Chen et al. uncovered the transcriptional, developmental, and functional heterogeneity of ILC1s and demonstrated the dynamic changes that occur in ILC1 subsets from different origins during ontogeny, which correspond to their different functional roles, facilitating adaptation to age-related immune demands.
Abstract
Type 1 innate lymphoid cells (ILC1s) represent the predominant population of liver ILCs and function as important effectors and regulators of immune responses, but the cellular heterogeneity of ILC1s is not fully understood. Here, single-cell RNA sequencing and flow cytometric analysis demonstrated that liver ILC1s could be dissected into Ly49E+ and Ly49E− populations with unique transcriptional and phenotypic features. Genetic fate-mapping analysis revealed that liver Ly49E+ ILC1s with strong cytotoxicity originated from embryonic non–bone marrow hematopoietic progenitor cells (HPCs), persisted locally during postnatal life, and mediated protective immunity against cytomegalovirus infection in newborn mice. However, Ly49E− ILC1s developed from BM and extramedullary HPCs after birth, gradually replaced Ly49E+ ILC1s in the livers with age, and contained the memory subset in recall response to hapten challenge. Thus, our study shows that Ly49E dissects liver ILC1s into two unique subpopulations, with distinct origins and a bias toward neonatal innate or adult immune memory responses.
Graphical Abstract
Introduction
Innate lymphoid cells (ILCs) can be categorized into groups 1, 2, and 3 based on differences in transcription factor expression and function (Spits et al., 2013; Vivier et al., 2018). Group 1 ILCs comprise conventional natural killer (cNK) cells and ILC1s (Riggan et al., 2019) and are characterized by the expression of pan-NK cell markers, such as NK1.1 and NKp46, and T-box expressed in T cells (T-bet) in mice (Weizman et al., 2017). Despite belonging to the same group, cNK cells and ILC1s differ in many aspects. In mice, ILC1s can be distinguished from cNK cells by cell surface marker expression, including CD49a, CXCR6, and CD200R1, and the lack of CD49b and Eomesodermin (Eomes) expression (Daussy et al., 2014; Gordon et al., 2012; Peng et al., 2013; Sojka et al., 2014; Weizman et al., 2017). Whereas cNK cells are circulating and widely distributed, ILC1s are tissue resident, with dramatic differences in frequency across tissues (O’Sullivan et al., 2016; Peng et al., 2013). Notably, liver ILC1s (also known as liver-resident NK cells) account for a relatively large proportion of all liver ILCs (Robinette et al., 2015), making the liver an important window for ILC1 study. Previous studies on liver ILC1s have shown their functional diversity and specificity, including immune memory potential (Paust et al., 2010; Peng et al., 2013; Weizman et al., 2019), immune regulatory effects (Zhou et al., 2019), and antiviral ability (Weizman et al., 2017). Whether this functional diversity and specificity reflect heterogeneity of liver ILC1s remains unclear.
Immune cells with different developmental origins might have discrete functions in health and disease. Studies on macrophages provide a typical example (Bajpai et al., 2018; Lavine et al., 2014). Divergent origins within CD8+ T cells (Smith et al., 2018) and ILC2s (Schneider et al., 2019) have also been recently reported. While ILC1s initially emerge in the fetal liver and exhibit a dynamically changed pool with age (Takeda et al., 2005), whether ILC1s with specific phenotype and discrete functions have different origins has not been defined.
In early life, adaptive immune responses are limited, and the innate immune system may play a larger role than during adulthood. Recent studies have revealed the accumulation of myeloid-derived suppressor cells (MDSCs; He et al., 2018) or γδT cells (Guo et al., 2018) in newborn mice, which play specific roles in both homeostatic maintenance and host defense. Interestingly, ILC1s are predominant over cNK cells during the embryonic and neonatal periods (Constantinides et al., 2015; Takeda et al., 2005). However, the roles of neonatal liver ILC1s during early life remain unexplored.
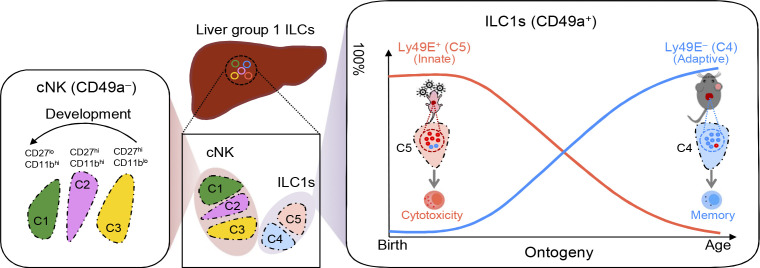
In this study, we found that murine liver ILC1s could be further subdivided into two subsets, which are distinguished by Ly49E expression. Ly49E+ ILC1s were predominantly present in the neonatal mouse liver but gradually decreased with age, whereas Ly49E− ILC1s showed the opposite trend. These two subsets also differ in hematopoietic origins and functionality. Specifically, Ly49E+ ILC1s arose from embryonic hematopoiesis and persisted in the liver after birth, whereas Ly49E− ILC1s were primarily derived from postnatal hematopoiesis. Compared with Ly49E− ILC1s, Ly49E+ ILC1s exhibited more cytotoxic potential and played an essential role in protecting newborn mice against viral infection, whereas adult Ly49E− but not Ly49E+ ILC1s displayed immune memory potential in the contact hypersensitivity (CHS) model. Thus, our findings showed that liver ILC1s are heterogeneously composed of at least two subpopulations with different hematopoiesis waves and distinct functional characteristics.
Results
Single-cell RNA sequencing (scRNA-seq) reveals transcriptional heterogeneity among liver group 1 ILCs
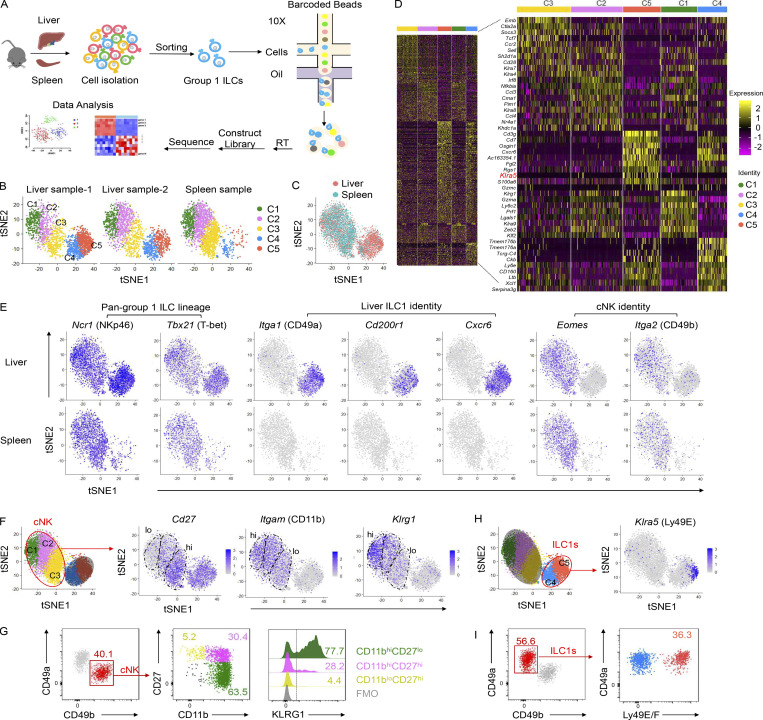
Group 1 ILCs comprise cNK cells and ILC1s. To delineate their heterogeneity, we sorted NK1.1+NKp46+ group 1 ILCs from the liver and spleen of adult C57BL/6 (B6) mice to perform scRNA-seq (Fig. 1 A). Approximately 2,000 hepatic or splenic cells were captured and passed quality control, with an average of 1,400 genes detected in each individual cell. Group 1 ILCs could be grouped into five main clusters (C1–C5) in two independent liver samples, whereas the splenic group 1 ILCs consisted predominantly of C1–C3, with a low frequency of C4 and the rare detection of C5 (Fig. 1, B and C). A comparison of the top 200 most variable genes in each cluster confirmed that each cluster was distinct from the others (Fig. 1 D). By analyzing the characteristic markers of cNK cells and ILC1s (Daussy et al., 2014; Paust et al., 2010; Tang et al., 2016; Weizman et al., 2017), we found that all clusters uniformly expressed the pan-group 1 ILC lineage markers Ncr1 (NKp46) and Tbx21 (T-bet; Fig. 1 E). C1–C3 expressed Eomes (Eomes) and Itga2 (CD49b; Fig. 1 E) and, therefore, might represent different subtypes of cNK cells. Based on expression of maturation-associated molecules (Castro et al., 2018; Chiossone et al., 2009), such as Itgam (CD11b), Cd27 (CD27), and Klrg1 (KLRG1), C1–C3 clusters likely represent the terminally mature stage (CD11bhiCD27loKLRG1+), intermediate stage (CD11bhiCD27hi), and immature stage (CD11bloCD27lo) of cNK cells, respectively (Fig. 1, F and G).
Figure 1.
Dissection and clustering of murine liver group 1 ILCs at steady state. (A) Overview of the study design. Group 1 ILCs (CD45+NK1.1+NKp46+CD3−CD19−) from the livers and spleens of naive WT C57BL/6 (B6) mice (6–8 wk old) were sorted for scRNA-seq. (B and C) t-SNE clustering plots of each hepatic and splenic group 1 ILC sample (B) and an overlay (C). Each point represents an individual cell. Hepatic and splenic group 1 ILCs could be grouped into five distinct clusters (C1–C5). Data were obtained from two independent experiments (liver) or one experiment (spleen). (D) Heatmap showing the expression levels of the top 200 genes (left) and top 10 genes (right) expressed in each liver group 1 ILC cluster (C1–C5). (E) Distribution of signature gene expression by the liver (top) and spleen (bottom) group 1 ILC clusters in t-SNE plots. (F–I) Distribution of Cd27, Itgam, Klrg1 (F), and Klra5 (H) expression in the t-SNE plot. Representative plots of CD27, CD11b, and KLRG1 expression on cNK cells (CD45+NK1.1+CD49b+CD49a−CD3−CD19−; G) and Ly49E/F expression on liver ILC1s (CD45+NK1.1+CD49a+CD49b−CD3−CD19−; I) of WT B6 mice (6–8 wk old). Fluorescence minus one (FMO) controls were gated on total cNK cells. Flow cytometric plots are representative of at least three independent experiments with n = 3–9 mice per experiment.
As shown in Fig. 1 E, Itga1 (CD49a), Cd200r1 (CD200R1), and Cxcr6 (CXCR6), which are the typical signature molecules associated with ILC1s, were enriched in the liver but rarely detected in the spleen and were detected only in C4 and C5 but not C1–C3, indicating that liver ILC1s represent a population distinct from cNK cells. By comparing the top 10 most variable genes in each cluster (Fig. 1 D), we found that Klra5 (Fig. 1 H), which encodes Ly49E, was specifically but not homogeneously expressed in C5. At the protein level, Ly49E/F was further confirmed to be expressed by a proportion of liver ILC1s (Fig. 1 I), likely representing C5 from the t-distributed stochastic neighbor embedding (t-SNE) map.
