their help with digitizing histology slide. Finally, we would like to thank the surgeons from the George Washington University Hospital, D. C., USA and Children’s National Hospital, D.C., USA, who participated in the study.
Funding:
Research reported in this article was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under award numbers 1R01EB020610, and R21EB024707. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests: Authors declare that they have no competing interests.
Data and materials availability:
All data are available in the main text or the supplementary materials.
References and Notes
- 1.Liow MHL, Chin PL, Pang HN, Tay DK-J, Yeo S-J, THINK surgical TSolution-One®(Robodoc) total knee arthroplasty, SICOT-J 3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rose PT, Nusbaum B, Robotic hair restoration, Dermatologic clinics 32, 97–107 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Perry TS, Profile: veebot [Resources_Start-ups], IEEE Spectrum 50, 23–23 (2013). [Google Scholar]
- 4.Kilby W, Dooley JR, Kuduvalli G, Sayeh S, Maurer CR Jr, The CyberKnife® robotic radiosurgery system in 2010, Technology in cancer research & treatment 9, 433–452 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Haidegger T, Autonomy for Surgical Robots: Concepts and Paradigms, IEEE Transactions on Medical Robotics and Bionics 1, 65–76 (2019). [Google Scholar]
- 6.Connor MJ, Dasgupta P, Ahmed HU, Raza A, Autonomous surgery in the era of robotic urology: friend or foe of the future surgeon?, Nature Reviews Urology 17, 643–649 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Weiser TG, Regenbogen SE, Thompson KD, Haynes AB, Lipsitz SR, Berry WR, Gawande AA, An estimation of the global volume of surgery: a modelling strategy based on available data, The Lancet 372, 139–144 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Patel SS, Patel MS, Mahanti S, Ortega A, Ault GT, Kaiser AM, Senagore AJ, Laparoscopic versus open colon resections in California: a cross-sectional analysis, The American Surgeon 78, 1063–1065 (2012). [PubMed] [Google Scholar]
- 9.Chapron C, Querleu D, Bruhat M-A, Madelenat P, Fernandez H, Pierre F, Dubuisson J-B, Surgical complications of diagnostic and operative gynaecological laparoscopy: a series of 29,966 cases., Human reproduction (Oxford, England) 13, 867–872 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Tsui C, Klein R, Garabrant M, Minimally invasive surgery: national trends in adoption and future directions for hospital strategy, Surgical endoscopy 27, 2253–2257 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Kang CY, Chaudhry OO, Halabi WJ, Nguyen V, Carmichael JC, Stamos MJ, Mills S, Outcomes of laparoscopic colorectal surgery: data from the Nationwide Inpatient Sample 2009, The American journal of surgery 204, 952–957 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Neville RF, Elkins CJ, Alley MT, Wicker RB, Hemodynamic comparison of differing anastomotic geometries using magnetic resonance velocimetry, Journal of Surgical Research 169, 311–318 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Williams CP, Rosen MJ, Jin J, McGee MF, Schomisch SJ, Ponsky J, Objective analysis of the accuracy and efficacy of a novel fascial closure device, Surgical innovation 15, 307–311 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Waseda M, Inaki N, Bermudez JT, Manukyan G, Gacek IA, Schurr MO, Braun M, Buess GF, Precision in stitches: Radius surgical system, Surgical endoscopy 21, 2056–2062 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Manilich E, Vogel JD, Kiran RP, Church JM, Seyidova-Khoshknabi D, Remzi FH, Key factors associated with postoperative complications in patients undergoing colorectal surgery, Diseases of the colon & rectum 56, 64–71 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Vignali A, Gianotti L, Braga M, Radaelli G, Malvezzi L, Di Carlo V, Altered microperfusion at the rectal stump is predictive for rectal anastomotic leak, Diseases of the colon & rectum 43, 76–82 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Oh PJ, Cheng N, Shah A, Montez J, Jarc A, Guo L, Gill IS, Hung AJ, Use of automated performance metrics to measure surgeon performance during robotic vesicourethral anastomosis and methodical development of a training tutorial, The Journal of urology 200, 895–902 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Yip M, Das N, in The Encyclopedia of MEDICAL ROBOTICS: Volume 1 Minimally Invasive Surgical Robotics, (World Scientific, 2019), pp. 281–313. [Google Scholar]
