Abstract
A “Leap-of-Faith” approach is used to treat patients with previously unknown ultrarare pathogenic mutations, often based on evidence from patients having dissimilar but more prevalent mutations. This uncertainty reflects the need to develop personalized prescreening platforms for these patients to assess drug efficacy before considering clinical trial enrollment. In this study, we report an 18-year-old patient with ultrarare Leigh-like syndrome. This patient had previously participated in two clinical trials with unfavorable responses. We established an induced pluripotent stem cell (iPSC)–based platform for this patient, and assessed the efficacy of a panel of drugs. The iPSC platform validated the safety and efficacy of the screened drugs. The efficacy of three of the screened drugs was also investigated in the patient. After 3 years of treatment, the drugs were effective in shifting the metabolic profile of this patient toward healthy control. Therefore, this personalized iPSC-based platform can act as a prescreening tool to help in decision-making with respect to patient’s participation in future clinical trials.
An iPSC-based personalized prescreening tool helps in selection of suitable clinical trial for patients with ultrarare diseases.
INTRODUCTION
In the case of inborn errors of metabolism, patients with previously unknown ultrarare variants differ with respect to underlying disease pathophysiology and mechanism of disease progression. This creates a group of patients predicted to have an uncertain response to a drug in a clinical trial, because the enrollment criteria for these clinical trials are mostly established through evaluation of patients with similar phenotypes but different mutations (1). This leads to delayed or imprecise treatment options for patients with ultrarare novel variants. Here, we report this 18-year-old patient with Leigh-like syndrome (LS-like) with compound heterozygous variants in the ECHS1 gene. One of the variants in this patient is novel, but there is a lack of information on both variants observed here. There is thus the scenario where new drug administration and clinical trial enrollment for this patient are fraught with uncertainty. Furthermore, drug trials need to be carried out sequentially and enough time needs to elapse between experimental drug testing periods for washout and recovery. This is problematic, where time is of the essence in the search for a possible effective treatment for hereditary metabolic disorders with still no effective treatment options. This “trial and error” approach is still ongoing for this patient. Two such attempts have been met with unfavorable responses: (i) α-tocopherol (Toco) was discontinued as the patient reported discomfort and (ii) the patient had participated in a clinical trial [RP103-MITO-001(NCT02023866)] involving cysteamine (Cys) salt but withdrew due to side effects. Therefore, a personalized prescreening tool that could help decide whether enrollment in a particular clinical trial with the assurance of best possible drug safety and efficacy would benefit this individual (and similarly other patients) with novel ultrarare mutations.
Induced pluripotent stem cells (iPSCs) have recently gained preeminence in disease modeling by recapitulating identical genetic and phenotypic aberrations (2), allowing demonstration of personalized medicine. Several single organ–specific disorders like Long-QT syndrome and Parkinson’s disease have been rigorously modeled through iPSC, providing better understanding of disease progression and drug efficacy testing platforms (3). Likewise, human iPSC–derived cardiomyocytes (CMs) have already been approved by the Food and Drug Administration (FDA) for safety and efficacy evaluations of new drugs (4). In the current study, we have demonstrated the development of an iPSC-based stable multisystem clinical trial selection platform for the patient with LS-like syndrome. We compared the LS-like patient with negative controls (healthy individuals) and a positive control (patient with classic LS). Leigh/Leigh-like syndrome (LS) is a rare group of progressive mitochondrial disorders of oxidative phosphorylation that involves multisystem abnormalities and heterogeneous manifestations. More than 100 mutations in mitochondrial and genetic DNA have been reported in LS, with novel variants being found regularly (5). With treatment options lacking, patients and physicians often explore experimental opportunities through enrollment in clinical trials or by sifting through the results of completed ones (6). However, the heterogeneity of mutations within patients with LS puts them at a risk of adverse drug response and uncertain clinical response. With scarcity of conventional models (cellular or animal), there is a wide preclinical to clinical gap to finding an optimal treatment for these patients.
The platform demonstrated here included iPSC-derived fibroblasts (representation of general cell population in the body), neural progenitor cells (NPCs) and mature neurons (neural system is the primary organ affected in many LS cases), and CMs (cardiac involvement is reported to lesser degree). Thereafter, this multisystem platform was used to assess the toxicity and efficacy of a panel of seven drugs (in clinical application or being tested in ongoing clinical trials) in affected iPSC-derived cells.
RESULTS
Case reports
Patient with LS-like phenotype (mitochondrial enoyl–coenzyme A hydratase short chain 1 deficiency)
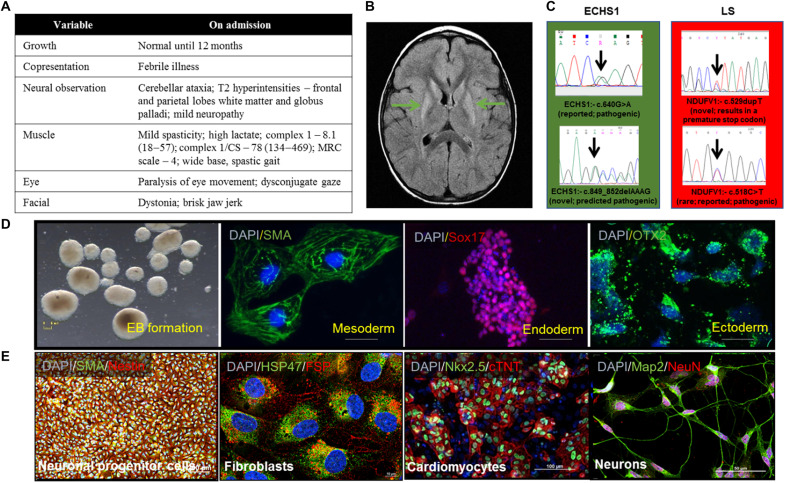
The patient is now 18 years of age. He was well with normal growth and development until 12 months of age when, after a febrile illness, he presented with cerebellar ataxia. The ataxia did not resolve and gradually worsened accompanied by dystonia with subsequent febrile illnesses. Baseline metabolic studies were unremarkable except for intermittently elevated lactate levels. Muscle histology was normal, but respiratory chain studies of isolated muscle mitochondria revealed low complex 1 activity and low complex 1/citrate synthase ratio (Fig. 1A). Mitochondrial and nuclear DNA testing revealed compound heterozygosity for two variants in ECHS1 c.518C>T (pAla173Val) and c.849_852delAAAG, causing a frameshift of 290 amino acids and a protein product lengthened by 25 amino acids. Both variants are predicted to be pathogenic. These biochemical and molecular results were consistent with mitochondrial enoyl–coenzyme A (CoA) hydratase short chain 1 (ECHS1) deficiency, an inborn error of valine metabolism [(EC4.2.1.17) OMIM 602292]. Brain magnetic resonance imaging (MRI) done shortly after presentation revealed patchy areas of T2 hyperintensity in deep white matter of the frontal and parietal lobes and basal ganglia. Follow-up brain MRI at 5 years of age revealed evidence of progressive disease in the basal ganglia (Fig. 1, A and B). Brain MR spectroscopy (MRS) was unremarkable. His clinical course has consisted of repeated exacerbations of ataxia and dystonia. The patient ultimately required mobility aids and became wheelchair bound at the age of 10 years. Expressive language was substantially impaired due to dystonia. Intellectual functioning was felt to be normal. He has been fed by gastronomy tube since 10 years of age due to oral dystonia and problems with swallowing and vomiting.
Fig. 1. Patient presentations and iPSC characterization.
(A) Table of presentations for ECHS1 patient at clinical visits. (B) Brain MRI scan of the ECHS1 patient showing T2 hyperintensity in deep white matter of the frontal and parietal lobes and basal ganglia (marked by arrows). (C) The genomic DNA of the iPSC clones were evaluated using target-specific Sanger sequencing to confirm mutations (n = 3). (D) Verification of trilineage differentiation potential of iPSC was carried out using spontaneous embryoid body (EB) culture. iPSC colonies were suspended in non-ES cell medium and allowed to form rotund structures in low attachment plates. The cells were allowed to spontaneously differentiate and plated onto gelatin-coated culture dishes on day 8. Immunostaining revealed cells from all three lineages: mesoderm, alpha-smooth muscle actin (SMA); endoderm, Sox17; and ectoderm, Otx2. Scale bars, 5 mm (EB), 10 μm (mesoderm), 100 μm (endoderm), and 15 μm (ectoderm) (n = 3). DAPI, 4′,6-diamidino-2-phenylindole. (E) iPSC differentiation to NPCs (Pax6 and Nestin), fibroblasts (HSP47 and FSP), CMs (Nkx2.5 and cardiac troponin), and neurons (Map2 and NeuN). Scale bars, 100 μm (NPCs), 100 μm (fibroblasts), 10 μm (CMs), and 50 μm (neurons) (n = 3). The “n” is representative of the biological replicates. The data are representative of three independent experiments.
Patient with classic LS (used as positive control)
This female toddler, previously well, presented at 26 months of age with an acute decrease in level of consciousness, respiratory failure requiring aggressive resuscitation with intubation and positive pressure ventilation, metabolic acidosis, and hyperammonemia. One and a half months before this acute presentation, this patient developed sudden onset of bilateral ptosis that resolved on its own and the patient was subsequently well until this fulminant presentation. Past medical history revealed she was a product of a normal pregnancy and had appropriate growth and development until this presentation. Family history was noncontributory. Biochemical investigations demonstrated lactic acidosis, and brain MRI showed leukodystrophy with increased signal in the basal ganglia. Brain MRS revealed an elevated lactate peak. DNA testing with mitochondrial encephalopathy/LS nuclear gene panel (GENEDx) revealed two variants in the NDUFV1 gene. The first pathogenic variant is c.529dupT(p.Y177Lfs*z2), and the second is c.640G>A(p.E214k). All other metabolic, microbiological, and serological studies were negative. The child never regained consciousness and died 2 weeks following her admission to hospital.
The patient with the classic characteristics of LS (positive control) provided an avenue to compare cellular similarities and differences with the patient with the LS-like phenotype. As there are no specific cellular or animal models available for this ultrarare novel variant to evaluate drug and dosage responses, a preclinical screening modality does not presently exist to decide upon a clinical trial or recommend a specific therapy. The platform here attempts to fill this gap.
Development and characterization of iPSC-based multisystem model
The peripheral blood mononuclear cells (PBMCs) from each patient and healthy controls were reprogrammed to iPSC, which were investigated for pluripotency markers-Oct4, Nanog, Sox2, SSEA4, Tra-1-81, and Tra-1-60 using immunostaining and polymerase chain reaction (PCR; fig. S1). The Sanger sequencing confirmed the presence of variants in the iPSC (Fig. 1C). To verify the functional pluripotency of iPSC, the embryoid bodies (EBs) were formed and assessed for trilineage differentiation potential toward mesoderm, endoderm, and ectoderm (Fig. 1D). The trilineage differentiation potential of iPSC was further characterized in our recently published studies (7, 8). iPSC differentiation into fibroblasts, NPCs, CMs, and neurons further verified their multilineage differentiation capabilities (Fig. 1E). Further, there was no mycoplasma contamination detected in generated iPSC lines as reported in our recent studies (7, 8). The presence of karyotypic abnormalities was investigated at intervals of 20 to 25 passages. None of the cell lines generated in the study reported any karyotypic abnormalities (fig. S2).