Ly49E expression dissects liver ILC1s into two distinct subsets
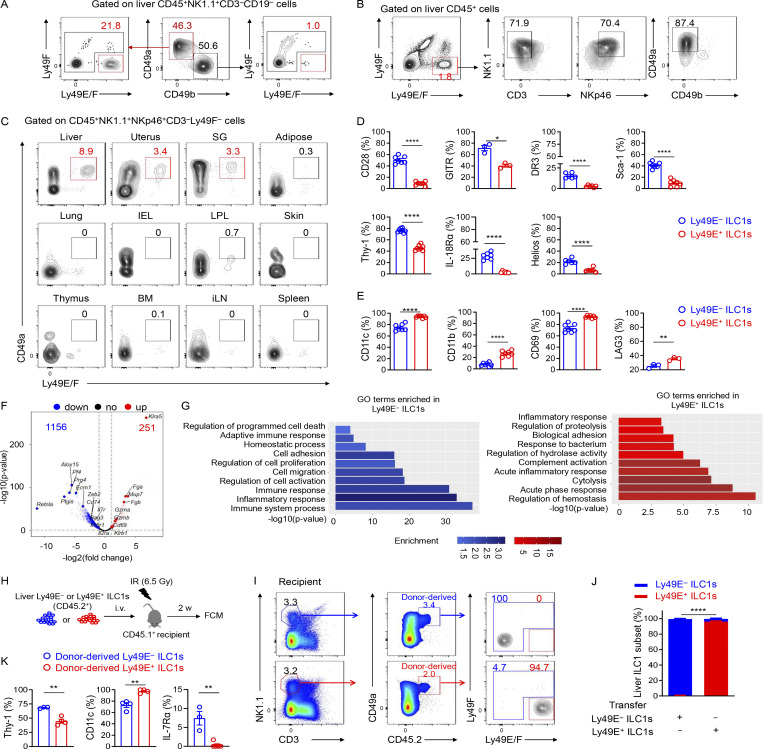
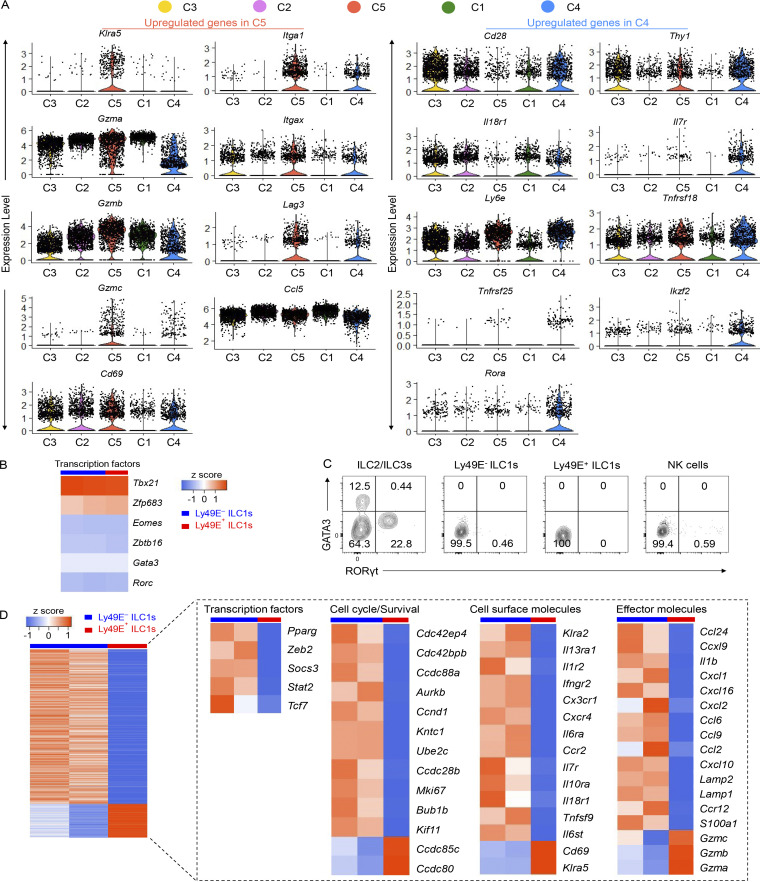
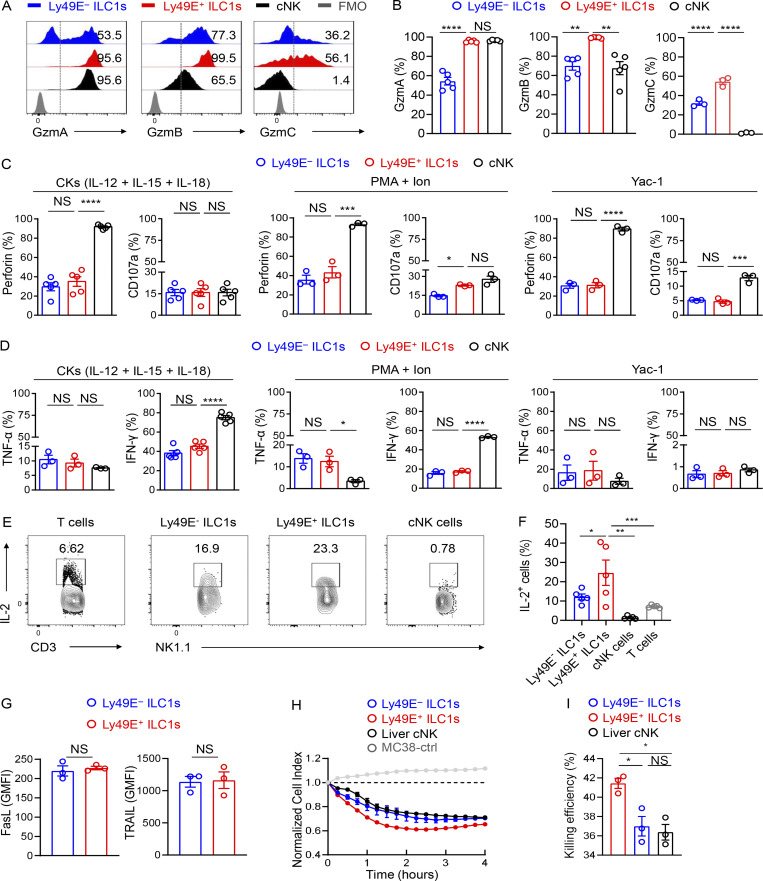
By using anti-Ly49E/F and anti-Ly49F mAbs to further examine Ly49E expression in the liver (no monospecific Ly49E mAb was available), we found that ∼20% of liver ILC1s expressed Ly49E, whereas Ly49E was rarely expressed on cNK cells (Fig. 2 A). Interestingly, gating Ly49E+ cells in liver leukocytes revealed that Ly49E was predominantly expressed by liver ILC1s rather than by other leukocytes (Fig. 2 B). Additionally, Ly49E expression by CD49a+NK1.1+NKp46+ ILC1s from multiple lymphoid and nonlymphoid tissues was undetectable or at much lower levels than liver ILC1s (Fig. 2 C). Further analysis revealed that Ly49E+ ILC1s expressed lower levels of Tnfrsf25 (DR3), Ly6e (Sca-1), Ikzf2 (Helios), Thy1 (Thy-1), Il18r1 (IL-18Rα), and costimulatory molecules, including Cd28 (CD28) and Tnfrsf18 (GITR), but higher levels of Itgax (CD11c), Itgam (CD11b), Cd69 (CD69), and Lag3 (LAG3) at both mRNA and protein levels, compared with Ly49E− ILC1s (Fig. S1 A and Fig. 2, D and E). Ly49E+ and Ly49E− ILC1s rarely expressed Gata3 (GATA3) and Rorc (RORγt), but highly expressed Tbx21 (T-bet) and Zfp683 (Hobit; Fig. S1, B and C), thus ruling out contamination of ILC2s and ILC3s. Further analysis of liver Ly49E+ and Ly49E− ILC1s using bulk RNA-seq showed that a total of 1,407 genes were identified as being differentially expressed by fold-changes >2.0 (Fig. 2 F). The differentially expressed genes include those encoding transcription factors, cell cycle regulators, cell surface receptors, and effector molecules (Fig. S1 D). Although gene enrichment analysis indicated that these two subsets were both enriched in genes related to immune and inflammatory responses, Ly49E+ ILC1s were more strongly enriched in genes associated with the acute immune response and cytolysis than Ly49E− ILC1s (Fig. 2 G), suggesting that Ly49E+ ILC1s might mediate an earlier immune response than Ly49E− ILC1s.
Figure 2.
Ly49E expression distinguishes two distinct clusters of liver ILC1s. (A) Representative plots of Ly49E expression on liver ILC1s (CD45+NK1.1+CD49a+CD49b−CD3−CD19−) and cNK cells (CD45+NK1.1+CD49b+CD49a−CD3−CD19−) of WT B6 mice (5–8 wk old). (B) Representative plots showing the expression of ILC1-lineage markers on Ly49E+CD45+ leukocytes from the livers of WT B6 mice (5–8 wk old). Data are representative of at least five independent experiments with n = 3–5 mice per experiment (A and B). (C) Representative plots of CD49a and Ly49E/F expression on CD45+NK1.1+NKp46+CD3−Ly49F− cells from the indicated tissues of WT B6 mice (5–8 wk old). Data are representative of at least three independent experiments with n = 1–4 mice per experiment. (D and E) Percentages of cells with higher expression levels of the indicated molecular signatures in liver Ly49E− ILC1s (D) or Ly49E+ ILC1s (E) of WT B6 mice (5–8 wk old). Data are pooled from one or two independent experiments with n = 3–4 mice. (F and G) Comparative transcriptome analysis between Ly49E+ or Ly49E− ILC1s from WT B6 mice (5–8 wk old) using bulk cell RNA-seq. Volcano plot (F) and GO Biological Processes (G) associated with the genes with >2.0-fold changes in expression are highlighted in red (higher in Ly49E+ ILC1s) or blue (higher in Ly49E− ILC1s). (H–K) 1–2 × 105 Ly49E+ or Ly49E− ILC1s were sorted from CD45.2+ WT mice (3–4 wk old) and adoptively transferred into sublethally irradiated CD45.1+ mice (6–8 wk old). 2 wk after transfer, CD45.2+ liver ILC1s in recipients were analyzed for Ly49E expression. (H) Schematic of the experimental design. (I) Representative plots showing Ly49E expression on donor-derived (CD45.2+) ILC1s in recipient liver. (J) Percentages of Ly49E+ or Ly49E− cells among host- or donor-derived liver ILC1s. Data are pooled from two (Ly49E+ ILC1s) or three (Ly49E− ILC1s) independent experiments with n = 7–9 mice (I and J). (K) Percentages of cells expressing the indicated molecular signatures among liver ILC1 subsets after 2 wk of adoptive transfer. Data are representative of at least two independent experiments with three to four recipient mice per group. Bar graphs show the mean ± SEM; an unpaired Student’s t test was used to compare experimental groups in D, E, J, and K; two-way ANOVA was used in I; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Figure S1.
Differentially expressed genes between two liver ILC1 subsets revealed by scRNA-seq and bulk RNA-seq. (A) Distribution of selected genes (upregulated in C4 or C5 cluster) within C1–C5 clusters from scRNA-seq analysis. (B) Heatmap showing the expression of selected ILC-featured transcription factors from bulk RNA-seq data of liver Ly49E+ ILC1s and Ly49E− ILC1s from WT B6 mice (5–8 wk old). (C) Representative plots of GATA3 and RORγt expression in ILC subsets of WT B6 mice (5–8 wk old). Flow cytometric plots are representative of two independent experiments with four to five mice per group. (D) Gene expression signature of the indicated cell populations revealed by bulk RNA-seq. Left heatmap shows total differential expressed genes with >2.0 fold-changes. Right heatmap shows genes that were manually selected from the left with different categories.
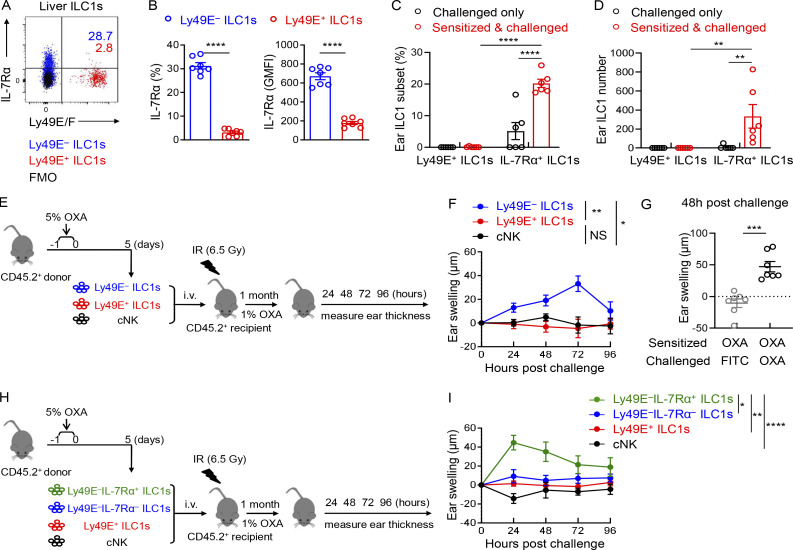
As shown in Fig. 2 F and Fig. S1, A and D, immune memory-associated molecules, such as Il18r1, Il7r, Il2ra, and Bag3 (Gillard et al., 2011; Kaech et al., 2003; Wang et al., 2018; Weizman et al., 2019), were expressed at higher levels by Ly49E− ILC1s. Interestingly, the expression of IL-7Rα (CD127), a key factor associated with ILC1 immune memory in CHS, required for long-term maintenance of liver memory ILC1s (Wang et al., 2018), was restricted to Ly49E− ILC1s at steady state (Fig. S2, A and B). After hapten sensitization and challenge, IL-7Rα+Ly49E− ILC1s but not Ly49E+ ILC1s accumulated in the inflamed ears (Fig. S2, C and D). Furthermore, only oxazolone (OXA)-sensitized Ly49E− ILC1s induced recipient ear increase swelling after the OXA challenge (Fig. S2, E and F), and this effect was antigen specific (Fig. S2 G). When OXA-sensitized liver ILC1s were devided into three populations based on Ly49E and IL-7Rα expression, only the ears of mice that received the sensitized IL-7Rα+Ly49E− ILC1s exhibited obvious swelling after challenge (Fig. S2, H and I). Altogether, these data showed that ILC1s with memory potential are enriched in the Ly49E− subset, based on their responses to the CHS model. However, the molecular basis for hapten recognition is not yet defined, and the receptors responsible for hapten recognition have not been identified.
Figure S2.
