Abstract
Molecular mechanisms of the non-structural protein 1 (NS1) in influenza A-induced pathological changes remain ambiguous. This study explored the pathogenesis of human infection by influenza A viruses (IAVs) through identifying human genes with codon usage bias (CUB) similar to NS1 gene of these viruses based on the relative synonymous codon usage (RSCU). CUB of the IAV subtypes H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2 was analyzed and the correlation of RSCU values of NS1 sequences with those of the human genes was calculated. The CUB of NS1 was uneven and codons ending with A/U were preferred. The ENC-GC3 and neutrality plots suggested natural selection as the main determinant for CUB. The RCDI, CAI and SiD values showed that the viruses had a high degree of adaptability to human. A total of 2155 human genes showed significant RSCU-based correlation (p < 0.05 and r > 0.5) with NS1 coding sequences and was considered as human genes with CUB similar to NS1 gene of IAV subtypes. Differences and similarities in the subtype-specific human protein–protein interaction (PPI) networks and their functions were recorded among IAVs subtypes, indicating that NS1 of each IAV subtype has a specific pathogenic mechanism. Processes and pathways involved in influenza, transcription, immune response and cell cycle were enriched in human gene sets retrieved based on the CUB of NS1 gene of IAV subtypes. The present work may advance our understanding on the mechanism of NS1 in human infections of IAV subtypes and shed light on the therapeutic options.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10709-022-00155-9.
Keywords: Influenza A virus, Codon usage bias, Nonstructural protein 1, Adaptation to host, Relative synonymous codon usage, NS1 human interactome
Introduction
Influenza A viruses (IAVs) are infectious agents that cause many epidemics and seasonal influenza pandemics (Chen et al. 2018; Zhou et al. 2015). The natural reservoirs of IAVs are aquatic wild birds and mammalians such as pigs, horses, swine (Koutsakos et al. 2019; Slusher et al. 2014; Urbaniak et al. 2014). Apart from H18N11 and H17N10, almost all subtypes of IAVs are grouped in avian influenza viruses (AIVs). The H1N1, H2N2 and H3N2 subtypes of IAVs are human-adapted subtypes due to their ability to α-2,6 sialylated glycan receptors in human epithelial cells in the upper respiratory tract (Cauldwell et al. 2014; Ge and Wang 2011; Imai et al. 2012; Jayaraman et al. 2011; Long et al. 2019; Mair et al. 2014; Makarova et al. 1999; Schäfer et al. 1993; Smith et al. 2009; Viswanathan et al. 2010). Since 1968, the H2N2 has stopped circulating in human but, like other H2 subtypes, continues to circulate in birds and sporadically in pigs. The H5N1, H7N7, H7N2, H7N3 and H9N2 subtypes, circulating in domestic poultry, are not adapted to human but have been reported to cause infections without sustained interhuman transmissions (Alonso and Diambra 2020). The H5, H6, H7, H9 and H10 AIV subtypes have crossed the species barrier to infect humans via infected poultry (Kessler et al. 2021) via acquiring the ability to infect human due mutations in hemagglutinin leading to increased affinity for α-2,6 sialylated glycan receptors in human (Mair et al. 2014). (Kessler et al. 2021)The IAV subtypes that have caused pandemics in humans are H1N1, H2N2, H3N2, H5N1, H7N9, H7N7 and H9N2, in descending order of the number of pandemic deaths in human (Koutsakos et al. 2019; Long et al. 2019; Philippon et al. 2020; Song and Qin 2020; Sutton 2018; Tanner et al. 2015; Wu et al. 2020; Yamaji et al. 2020). The IAV subtypes are potential causal agents of pandemic in the human population in the future. However, (Gao et al. 2013; Koopmans et al. 2004; Marinova-Petkova et al. 2017; Meineke et al. 2019; Skowronski et al. 2006; Song and Qin 2020; Terebuh et al. 2018; Tong et al. 2018; Tweed et al. 2004; Um et al. 2021; Westenius et al. 2018) current management of human influenza A is limited to vaccines and antiviral treatments targeting viral proteins (Uyeki et al. 2019). Due to their rapid mutation capacity, IAVs have a strong evasion mechanism to the host immune system (Chang et al. 2020; Huang et al. 2021; Li et al. 2012; Peacock et al. 2017; Peng et al. 2014; Sitaras et al. 2014), thus reducing the effectiveness of antivirals targeting viral proteins. To counteract these problems, the development of drugs targeting the virus-host interactions that promote IAV infection and replication is necessary. Thus, analyzing the implications of virulence factors on host processes such as gene expression is crucial.
Among the promising targets of the treatment and prevention of influenza, the non-structural protein 1 (NS1) is a multifaceted protein majorly known for its role in evasion of the host immune system and RNA binding and is considered to be the main viral weapon for combating the innate immune response and promoting viral multiplication in the hostile environment of the host (Han et al. 2019; Zhang et al. 2019). NS1 is a key regulator in the replication and pathogenesis of avian influenza (Rosário-Ferreira et al. 2020). Studies have shown that NS1 interacts with a variety of host proteins to participate in viral replication, gene transcription regulation and induction of cell pathological changes, etc. (Nemeroff et al. 1998; Nogales et al. 2018). In host cells, NS1 of H3N2 can interact with nucleoli to regulate the stress related to cell cycle progression based on the rRNA gene promoter (Yan et al. 2017). Moreover, the NS1 protein of IAV subtype H5N1 interacts with the heat shock protein Hsp90 and is involved in the apoptosis induced by IAV via caspase cascade (Zhang et al. 2011). It was also reported that mammalian adaptation of avian influenza A virus implicates gradual shift in NS1 and that interplay among immune evasion and virus evolution is fundamental in IAV mammalian adaptation (Chauché et al. 2018).
The uneven usage of the synonymous codons encoding the same amino acids is also called codon usage bias (CUB), which is widespread in biology (Deng et al. 2020). CUB is mainly determined by codon in the third position of nucleic acid and CUB analysis has been widely used in virus genome studies and contributes to the understanding of the shifts of the evolution and genetics in the virus (Cutter et al. 2006). Liu et al. (2011) found that the synonymous codon usage pattern is a mixture of coincidence and antagonism to that of the host cells. Viruses tend to use different codon usage patterns to improve overall virus survival and evade the host immune system (Moratorio et al. 2013). Previous studies indicated that similarities of relative synonymous codon usage (RSCU) of genes between pathogens and their host can impact gene expression in the host and that the pathogens depend on their hosts for their multiplication (Alonso and Diambra 2020; Jitobaom et al. 2020; Kula et al. 2018; Michely et al. 2013; Miller et al. 2017; Nambou and Anakpa 2020; Pepin et al. 2008). Recently, codon usage similarities between viral genes and human genes has been demonstrated and indicated that the infection (viral proteins engaging the host cell proteins) may induce pathological effects as a result of the expression and translation of RNA transcripts and the effects of proteins from these transcripts (Maldonado et al. 2021). Translational adaptation of human viruses to the tissues they infect has been also demonstrated and indicated that viruses adapt their codon usage toward their tropism and hijack the host defense machinery (Hernandez-Alias et al. 2021). Based on this evolution of molecular compatibility among the pathogens and their hosts, we can retrieve key information for infectious diseases, especially the translation effects more likely affected by the evolution of CUB. Thus, the co-evolution analysis based on CUB similarities of viral genes and host genes represents a way to identify host genes that may have transcripts dysregulated in response to infection. Previously, based on RSCU similarity analysis, we identified key genes involved in COVID-19 and predicted potential therapeutics for this disease (Nambou and Anakpa 2020). Proceeding so will open new avenues for understanding the pathogenesis of human infection by the IAV subtypes; however, there is no report of such analysis regarding IAV. Thus, analyzing the similarity between RSCU values of virulence genes of IAV and human gene RSCU values is potential in providing novel significant information on the pathogenesis of IAV. Because of its multifarious implications in the infectious pathogenesis, revealing the CUB of the NS1 protein will help understand the evolution and adaptability of IAVs to human as well as cellular and molecular changes induced by these viruses in human (Luo et al. 2020). However, few studies have reported the codon usage pattern of the NS1 protein of the IAV subtypes and the genome-wide human genes with CUB similar to that of this viral molecule has yet to be exhaustively scrutinized.
In this study, we downloaded NS1 coding sequences of the IAV subtypes H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2 from NCBI. The codon usage was studied to explore the adaptability and CUB of NS1 coding sequences between these IAV subtypes and human, providing a basis for a better understanding of the evolution and human adaptability of IAV subtypes. Further, RSCU-based correlation analysis allowed the identification of the human genes with CUB similar to that of NS1 gene and their functional roles.