Patients’ clinical characteristics were reproduced in iPSC model
To demonstrate establishment and clinical relevance of the iPSC-based cellular model, the most commonly observed clinical presentations in patients were equated with reported parameters at the cellular level (Fig. 2A) and evaluated as follows
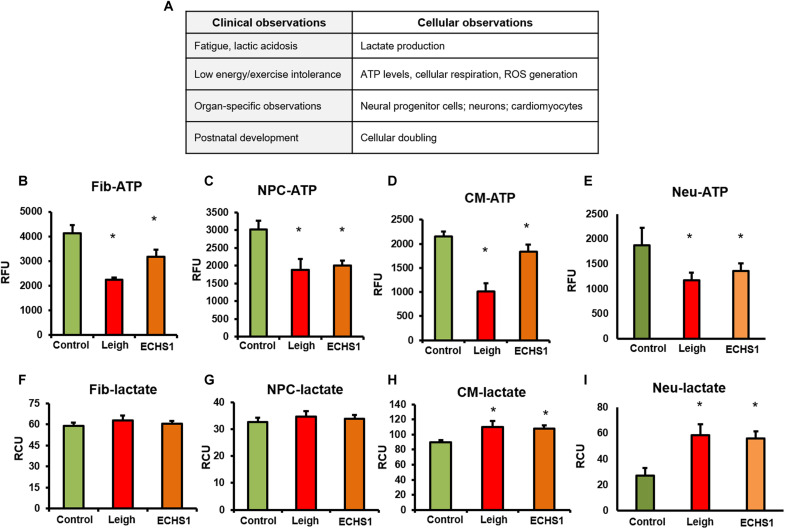
Fig. 2. Disease characteristics in patient and iPSC-derivatives.
(A) Comparison of clinical observations of patient versus equivalent cellular parameters; functional parameters similar to clinical observations investigated in fibroblasts, NPCs, CMs, and neurons. (B to E) Total ATP production in all four cell types—fibroblasts (Fib), NPCs, CMs, and neurons—was decreased in patients with LS and ECHS1 deficiency compared to healthy control (n = 8). RFU, relative fluorescence units. (F and G) Lactate levels did not change in fibroblasts (n = 5) and NPCs (n = 6) in both patients with LS and ECHS1 deficiency compared to control. (H and I) In CMs and neurons, there was a significant increase in lactate production in LS and ECHS1 patients compared to healthy control (n = 6). The “n” is representative of the biological replicates and was analyzed by unpaired t test. The data are representative of three independent experiments and presented as means ± SD; *P < 0.05 in comparison to control. RCU, relative colorimetric units.
Cellular adenosine triphosphate and lactate analyses
The down-regulation of adenosine triphosphate (ATP) levels in patients with LS and ECHS1 deficiency is a typical characteristic (5, 9–11), including patients considered for this study. We found a significant reduction in ATP levels in iPSC-derived fibroblasts, NPCs, CMs, and neurons of the LS and ECHS1 (LS-like) patients compared to healthy controls (Fig. 2, B to E). When wild-type (WT) ECHS1 protein was expressed ectopically in iPSC-derived fibroblasts of ECHS1 patient using an expression vector, there was a significant increase in ATP levels compared to ECHS1 variant fibroblasts (fig. S3A). The expression of WT pECHS1 protein was confirmed by immunoblotting (fig. S3B).
Lactate accumulation in blood, urine, and/or cerebrospinal fluid is a hallmark of LS and other mitochondrial disorders (12, 13). Both ECHS1 patient and positive control (LS patient) were presented with lactic acidosis. However, the lactate levels in fibroblasts and NPCs were not significantly different in the patients’ and in control cells (Fig. 2, F and G). CMs and neurons displayed a significant increase in lactate levels in both ECHS1 and LS patient cells compared to healthy control CMs and neurons (Fig. 2, H and I). This accumulation of lactate in CMs and neurons could be due to problem in transfer of electrons down the respiratory chain, which is commonly observed in metabolically active cells, e.g., CMs and neurons in patients with mitochondrial disorders (14, 15).
Mitochondrial membrane potential
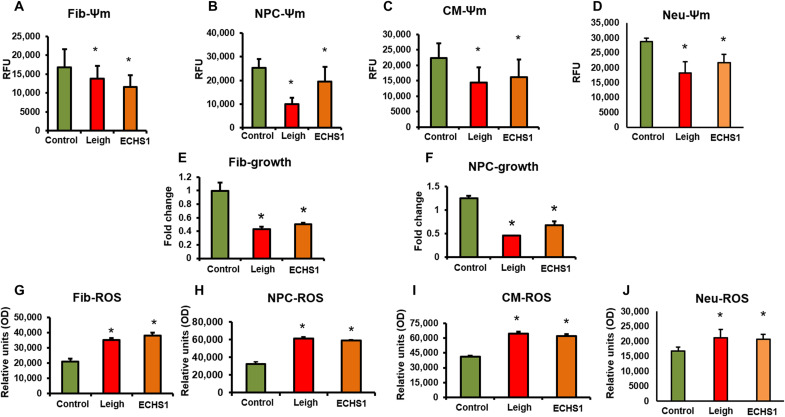
Mitochondrial membrane potential (Ψm) serves as a key indicator of the mitochondrial health, integrity of mitochondrial structure, and function (16, 17). We found a significant decrease in Ψm in fibroblasts, NPCs, CMs, and neurons from the ECHS1 and LS patients compared to control group (Fig. 3, A to D). The decrease in Ψm normally is underlined by the loss of optimal mitochondrial fusion, largely observed in patients with mitochondrial disorders. Furthermore, when WT ECHS1 protein was ectopically expressed in fibroblasts of ECHS1 patient using an expression vector, there was a significant improvement in Ψm levels compared to ECHS1 variant fibroblasts (fig. S3C).
Fig. 3. Disease characteristics in iPSC-derivatives.
(A to D) Mitochondrial membrane potential (Ψm) was measured by tetramethylrhodamine ethyl ester assay. The membrane potential decreased in all four cell types—fibroblasts, NPCs, CMs, and neurons—from patients with LS and ECHS1 deficiency compared to healthy control (n = 20). (E and F) Cellular growth was assessed through cell doubling count. Briefly, cells were plated after being stained with CytoPainter. The cell number was counted after 2 days. The data are presented as the ratio of day 2 count versus day 0 count. The fibroblasts and NPCs from the LS and ECHS1 patients had severely retarded growth compared to healthy control (n = 15). (G to J) Reactive oxygen species (ROS) production as measured using an H2DCFDA dye increased in all four cell types—fibroblasts, NPCs, CMs, and neurons—from patients with LS and ECHS1 deficiency compared to healthy control (n = 12). OD, optical density. The “n” is representative of the biological replicates and was analyzed by unpaired t test. The data are representative of three independent experiments. Data are represented as means ± SD; *P < 0.05 in comparison to control.
Cellular growth
Growth retardation, muscle loss, and loss of body weight in general are present in patients with mitochondrial disorders (18, 19). To relate to these clinical presentations, we measured the cellular growth. There was a significant reduction in growth rate of fibroblasts and NPCs from the LS and ECHS1 patients compared to control cells from healthy individuals (Fig. 3, E and F).
Reactive oxygen species
Strong association of reactive oxygen species (ROS) generation with impaired mitochondrial function has been reported (20). ROS scavenging has been defined as a central goal for numerous therapeutic options in combating mitochondrial disorders (21, 22). ROS generation showed a significant increase in fibroblasts, NPCs, CMs, and neurons from the LS and the ECHS1 patients compared to healthy controls (Fig. 3, G to J). On the other hand, when WT ECHS1 protein was expressed in fibroblasts of ECHS1 patient using an expression vector, there was a significant decrease in ROS levels compared to ECHS1 variant fibroblasts (fig. S3D).
Cellular bioenergetics or mitochondrial respiration analyses
To determine mitochondrial respiration, oxygen consumption rate (OCR) was measured using Seahorse XFe24 analyzer. The basal OCR is the amount of oxygen consumption that is linked to ATP synthesis in the mitochondria. We found a significant down-regulation in OCR in both patients’ iPSC-fibroblasts, CMs, and neurons compared to control (fig. S4, A to I). The maximal respiration and spare respiratory capacity showed a significant decrease in fibroblasts, CMs, and neurons from the ECHS1 and Leigh patients compared to control group (fig. S4, D to I). These parameters are strong indicators for mitochondrial dysfunction in iPSC-derived cells in both ECHS1 patient and positive control (LS patient). When WT ECHS1 protein was expressed in iPSC-derived fibroblasts of ECHS1 patient using an expression vector, there was a significant improvement in OCR, basal, maximal, and spare respiratory capacity (fig. S5, A to D).
Functional and phenotypic analysis of iPSC-derived neurons and CMs
Inborn mitochondrial disorders including LS are associated with neuronal and cardiac complications. This leads to impaired development and functioning of neurons and CMs. In the current study, functional characterization of iPSC-derived neurons and CMs was performed by microelectrode array (MEA) system and by calcium imaging. The postdifferentiation maturation of neurons was followed for a period of 6 weeks with weekly data acquisition. Our data demonstrate that ECHS1 iPSC-derived neurons tend to retain neural progenitor phenotype compared to control cells, indicating a delay in maturation of these cells. Neurons are known to form electrical networks, and their activity can be seen in the form of network bursts. The neuronal activity in control cells was observed from the second week of maturation, whereas neuronal activity in ECHS1 cells was observed from week 3 onward as observed in terms of firing rate and burst frequency (fig. S6, A to D). Furthermore, neuronal bursts (demarcated in blue squares) observed in the control neurons were significantly higher than ECHS1 and Leigh neurons (fig. S6, E to G). The calcium handling analysis further demonstrated that neurons from iPSC of ECHS1 patient tend to retain their neural progenitor phenotype. We observed a stagnant calcium flux in ECHS1 cells when compared with healthy control iPSC neurons, where a sharp peak in fluorescence was observed (fig. S7, A and B).
The functional characterization of CMs as performed by the MEA system demonstrated an increase in corrected field potential (cFPD) in ECHS1 and Leigh CMs compared to control group (fig. S8). cFPD is a very good indicator of abnormal beating of CMs and arrhythmia. Furthermore, we also observed a decline in the spike amplitude, contractility, and conduction velocity of electrical propagation in ECHS1 and Leigh CMs compared to healthy controls (fig. S8, A to E). The calcium handling analysis of CMs showed a delayed action potential for both Leigh and ECHS1 CMs compared to control cells (fig. S8, F to I). This phenotypic cellular and functional analysis on the iPSC-based multisystem model further validates its potential in recapitulating patient pathologies in a dish.