Adult Ly49E− ILC1s have the capacity to confer memory responses in the CHS model. (A) Representative dot plots showing Ly49E/F and IL-7Rα expression on liver ILC1 subsets from WT B6 mice (6–8 wk old). Data are representative of at least two independent experiments with three to four mice per experiment. (B) Percentages and GMFI of IL-7Rα expression among liver Ly49E+ or Ly49E− ILC1s from B6 mice (6–8 wk old). Data are pooled from two independent experiments with seven mice per experiment. (C and D) WT B6 mice (6–8 wk old) were abdominally sensitized with 5% OXA and challenged with 1% OXA on the ears 5 d later (sensitized and challenged). The control mice were only challenged with 1% OXA on the ears without prior abdominal sensitization (challenged only). 48 h after 1% OXA challenge, the ear cell suspensions of two groups were prepared for flow cytometric analysis. Statistical analysis showing the percentages (C) and absolute numbers (D) of Ly49E+ or Ly49E−IL-7Rα+ ILC1s in mouse ears. Ear ILC1s were gated as CD3−CD19−NK1.1+NKp46+CD49a+. Data were pooled from two independent experiments with n = 6 mice per group. (E and F) Sublethally irradiated WT B6 mice (6–8 wk old) received 5 × 104 Ly49E+ ILC1s, Ly49E− ILC1s, or liver cNK cells obtained from 5% OXA-sensitized WT B6 mice (4 wk old) and were challenged 1 mo later with 1% OXA on the right ears and solvent control on the left ears. Schematic showing the experimental design (E). Ear swelling of recipient mice was measured 24–96 h after challenge (F). Data were pooled from three independent experiments with n = 5–7 mice per group. (G) Sublethally irradiated WT B6 mice (6–8 wk old) received OXA-sensitized Ly49E− ILC1s and were challenged with 1% OXA on the right ears or 0.5% FITC on the left ears 1 mo later. Ear swelling of recipient mice was measured 48 h after challenge. Data were pooled from two independent experiments with n = 7 mice per group. (H and I) Sublethally irradiated WT B6 mice (6–8 wk old) received 5 × 104 Ly49E+ ILC1s, Ly49E−IL-7Rα− ILC1s, Ly49E−IL-7Rα+ ILC1s, or liver cNK cells from OXA-sensitized WT B6 mice (4 wk old) and were challenged 1 mo later with 1% OXA on the right ears or solvent control on the left ears. Schematic showing the experimental design (H). Ear swelling was measured 24–96 h after challenge of recipient mice (I). Data were pooled from four independent experiments with n = 6–14 mice per group. Bar graphs show the mean ± SEM; unpaired, two-tailed Student’s t test were used in B and G, two-way ANOVA in C, D, F, and I; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To explore whether liver Ly49E+ and Ly49E− ILC1s represent different maturation stages like cNK cells, we performed adoptive transfer experiments (Fig. 2 H). In line with previous reports (Filtjens et al., 2013), donor ILC1 subsets could be found in the recipient liver, and they maintained Ly49E-positive or -negative status after transfer (Fig. 2, I and J). Consistent with the differences between Ly49E+ and Ly49E− ILC1s at steady state (Fig. 2, D and E; and Fig. S2 A), flow cytometric analysis revealed that the transferred Ly49E+ ILC1s retained higher CD11c expression and lower expression of Thy-1 and IL-7Rα than transferred Ly49E− ILC1s (Fig. 2 K). These results suggest that liver Ly49E+ and Ly49E− ILC1s represent phenotypically stable subsets rather than developmental intermediates.
Ly49E+ and Ly49E− subsets originate from different hematopoietic waves
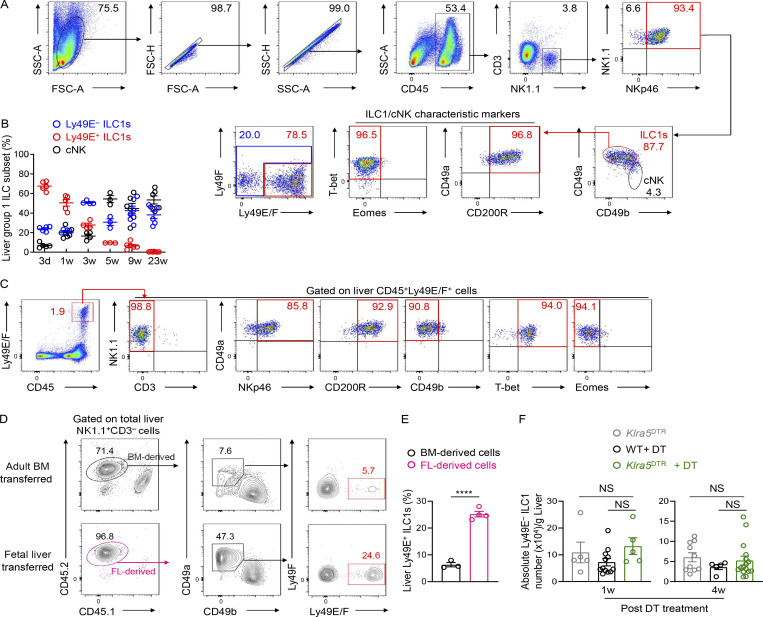
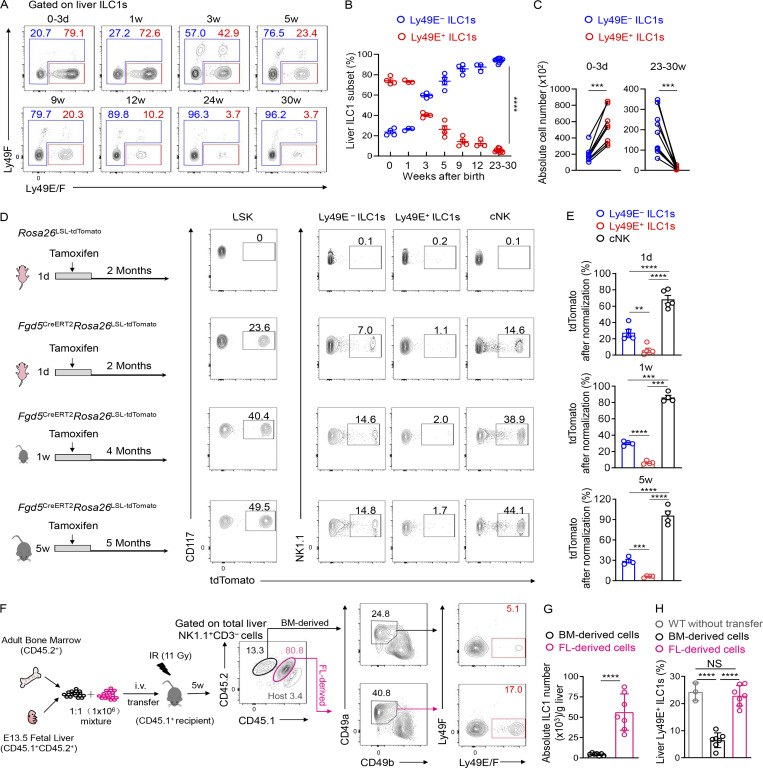
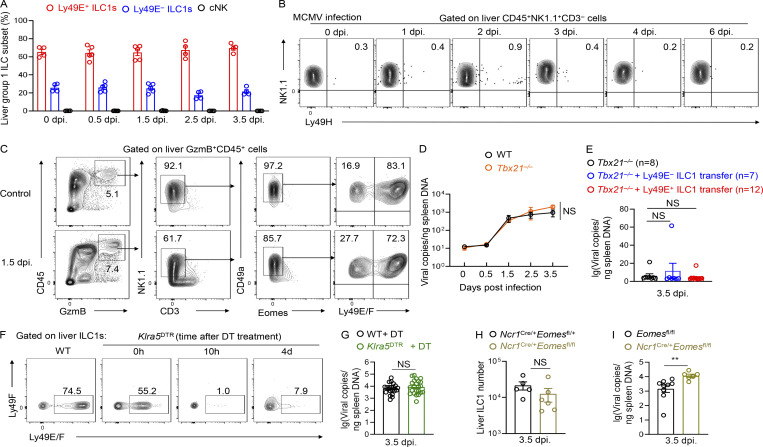
Studies have revealed that, during early life, a large fraction of liver group 1 ILCs express Ly49E and ILC1s predominate over cNK cells (Constantinides et al., 2015; Filtjens et al., 2013; Stevenaert et al., 2003; Takeda et al., 2005; Van Beneden et al., 2001). We consistently found that nearly 90% of group 1 ILCs in the neonatal liver expressed CD49a, T-bet, and CD200R1 and lacked Eomes expression, suggesting their ILC1 identity (Fig. S3 A). In contrast, CD49a−CD49b+ cNK cells accounted for <10% of the total group 1 ILCs in the neonatal liver (Fig. S3, A and B). Moreover, the majority (>70%) of liver ILC1s in neonatal mice were Ly49E-positive (Fig. 3, A and B). Neonatal ILC1s, rather than cNK cells or T cells, represented the predominant Ly49E-expressing leukocytes in the neonatal liver (Fig. S3 C). While the frequency and number of Ly49E+ ILC1s decreased with age, Ly49E− ILC1s showed an opposite trend (Fig. 3, A–C).
Figure S3.
The neonatal liver is enriched with Ly49E+ ILC1s that are rarely replenished after birth. (A) Representative plots showing the gating strategy and characteristics of neonatal liver group 1 ILCs (0–3 d old). (B) Percentages of Ly49E+ ILC1s, Ly49E− ILC1s, and cNK cells among liver group 1 ILCs from WT mice at different ages. Data for each time point are representative of three independent experiments with n = 3–6 mice. (C) Flow cytometric plots showing Ly49E expression among CD45+ leukocytes from neonatal liver (0–3 d old). Data are representative of at least three independent experiments. (D and E) Lethally irradiated recipient CD45.1+ mice received donor CD45.2+ BM cells (6–8 wk old) or CD45.2+ fetal liver cells (E13.5–15.5). 1 mo later, the livers of recipient mice were harvested for flow cytometric analysis of Ly49E expression on ILC1s (D). Percentages of BM- or fetal liver–derived Ly49E+ cells among donor-derived ILC1s (E). Data are representative of two independent experiments with n = 3 or 4 mice. (F) Absolute cell numbers of Ly49E− ILC1s in the liver of WT and Klra5DTR mice at 1 or 4 wk after intrahepatic (i.h.) treatment with DT once during the neonatal period (0–3 d old). Data were pooled from two or three independent experiments with n = 5–16 mice. Bar graphs show the mean ± SEM; unpaired Student’s t test was used in E, and one-way ANOVA followed by Dunnett’s multiple comparison test in F; ****, P < 0.0001.
Figure 3.
BM hematopoiesis contributes little to the Ly49E+ ILC1 pool. (A) Representative plots showing Ly49E expression on liver ILC1s from WT B6 mice at different ages. Data are representative of three independent experiments with n = 3–9 mice per condition. (B) Percentages of Ly49E+ or Ly49E− cells among liver ILC1s at different ages. Data for each time point are representative of three independent experiments with n = 3–9 mice. (C) Absolute cell numbers of ILC1 subsets per liver from WT B6 mice at ages 0–3 d or 23–30 wk. Data for 0–3 d are pooled from two independent experiments with n = 8 mice. Data for 23–30 wk are pooled from three independent experiments with n = 10 mice. (D and E) Fate-mapping analysis of neonatal (1 d and 1 wk) and adult (5 wk) Fgd5+ LSK cell–derived group 1 ILCs. Representative plots show tdTomato expression on BM LSK cells and liver group 1 ILC subsets from Fgd5CreERT2Rosa26tdTomato mice exposed to tamoxifen at different ages (D). Labeling efficiency of Ly49E− ILC1s, Ly49E+ ILC1s, and cNK cells normalized against LSK cells (E). Data are pooled from at least three independent experiments (1 d: n = 5; 1 wk: n = 4; 5 wk: n = 4). (F–H) Representative plots showing Ly49E expression on liver ILC1s from lethally irradiated recipient CD45.1+ mice that received the adoptive transfer of donor CD45.2+ BM (6–8 wk old) and CD45.1+CD45.2+ fetal liver (E13.5 fetal liver [FL]) MNCs (mixed at 1:1; F). Absolute cell numbers of donor BM- or FL-derived ILC1s in recipient livers (G). Percentages of donor BM- or FL-derived Ly49E+ cells among donor-derived ILC1s (H). Data are representative of at least two independent experiments with n = 3 or 7 mice per group. Bar graphs show the mean ± SEM; two-way ANOVA was used in B, paired Student’s t test in C, one-way ANOVA followed by Dunnett’s multiple comparison test in E and H, and unpaired Student’s t test in G; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To explore whether Ly49E+ and Ly49E− ILC1s originated from different hematopoietic waves, we established an inducible fate-mapping model (Fgd5CreERT2Rosa26tdTomato), in which Fgd5 is specifically expressed in hematopoietic stem cells (HSCs) but not in lineage-restricted progenitors and mature immune cells (Gazit et al., 2014; Sawen et al., 2018); thus, HSCs and their descendants could be labeled upon tamoxifen administration. Fgd5CreERT2Rosa26tdTomato mice were administered with tamoxifen at the ages of 1 d, 1 wk, and 5 wk (Fig. 3, D and E). Several months after tamoxifen treatment, bone marrow (BM) HSCs, characterized as Lin−Sca-1+c-kit+ (LSK) cells, were substantially labeled (Fig. 3, D and E), confirming that LSK cells and their descendants could be successfully fate-mapped. After normalization, we found that neonatal (1 d and 1 wk) HSCs contributed to nearly all cNK cells, ∼30% of Ly49E− ILC1s, and rare (<7%) Ly49E+ ILC1s (Fig. 3, D and E). Similarly, upon tamoxifen exposure at the age of 5 wk, most liver cNK cells (∼95%) were fate-mapped after normalization, whereas liver Ly49E+ ILC1s remained unlabeled, and Ly49E− ILC1s exhibited an intermediate expression of tdTomato (Fig. 3, D and E), suggesting little contribution of adult BM HSCs to the Ly49E+ ILC1 pool. To further assess the contributions of adult BM hematopoiesis, we generated chimeric mice transplanted with BM and fetal liver cells (Fig. 3 F). BM cells generated fewer liver ILC1s than fetal liver cells (Fig. 3 G), alongside very low expression of Ly49E on BM-derived ILC1s (Fig. 3, F and H). Similar results were observed even in a noncompetitive setting (Fig. S3, D and E).
