Summary
The dynamic nature of sleep in many animals suggests distinct stages which serve different functions. Genetic sleep induction methods in animal models provide a powerful way to disambiguate these stages and functions, although behavioral methods alone are insufficient to accurately identify what kind of sleep is being engaged. In Drosophila, activation of the dorsal fan-shaped body (dFB) promotes sleep, but it remains unclear what kind of sleep this is, how the rest of the fly brain is behaving, or if any specific sleep functions are being achieved. Here, we developed a method to record calcium activity from thousands of neurons across a volume of the fly brain during spontaneous sleep and compared this to dFB-induced sleep. We found that spontaneous sleep typically transitions from an active ‘wake like’ stage to a less active stage. In contrast, optogenetic activation of the dFB promotes sustained wake-like levels of neural activity, even though flies become unresponsive to mechanical stimuli. When we probed flies with salient visual stimuli, we found that the activity of visually responsive neurons in the central brain was blocked by transient dFB activation, confirming an acute disconnect from the external environment. Prolonged optogenetic dFB activation nevertheless achieved a key sleep function, by correcting visual attention defects brought on by sleep deprivation. These results suggest that dFB activation promotes a distinct form of sleep in Drosophila, where brain activity appears similar to wakefulness, but responsiveness to external sensory stimuli is profoundly suppressed.
Keywords: Sleep stages, optogenetics, REM sleep, visual attention, calcium imaging, brain
Graphical Abstract
eTOC Blurb
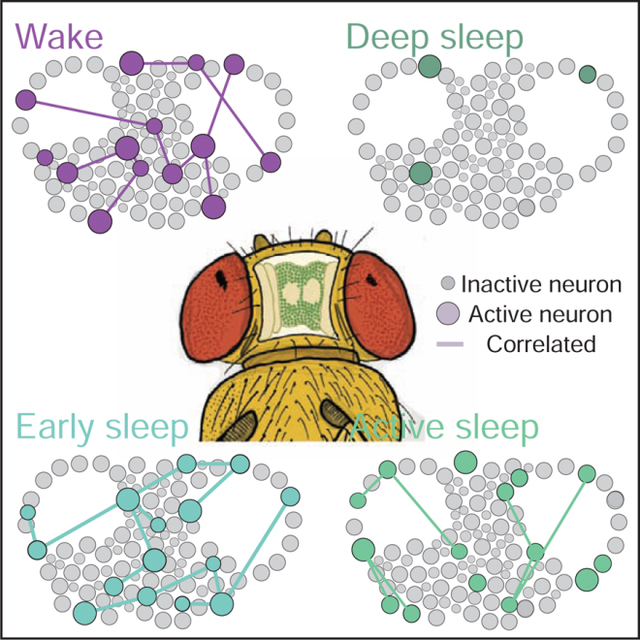
Tainton-Heap et al. track calcium activity in neurons across the fly brain to compare spontaneous and optogenetically induced sleep. They uncover an active, wake-like sleep stage as well as a less active deep sleep stage. Induced sleep appears to promote wake-like levels of brain activity, while suppressing responsiveness to external stimuli.
Introduction
Sleep in one form or another has been documented in most animals, suggesting that it serves conserved and important functions across species [1–3]. Extended bouts of behavioral quiescence are typically the first criteria sought in all sleep studies, with experiments often probing additional sleep criteria such as animal posture, increased arousal thresholds, homeostatic regulation, and specific neural correlates. However, sleep is not just a single phenomenon, and different animals could be manifesting distinct forms of sleep [4]. Since most sleep is evidenced by behavioral quiescence, this criterion alone is probably insufficient to discriminate between distinct forms of sleep, and consequently between potentially different sleep functions. While it has long been accepted that distinct sleep stages such as slow-wave sleep (SWS) and rapid eye movement (REM) sleep exist in mammals and birds [5–7], only recently has the possibility of sleep stages been explored in other animals, such as reptiles [8], fish [9], and invertebrates such as cuttlefish [10]. To be able to properly understand sleep biology requires disambiguating various forms of sleep, as these may be accomplishing different functions that could be confused if grouped together simply under behavioral quiescence alone.
Over the past two decades, the fruit fly Drosophila melanogaster has become an important model for understanding sleep biology and functions [11]. Original hypotheses regarding potential sleep functions, such as the idea that sleep rescales synaptic weights [12], became more readily testable in this model [13–15] after it was understood that flies indeed require sleep in a similar way as other animals [16, 17]. However, sleep research in Drosophila and other invertebrate models such as the roundworm Caenorhabditis elegans [18, 19] have generally considered sleep as a single, unitary phenomenon linked primarily to extended behavioral quiescence.
Although sleep is typically studied as a spontaneous transition that follows predictably after extended wakefulness, the recent development of genetic techniques to induce sleep on demand in animal models such as Drosophila have provided novel approaches to manipulating sleep as an experimental variable. Notably, sleep-promoting neurons were discovered in the central brain of Drosophila that, when activated, rapidly produced a sleep-like state that is conducive to memory consolidation [20] and that promotes behavioral plasticity [21], which are both proposed sleep functions [22]. These sleep-promoting neurons in the fly brain project to a region in the central complex called the dorsal fan-shaped body (dFB), and were found to form a homeostatically-regulated circuit that becomes more active when sleep pressure (i.e., extended wakefulness) accrues above a set threshold, determined in part by the membrane potential of the dFB neurons [23–25]. Together, these experiments provide compelling evidence that neurons in the dFB promote sleep in flies, and therefore that sleep functions can be studied by manipulating these neurons experimentally [21, 23, 26–28]. It remains unclear however what kind of sleep is being engaged when sleep-promoting neurons in Drosophila are being activated (by using, increasingly, the dFB-expressing strain R23E10 [29]), or how the rest of the fly brain is behaving during genetically induced sleep. We employed two-photon imaging of a calcium reporter localized to neural soma to track fly brain activity and connectivity during spontaneous sleep and wake. We then compared these results with optogenetically-induced sleep to determine if activation of the dFB might be promoting a distinct sleep stage.
Results
Wake and sleep both include epochs of high and low neural activity
Previous electrophysiological studies employing local field potential (LFP) recordings in Drosophila identified distinct forms of electrical activity during sleep [26, 30, 31], however it remained unclear how individual neurons behaved across the fly brain. To sample neural activity during sleep and wake, we developed a calcium imaging preparation to track fluorescence traces of thousands of individual neurons over several hours in behaving animals, employing 2-photon microscopy (Figure 1A, top: Figure S1A). We first investigated spontaneous sleep in flies expressing pan-neuronal GCaMP6f [32] under the control of a nuclear-localized sequence (nls). Flies were sleep deprived for 24 hours prior to imaging, to facilitate the occurrence of daytime sleep episodes [23]. Flies were positioned on an air-supported ball and filmed to monitor their behavior (Figure 1A, bottom). Simultaneously, the brain was perfused with oxygenated extracellular fluid while calcium activity was recorded over 18 optical slices of the central brain at 3.3Hz (Figure 1B, Figure S1B). Neural activity was extracted from regions of interest (ROIs) that corresponded to cell bodies (Figure 1C), and active neurons were identified and their activity levels were tracked throughout an experiment (Figure 1D,E; Figure S1C; Video S1). Additionally, we measured temporal correlations among active neurons, to estimate functional connectivity [33] in the fly brain during wake and sleep (Figure 1F; Figure S1D–G).
Figure 1: Neural activity during wake and spontaneous sleep stages.
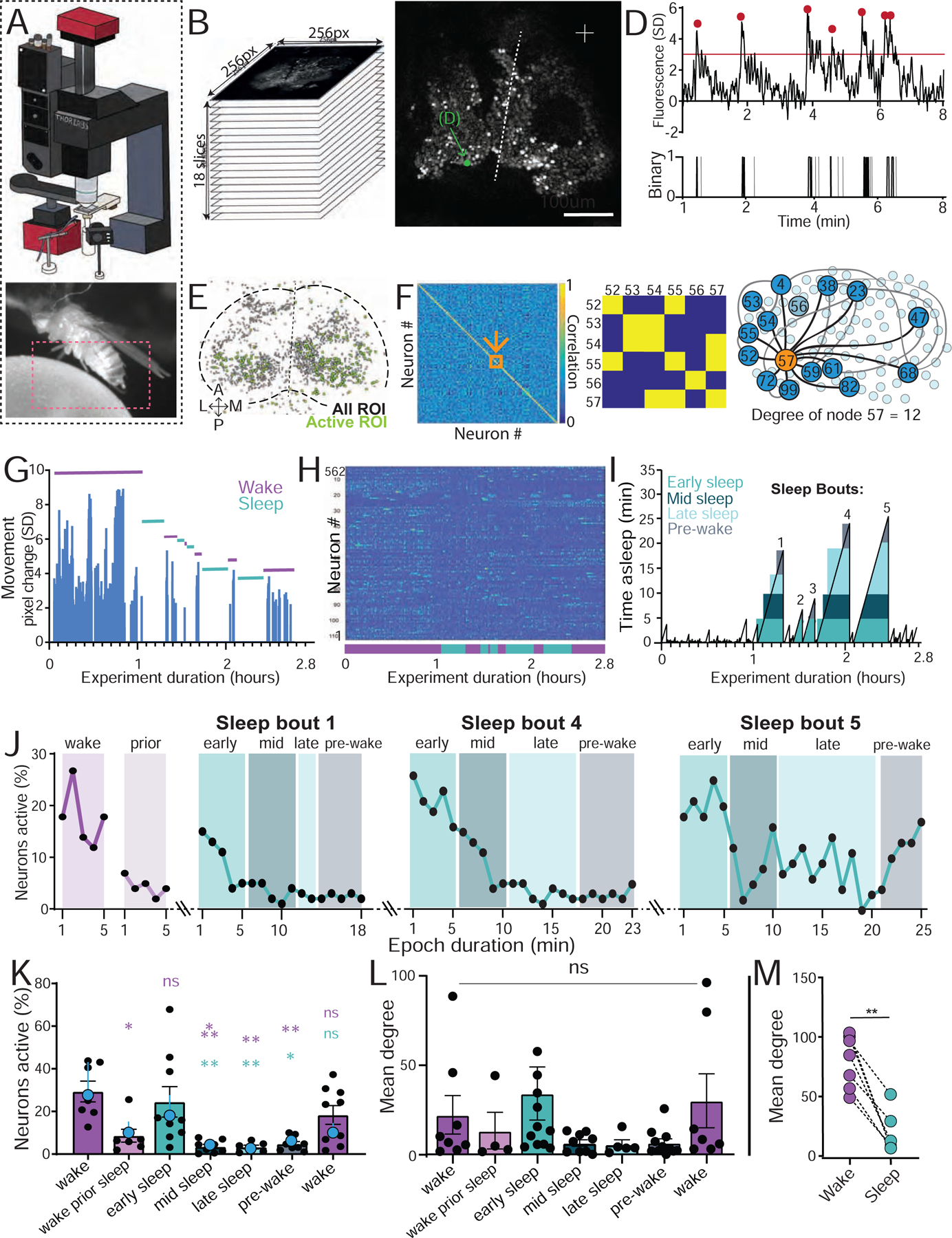
A) Top: microscopy setup (see Figure S1A,B). Bottom: temporally-matched behavior captured from fly legs and abdomen (red box). B) Brain imaging across 18 z-slices, over ~100μm depth from the top of the brain. C) Example GCaMP6f expression. Arrow: a single neuronal soma. D) Activity trace of single neuron indicated in C. Activity events are indicated (red dots, top), alongside the corresponding binary data (bottom; gray traces did not meet criteria. See Figure S1C). E) A collapsed mask from one fly of neurons found to be active (green) in C alongside all identified regions of interest (ROIs, gray). F) Left: adjacency matrix of pairwise correlations between all active neurons in E. Middle: significant correlations (yellow) were identified (See Figure S1D–G). Right: mean degree was calculated by averaging the number of significant correlations among all active neurons. G) Movement data from an example fly. Sleep (green bars) was determined by >5min immobility. H) Corresponding active neurons during the behavioral trace in G. I) Sleep epochs in the same sample fly, corresponding to panels G and H, with temporally segmented sleep bouts: early sleep (0–5 min), mid sleep (5 – 10 min), late sleep (< 10 min), and prior to wake (5 min to wake). J) Neural activity (% neurons active) during 3 sleep bouts indicated in (I), compared to wake and wake prior to sleep. K) Average neural activity (% active neurons ± sem) for successive 5 min epochs of wake and sleep. Blue datapoints indicates average (± sem) for the fly used as an example in G-J. L) Average connectivity (mean degree ± sem) for the data in K. M) Mean degree for the data in L combining all sleep epochs per fly, compared to wake epochs of similar duration. For K and L, statistical tests are one-way ANOVAs with Dunnett’s multiple comparison correction. ns = not significant, * = p < 0.05, *** = p < 0.001. For M, statistical tests is Wilcoxon signed-rank test, ** = p < 0.01. n=7 flies. See also Figure S1 and Video S1.