As demonstrated in the above sections, most of the commonly observed clinical presentations of LS-like and LS pertaining to ATP levels, lactate, mitochondrial respiration, Ψm, ROS levels, and cellular phenotypic alterations were reproduced in iPSC-derived cells from the ECHS1 patient in the current study. Maintaining ECHS1 protein levels in iPSC-derived fibroblasts of ECHS1 patient using an expression vector improved cellular bioenergetics, as well as the biochemical and molecular behavior of these cells. Therefore, these data establish the pathogenicity of ECHS1 mutation. Further, these results provide evidence for establishment of iPSC-based multisystem model for LS-like (ECHS1) to test the safety and efficacy of a panel of drugs.
Drug panel testing
The panel of seven drugs tested in iPSC-based multisystem platform was as follows: two drugs commonly administered to patients with mitochondrial disorders [coenzyme Q10 and α-lipoic acid (ALA)], two drugs discontinued in the ECHS1 patient because of gastrointestinal discomfort (Toco and Cys), two drugs in different stages of clinical trials (rapamycin (Rapa; NCT03747328) and elamipretide (Elam), and riboflavin (Ribo) is a dietary supplement that has shown effectiveness for several mitochondrial disorders. The doses of different drugs were decided on the basis of previous literature to reflect a possible wide range of administration (23–32).
1) Coenzyme Q10 [ubiquinol (Ubi)] is an essential electron carrier in the mitochondrial chain. It is highly beneficial in reducing oxidative damage. Coenzyme Q10, especially in its Ubi form, is electron-rich and has been found to be highly soluble in water.
2) ALA: ALA is a naturally occurring antioxidant typically synthesized in body and supplemented through diet (33). ALA is readily soluble in both water and fat. This facilitates tissue wide distribution of ALA after administration.
3) Toco: Toco is a form of vitamin E and a ROS scavenger. At a very high dose, Toco is known to be toxic (25). Toco is fat soluble in nature.
4) Cys hydrochloride: Cys is a water soluble aminothiol, synthesized in body, and promotes glutathione production (potent antioxidant). This drug is presently being tested in an open-label phase 2 trial (RP-103-MITO-001) for inherited mitochondrial syndromes.
5) Rapa: Rapa has been shown to improve motor endurance and correct muscle abnormalities during mitochondrial myopathy. It is presently being evaluated in clinical trials for various mitochondrial disorders (32).
6) Elam (SS-31, MTP-131, and Bendavia): Elam is a mitochondria-targeting tetrapeptide antioxidant; it is known to interact with cardiolipin that helps in stabilizing mitochondrial membrane potential (34).
7) Ribo (vitamin B2): Ribo is essential vitamin of the vitamin B family. It protects cells from oxidative stress by interacting with flavoenzymes, which, in turn, are important in mitochondria-based energy production. Ribo is supplemented through numerous food options and has low solubility in water.
In silico analysis of the drug panel
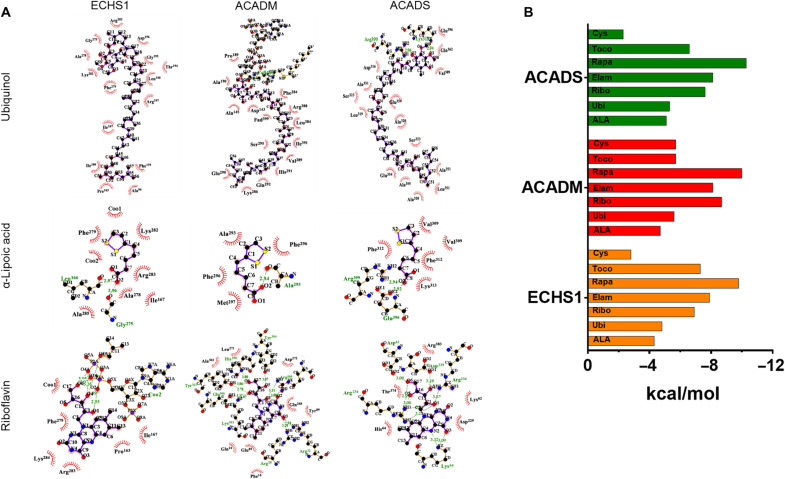
Before investigating the safety and efficacy of the abovementioned drug panel in iPSC-based models, we performed in silico analysis to predict the effectiveness of these drugs. The ECHS1 deficiency causes a defect in β-oxidation and valine degradation, which leads to accumulation of reactive metabolites including methacrylyl-CoA and acrylyl-CoA in patients. The toxic effects of these accumulated metabolites are reported to be important contributors to clinical pathophysiology of these patients (9, 35). We performed molecular docking studies of all the seven drugs being tested in the current study for their interactions with enzymes that can regulate the accumulation of the abovementioned toxic metabolites. These enzymes included ECHS1 (mitochondrial) by itself; ACADM (medium chain–specific acyl-CoA dehydrogenase, mitochondrial), which can use methacrylyyl-CoA as its substrate; and ACADS (short chain–specific acyl-CoA dehydrogenase, mitochondrial) that uses acrylyl-CoA as its substrate. Our in silico analysis demonstrated favorable binding affinities of all seven drugs with these enzymes (Fig. 4 and fig. S9).
Fig. 4. Evaluation of drug interaction with ECHS1 and related enzymes.
In silico analysis was performed to assess the interaction of Ubi, ALA, Ribo, Toco, Cys, Rapa, and Elam with ECHS1 (mitochondrial), ACADM (mitochondrial), and ACADS (mitochondrial). (A) The figure depicts molecular interaction of Ubi, ALA, and Ribo with ECHS1, ACADM, and ACADS, respectively, using docking analysis. The analysis in this figure show visualization of the top docking site/pockets where the drug interacted with respective target molecules to understand multitudinous interactions among drugs and their molecular binding sites using LigPlot+ software. The interaction of Toco, Cys, Rapa, and Elam with these enzymes is presented in fig. S9. (B) The histogram demonstrates binding energy during molecular interactions for all the drugs against ECHS1, ACADM, and ACADS, respectively. To evaluate the protein-ligand binding at the molecular level, the binding energy was expressed in terms of kcal/mol (kilocalorie per mole); these units define the energy density (binding energy) of the interaction between the two molecules in question. The stability of an interaction is inversely proportional to their binding energy, which is represented in (B). Hence, if two interacting molecules have a lower binding energy, then the probability of their interaction increases.
In addition, the binding affinities of all seven drugs with different protein targets involved in cellular bioenergetics and ROS scavenging were tested. The targets selected for this analysis were glutamate aspartate transporter (GLAST), which is responsible to maintain the malate aspartate shuttle; cytochrome P450 family 2 subfamily B6 proteins that breaks down arachidonic acid into epoxyeicosatrienoic acid (EET) molecules; glutathione peroxidase, which can regulate oxidized and reduced form of glutathione, thereby influencing cellular ROS scavenging potential; and farnesyl pyrophosphate synthase (FPPS), an enzyme that affects both cellular bioenergetics and ROS scavenging potential (figs. S10 and S11). Our data demonstrate that the binding energies of all the drugs with the exception of Cys showed very good affinity with the above targets (figs. S10 and S11). These in silico analyses suggested that the drugs selected in our study have the ability to influence metabolic changes in ECHS1 patient and may improve patient condition by influencing both cellular bioenergetics and ROS scavenging.
Assessment of safety and efficacy of drug panel in iPSC-based models
The safety of the drug panel was assessed by measuring cytotoxicity through the release of lactate dehydrogenase (LDH). None of the cell types (fibroblasts, NPCs, CMs, and neurons) showed any cytotoxicity to all these drugs at doses tested in this study (fig. S12, A to D). These results are consistent with the available reports from different animal studies (31) and clinical trials conducted so far (34, 36–38).
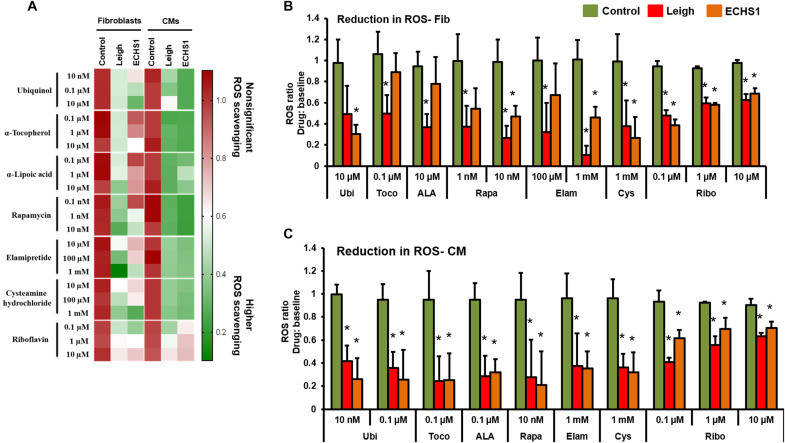
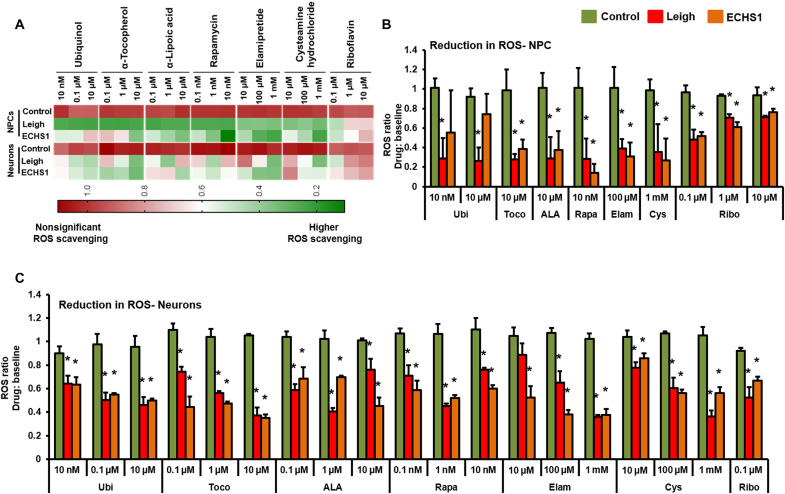
The current therapeutic options to combat different mitochondrial disorders focus on scavenging ROS and improvement in mitochondrial membrane potential (39–42). Therefore, in the current study, the efficacy of different drugs was assessed by determining the ability of individual drugs to quench ROS and restore Ψm to normal levels. The treatment with all seven drugs showed significantly reduced ROS generation in different cell types for the ECHS1 patient and positive control (LS patient; Figs. 5, A to C, and 6, A to C). Ubi (10 μM) was effective at best in ECHS1 fibroblasts, while LS fibroblasts showed a nonsignificant reduction (Fig. 5B). Toco (0.1 μM), ALA (10 μM), Rapa (1 nM), and Elam (100 μM) on the other hand showed significant reduction in the ROS measured in comparison to respective baselines in the LS fibroblasts, while a trend of nonsignificant ROS scavenging was seen in ECHS1 fibroblasts. Rapa (10 nM), Elam (1 mM), and Cys (1 mM) showed marked scavenging of ROS in fibroblasts of both patients. The reduction in ROS was also recorded for NPCs and neurons of both LS and ECHS1 origin, albeit in a different fashion in comparison to fibroblasts (Fig. 6, A to C). The need for inclusion of multiple cell platforms for drug evaluation becomes apparent here, when similar concentrations of the drugs display different efficacies of ROS reduction in various cell types. Different concentrations of Ubi were not effective in down-regulating ROS levels in the LS fibroblasts, whereas ECHS1 fibroblasts responded well (Fig. 5B). Ubi had an opposite effect on NPCs, e.g., at 10 nM and 10 μM, Ubi showed significant reduction in the ROS measured in LS NPCs but not in ECHS1 NPCs (Fig. 6B). The underlying mechanism for this differential effect in fibroblasts and NPCs here might be due to variations in drug uptake and/or processing pathway between the cell types. The remainder of the drugs—Toco (10 μM), ALA (10 μM), Rapa (10 nM), Elam (100 μM), and Cys (1 mM)—showed significant reduction in ROS levels compared to baseline in NPCs of both patients (Fig. 6B). Ribo treatment at all three doses (0.1, 1, and 10 μM) showed reduction in ROS levels in all the cell types—fibroblasts, NPCs, CMs, and neurons (Figs. 5, A to C, and 6, A to C). The CMs and neurons from both patients were the most receptive to all the drugs with significant reduction in ROS levels across all the drugs (Figs. 5C and 6C).