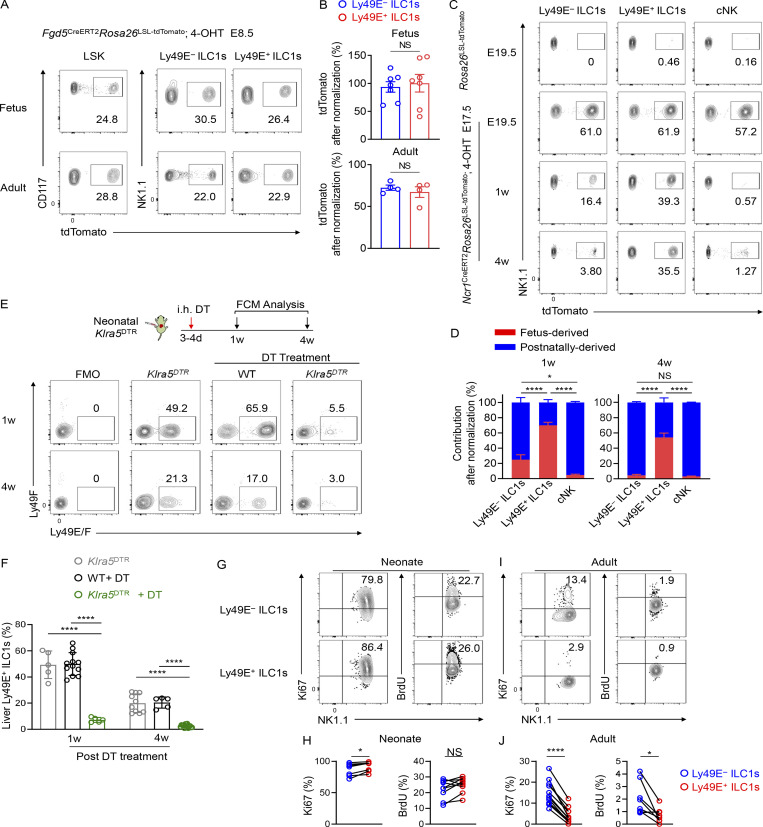
The finding that fetal liver cells have the potential to generate Ly49E+ ILC1s (Fig. 3, F–H) implies the importance of embryonic hematopoiesis for Ly49E+ ILC1 development. Before the emergence of HSCs, Fgd5 is expressed by hemogenic endothelial cells during the early embryonic life (Gazit et al., 2014; Sawen et al., 2018). In Fgd5CreERT2Rosa26tdTomato embryos pulsed with 4-hydroxytamoxifen (4-OHT) at embryonic day 8.5 (E8.5), when hematopoiesis primarily occurs in the yolk sac (YS), fetal liver LSK cells expressing tdTomato were observed at E16.5 (Fig. 4, A and B). Fetal liver Ly49E+ and Ly49E− ILC1s were also fate-mapped, suggesting that they originate from early embryonic hematopoiesis (Fig. 4, A and B). Considerable proportions of adult LSK cells (∼31%), Ly49E− ILC1s (∼22%), and Ly49E+ ILC1s (∼20%) remained fate-mapped, suggesting that embryonic progenitors could ultimately generate adult liver ILC1 subsets (Fig. 4, A and B).
Figure 4.
Ly49E+ ILC1s are primarily derived from embryonic hematopoiesis. (A and B) tdTomato expression on fetal liver LSK cells (E14.5), ILC1 subsets (E16.5), adult BM LSK cells, and adult liver ILC1 subsets from Fgd5CreERT2Rosa26tdTomato mice pulsed with 4-OHT at E8.5 (A). Labeling efficiency of Ly49E− ILC1s and Ly49E+ ILC1s in A were normalized to LSK cells (B). Data are pooled from at least three experiments (fetus: n = 7; adult: n = 4). (C and D) tdTomato expression on liver Ly49E− ILC1s, Ly49E+ ILC1s, and cNK cells from E19.5, 1-wk-old, and 4-wk-old Ncr1CreERT2Rosa26td-Tomato mice pulsed with 4-OHT at E17.5, Rosa26td-Tomato mice were used as controls (C). Contribution of fetus-derived Ly49E− ILC1s, Ly49E+ ILC1s, and cNK cells were calculated as labeling efficiency of the cells from Ncr1CreERT2Rosa26tdTomato mice at 1 or 4 wk relative to that at E19.5, and the unlabeled cells were considered postnatally derived (D). Data are pooled from at least three independent experiments (E19.5: n = 4; 1 wk: n = 5; 4 wk: n = 5). (E and F) Representative plots (E) and percentages (F) of Ly49E+ cells among liver ILC1s from WT and Klra5DTR mice 1 or 4 wk after intrahepatic (i.h.) treatment with DT once during the neonatal period. Data are pooled from two or three independent experiments with n = 5 or 8 (Klra5DTR mice), n = 8 or 15 (DT-treated WT mice), or n = 7 or 12 (DT-treated Klra5DTR mice). (G–J) Representative plots (G and I) and percentages (H and J) of Ki67+ or BrdU+ cells among liver ILC1 subsets in neonatal (0–3 d old) and adult (6–8 wk old) B6 mice. Data are pooled from two or three independent experiments (neonate: n = 7 or 4; adult: n = 12 or 8). Bar graphs show the mean ± SEM; unpaired Student’s t test was used in B, one-way ANOVA in D and F, paired Student’s t test in H and J; *, P < 0.05; ****, P < 0.0001.
We wondered whether embryonic hematopoiesis-derived ILC1s self-maintained or were generated from BM HSC-independent progenitors during adulthood. Using an inducible fate-mapping model, based on Ncr1 (NKp46), we found that Ly49E− ILC1s, Ly49E+ ILC1s, and cNK cells could all be labeled with high and comparable efficiencies in the E19.5 fetal liver pulsed with 4-OHT at E17.5; however, the proportions of labeled Ly49E− ILC1s and cNK cells decreased dramatically after birth (Fig. 4 C). Fetus-derived Ly49E− ILC1s accounted for only 25% of liver Ly49E− ILC1s in 1-wk-old mice and almost disappeared in 4-wk-old mice, suggesting that fetus-derived Ly49E− ILC1s were gradually replaced by unlabeled postnatal progenitor-generated Ly49E− ILC1s (Fig. 4, C and D). In contrast, fetus-derived Ly49E+ ILC1s remained detectable at a relatively high frequency after birth (Fig. 4, C and D), suggesting that they could self-maintain into adulthood. To verify this finding, Ly49E+ cells were depleted by a single injection of diphtheria toxin (DT) into newborn Klra5DTR mice, which did not affect the numbers of Ly49E− ILC1s (Fig. S3 F). Ly49E+ ILC1s could not be generated even at the age of 4 wk in these mice (Fig. 4, E and F), suggesting that postnatal hematopoiesis was unable to efficiently give rise to liver Ly49E+ ILC1s.
We then detected the proliferation ability of ILC1 subsets by examining the proliferation marker Ki67 in vitro and the incorporation of the thymidine analog BrdU in vivo. Both Ly49E− and Ly49E+ ILC1s actively proliferated during neonatal life (Fig. 4, G and H), whereas Ly49E+ ILC1s proliferated at a significantly lower rate than Ly49E− ILC1s during adult life (Fig. 4, I and J), suggesting a quiescent state of Ly49E+ ILC1s after birth. Taken together, these data reveal that embryonic hematopoiesis-derived Ly49E+ ILC1s contribute to the majority of the Ly49E+ ILC1 pool in the adult liver, whereas liver Ly49E− ILC1s are gradually replenished by hematopoietic progenitors after birth.
Ly49E marks a highly cytotoxic subset of liver ILC1s
The differences in origins and transcriptional profiles led us to wonder whether liver Ly49E+ ILC1s served distinct immunologic functions. The transcriptional profile suggested their cytotoxic potential (Fig. 2, F and G; and Fig. S1, A and D). Flow cytometric analysis revealed that resting Ly49E+ ILC1s expressed higher levels of granzyme family proteins, such as granzyme A (GzmA), GzmB, and GzmC, than resting Ly49E− ILC1s (Fig. S4, A and B). Notably, the expression levels of GzmB and GzmC in Ly49E+ ILC1s were even higher than those in liver cNK cells (Fig. S4, A and B). Similar results were obtained upon stimulation with IL-12, IL-15, and IL-18 (Fig. 5, A and B); PMA plus ionomycin (Fig. 5 C); or coculture with Yac-1 cells (Fig. 5 D). Meanwhile, Ly49E+ and Ly49E− ILC1s expressed similar levels of perforin, CD107a, IFN-γ, and TNF-α (Fig. S4, C and D). Notably, both Ly49E+ and Ly49E− ILC1s could produce IL-2, with the former showing more robust potential (Fig. S4, E and F). NK1.1+ cells are capable of using death ligands such as TRAIL to induce cytotoxicity (Arase et al., 1995; Kayagaki et al., 1999). However, there was no difference in FasL and TRAIL expression between Ly49E+ and Ly49E− ILC1s (Fig. S4 G). To ascertain the cytotoxic capacity of Ly49E+ ILC1s, we performed flow-based killing assays. Cytokine-activated group 1 ILC subsets were used as effectors against the Yac-1 cells. Ly49E+ ILC1s lysed YAC-1 cells more efficiently than both Ly49E− ILC1s and cNK cells (Fig. 5, E and F). Additionally, Ly49E+ ILC1s could also efficiently lyse MC38 tumor cells (Fig. S4, H and I). Overall, these results reveal that Ly49E+ ILC1s are more potently cytotoxic than Ly49E− ILC1s.
Figure S4.
Comparison of effector functions between Ly49E+ and Ly49E− ILC1s. (A and B) Representative histograms (A) and percentages (B) of cells expressing the indicated intracellular functional molecules (GzmA, GzmB, and GzmC) in liver group 1 ILC subsets of WT B6 mice (5–8 wk old) at steady state. Data are representative of two or three independent experiments with n = 3–5 mice per experiment. (C and D) Percentages of cells expressing perforin and CD107a (C) or TNF-α and IFN-γ (D) among liver group 1 ILC subsets of WT B6 mice (5–8 wk old) after stimulation for 18 h with 10 ng/ml IL-12, 10 ng/ml IL-15, and 50 ng/ml IL-18, 4 h with PMA plus ionomycin (PMA + Ion), or with Yac-1 cells. Data are representative of two or three independent experiments with n = 3 mice per experiment. (E and F) Representative plots (E) and percentages (F) of cells expressing IL-2 in liver T cells and group 1 ILC subsets of WT B6 mice (5–8 wk old) after stimulation for 4 h with PMA and Ion. Data are representative of two independent experiments with n = 5 mice per experiment. (G) GMFI of cells expressing FasL and TRAIL among Ly49E+ or Ly49E− ILC1s derived from WT B6 mice (5–8 wk old). Data are representative of two independent experiments with n = 3 mice per experiment. (H and I) Real-time cytotoxicity assays were performed using liver group 1 ILC subsets from WT B6 mice (3–6 wk old) as effector cells and MC38 tumor cells as the target cells, with an effector-target (E:T) ratio of 10:1. The cell index during 4 h (H) and percentages of killing efficiency at the 4-h time point (I) after incubation with effector cell. Data are representative of two independent experiments with n = 3 samples per group. Bar graphs show the mean ± SEM; one-way ANOVA followed by Dunnett’s multiple comparison test was used in B–D, F and I, and unpaired Student’s t test in G; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure 5.
Ly49E+ ILC1s are effective in cytotoxicity. (A and B) Representative histograms (A) and percentages (B) of cells expressing the indicated intracellular functional molecules (GzmA, GzmB, and GzmC) in liver group 1 ILC subsets of WT B6 mice (5–8 wk old) after stimulation for 18 h with 10 ng/ml IL-12, 10 ng/ml IL-15, and 50 ng/ml IL-18. Fluorescence minus one (FMO) control was gated on total group 1 ILCs (A). (C and D) Percentages of cells expressing the indicated molecules among the liver group 1 ILC subsets for 4 h with PMA plus ionomycin (PMA + Ion; C) or with Yac-1 cells (D). Data are representative of two or three independent experiments with n = 3–6 mice per experiment (A–D). (E and F) Flow-based killing assays were performed by using liver group 1 ILC subsets of WT B6 mice (3–6 wk old) that were preactivated with IL-12, IL-15, and IL-18 for 18 h as effector cells and Yac-1 tumor cells as the target cells, with an effector–target (E:T) ratio of 10:1. Representative plots (E) and percentages (F) of 7-AAD–positive target cells after incubation with effector cells. Data are pooled from two independent experiments with n = 14–30 samples per group. Bar graphs show the mean ± SEM; one-way ANOVA, followed by Dunnett’s multiple comparison test, was used to compare the experimental groups in B–D and F; **, P < 0.01; ****, P < 0.0001.