Materials and methods
Sequence data
The complete human genome was obtained from GENCODE (https://www.gencodegenes.org/human/). The nucleotides sequences of IAV NS1 protein sequences were downloaded from the NCBI database after searching with specific keywords. After manual curation, only NS1 protein sequences were included. The IAV types included were those having been reported to cause human infections with number of NS1 sequences available higher than 1000. Thus, H1N1 (45,217 NS1 sequences), H3N2 (52,887 NS1 sequences), H3N8 (4553 NS1 sequences), H5N1 (7486 NS1 sequences), H5N2 (2894 NS1 sequences), H5N8 (1958 NS1 sequences), H7N9 (3957 NS1 sequences) and H9N2 (9375 NS1 sequences) were finally included in this study.
Relative synonymous codon usage (RSCU) analysis
According to the equation by Sharp and Li (1986), we calculated the relative synonymous codon usage (RSCU) values among different codons of IAV nonstructural protein 1 and human gene sequences using the R packages “vhcub” (Anwar et al. 2019). RSCU refers to the probability for a codon to be used between synonymous codons of corresponding amino acids. When RSCU < 1, it means that the codon is used less; conversely, when RSCU > 1, it means that the codon is used more. When RSCU = 1, there is no bias in the use of the codon, with RSCU value of > 1.6 and < 0.6 representing "overrepresented" and "underrepresented" codons, respectively. For calculation of RSCU, please refer to the work by He et al. (2019).
Principal component analysis (PCA)
PCA is a common multivariate statistical method, which can transform the dimension of multiple indicators into several comprehensive indicators. In our study, PCA was performed and visualized using the R packages “ade4”, “ggplot2” and “ggrepel” for analyzing the codon usage of NS1 sequences in different IAV subtypes based on the RSCU values.
Effective number of codons (ENC) analysis
ENC reflects the degree of preference for the unbalanced use of synonymous codons in the codon family. ENC was used to assess the CUB of the IAV NS1 coding sequences using the R package “vhcub” (Anwar et al. 2019). The codon with high expression usually has high codon bias and relatively small ENC. The gene codon with low expression has low codon bias and relatively high ENC. Therefore, ENC can be used to measure the expression level of endogenous genes. The ENC measures CUB for a gene, ranging from 20 (high CUB; a gene uses only one codon) to 61 (no bias; synonymous codons are used equally).
ENC-plot analysis
ENC-plot is based on ENC as the abscissa and GC3 (GC content of the third codon) as the ordinate of a scatter diagram, and can be used to evaluate the role of mutation pressure in codon bias. If mutation pressure only acts on the choice of codon bias, the points in the graph will appear on the top of the expected ENC curve. On the contrary, there may be other factors, such as natural selection pressure, involved. ENC-plot was performed using the R package “vhcub” (Anwar et al. 2019).
Parity rule 2 (PR2) analysis
The PR2 plot was used to explore the effects of mutation pressure and natural selection pressure on the usage of a single gene codon. The figure was drawn with A3/(A3 + U3) as the vertical coordinate and G3/(G3 + C3) as the horizontal coordinate. The central point of the figure is where A = U (0.5) and C = G (0.5), representing the equal role of mutation pressure and natural selection pressure. PR2 plot was performed using the R package “vhcub” (Anwar et al. 2019).
Codon adaptation index (CAI) analysis
The codon adaptation index (CAI) is a parameter, which is used to measure the relative adaptation of viruses and their potential hosts. CAI values range from 0 to 1. A sequence with high CAI is believed to have a strong adaptability to its hosts. CAI analysis was performed by R package “vhcub” (Anwar et al. 2019).
Neutrality analysis
Neutrality plot (GC12 values against GC3 values) was used to evaluate the effects of mutation pressure and natural selection on codon usage. The slope of the regression curve between GC12 and GC3 content represents the mutation pressure (Jia et al. 2015; Sueoka 1988). When the slope = 1.0, there is no natural selection pressure or the natural selection pressure is weak (Jia et al. 2015; Sueoka 1988). On the contrary, when the regression curve deviates from the diagonal, natural selection has a significant impact on the usage of codons (Jia et al. 2015; Sueoka 1988). The GC12 was calculated as the average of GC1 and GC2 from each sequence. The visualization of the neutrality plots was performed using the GraphPad Prism software (GraphPad Software, San Diego, California USA, www.graphpad.com).
Relative codon deoptimization index (RCDI) analysis
The relative codon deoptimization index (RCDI) value is a parameter of codon deoptimization to measure the similarity of codons used by a given gene over the reference genome. The higher genetic similarity between the virus and the host is, the higher the translation ratio is, and the closer the RCDI value is to 1 (Mueller et al. 2006). The lower RCDI value indicates higher adaptability between the virus and the host. However, high RCDI may indicate that some genes are expressed during the latency phase or show low replication levels. RCDI analysis was performed with the “vhcub” (Anwar et al. 2019) library in R.
Similarity index
SiD was used to measure the effect of host CUB on IAV NS1. The value of SiD is between 0 and 1. Higher SiD values indicates that the host has a decisive influence on codon usage. SiD value was calculated using the R package “vhcub” (Anwar et al. 2019).
Synonymous codon usage order (SCUO) analysis
The SCUO (0 ⩽ SCUO ⩽ 1) is the synonymous CUB for the whole sequence. A higher SCUO indicates a higher CUB in the sequence. SCUO was analyzed using the R package “vhcub” (Anwar et al. 2019).
Identification of human genes with CUB similar to NS1 CUB
To disclose human genes probably affected by the transcription of NS1, we calculated the Pearson correlation between the human genes and the NS1 sequences from the IAV subtypes based on the RSCU values. The correlation analysis was performed using the Hmisc library in R. The human genes which were correlated with NS1 sequences with |r|> 0.5 and p < 0.05 were considered as those potentially affected by the NS1 transcription.
Protein–protein interaction network of human genes with CUB similar to that of NS1 gene of IAV subtypes
To construct the protein–protein interaction (PPI) network of the human interactome of NS1 gene of the IAV subtypes, we selected the human genes correlated with IAV NS1 from each subtype as the input list for analysis in Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/). The basic settings of analysis chosen were: full network for the network type; confidence for the meaning of network edges; the active interaction sources included textmining, experiments, databases, co-expression, neighborhood, gene fusion and co-occurrence; and the minimum required interaction score at high confidence was 0.7. The obtained PPI networks were exported for visualization in Cytoscape 3.4.0 software (Cytoscape Consortium, SanDiego, CA, USA) and the MCODE plugin was used for the extraction of PPI clusters with hub genes based on the following settings: degree cut-off ≥ 2, node score cut-off ≥ 0.2, K-core ≥ 2, and max depth = 100.
Functional enrichment analysis
The functional analysis of the genes was performed using the TCGAanalyze_EAcomplete function in the TCGAbiolinks library in R. The enrichment terms with adjusted p value of < 0.05 were retained as significantly enriched terms. The visualization was performed with the same library using the function TCGAvisualize_EAbarplot.
Statistical analysis
The GraphPad Prism version 9.0.0 for Windows (GraphPad Software, San Diego, California USA, www.graphpad.com) was used for statistical processing and the one-way ANOVA followed by Tukeys multiple comparison posttest was used to compare the differences in ENC, RCDI, and CAI values between IAV subtypes. The adjusted p-values < 0.05 indicated statistically significant differences among subtypes. The p-values among groups were represented by an inset network on the figures.
Results
A- and U-ending codons are preferred in NS1 coding sequences of IAV subtypes
As shown in Fig. 1, the results of RSCU analysis were used to estimate the codon usage patterns of NS1 coding sequences from different subtypes of IAV. The NS1 coding sequences tended to use codons with A-/U-ending. For H1N1, 27 preferred codons among which 19 had A-/U-ending and9 were highly used with A-U-ending were identified. For H3N2, 28 preferred codons were identified among which 22 had A-/U-ending and 12 were highly used with 10 A-/U-ending. For H3N8, 27 codons were preferred, among which 19 had A-/U-ending and 10 were highly used with 9 of them having A-U-ending codons. For H5N1, 27 preferred codons, with 16 of them having A-/U-ending were identified; 13 of them were highly used and 9 had A-/U-ending. For H5N2, 28 preferred codons among which 19 had A-/U-endingand 10 were highly used with 9 having A-U-ending were identified. For H5N8, 29 preferred codons among which 19 had A-/U-ending and 10 were highly used were identified. For H7N9, 26 preferred codons among which 21 had A-/U-ending and 12 (10 codons with A-/U-ending) were highly used were identified. For H7N9, 27 codons were preferred, and among them, 20 codons had A-/U-endingwhile 11 were highly used codons.