Fig. 5. Effect of drug panel on ROS in fibroblasts and CMs.
Efficacy of a panel of seven drugs—coenzyme Q10 (Ubi), ALA, Toco, Cys, Rapa, Elam, and Ribo—was evaluated by determining the ability of individual drugs to scavenge ROS in iPSC-derived fibroblasts and CMs. (A) Clustered heatmap for dosage analyses of different drugs. Each row represents data for a specific drug dose, and each set of columns represents data from different cells (fibroblasts and CMs) from healthy control, LS, and ECHS1 patients. All values are represented as ratio of ROS scavenging with drug: baseline. Color representation: green is higher, and red is lower/no ROS scavenging. (B) Drug doses with significant ROS scavenging in fibroblasts (n = 3). (C) Drug doses with significant ROS scavenging in CMs (n = 3). The “n” is representative of the biological replicates and was analyzed by one-way analysis of variance (ANOVA). The data are representative of three independent experiments. Data are represented as means ± SD; *P < 0.05 in comparison to control.
Fig. 6. Effect of drug panel on ROS on NPCs and neurons.
Efficacy of a panel of seven drugs—Coenzyme Q10 (Ubi), ALA, Toco, Cys, Rapa, Elam, and Ribo was evaluated by determining the ability of individual drugs to scavenge ROS in different iPSC derivatives: NPCs and neurons. (A) Clustered heatmap for dosage analyses of different drugs. Each set of column represents data for a specific drug dose and each row represents data from different cells (NPCs and neurons) from healthy control, LS, and ECHS1 patients. All values are represented as ratio of ROS scavenging with drug: baseline. Color representation: green is higher, and red is lower/no ROS scavenging. (B) Drug doses with significant ROS scavenging in NPCs (n = 3). (C) Drug doses with significant ROS scavenging in neurons (n = 3). The “n” is representative of the biological replicates and was analyzed by one-way ANOVA. The data are representative of three independent experiments. Data are represented as means ± SD. *P < 0.05 in comparison to control.
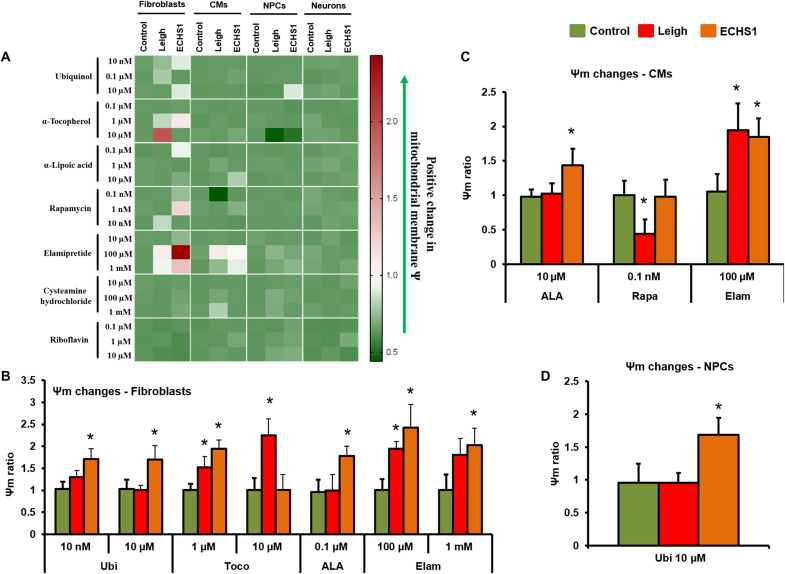
Mitochondrial membrane potential is associated with energy storage, optimal mitochondrial fusion dynamics, and a requirement for ideal ATP generation, thus making it a good indicator of the mitochondrial function. Ψm showed a broad range of responses, with most drug concentrations showing nonsignificant changes in CMs, NPCs, and neurons (Fig. 7A and fig. S13A). Treatment with Ubi improved Ψm in fibroblasts from ECHS1 (10 nM and 10 μM) patient (Fig. 7B). Ubi displayed no changes in Ψm in CMs of both patients and significantly increased Ψm of ECHS1 NPCs (10 μM; Fig. 7, A and D). Toco showed a significant increase in Ψm in LS fibroblasts (1 and 10 μM; Fig. 7B). Similarly, Toco increased Ψm in ECHS1 fibroblasts (1 μM; Fig. 7B). ALA treatment showed no changes in any of the LS cell types; however, there was a significant improvement in Ψm in ECHS1 fibroblasts (0.1 μM; Fig. 7B) and ECHS1 CMs (10 μM; Fig. 7C). Cys treatment in both LS and ECHS1 did not register any change in all cell types compared to control groups (Fig. 7A). Rapa treatment significantly reduced Ψm in LS CMs (0.1 nM; Fig. 7C). In ECHS1 cells, Rapa did not register any significant changes in fibroblasts, CMs, or NPCs (Fig. 7, B to D). Ribo treatment did not show any improvement in Ψm in all the cell types at all three (0.1, 1, and 10 μM) doses (fig. S13B). Elam was found to be beneficial in improving Ψm in cells from both patients. The cells that responded positively to Elam include fibroblasts and CMs from both the LS and ECHS1 patients at a dose of 100 μM (Fig. 7, B and C). Further, Elam at 1 mM dose showed positive trend with LS fibroblasts and significant improvement in the Ψm of ECHS1 patient fibroblasts (Fig. 7B).
Fig. 7. Effect of drug panel on mitochondrial membrane potential.
Efficacy of a panel of seven drugs—coenzyme Q10 (Ubi), ALA, Toco, Cys, Rapa, Elam, and Ribo—was evaluated by determining the effects of individual drugs on mitochondrial membrane potential (Ψm) in different iPSC derivatives—fibroblasts, NPCs, CMs, and neurons. (A) Clustered heatmap for drug dosage analyses of different drugs. Each row represents data for a specific drug dose, and each set of columns represents data from different cells (fibroblasts, NPCs, CMs, and neurons) from healthy control, LS, and ECHS1 patients. All values are represented in ratio of Ψm drug: baseline. The color changing from low (green) to high (red) correspond to improvement in Ψm. (B) Drug doses with significant mitochondrial membrane potential changes in fibroblasts (n = 3). (C) Drug doses with significant mitochondrial membrane potential changes in CMs (n = 3). (D) Drug doses with significant mitochondrial membrane potential changes in NPCs (n = 3). The “n” is representative of the biological replicates and was analyzed by one-way ANOVA. The data are representative of three independent experiments. Data are represented as means ± SD; *P < 0.05 in comparison to control.
Drug treatment in iPSC derivatives improved cellular phenotype and mitochondrial respiration
In the next experiment, the efficacy of the most effective drugs from the above studies was investigated for the ability of these drugs to improve the cellular phenotype, thereby providing us an understanding of their clinical efficacy in the patient. We administered Ubi, ALA, Ribo, and Elam (10 μM) for 7 days and tested their potential in improving neuronal and cardiac cell electrophysiology, Ca2+ handling properties, and mitochondrial respiration. In ECHS1 neurons, administration of both Ubi and ALA demonstrated promise in improving neuronal activity in terms of mean firing rate and burst frequency (fig. S14, A and B). The treatment with Ribo markedly improved calcium flux and handling capacity of ECHS1 neurons (fig. S15, A and B). Similarly, in ECHS1 CMs, there was a trend toward improvement in contractility and conduction velocity of electrical propagation after treatment with Ubi, ALA, and Ribo (fig. S16, A to D). Ribo was also able to significantly improve calcium handling of the ECHS1 CMs (fig. S16, E to H). These data suggest that treatment with Ubi, ALA, and Ubi resulted in improvement in neuronal and CM function. Furthermore, our seahorse analysis demonstrated that in ECHS1 neurons, after treatment with Ubi and ALA, there was a trend toward improvement in maximal and spare mitochondrial respiratory capacity (fig. S17). In ECHS1 CMs, treatment with Ubi caused a significant improvement in maximal and spare mitochondrial respiratory capacity, and there was a trend toward improvement in these parameters in CMs with ALA treatment (fig. S18). In fibroblasts, Ubi and ALA showed a significant improvement in maximal and spare respiratory capacity, whereas Ribo treatment was able to improve basal and maximal respiratory capacity (fig. S19). Treatment with these three drugs did not cause any significant change in the proliferation of ECHS1 fibroblasts (fig. S20). These data suggest that Ubi, ALA, and Ribo were very effective in improving mitochondrial respiration, cellular function, and phenotype in iPSC-derived fibroblasts, CMs, and neurons of ECHS1 patient, whereas treatment with Elam did not show any significant effect, but there was a trend toward improvement in cellular function.