Epochs of wake and sleep were identified based on the filmed behavior of individual flies (Figure 1G), with sleep initially defined as >5 min immobility [17]. From this, the corresponding neural activity traces were identified (Figure 1H). As a preliminary analysis of differences between wake and sleep, we calculated the average activity (% neurons active out of total ROIs, see STAR Methods) for wake versus sleep. These were not significantly different (average % active wake = 24.05 ± 4.25, average % active sleep = 23.21% ± 9.790; n=7, p = 0.6875, Mann–Whitney U test). To investigate this brain readout more closely, we partitioned sleep epochs into successive 5 min bins [26] and compared these to waking epochs (Figure 1I). We observed considerable variability in neural activity across a sleep bout as well as between different sleep bouts (Figure 1J), with early sleep (0–5min) typically resembling average waking activity (Figure 1J,K). Interestingly, we observed that in the 5 minutes immediately preceding sleep, activity levels drop even though the fly is still behaviorally ‘awake’ (wake prior sleep, Figure J,K). After the last detected movement, neural activity reverts to average wake-like levels during early (0–5 min) sleep and then decreases as sleep progresses, where during late sleep (>10 min) neural activity levels are typically lowest (but not always, see sleep bout 5, Figure 1J). Once a fly wakes up, neural activity resembles average wakefulness again (Figure 1K). We also calculated neural connectivity (mean degree of active neurons, Figure 1F; Figure S1D–G) across these different sleep stages, and found that they mirrored neural activity levels (Figure 1L). However, when we combined all sleep epochs and compared them to equivalent wake durations per fly, we found significantly decreased connectivity among active neurons during sleep compared to wake (Figure 1M).
Our imaging analysis tracked the activity of identified ROIs over extended periods of time (Figure 2A–D, and see STAR Methods), allowing us to ask if different sleep stages recruited overlapping or distinct groups of neurons, compared to wake (Figure 2E). To establish a baseline for neural turnover, we investigated the overlap between three successive 5 min waking epochs (Figure 2C,D) across all of our flies, and found that this was 23% on average (Figure 2D, F). This level of overlap is partly a consequence of our criteria for calling a neuron active (Figure S1C), which was set at 3 standard deviations (dotted line in Figure 2B,C; Figure S2A,B) and constrained by the calcium signal properties (Figure S2C–F). Wake immediately prior to sleep was not significantly different than the waking average, showing 16% overlap with any random 5 min wake (Figure 2F). Transition into early ‘active’ sleep also showed a similar overlap with wake, at 18%. In contrast, the fewer neurons that remained active during late sleep were largely comprised of cells that were also observed to be active during wake (60%, significantly more than the waking average, Figure 2F). In contrast, early and late sleep were almost completely non-overlapping (5%). Finally, wake immediately prior to sleep had significantly more neurons in common with early sleep (31%) than late sleep (4%). Together, these comparisons show that different sleep stages not only comprise different cell numbers, but also largely different cell identities. One caveat to these findings, however, is overlap may be less likely between epochs involving fewer active neurons (e.g., late sleep and wake prior to sleep).
Figure 2: Neural identity overlaps during wake and spontaneous sleep stages.
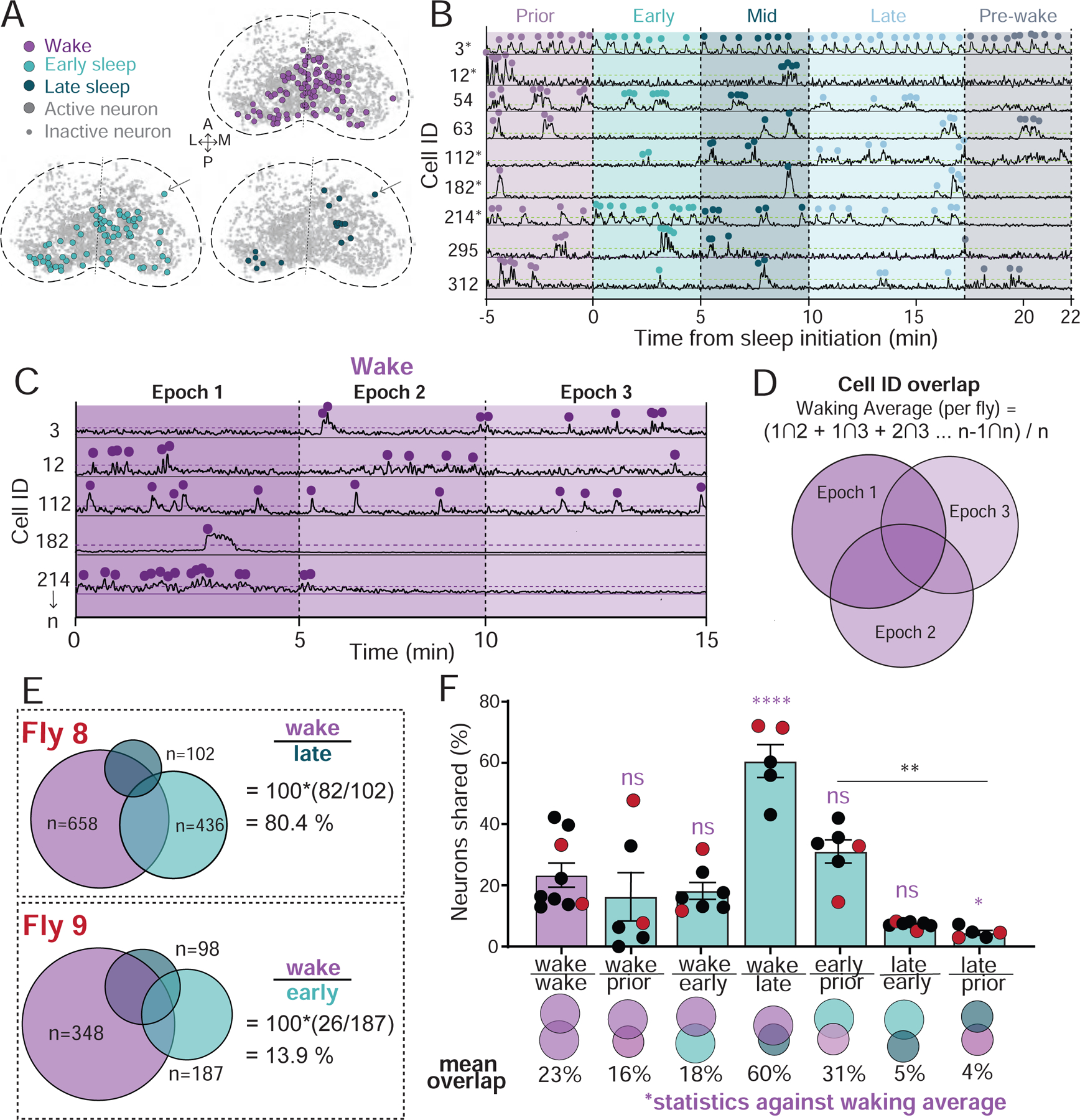
A) Collapsed masks of neurons active during wake, early sleep, and late sleep in neurons from Figure 1E. Active neurons met activity criteria in other epochs. Inactive neurons are ROIs that were identified through segmentation, but never met activity criteria. Arrow: an active neuron tracked across epochs. B) Example neural activity traces for neurons during a sleep bout. Dotted green line: activity threshold (colored dots; see Figure S2A,B). *, same neuron as in C. C) Example neural activity across three successive 5 min epochs of wakefulness. Dotted purple line: activity threshold (colored dots). D) Calculation of the average overlap in neural activity during wake (waking average). Neurons indicated are the same as in B and C. E) Overlaps in neural identities during wake and sleep stages in two example flies. The number of active neurons in each is indicated. % overlap = 100*((#stage 1 ∩ #stage 2) / #stage2). F) Overlap in neural identities compared to the waking average (% overlap ± sem). Flies from E are indicated with red dots. One-way ANOVAs with Dunnett’s multiple comparison correction. ns = not significant, * = p < 0.05, ** = p < 0.01, **** = p < 0.0001. Waking average, n = 9; all other data, n = 7. See also Figure S2.
Short bouts of immobility (1–5min) are active sleep bouts
Sleep in Drosophila is typically assessed by quiescence lasting at least 5 minutes, which has been associated with increased arousal thresholds [17]. Yet, average arousal thresholds for freely-walking flies can already be significantly higher after only 1 minute of immobility [31], suggesting that a form of sleep might already be engaged much earlier. We wondered whether this could be the early ‘active’ sleep we have identified above. To investigate this possibility, we identified all 1–5 minute immobility epochs in our dataset, which had not qualified as sleep according to the 5-minute criterion (Figure 3A, arrows). We compared the % active cells in these brief immobility epochs with the % active in immediately preceding waking epochs. Because the duration of immobility was variable (1–5min), we matched the preceding waking epoch in time (Figure 3B) and found these matched comparisons across our datasets (Figure 3C,D). Surprisingly, we found that 1–5min bouts of immobility resembled early ‘active’ sleep, with wake-like levels of neural activity immediately preceded by lower levels of neural activity (Figure 3E). Although fewer cells were active in the waking period immediately preceding, a significant percentage (40%) remained active during the subsequent ‘short sleep’ bout (Figure 3F). We next questioned what the neural overlap was between short sleep and early sleep, and found this to be 53%, significantly higher than the overlap with wake (Figure 3F). Taken together, these observations suggest that 1–5 min bouts of immobility are similar to the ‘active’ sleep stage most evident at the beginning of an extended sleep bout.
Figure 3: Short sleep bouts.