Ly49E+ ILC1s confer protection against murine CMV (MCMV) infections in neonatal mice
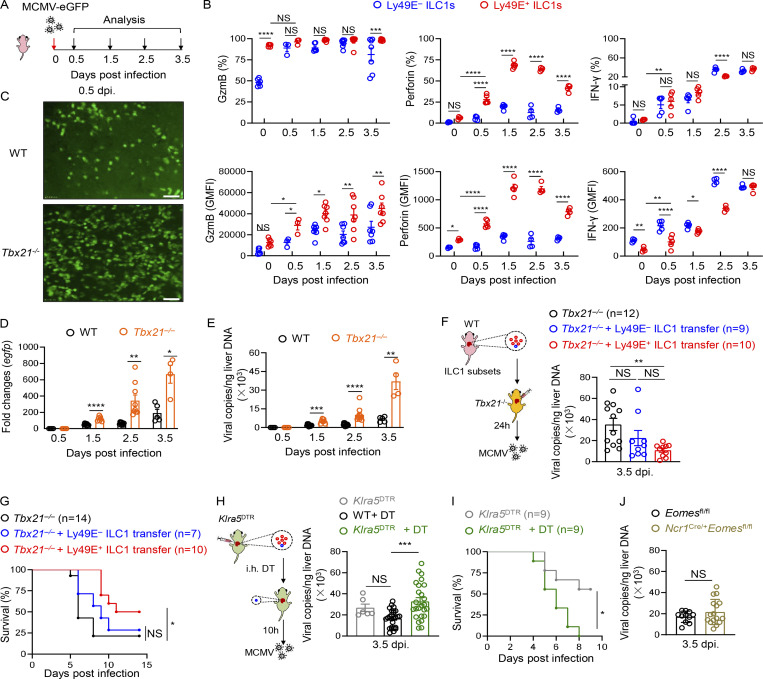
The enrichment and functionality of Ly49E+ ILC1s in neonates prompted us to explore their roles in immune defense in early life. Because CMV is a hepatotropic virus associated with serious health problems in neonates (Schleiss, 2013), we infected neonatal mice with recombinant MCMV expressing eGFP (Pan et al., 2018; Fig. 6 A). Although MCMV infection can induce cNK cell expansion in adult mice (Sun et al., 2009), cNK cells were rarely present in the neonatal liver after MCMV infection, at a much lower frequency than Ly49E+ ILC1s (Fig. S5 A). MCMV infection induced the rapid response of liver Ly49E+ ILC1s in neonates, as evidenced by the increased expression of GzmB, perforin, and IFN-γ as early as 0.5 d post infection (dpi) compared with those in uninfected mice (0 dpi; Fig. 6 B), and expression levels of these molecules remained high until 3.5 dpi. Upon MCMV infection, Ly49E− ILC1s also upregulated GzmB expression, with similar percentages of GzmB+ cells but lower GzmB geometric mean fluorescence intensity (GMFI) compared with Ly49E+ ILC1s (Fig. 6 B). The percentages and GMFI of perforin were lower in Ly49E− ILC1s than Ly49E+ ILC1s (Fig. 6 B). However, there was no increase in the percentage of IFN-γ+ cells, and the GMFI of IFN-γ was even lower in Ly49E+ ILC1s compared with Ly49E− ILC1s (Fig. 6 B). Notably, Ly49E+ ILC1s were the primary lymphocytes expressing GzmB after MCMV infection, as Ly49E− ILC1s rarely coexisted (Fig. S5 C). In adult mice, cNK cells mediate essential resistance to MCMV infection through the m157–Ly49H axis (Arase et al., 2002; Brown et al., 2001; Dokun et al., 2001; Smith et al., 2002). Interestingly, in our setting, neonatal liver NK1.1+ ILCs did not express high levels of Ly49H either under steady state or after MCMV infection (Fig. S5 B), raising a possibility that Ly49E+ ILC1s counteract MCMV in a Ly49H-independent manner.
Figure 6.
Ly49E+ ILC1s play a role in the host defense against MCMV infection during early life. (A and B) Schematic showing the experimental design used for MCMV infections in neonatal mice within 3 d of age (A). Percentages and GMFI of GzmB, perforin, or IFN-γ expressed by Ly49E+ or Ly49E− ILC1s derived from neonatal mice at different time points after infection with MCMV-eGFP (B). Data presented for each time point are representative of at least three independent experiments with n = 3–5 mice. (C) Representative images of the intact liver of neonatal WT and Tbx21−/− mice 0.5 dpi with MCMV-eGFP; scale bar = 100 μm. (D and E) egfp expression (D) and viral copies (E) in the livers of neonatal Tbx21−/− and WT mice were measured 0.5, 1.5, 2.5, and 3.5 dpi with MCMV-eGFP. Data are pooled from two independent experiments with n = 4–12 mice per group. (F and G) 1–1.5 × 105 neonatal Ly48E+ or Ly49E− ILC1s were transferred into neonatal Tbx21−/− mice 1 d before MCMV-eGFP infection. Liver viral copies were measured 3.5 dpi (F). Data were pooled from three independent experiments with n = 9–12 mice per group. Survival of neonatal Tbx21−/− mice that received Ly49E+ or Ly49E− ILC1s followed by MCMV infection (G). Data are pooled from three independent experiments with n = 7–14 mice per group. (H and I) Neonatal Klra5DTR mice were administered DT 10 h before MCMV-eGFP infection. Liver viral copies were measured 3.5 dpi. MCMV-infected Klra5DTR littermates and DT-treated WT mice were as controls (H). Data were pooled from three independent experiments with n = 6–26 mice per group. Survival of neonatal Klra5DTR mice were shown (I). Data are pooled from two independent experiments with n = 9 mice per group. (J) Viral titers in the livers of neonatal Ncr1Cre/+Eomes fl/fl and Eomes fl/fl mice were measured 3.5 dpi with MCMV-eGFP. Data are pooled from three independent experiments with n = 11–16 mice per group. Bar graphs show the mean ± SEM; two-way ANOVA was used in B, D, and E, one-way ANOVA followed by Dunnett’s multiple comparison test in F and H, unpaired two-tailed Student’s t test in J, and log-rank (Mantel–Cox) test in G and I; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S5.
Ly49E+ ILC1s mediate protective responses against MCMV infection in neonatal mice. (A) Percentages of Ly49E+ ILC1s, Ly49E− ILC1s, and cNK cells among liver group 1 ILCs from neonatal WT mice within 3 d of age after infection with MCMV-eGFP. Data for each time point are representative of three independent experiments with n = 4–5 mice. (B) Representative plots showing Ly49H expression on neonatal liver group 1 ILCs after MCMV-eGFP infection. (C) Flow cytometric analysis of GzmB expression in CD45+ leukocytes from neonatal livers before and after MCMV-eGFP infection. (D) Viral copies in the spleen of neonatal Tbx21−/− and WT mice were measured 0.5, 1.5, 2.5, and 3.5 dpi with MCMV-eGFP. Data are representative of at least three independent experiments with n = 3 mice per group. (E) 1–1.5 × 105 neonatal Ly48E+ or Ly49E− ILC1s were transferred into neonatal Tbx21−/− mice 1 d before MCMV-eGFP infection. Spleen viral copies were measured 3.5 dpi. Data were pooled from two independent experiments with n = 7–12 mice per group. (F) The depletion effect of Klra5DTR mice after DT treatment. Plots are representative of at least three independent experiments. (G) Neonatal Klra5DTR mice were administered DT 10 h before MCMV-eGFP infection. Spleen viral copies were measured 3.5 dpi. Data were pooled from three independent experiments with 21–23 mice. (H and I) 0–3-d-old neonatal Ncr1Cre/+Eomesfl/+ and Ncr1Cre/+Eomesfl/fl mice were infected with MCMV-eGFP. The absolute cell number of liver ILC1s (H) and viral titers in the spleen (I) were measured 3.5 d later. **, P < 0.01. Data are representative of three independent experiments with five to nine mice per experiment. Bar graphs show the mean ± SEM; two-way ANOVA was used in D and E, unpaired two-tailed Student’s t test in G–I.
To explore whether neonatal liver Ly49E+ ILC1s play a role in the innate defense against MCMV, we used Tbx21−/− mice, which are deficient in liver ILC1s (Daussy et al., 2014; Gordon et al., 2012; Zhou et al., 2019). We found larger eGFP-positive areas in Tbx21−/− liver after MCMV infection (Fig. 6 C). Higher egfp expression and increased viral DNA copies after MCMV infection were observed in the neonatal liver at different time points in Tbx21−/− mice compared with control mice (Fig. 6, D and E), but no difference was observed in spleen (Fig. S5 D), indicating the early local protection of liver ILC1s against MCMV. To confirm the specific protective roles of Ly49E+ ILC1s, Ly49E+ ILC1s were adoptively transferred into neonatal Tbx21−/− mice before infection. Decreased viral copies were observed in the liver of Ly49E+ ILC1-transferred mice (Fig. 6 F). Although there was a slight decrease of viral loads in Ly49E− ILC1-transferred mice (Fig. 6 F), their survival was not altered compared with those without transfer (Fig. 6 G). In contrast, transfer of Ly49E+ ILC1s prolonged the survival time (Fig. 6 G).
Because T-bet can also regulate differentiation of cNK cells and T cells (Lazarevic et al., 2013), we specifically depleted Ly49E+ cells, which were mainly Ly49E+ ILC1s, by using Klra5DTR mice via DT injection (Fig. S5 F). We found that Ly49E+ cell depletion consistently rendered the neonatal liver more susceptible to MCMV infection (Fig. 6 H) and shortened its survival time (Fig. 6 I); however, the absence or replenishment of Ly49E+ ILC1s had no obvious impact on viral copies in the spleen (Fig. S5 G). Viral copies in the liver of neonatal Ncr1Cre/+Eomesfl/fl mice, in which ILC1s were not affected (Fig. S5 H) but cNK cells were reduced (Weizman et al., 2017; Zhou et al., 2019), did not differ from those in control mice (Fig. 6 I), but were higher in the spleen of Ncr1Cre/+Eomesfl/fl mice (Fig. S5 I). Taken together, these findings demonstrated that Ly49E+ ILC1s are important effectors that confer protection against MCMV infection during the neonatal stage.
Discussion
Group 1 ILCs consist of cNK cells and ILC1s. Although cNK cells have been extensively studied for decades, research on ILC1s remains in its infancy. In this study, the scRNA-seq analysis revealed heterogeneity within both cNK and ILC1 subsets. cNK cells could be further divided into three clusters, which corresponded to different developmental stages. Ly49E expression could dissect ILC1s into two subsets: Ly49E+ ILC1s and Ly49E− ILC1s, which originated from different hematopoietic waves. Specifically, Ly49E+ ILC1s were primarily dependent on embryonic hematopoiesis and self-maintenance after birth, whereas Ly49E− ILC1s primarily originated from postnatal hematopoiesis. Functionally, Ly49E+ ILC1s were highly cytotoxic and sensitive to external stimuli, exhibiting typical characteristics of innate immune cells. With increasing age, liver ILC1s gradually became predominated by the Ly49E− population, which expressed memory-associated molecules and displayed memory potential in the CHS model. Overall, our study depicted the landscape of liver group 1 ILC composition, highlighting the previously unrecognized heterogeneity of liver ILC1s.
Newly developed single-cell profiling technologies provide valuable tools for dissecting cellular heterogeneity of ILC1s at high resolution. However, differing sample preparation process, technical platforms, and data analysis methods might yield different cell clustering results (Kharchenko, 2021; Papalexi and Satija, 2018). For instance, McFarland et al. (2021) recently showed that liver ILC1s were just grouped as one cluster by scRNA-seq. In contrast, other studies show that liver ILC1s can be subdivided into several subpopulations by quantitative single-cell RT-PCR or scRNA-seq analysis (Park et al., 2019; Perchet et al., 2018). Recently, Friedrich et al. (2021) separated liver ILC1s into two subsets by using IL-7Rα and reported that IL-7Rα+ ILC1s were precursors of IL-7Rα− ILC1s, the latter of which highly expressed cytotoxic molecules. Consistently, our study also showed high cytotoxic potential of Ly49E+ ILC1s, which partially overlap with IL-7Rα− ILC1s, as evidenced by negative expression of IL-7Rα on Ly49E+ ILC1s. However, IL-7Rα was heterogeneously expressed by Ly49E− ILC1s, and there was no conversion between Ly49E+ ILC1s and Ly49E− ILC1s in our study. These findings raise a possibility that the differentiation from IL-7Rα+ ILC1s into IL-7Rα− ILC1s might be confined to Ly49E− cell populations.
The layered ontogeny of lymphocytes has recently been reported, such as CD8+ T cells (Smith et al., 2018) and ILC2s (Schneider et al., 2019); however, the distinction of lymphocytes with different origins remains challenging. Using Fgd5- or Ncr1-based inducible fate-mapping models, Ly49E separates liver ILC1s into two subsets with different hematopoietic waves. While liver Ly49E+ ILC1s mainly originate through embryonic hematopoiesis and self-maintain into adulthood, Ly49E− ILC1s have heterogeneous developmental origins. Fetus-derived Ly49E− ILC1s can be replaced continuously by postnatal hematopoietic progenitors. However, the fate-mapping model indicated that postnatal BM hematopoiesis contributed only ∼30% of Ly49E− ILC1s. Because the adult liver contains hematopoietic progenitors with ILC1 development potential (Bai et al., 2021), the rest of the Ly49E− ILC1s might develop from local hematopoietic progenitors during postnatal life. In contrast with Ly49E− ILC1s, the majority of liver Ly49E+ ILC1s originate through embryonic hematopoiesis and self-maintain into adulthood. A recent study reported that erythromyeloid progenitors (EMPs) in the YS gave rise to potently cytotoxic fetal NK cells (Dege et al., 2020). However, the EMP progenies in that study were not precisely defined for their ILC1 or cNK cell identities; therefore, further investigations should examine whether YS EMPs contribute to Ly49E+ ILC1s.
In addition to liver ILC1s, Ly49E was expressed by a minor fraction of NK1.1+NKp46+ cells in the uterus and the salivary gland (SG). A recent study (McFarland et al., 2021) showed negative expression of IL-7Rα on uterus and SG NK1.1+NKp46+ cells, which is similar to liver Ly49E+ ILC1s depicted in this study. However, their single-cell analysis revealed one tissue-specific metacell shared by the uterus and SG, but no tissue-specific metacells commonly exist between the liver and uterus/SG. The researchers found that the shared SG-uterine metacell expressed Eomes and Tcf7, both of which were expressed at low levels of liver ILC1s. These results suggest that, despite Ly49E expression across these tissues, it is likely that Ly49E+NK1.1+NKp46+ cells in the liver and uterus/SG might be different in transcription factor expression. Nevertheless, it would be of interest to investigate whether Ly49E can be commonly used to mark embryonic hematopoiesis-dependent ILC1s/NK cells.