Assessment of efficacy of drug panel in ECHS1 patient
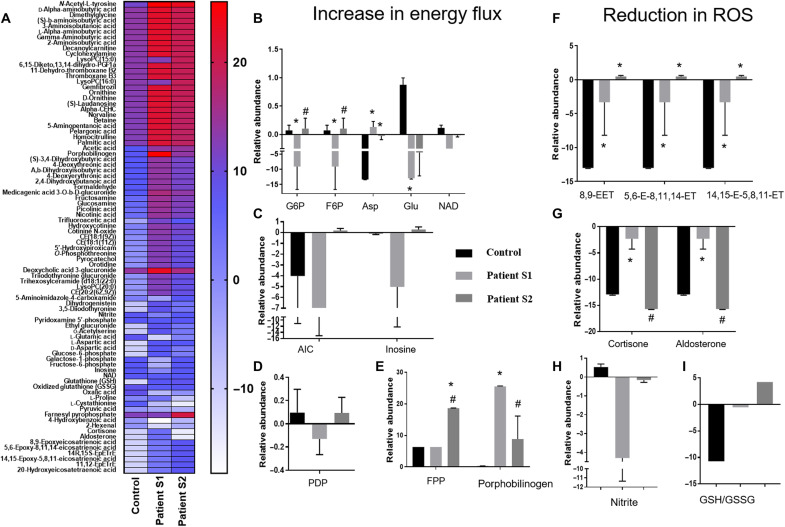
In the next step, as a proof of concept and to further validate the efficacy of our iPSC-based platform, we investigated metabolomic profiles of the ECHS1 patient who was administered a combination of Ubi, ALA, and Ribo. We performed nontargeted metabolomics [using a liquid chromatography-quadrupole time of flight/mass spectrometry (LC-QTOF/MS) system] in the patient’s blood cells, before and after 3 years of drugs administration and compared with healthy control. After administration of these drugs, despite no evidence for substantial clinical improvement, there has been no clinical deterioration and patient remains very stable. However, we observed a significant improvement in the metabolic profile of the patient after supplementation of these drugs (Fig. 8). A total of 200 metabolites were detected in our assay using two different column separations (C18 and SB). We observed a shift in the metabolomic profile of ECHS1 patient toward healthy control after treatment with Ubi, ALA, and Ribo (Fig. 8 and fig. S21, A and B). On closer observation, there were 85 different metabolites with significant changes within the groups (Fig. 8). Among these, there were a group of 18 metabolites (highlighted as histograms in Fig. 8, B to I) that showed significant improvement after treatment. These were metabolites from glycolysis and malate aspartate shuttle—pyridoxal-5 phosphate, a cofactor in many metabolic enzymes; porphobilinogen; farnesyl pyrophosphate; inosine; and 5-aminoimidazole-4-carboxamide—that show an improvement toward patient’s overall energy flux (Fig. 8, B to E). Further, metabolites including reduced and oxidized glutathione, EETs, nitrite, and hormones such as cortisone and aldosterone reflect an improvement in patients’ overall ROS levels (Fig. 8, F to I), which is also evident from our enrichment analysis that shows affected metabolic pathways (fig. S21C). The overall metabolic profile of the patient after 3 years of oral treatment with these drugs showed a correction in term of total energy flux and reduction in ROS levels, which is similar to what was indicated in our in silico analysis of these drugs and observed in our screening by iPSC-based models.
Fig. 8. Metabolomic analysis of patient PBMCs before and after the treatment.
Patient PBMCs were analyzed for the presence of metabolites and their relative abundance using mass spectrometry before (patient sample, S1) and after (patient sample, S2) the drug treatment. These profiles were compared to the metabolomic profile of a healthy individual (control). The metabolites were broadly screened for their association in modulating cellular energy flux and ROS levels. (A) The figure shows the heatmap of significantly altered metabolites in patient PBMCs. (B) Histograms showing metabolites involved in energy metabolism such as glucose-6-phosphate (G6P), fructose-6-phosphate (F6P), aspartate (Asp), glutamate (Glu), and nicotinamide adenine dinucleotide (NAD). (C) Relative abundance of 5-aminoimidazole-4-carboxamide (AIC) and inosine metabolites in control, S1, and S2 groups. (D) Histograms showing comparative relative abundance of pyridoxamine 5-phosphate (PDP). (E) Relative abundance of farnesyl pyrophosphate (FPP) and porphobilinogen. (F) Relative abundance of EET intermediates involved in ROS reduction related to arachidonic acid metabolism. (G) Relative abundance of cortisone and aldosterone. (H) Relative abundance of nitrite. (I) Relative abundance of reduced (GSH) to oxidized (GSSG) glutathione GSH/GSSG ratio. The “n” is representative of technical replicates and was analyzed by two-way ANOVA. Data are represented as means ± SD; *P < 0.05 in comparison to control and #P < 0.05 in comparison to patient sample, S1.
Furthermore, ECHS1 deficiency is reported to cause increased acylcarnitine levels and accumulation of short-chain fatty acid derivatives (9). In the patient’s blood sample, we observed a significant accumulation of metabolites such as decanoylcarnitine, aminobutyric acid, and other butyric acid derivatives including 3,4-dihydroxybutanoic acid that are known to cause neurological symptoms (43, 44). After treatment with the abovementioned three drugs (Ubi, ALA, and Ribo), there was a decreasing trend in the levels of these metabolites (fig. S22). All these observations verify the validity of our iPSC-based personalized platform to assess drug safety and efficacy before considering these patients for enrollment in any particular clinical trial.
DISCUSSION
When drugs reach phase 2 clinical trials, they would have already gone through several rigorous quality control steps. The primary and secondary outcomes in healthy controls from phase 0/1 trials help inform a guided dosage range of the experimental regime, which, despite these controlled approaches, have a likelihood of low efficacy in a clinical trial (45). The unique characteristics of the enrolled patient that might confound a clinical trial are patient-specific pathophysiology (46), disease progression (47), enrichment of drug response to certain treatment-sensitive subtypes (48), and, underlining all these, the unaddressed genetic heterogeneity (49, 50). These situations are further compounded when novel pathogenic variants exist (51). The nonavailability of relevant preclinical models for ultrarare and novel mutations leads to uncertainty for the inclusion of these patients in a clinical trial for approved drugs (52). However, an iPSC-based personalized clinical trial selection platform as presented in this study can act as an auxiliary system to phase 2 trials, wherein one could specifically challenge those molecules that have undergone rigorous testing in patients with similar phenotypes but dissimilar mutations.
In the current study, the iPSC multisystem platform allowed rapid testing of available drugs in ECHS1 patient–specific cells to find out the safest and most efficient treatment options. Patient-specific iPSC derivatives permitted the detection of cellular level changes not only in those cells where the disease manifestation was expected (iPSC-NPCs and neurons) but also in CMs that were not apparent in the patient (but reported in other patients) at the organ (heart) level but could present at an older age (6). In the current study, the disease pathogenicity of ECHS1 mutation was confirmed by observing improvement in cellular phenotype upon introduction of WT ECHS1 protein in iPSC-derived fibroblasts of ECHS1 patient. Further, compromise in cellular functions and physiological differences (through investigation of ROS, Ψm, and mitochondrial respiration) provided a comprehensive tool to test the toxicity and efficacy of various experimental drugs in ECHS1 patient–specific cells.
In case of drugs that were administered to the ECHS1 patient, all of these showed efficacy in reducing ROS across different iPSC-derived cell types. Ubi showed beneficial effects in NPCs and fibroblasts, Ribo was able to reduce ROS in all cell types, while ALA showed significant improvement in Ψm in fibroblasts and CMs. This makes a stronger case for the continuation of usage of combination of these drugs for the ECHS1 patient. Of the two drugs that were discontinued for the patient, Toco showed a concerning reduction trend in Ψm of ECHS1 NPCs, while Cys showed no marked difference in the cell types tested. While Toco can be ruled out of future treatment strategies for the ECHS1 patient, the discomfort caused by Cys can be presumably attributed to its dynamics in cells of other lineages than the ones studied here or due to physiological parameters other than LDH, ROS, and Ψm. Further, our in silico analysis also demonstrated that Ubi, ALA, and Ribo could be beneficial against ECHS1 deficiency and can have positive influence on cellular bioenergetics and ROS scavenging. In our in vitro studies, all three drugs—Ubi, ALA, and Ribo—showed a significant potential in improving mitochondrial respiration in the ECHS1 neurons, CMs, and fibroblasts, respectively. We also observed improvements in electrophysiology and calcium handling properties of ECHS1 neurons and CMs after treatment with Ubi, ALA, and Ribo. Furthermore, the ability of all three drugs in scavenging ROS in iPSC-derived cells reflects their affinity toward cytochrome P450 B6 enzyme family. Similarly, these drugs demonstrated binding affinities for GLAST and FPPS, which could regulate and are essential for maintaining cellular bioenergetics.
In literature, specific treatments for LS have been reported (with varying successes) only for a few mutations causing LS or LS-like disorder, specifically with mutations in the following genes: SLC19A3, BTD, PDSS2, ETHE1, PDHA1, and TPK1 (53, 54). Therefore, for the rest of the patients with variants, the consensus standards for the ongoing care of diagnosed patients include endorsing varying vitamin “cocktails,” exercise, and diet regimen changes. Of the recently concluded and ongoing clinical trials that were available to be tested for our LS-like patient included these drugs—Elam, cys bitartate, Epi-743, KH176, and ABI-0029 (Rapa). With so many choices and having no information on the respective drug responses in a model with specific pathological variants, the patient faced steep challenges in participating in any of these clinical trials. These challenges were geographical (significant amount of travel), financial (cost of relocation along with family), and emotional (temporarily leaving the institution, support network, and personal medical care). Notably, here is the fact that out of the numerous options, our patient was enrolled for the Cys trial, which had to be discontinued because of side effects. This restricted the patient’s choice to participate in other trials that may have fared better results. iPSC-based clinical trial selection platforms as demonstrated in the current study can be applied to these patient-specific scenarios where time and financial lenience in developing the platform allows efficient decision-making.
In extension with the abovementioned observations and facts, a mode of action for the ECHS1 patients with the LS-like syndrome was established: The clinically safe FDA-approved effective drugs—Ubi, ALA, and Ribo—were administered to the patient and showed significant clinical improvement in patient’s metabolic profile, which was also reflected in the improvement observed in the phenotypes of the iPSC derivatives. Therefore, the outcome of current study validates the effectiveness of iPSC-based personalized prescreening tool for patients with ultrarare variants. The intention behind creating this platform is to provide a selection of effective drugs against respective pathologies and transfer this knowledge back to the clinicians and patients, which will help them take an informed clinically safe and effective decision. This iPSC-based tool may aid patients with ultrarare disorders in choosing an effective and safe clinical trial.