Fig. 1.
The relative synonymous codon usage (RSCU) values of the NS1 coding sequences of IAV subtypes
ENC of the NS1 coding sequences of IAV subtypes
ENC was used to describe the CUB of NS1 coding sequences of IAV subtypes. A smaller ENC value indicates a stronger CUB. The boxplot showing the ENC values of NS1 coding sequences of IAV subtypes was depicted in Fig. 2. Statistical analysis indicated significant differences among the mean ENC values of pairwise-compared IAV subtypes (Additional File S1). The mean ENC value of NS1 sequences of H1N1 (56.07 ± 2.11) was the highest, followed by those of H3N2 (52.01 ± 1.54), H3N8 (51.56 ± 1.27), H5N2 (51.34 ± 1.97), H5N8 (51.02 ± 1.79), H5N1 (49.98 ± 2.35), H9N2 (49.61 ± 2.11) and H7N9 (48.81 ± 2.23). These results indicated that the CUB of NS1 from H7N9 was the highest, followed by H7N9, H5N1, H5N8, H5N2, H3N8, H3N2 and H1N1, in this order.
Fig. 2.

The effective number of codons (ENC) values of NS1 of IAV subtypes. The egdes of the inset network indicates the distribution of adjusted p-values among the compared node groups of IAV subtypes
Trends in codon usage variations
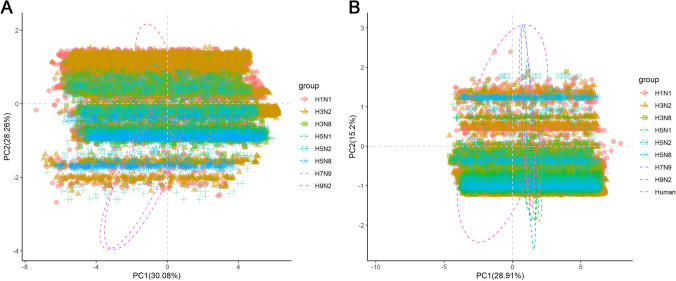
PCA, a common multivariate statistical method, is used to estimate the relationship between samples and variables. In this study, PCA was used to evaluate the usage of synonymous codons in NS1 coding sequences of IAV subtypes. The first two principal components of the NS1 coding sequences accounted for 30.08% and 28.26%, respectively (Fig. 3A). These values reflected that the first two principal components are the main influencing factors for the codon usage of NS1 coding sequences of IAV subtypes. In addition, PCA was clustered into two categories based on NS1 coding sequences of IAV subtypes, among which some individual points were overlapped, suggesting that these points may have the same codon usage trend. Moreover, Fig. 3B demonstrated that the points of NS1 coding sequences of IAV subtypes were covered by the point of gene sequences from Homo sapiens, indicating that NS1 coding sequences of these viruses may share similar codon usage patterns with their human host. The individual overlap of each virus with H. sapiens was as depicted in Supplementary Figure S1.
Fig. 3.
Principal component analysis (PCA) for NS1 of IAV subtypes based on the RSCU values. A PCA based on the RSCU values of NS1 of IAV subtypes; B PCA based on the RSCU values of NS1 of IAV subtypes and RSCU values of all genes of H. sapiens
ENC-plot analysis
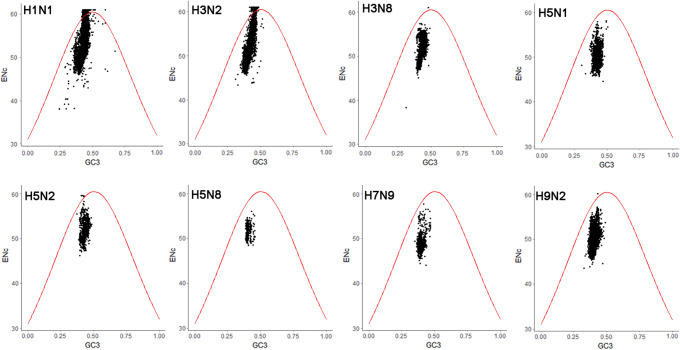
ENC-plots (ENC vs GC3) were generated to explore factors that affect the codon usage of the NS1 coding sequences of IAV subtypes. If codon bias is only affected by natural selection pressure, all points fall below the standard curve. However, points above the standard curve indicate that codon usage is affected by mutation pressure. The points representing the ENC values of NS1 coding sequences of IAV subtypes H7N9, H5N8, and H5N1 were exclusively below the expected ENC curve, suggesting that natural selection pressure is the main factor affecting the codon usage of NS1 in these viruses (Fig. 4). For H5N2, H3N8, H3N2, H9N2 and H1N1, the points were on both sides of the expected ENC curve (Fig. 4), demonstrating that the NS1 coding sequences of H5N2, H3N8, H3N2, H9N2 and H1N1 may be affected by both natural selection and mutation pressure.
Fig. 4.
ENC-plot analysis of NS1 of IAV subtypes. ENC-plot analysis of the NS1, with ENC, plotted against GC3s of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9, and H9N2 was generated. The red curve represents the expected ENC curve when the CUB is determined by the GC3s composition only. The black dots represent the distribution of ENC values of NS1of AIV subtypes
Neutrality plot
In order to determine the levels of natural selection pressure and mutation pressure in codon usage of NS1 coding sequences of IAV subtypes, we performed neutrality plot analysis. As shown in Fig. 5A, Pearson correlation analysis indicated significant negative correlations between the GC12 and the GC3 values of NS1 coding sequences in the IAV subtypes H1N1 (r = -0.28, p = 0.00), H5N2 (r = -0.24, p = 3.22 ) and H5N8 (r = -0.08, p = 0.008). On the contrary significant positive correlations were recorded for H3N2 (r = 0.30, p = 0.00), H3N8 (0.12, p = 3.18), H5N1 (r = 0.05, p = 0.004), H7N9 (r = 0.51, 4.48) and H9N2 (0.21, 1.60). Moreover, as shown in Fig. 5B, the slopes of simple linear regression analysis were -0.11, 0.15, 0.09, 0.03, -0.19, -0.04, 0.27 and 1.19 for H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2, respectively. These results indicated that the levels of natural selection in the codon usage of NS1 coding sequences in H1N1, H3N2, H3N8, H5N1, H5N2, H5N8 and H7N9 were respectively 89%, 85%, 91%, 97%, 81%, 96% and 73%. For H9N2, the slope was higher than 1, indicating exclusive neutrality. These results indicated that natural selection was the main factor affecting the codon usage of NS1 coding sequences in H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2 while mutation pressure was a minor influencing factor.
Fig. 5.
Neutrality plot analysis of GC3s against GC12s for all the coding sequences based on NS1 of IAV subtypes. A Pearson correlation analysis between the GC3s and GC12s of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9, and H9N2. B Neutrality plots of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9, and H9N2
Parity analysis
PR2 bias plot was used to analyze whether CUB exists in NS1 coding sequences of IAV subtypes. When there is no deviation from natural selection and mutation, the points fall in the middle of the graph, and the coordinates of this middle point is (0.5, 0.5). Figure 6 showed that most of the points fall in the upper right quadrant of the PR2 plots from each of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2 viruses, indicating that A3s were more than U3s while G3s were more than C3s, which demonstrated that the codon usage pattern of NS1 in the IAV subtypes was affected by mutation and natural selection pressure.
Fig. 6.