- 19.Yang G-Z, Cambias J, Cleary K, Daimler E, Drake J, Dupont PE, Hata N, Kazanzides P, Martel S, Patel RV, Medical robotics—Regulatory, ethical, and legal considerations for increasing levels of autonomy, Science Robotics 2, 8638 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Rosenberg LB, in Virtual Reality Annual International Symposium, 1993., 1993 IEEE, (IEEE, 1993), pp. 76–82. [Google Scholar]
- 21.Marinho MM, Harada K, Morita A, Mitsuishi M, SmartArm: Integration and validation of a versatile surgical robotic system for constrained workspaces, The International Journal of Medical Robotics and Computer Assisted Surgery 16, e2053 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Marinho MM, Harada K, Deie K, Ishimaru T, Mitsuishi M, SmartArm: Suturing Feasibility of a Surgical Robotic System on a Neonatal Chest Model, arXiv preprint arXiv:2101.00741 (2021). [Google Scholar]
- 23.Song C, Ma X, Xia X, Chiu PWY, Chong CCN, Li Z, A robotic flexible endoscope with shared autonomy: a study of mockup cholecystectomy, Surg Endosc 34, 2730–2741 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Shademan A, Decker RS, Opfermann JD, Leonard S, Krieger A, Kim PCW, Supervised autonomous robotic soft tissue surgery, Science Translational Medicine 8, 337ra64–337ra64 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Chen TP, Pickett M, Dehghani H, Buharin V, Stolyarov R, DeMaio E, Oberlin J, Chen T, O’Shea L, Galeotti J, Calef T, Kim PC, Demonstration of Fully Autonomous, Endoscopic Robot-Assisted Closure of Ventral Hernia using Smart Tissue Autonomous Robot (STAR) in preclinical Porcine Models (SAGES Emerging Technology Session, 2020). [Google Scholar]
- 26.Hu D, Gong Y, Seibel EJ, Sekhar LN, Hannaford B, Semi-autonomous image-guided brain tumour resection using an integrated robotic system: A bench-top study, Int J Med Robot 14 (2018), doi: 10.1002/rcs.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murali A, Sen S, Kehoe B, Garg A, McFarland S, Patil S, Boyd WD, Lim S, Abbeel P, Goldberg K, Learning by observation for surgical subtasks: Multilateral cutting of 3d viscoelastic and 2d orthotropic tissue phantoms, in 2015 IEEE International Conference on Robotics and Automation (ICRA), (IEEE, 2015), pp. 1202–1209. [Google Scholar]
- 28.Richter F, Shen S, Liu F, Huang J, Funk EK, Orosco RK, Yip MC, Autonomous robotic suction to clear the surgical field for hemostasis using image-based blood flow detection, IEEE Robotics and Automation Letters 6, 1383–1390 (2021). [Google Scholar]
- 29.Martin JW, Scaglioni B, Norton JC, Subramanian V, Arezzo A, Obstein KL, Valdastri P, Enabling the future of colonoscopy with intelligent and autonomous magnetic manipulation, Nat Mach Intell 2, 595–606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijsman PJM, Broeders IAMJ, Brenkman HJ, Szold A, Forgione A, Schreuder HWR, Consten ECJ, Draaisma WA, Verheijen PM, Ruurda JP, Kaufman Y, First experience with THE AUTOLAPTM SYSTEM: an image-based robotic camera steering device, Surg Endosc 32, 2560–2566 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Tanwani AK, Sermanet P, Yan A, Anand R, Phielipp M, Goldberg K, Motion2vec: Semi-supervised representation learning from surgical videos, in 2020 IEEE International Conference on Robotics and Automation (ICRA), (IEEE, 2020), pp. 2174–2181. [Google Scholar]
- 32.Luongo F, Hakim R, Nguyen JH, Anandkumar A, Hung AJ, Deep learning-based computer vision to recognize and classify suturing gestures in robot-assisted surgery, Surgery (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedram SA, Shin C, Ferguson PW, Ma J, Dutson EP, Rosen J, Autonomous Suturing Framework and Quantification Using a Cable-Driven Surgical Robot, IEEE Transactions on Robotics (2020). [Google Scholar]
- 34.Varier VM, Rajamani DK, Goldfarb N, Tavakkolmoghaddam F, Munawar A, Fischer GS,Collaborative Suturing: A Reinforcement Learning Approach to Automate Hand-off Task in Suturing for Surgical Robots, in 2020 29th IEEE International Conference on Robot and Human Interactive Communication (RO-MAN), (IEEE, 2020), pp. 1380–1386. [Google Scholar]
- 35.Attanasio A, Scaglioni B, De Momi E, Fiorini P, Valdastri P, Autonomy in surgical robotics, Annual Review of Control, Robotics, and Autonomous Systems 4, 651–679 (2021). [Google Scholar]