Although Ly49E was identified as an inhibitory receptor, its role under pathophysiological conditions remains controversial. Studies have shown that Ly49E is not involved in several pathophysiological situations, including malaria infection (Filtjens et al., 2014), chemically induced colitis (Van Acker et al., 2016), and different tumors (Filtjens et al., 2016b). However, young Ly49E-KO mice could control the parasite infection better (Filtjens et al., 2016a). Although Ly49E activation by uPA, a non-MHC class I ligand (Van Den Broeck et al., 2008), could inhibit the effector functions of NK cell lines or fetal thymic NK cells, freshly isolated liver group 1 ILCs from Ly49E-deficient and WT mice did not differ in cytotoxic capacity in the presence of uPA (Filtjens et al., 2013). Because of these contradictory observations, whether uPA represents a high-affinity ligand for Ly49E remains unclear. Previously, an NKp46−CD8αα−Ly49E+ population has been identified in small intestinal intraepithelial lymphocytes (IELs; Van Acker et al., 2017), but the functional role of Ly49E in this subset was unknown. Our study revealed that Ly49E+ ILC1s played a protective role against early viral infections; however, whether Ly49E is directly involved in the antiviral process requires further investigation.
The phenotypic and functional characterization of human ILC1s remain controversial. Studies have reported that fetal CD56+ group 1 ILCs are highly responsive to cytokines IL-12 and IL-18, which could be rapidly secreted by myeloid cells through the activation of pattern recognition receptors upon infection, yielding increased cytotoxicity and cytokine production relative to their adult counterparts (Ivarsson et al., 2013). Therefore, human CD56+ group 1 ILCs might be actively involved in the host defense during early life, in a manner similar to neonatal murine Ly49E+ ILC1s. Despite the lack of any identified Ly49E homolog in humans, transcriptomic analysis revealed that mouse Ly49E+ ILC1s and human fetal liver NK or ILC1s shared common genetic features, such as Gzm (NKG7, GNLY related), Klrd1 (KLRD1), and Cd69 (CD69; Bennstein et al., 2020; Popescu et al., 2019), suggesting the potential for a similar fetus-derived ILC1 population in humans.
Because of the lack of an experienced adaptive response, newborn infants rely heavily on the innate immune system to confer host protection against invading pathogens. CMV is a common virus that typically results in an asymptomatic infection in healthy individuals but can cause severe illness in neonates who experience congenital or perinatal infections (Swanson and Schleiss, 2013). Compelling evidence has been provided to support the necessity of cNK cells to fight against CMV infections in adults (Lanier, 2008); however, cNK cells are rarely present in neonatal mice, suggesting that they may make less contribution during the neonatal stage than during adulthood. In the present study, we detected increased MCMV DNA copies in neonatal Tbx21−/− mice, in which ILC1s were absent, and the adoptive transfer of Ly49E+ ILC1s, but not Ly49E− ILC1s, reduced viral loads and prolonged survival, suggesting an important role for Ly49E+ ILC1s in the host defense against viral infection during early ontogeny.
Previous studies have shown that proinflammatory cytokines, such as IL-12, IL-18, IFN-α, and IFN-β, are required for regulation of NK cell or ILC1 activation and responses to viral infection through the induction of IFN-γ or cytotoxicity during infection (Biron and Tarrio, 2015; Nguyen et al., 2002; Weizman et al., 2017). Further investigations are required to explore whether these cytokines are also essential for activation of neonatal Ly49E+ ILC1s during MCMV infection. Furthermore, previous studies have revealed that granzymes and perforin are critical to the control of MCMV infection (Andoniou et al., 2014; Fehniger et al., 2007), especially during the acute infection phase (Riera et al., 2000). Cytotoxicity is particularly important for Ly49H-dependent MCMV control (Pak-Wittel et al., 2015). Both liver Ly49E+ and Ly49E− ILC1s have cytotoxic capabilities, and the former subset is more potently cytotoxic. Moreover, cell numbers of Ly49E− ILC1s are far fewer than Ly49E+ ILC1s in the neonatal liver, and liver GzmB+ leukocytes in neonatal mice during MCMV infection are mainly confined to Ly49E+ ILC1s. Therefore, it would be of interest to investigate whether the cytotoxic capability of Ly49E+ ILC1s or other specific molecular mechanisms contribute to the viral control during early life. However, the decline in Ly49E+ ILC1s proportions with age suggests that this subset may be less important for antiviral immunity during adulthood, and their replacement with Ly49E− ILC1s may play a compensatory role. A recent study revealed that during MCMV infection, liver ILC1s upregulate IL-18Rα expression, and IL-18Rα+ ILC1s were able to confer memory responses during secondary MCMV infections (Weizman et al., 2019). We found that IL-18Rα was preferentially expressed by Ly49E− ILC1s, raising the possibility that Ly49E− ILC1s may also participate in MCMV-induced memory responses in adults.
Taken together, our findings uncover the transcriptional, developmental, and functional heterogeneity of ILC1s and provide additional evidence to support the diversity of ILC1s in different tissues. Our study also demonstrates the dynamic changes that occur in ILC1 subsets from different origins during ontogeny, which correspond to their different functional roles, facilitating adaptation to age-related immune demands.
Materials and methods
Mice
All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Science and Technology of China. WT B6 mice were purchased from the Shanghai Experimental Animal Center. Tbx21−/− (T-bet–deficient) mice, CD45.1+ mice, and Fgd5CreERT2 mice were obtained from Jackson Laboratory. Ncr1Cre/+Eomesfl/fl mice were provided by Zhongjun Dong (Tsing Hua University). Rosa26LSL-tdTomato and Ncr1CreERT2 mice were obtained from the Shanghai Model Organisms Center. Klra5DTR mice were generated by Biocytogen Co. All mice were generated on or backcrossed to the B6 background for three or more generations. Various ages of both male and female mice were used. Mice were sex- and age-matched for all adult-related experiments; mice were age-matched for all neonate-related experiments. All mice were housed in a specific pathogen–free facility with unrestricted access to food and water, in compliance with the guidelines for the use of experimental animals at the University of Science and Technology of China. Experiments were conducted in accordance with approved institutional protocols.
Virus and infection
MCMV-eGFP (provided by Zhikang Qian; Pan et al., 2018) was propagated on primary MEFs (Brune et al., 2001) derived from WT B6 mice, and viral titers were determined using a 96-well plate. Briefly, cell-free media from the virus-infected cultures were collected to determine the virus titers using the 50% tissue culture infectious dose (TCID50) assay in MEFs. To measure viral copies, experimental neonatal mice were infected i.p. with 5 × 106 TCID50 MCMV in 0.01–0.03 ml DMEM per g body weight. To assess survival, experimental neonatal mice were infected i.p. with 2 × 106 TCID50 MCMV in 0.01–0.03 ml per g body weight.
Virus quantification
MCMV viral titers were determined as previously described (Weizman et al., 2017). The DNeasy Blood and Tissue Kit (Qiagen) was used for DNA isolation from the liver or spleen. After isolation, DNA concentrations were measured using a Nanodrop (Thermo Fisher Scientific). Quantitative real-time PCR was performed using 10 ng DNA, a supermix containing SYBR Green (Takara), and primers specific to MCMV IE-1 DNA (forward: 5′-AGCCACCAACATTGACCACGCAC-3′; reverse: 5′-GCCCCAACCAGGACACACAACTC-3′) and egfp (forward: 5′-GAAGAACGGCATCAAGGTG-3′; reverse: 5′-CTGGGTGCTCAGGTAGTGG-3′). Copy numbers were determined by comparing the Cq values to a standard curve generated using known dilutions of IE-1 plasmid and normalized relative to the total DNA content. Actin was used as a housekeeping gene for data normalization.
Cell isolation
Isolation of liver MNCs and splenocytes was performed as previously described (Wang et al., 2006). Briefly, livers were removed and pressed through a 200-gauge mesh, and liver MNCs were collected after 40 and 70% Percoll density gradient centrifugation. The spleens were removed and pressed through a 200-gauge mesh, and splenocytes were obtained by lysing the erythrocytes with red cell lysis buffer (BioLegend). BM cells were obtained by flushing femurs and then lysing the erythrocytes with red cell lysis buffer. Cells from the thymus and inguinal LNs (iLNs) were obtained by mechanical disruption.
The lung, SG, uterus, and adipose and ear skin tissues were cut into pieces and digested in RPMI 1640 (Hyclone) with 0.1% collagenase IV, 0.25 mg/ml DNase I, and 10% FBS (Gibco BRL) with shaking at 37°C for 40 min. The supernatant was collected and purified by Percoll gradient centrifugation.
IELs were isolated as described previously (Ivanov et al., 2006). Briefly, small intestine mesentery and Peyer’s patches were excised and gently washed to remove the luminal contents. The intestines were cut into pieces and placed into a 50-ml Erlenmeyer flask. The intestinal pieces were incubated in 20 ml IMDM (Hyclone) supplemented with 5% FBS, 5 mM EDTA, and 15 mM Hepes and rotated at 200 rpm for 30 min at 37°C. This procedure was repeated twice. After each rotation, the supernatants containing IELs were collected. The intestine was washed, minced, and incubated with IMDM, 10% FBS, 0.1% collagenase IV, and 0.25 mg/ml DNase I with shaking at 37°C for 30 min. This procedure was repeated twice. After each round of rotation, the supernatants containing lamina propria–associated lymphocytes were collected. The supernatants were filtered through a 200-gauge steel mesh, and IELs and lamina propria–associated lymphocytes were collected after Percoll gradient centrifugation.
BrdU incorporation
Mice were injected i.p. with BrdU (1 mg for adult mice or 0.1 mg for neonatal mice) 1 d before harvest. BrdU staining was performed according to the FITC BrdU Flow Kit (BD Biosciences) instructions.
Antibody staining and flow cytometry
The isolated cells were incubated with rat serum to block Fc receptors for 30 min at 4°C, followed by staining with fluorescently labeled mAbs (Table S1) against surface molecules. For intracellular cytokine staining, cells were stimulated with 30 ng/ml PMA (Sigma-Aldrich) and 1 µg/ml ionomycin (Sigma-Aldrich) or incubated with Yac-1 cells (effector/target ratio of 3:1) for 4 h, and 2 µg/ml monensin (Sigma-Aldrich) was added at the beginning of the stimulation procedure. For cytokine stimulation, the cells were stimulated with 10 ng/ml muIL-12, 10 ng/ml muIL-15 (Peprotech), and 50 ng/ml muIL-18 (R&D Systems) for 18 h, and 2 µg/ml monensin was added during the last 4 h of stimulation. CD107a was stained during stimulation. After surface staining, the cells were fixed, permeabilized using a Foxp3/Transcription Factor Staining Buffer Set (eBioscience), and stained with mAbs against intracellular molecules. All data were collected using a flow cytometer (LSR II and LSRFortessa; BD Biosciences) and analyzed with FlowJo software (TreeStar).
Cell sorting and adoptive transfer
A FACS Aria cell sorter (BD Biosciences) was used to purify cNK cells (CD3−NK1.1+CD49b+CD49a−), Ly49E+ ILC1s (CD3−NK1.1+CD49a+CD49b−Ly49F−Ly49E+), Ly49E− ILC1s (CD3−NK1.1+CD49a+ CD49b−Ly49E−), IL-7Rα+Ly49E− ILC1s (CD3−NK1.1+CD49a+CD49b−Ly49E− IL-7Rα+), and IL-7Rα−Ly49E− ILC1s (CD3−NK1.1+CD49a+CD49b−Ly49E− IL-7Rα−). The purity of the sorted cell populations was >90%, as verified after sorting by flow cytometry. For the in vivo study, purified cells were transferred i.v. into sublethally irradiated (6.5 Gy administered 1 d before adoptive transfer) adult recipient mice, which were harvested 2 wk later or challenged 1 mo later with OXA. For some neonatal experiments, purified cells were transferred into neonatal Tbx21−/− mice by intrahepatic injection. Mice were infected with MCMV 1 d later and harvested 3.5 dpi or assessed for survival.
Lineage tracing of HSCs and group 1 ILCs
For fate-mapping analysis of HSCs, Fgd5CreERT2 mice were crossed with Rosa26-LSL-tdTomato reporter mice. To label the HSCs of newborns (1 d), 2.5 mg tamoxifen (MedChemExpress) was administered to dams by oral gavage three times within 24 h. To mark neonatal (1 wk) and adult (5 wk) HSCs, mice were injected i.p. with 500 µg tamoxifen (a single dose) and 2.5 mg tamoxifen (every 24 h for five consecutive days), respectively. To induce the labeling of YS Fgd5+ progenitors, pregnant females were injected i.p. with 1.25 mg 4-OHT (MedChemExpress) and 0.75 mg progesterone (MedChemExpress) on E8.5. For the induction of embryonic Ncr1 fate-mapping, Ncr1CreERT2 males were crossed with Rosa26-LSL-tdTomato females, followed by i.p. injection of pregnant mice with 1.5 mg 4-OHT and 1 mg progesterone on E17.5. Because 4-OHT treatment interferes with natural delivery, pups were delivered by cesarean section and fostered with lactating ICR females.
DT-mediated Ly49E+ cell depletion
DT (List Biological Laboratories) was stored at −80°C. Neonatal Klra5DTR or WT mice were injected i.p. with a single dose of DT (4 ng/g body weight).
CHS
On days 0 and 1, WT mice were sensitized by painting the shaved abdominal skin with 50 μl 5% OXA (Sigma-Aldrich) in a solution of acetone/methanol (1:1). After 5 d, these mice were challenged by painting the skin of their ears with 20 μl 1% OXA for ear-based ILC1 analysis.