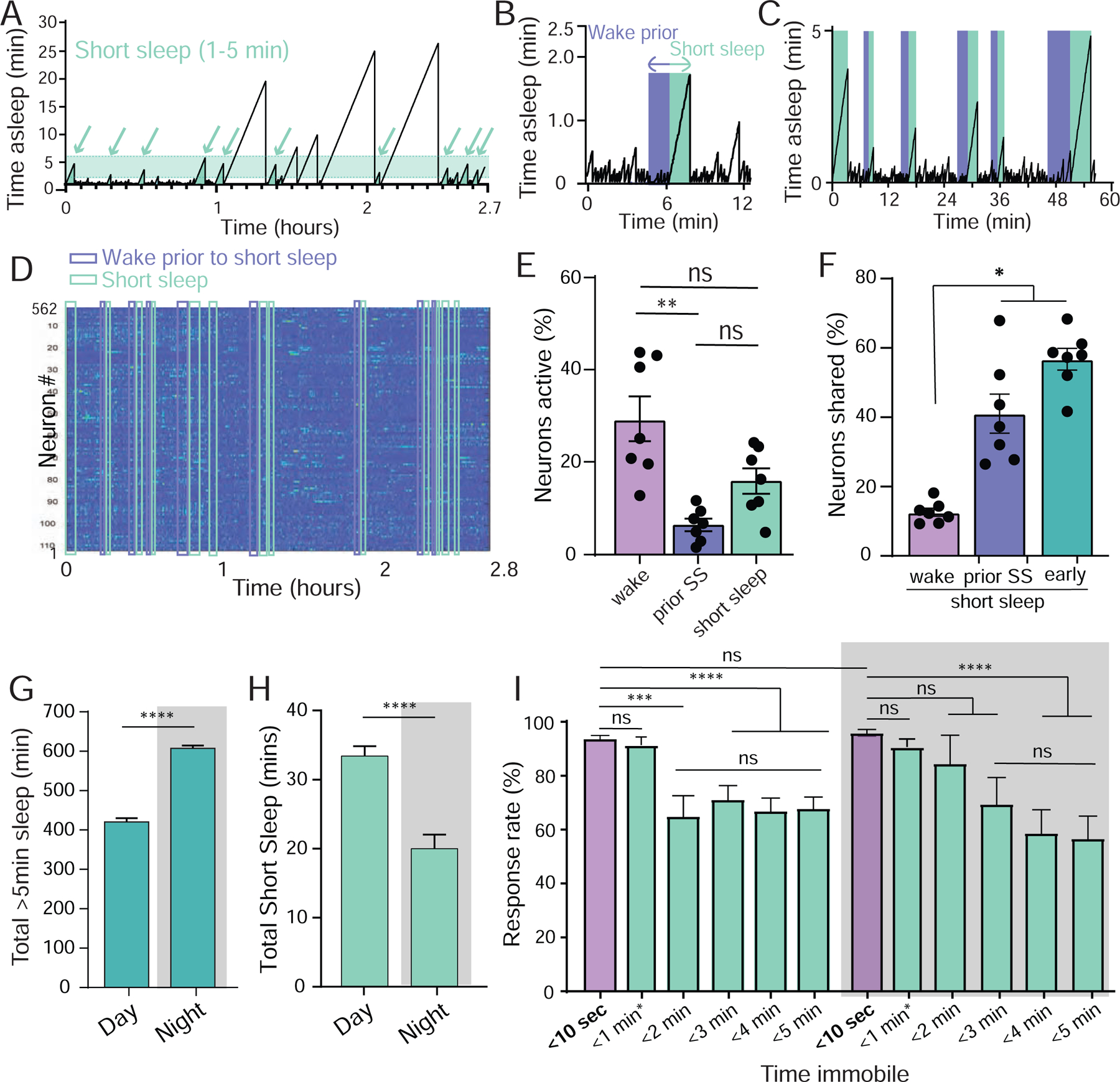
A) Short sleep (shaded, 1–5min) epochs (arrows, from same fly as in Figure 1I). B,C) For every 1–5min epoch identified (green), an immediately preceding ‘prior wake’ epoch of equal length (1–5min) was identified and used for further comparisons (purple). D) Neural activity across all active ROIs for the duration of the experiment in A, with all short sleep and corresponding prior wake epochs indicated. E) Neural activity (% neurons active ± sem) in short sleep (green) compared to wake prior to sleep (purple) and the waking average (lavender). F) Significantly more active neurons are shared between short sleep and early sleep, and between short sleep and prior sleep, than between short sleep and wake. E, Wilcoxon matched-pairs signed rank test, * = p < 0.05; F, one-way ANOVA with Bonferroni multiple comparison correction. ns = not significant, * = p < 0.05, ** = p < 0.01. n=7 flies. G) Average total sleep (± sem) for day and night in freely-walking flies, based on >5min immobility criterion. H) Average total 1–5min sleep (± sem) for day and night in the same flies as in G. I. Average behavioral responsiveness (% ± sem) to a vibration stimulus during day and night as a function of prior immobility duration, for different durations (green) compared to briefly quiescent flies (<10 sec, lavender). Flies are the same as in G and H. <1min* epoch includes <10 sec. For G and H, test = Wilcoxon signed-rank test; for I, test is One-way ANOVA with Dunnett’s multiple comparison correction. ns = not significant, *** = p < 0.001, **** = p < 0.0001.
Our experiments so far were performed on tethered flies that were prepared for brain imaging. We were curious how common 1–5 min short sleep bouts were in freely-walking flies maintained in a more traditional setting for sleep experiments in Drosophila, namely small glass tubes [16, 17]. We used the Drosophila ARousal Tracking (DART, [34]) system to track sleep (>5 min immobility) in freely-walking flies (Figure 3G) and compared this to short (1–5 min) bouts of immobility in the same animals. We found that flies display a cumulative total of ~30 minutes of 1–5 min short sleep bouts during the day and 20 minutes at night (Figure 3H). Whether these short sleep bouts represent aborted longer sleep bouts (e.g., flies were woken up before 5 min) is unclear. However, it is clear that average behavioral responsiveness is already significantly lower within the first 5 minutes of quiescence (Figure 3I), suggesting that 1–5 min bouts of immobility could be a form of ‘active’ sleep.
dFB-induced sleep resembles wakefulness
Sleep in Drosophila is promoted by activating neurons that project to the dFB [20, 23–25], and transient activation of these neurons using thermogenetic or optogenetic tools rapidly induces sleep in flies [26, 27, 35]. This suggests that dFB neurons might be responsible for the promoting the early sleep stage and short sleep bouts we have identified in our brain imaging experiments. We therefore next questioned what kind of sleep dFB activation is promoting, and if it indeed resembles an early spontaneous sleep stage. We used an optogenetic strategy to activate R23E10-Gal4 neurons (Figure 4A), which express primarily in the dFB [29]. Flies were fed all-trans retinal (ATR) to activate a red-shifted channelrhodopsin (CsChrimson [36]) expressed in these neurons, and thereby induce flies to sleep by exposing their brain to red light (Figure 4B, Figure S1A, and see STAR Methods). Simultaneous calcium imaging and behavioral analysis in R23E10-Gal4>UAS-CsChrimson-mVenus>UAS-GCaMP6f flies confirmed that red-light activation of this circuit rendered flies quiescent, while also eliciting neural responses in these cells (Figure 4C–E). To confirm that dFB activated flies in our brain recording preparation were indeed sleeping, we probed a separate group of flies in the same preparation for their arousability, by using air puff stimuli (Figure 4F). This elicited a robust and reliable behavioral response from awake flies, whereas the same flies were mostly unresponsive during dFB activation (Figure 4F, red; see Video S2 for an example of a responsive fly). This decreased level of arousability lasted the duration of the optogenetic activation, reaching a nadir of non-responsiveness after five minutes of dFB activation (Figure 4F). When optogenetic activation was stopped, flies rapidly awakened (Figure 4E) and their responsiveness returned to baseline levels (Figure 4F).
Figure 4: dFB-induced sleep resembles wakefulness.
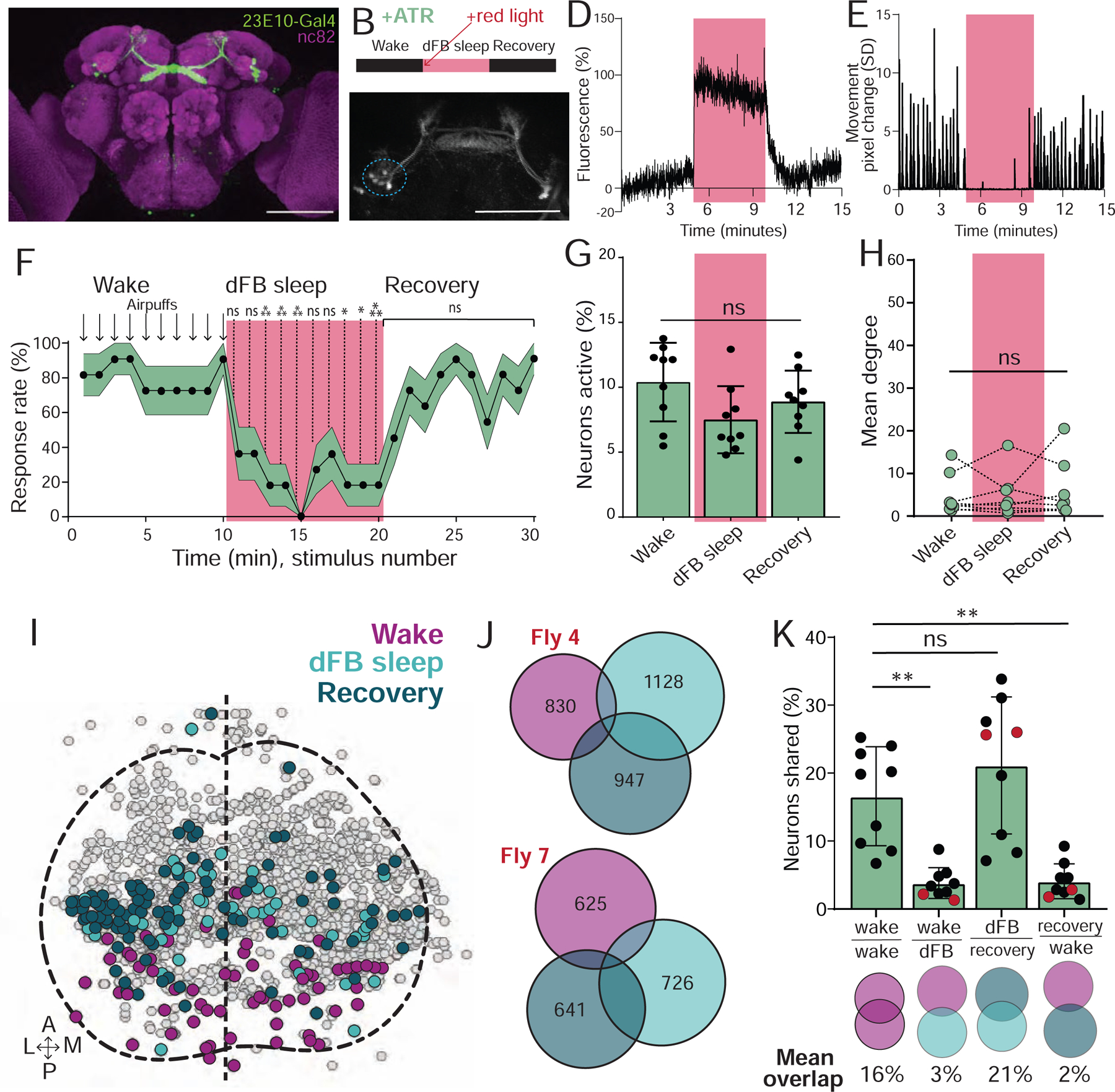
A) R23E10:Gal4-UAS:GFP expression (green). Scalebar=100μm. B) Experimental sequence. C) R23E10:Gal4-UAS:CsChrimson; UAS:GCaMP6f imaging during optogenetic activation. Scalebar=100μm. D) Optogenetic activation (red) led to an increase in GCaMP6f fluorescence in dorsal fan-shaped body (dFB) soma (circled blue in C). E) Corresponding movement trace from fly in D. F) Behavioral responsiveness (% ± sem) of flies to air puffs, before, during (red), and after optogenetic activation of the dFB. G) % neurons active (± SD) in UAS:Chrimson / X ; Nsyb:LexA/+ ; LexOp:nlsGCaMP6f / R23E10:Gal4 flies did not change during optogenetic activation of the dFB (red) in ATR-fed flies, compared to baseline wake and recovery. H) Mean degree did not change during optogenetic activation of the dFB (red), compared to baseline wake and recovery. I) Example collapsed mask of neurons active during waking, dFB sleep, and recovery. J) Overlaps between these three neural groups in two example flies. Numbers indicate active neurons within each condition. K) Analysis of the overlapping neurons between conditions. Red dots indicate flies shown in J. F: 2-way ANOVA with Dunnett’s multiple comparison test. *=p<0.05, **=p<0.01, ***=p<0.001; n=11 flies. G, H, and K: one-way ANOVA with Kruskal-Wallis multiple comparison test, ns = non-significant; n = 9 flies. See also Figure S3 and Video S2.