- 36.Marecik SJ, Chaudhry V, Jan A, Pearl RK, Park JJ, Prasad LM, A comparison of robotic, laparoscopic, and hand-sewn intestinal sutured anastomoses performed by residents, The American journal of surgery 193, 349–355 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Leonard S, Wu KL, Kim Y, Krieger A, Kim PC, Smart tissue anastomosis robot (STAR): A vision-guided robotics system for laparoscopic suturing, IEEE Transactions on Biomedical Engineering 61, 1305–1317 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Le HN, Nguyen H, Wang Z, Opfermann J, Leonard S, Krieger A, Kang JU, Demonstration of a laparoscopic structured-illumination three-dimensional imaging system for guiding reconstructive bowel anastomosis, Journal of biomedical optics 23, 056009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Decker RS, Shademan A, Opfermann JD, Leonard S, Kim PC, Krieger A, Biocompatible near-infrared three-dimensional tracking system, IEEE Transactions on Biomedical Engineering 64, 549–556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronneberger O, Fischer P, Brox T, U-net: Convolutional networks for biomedical image segmentation, in International Conference on Medical image computing and computer-assisted intervention, (Springer, 2015), pp. 234–241. [Google Scholar]
- 41.Long J, Shelhamer E, Darrell T, Fully convolutional networks for semantic segmentation, in Proceedings of the IEEE conference on computer vision and pattern recognition, (2015), pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Bo L, Fuxin L, Adaptive wing loss for robust face alignment via heatmap regression, in Proceedings of the IEEE/CVF International Conference on Computer Vision, (2019), pp. 6971–6981. [Google Scholar]
- 43.Yu Y, Si X, Hu C, Zhang J, A review of recurrent neural networks: LSTM cells and network architectures, Neural computation 31, 1235–1270 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Kam M, Saeidi H, Wei S, Opfermann JD, Leonard S, Hsieh MH, Kang JU, Krieger A, Semi-autonomous Robotic Anastomoses of Vaginal Cuffs Using Marker Enhanced 3D Imaging and Path Planning, in Medical Image Computing and Computer Assisted Intervention – MICCAI 2019, Lecture Notes in Computer Science. Shen D, Liu T, Peters TM, Staib LH, Essert C, Zhou S, Yap P-T, Khan A, Eds. (Springer International Publishing, Cham, 2019), pp. 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saeidi H, Ge J, Kam M, Opfermann JD, Leonard S, Joshi AS, Krieger A, Supervised Autonomous Electrosurgery via Biocompatible Near-Infrared Tissue Tracking Techniques, IEEE Transactions on Medical Robotics and Bionics 1, 228–236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saeidi H, Le HND, Opfermann JD, Leonard S, Kim A, Hsieh MH, Kang JU, Krieger A, Autonomous laparoscopic robotic suturing with a novel actuated suturing tool and 3D endoscope, in 2019 International Conference on Robotics and Automation (ICRA), (2019), pp. 1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kröger T, Opening the door to new sensor-based robot applications—The Reflexxes Motion Libraries, in 2011 IEEE International Conference on Robotics and Automation, (2011), pp. 1–4. [Google Scholar]
- 48.Smits R, Bruyninckx H, Aertbeliën E, KDL: Kinematics and dynamics library (2011). [Google Scholar]
- 49.Hennersperger C, Fuerst B, Virga S, Zettinig O, Frisch B, Neff T, Navab N, Towards MRI-Based Autonomous Robotic US Acquisitions: A First Feasibility Study, IEEE Transactions on Medical Imaging 36, 538–548 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Bruyninckx H, Soetens P, Koninckx B, The real-time motion control core of the Orocos project, in 2003 IEEE international conference on robotics and automation (Cat. No. 03CH37422), (IEEE, 2003), vol. 2, pp. 2766–2771. [Google Scholar]
- 51.Schreiber G, Stemmer A, Bischoff R, the fast research interface for the kuka lightweight robot, in IEEE Workshop on Innovative Robot Control Architectures for Demanding (Research) Applications How to Modify and Enhance Commercial Controllers (ICRA 2010), (Citeseer, 2010), pp. 15–21. [Google Scholar]
- 52.Spong MW, Hutchinson S, Vidyasagar M, Robot modeling and control (John Wiley & Sons, 2020). [Google Scholar]
- 53.Forkman J, Estimator and tests for common coefficients of variation in normal distributions, Communications in Statistics—Theory and Methods 38, 233–251 (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Enhanced autonomous intestinal anastomosis.
Materials and Methods
Fig. S1. Detailed block diagram of the overall control strategy.
Fig. S2. Ex-vivo experiment testbeds.
Fig. S3. Examples of suture metrics and animal procedures.
Fig. S4. Suture (re)planning workflow and deformation tracking.
Table S1. Quantitative geometry quality of ex vivo end-to-end anastomosis.
Data Availability Statement
All data are available in the main text or the supplementary materials.