The PR2 plots of NS1 of IAV subtypes. The AT [A3%/(A3% + T3%)] and GC [G3%/ (G3% + C3%)] bias of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9, and H9N2 were plotted. The center of the plot (both the coordinates is 0.5) indicates a position where there is no bias
Codon usage adaptation of NS1 of the subtypes of IAV to human
In order to explore the adaptability and codon usage optimization of NS1 of the IAV subtypes and their human host, we calculated CAI, RCDI and SiD to analyze the adaptability of codons. The CAI values of NS1 in the IAV subtypes were 0.57 ± 0.015, 0.59 ± 0.011, 0.58 ± 0.015, 0.6 ± 0.024, 0.58 ± 0.012, 0.59 ± 0.011, 0.59 ± 0.012, 0.59 ± 0.014 for H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2, respectively (Fig. 7A). Significant differences in CAI were found among the pairwise comparisons of the eight viruses (Additional File S1). These values indicated a high degree of adaptation between the IAV subtypes and human. Furthermore, we analyzed the role of CUB in gene expression. The average value of RCDI was the highest for H7N9 (3.25 ± 0.17), followed by H9N2 (3.22 ± 0.19), H5N8 (3.15 ± 0.13), H3N2 (3.1 ± 0.14), H1N1 (3.06 ± 0.17), H5N1 (3.01 ± 0.19), H5N2 (2.95 ± 0.18) and H3N8(2.92 ± 0.21), indicating that the codon usage deoptimization occurred in this decreasing order (Fig. 7B). Significant differences in RCDI were found among the pairwise comparisons of the eight viruses (Additional File S1). Significant differences in SCUO values were also found among the eight IAV subtypes (Fig. 7C, Additional File S1). In addition, SiD analysis was used to assess the influence of the human on codon usage patterns. We found that SiD values were the highest in H1N1 (0.4919), suggesting that the human exerts more effects on this virus (Fig. 7D). However, the RCDI and SiD values were not significantly different among the studied IAV subtypes. In addition, a negative correlation was recorded between the CAI values of NS1 of the eight studied IAV subtypes and the values of ENC and GC3 (Fig. 7E), which indicated that the highly expressed genes had a superior scale of CUB and preferred the codons with A or U at the synonymous position. Positive correlations were also recorded between CAI, RCDI and SCUO values (Fig. 7E).
Fig. 7.
Adaptation analysis of NS1 of IAV subtypes. A The codon adaptation index (CAI) analysis of IAV NS1. B The RCDI values of IAV NS1. C The SCUO analysis of IAV subtypes. D The similarity index (SiD) analysis of IAV NS1. E Heatmap showing the Pearson correlation between the ENC, CAI, RCDI and SCUO values of NS1 coding sequences of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9, and H9N2. The egdes of the inset network indicates the distribution of adjusted p-values among the compared node groups of IAV subtypes in A–C. Statistical analysis was not applicable to D
Identification of human genes with CUB similar to NS1 CUB and their functional roles
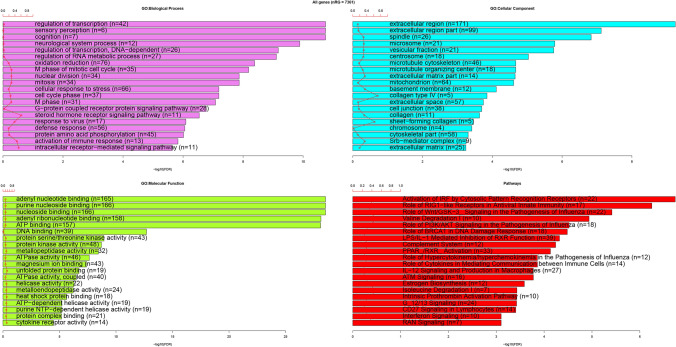
In order to identify human genes with CUB similar to CUB of NS1 gene of the IAV viral subtypes, Pearson correlation based on RSCU values was calculated among human genes and NS1 sequences. We obtained 2147 human genes correlated to NS1 with |r|> 0.5 and p < 0.05 (Additional File S2), which were considered as human genes with CUB similar to CUB of NS1 gene; RSCU values of all of these genes were positively correlated with RSCU values of NS1 gene (Additional File S2). Functional enrichment (Fig. 8) results showed that the human genes with CUB similar to CUB of NS1 were enriched in the biological processes including regulation of transcription (77 genes), DNA-dependent regulation of transcription (26 genes), regulation of RNA metabolic process (27 genes), cellular response to stress (66 genes), defense response (56 genes), response to virus (17 genes), activation of immune response (13 genes), oxidation reduction (76 genes) cell cycle phase (37 genes) and M phase of mitotic cell cycle ( 35 genes). For cellular component, extracellular region (171 genes) was the most enriched term whereas in the category of molecular function, adenyl nucleotide binding (165 genes) was the most represented term. In pathway enrichment analysis, we found that pathways such as “activation of IRF by cytosolic pattern recognition receptors” (22 genes), “role of RIG1-like receptors in antiviral innate immunity” (17 genes), “role of Wnt/GSK-3”_signaling in the pathogenesis of influenza (22 genes). The pathways related to influenza such as “role of PI3K/AKT signaling in the pathogenesis of influenza” (18 genes) and “role hypercytokenemia/hyperchemokinemia in the pathogenesis of influenza” (12 genes) were also enriched. Immune-related pathways such as “complement system”, “PPAR/RXR activation”, “role of cytokines in mediating communication between immune cells” (14 genes), “CD27 signaling in lymphocytes”, and “interferon signaling” (10 genes) were also significantly enriched.
Fig. 8.
Functional enrichment analysis of human genes with CUB similar to that of NS1 gene of IAV subtypes. The GO and pathway functional enrichment analysis was performed and the top 10 terms were visualized for each GO categories and pathway category
Protein–protein interaction network prediction and identification of hub clusters of the human genes with CUB similar to that of NS1 of IAV subtypes and their functional roles
In order to explore whether proteins corresponding to human genes with CUB similar to NS1 gene of IAV subtypes interact with each other, we selected the top-2000 genes from these genes for PPI network construction (Supplementary Figure S2). The network was composed of 1877 nodes with 12,707 edges, and the average number of neighbors was 13.578 (Fig. 9A). To uncover hub genes from this network, we performed network clustering analysis using the MCODE plugin and identified 46 clusters. The hub clusters with the highest score (cluster1, score = 31.273) was composed of 34 nodes and 516 edges (Fig. 9A). Further analysis indicated that the hub genes in cluster1 were enriched in the biological process of nuclear division (12 genes), mitosis (12 genes), M phase of mitotic cell cycle (12 genes), M phase (11 genes), regulation of cell cycle (11 genes) and organelle fission (9 genes) (Fig. 9B). The most enriched cellular components of the hub genes (cluster 1) were spindle (10 genes), microtubule cytoskeleton (11 genes) and condensed chromosome (6 genes) while the most significant molecular functions were ATP binding (9 genes) and adenyl ribonucleotide binding (9 genes) (Fig. 9B). The most significant pathways were “role of CHK proteins in cell cycle checkpoint control” (5 genes), ATM signaling (5 genes) and mitotic roles of polo-like kinase (5 genes) (Fig. 9B).
Fig. 9.
Hub gene subnetwork extracted from the protein–protein interaction (PPI) network of the human genes with CUB similar to that of NS1 of IAV subtypes. A The PPI network based on the top-2000 (based on Pearson correlation) human genes with CUB similar to that of NS1 of IAV subtypes was constructed and the MCODE plugin was used for extracting the hub gene subnetworks and the hub gene cluster with the highest score was visualized. B Functional enrichment analysis of hub genes extracted from the human genes with CUB similar to that of NS1 of IAV subtypes. The GO and pathway functional enrichment analysis was performed and the top 10 terms were visualized for each GO categories and pathway category
Similarities and differences of the human protein–protein interaction networks of NS1 coding sequences from different IAV subtypes
In order to discover the dissimilarities and the commonalities between the IAV subtypes H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2, we retrieved the human genes interacting with each of the IAV subtypes. The intersection analysis indicated important intersection genes between the list of genes from the eight IAV subtypes (Fig. 10A). Genes specific to each of the IAV subtypes were also identified (Fig. 10A). The hub subnetworks from the PPI networks constructed from human genes with CUB similar to CUB of NS1 gene of each IAV subtype were identified and depicted in Fig. 10B. These results indicated that though similarities were found in the interaction networks of IAV subtypes, some specific genes were associated to each IAV subtype, which may explain the differences in the response of human to each type of viral infections. The biological processes and pathways enriched from the list of genes from each subtype was summarized in Additional File S3 and indicated differences and similarities among the IAV subtypes.
Fig. 10.
Similarities and differences of the PPI networks of human genes with CUB similar to that of NS1 coding sequences from different IAV subtypes. A Intersection analysis of the different PPI networks of human genes with CUB similar to that of NS1 coding sequences from each IAV subtype. B Networks of hub genes of the different PPI networks of human genes with CUB similar to that of NS1 coding sequences from each IAV subtype
Discussion
The influenza A virus NS1 protein is a key viral protein that plays a key role in immune escape and the weakening of immune system in the host (Fernandez-Sesma 2007). Herein, starting from the CUB analysis, we aimed to study the factors influencing the expression of NS1 gene of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2 during adaptation to the human host and reveal the human genes whose expression would be regulated by the expression of NS1 in the human host. We first computed the CUB indices and subsequently analyzed the correlation between the RSCU values of human genes and those of different NS1 coding sequences of IAV subtypes H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2. The NS1 gene of IAV subtypes had high CAI values and codons ending with A/U were preferentially used. In addition, natural selection was found as the most determinant factor for CUB. The adaptation indices based on the NS1 coding sequences indicated a high degree of adaptability of different IAV subtypes to the host. Most importantly, we identified the human genes with CUB similar to that of NS1 gene and found that they were majorly involved in biological processes related to transcription and cellular response to stress; these genes were also involved in multiple pathways including those related to influenza, response to virus, defense response and immune response. This is the first study to reveal human genes with CUB similar to CUB of NS1 gene of IAV subtypes. The present findings may help develop vaccines and therapeutics for influenza A.