MATERIALS AND METHODS
Patient recruitment and blood collection
The study was approved by the “Research Ethics Board” (HS18974) of the University of Manitoba. The patients with Leigh and LS-like syndrome were diagnosed at the Children’s Hospital Research Institute of Manitoba. The informed consents were obtained in accordance with the institutional policy. Blood (10 ml) was collected from a patient with LS-like syndrome harboring compound heterozygous mutations in the ECHS1 gene, a deceased patient with LS harboring compound heterozygous mutations in the NDUFV1 gene, and four sex-matched controls. The PBMCs were extracted using the Lympholyte-H Kit (Cedarlane, Canada). All the protocols and procedures were approved by the University of Manitoba’s Health Research Ethics Board. Before blood collection, written informed consent from all the patients’ parents and healthy individuals (controls) was obtained.
iPSC generation and maintenance
The PBMCs were reprogrammed to iPSC using a commercial kit (CytoTune-iPS 2.0 Sendai Reprogramming Kit, Thermo Fisher Scientific, lot no. L2120016). Briefly, PBMCs (4 × 105 cells) were cultured in a six-well plate for 24 hours and then transduced with Sendai virus. The multiplicity of infection for each of the three viruses in the kit was KOS:c-MYC:KlF4 at 5:5:3. After 4 days, cells were plated onto Geltrex (Gibco)–coated plates and cultured in a mixture of PBMC medium (Life Technologies) and TeSR-E8 (STEMCELL Technologies Inc.) medium at a ratio of 1:1. When cells showed transformation and started attaching to the Geltrex, the cell culture medium was replaced with TeSR-E8. After a successful adaptation to feeder-free conditions of at least three iPSC colonies from each patient and control, the cells were expanded in TeSR-E8 medium using manual gridding protocol.
Assessment of pluripotency of iPSC
The pluripotency of generated iPSC was evaluated by immunostaining the cells with different markers (Oct4A, SOX2, NANOG, SSEA4, TRA-1-60, and TRA-1-81) using a commercial kit (StemLight Pluripotency Antibody Kit, Cell Signaling Technology, catalog no. 9656S). Briefly, the iPSCs were fixed with 4% paraformaldehyde and permeabilized with ice-cold methanol for 10 min at −20°C and then rinsed with phosphate-buffered saline (PBS) for 5 min. After blocking with 5% bovine serum albumin solution [containing 0.3% Triton X-100 (w/v)] for 60 min, the cells were probed with primary antibody (1:500) for 12 hours. After washing with PBS, the iPSC was incubated with fluorochrome-conjugated secondary antibody (1:1000) for 2 hours. The nuclei were stained with 4′,6-diamidino-2-phenylindole. The iPSCs were washed with PBS and examined under fluorescent microscope (Cytation 5, BioTek Instruments).
The pluripotency of iPSC was also evaluated by PCR. Total RNA was isolated using a column extraction kit (Bio-Rad, no. 7326820). Two micrograms of total RNA was converted to cDNA using a cDNA synthesis kit (Life Technologies, no. 4374966) as per the manufacturer’s protocol. Gene expression of pluripotency markers including Oct4, Sox2, and Nanog was performed using SYBR Green Supermix (Bio-Rad, no. 1725271) and Bio-Rad C-1000 Touch. The detail of primers used for the PCR is listed in table S1.
EB formation
To prepare EBs, the colonies were carefully lifted following dispase treatment and suspended in Nunclon Sphera low attachment dishes (Thermo Fisher Scientific). The EBs were cultured in a medium that was prepared using high-glucose Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal bovine serum, nonessential amino acids, β-mercaptoethanol, l-glutamine, and penicillin/streptomycin (Life Technologies). At day 8, the surviving EBs were plated onto gelatin-coated dishes. The differentiation potential of generated EBs toward the three germ layers—ectoderm, mesoderm, and endoderm—was measured using a commercial kit (STEMdiff Trilineage Differentiation Kit, STEMCELL Technologies Inc.).
Fibroblast differentiation
Once EBs were plated after 4 days of suspension culture, spontaneous fibroblasts were allowed to grow out and visually assessed. The fibroblasts were manually dissected from the culture plate and stained for HSP47 (catalog no. AB77609, Abcam) and fibroblast surface protein (FSP) (catalog no. AB11333, Abcam). The cell populations that were positive for both markers were used for further studies.
NPC differentiation
The iPSC-derived EBs were treated with SB 431542 (a transforming growth factor–β/Smad inhibitor; Torcis) for 2 days, followed by plating on polyornithine-coated plates on day 5. The neural rosettes, which appeared at days 7 to 10 onward, were excised and grown on polyornithine-coated plates in Neurocell medium (Wisent Bioproducts). The immunostaining was carried out after passage 3 (using Accutase) with Pax6 (catalog no. sc-81649, Santa Cruz Biotechnology) and nestin (catalog no. sc-23927, Santa Cruz Biotechnology).
CM differentiation
The iPSCs (>p20) were passaged using EDTA and grown until the cells reached ∼75% confluency. The medium was changed to CDM3, which consists of RPMI 1640 (Life Technologies), Oryza sativa–derived recombinant human albumin (500 μg ml−1; Sigma-Aldrich), and l-ascorbic acid 2-phosphate (213 μg ml−1; Sigma-Aldrich). Medium was changed every other day (48 hours). At days 0 to 2, medium was supplemented with 6 μM glycogen synthase kinase 3-β inhibitor CHIR99021 (APExBio). On day 2, the medium was changed to CDM3 supplemented with 2 μM Wnt inhibitor Wnt-C59 (Calbiochem Research Biochemicals). The medium was changed on day 4 and every other day for CDM3 cultures. The contracting cells were observed from day 7. At day 10, the medium was changed to CDM3L, which consists of RPMI 1640 no glucose (Life Technologies), recombinant human albumin (500 μg ml−1), and l-ascorbic acid 2-phosphate (213 μg ml−1) supplemented with 4 mM l-lactic acid (Sigma-Aldrich). CM formation was further confirmed by staining for NKX2.5 (Cell Signaling Technology no. 8792S) and cardiac troponin (catalog no. AB8295, Abcam).
Neuron differentiation
The iPSCs were differentiated into forebrain neurons using a combination of the following commercially available kits: STEMdiff SMADi Neural Induction Kit (catalog no. 08581, STEMCELL Technologies), STEMdiff Neural progenitor medium (catalog no. 05833, STEMCELL Technologies), STEMdiff Forebrain Neuron Differentiation Kit (catalog no. 08600, STEMCELL Technologies), and STEMdiff Forebrain Neuron Maturation Kit (catalog no. 08605, STEMCELL Technologies). Briefly, iPSCs were plated as single cells on Matrigel-coated plates. When the cells were at >80% confluency, the SMADi neural induction medium (prepared as per the manufacturer’s directions) was added to the single-cell culture and replenished every alternate day until day 18. The obtained NPCs were passaged on days 6, 12, and 18. Thereafter, NPCs were cultured and expanded in neural progenitor medium and used for differentiation as needed. On day 19, the NPCs were supplemented with Forebrain Neuron Differentiation medium (prepared as per the manufacturer’s directions), and the medium was replenished daily until day 25. From day 25 onward, the cells were maintained in Forebrain Neuron Maturation medium (prepared as per the manufacturer’s directions) and observed for mature neuron morphology. The mature neurons were immunostained for microtubule-associated protein 2 (MAP2) and neuronal nuclear protein (NeuN).
Confirmation of patient mutations in iPSC
To confirm the presence of patient mutations in iPSC, Sanger sequencing was performed. The specific primers flanking the compound heterozygous mutations were prepared (table S1). The products were limited to a size range of 100 to 300 base pairs. The genomic DNA from iPSC colonies was isolated using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific). The AmpliTaq Gold DNA polymerase with buffer II and MgCl2 (Thermo Fisher Scientific) was used for the PCR reaction. The PCR products were processed for Sanger sequencing at TCAG DNA/Sequencing Facility at the Centre for Applied Genomics, Toronto. The resulting files were evaluated through the Chromas Software.
ATP measurement
The extracellular ATP was quantified in iPSC-fibroblasts, NPCs, CMs, and neurons using a luminescent ATP detection assay kit (catalog no. ab113849, Abcam). A total of 105 cells were plated into each well of a 96-well plate in the complete respective medium of each cell type. The drugs, where required, were added for a 24-hour period. At the end of the treatment, the plate was centrifuged at 1000 rpm for 5 min to pellet-floating cells. The cells were lysed using a detergent (provided in the kit) and incubated with the substrate solution (at room temperature) for 5 min in an orbital shaker at 650 rpm and then for 10 min in the dark. The ATP standards were loaded on the same plate as references for quantification. The quantification of plates was performed with the luminescence detection module of the Cytation 5 plate reader (BioTek Instruments).
Lactate assay
The lactate levels were detected in iPSC-fibroblasts, NPCs, CMs, and neurons using a commercially available kit (L-Lactate Assay Kit, catalog no. ab65330, Abcam). The cells were plated into a 96-well plate (105 cells per well) in complete medium for each cell type. The drug treatment, where required, was for a 24-hour period. A reaction mix composed of cell lysates was prepared according to the manufacturer’s instructions. Briefly, the cells were suspended in 4× lactate assay buffer, followed by homogenization. The resultant supernatant was collected after centrifugation, and the manufacturer-provided reaction mix was added to the samples and incubated for 30 min. The quantification was performed with Cytation 5 plate reader (BioTek Instruments) at a wavelength of 570 nm. The lactate concentrations were calculated from the standard curve (ranging from 0 to 10 nmol per well) generated from the lactate standards according to the manufacturer’s instructions.
Mitochondrial membrane potential measurement
The mitochondrial membrane potential (Ψm) was measured in iPSC-fibroblasts, NPCs, CMs, and neurons using tetramethylrhodamine ethyl ester (TMRE) assay. Briefly, the cells were plated in a 96-well plate and then incubated for 10 min with 50 μM FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone), a protonophore that collapses the Ψm, as a negative control. TMRE (1 μM final concentration) was then added for 30 min, cells were further collected and washed, and fluorescence was read using Cytation 5 plate reader (BioTek Instruments) at an excitation wavelength of 549 nm and an emission wavelength of 575 nm.
Cellular growth count
Cellular growth was assessed through cell doubling count. Briefly, fibroblasts and NPCs were plated after staining with CytoPainter (catalog no. 138891, Abcam). The cell number was counted after 2 days using Cytation 5 plate reader (BioTek Instruments). The data are presented as ratio of day 2 count versus plating number (1 × 104).
ROS measurement
The ROS formation was measured in fibroblasts, NPCs, CMs, and neurons using a dye, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA). Briefly, the cells (1 × 106) were plated in a 96-well plate, washed with PBS, and then incubated with 5 μM H2DCFDA and Hoechst (0.5 μg/ml) in the respective medium. The fluorescence signal was recorded using Cytation 5 plate reader (BioTek Instruments) at an excitation wavelength of 485 nm and an emission wavelength of 535 nm.
