Abstract
Cells responding to DNA damage implement complex adaptive programs that often culminate in one of two distinct outcomes: apoptosis or senescence. To systematically identify factors driving each response, we analyzed human IMR-90 fibroblasts exposed to increasing doses of the genotoxin etoposide and identified SRC as a key kinase contributing early to this dichotomous decision. SRC was activated by low but not high levels of etoposide. With low DNA damage, SRC-mediated activation of p38 critically promoted expression of cell survival and senescence proteins, while SRC-mediated repression of p53 prevented a rise in proapoptotic proteins. With high DNA damage, failure to activate SRC led to elevation of p53, inhibition of p38, and apoptosis. In mice exposed to DNA damage, pharmacologic inhibition of SRC prevented the accumulation of senescent cells in tissues. We propose that inhibiting SRC could be exploited to favor apoptosis over senescence in tissues to improve health outcomes.
The kinase SRC emerges as an early vital driver of cell senescence in response to sublethal DNA damage.
INTRODUCTION
Senescent cells arise in tissues responding to sublethal damage to cellular components. They are characterized by morphological changes, indefinite cell cycle arrest with increased expression of cyclin-dependent kinase inhibitors p21 (CDKN1A) and p16 (CDKN2A), elevated resistance to apoptosis, and metabolic alterations including increased lysosomal function and activation of a senescence-associated β-galactosidase enzyme (SA-β-Gal) (1). They also actively secrete proinflammatory molecules and tissue-remodeling factors, a trait known as the senescence-associated secretory phenotype (SASP) (1). Senescent cells are beneficial for physiological processes such as tissue repair, embryonic development, and cancer prevention; however, an excessive accumulation of senescent cells can be detrimental to tissue function, disrupt organ homeostasis, and contribute to disease development (2). Accordingly, the increased presence of senescent cells in aging tissues and organs has been linked to many age-associated physiologic declines and chronic diseases (3). Recently, therapeutic strategies (“senotherapies”) aiming to eliminate senescent cells in tissues (“senolysis”) have been developed, but their precise mechanisms of action are not fully understood and their efficacy in clinical settings is still being investigated (4). An alternative emerging strategy to reduce the senescent cell compartment for health benefit aims to prevent the buildup of senescent cells in tissues before they become detrimental.
Although DNA-damaging stimuli can trigger apoptotic cell death, they can also be potent inducers of senescence. Whether the cell undergoes apoptosis or senescence is strongly influenced by the cell type and the extent and nature of the genotoxic injury (5). In this regard, senescent cells represent a state of increased resistance to apoptotic cell death, with elevated activity of prosurvival pathways such as those elicited by the BCL2 family of antiapoptotic proteins (6), although some proapoptotic effectors are also increased in senescence (7). The specific molecular determinants of the decision between apoptosis and senescence have not been identified. Given that senolytic agents generally operate by suppressing the antiapoptotic defense of senescent cells, identifying damage-sensing signaling pathways that direct the early cellular response toward apoptosis or senescence can uncover senescent cell vulnerabilities and help design effective senotherapies.
Given the limited knowledge of the molecular effectors that direct response pathways toward senescence or apoptosis, we sought to study systematically these two responses in the same cell type, changing only the magnitude of damage. We hypothesized that by uncovering signaling differences between cells committed to either senescence or apoptosis, we might identify critical factors tilting cells to one response or the other. In this study, we identified the kinase SRC as a key protein that was activated in human IMR-90 fibroblasts undergoing senescence in response to DNA damage and therefore surviving the genotoxic injury. By contrast, IMR-90 cells undergoing cell death showed minimal SRC activation. Signaling through SRC is aberrantly activated in many malignant cells often via upstream receptor tyrosine kinases (RTKs) (8), and SRC modulates several major signaling pathways, including mitogen-activated protein kinases (MAPKs), phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT1), signal transducers and activators of transcription (STATs), and signaling through p53 (TP53), all central to the apoptotic and senescence programs (9, 10). In the paradigm uncovered here, SRC activation by low levels of the DNA-damaging agent etoposide caused cells to undergo senescence and survival, with reduced p53 signaling and hence lower levels of PUMA (p53–up-regulated modulator of apoptosis; a transcriptional target of p53) and activated MAPK p38 signaling, leading to increased abundance of the prosurvival protein BCL2L2. Moreover, commitment to senescence included activation of focal adhesion kinase (FAK) by integrins and an epithelial-to-mesenchymal transition (EMT)–like molecular program that reinforced the senescent phenotype. By contrast, diminished SRC activity in response to high levels of etoposide-triggered DNA damage led to decreased p38 function and increased p53 function, promoting apoptosis. In tissues of mice treated with DNA-damaging agents, the accumulation of senescent cells was reduced by pharmacologic inhibition of SRC. We propose that SRC inhibition might be exploited for promoting apoptosis of senescent cells in aged or damaged tissues to improve health span and disease outcomes.