To measure brain activity during dFB-induced sleep, we expressed an nlsGCaMP6f reporter in all neurons (R23E10-Gal4>UAS-Chrimson88-tdTomato;Nsyb-LexA>LexOp-nlsGCaMP6f) in flies that had been fed ATR prior to experiments. This allowed simultaneous imaging of pan-neuronal calcium activity, as shown in Figure 1, alongside optogenetic activation of the dFB. We measured the activity of individual neurons during wakefulness, during dFB activation, and after the red light was turned off. We observed no effect of red light alone on behavior, neural activity, or connectivity in control (-ATR) flies (Figure S3A–C). When we activated the dFB in ATR-fed flies, we found that the quiescence and loss of responsiveness observed in the first 5 minutes of induced sleep was not associated with any significant change in average neural activity levels in the brain (Figure 4G). Further, 5 minutes of dFB sleep induction did not promote a subsequent sleep stage after activation was turned off (Figure 4G). Since neural activity typically decreases after 5 minutes of spontaneous sleep (Figure 1J,K), we conducted a second set of experiments where we induced sleep for 15 minutes, to determine if extended dFB activation promoted an eventual sleep stage with decreased neural activity. However, neural activity levels remained no different from waking even after 15 minutes of induced sleep (Figure S3D). There was also no change in the connectivity of active neurons during induced sleep, compared to baseline waking (Figure 4H, Figure S3E). Next, we determined the proportion of active neurons that were shared between induced sleep and wake (Figure 4I–K). We found little overlap (2–3%) between 5min of dFB-induced sleep and wake, but more overlap between induced sleep and the 5 min of wake post-sleep (recovery, 21%) (Figure 4K). In conclusion, dFB-induced sleep bears some hallmarks of early spontaneous sleep, although the almost complete turnover of neural identities suggests that neural dynamics are not entirely similar.
dFB-induced sleep corrects attention defects following sleep deprivation
Our brain imaging results suggest that optogenetic activation of the dFB promotes ‘active’ sleep, but whether this achieves any specific sleep function remains unclear [27]. We have recently shown that sleep deprivation impairs visual attention in Drosophila [37]. We therefore next investigated if prolonged dFB activation might reverse this behavioral defect. To measure visual attention in Drosophila, we used an open field arena paradigm [38] where individual flies walked towards a target fixation stimulus in the presence of a distracting moving grating (Figure 5A). We tested visual attention before and after 10 hours of daytime dFB activation (Figure 5B). Consistent with our previous study [37], we observed that 24 hours of sleep deprivation resulted in increased distractibility (Figure 5C; pre, -Red light +SD; Figure 5D, top). This effect persisted when flies were subjected to an additional 10 hours of daytime sleep deprivation (Figure 5C; post, +SD (long)). To confirm that the impaired attention phenotype observed following 24 hours of sleep deprivation was indeed associated with lost sleep, we measured recovery sleep in siblings (Figure S4A–C). We found that 10 hours of dFB-induced sleep (see Figure S4D–G) significantly improved visual attention (Figure 5C, post: +Red light + SD; Figure 5D, bottom) compared to extended sleep-deprived controls (post: +SD (long)), and was not different than flies that were never sleep deprived (post: -Red light). In contrast, flies that were only allowed spontaneous daytime sleep after sleep deprivation (post: -Red light + SD) trended to improved visual attention but were not significantly different than extended sleep-deprived controls. To investigate if induced sleep affected other behaviors, we measured walking speed and optomotor responsiveness in the same animals, and found that these were not affected (Figure S4D,E).
Figure 5. dFB-induced sleep corrects visual attention deficits following sleep deprivation.
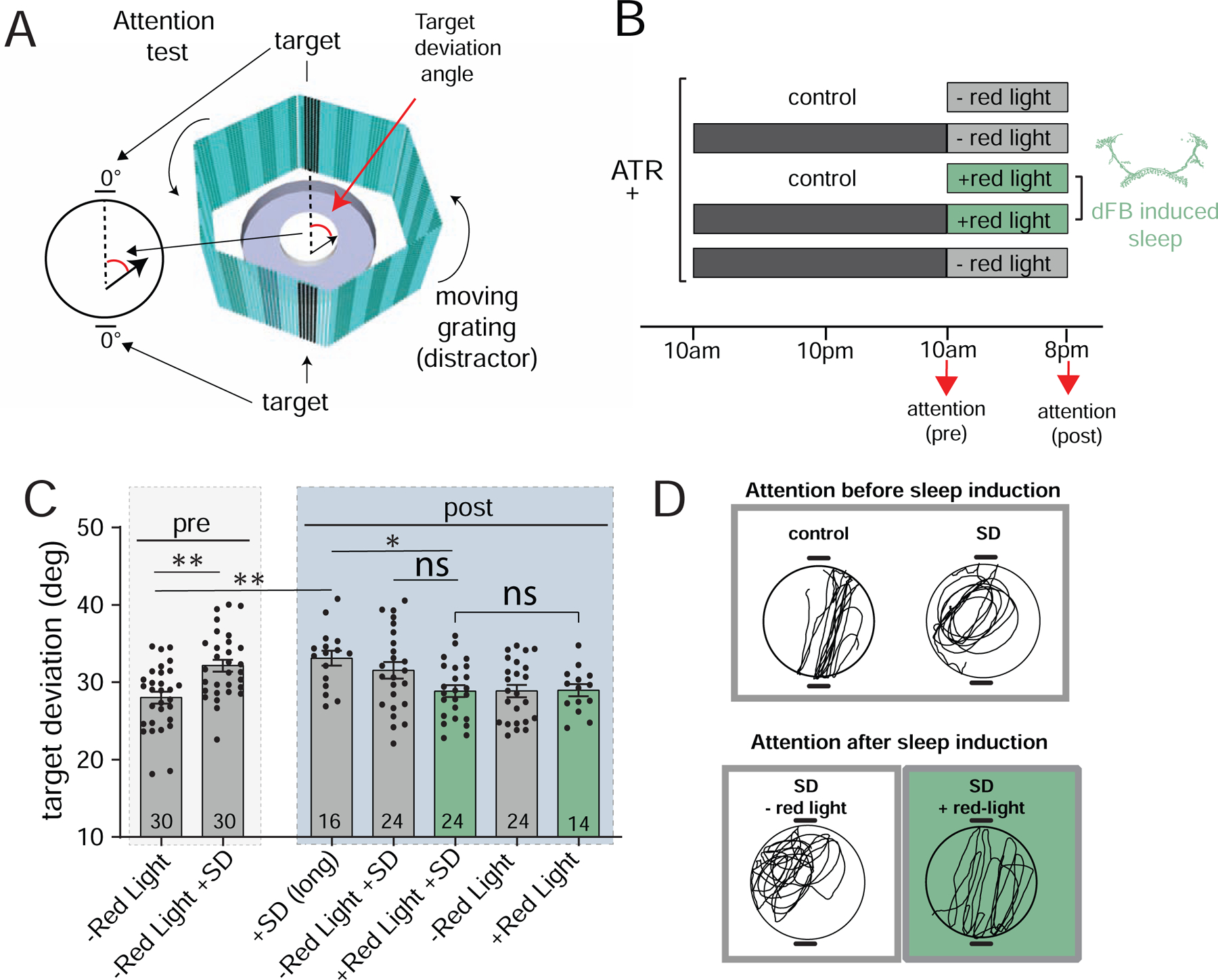
A) Diagram of the visual attention paradigm. Flies walking central platform are presented with two opposing flickering dark bars (targets) and a moving cyan grating in the background (distractor). Inset: attention is measured by the angle between the fly’s trajectory (black arrow) and the closest target (target deviation). B) Protocol for behavioral testing (all experiments with w+; R23E10-Gal4/+>UAS-CsChrimson/+ flies). Attention (pre & post) indicate when flies were tested for visual attention. C) Target deviation (degrees ± sem) before (pre) and after (post) dFB-induced sleep, for all of the conditions outlined in B. Fly numbers are indicated, pooled across 2–3 experiments each. D) Example traces of walking paths of flies immediately following sleep deprivation (SD) compared to non sleep-deprived controls (top), and after subsequent sleep induction (SD +red light) compared to controls (SD – red light). Statistical tests for C is one-way ANOVA with Dunnett’s multiple comparison correction. * = p<0.05, ** = p<0.01, *** = p<0.001. See also Figure S4.
dFB-induced sleep results in a loss of responses in visually-responsive neurons
We showed earlier that acute dFB sleep induction renders a fly profoundly less responsive to mechanical stimuli (Figure 4F, Figure S4D–G), however neural activity appears indistinguishable from wakefulness (Figure 4G,H; Figure S3D,E), and therefore most resembles early spontaneous sleep (Figure 1) and short sleep bouts (Figure 3). Having not excluded that acute activation of this Gal4 circuit drives a semi-paralyzed waking state, we next probed responsiveness levels in the brain itself, by investigating a sensory modality more relevant to our attention experiments: vision. Unlike many sleeping animals, insects cannot shut their eyes, so suppression of neural responses to visual stimuli during sleep in bees or flies (e.g. [39, 40]) must stem entirely from altered brain dynamics. We therefore next asked if dFB sleep suppresses visual responses in the fly brain.
To probe visual responsiveness in the fly brain, we used a flickering ultraviolet (UV, 405nm) stimulus (see STAR Methods), which is highly salient to flies [41]. We first confirmed that the UV light elicited robust local-field potential (LFP) responses in the fly optic lobes (Figure S5A–C). To measure visual responsiveness in the fly brain during sleep and wake, we next tracked neural activity throughout the course of the experiment, which we achieved by utilizing the same preprocessing pipeline as before (Figure S1), paired with k-means clustering to identify neural responses (Figure S5D–G, and see STAR Methods). We recorded calcium activity during presentation of the visual stimulus (V1), during baseline wakefulness, and during dFB sleep (Figure 6A). Crucially, during dFB sleep the same visual stimulus that was shown in wakefulness was presented to the fly a second time (V2). We observed different categories of neurons (Figure 6B): some responded to the visual stimulus (Figure 6C, top), some responded to dFB activation (Figure 6C, bottom), some were spontaneously active during sleep or wake, and some were active across these different categories (individual traces in Figure 6D).
Figure 6. Visual responses in the brain are lost during dFB sleep.