On days 0 and 1, the WT mice were sensitized by painting their shaved abdominal skins with 50 μl 5% OXA or 0.5% FITC (Sigma-Aldrich) in a solution of acetone/dibutyl phthalate (1:1). After 5 d, these mice were used as donors for adoptive transfer of purified liver group 1 ILC subsets. Recipient mice were challenged by painting the skin of their right ears with 20 μl 1% OXA or 20 μl 0.5% FITC; left ears were painted with solvent as controls. Ear thickness was measured every 24 h using a micrometer.
To evaluate acute hapten-induced irritation, background swelling was measured as the swelling measured in naive mice treated with the same solvent. Hapten-specific ear swelling was calculated as follows: (thickness of the hapten-treated ear − thickness of the control ear) − background swelling.
Cytotoxicity assay
Flow-based killing assays were performed as previously described (Grossman et al., 2004). Briefly, target Yac-1 cells were labeled at 37°C for 10 min with 200 nM CFSE (Sigma-Aldrich). Labeled target cells (1 × 104) were added to 96-well round-bottom plates along with purified liver group 1 ILC subsets (1 × 105) in complete RPMI for 4 h. Immediately before analysis, 0.5 mg/ml (final concentration) of 7-amino-actinomycin D (7-AAD; eBioscience) was added to each sample. Cells positive for both 7-AAD and CFSE were considered lysed.
Real-time cytotoxicity assays were monitored using an xCELLigence Real-Time Cell Analyzer-Multiple Plate system (Roche Applied Science). To obtain an equilibrium of E-Plate 16 (Roche Applied Science), 50 μl complete RPMI was added to each well. Then, 1 × 104 MC38 cells (target cells) in 100 μl culture medium per well were plated into E-Plate 16, which was subsequently placed into the Real-Time Cell Analyzer-Multiple Plate instrument at 37°C with 5% CO2. 28 h later, 1 × 105 liver group 1 ILC subsets (effectors) in 100 μl culture medium per well were added. The cell index represented changes in electrical impedance and reflected the number of unkilled target cells on biocompatible microelectrode surfaces. For real-time monitoring, the cell index was read automatically every 15 min. Cell index data in each group represented the mean value from three wells.
scRNA-seq and graphic display of scRNA-seq data
Group 1 ILCs from the liver and spleen (CD45+ NK1.1+NKp46+ CD3−CD19−) were sorted using a FACS Aria cell sorter (BD Biosciences). Single-cell barcode scRNA-seq libraries of the purified group 1 ILCs were generated using Chromium Single Cell 3′ Library (v2; 10x Genomics), according to the manufacturer’s instructions, and scRNA-seq libraries were sequenced with a HiSeq X Ten system (Illumina).
Data were mapped to the mouse genome mm10 using Cell Ranger 2.1.1 (10x Genomics). Raw gene expression data were converted to a Seurat object using Seurat R (v3.0; Butler et al., 2018) for further analyses. The Seurat object was first generated using the following criteria: each gene was expressed by ≥50 cells, and ≥500 genes were expressed per cell. We further filtered out cells that had unique feature counts >3,000 or <500 and >4% mitochondrial counts. Variable genes were selected based on average expression and dispersion. Principal component analysis was performed using variable genes. Clusters and t-SNE plots were generated based on selected principal component analysis dimensions. Marker genes were identified by the Seurat function FindVariableFeatures. Scaled expression data of the top 200 and top 10 marker genes for each cluster were used to generate heatmaps.
Bulk RNA-seq and transcriptomic analysis
Liver Ly49E+ or Ly49E− ILC1s from WT adult mice were sorted, and RNA was isolated using TRIzol (Thermo Fisher Scientific) and Direct-zol RNA Miniprep kit (Zymo Research Corp). RNA amplification and RNA-seq were performed by Novogene Bioinformatics Technology Co.
Differential expression analysis was executed with edgeR (v3.26.8). Genes were considered to be differentially expressed between two groups (one sample for Ly49E+ group versus two samples for Ly49E− group) if their fold-changes were >2.0 and adjusted P values were <0.01. Gene Ontology (GO) analysis was performed using GOrilla ranked lists analysis (Eden et al., 2009). The full list of enriched GO terms and volcano plots were generated for differentially expressed genes with >2.0-fold differences in expression between adult Ly49E+ and Ly49E− ILC1s.
Statistical methods
Statistical analyses were performed using GraphPad Prism v6.0 and v8.0. For graphs, data are presented as the mean ± SEM; P values were calculated using one- or two-way ANOVA or Student’s t test (Welch’s correction used for unequal variances). Significance was defined as follows: NS, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001; n in animal experiments refers to numbers of animals per experimental group.
Online supplemental material
Fig. S1 shows differentially expressed genes between liver ILC1 subsets. Fig. S2 shows that Ly49E− ILC1s rather than Ly49E+ ILC1s specifically express IL-7Rα and exhibit memory potential in the CHS model. Fig. S3 shows Ly49E+ ILC1s in neonatal liver and little contribution of adult BM hematopoiesis to the Ly49E+ ILC1 pool. Fig. S4 shows the expression of granzymes and other functional molecules and cytotoxicity against MC38 cells in Ly49E+ and Ly49E− ILC1s. Fig. S5 shows liver group 1 ILC composition and the expression of Ly49H and GzmB in Ly49E+ ILC1s during MCMV infection, and also shows the viral loads in spleens of Tbx21−/− mice adoptively transferred with ILC1 subsets or not, Klra5DTR mice or Ncr1Cre/+Eomesfl/fl mice after MCMV infection. Table S1 lists antibodies and fluorochromes.
Supplementary Material
lists antibodies and fluorochromes.
Acknowledgments
We thank Prof. Zhikang Qian (University of Chinese Academy of Sciences, Shanghai, China) for providing MCMV-eGFP.
This work was supported by the Natural Science Foundation of China (#81788101, 92042305, 81922029, 31900637, 81821001), National Key R&D Program of China (2020YFA0804100, 2019YFA0508502/3), and CAMS Innovation Fund for Medical Sciences 2019-I2M-5-073.
Author contributions: Y. Chen, X. Wang, H. Peng, and Z. Tian initiated and designed the research. Y. Chen, B. Li, and X. Hao analyzed the RNA-seq data. Y. Chen and X. Wang performed the animal experiments and analyzed and interpreted results. Y. Chen, X. Wang, H. Peng, and Z. Tian wrote the manuscript. R. Sun, H. Wei, and K. Qu contributed to discussions of results. W. Tao and S. Zhu contributed to MCMV infection experiments.
Data availability
Murine bulk RNA-seq and scRNA-seq data are deposited under GEO accession number GSE171851. All other data needed to evaluate the conclusions in the paper are present in the paper or the online supplemental material.
References
- Andoniou, C.E., Sutton V.R., Wikstrom M.E., Fleming P., Thia K.Y.T., Matthews A.Y., Kaiserman D., Schuster I.S., Coudert J.D., Eldi P., et al. 2014. A natural genetic variant of granzyme B confers lethality to a common viral infection. PLoS Pathog. 10:e1004526. 10.1371/journal.ppat.1004526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arase, H., Arase N., and Saito T.. 1995. Fas-mediated cytotoxicity by freshly isolated natural killer cells. J. Exp. Med. 181:1235–1238. 10.1084/jem.181.3.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arase, H., Mocarski E.S., Campbell A.E., Hill A.B., and Lanier L.L.. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 296:1323–1326. 10.1126/science.1070884 [DOI] [PubMed] [Google Scholar]
- Bai, L., Vienne M., Tang L., Kerdiles Y., Etiennot M., Escaliere B., Galluso J., Wei H., Sun R., Vivier E., et al. 2021. Liver type 1 innate lymphoid cells develop locally via an interferon-gamma-dependent loop. Science. 371:eaba4177. 10.1126/science.aba4177 [DOI] [PubMed] [Google Scholar]
- Bajpai, G., Schneider C., Wong N., Bredemeyer A., Hulsmans M., Nahrendorf M., Epelman S., Kreisel D., Liu Y., Itoh A., et al. 2018. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat. Med. 24:1234–1245. 10.1038/s41591-018s4150059-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennstein, S.B., Weinhold S., Manser A.R., Scherenschlich N., Noll A., Raba K., Kogler G., Walter L., and Uhrberg M.. 2020. Umbilical cord blood-derived ILC1-like cells constitute a novel precursor for mature KIR+NKG2A- NK cells. eLife. 9:e55232. 10.7554/eLife.55232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron, C.A., and Tarrio M.L.. 2015. Immunoregulatory cytokine networks: 60 years of learning from murine cytomegalovirus. Med. Microbiol. Immunol. 204:345–354. 10.1007/s00430-015s0040412-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M.G., Dokun A.O., Heusel J.W., Smith H.R., Beckman D.L., Blattenberger E.A., Dubbelde C.E., Stone L.R., Scalzo A.A., and Yokoyama W.M.. 2001. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 292:934–937. 10.1126/science.1060042 [DOI] [PubMed] [Google Scholar]
- Brune, W., Hengel H., and Koszinowski U.H.. 2001. A mouse model for cytomegalovirus infection. Curr. Protoc. Immunol. Chapter 19:Unit 19.7. 10.1002/0471142735.im1907s43 [DOI] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, W., Chelbi S.T., Niogret C., Ramon-Barros C., Welten S.P.M., Osterheld K., Wang H., Rota G., Morgado L., Vivier E., et al. 2018. The transcription factor Rfx7 limits metabolism of NK cells and promotes their maintenance and immunity. Nat. Immunol. 19:809–820. 10.1038/s41590-018s4150144-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiossone, L., Chaix J., Fuseri N., Roth C., Vivier E., and Walzer T.. 2009. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 113:5488–5496. 10.1182/blood-2008b10-187179 [DOI] [PubMed] [Google Scholar]
- Constantinides, M. G., Gudjonson H., McDonald B. D., Ishizuka I. E., Verhoef P. A., Dinner A. R., and Bendelac A.. 2015. PLZF expression maps the early stages of ILC1 lineage development. J. Immunol. 112:5123–5128. 10.1073/pnas.1423244112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy, C., Faure F., Mayol K., Viel S., Gasteiger G., Charrier E., Bienvenu J., Henry T., Debien E., Hasan U.A., et al. 2014. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 211:563–577. 10.1084/jem.20131560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dege, C., Fegan K.H., Creamer J.P., Berrien-Elliott M.M., Luff S.A., Kim D., Wagner J.A., Kingsley P.D., McGrath K.E., Fehniger T.A., et al. 2020. Potently cytotoxic natural killer cells initially emerge from erythro-myeloid progenitors during mammalian development. Dev. Cell. 53:229–239.e7. 10.1016/j.devcel.2020.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokun, A.O., Kim S., Smith H.R., Kang H.S., Chu D.T., and Yokoyama W.M.. 2001. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2:951–956. 10.1038/ni714 [DOI] [PubMed] [Google Scholar]
- Eden, E., Navon R., Steinfeld I., Lipson D., and Yakhini Z.. 2009. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 10:48. 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehniger, T.A., Cai S.F., Cao X., Bredemeyer A.J., Presti R.M., French Anthony R., and Ley T.J.. 2007. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 26:798–811. 10.1016/j.immuni.2007.04.010 [DOI] [PubMed] [Google Scholar]
- Filtjens, J., Coltel N., Cencig S., Taveirne S., Van Ammel E., Van Acker A., Kerre T., Matthys P., Taghon T., Vandekerckhove B., et al. 2016a. The Ly49E receptor inhibits the immune control of acute Trypanosoma cruzi infection. Front. Immunol. 7:472. 10.3389/fimmu.2016.00472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filtjens, J., Foquet L., Taveirne S., Van Ammel E., Vanhees M., Van Acker A., Kerre T., Taghon T., Vandekerckhove B., Plum J., et al. 2014. Contribution of the Ly49E natural killer receptor in the immune response to Plasmodium berghei infection and control of hepatic parasite development. PLoS One. 9:e87463. 10.1371/journal.pone.0087463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filtjens, J., Keirsse J., Van Ammel E., Taveirne S., Van Acker A., Kerre T., Taghon T., Vandekerckhove B., Plum J., Van Ginderachter J.A., and Leclercq G.. 2016b. Expression of the inhibitory Ly49E receptor is not critically involved in the immune response against cutaneous, pulmonary or liver tumours. Sci. Rep. 6:30564. 10.1038/srep30564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filtjens, J., Taveirne S., Van Acker A., Van Ammel E., Vanhees M., Kerre T., Taghon T., Vandekerckhove B., Plum J., and Leclercq G.. 2013. Abundant stage-dependent Ly49E expression by liver NK cells is not essential for their differentiation and function. J. Leukoc. Biol. 93:699–711. 10.1189/jlb.0812378 [DOI] [PubMed] [Google Scholar]