The CUB analysis suggested that the usage of codons of NS1 coding sequences of IAV subtypes is mainly influenced by natural selection. The RSCU analysis showed that the A-/U-ending codons were those preferred in the coding sequences of NS1 of IAV subtypes, suggesting that NS1 gene of IAV subtypes is less affected by mutation pressure. The results were supported by the findings of Luo et al. (2020). All IAV viruses isolated from humans tend to eliminate GC and CpG components during long-term adaptation to avoid activating the anti-viral immune response (Greenbaum and Ghedin 2015; Luo et al. 2020). The ENC-plot, neutrality plot and PR2 analyses clearly indicated that codon usage of NS1 coding sequences of IAV subtypes is influenced by natural selection in a broader extent and mutation pressure in a minor degree. We also found that the NS1 coding sequences of H1N1 and H3N2 were greatly affected by mutation pressure compared to other IAV subtypes, which may increase their pathogenicity compared to other subtypes and this was supported by the findings that mutations in the NS1 region of a virus will affect the stability and function of the protein (Solórzano et al. 2005). The NS1 protein plays an important role in the process of innate immunity against the host, allowing the IAV to replicate more effectively in the host cell (Fernandez-Sesma et al. 2006; Monteagudo et al. 2019). The virus relies on the host cellular components and metabolism to complete its own replication and assembly. The codon usage pattern of viruses reflects the evolution of viruses to improve their survival and fitness in the host (He et al. 2017; Hu et al. 2011; Luo et al. 2019; Taylor et al. 2017). Therefore, comparing the CUB of IAV helps improve our understandings on the evolutionary process of the virus. We found that the ENC value of the NS1 coding sequences of IAV subtypes was higher than 35, indicating lower CUB. Lower CUB in RNA viruses can avoid the competition on synthetic resources between the virus and the host cells so as to improve the efficiency of viral replication (Butt et al. 2016). NS1 is a nonstructural protein of IAV that plays a pivotal role in the early life cycle of the virus. Moreover, we found that the mean ENC values between the eight subtypes was significant, indicating that the codon usage pattern of these subtypes differed significantly from each other. Numerous studies have reported that the relationship between the virus and the host affects the occurrence, dynamics, and evolution of infectious diseases (Gibbs et al. 2010; Irwin et al. 2012; Puigbò et al. 2010; Rodelo-Urrego et al. 2013; Torres-Pérez et al. 2011). CAI values showed that the NS1 coding sequences of IAV subtypes had strong adaptability to human. The high adaptation of NS1 gene of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2 to human indicated the shaping of their codon usage profiles for improved adaptation to those of human, thus achieving their increased replication rate, which indicates high pathogenicity of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2 in humans. The results of CAI was supported by RCDI analysis because low RCDI may indicate high adaptability of both the virus and the host (Perez-Padilla et al. 2009). However, we found that there are differences in the RCDI values among the eight IAV subtypes, among which the RCDI values of H1N1 and H3N2 were the highest, suggesting that both viruses are better adapted to human. The low RCDI values indicated high similarity and adaptation of codon usage among NS1 gene of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2 and human, allowing these IAV subtypes to express NS1 gene efficiently with an elevated replication rate. The SiD value of NS1 coding sequences of IAV subtypes was about 0.49, indicating that the human is less affected by the usage of NS1 codons. The results were verified by neutrality plot and ENC-plot analysis. Therefore, in this study, we found that IAV NS1 showed a certain adaptability to human, which may be beneficial to IAV infection of human populations. The SiD value of NS1 coding sequences of H1N1 was the highest, suggesting the similarity of the genome between NS1 of H1N1 and human, which explains why H1N1 is reported to be a common human influenza A in the IAV subtypes. The high SiD value of H1N1 NS1 sequences may reflect the length of time the virus has circulated in human hosts (about 1918–2009) compared to the other viruses (H3N2 from the 1960s to present). The major factors shaping CUB in various species are natural selection and mutational pressure associated with genetic drift. In the present work, natural selection was noted as the main factor shaping the codon usage of NS1 coding sequences in H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2. This might reflect the length of time the viruses have evolved with the human. To the best of our knowledge, a systematic analysis of the evolutionary adaptation of NS1 gene of IAV subtypes to human has not been performed so far. Thus, our study is the first to explore this area and our results may be essential for the development of vaccines against these viruses (Baker et al. 2015). Previous studies indicated that codon usage adaptation of NS1 gene in human was involved in the spread of the pandemic Zika virus (de Melo Freire et al. 2015; Freire et al. 2018).
Up to date, the human interactome of virulence genes of IAV is not elucidated. Numerous studies have experimentally revealed proteins that can be affected by the IAV subtypes. For example, it was demonstrated that IAV NS1 hinders the activation of NF-κB, alpha/beta interferon, Jun N-terminal kinase and AP-1 transcription factors (Ludwig et al. 2002; Wang et al. 2000). However, the effect of NS1 in gene regulation in infected human remains to be explored. To date, the use of high throughput approaches in the identification of the effect of IAV subtypes would improve our knowledge on the molecular mechanism of these viruses or their virulence factors. However, studies based on those approaches are limited. Here, using the RSCU correlative approach, we uncovered human genes that could be significantly correlated with the NS1 gene of IAV subtypes H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2. This is the first study to uncover human genes with CUB similar to CUB of NS1 gene of IAV subtypes on a large scale. It is worth noting that the similarity in CUB may be the need for the host cell to rapidly produce the proteins (translation) when the transcript is present. The identification of these human genes with CUB similar to CUB of NS1 gene will be useful for diagnostic, prognostic and therapeutic purposes in human cases of infection by IAV subtypes. Moreover, PPI network construction indicated that human proteins in the network strongly interacted with each other, indicating that protein encoded by human genes with CUB similar to that of NS1 gene of H1N1, H3N2, H3N8, H5N1, H5N2, H5N8, H7N9 and H9N2 are involved in coordinated functions leading to pathological changes in the body. Thus, we analyzed the functional role of these human genes and found that human genes in the interactome were involved in the regulation of transcription. Previous studies have indicated that transcription of genes in the host is affected by pathogens (Kainulainen et al. 2014; Terasaki et al. 2016). Our results corroborated with these findings. Recent single cell RNA sequencing analysis indicated that the complexity of the expression profile of viral genes can lead to multifarious host transcriptional responses during the influenza A virus infection (Sun et al. 2020). Other studies have indicated that influenza virus infection can affect cellular transcription by altering the distribution of the polymerase II on cellular genes, which leads to host shut-off (Engelhardt and Fodor 2006). Thus, the modulation of genes involved in transcription or the development of therapeutics based on these genes will be vital for treating human infections caused by the IAV subtypes. Processes such as “cellular response to stress”, “defense response”, “response to virus”, “activation of immune response”, “oxidation reduction” and “cell cycle phase” were also predicted to be functionally altered by the NS1 of IAV subtypes. These observations confirmed the multifunctional role of NS1 gene especially its involvement in the immune and weakening of host defense following viral infection. Some of these processes have been reported to be affected by IAVs in past studies (Jang et al. 2015; Josset et al. 2008). Moreover, we found that the pathways enriched largely included those associated with influenza (eg. “role of Wnt/GSK-3″_signaling in the pathogenesis of influenza”, “role of PI3K/AKT signaling in the pathogenesis of influenza” and “role hypercytokenemia/hyperchemokinemia in the pathogenesis of influenza”), further confirming the credibility of our approach that the similarity of RSCU of parasite and host may affect the diverse processes in the host and drive the subsequent pathological conditions. The pathological conditions induced by the NS1 gene is probably driven by abnormal regulation of cell cycle as demonstrated by the enrichment of the hub genes of the human PPI network of NS1 of IAV subtypes in processes such as nuclear division, mitosis, M phase of mitotic cell cycle and the pathway of “role of CHK proteins in cell cycle checkpoint control”. Thus, our study not only revealed pathways already known to be affected by NS1 gene of IAV subtypes, but also disclosed new pathways and processes that could be modulated in future, thus enhancing our understanding of the molecular mechanism of IAV infections. The obtained data will also help develop appropriate diagnostic approaches and therapeutics for the infected patients. In our previous work, we identified the human genes whose RSCU values are correlated with those of the genes of respiratory coronaviruses (Nambou and Anakpa 2020). Overlapping of these coronavirus-related human genes (Nambou and Anakpa 2020) with the human genes correlated with NS1 gene of IAV subtypes, we identified 1723 common genes; the functional enrichment of these common genes indicated their involvement mainly in defense response (56 genes) and immune-related processes and pathways (Supplementary Figure S3). These observations indicated that the NS1 gene of IAV subtypes may induce some pathogenetic mechanisms similar to those of other respiratory viruses, namely the coronaviruses.