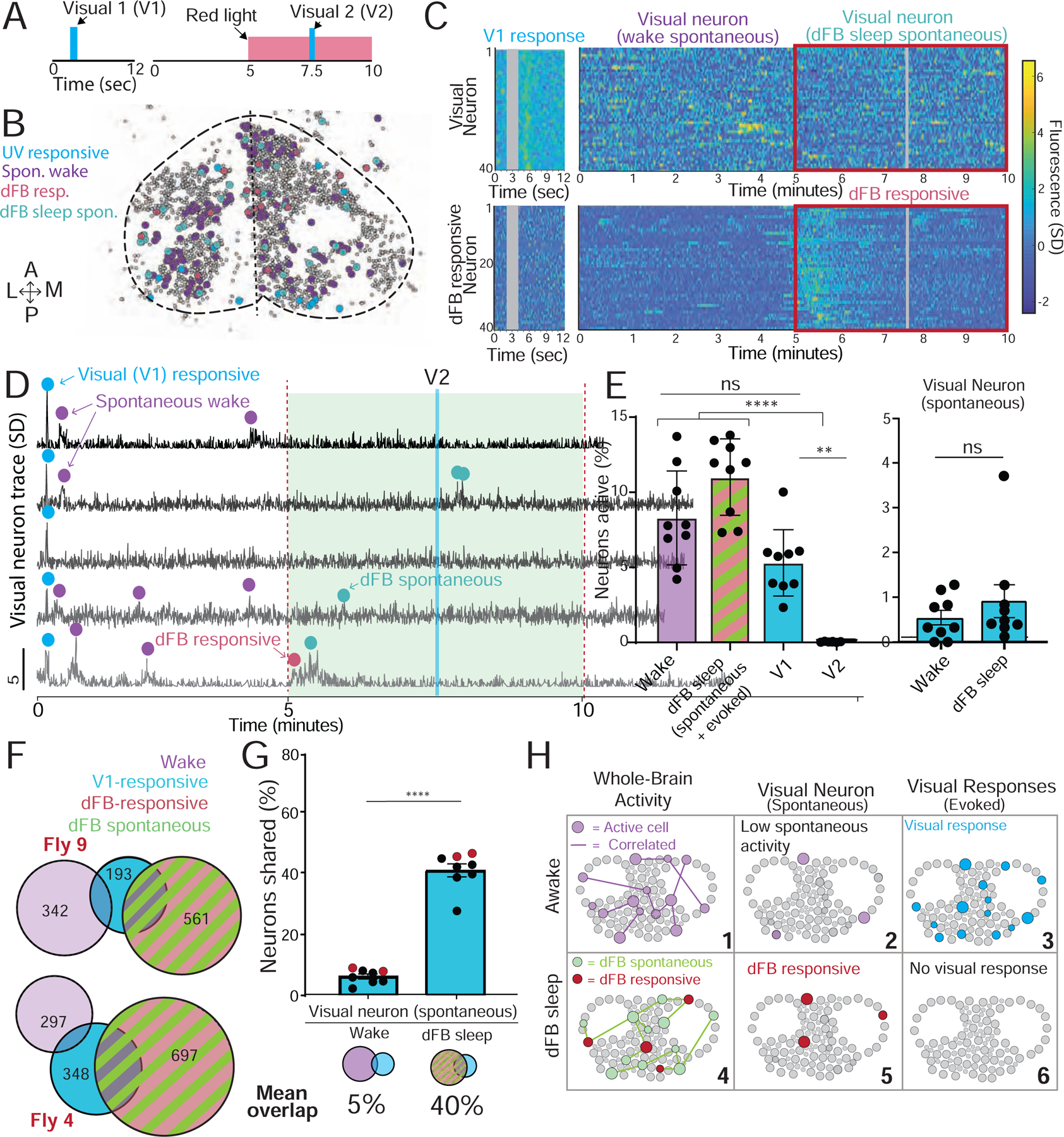
A) A 2-second ultraviolet (UV) stimulus (V1) was used to probe visual responses in the brain of w+ ; UAS:CsChrimson / X ; Nsyb:LexA/+ ; LexOp:nlsGCaMP6f / R23E10:Gal4 flies during wakefulness and during dFB sleep (V2). B) A collapsed mask of spontaneous baseline neurons (purple), dFB-responsive neurons (red), spontaneous dFB sleep neurons (green), and visually responsive neurons (blue) in an example fly. C) Top left: visually responsive neurons in an example fly. Top right: spontaneous activity in the same neurons during wake and dFB sleep (red box). Bottom left: data from the same fly for dFB responsive neurons presented with the visual stimulus. Bottom right: the same dFB responsive neurons during wake and dFB sleep (red box). Gray bar: timing of visual stimulus. D) Calcium activity traces of five neurons found to be responsive to the first visual stimulus during wake (V1, blue dot); purple, spontaneous waking activity; green, spontaneous activity during dFB activation; red, response directly driven by activation of the dFB. E) Left: % active neurons during wake (purple), dFB sleep (green/red) and following visual stimulation (cyan). Right: spontaneous activity of visual neurons during both wake and dFB sleep. F) Overlapping active neurons between V1 (blue), wake (purple) and dFB sleep (red/green) in two example flies. The number of active neurons per group is indicated. G) V1 neurons had a significantly higher overlap across dFB sleep-active neurons compared to spontaneous wake neurons. Red dots indicate the flies shown in F. H) Cartoon of dFB sleep effects on spontaneous and evoked activity. (6). Tests = one-way ANOVA with Friedman multiple comparison test. ns = non-significant, * = p<0.05, ** =p<0.01, **** =p<0.0001. n = 9 flies. Error bars in E are SD, error bars in G are sem. See also Figure S5 and S6.
Consistent with our other experiments, dFB-induced sleep did not decrease neural activity in the fly brain (Figure 6E, left: dFB sleep (spontaneous + evoked); Figure 6C, red boxes). Nevertheless, neurons that responded to the UV stimulus during wakefulness almost never responded to the UV stimulus during dFB-induced sleep (Figure 6E, left: V1 vs V2; Figure 6C, top). In control (-ATR) flies, visual responses were robust during the red-light stimulus, confirming that this loss of visual responses was not a red-light effect (Figure S5D–H). Visually responsive neurons nevertheless maintained a low level of spontaneous activity during wakefulness and dFB sleep (Figure 6E, right), indicating that these neurons were not completely silenced. Rather, they were completely disconnected from the visual stimulus. Comparing neural identities across our experimental conditions (Figure 6F), we found that 40% of visually responsive neurons were also dFB-active neurons (Figure 6G, and see example in Figure 6D, bottom trace), which was significantly more than the average overlap with wake-active neurons (5%).
In recent electrophysiological studies we found that the dFB produces theta-like (7–10Hz) oscillatory activity when activated [26, 35]. We therefore replicated our last experiment employing a more physiologically relevant sinusoidal 7Hz red-light activation regime (see STAR Methods) in a different set of flies and found the same result: dFB activation abolishes visual responsiveness and ~40% of visually responsive neurons are also dFB active (Figure S6A–D).
Together, our imaging results present the following scenario for dFB-induced sleep. During wakefulness, the brain maintains an average level of activity through populations of neurons that are highly dynamic, with correlated activity among these neurons (Figure 6H–1). During wake, visually responsive neurons in the central brain display low spontaneous activity (Figure 6H–2), however visual responses are robust (Figure 6H–3). During dFB-induced sleep, spontaneous activity and connectivity are not different from wakefulness (Figure 6H–4). However, increased activity in dFB-responsive neurons (Figure 6H–5) suppresses visual responsiveness in the central brain (Figure 6H–6).
Discussion
To be able to experimentally switch sleep on and off ‘at will’ in flies is a compelling approach towards uncovering conserved biological processes underlying the diverse functions of sleep [11]. Accordingly, the discovery of sleep-promoting neurons in the dorsal fan-shaped body (dFB) of Drosophila [20] has led to understanding how homeostat-like neurons in the fly brain might integrate sleep pressure signals from other circuits to activate a ‘sleep switch’ [23–25, 35, 42]. On the other hand, neurons in the fly dFB appear to regulate diverse behaviors during wakefulness more generally, such as responses to visual and mechanical stimuli [26, 35, 43, 44], hinting that they involve more than just sleep-regulatory neurons. Indeed, potentially overlapping roles between sleep and wake seems intuitive, as sleep by definition requires altered arousal thresholds that logically begin during waking. If a sleep switch also serves more broadly as a perceptual switch [4, 35], then one could question if dFB-activated flies are really asleep. When optogenetic activation of the dFB is stopped, flies wake up almost immediately, which is not what might be expected following spontaneous sleep onset. Additionally, we have shown elsewhere that flies remain more responsive during earlier stages of spontaneous sleep compared to later stages [31, 34], yet dFB activation rapidly renders flies deeply unresponsive. These discrepancies with spontaneous sleep highlight that current optogenetic approaches to activating circuits may not always be physiologically accurate; it seems unlikely for example that all dFB neurons would be activated simultaneously during spontaneous sleep. This may explain differences observed between early spontaneous sleep and dFB-induced sleep, which we propose are related. Future spontaneous sleep imaging experiments specifically labelling dFB neurons (or other candidate neurons) should resolve the exact pattern of cells involved.
If dFB-induced sleep resembles wakefulness, then why are flies unresponsive to mechanical and visual stimuli? The observation that R23E10-Gal4 also expresses in non-dFB circuits such as descending or ventral nerve cord neurons [29] may explain some of the observed behavioral effects, but does not explain loss of visual responsiveness in central brain neurons. Perhaps synchronized activity in the dFB [26, 35] causes brain activity in other circuits to be reorganized in a way that is not conducive to processing external stimuli, and this may simultaneously promote a neurochemical environment that optimize internally-driven homeostatic processes. One caveat of our brain imaging approach is that it focused on neuronal cell bodies, and it remains unclear what aspect of neuronal signaling is represented by the peaks of calcium activity we tracked through time. It could be that other neuronal compartments become less active or less correlated during dFB sleep, or that longer (>15min) recordings might reveal decreased neural activity during prolonged dFB activation, as in our visual attention experiments.
Our calcium imaging experiments support the view that spontaneous sleep in Drosophila is dynamic [26, 31, 34, 45]. We uncovered different sleep stages, including an early ‘wake-like’ active stage and a later less active ‘deep’ sleep stage. Interestingly, ‘wake-like sleep’ is often preceded by ‘sleep-like wake’ (consistent with an earlier study [46]), showing that either behavioral state can be confounded at the level of brain activity. This paradox was evident even for shorter sleep bouts (1–5min), which were quite common in our imaging experiments and associated with increased arousal thresholds in freely-walking animals. This suggests that ‘active’ sleep functions, which could involve optimizing attentional processes, might also be engaged during these shorter bouts of quiescence that never transform into deep sleep. Although short sleep bouts only amounted to about an hour total over a day and night in wild-type flies, together with any ‘active’ sleep occurring within longer (>5min) sleep bouts this could add up to a significant proportion of sleep time in Drosophila. The total amount and distribution of ‘active’ sleep occurring in flies remains to be determined, although this could depend on homeostatic regulation [37] as well as waking experience [47].
In mammals and birds, sleep stages are formally characterized by their electrophysiological correlates: slow wave sleep (SWS) presents with high amplitude 1–4Hz brain oscillations, whereas during rapid eye movement (REM) sleep, brain activity looks more similar to an awake brain [48, 49]. REM sleep thus qualifies as sleep largely on the observation that behavioral responsiveness to the outside world is suppressed, i.e., that arousal thresholds are elevated [50, 51]. It is also worth noting that arousal thresholds in mammals can be as high during REM sleep as so-called ‘deep’ sleep [50–53]. For these reasons, REM sleep has also been termed ‘paradoxical’ sleep, which is its original label [54, 55].
Sleep functions probably predate the idiosyncratic electrophysiological signatures that happen to be associated with them in mammals. Thus, SWS has been associated with cellular processes such as growth, stress regulation, metabolite clearance, and synaptic homeostasis [56–58]. In contrast, REM sleep has been associated with circuit-level homeostatic processes such as emotional or motor learning [59–62]. It seems likely that the need to curate cellular processes while animals are behaviorally quiescent predates the advent of SWS, as suggested by findings that even brainless nematodes become briefly quiescent to satisfy some of these ancient biological needs [18]. Decreased neural activity during drug-induced ‘deep’ sleep in flies [21, 26] suggests a conserved GABA-driven regulatory mechanism that promotes prolonged neural quiescence, which may be functionally related to the synchronized ‘down-states’ enabled by SWS in mammals and birds [63–65]. In contrast, the rapid eye movements that are associated with REM sleep in certain vertebrates may be less relevant to this eponymous sleep stage than the observation that the brain is essentially awake, but selectively unresponsive to the outside world [51]. Our work suggests that some sleep stages and functions may require the fly brain to remain active but disconnected from the outside world. Whether this form of sleep in flies is similar to paradoxical sleep stages in other animals should be determined by further evaluating the functions being served, and if these differ from conserved deep sleep functions.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources can be directed and fulfilled by the Lead Contact, Bruno van Swinderen (b.vanswinderen@uq.edu.au).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The data underpinning the publication is stored in the University of Queensland Research Data Management (UQRDM) via UQ eSpace, the institutional data storage repository of the University of Queensland (UQ).The metadata in UQ eSpace is indexed by common search engines (e.g., Google) as well as by Research Data Australia (the national data discovery platform) and also by Data Citation Index. Matlab analysis scripts used in this study are available from the Lead Contact upon request.
Experimental Model and Subject Details:
Animals:
Drosophila melanogaster flies were reared in plastic vials on standard yeast-based medium under a 12:12 light/dark (8 AM:8 PM) cycle and maintained at 25°C with 50% humidity. Adult, 3–5 day-old female flies were used for all experiments and randomly assigned to experimental groups. Fly lines used for behavioral experiments were R23E10-Gal4 (attp2; Bloomington 49032; Bloomington Drosophila Stock Center, Bloomington, Indiana), UAS-CsChrimson-mVenus (attp18; Bloomington 55134; Provided by Janelia Research Campus, Ashburn, Virginia), and Canton-S (Bloomington 64349). For 2-photon experiments, flies with the genotype 10XUAS-Chrimson88-tdTomato (attp18) / +: LexAop-nlsGCaMP6f (VIE-260b); kindly provided by Barry J. Dickson) / +: Nsyb-LexA (attP2), LexAop-PAGFP (VK00005) / R23E10-Gal4 were used. Optogenetically-manipulated fly lines were maintained on food containing 0.2mM all-trans retinal (ATR; Merck, Darmstadt, Germany) for 24 hours prior to assay to allow for sufficient consumption.