- Friedrich, C., Taggenbrock R.L.R.E., Doucet-Ladeveze R., Golda G., Moenius R., Arampatzi P., Kragten N.A.M., Kreymborg K., Gomez de Agüero M., Kastenmüller W., et al. 2021. Effector differentiation downstream of lineage commitment in ILC1s is driven by Hobit across tissues. Nat. Immunol. 22:1256–1267. 10.1038/s41590-021-01013-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit, R., Mandal P.K., Ebina W., Ben-Zvi A., Nombela-Arrieta C., Silberstein L.E., and Rossi D.J.. 2014. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J. Exp. Med. 211:1315–1331. 10.1084/jem.20130428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillard, G.O., Bivas-Benita M., Hovav A.H., Grandpre L.E., Panas M.W., Seaman M.S., Haynes B.F., and Letvin N.L.. 2011. Thy1+ NK [corrected] cells from vaccinia virus-primed mice confer protection against vaccinia virus challenge in the absence of adaptive lymphocytes. PLoS Pathog. 7:e1002141. 10.1371/journal.ppat.1002141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, S.M., Chaix J., Rupp L.J., Wu J., Madera S., Sun J.C., Lindsten T., and Reiner S.L.. 2012. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 36:55–67. 10.1016/j.immuni.2011.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman, W.J., Verbsky J.W., Barchet W., Colonna M., Atkinson J.P., and Ley T.J.. 2004. Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity. 21:589–601. 10.1016/j.immuni.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Guo, X.-z.J., Dash P., Crawford J.C., Allen E.K., Zamora A.E., Boyd D.F., Duan S., Bajracharya R., Awad W.A., Apiwattanakul N., et al. 2018. Lung γδ T cells mediate protective responses during neonatal influenza infection that are associated with type 2 immunity. Immunity. 49:531–544.e6. 10.1016/j.immuni.2018.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y.-M., Li X., Perego M., Nefedova Y., Kossenkov A.V., Jensen E.A., Kagan V., Liu Y.-F., Fu S.-Y., Ye Q.-J., et al. 2018. Transitory presence of myeloid-derived suppressor cells in neonates is critical for control of inflammation. Nat. Med. 24:224–231. 10.1038/nm.4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., and Littman D.R.. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Ivarsson, M.A., Loh L., Marquardt N., Kekäläinen E., Berglin L., Björkström N.K., Westgren M., Nixon D.F., and Michaëlsson J.. 2013. Differentiation and functional regulation of human fetal NK cells. J. Clin. Invest. 123:3889–3901. 10.1172/JCI68989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech, S.M., Tan J.T., Wherry E.J., Konieczny B.T., Surh C.D., and Ahmed R.. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191–1198. 10.1038/ni1009 [DOI] [PubMed] [Google Scholar]
- Kayagaki, N., Yamaguchi N., Nakayama M., Takeda K., Akiba H., Tsutsui H., Okamura H., Nakanishi K., Okumura K., and Yagita H.. 1999. Expression and function of TNF-related apoptosis-inducing ligand on murine activated NK cells. J. Immunol. 163:1906–1913 [PubMed] [Google Scholar]
- Kharchenko, P.V. 2021. The triumphs and limitations of computational methods for scRNA-seq. Nat. Methods. 18:723–732. 10.1038/s41592-021-01171-x [DOI] [PubMed] [Google Scholar]
- Lanier, L.L. 2008. Evolutionary struggles between NK cells and viruses. Nat. Rev. Immunol. 8:259–268. 10.1038/nri2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine, K.J., Epelman S., Uchida K., Weber K.J., Nichols C.G., Schilling J.D., Ornitz D.M., Randolph G.J., and Mann D.L.. 2014. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc. Natl. Acad. Sci. USA. 111:16029–16034. 10.1073/pnas.1406508111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic, V., Glimcher L.H., and Lord G.M.. 2013. T-bet: A bridge between innate and adaptive immunity. Nat. Rev. Immunol. 13:777–789. 10.1038/nri3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland, A.P., Yalin A., Wang S.Y., Cortez V.S., Landsberger T., Sudan R., Peng V., Miller H.L., Ricci B., David E., et al. 2021. Multi-tissue single-cell analysis deconstructs the complex programs of mouse natural killer and type 1 innate lymphoid cells in tissues and circulation. Immunity. 54:1320–1337.e4. 10.1016/j.immuni.2021.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, K.B., Salazar-Mather T.P., Dalod M.Y., Van Deusen J.B., Wei X.Q., Liew F.Y., Caligiuri M.A., Durbin J.E., and Biron C.A.. 2002. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J. Immunol. 169:4279–4287. 10.4049/jimmunol.169.8.4279 [DOI] [PubMed] [Google Scholar]
- O’Sullivan, T.E., Rapp M., Fan X., Weizman O.-E., Bhardwaj P., Adams N.M., Walzer T., Dannenberg A.J., and Sun J.C.. 2016. Adipose-resident group 1 innate lymphoid cells promote obesity-associated insulin resistance. Immunity. 45:428–441. 10.1016/j.immuni.2016.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak-Wittel, M.A., Yang L., Schreiber R.D., Piersma S.J., Yokoyama W.M., and Parikh B.A.. 2015. Dual requirement of cytokine and activation receptor triggering for cytotoxic control of murine cytomegalovirus by NK cells. PLoS Pathog. 11:e1005323. 10.1371/journal.ppat.1005323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, D., Han T., Tang S., Xu W., Bao Q., Sun Y., Xuan B., and Qian Z.. 2018. Murine cytomegalovirus protein pM91 interacts with pM79 and is critical for viral late gene expression. J. Virol. 92:e00675-18. 10.1128/JVI.00675-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalexi, E., and Satija R.. 2018. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 18:35–45. 10.1038/nri.2017.76 [DOI] [PubMed] [Google Scholar]
- Park, E., Patel S., Wang Q., Andhey P., Zaitsev K., Porter S., Hershey M., Bern M., Plougastel-Douglas B., Collins P., et al. 2019. Toxoplasma gondii infection drives conversion of NK cells into ILC1-like cells. eLife. 8:e47605. 10.7554/eLife.47605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paust, S., Gill H.S., Wang B.Z., Flynn M.P., Moseman E.A., Senman B., Szczepanik M., Telenti A., Askenase P.W., Compans R.W., and Von Andrian U.H.. 2010. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat. Immunol. 11:1127–1135. 10.1038/ni.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, H., Jiang X., Chen Y., Sojka D.K., Wei H., Gao X., Sun R., Yokoyama W.M., and Tian Z.. 2013. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Invest. 123:1444–1456. 10.1172/JCI66381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchet, T., Petit M., Banchi E.G., Meunier S., Cumano A., and Golub R.. 2018. The notch signaling pathway is balancing type 1 innate lymphoid cell immune functions. Front. Immunol. 9:1252. 10.3389/fimmu.2018.01252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu, D.M., Botting R.A., Stephenson E., Green K., Webb S., Jardine L., Calderbank E.F., Polanski K., Goh I., Efremova M., et al. 2019. Decoding human fetal liver haematopoiesis. Nature. 574:365–371. 10.1038/s41586-019s4151652-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera, L., Gariglio M., Valente G., Mullbacher A., Museteanu C., Landolfo S., and Simon M.M.. 2000. Murine cytomegalovirus replication in salivary glands is controlled by both perforin and granzymes during acute infection. Eur. J. Immunol. 30:1350–1355. [DOI] [PubMed] [Google Scholar]
- Riggan, L., Freud A.G., and O’Sullivan T.E.. 2019. True detective: Unraveling group 1 innate lymphocyte heterogeneity. Trends Immunol. 40:909–921. 10.1016/j.it.2019.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette, M.L., Fuchs A., Cortez V.S., Lee J.S., Wang Y., Durum S.K., Gilfillan S., and Colonna M.; Immunological Genome Consortium . 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16:306–317. 10.1038/ni.3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawen, P., Eldeeb M., Erlandsson E., Kristiansen T.A., Laterza C., Kokaia Z., Karlsson G., Yuan J., Soneji S., Mandal P.K., et al. 2018. Murine HSCs contribute actively to native hematopoiesis but with reduced differentiation capacity upon aging. eLife. 7:e41258. 10.7554/eLife.41258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiss, M.R. 2013. Cytomegalovirus in the neonate: Immune correlates of infection and protection. Clin. Dev. Immunol. 2013:501801. 10.1155/2013/501801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C., Lee J., Koga S., Ricardo-Gonzalez R.R., Nussbaum J.C., Smith L.K., Villeda S.A., Liang H.-E., and Locksley R.M.. 2019. Tissue-resident group 2 innate lymphoid cells differentiate by layered ontogeny and in situ perinatal priming. Immunity. 50:1425–1438.e5. 10.1016/j.immuni.2019.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.R.C., Heusel J.W., Mehta I.K., Kim S., Dorner B.G., Naidenko O.V., Iizuka K., Furukawa H., Beckman D.L., Pingel J.T., et al. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA. 99:8826–8831. 10.1073/pnas.092258599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N.L., Patel R.K., Reynaldi A., Grenier J.K., Wang J., Watson N.B., Nzingha K., Yee Mon K.J., Peng S.A., Grimson A., et al. 2018. Developmental origin governs CD8+ T cell fate decisions during infection. Cell. 174:117–130.e14. 10.1016/j.cell.2018.05.029 [DOI] [PubMed] [Google Scholar]
- Sojka, D.K., Plougastel-Douglas B., Yang L., Pak-Wittel M.A., Artyomov M.N., Ivanova Y., Zhong C., Chase J.M., Rothman P.B., Yu J., et al. 2014. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 3:e01659. 10.7554/elife.01659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits, H., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. 2013. Innate lymphoid cells—a proposal for uniform nomenclature. Nat. Rev. Immunol. 13:145–149. 10.1038/nri3365 [DOI] [PubMed] [Google Scholar]
- Stevenaert, F., Van Beneden K., De Creus A., Debacker V., Plum J., and Leclercq G.. 2003. Ly49E expression points toward overlapping, but distinct, natural killer (NK) cell differentiation kinetics and potential of fetal versus adult lymphoid progenitors. J. Leukoc. Biol. 73:731–738. 10.1189/jlb.0902443 [DOI] [PubMed] [Google Scholar]
- Sun, J.C., Beilke J.N., and Lanier L.L.. 2009. Adaptive immune features of natural killer cells. Nature. 457:557–561. 10.1038/nature07665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, E.C., and Schleiss M.R.. 2013. Congenital cytomegalovirus infection: new prospects for prevention and therapy. Pediatr. Clin. N. Am. 60:335–349. 10.1016/j.pcl.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, K., Cretney E., Hayakawa Y., Ota T., Akiba H., Ogasawara K., Yagita H., Kinoshita K., Okumura K., and Smyth M.J.. 2005. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood. 105:2082–2089. 10.1182/blood-2004blo08-3262 [DOI] [PubMed] [Google Scholar]
- Tang, L., Peng H., Zhou J., Chen Y., Wei H., Sun R., Yokoyama W.M., and Tian Z.. 2016. Differential phenotypic and functional properties of liver-resident NK cells and mucosal ILC1s. J. Autoimmun. 67:29–35. 10.1016/j.jaut.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Van Acker, A., Gronke K., Biswas A., Martens L., Saeys Y., Filtjens J., Taveirne S., Van Ammel E., Kerre T., Matthys P., et al. 2017. A murine intestinal intraepithelial NKp46-negative innate lymphoid cell population characterized by group 1 properties. Cell Rep. 19:1431–1443. 10.1016/j.celrep.2017.04.068 [DOI] [PubMed] [Google Scholar]
- Van Acker, A., Louagie E., Filtjens J., Taveirne S., Van Ammel E., Kerre T., Elewaut D., Taghon T., Vandekerckhove B., Plum J., and Leclercq G.. 2016. The role of Ly49E receptor expression on murine intraepithelial lymphocytes in intestinal cancer development and progression. Cancer Immunol. Immunother. 65:1365–1375. 10.1007/s00262-016s0021894-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beneden, K., Stevenaert F., De Creus A., Debacker V., De Boever J., Plum J., and Leclercq G.. 2001. Expression of Ly49E and CD94/NKG2 on fetal and adult NK cells. J. Immunol. 166:4302–4311. 10.4049/jimmunol.166.7.4302 [DOI] [PubMed] [Google Scholar]
- Van Den Broeck, T., Stevenaert F., Taveirne S., Debacker V., Vangestel C., Vandekerckhove B., Taghon T., Matthys P., Plum J., Held W., et al. 2008. Ly49E-dependent inhibition of natural killer cells by urokinase plasminogen activator. Blood. 112:5046–5051. 10.1182/blood-2008-06-164350 [DOI] [PubMed] [Google Scholar]
- Vivier, E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. 2018. Innate lymphoid cells: 10 years on. Cell. 174:1054–1066. 10.1016/j.cell.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Wang, J., Sun R., Wei H., Dong Z., Gao B., and Tian Z.. 2006. Poly I:C prevents T cell-mediated hepatitis via an NK-dependent mechanism. J. Hepatol. 44:446–454. 10.1016/j.jhep.2005.08.015 [DOI] [PubMed] [Google Scholar]
- Wang, X., Peng H., Cong J., Wang X., Lian Z., Wei H., Sun R., and Tian Z.. 2018. Memory formation and long-term maintenance of IL-7Rα+ ILC1s via a lymph node-liver axis. Nat. Commun. 9:4854. 10.1038/s41467-018-07405-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weizman, O.-E., Adams N.M., Schuster I. S., Krishna C., Pritykin Y., Lau C., Degli-Esposti M.A., Leslie C.S., Sun J.C., and O’Sullivan T.E.. 2017. ILC1 confer early host protection at initial sites of viral infection. Cell. 171:795–808.e12. 10.1016/j.cell.2017.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weizman, O.-E., Song E., Adams N.M., Hildreth A.D., Riggan L., Krishna C., Aguilar O.A., Leslie C.S., Carlyle J.R., Sun J.C., and O'Sullivan T.E.. 2019. Mouse cytomegalovirus-experienced ILC1s acquire a memory response dependent on the viral glycoprotein m12. Nat. Immunol. 20:1004–1011. 10.1038/s41590-019s4150430-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Peng H., Li K., Qu K., Wang B., Wu Y., Ye L., Dong Z., Wei H., and Tian Z.. 2019. Liver-resident NK cells control antiviral activity of hepatic T cells via the PD-1-PD-L1 axis. Immunity. 50:403–417.e4. 10.1016/j.immuni.2018.12.024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists antibodies and fluorochromes.
Data Availability Statement
Murine bulk RNA-seq and scRNA-seq data are deposited under GEO accession number GSE171851. All other data needed to evaluate the conclusions in the paper are present in the paper or the online supplemental material.