Our study presents some limitations. We only included IAV subtypes with the number of NS1 coding sequences higher than 1000 in order to keep statistical power; doing so, the other IAV subtypes were excluded from the study, which may lead to some information loss. In addition, we did not include the IAV subtypes that had not infected human, including these viruses may be informative for predicting their effect in case of human infection by these viruses. Furthermore, comparative analysis of the co-adaptation of NS1 gene of IAV subtypes and human to that of other natural reservoirs and intermediate hosts was not performed. Performing such analysis would help understand the differential adaptation of IAV subtypes to their hosts. Moreover, due to the huge number of analyses performed, we did not compare the results from allele A and allele B of NS1 gene, which will be addressed in a future study. In the future, we will also update our research based on experimental and public data and verify our results through in vivo and in vitro experiments.
In sum, our study showed that the CUB of the NS1 coding sequences of IAV subtypes is highly conserved and subtype-specific though similarities were found. The codon usage pattern is mainly affected by the pressure of natural selection and mutation pressure in a lesser extent. Additionally, IAV NS1 showed good adaptability to the host. The findings are vital for understanding the CUB of IAV NS1 and the factors that affect the evolutionary process of IAV, and may contribute to the development of the IAV vaccine. We also identified the human genes with CUB similar to CUB of NS1 gene of IAV subtypes and their function, which may shed light in the pathogenesis of human infection by these viruses and may help develop adequate treatments for IAV-infected patients. The functional role of the predicted human genes needs to be verified by other methods, especially at the translational regulation level.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (XLS 100 kb) Additional File S1. Results of one-way ANOVA analysis of ENC, CAI, RCDI and SCUO among the IAV subtypes.
Supplementary file2 (CSV 268 kb) Additional File S2. The human genes with CUB similar to that of NS1 gene. The Pearson correlation between NS1 gene and human genes was calculated based on the RSCU values. Genes showing a correlation of |r|>0.5 and p<0.05 were considered as belonging to the human genes with CUB similar to that of NS1 gene based on RSCU.
Supplementary file3 (XLS 203 kb) Additional File S3 Biological processes and pathways enriched from the human interactome of NS1 coding sequences of each IAV subtype.
Supplementary file4 (TIF 4256 kb) Supplementary Figure S1. Principal component analysis indicating the overlap of the RSCU values of each of IAV subtypes with that of human.
Supplementary file5 (TIF 20965 kb) Supplementary Figure S2. Protein-Protein interaction network of the human genes with CUB similar to that of the NS1 coding sequences of IAV subtypes.
Supplementary file6 (TIF 21285 kb) Supplementary Figure S3. Functional enrichment analysis of human genes with CUB similar to respiratory coronaviruses as well as the NS1 coding sequences of IAV subtypes.
Acknowledgements
Not applicable.
Author contributions
KN conceived, designed, analyzed the data and wrote the manuscript. MA and YT made substantial contributions to the acquisition, analysis, interpretation of data and proofread the manuscript critically for important intellectual content. All the authors approved the version to be published, and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was not supported by funding organizations.
Data availability
All data generated or analyzed during this study are included in this published article.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Komi Nambou, Email: noelnambou@yahoo.fr.
Yin Selina Tong, Email: biochem_selina0522@hotmail.com.
References
- Alonso AM, Diambra L. SARS-CoV-2 codon usage bias downregulates host expressed genes with similar codon usage. Front Cell Dev Biol. 2020;8:831. doi: 10.3389/fcell.2020.00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar AM, Soudy M, Mohamed R. vhcub: virus-host codon usage co-adaptation analysis. F1000Research. 2019;8:2137. doi: 10.12688/f1000research.21763.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SF, Nogales A, Martínez-Sobrido L. Downregulating viral gene expression: codon usage bias manipulation for the generation of novel influenza A virus vaccines. Futur Virol. 2015;10:715–730. doi: 10.2217/fvl.15.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AM, Nasrullah I, Qamar R, Tong Y. Evolution of codon usage in Zika virus genomes is host and vector specific. Emerg Microbes Infect. 2016;5:e107. doi: 10.1038/emi.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauldwell AV, Long JS, Moncorgé O, Barclay WS. Viral determinants of influenza A virus host range. J Gen Virol. 2014;95:1193–1210. doi: 10.1099/vir.0.062836-0. [DOI] [PubMed] [Google Scholar]
- Chang P, Sealy JE, Sadeyen JR, Bhat S, Lukosaityte D, Sun Y, Iqbal M. Immune escape adaptive mutations in the H7N9 avian influenza hemagglutinin protein increase virus replication fitness and decrease pandemic potential. J Virol. 2020;94:e00216–e00220. doi: 10.1128/JVI.00216-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauché C, Nogales A, Zhu H, Goldfarb D, Ahmad Shanizza AI, Gu Q, Parrish CR, Martínez-Sobrido L, Marshall JF, Murcia PR. Mammalian adaptation of an avian influenza A virus involves stepwise changes in NS1. J Virol. 2018;92:e01875–e01917. doi: 10.1128/JVI.01875-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu S, Goraya MU, Maarouf M, Huang S, Chen JL. Host immune response to influenza A virus infection. Front Immunol. 2018;9:320. doi: 10.3389/fimmu.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Wasmuth JD, Blaxter ML. The evolution of biased codon and amino acid usage in nematode genomes. Mol Biol Evol. 2006;23:2303–2315. doi: 10.1093/molbev/msl097. [DOI] [PubMed] [Google Scholar]
- de Melo Freire CC, Iamarino A, de Lima Neto DF, de Andrade Zanotto PM (2015) Spread of the pandemic Zika virus lineage is associated with NS1 codon usage adaptation in humans. BioRxiv 032839.