Method Details:
Behavioral analysis:
Sleep behavior was calculated with the Drosophila ARousal Tracking system (DART) as previously described [34]. Prior to analysis, 3–5 day-old virgin females were collected and loaded individually into 65 mm glass tubes (Trikinetics, Waltham, MA) that were plugged at one end with standard fly food, containing either vehicle or 0.2mM ATR. Controls were placed onto standard food and housed under identical conditions as the experimental groups. The tubes were placed onto platforms (6 total platforms, 17 tubes per platform, up to 102 flies total) for filming. Flies were exposed to ultra-bright red LED (617 nm Luxeon Rebel LED, Luxeon Star LEDs, Ontario, Canada) which produced 0.7mW/mm2 at a distance of 4–5 cm from the fly for the duration of the experiment for optogenetic activation. Sleep behavior was probed with hourly exposure to a train of 5 vibrational stimuli (1.2 g), each lasting 200 ms and spaced 800 ms apart. Fly movement in response to this stimulus was recorded, using 3mm as a movement threshold. A decreasing percentage of responsive flies is associated with increased sleep intensity [31, 34]. Sleep intensity was measured as the proportion of immobile flies that responded to these stimuli. For >5min sleep experiments, flies were determined to have responded if they moved by a threshold of at least 3 mm (~3 body lengths) within the minute following the stimulus. Significance was determined by performing Mann-Whitney U tests between groups, following a test for normality. For 1–5 min sleep experiments, flies were determined to have responded to the hourly stimulus if they moved by a threshold of at least 3 mm (~3 body lengths) within 10s following the stimulus. Significance for short sleep responsiveness data was determined using One-way ANOVA with Dunnett’s multiple comparison correction.
Sleep deprivation and visual attention paradigm:
Flies were sleep deprived (SD) with the use of the previously described Sleep Nullifying Apparatus (SNAP) [37]. For behavioral testing, female progeny from a w+; 23E10-gal4 and w1118; UAS-CsChrimson cross had their wings clipped while anesthetized by CO2, at least 3 days prior to the experiment. Flies were housed in vials of up to ~40 females and ~5 males up until 24 hrs before the experiment, when they were divided into different experimental groups of ~12 females and 1 male and were transferred to 0.2mM ATR food. For testing for sleep rebound, sibling-matched flies were placed into 65 mm glass tubes (Trikinetics), containing either standard food media or 0.2mM ATR food, and monitored using the DART platform. Flies were sleep deprived on the SNAP device for 24hrs as previously described [37], while control flies were allowed to rest. Immediately following sleep deprivation (at 10am), some sleep-deprived and control flies were tested for visual attention, while some were transferred to a red light exposure condition for 10 hrs sleep induction. Following sleep induction, these flies were tested for visual attention (at 8pm), alongside controls that had been allowed to rest normally without sleep induction, in addition to a group that had undergone a long sleep deprivation protocol (34 hrs, until the 8pm test). Visual attention was measured by tracing the walking paths of flies responding to competing visual stimuli (7 Hz flickering targets, overlayed against a 3 Hz grating of speed 54°s −1 in the background) as described previously [37] CeTran (3.4) software [38] was used to calculate the target deviation angle and the walking speed of individual flies, in addition to custom scripts in R programming language used to calculate the optomotor response (the turning angle of the fly in °s −1 in the direction of the grating movement). Prism software was used to test for normality of data, and to perform one-way ANOVAs with Dunnett’s multiple comparison correction to test for significant differences between groups. To test for a sleep rebound resulting from the sleep deprivation paradigm, flies were recorded for sleep behavior for six consecutive hours post-sleep deprivation, compared to handling controls. All analysis was performed with DART software, as described above.
Two-photon imaging:
2-Photon Imaging was performed using a ThorLabs Bergamo series 2 multiphoton microscope. A Ti:Sapphire laser (Mai Tai Deepsee, Spectra Physics) tuned to 920nm was used as an excitation source, and laser power was controlled with a pockels cell with an electro-optic modulator (Conoptics). Z-movement of the imaging objective (Nikon CFI APO 40XW NIR) was performed with a Piezo controlled Focus motor with 400μm of movement. Fluorescence was detected with a High Sensitivity GaAsP PMT (ThorLabs, PMT2000). GCaMP fluorescence was filtered through the microscope with a 594 dichroic beam splitter and a 525/25nm band pass filter. A 617nm LED at 0.31mA/mm2 was delivered through the objective for activation of CsChrimson [36].
For two-photon experiments, flies were secured to a custom-built holder. Extracellular fluid (ECF) containing 103 NaCl, 10.5 trehalose, 10 glucose, 26 NaHCO3, 5 C6H15NO6S, 5 MgCl2 (hexa-hydrate), 2 Sucrose, 3 KCl, 1.5 CaCl (dihydrate), and 1 NaH2PO4 (in mM) at room temperature was used to fill a chamber over the head of the fly. The brain was accessed by removing the cuticle of the fly with forceps, and the perineural sheath was removed with a microlance. Flies were allowed to recover from this for one hour before commencement of experiments. For experiments in Figure 1 and Figure S2, dissections were performed through the cuticle on the top of the head, of flies that had been sleep deprived for the 24 hours prior.
Imaging was performed across 18 z-slices, separated by 6μm, with two additional flyback frames. The entire nlsGCaMP6f signal was located within a 256 x 256px area, corresponding to 667 x 667μm. Fly behavior was recorded with a Firefly MV 0.3MP camera (FMVU-03MTM-CS, FLIR Systems), which was mounted to a 75mm optical lens and an infrared filter. Camera illumination was provided by a custom-built infrared array consisting of 24 3mm infrared diodes. Behavioral data was collected for the duration of all experiments. For optogenetic experiments in either experimental or control animals, either a 15 or 45 minute imaging timeframe was used. For the 15-minute experiments, this was comprised of 5 minutes of baseline activity, followed by five minutes of CsChrimson activation with 0.01mW of 617nm light (a block activation was used in experiments displayed in Figure 6, and a 7Hz sine wave light stimulus was used for experiments displayed in Figure S6), and followed by five minutes of recovery. For the 45-minute experiments in Figure S3D–E, this was comprised of 15 minutes of baseline activity, followed by 15 minutes of CsChrimson activation as described above, and 15 minutes of recovery. For further processing, these segments were divided into three five-minute long segments each for baseline, CsChrimson activation and recovery. Baseline and recovery data were averaged per fly and compared to the three successive 5 min dFB sleep epochs.
Behavioral responsiveness probing in tethered flies:
For probing behavioral responsiveness, flies were subjected to a 50ms long, 10psi air puff stimulus, which was generated using a custom-built apparatus and delivered through a 3mm-diameter tube onto the front of the fly. Flies were subjected to 10 pre-stimuli at a rate of one puff/minute, to characterize the baseline response rate. dFB sleep was then induced for 10 minutes, where a further 10 air puff stimuli were delivered, at a rate of one puff/minute. Following this, red light activation was stopped and a further 10 ‘recovery’ air puffs were applied. Behavioral responses to the air puff were noted as a ‘yes’ (1) or ‘no’ (0), which were characterized as the fly rapidly walking on the ball immediately following the air puff. For statistical analysis, the pre-condition was compared to the 10 minutes of activation, or 10 minutes of the recovery condition.
Optic lobe LFP recordings of visual UV stimulus:
Flies were tethered and prepared as described for the 2photon imaging. Recordings were performed by using a glass electrode (7–9 GΩ) with a field effect transistor (FETs) for amplification of small currents. Data was further amplified (X1000) and filtered between 0.1Hz and 5kHz (A-M Systems Model 1700), digitized (Axon Digidata 1400 A Digitizer) and sampled at 1000Hz with the acquisition software AxoGraph 1.4.4 (Axon Instruments). For visual stimulation we used a single UV LED diode with an intensity of 10mW, which is comparable to the 2photon experiments. The visual stimulation was controlled via a custom written AxoGraph script. The visual stimulus was a 7Hz UV flicker with (25ms on, 75ms off, 10mW of power). Each trial was recorded for a duration of 180s with a 60s baseline, a 60s stimulation period, and a 60s recovery. Every animal was tested for consecutive 5–6 trials with 30 seconds in between trials.
Visual responsiveness:
Flies were tethered as described above, using both dFB-sleep and control flies. For measuring visual responsiveness, the addition of a UV-LED emitting 10mW of 405nm light was placed in the frontal visual field of the fly. Initially, a 12 second recording was performed, where a 2 second flickering 7Hzvisual stimulus was presented to the fly. Following this, a 15-minute recording session followed, consisting of 5 minutes of baseline condition, 5 minutes of red light (same properties as described above) and 5 minutes of recovery. During the red light stimulus, an additional 2 second 7Hz UV-light visual stimulus was presented to the fly, 2.5minutes into the red light delivery.
Spontaneous sleep recordings and analysis of behavioral data:
Flies with the genotype UAS:Chrimson / X ; Nsyb:LexA/+ ; LexOp:nlsGCaMP6f/+ were tethered as described above, with a window cut in through the top of the head. Simultaneous 2-photon and behavioral imaging were acquired for a minimum of 2 hours per fly, with a maximum of 2.8 hours. Behavioral data for each fly was analyzed using a custom-written MatLab code that measured the change in greyscale pixel values occurring over the legs of the fly on the ball over the entire time series. Data was temporally segmented into short sleep (1–5min inactivity), sleep (>5 minutes inactivity) and wake (<1min inactivity). These experimental segments were further divided into wake prior to sleep (5 minutes immediately prior to the start of immobility), early sleep (<5min), mid sleep (5–10min), late sleep (>10min) and sleep prior to wake (5 minutes immediately prior to a fly resuming movement). The temporal locations of these epochs weres used to extract the corresponding 2-photon imaging data.
Quantification and Statistical Analysis:
LFP Analysis:
AxoGraph data files were first converted into Matlab files. A custom written Matlab script was used for the spectral analysis of the LFP data using the Matlab FieldTrip toolbox. Data was first filtered to exclude 50Hz line noise (iirnotch.m, filter.m) and subsequently filtered between 0.2–100Hz using a second order butterworth filter (butter.m, filter.m). To generate the spectrogram we used a frequency resolution of 0.01Hz for a frequency band between 6–8Hz and a time resolution of 1000Hz for the duration of one trial (180s) and used the ft_specest_wavelet.m function. Data for each trial was then normalized by averaging the power for the first 58 seconds and divide the whole spectrum by the baseline average. We first averaged all trials per fly and then averaged this data for all flies and normalized the resulting data between 0 and 1. For the power spectrum we divided the filtered data into baseline and UV stimulation and computed Fourier transformations for both datasets (fft.m) and calculated the resulting power spectrum (Powerspectrum = 2*dt^2/T * FourierTransform.*conj(FourierTransform), dt=step size(1/1000), T=60s) and transformed the power to log10 for better representation. Again, we first averaged all trials per fly and then averaged this data for all flies.
Imaging analysis:
Preprocessing of images was carried out using custom written Matlab scripts and ImageJ. To remove X-Y motion artefacts, a reference slice (at the middle timepoint) in each experimental segment (baseline, treatment, recovery for dFB and THIP experiments, sleep and wake for spontaneous sleep experiments) was computed separately. Second, in each experimental segment, the images (per z-slice) were registered to the corresponding reference slices (per z-slice) mentioned above. Image registration was achieved using efficient sub-pixel image registration by cross-correlation [69].
Each z-slice in a volume (18 z-slices and 2 flyback slices) is acquired at a slightly different time point compared to the rest of the slices. Hence to perform volume (x,y,z) analysis of images, all the slices within a volume need to be adjusted for timing differences. This was achieved by using the 9th z-slice as the reference slice and temporal interpolation was performed for all the other z-slices using ‘sinc’ interpolation. The timing correction approach implemented here is conceptually similar to the methods using in fMRI for slice timing correction.
A standard deviation projection of the entire time series was used for watershed segmentation with the ‘Morphological segmentation’ ImageJ plugin [70]. Using a custom-written MatLab (Mathworks) code, the mean fluorescent value of all pixels within a given ROI were extracted for the entire time series. This resulted in an n x t array for each slice of each experiment, where n refers to the number of neurons in each Z-slice, and t refers to the length of the experiment in time frames. These greyscale values were z-scored for each neuron, and the z-scored data was transformed into a binary matrix where a value of > 3 standard deviations of the mean was allocated a ‘1’, and every value < 3 standard deviations was allocated a ‘0’. To determine whether a neuron fired during the entire time series, a rolling sum of the binary matrix was performed, where ten consecutive time frames were summed together. If the value of any of these summing events was greater or equal to seven (indicating a fluorescent change of > 3 standard deviations in 7/10 time frames), a neuron was deemed to be active.