Measurement of cellular bioenergetics
The cellular bioenergetics was determined using the extracellular flux (XFe24) analyzer (Seahorse Bioscience). The mean basal respiration was determined by recording OCR measurements before adding inhibitors or activators. The cells were seeded in XF Cell Culture Microplates. On the day of the experiment, the medium was changed to bicarbonate-free DMEM containing glucose (Sigma-Aldrich), l-glutamine, and sodium pyruvate (Sigma-Aldrich). The pH was adjusted to 7.0 to 7.4. The cells were incubated at 37°C for 1 hour before starting the assay. First, the baseline OCR was measured. Proton leak was measured by the addition of 1 μM oligomycin. Maximal capacity respiration was determined by the sequential injection of FCCP (2 μM). Nonmitochondrial respiration was measured by the addition of 1 μM rotenone and 1 μM antimycin A. Data were corrected for the nonmitochondrial respiration and normalized to the total protein concentration measured by Bradford method. Data are presented as picomole of oxygen per minute per milligram of protein.
Drug challenges in iPSC-derived cells
Ubi (catalog no. 1705334, Sigma-Aldrich) stock was prepared in acetone solution, and working concentrations were prepared in respective medium. Toco (T3251, Sigma-Aldrich) stock was prepared in ethanol solution, and working concentrations were prepared in respective medium. ALA (07039, Sigma-Aldrich) stock was prepared in ethanol solution, and working concentrations were prepared in respective medium. Cys hydrochloride (22193-25 CA, Cayman Chemicals) stock was prepared in water solution, and working concentrations were prepared in respective medium. Rapa (catalog no. AAJ62473MF, Sigma-Aldrich) stock was prepared in dimethyl sulfoxide (DMSO) solution, and working concentrations were prepared in respective medium. Elam (HY-P0125, MedChemExpress) stock was prepared in DMSO solution, and working concentrations were prepared in respective medium. Ribo (catalog no. 132350250, Acros Organics) stock was prepared in 0.1 N NaOH solution, and working concentrations were prepared in respective medium. For cytotoxicity studies, ROS measurement, and mitochondrial membrane assessment, the drugs were administered for 24 hours. For MEA analysis, calcium handling assessment, and Seahorse analysis, Ubi, ALA, Ribo, and Elam (10 μM) were used to treat neurons, CMs, and fibroblasts for 7 days to mimic long-term treatment in patient.
LDH assay
The cytotoxicity caused by drugs in fibroblasts, NPCs, CMs, and neurons was measured by detecting LDH release from the cells using the LDH Cytotoxicity Detection Kit (Clontech Laboratories). The cells were plated at 104 cells per well in a 96-well plate and treated with the maximum dosage of respective drugs (10 μM Ubi, 10 μM Toco, 10 nM Rapa, 1 mM Elam, 1 mM Cys HCl, 100 μM ALA, and 10 μM Ribo) used in the study. After 3 days of culture, drug-mediated toxicity was assessed by LDH release from the affected cells using Cytation 5 plate reader (BioTek Instruments).
MEA analysis
MEA plates were coated with Geltrex in DMEM F12 (1:100). The neurons and CMs (40,000 cells per well) were seeded in 24-well MEA plates, covering all 16 electrodes uniformly. The data were recorded for neurons and CMs on Maestro Edge MEA System (Axion Biosystems Inc.) using Axis Navigator 3.5.1 software. The neural metric tool was used to generate raster plots for neurons. In these plots, each line is a spike that represents firing of neurons, and a group of five continuous spikes makes up a burst that is represented by a blue line in the figure. A series of bursts from neighboring neurons make up a network burst represented by the activity in the purple box in each raster plot. Similarly, the recordings for CMs were analyzed using a cardiac metric tool for field potential and contractility.
Calcium signaling analysis
Calcium flux was evaluated using Fluo-4 AM dye for both neurons and CMs by following the manufacturer’s protocol. Briefly, 3 μM Fluo-4 AM solution in Hanks’ balanced salt solution (HBSS) was loaded in the respective cells. After incubation in the dark at room temperature for 45 min, the cells were washed in HBSS and further incubated at 37°C in the dark for de-esterification for 20 min. Neurons were evoked using a buffer containing 10 mM KCl after 5 min of baseline recording for an additional 10 min. The calcium imaging in CMs was performed by recording 30-s video loops. Imaging was performed using the Nikon Ti2 inverted fluorescent microscope. For CMs, data were collected from 10 different regions of interest per loop, and 80 to 130 beats were analyzed and plotted for every group of CMs. Similar analysis was performed for neurons. The data were analyzed using the Clampfit 10.7.0.3 and Nikon AR software.
In silico analysis
Protein database structures [Protein Data Bank (PDB)] for the respective targets [ACADM, mitochondrial (PDB ID: 1EGC); ACADS, mitochondrial (PDB ID: 2VIG); Enoyl-CoA hydratase, mitochondrial (PDB ID: 2HW5); cytochrome P450 family 2 subfamily B6 (PDB ID: 5uda); FPPS (PDB ID: 4xqr); GLAST (PDB ID: 4p60); and glutathione peroxidase (PDB ID: 2f8a)] were retrieved. The structure of ALA was retrieved from PubChem (PubChem CID:864), and the structure of Ubi was constructed using the Frog2 software, using the SMILES formula [CC1 = C(C(=C(C(=C1O)OC)OC)O)CC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)C] procured from the PubChem (PubChem CID:9962735). AutoDock MGL tools version 1.5.6. were used to prepare molecular targets and drugs for docking. AutoDock Vina version 1.1.2. was used for the docking analysis. A comparative analysis of the obtained binding energies (kilocalorie per mole) was performed. Furthermore, PyMol version 2.4.1 was used to visualize the top docking site/pockets where the drug interacted with the target molecule. Last, to analyze the multitudinous interactions among the drugs and the molecular target binding sites, LigPlus software was used.
Metabolomics of patient samples
A total of 2 million PBMCs were used for sample preparation. The extracted samples were dried using a nitrogen evaporator. The dried samples were reconstituted and used for analysis in LC-QTOF/MS system. The analysis was performed on a Rapid Resolution HPLC system (1290 Infinity Agilent Ltd., Santa Clara, CA) that was coupled to a 6538 UHD Accurate LC-QTOF-MS (Agilent Technologies, Santa Clara, CA) with dual electrospray ionization (ESI) source (Agilent Technologies, Santa Clara, CA). The sample acquisition was performed in both positive (+) and negative (−) ESI modes. Metabolites were annotated using Metlin Database. These metabolites were then fed into MetaboAnalyst online tool for enrichment and pathway analysis. The heatmaps and histograms were generated using GraphPad Prism 7 (San Diego, CA).
ECHS1 expression in cells
WT ECHS1 protein was ectopically expressed in iPSC-derived fibroblasts of ECHS1 patient using an expression vector (OriGene no. RC200369). The expression vector (5 μg) was transfected using Lipofectamine 3000 reagent (Life Technologies, no. L3000008) as per the manufacturer’s protocol. The transfected fibroblasts were selected using G418 (Life Technologies, no. 11811023) at 300 μg/ml in DMEM high-glucose media (Gibco, no. 10569010). The expression of WT pECHS1 protein was confirmed by immunoblotting against its DDK tag using anti-DDK antibody (OriGene, no. TA50011100).
Cell proliferation assay
The proliferation of fibroblasts was assessed using a WST1 assay kit (BioVision Inc., no. K301-500) using the manufacturer’s instructions. Briefly, 5000 cells were seeded in 96-well cell culture plates. After respective treatments, 10 μl of WST1 reagent was added to the cells and incubated for 60 min at 37°C in a CO2 incubator. The absorbance of the plate was read at 450 nm.
Statistical analysis
Data are presented as means ± SD/SEM as indicated in respective figure legends. The continuous variable between two independent groups was compared using the Student’s t test. Two or more groups with one variable were analyzed using one-way analysis of variance (ANOVA) with multiple comparison analysis using Tukey’s post hoc test. Two or more groups with two variables were analyzed using two-way ANOVA with Tukey’s post hoc test for multiple comparison analysis where appropriate. Unless noted otherwise, adjusted P values of <0.05 were considered statistically significant. All statistical analysis was performed using GraphPad Prism 7.
Acknowledgments
We thank E. Bloomfield of Children’s Hospital Research Institute of Manitoba, Winnipeg in assisting with human blood sample collection. We thank B. Cohen of Children’s Hospital of Akron, Ohio for helpful discussions. The assistance of J. Wrogemann in providing the brain MRI image is greatly appreciated.
Funding: This work was supported by a grant from the Canadian Institutes of Health Research (MOP142265 to S.D.).
Author contributions: The study was conceptualized and designed by G.L.S., A.S., C.R.-G., and S.D. G.L.S., A.S., N.S., W.Y., and K.N.A. carried out the experiments and acquired the data. G.L.S., A.S., and S.D. interpreted the data and performed statistical analysis. M.R.A. and P.F. carried out Seahorse analysis, interpreted the data, and performed statistical analysis. M.A. carried out metabolomics analysis, interpreted the data, and performed statistical analysis. E.V. and A.K. performed in silico analysis. C.R.-G. recruited patients and healthy controls for the study. G.L.S., A.S., C.R.-G., and S.D. drafted the manuscript. All authors have read and approved the final manuscript.