- Deng Y, de Lima HF, Kalfon J, Chu D, von der Haar T. Hidden patterns of codon usage bias across kingdoms. J R Soc Interface. 2020;17:20190819. doi: 10.1098/rsif.2019.0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt OG, Fodor E. Functional association between viral and cellular transcription during influenza virus infection. Rev Med Virol. 2006;16:329–345. doi: 10.1002/rmv.512. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sesma A. The influenza virus NS1 protein: inhibitor of innate and adaptive immunity. Infect Disord Drug Targets. 2007;7:336–343. doi: 10.2174/187152607783018754. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sesma A, Marukian S, Ebersole BJ, Kaminski D, Park MS, Yuen T, Sealfon SC, García-Sastre A, Moran TM. Influenza virus evades innate and adaptive immunity via the NS1 protein. J Virol. 2006;80:6295–6304. doi: 10.1128/JVI.02381-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire CCM, Palmisano G, Braconi CT, Cugola FR, Russo FB, Beltrão-Braga PC, Iamarino A, Lima Neto DF, Sall AA, Rosa-Fernandes L, Larsen MR, Zanotto PMA. NS1 codon usage adaptation to humans in pandemic Zika virus. Mem Inst Oswaldo Cruz. 2018;113:e170385. doi: 10.1590/0074-02760170385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Ge S, Wang Z. An overview of influenza A virus receptors. Crit Rev Microbiol. 2011;37:157–165. doi: 10.3109/1040841X.2010.536523. [DOI] [PubMed] [Google Scholar]
- Gibbs AJ, Fargette D, García-Arenal F, Gibbs MJ. Time–the emerging dimension of plant virus studies. J Gen Virol. 2010;91:13–22. doi: 10.1099/vir.0.015925-0. [DOI] [PubMed] [Google Scholar]
- Greenbaum BD, Ghedin E. Viral evolution: beyond drift and shift. Curr Opin Microbiol. 2015;26:109–115. doi: 10.1016/j.mib.2015.06.015. [DOI] [PubMed] [Google Scholar]
- Han CW, Jeong MS, Jang SB. Structure and function of the influenza A virus non-structural protein 1. J Microbiol Biotechnol. 2019;29:1184–1192. doi: 10.4014/jmb.1903.03053. [DOI] [PubMed] [Google Scholar]
- He W, Zhang H, Zhang Y, et al. Codon usage bias in the N gene of rabies virus. Infect Genet Evol. 2017;54:458–465. doi: 10.1016/j.meegid.2017.08.012. [DOI] [PubMed] [Google Scholar]
- He Z, Gan H, Liang X. Analysis of synonymous codon usage bias in potato virus M and its adaption to hosts. Viruses. 2019;11:752. doi: 10.3390/v11080752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Alias X, Benisty H, Schaefer MH, Serrano L. Translational adaptation of human viruses to the tissues they infect. Cell Rep. 2021;34:108872. doi: 10.1016/j.celrep.2021.108872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JS, Wang QQ, Zhang J, et al. The characteristic of codon usage pattern and its evolution of hepatitis C virus. Infect Genet Evol. 2011;11:2098–2102. doi: 10.1016/j.meegid.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Huang MT, Zhang S, Wu YN, et al. Dual R108K and G189D Mutations in the NS1 protein of A/H1N1 influenza virus counteract host innate immune responses. Viruses. 2021;13:905. doi: 10.3390/v13050905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin NR, Bayerlová M, Missa O, Martínková N. Complex patterns of host switching in New World arenaviruses. Mol Ecol. 2012;21:4137–4150. doi: 10.1111/j.1365-294X.2012.05663.x. [DOI] [PubMed] [Google Scholar]
- Jang HJ, Lee HJ, Kang KS, et al. Molecular responses to the influenza A virus in chicken trachea-derived cells. Poult Sci. 2015;94:1190–1201. doi: 10.3382/ps/pev033. [DOI] [PubMed] [Google Scholar]
- Jayaraman A, Pappas C, Raman R, et al. A single base-pair change in 2009 H1N1 hemagglutinin increases human receptor affinity and leads to efficient airborne viral transmission in ferrets. PLoS ONE. 2011;6:e17616. doi: 10.1371/journal.pone.0017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Liu S, Zheng H, et al. Non-uniqueness of factors constraint on the codon usage in Bombyx mori. BMC Genomics. 2015;16:356. doi: 10.1186/s12864-015-1596-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitobaom K, Phakaratsakul S, Sirihongthong T, et al. Codon usage similarity between viral and some host genes suggests a codon-specific translational regulation. Heliyon. 2020;6:e03915. doi: 10.1016/j.heliyon.2020.e03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L, Frobert E, Rosa-Calatrava M. Influenza A replication and host nuclear compartments: many changes and many questions. J Clin Virol. 2008;43:381–390. doi: 10.1016/j.jcv.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Kainulainen M, Habjan M, Hubel P, Busch L, Lau S, Colinge J, Superti-Furga G, Pichlmair A, Weber F. Virulence factor NSs of rift valley fever virus recruits the F-box protein FBXO3 to degrade subunit p62 of general transcription factor TFIIH. J Virol. 2014;88:3464–3473. doi: 10.1128/JVI.02914-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S, Harder TC, Schwemmle M, Ciminski K. Influenza A viruses and zoonotic events-are we creating our own reservoirs? Viruses. 2021;13:2250. doi: 10.3390/v13112250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M, Wilbrink B, Conyn M, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet (london, England) 2004;363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- Koutsakos M, Kedzierska K, Subbarao K. Immune responses to avian influenza viruses. J Immunol (baltimore, Md : 1950) 2019;202:382–391. doi: 10.4049/jimmunol.1801070. [DOI] [PubMed] [Google Scholar]
- Kula A, Saelens J, Cox J, et al. The evolution of molecular compatibility between bacteriophage ΦX174 and its host. Sci Rep. 2018;8:8350. doi: 10.1038/s41598-018-25914-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Shao TJ, Yu XF, et al. Molecular evolution of HA gene of the influenza A H1N1 pdm09 strain during the consecutive seasons 2009–2011 in Hangzhou, China: several immune-escape variants without positively selected sites. J Clin Virol. 2012;55:363–366. doi: 10.1016/j.jcv.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Liu YS, Zhou JH, Chen HT, Ma LN, Pejsak Z, Ding YZ, Zhang J. The characteristics of the synonymous codon usage in enterovirus 71 virus and the effects of host on the virus in codon usage pattern. Infect Genet Evol. 2011;11:1168–1173. doi: 10.1016/j.meegid.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Mistry B, Haslam SM, Barclay WS. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol. 2019;17:67–81. doi: 10.1038/s41579-018-0115-z. [DOI] [PubMed] [Google Scholar]
- Ludwig S, Wang X, Ehrhardt C, et al. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J Virol. 2002;76:11166–11171. doi: 10.1128/JVI.76.21.11166-11171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Li Y, Yu S, et al. Better fit of codon usage of the polymerase and nucleoprotein genes to the chicken host for H7N9 than H9N2 AIVs. J Infect. 2019;79:174–187. doi: 10.1016/j.jinf.2019.05.012. [DOI] [PubMed] [Google Scholar]
- Luo W, Tian L, Gan Y, et al. The fit of codon usage of human-isolated avian influenza A viruses to human. Infect Genet Evolut. 2020;81:104181. doi: 10.1016/j.meegid.2020.104181. [DOI] [PubMed] [Google Scholar]
- Mair CM, Ludwig K, Herrmann A, Sieben C. Receptor binding and pH stability—how influenza A virus hemagglutinin affects host-specific virus infection. Biochem Biophys Acta. 2014;1838:1153–1168. doi: 10.1016/j.bbamem.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Makarova NV, Kaverin NV, Krauss S, Senne D, Webster RG. Transmission of Eurasian avian H2 influenza virus to shorebirds in North America. J Gen Virol. 1999;80(Pt 12):3167–3171. doi: 10.1099/0022-1317-80-12-3167. [DOI] [PubMed] [Google Scholar]
- Maldonado LL, Bertelli AM, Kamenetzky L. Molecular features similarities between SARS-CoV-2, SARS, MERS and key human genes could favour the viral infections and trigger collateral effects. Sci Rep. 2021;11:4108. doi: 10.1038/s41598-021-83595-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova-Petkova A, Laplante J, Jang Y, et al. Avian influenza A(H7N2) virus in human exposed to sick cats, New York, USA, 2016. Emerg Infect Dis. 2017;23:2046–2049. doi: 10.3201/eid2312.170798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meineke R, Rimmelzwaan GF, Elbahesh H. Influenza virus infections and cellular kinases. Viruses. 2019;11:171. doi: 10.3390/v11020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michely S, Toulza E, Subirana L, et al. Evolution of codon usage in the smallest photosynthetic eukaryotes and their giant viruses. Genome Biol Evol. 2013;5:848–859. doi: 10.1093/gbe/evt053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JB, Hippen AA, Wright SM, Morris C, Ridge PG (2017) Human viruses have codon usage biases that match highly expressed proteins in the tissues they infect. Biomed Genet Genomics