To determine the activity of neurons during individual treatments, the same summing protocol was performed but only within the time frame where that treatment occurred (i.e. baseline activity was only calculated based on the first five minutes of recording). For THIP sleep experiments, the five minutes occurring after an initial 30 seconds of behavioral inactivity were used. After identifying firing neurons for each condition, the percentage of active neurons was calculated across the volume by taking the number of active neurons and dividing it by the total number of neurons (defined as the number of ROIs that are <15pixels (the approximate size of a neuron in our imaging setup)).
To perform graph-theory analysis, all active neurons identified above were concatenated into a single matrix of all active neurons, separated into individual treatment conditions for each fly. Neural traces were correlated to each other, creating an adjacency matrix of correlation coefficients. To determine a threshold for significant correlations, neural traces were temporally shuffled separately (see Figure S1) and were these temporally-unmatched traces were correlated to each other. This process was repeated 1000 times, resulting in 1000 adjacency matrices of correlation values for temporally shuffled data. All correlation values were concatenated together, and the 99th percentile value of the correlation coefficients was used as a significant threshold for temporally aligned correlation values. This value was used to transform the original correlation matrix into a binary matrix, representing only significant correlations between neurons. These binary matrices were then treated as a graph, where each neuron represented a node, and significant correlations between matrices were represented as edges. To calculate the mean degree of a graph, the binary adjacency matrix was summed, and the mean of this was extracted.
Convolution of calcium signals:
To create an average nls-GCaMP6f response kernal, 70 randomly chosen activity events (as identified by the thresholds outlined above) in 10 flies were averaged together across an 8-second window. Additionally, four binary vectors were created, spanning 120 seconds with action potential events incorporated from 30–60s, with the firing rates 1Hz, 0.5Hz, 0.2Hz and 0.1Hz. Using the MatLab function “conv”, the kernel and one of the binary vectors were convolved together, resulting in the equivalent multiplication of the two polynomial vectors. This was repeated for each binary vector, resulting in a convolved signal for each firing rate. This convolved signal represents potential GCaMP6f dynamics, based on the kinematics of the GCaMP6f response and the likely firing rates of a given neuron.
Comparison of neural populations during sleep:
To determine whether neurons employed during sleep are different to those during wake, we first extracted the neuronal IDs of all neurons active during 5 minutes of the sleep stages described below. For spontaneous sleep recordings, this consisted of the neurons active during 5 minutes of wake, wake prior to sleep, early, mid and late sleep. For dFB experiments, the identities of neurons during wake, treatment and recovery were collated. For short sleep epochs in spontaneous sleep experiments where the length of a sleep bout was <5min, the IDs were compared to a time-locked period of wake prior to sleep. To serve as a control for the baseline level of overlap in neural IDs, for the spontaneous sleep dataset, in the same animals, we took instances where there was 15 minutes of continuous wake and segmented this into three five-minute epochs. The level of overlap between these epochs was calculated and used as a ‘waking average’ to compare all condition overlaps against. For dFB-activation experiments, a waking average dataset was collected by imaging and comparing the overlaps between 15 min of wake, in a separate group of flies with the genotype 10XUAS-Chrimson88-tdTomato/X: LexAop-nlsGCaMP6f / +: Nsyb-LexA, LexAop-PAGFP / R23E10-Gal4 flies. The identities of neurons found as active within each epoch were compared, resulting in an average percentage of neurons shared across wake epochs. This was then repeated to compare the identities of neurons active in all behavioral states described above.
Analysis of visual responsiveness during 2-photon imaging experiments:
To analyze both dFB-activation and visual responsiveness data, experiments were preprocessed as described above, to identify active neurons. To identify neurons responsive to the first visual stimulus, in MatLab v2018b, neurons found to be active during the first visual recording underwent a linear regression, where the regressor consisted of the onset of the UV light stimulus. Visually responsive neurons were identified as having a R2 value of >0.1 to the regressor. To identify the nature of R23E10-activation responsive neurons, the traces of neurons active during the red light stimulus were clustered using K-means clustering into 36 components, which was identified as the optimal number of components through both the optimal-K Matlab function, as well as the elbow method (a rudimentary estimator of the relationship between the number of components and the explained variability of those components). To identify which of these components were likely to be driven by either R23E10 activity, or the second visual response, a multivariate linear regression was performed against 4 regressors, consisting of both the onset and offset of the red light stimulus, the UV stimulus, as well as a regressor that represented neurons that were ‘on’ for the duration of the red light. Any component with an R2 value > 0.1 to any of these regressors was identified as responsive. Additionally, components were required to belong to >80% of flies for them to be included in any analysis. Visual responsive experiments were repeated in control (-ATR) flies with the same genotype, to ensure that any observed changes were not an artefact of red light exposure. These were analyzed the same as described above to identify visually responsive neurons (Figure S6).
Statistical analyses:
Data was analyzed and graphed using Graphpad Prism. All data was checked for Gaussian distribution using a D’Agostino-Pearson normality test prior to statistical testing. Specific descriptions of statistical tests used can be found in the figure legends.
Supplementary Material
Genotype of fly UAS:Chrimson / X ; Nsyb:LexA/+ ; LexOp:nlsGCaMP6f/+. One optical plane is shown, sped 25X. Scale bar = 100μm.
Genotype of fly is R23E10-Gal4>UAS-CsChrimson-mVenus>UAS-GCaMP6f fly, exposed to red light.
Key Resources Table:
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| NaCl | Sigma-Aldrich | S7653-1KG |
| trehalose | Sigma-Aldrich | T9531-100G |
| glucose | Chem Supply | 9326410004041 |
| NaHCO3 | Ajax Finechem pty ltd | 3964-500G |
| MgCl2 (hexa-hydrate) | EMD Millipore Corp | 442615-500GM |
| Sucrose | Chem Supply | SA030-500G |
| KCl | Sigma-Aldrich | P9333-1KG |
| CaCl (dihydrate) | Sigma-Aldrich | 223506-500G |
| NaH2PO4 | Sigma-Aldrich | RDD007 |
| C6H15NO6S | Sigma-Aldrich | T1375 |
| all-trans retinal | Sigma-Aldrich | R2500-1G |
| Deposited Data | ||
| 1) Spontaneous sleep 2p + behavior 2) dFB sleep 2p (short and long) + controls + behavior 3) dFB sleep and vision 2p + controls + behavior 4) DART 5) Buridans (visual attention) 6) Local Field Potential 7) Statistics files |
The University of Queensland data repository | https://espace.library.uq.edu.au/ |
| Experimental Models: Organisms/Strains | ||
| R23E10-Gal4 (attp2) | Bloomington Drosophila Stock Center. [66] | Bloomington ID 49032 |
| UAS-Chrimson (P[20xUAS-IVS-CsChrimson.mVenus]attp18) | Vivek Jayaraman, JFRC [36] | Bloomington ID 55134 |
| lexAop-Syn21-nlsGCaMP6f-p10 in VIE-260b | This paper (provided by Barry J. Dickson) | N/A |
| 10xUAS-Chrimson88-tdT(attP18) | [67] | N/A |
| Nsyb-LexAp65(attP2) | [68] | N/A |
| 13XLexAop2-IVS-Syn21-mPA-p10 (VK00005) | [68] | N/A |
| Software and Algorithms | ||
| Drosophila ARousal Tracking (DART) system | BFK-LAB [34] |
http://www.bfklab.com/dart |
| MatLab analysis scripts | This paper | https://espace.library.uq.edu.au/ |
| CeTran (3.4) software | Buritracker | http://buridan.sourceforge.net/ |
| MatLab v2018b | Mathworks | https://au.mathworks.com/products/get-matlab.html |
| Prism 8.1.2 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagej.net |
| FiJi | FijiSc [70] | https://imagej.net/Fiji |
| ThorLabs Imaging software v3.0 | Thorlabs | https://www.thorlabs.com/navigation.cfm?Guide_ID=2191 |
| AxoGraph 1.4.4 | Axon Instruments | https://axograph.com/index.php?option=com_content&view=article&id=12&Itemid=136 |
| Other | ||
| 65 mm glass tubes | Trikinetics | PGT7x65 |
| 617 nm Luxeon Rebel LED | Luxeon Star LEDs, Ontario, Canada | MR-H2060-10S |
| Sleep Nullifying Apparatus (SNAP) | [37] | N/A |
| LED arena for visual attention experiments | [37] | N/A |
| ThorLabs Bergamo series 2 multiphoton microscope | ThorLabs | BERGAMO |
| Ti:Sapphire laser (920nm tuned) | Spectra Physics | Mai Tai Deepsee |
| High Speed Pockels Cell Module | Thorlabs | BCM-PCA100 |
| Galvo-resonant scanner | Thorlabs | OPX1100 |
| Nikon CFI APO 40XW NIR objective | Thorlabs | N40X-NIR |
| Piezo Focus Motor | Thorlabs | PI-P725 |
| High Sensitivity GaAsP PMT | ThorLabs | PMT2000 |
| 594 dichroic beam splitter | ThorLabs | 22-0104 |
| 680 nm Shortpass Dichroic | Thorlabs | 22-0312 |
| 525/25nm band pass filter | ThorLabs | 22-0045 |
| 617nm LED | ThorLabs | M617F1 |
| 405nm LED | ThorLabs | M405L4 |
| 4 channel LED driver | Thorlabs | DC4104 |
| Firefly MV 0.3MP camera | FLIR Systems | FMVU-03MTM-CS |
| Custom air puff generator | This paper | N/A |
| 1400 A Digitizer | Axon Digidata | N/A |
| Custom sine wave generator | This paper | N/A |
Highlights:
Calcium imaging reveals distinct spontaneous sleep stages in the fly brain
Short bouts of quiescence in Drosophila may involve an active sleep stage
Optogenetic activation of a sleep switch promotes wake-like brain activity
Prolonged activation of the sleep switch corrects visual attention defects
Acknowledgements
We thank members of the van Swinderen lab for critical discussions about the work, and Melvyn Yap for help with electrophysiological validation of the UV stimulus. We thank Barry Dickson for providing us with the LexAop-nlsGCaMP6f (VIE-260b) Drosophila line, and Sridhar Jagannathan for providing us with custom-written Matlab code for performing the slice timing correction. This study was funded NHMRC GNT1065713 (to BvS) and NIH R01 grant NS076980–01 (to PJS and BvS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
Declaration of interests
The authors declare no competing interests.