Competing interests: The authors declare that they have no competing interests. The authors declare no other relationships or activities that could appear to have influenced the submitted work. Ethical approval was given by the Research Ethics Board (HS18974) of the University of Manitoba.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S22
Table S1
REFERENCES AND NOTES
- 1.Renfro L. A., Sargent D. J., Statistical controversies in clinical research: Basket trials, umbrella trials, and other master protocols: A review and examples. Ann. Oncol. 28, 34–43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamazaki T., El Rouby N., Fredette N. C., Santostefano K. E., Terada N., Concise review: Induced pluripotent stem cell research in the era of precision medicine. Stem Cells Dayt. Ohio 35, 545–550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meseguer-Ripolles J., Khetani S. R., Blanco J. G., Iredale M., Hay D. C., Pluripotent stem cell-derived human tissue: Platforms to evaluate drug metabolism and safety. AAPS J. 20, 20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baghbaderani B. A., Tian X., Neo B. H., Burkall A., Dimezzo T., Sierra G., Zeng X., Warren K., Kovarcik D. P., Fellner T., Rao M. S., cGMP-manufactured human induced pluripotent stem cells are available for pre-clinical and clinical applications. Stem Cell Rep. 5, 647–659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruhoy I. S., Saneto R. P., The genetics of Leigh syndrome and its implications for clinical practice and risk management. Appl. Clin. Genet. 7, 221–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sofou K., De Coo I. F. M., Isohanni P., Ostergaard E., Naess K., De Meirleir L., Tzoulis C., Uusimaa J., De Angst I. B., Lönnqvist T., Pihko H., Mankinen K., Bindoff L. A., Tulinius M., Darin N., A multicenter study on Leigh syndrome: Disease course and predictors of survival. Orphanet J. Rare Dis. 9, 52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sequiera G. L., Rockman-Greenberg C., Dhingra S., Generation of human induced pluripotent stem cell (hiPSC) line UOMi001-A from a patient with Leigh-like syndrome harbouring compound heterozygous variants in ECHS1 gene. Stem Cell Res. 48, 101934 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Sequiera G. L., Rockman-Greenberg C., Dhingra S., Induced pluripotent stem cell line UOMi002-A from a patient with Leigh syndrome with compound heterozygous mutations in the NDUFV1 gene. Stem Cell Res. 48, 101964 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Haack T. B., Jackson C. B., Murayama K., Kremer L. S., Schaller A., Kotzaeridou U., de Vries M. C., Schottmann G., Santra S., Büchner B., Wieland T., Graf E., Freisinger P., Eggimann S., Ohtake A., Okazaki Y., Kohda M., Kishita Y., Tokuzawa Y., Sauer S., Memari Y., Kolb-Kokocinski A., Durbin R., Hasselmann O., Cremer K., Albrecht B., Wieczorek D., Engels H., Hahn D., Zink A. M., Alston C. L., Taylor R. W., Rodenburg R. J., Trollmann R., Sperl W., Strom T. M., Hoffmann G. F., Mayr J. A., Meitinger T., Bolognini R., Schuelke M., Nuoffer J.-M., Kölker S., Prokisch H., Klopstock T., Deficiency of ECHS1 causes mitochondrial encephalopathy with cardiac involvement. Ann. Clin. Transl. Neurol. 2, 492–509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R. Ganetzky, C. Stojinski, in GeneReviews®, M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. Bean, K. Stephens, A. Amemiya, Eds. (University of Washington, Seattle, Seattle (WA), 1993). [PubMed] [Google Scholar]
- 11.Houštěk J., Pícková A., Vojtíšková A., Mráček T., Pecina P., Ješina P., Mitochondrial diseases and genetic defects of ATP synthase. Biochim. Biophys. Acta BBA Bioenerg. 1757, 1400–1405 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hommes F. A., Polman H. A., Reerink J. D., Leigh’s encephalomyelopathy: An inborn error of gluconeogenesis. Arch. Dis. Child. 43, 423–426 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Detre J. A., Wang Z., Bogdan A. R., Gusnard D. A., Bay C. A., Bingham P. M., Zimmerman R. A., Regional variation in brain lactate in leigh syndrome by localized 1H magnetic resonance spectroscopy. Ann. Neurol. 29, 218–221 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Holmgren D., Wåhlander H., Eriksson B. O., Oldfors A., Holme E., Tulinius M., Cardiomyopathy in children with mitochondrial disease; clinical course and cardiological findings. Eur. Heart J. 24, 280–288 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Parikh S., Goldstein A., Koenig M. K., Scaglia F., Enns G. M., Saneto R., Anselm I., Cohen B. H., Falk M. J., Greene C., Gropman A. L., Haas R., Hirano M., Morgan P., Sims K., Tarnopolsky M., Van Hove J. L. K., Wolfe L., DiMauro S., Diagnosis and management of mitochondrial disease: A consensus statement from the Mitochondrial Medicine Society. Genet. Med. 17, 689–701 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zorova L. D., Popkov V. A., Plotnikov E. Y., Silachev D. N., Pevzner I. B., Jankauskas S. S., Babenko V. A., Zorov S. D., Balakireva A. V., Juhaszova M., Sollott S. J., Zorov D. B., Mitochondrial membrane potential. Anal. Biochem. 552, 50–59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman J. R., Nunnari J., Mitochondrial form and function. Nature 505, 335–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmieri L., Alberio S., Pisano I., Lodi T., Meznaric-Petrusa M., Zidar J., Santoro A., Scarcia P., Fontanesi F., Lamantea E., Ferrero I., Zeviani M., Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum. Mol. Genet. 14, 3079–3088 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Koenig M. K., Presentation and diagnosis of mitochondrial disorders in children. Pediatr. Neurol. 38, 305–313 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi G., Cortopassi G., Oxidative stress in inherited mitochondrial diseases. Free Radic. Biol. Med. 88, 10–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin M. T., Beal M. F., Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Hirano M., Emmanuele V., Quinzii C. M., Emerging therapies for mitochondrial diseases. Essays Biochem. 62, 467–481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddux B. A., See W., Lawrence J. C., Goldfine A. L., Goldfine I. D., Evans J. L., Protection against oxidative stress—Induced insulin resistance in rat L6 muscle cells by micromolar concentrations of α-lipoic acid. Diabetes 50, 404–410 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Kim K.-H., Lee B., Kim Y.-R., Kim M.-A., Ryu N., Jung D. J., Kim U.-K., Baek J.-I., Lee K.-Y., Evaluating protective and therapeutic effects of alpha-lipoic acid on cisplatin-induced ototoxicity. Cell Death Dis. 9, 827 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kappus H., Diplock A. T., Tolerance and safety of vitamin E: A toxicological position report. Free Radic. Biol. Med. 13, 55–74 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Hekimi S., Understanding ubiquinone. Trends Cell Biol. 26, 367–378 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Sabbah H. N., Gupta R. C., Singh-Gupta V., Zhang K., Effects of elamipretide on skeletal muscle in dogs with experimentally induced heart failure. ESC Heart Fail. 6, 328–335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatfield K. C., Sparagna G. C., Chau S., Phillips E. K., Ambardekar A. V., Aftab M., Mitchell M. B., Sucharov C. C., Miyamoto S. D., Stauffer B. L., Elamipretide improves mitochondrial function in the failing human heart. JACC Basic Transl. Sci. 4, 147–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao Z., Choo Y. S., Lesort M., Cystamine and cysteamine prevent 3-NP-induced mitochondrial depolarization of Huntington’s disease knock-in striatal cells. Eur. J. Neurosci. 23, 1701–1710 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Johnson S. C., Yanos M. E., Kayser E.-B., Quintana A., Sangesland M., Castanza A., Uhde L., Hui J., Wall V. Z., Gagnidze A., Oh K., Wasko B. M., Ramos F. J., Palmiter R. D., Rabinovitch P. S., Morgan P. G., Sedensky M. M., Kaeberlein M., mTOR Inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Civiletto G., Dogan S. A., Cerutti R., Fagiolari G., Moggio M., Lamperti C., Benincá C., Viscomi C., Zeviani M., Rapamycin rescues mitochondrial myopathy via coordinated activation of autophagy and lysosomal biogenesis. EMBO Mol. Med. 10, , 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almannai M., El-Hattab A. W., Ali M., Soler-Alfonso C., Scaglia F., Clinical trials in mitochondrial disorders, an update. Mol. Genet. Metab. 131, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parikh S., Saneto R., Falk M. J., Anselm I., Cohen B. H., Haas R., A modern approach to the treatment of mitochondrial disease. Curr. Treat. Options. Neurol. 11, 414–430 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garone C., Viscomi C., Towards a therapy for mitochondrial disease: An update. Biochem. Soc. Trans. 46, 1247–1261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters H., Buck N., Wanders R., Ruiter J., Waterham H., Koster J., Yaplito-Lee J., Ferdinandusse S., Pitt J., ECHS1 mutations in Leigh disease: A new inborn error of metabolism affecting valine metabolism. Brain J. Neurol. 137, 2903–2908 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Avula S., Parikh S., Demarest S., Kurz J., Gropman A., Treatment of mitochondrial disorders. Curr. Treat. Options. Neurol. 16, 292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karaa A., Kriger J., Grier J., Holbert A., Thompson J. L. P., Parikh S., Hirano M., Mitochondrial disease patients’ perception of dietary supplements’ use. Mol. Genet. Metab. 119, 100–108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enns G. M., Cohen B. H., Clinical trials in mitochondrial disease: An update on EPI-743 and RP103. J. Inborn Errors Metab. Screen. 5, 1–13 (2017). [Google Scholar]
- 39.Nissanka N., Moraes C. T., Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease. FEBS Lett. 592, 728–742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craven L., Alston C. L., Taylor R. W., Turnbull D. M., Recent advances in mitochondrial disease. Annu. Rev. Genomics Hum. Genet. 18, 257–275 (2017). [DOI] [PubMed] [Google Scholar]
- 41.D. R. Thorburn, J. Rahman, S. Rahman, in GeneReviews®, M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. Bean, K. Stephens, A. Amemiya, Eds. (University of Washington, Seattle, 1993). [Google Scholar]
- 42.Iannetti E. F., Smeitink J. A. M., Willems P. H. G. M., Beyrath J., Koopman W. J. H., Rescue from galactose-induced death of Leigh Syndrome patient cells by pyruvate and NAD +. Cell Death Dis. 9, 1135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shinka T., Inoue Y., Ohse M., Ito A., Ohfu M., Hirose S., Kuhara T., Rapid and sensitive detection of urinary 4-hydroxybutyric acid and its related compounds by gas chromatography-mass spectrometry in a patient with succinic semialdehyde dehydrogenase deficiency. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 776, 57–63 (2002). [DOI] [PubMed] [Google Scholar]
- 44.An M., Gao Y., Urinary biomarkers of brain diseases. Genomics Proteomics Bioinformatics 13, 345–354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fogel D. B., Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: A review. Contemp. Clin. Trials Commun. 11, 156–164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu P.-P., Xie Y., Meng X.-Y., Kang J.-S., History and progress of hypotheses and clinical trials for Alzheimer’s disease. Signal Transduct. Target. Ther. 4, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGhee D. J. M., Ritchie C. W., Zajicek J. P., Counsell C. E., A review of clinical trial designs used to detect a disease-modifying effect of drug therapy in Alzheimer’s disease and Parkinson’s disease. BMC Neurol. 16, 92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon N., Simon R., Adaptive enrichment designs for clinical trials. Biostatistics 14, 613–625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baird R. D., Caldas C., Genetic heterogeneity in breast cancer: The road to personalized medicine? BMC Med. 11, 151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su X. W., Broach J. R., Connor J. R., Gerhard G. S., Simmons Z., Genetic heterogeneity of amyotrophic lateral sclerosis: Implications for clinical practice and research. Muscle Nerve 49, 786–803 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Catenacci D. V. T., Next-generation clinical trials: Novel strategies to address the challenge of tumor molecular heterogeneity. Mol. Oncol. 9, 967–996 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Issa A. M., Thorogood A., Joly Y., Knoppers B. M., Accelerating evidence gathering and approval of precision medicine therapies: The FDA takes aim at rare mutations. Genet. Med. 21, 542–544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.S. Rahman, D. Thorburn, in GeneReviews®, M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. Bean, K. Stephens, A. Amemiya, Eds. (University of Washington, Seattle, 1993). [Google Scholar]
- 54.Distelmaier F., Haack T. B., Wortmann S. B., Mayr J. A., Prokisch H., Treatable mitochondrial diseases: Cofactor metabolism and beyond. Brain 140, e11 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S22
Table S1