- Monteagudo PL, Muñoz-Moreno R, Fribourg M, et al. Differential modulation of innate immune responses in human primary cells by influenza A viruses carrying human or avian nonstructural protein 1. J Virol. 2019;94:e00999–e01019. doi: 10.1128/JVI.00999-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moratorio G, Iriarte A, Moreno P, Musto H, Cristina J. A detailed comparative analysis on the overall codon usage patterns in West Nile virus. Infect Genet Evol. 2013;14:396–400. doi: 10.1016/j.meegid.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambou K, Anakpa M. Deciphering the co-adaptation of codon usage between respiratory coronaviruses and their human host uncovers candidate therapeutics for COVID-19. Infect Genet Evolut. 2020;85:104471. doi: 10.1016/j.meegid.2020.104471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/S1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. Modulation of innate immune responses by the influenza A NS1 and PA-X proteins. Viruses. 2018;10:708. doi: 10.3390/v10120708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock TP, Benton DJ, James J, et al. Immune escape variants of H9N2 influenza viruses containing deletions at the hemagglutinin receptor binding site retain fitness in vivo and display enhanced zoonotic characteristics. J Virol. 2017;91:5. doi: 10.1128/JVI.00218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Zou Y, Li H, Li K, Jiang T. Inferring the antigenic epitopes for highly pathogenic avian influenza H5N1 viruses. Vaccine. 2014;32:671–676. doi: 10.1016/j.vaccine.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Pepin KM, Domsic J, McKenna R. Genomic evolution in a virus under specific selection for host recognition. Infect Genet Evol. 2008;8:825–834. doi: 10.1016/j.meegid.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Perez-Padilla R, de la Rosa-Zamboni D, Samuel PDL, et al. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N Engl J Med. 2009;361:680–689. doi: 10.1056/NEJMoa0904252. [DOI] [PubMed] [Google Scholar]
- Philippon DAM, Wu P, Cowling BJ, Lau EHY. Avian influenza human infections at the human-animal interface. J Infect Dis. 2020;222:528–537. doi: 10.1093/infdis/jiaa105. [DOI] [PubMed] [Google Scholar]
- Puigbò P, Aragonès L, Garcia-Vallvé S. RCDI/eRCDI: a web-server to estimate codon usage deoptimization. BMC Res Notes. 2010;3:87. doi: 10.1186/1756-0500-3-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodelo-Urrego M, Pagán I, González-Jara P, et al. Landscape heterogeneity shapes host-parasite interactions and results in apparent plant-virus codivergence. Mol Ecol. 2013;22:2325–2340. doi: 10.1111/mec.12232. [DOI] [PubMed] [Google Scholar]
- Rosário-Ferreira N, Preto AJ, Melo R, Moreira IS, Brito RM. The central role of non-structural protein 1 (NS1) in influenza biology and infection. Int J Mol Sci. 2020;21:1511. doi: 10.3390/ijms21041511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer JR, Kawaoka Y, Bean WJ, Süss J, Senne D, Webster RG. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194:781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sitaras I, Kalthoff D, Beer M, Peeters B, de Jong MC. Immune escape mutants of Highly Pathogenic Avian Influenza H5N1 selected using polyclonal sera: identification of key amino acids in the HA protein. PLoS ONE. 2014;9:e84628. doi: 10.1371/journal.pone.0084628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski DM, Tweed SA, Petric M, Booth T, Li Y, Tam T. Human illness and isolation of low-pathogenicity avian influenza virus of the H7N3 subtype in British Columbia, Canada. J Infect Dis. 2006;193:899–900. doi: 10.1086/500219. [DOI] [PubMed] [Google Scholar]
- Slusher MJ, Wilcox BR, Lutrell MP, et al. Are passerine birds reservoirs for influenza A viruses? J Wildl Dis. 2014;50:792–809. doi: 10.7589/2014-02-043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Bahl J, Vijaykrishna D, et al. Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci USA. 2009;106:11709–11712. doi: 10.1073/pnas.0904991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solórzano A, Webby RJ, Lager KM, Janke BH, García-Sastre A, Richt JA. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J Virol. 2005;79:7535–7543. doi: 10.1128/JVI.79.12.7535-7543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Qin K. Human-infecting influenza A (H9N2) virus: a forgotten potential pandemic strain? Zoonoses Public Health. 2020;67:203–212. doi: 10.1111/zph.12685. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci USA. 1988;85:2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Vera JC, Drnevich J, Lin YT, Ke R, Brooke CB. Single cell heterogeneity in influenza A virus gene expression shapes the innate antiviral response to infection. PLoS Pathog. 2020;16:e1008671. doi: 10.1371/journal.ppat.1008671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton TC. The pandemic threat of emerging H5 and H7 avian influenza viruses. Viruses. 2018;10:461. doi: 10.3390/v10090461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner WD, Toth DJ, Gundlapalli AV. The pandemic potential of avian influenza A(H7N9) virus: a review. Epidemiol Infect. 2015;143:3359–3374. doi: 10.1017/S0950268815001570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TL, Dimitrov KM, Afonso CL. Genome-wide analysis reveals class and gene specific codon usage adaptation in avian paramyxoviruses 1. Infect Genet Evol. 2017;50:28–37. doi: 10.1016/j.meegid.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Terasaki K, Ramirez SI, Makino S. Mechanistic insight into the host transcription inhibition function of Rift Valley fever virus NSs and its importance in virulence. PLoS Negl Trop Dis. 2016;10:e0005047. doi: 10.1371/journal.pntd.0005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terebuh P, Adija A, Edwards L, et al. Human infection with avian influenza A(H7N2) virus-Virginia, 2002. Influenza Other Respir Viruses. 2018;12:529–532. doi: 10.1111/irv.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong XC, Weng SS, Xue F, Wu X, Xu TM, Zhang WH. First human infection by a novel avian influenza A(H7N4) virus. J Infect. 2018;77:249–257. doi: 10.1016/j.jinf.2018.06.002. [DOI] [PubMed] [Google Scholar]
- Torres-Pérez F, Palma RE, Hjelle B, Holmes EC, Cook JA. Spatial but not temporal co-divergence of a virus and its mammalian host. Mol Ecol. 2011;20:4109–4122. doi: 10.1111/j.1365-294X.2011.05241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweed SA, Skowronski DM, David ST, et al. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis. 2004;10:2196–2199. doi: 10.3201/eid1012.040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um S, Siegers JY, Sar B, et al. Human infection with avian influenza A(H9N2) Virus, Cambodia, February 2021. Emerg Infect Dis. 2021;27:2742–2745. doi: 10.3201/eid2710.211039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak K, Kowalczyk A, Markowska-Daniel I. Influenza A viruses of avian origin circulating in pigs and other mammals. Acta Biochim Pol. 2014;61:433–439. doi: 10.18388/abp.2014_1861. [DOI] [PubMed] [Google Scholar]
- Uyeki TM, Bernstein HH, Bradley JS, et al. Clinical practice guidelines by the Infectious Diseases Society of America: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal influenzaa. Clin Infect Dis. 2019;68:895–902. doi: 10.1093/cid/ciy874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan K, Koh X, Chandrasekaran A, et al. Determinants of glycan receptor specificity of H2N2 influenza A virus hemagglutinin. PLoS ONE. 2010;5:e13768. doi: 10.1371/journal.pone.0013768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, García-Sastre A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol. 2000;74:11566–11573. doi: 10.1128/JVI.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenius V, Mäkelä SM, Julkunen I, Österlund P. Highly pathogenic H5N1 influenza a virus spreads efficiently in human primary monocyte-derived macrophages and dendritic cells. Front Immunol. 2018;9:1664. doi: 10.3389/fimmu.2018.01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Xiao L, Li L. Research progress on human infection with avian influenza H7N9. Front Med. 2020;14:8–20. doi: 10.1007/s11684-020-0739-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji R, Saad MD, Davis CT, et al. Pandemic potential of highly pathogenic avian influenza clade 2344 A(H5) viruses. Rev Med Virol. 2020;30:e2099. doi: 10.1002/rmv.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Du Y, Wang G, Li K. Non-structural protein 1 of H3N2 influenza A virus induces nucleolar stress via interaction with nucleolin. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-18087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yang Y, Zhou X, et al. The NS1 protein of influenza A virus interacts with heat shock protein Hsp90 in human alveolar basal epithelial cells: implication for virus-induced apoptosis. Virol J. 2011;8:181. doi: 10.1186/1743-422X-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Xie Y, Muñoz-Moreno R, et al. Structural basis for influenza virus NS1 protein block of mRNA nuclear export. Nat Microbiol. 2019;4:1671–1679. doi: 10.1038/s41564-019-0482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Li X, Liu J, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 (XLS 100 kb) Additional File S1. Results of one-way ANOVA analysis of ENC, CAI, RCDI and SCUO among the IAV subtypes.
Supplementary file2 (CSV 268 kb) Additional File S2. The human genes with CUB similar to that of NS1 gene. The Pearson correlation between NS1 gene and human genes was calculated based on the RSCU values. Genes showing a correlation of |r|>0.5 and p<0.05 were considered as belonging to the human genes with CUB similar to that of NS1 gene based on RSCU.
Supplementary file3 (XLS 203 kb) Additional File S3 Biological processes and pathways enriched from the human interactome of NS1 coding sequences of each IAV subtype.
Supplementary file4 (TIF 4256 kb) Supplementary Figure S1. Principal component analysis indicating the overlap of the RSCU values of each of IAV subtypes with that of human.
Supplementary file5 (TIF 20965 kb) Supplementary Figure S2. Protein-Protein interaction network of the human genes with CUB similar to that of the NS1 coding sequences of IAV subtypes.
Supplementary file6 (TIF 21285 kb) Supplementary Figure S3. Functional enrichment analysis of human genes with CUB similar to respiratory coronaviruses as well as the NS1 coding sequences of IAV subtypes.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.