References
- 1.Campbell SS, and Tobler I (1984). Animal sleep: A review of sleep duration across phylogeny. Neuroscience & Biobehavioral Reviews 8, 269–300. [DOI] [PubMed] [Google Scholar]
- 2.Capellini I, Barton RA, McNamara P, Preston BT, and Nunn CL (2008). Phylogenetic analysis of the ecology and evolution of mammalian sleep. Evolution 62, 1764–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keene AC, and Duboue ER (2018). The origins and evolution of sleep. J Exp Biol 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirszenblat L, and van Swinderen B (2015). The Yin and Yang of Sleep and Attention. Trends Neurosci 38, 776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sterman MB, Knauss T, Lehmann D, and Clemente CD (1965). Circadian sleep and waking patterns in the laboratory cat. Electroencephalogr Clin Neurophysiol 19, 509–517. [DOI] [PubMed] [Google Scholar]
- 6.Tobler I, and Borbély AA (1988). Sleep and EEG spectra in the pigeon (Columba livia) under baseline conditions and after sleep deprivation. J Comp Physiology A 163, 729–738. [Google Scholar]
- 7.Rattenborg NC, van der Meij J, Beckers GJL, and Lesku JA (2019). Local Aspects of Avian Non-REM and REM Sleep. Front Neurosci 13, 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shein-Idelson M, Ondracek JM, Liaw HP, Reiter S, and Laurent G (2016). Slow waves, sharp waves, ripples, and REM in sleeping dragons. Science 352, 590–595. [DOI] [PubMed] [Google Scholar]
- 9.Leung LC, Wang GX, Madelaine R, Skariah G, Kawakami K, Deisseroth K, Urban AE, and Mourrain P (2019). Neural signatures of sleep in zebrafish. Nature 571, 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iglesias TL, Boal JG, Frank MG, Zeil J, and Hanlon RT (2019). Cyclic nature of the REM sleep-like state in the cuttlefish Sepia officinalis. J Exp Biol 222. [DOI] [PubMed] [Google Scholar]
- 11.Kirszenblat L, and van Swinderen B (2019). Sleep in Drosophila. Handbook of Sleep Research. Ed. Dringenberg Hans. (Elsevier B.V., 2019–01-01). [Google Scholar]
- 12.Tononi G, and Cirelli C (2003). Sleep and synaptic homeostasis: a hypothesis. Brain Res Bull 62, 143–150. [DOI] [PubMed] [Google Scholar]
- 13.Donlea JM, Ramanan N, and Shaw PJ (2009). Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324, 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilestro GF, Tononi G, and Cirelli C (2009). Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bushey D, Tononi G, and Cirelli C (2011). Sleep and synaptic homeostasis: structural evidence in Drosophila. Science 332, 1576–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, and Pack AI (2000). Rest in Drosophila is a sleep-like state. Neuron 25, 129–138. [DOI] [PubMed] [Google Scholar]
- 17.Shaw PJ, Cirelli C, Greenspan RJ, and Tononi G (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837. [DOI] [PubMed] [Google Scholar]
- 18.Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, and Pack AI (2008). Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451, 569–572. [DOI] [PubMed] [Google Scholar]
- 19.Nichols ALA, Eichler T, Latham R, and Zimmer M (2017). A global brain state underlies C. elegans sleep behavior. Science 356, eaam6851 [DOI] [PubMed] [Google Scholar]
- 20.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, and Shaw PJ (2011). Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332, 1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dissel S, Angadi V, Kirszenblat L, Suzuki Y, Donlea J, Klose M, Koch Z, English D, Winsky-Sommerer R, van Swinderen B, et al. (2015). Sleep restores behavioral plasticity to Drosophila mutants. Curr Biol 25, 1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tononi G, and Cirelli C (2014). Sleep and the price of plasticity: from synaptic and cellular homeostasis to memory consolidation and integration. Neuron 81, 12–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donlea JM, Pimentel D, and Miesenbock G (2014). Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81, 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pimentel D, Donlea JM, Talbot CB, Song SM, Thurston AJF, and Miesenbock G (2016). Operation of a homeostatic sleep switch. Nature 536, 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donlea JM, Pimentel D, Talbot CB, Kempf A, Omoto JJ, Hartenstein V, and Miesenböck G (2018). Recurrent Circuitry for Balancing Sleep Need and Sleep. Neuron 97, 378–389.e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yap MHW, Grabowska MJ, Rohrscheib C, Jeans R, Troup M, Paulk AC, van Alphen B, Shaw PJ, and van Swinderen B (2017). Oscillatory brain activity in spontaneous and induced sleep stages in flies. Nat Comm 8, 1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dag U, Lei Z, Le JQ, Wong A, Bushey D, and Keleman K (2019). Neuronal reactivation during post-learning sleep consolidates long-term memory in Drosophila. eLife 8, e42786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanhope BA, Jaggard JB, Gratton M, Brown EB, and Keene AC (2020). Sleep Regulates Glial Plasticity and Expression of the Engulfment Receptor Draper Following Neural Injury. Curr Biol 30, 1092–1101.e1093. [DOI] [PubMed] [Google Scholar]
- 29.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Reports 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nitz DA, van Swinderen B, Tononi G, and Greenspan RJ (2002). Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol 12, 1934–1940. [DOI] [PubMed] [Google Scholar]
- 31.van Alphen B, Yap MH, Kirszenblat L, Kottler B, and van Swinderen B (2013). A dynamic deep sleep stage in Drosophila. Journal Neurosci 33, 6917–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sporns O (2018). Graph theory methods: applications in brain networks. Dial Clin Neurosci 20, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faville R, Kottler B, Goodhill GJ, Shaw PJ, and van Swinderen B (2015). How deeply does your mutant sleep? Probing arousal to better understand sleep defects in Drosophila. Sci Reports 5, 8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Troup M, Yap MHW, Rohrscheib C, Grabowska MJ, Ertekin D, Randeniya R, Kottler B, Larkin A, Munro K, Shaw PJ, et al. (2018). Acute control of the sleep switch in Drosophila reveals a role for gap junctions in regulating behavioral responsiveness. eLife 7, e37105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, et al. (2014). Independent optical excitation of distinct neural populations. Nat Methods 11, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirszenblat L, Ertekin D, Goodsell J, Zhou Y, Shaw PJ, and van Swinderen B (2018). Sleep regulates visual selective attention in Drosophila. J Exp Biol 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colomb J, Reiter L, Blaszkiewicz J, Wessnitzer J, and Brembs B (2012). Open source tracking and analysis of adult Drosophila locomotion in Buridan’s paradigm with and without visual targets. PLoS One 7, e42247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaiser W, and Steiner-Kaiser J (1983). Neuronal correlates of sleep, wakefulness and arousal in a diurnal insect. Nature 301, 707–709. [DOI] [PubMed] [Google Scholar]
- 40.van Swinderen B, and Greenspan RJ (2003). Salience modulates 20–30 Hz brain activity in Drosophila. Nat Neurosci 6, 579–586. [DOI] [PubMed] [Google Scholar]
- 41.Heisenberg M, and Buchner E (1977). The role of retinula cell types in visual behavior of Drosophila melanogaster. J Comp Physiol 117, 127–162. [Google Scholar]
- 42.Liu S, Liu Q, Tabuchi M, and Wu MN (2016). Sleep Drive Is Encoded by Neural Plastic Changes in a Dedicated Circuit. Cell 165, 1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan Y, Zhou Y, Guo C, Gong H, Gong Z, and Liu L (2009). Differential roles of the fan-shaped body and the ellipsoid body in Drosophila visual pattern memory. Learn Mem 16, 289–295. [DOI] [PubMed] [Google Scholar]
- 44.Weir PT, and Dickinson MH (2015). Functional divisions for visual processing in the central brain of flying Drosophila. Proc Natl Acad Sci USA 112, E5523–E5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiggin TD, Goodwin PR, Donelson NC, Liu C, Trinh K, Sanyal S, and Griffith LC (2020). Covert sleep-related biological processes are revealed by probabilistic analysis in Drosophila. Proc Natl Acad Sci U S A 117, 10024–10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Swinderen B, Nitz DA, and Greenspan RJ (2004). Uncoupling of brain activity from movement defines arousal States in Drosophila. Curr Biol 14, 81–87. [PubMed] [Google Scholar]
- 47.Kirszenblat L, Yaun R, and van Swinderen B (2019). Visual experience drives sleep need in Drosophila. Sleep 42, zsz102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dement W, and Kleitman N (1957). Cyclic variations in EEG during sleep and their relation to eye movements, body motility, and dreaming. Clin Neurophysiol 9, 673–690. [DOI] [PubMed] [Google Scholar]
- 49.Peever J, and Fuller PM (2017). The Biology of REM Sleep. Curr Biol 27, R1237–R1248. [DOI] [PubMed] [Google Scholar]
- 50.Ermis U, Krakow K, and Voss U (2010). Arousal thresholds during human tonic and phasic REM sleep. J Sleep Res 19, 400–406. [DOI] [PubMed] [Google Scholar]
- 51.Andrillon T, and Kouider S (2020). The vigilant sleeper: neural mechanisms of sensory (de)coupling during sleep. Curr Op Physiol 15, 47–59. [Google Scholar]
- 52.Dillon RF, and Webb WB (1965). Threshold of Arousal from “Activated” Sleep in the Rat. J Comp Physiol Psychol 59, 446–447. [DOI] [PubMed] [Google Scholar]
- 53.Siegel J, and Langley TD (1965). Arousal threshold in the cat as a function of sleep phase and stimulus significance. Experientia 21, 740–741. [DOI] [PubMed] [Google Scholar]
- 54.Jouvet M, Michel F, and Courjon J (1959). [On a stage of rapid cerebral electrical activity in the course of physiological sleep]. C R Seances Soc Biol Fil 153, 1024–1028. [PubMed] [Google Scholar]
- 55.Siegel JM (2011). REM sleep: a biological and psychological paradox. Sleep Med Rev 15, 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Born J, Muth S, and Fehm HL (1988). The significance of sleep onset and slow wave sleep for nocturnal release of growth hormone (GH) and cortisol. Psychoneuroendocrinology 13, 233–243. [DOI] [PubMed] [Google Scholar]
- 57.Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O’Donnell J, Christensen DJ, Nicholson C, Iliff JJ, et al. (2013). Sleep drives metabolite clearance from the adult brain. Science 342, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Vivo L, Bellesi M, Marshall W, Bushong EA, Ellisman MH, Tononi G, and Cirelli C (2017). Ultrastructural evidence for synaptic scaling across the wake/sleep cycle. Science 355, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker MP, and Stickgold R (2004). Sleep-dependent learning and memory consolidation. Neuron 44, 121–133. [DOI] [PubMed] [Google Scholar]
- 60.Wagner U, Gais S, and Born J (2001). Emotional memory formation is enhanced across sleep intervals with high amounts of rapid eye movement sleep. Learn Mem 8, 112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ravassard P, Hamieh AM, Joseph MA, Fraize N, Libourel PA, Lebarillier L, Arthaud S, Meissirel C, Touret M, Malleret G, et al. (2016). REM Sleep-Dependent Bidirectional Regulation of Hippocampal-Based Emotional Memory and LTP. Cerebral Cortex 26, 1488–1500. [DOI] [PubMed] [Google Scholar]
- 62.Peters KR, Smith V, and Smith CT (2007). Changes in sleep architecture following motor learning depend on initial skill level. J Cog Neurosci 19, 817–829. [DOI] [PubMed] [Google Scholar]
- 63.Boucetta S, Cisse Y, Mainville L, Morales M, and Jones BE (2014). Discharge profiles across the sleep-waking cycle of identified cholinergic, GABAergic, and glutamatergic neurons in the pontomesencephalic tegmentum of the rat. J Neurosc 34, 4708–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Meij J, Martinez-Gonzalez D, Beckers GJL, and Rattenborg NC (2019). Intra-”cortical” activity during avian non-REM and REM sleep: variant and invariant traits between birds and mammals. Sleep 42. [DOI] [PubMed] [Google Scholar]
- 65.Bazhenov M, Timofeev I, Steriade M, and Sejnowski TJ (2002). Model of thalamocortical slow-wave sleep oscillations and transitions to activated States. J Neurosci 22, 8691–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA 105, 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strother JA, Wu S-T, Wong AM, Nern A, Rogers EM, Le JQ, Rubin GM, and Reiser MB (2017). The Emergence of Directional Selectivity in the Visual Motion Pathway of Drosophila. Neuron 94, 168–182.e110. [DOI] [PubMed] [Google Scholar]
- 68.Pfeiffer BD, Truman JW, and Rubin GM (2012). Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci USA 109, 6626–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guizar-Sicairos M, Thurman ST, and Fienup JR (2008). Efficient subpixel image registration algorithms. Optics Letters 33, 156–158. [DOI] [PubMed] [Google Scholar]
- 70.Legland D, Arganda-Carreras I, and Andrey P (2016). MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32, 3532–3534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotype of fly UAS:Chrimson / X ; Nsyb:LexA/+ ; LexOp:nlsGCaMP6f/+. One optical plane is shown, sped 25X. Scale bar = 100μm.
Genotype of fly is R23E10-Gal4>UAS-CsChrimson-mVenus>UAS-GCaMP6f fly, exposed to red light.
Data Availability Statement
The data underpinning the publication is stored in the University of Queensland Research Data Management (UQRDM) via UQ eSpace, the institutional data storage repository of the University of Queensland (UQ).The metadata in UQ eSpace is indexed by common search engines (e.g., Google) as well as by Research Data Australia (the national data discovery platform) and also by Data Citation Index. Matlab analysis scripts used in this study are available from the Lead Contact upon request.