RESULTS
Following DNA damage, SRC is activated in fibroblasts committed to senescence and survival
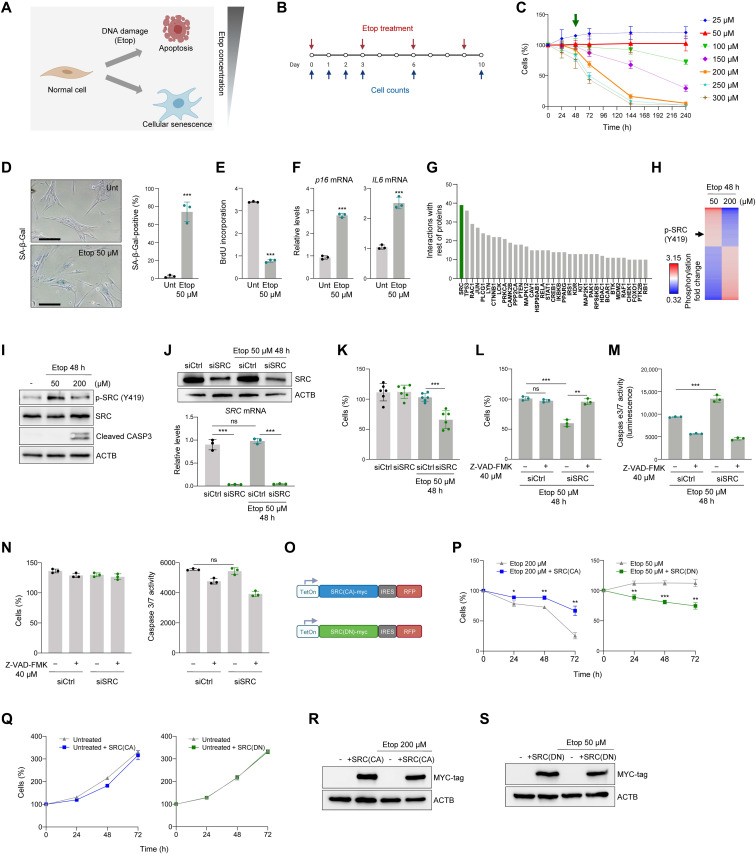
To systematically investigate the signaling pathways that govern whether cells undergo senescence or apoptosis, we first determined the thresholds of damage that triggered each cell response. We tested a range of doses of the DNA-damaging agent etoposide (25 to 300 μM) in proliferating human diploid IMR-90 fibroblasts, refreshing etoposide every 3 days (Fig. 1, A and B), and measured cell viability through up to 10 days (Fig. 1C). We focused on 50 and 200 μM doses of etoposide for subsequent experiments, as both doses led to similar cell survival by 48 hours; thereafter, 200 μM etoposide treatment caused cell death, while cells treated with 50 μM etoposide maintained viability and reached senescence (below). Treatment with 200 μM etoposide caused apoptosis, as it was rescued by the addition of Z-VAD-FMK (fig. S1, A and B), a broad-spectrum inhibitor of caspases [the proteases that execute the apoptotic program (11)]. Thus, we were able to compare etoposide-induced senescence (50 μM) and apoptosis (200 μM) side by side.
Fig. 1. Prosurvival signaling in senescence-committed cells implicates active SRC.
(A and B) Strategy (A) and timeline (B) to assess the threshold of etoposide (Etop) triggering IMR-90 cell senescence or apoptosis. (C) Percent live cells, assessed by direct cell counts, following etoposide treatment as in (B); green arrow, time selected for subsequent analyses. (D) SA-β-Gal staining 10 days after no treatment or 50 μM etoposide. Scale bar, 100 μm. Right, percent SA-β-Gal–positive cells. (E and F) BrdU incorporation (E) and RT-qPCR analyses of p16 and IL6 mRNA levels, normalized to ACTB mRNA levels (F), in cells processed as in (B). (G) STRING analysis of proteins differentially phosphorylated 48 hours after 50 or 200 μM etoposide treatments; graph, number of protein interactions for each protein indicated. (H) Heatmap representation of protein sites with significantly changed phosphorylation after 50 and 200 μM etoposide treatment (n = 2 per condition). Arrow, SRC(Y419) phosphorylation. (I) Western blot analysis of the indicated proteins; ACTB, loading control. (J) Western blot analysis of SRC levels (top) and RT-qPCR analysis SRC mRNA levels (bottom) in IMR-90 fibroblasts treated as shown. (K) IMR-90 fibroblasts were transfected with siCtrl or siSRC, treated as indicated, and live cells were counted 48 hours later. (L to N) Forty-eight hours after siRNA transfections, cells were treated with caspase inhibitor Z-VAD-FMK and 50 μM etoposide; 48 hours later, the remaining cells were counted (L) and caspase 3/7–dependent activity was assessed (M); cell viability and caspase 3/7 activity without etoposide exposure (N). (O to S) Constitutively active SRC(CA) and dominant-negative SRC(DN) lentiviral constructs (O) were transduced and induced by doxycycline (1 μg/ml) in IMR-90 cells treated with 200 or 50 μM etoposide (P) or proliferating (Q); cell viability was assessed as in (C). Western blot analysis of myc-tagged SRC mutants after 72 hours in doxycycline (R and S). Data in (C) to (F), (J) to (N), (P), and (Q) are means ± SD (n = 3 experiments); significance (*P < 0.05, **P < 0.01, ***P < 0.001) using two-tailed Student’s t test. ns, not significant.
To study whether 50 μM etoposide triggered cell senescence, we measured several well-established senescence markers. First, we assessed SA-β-Gal activity in IMR-90 cells that were either left untreated or treated with 50 μM etoposide for 10 days (Fig. 1D); as shown, 50 μM etoposide increased SA-β-Gal activity, yielding ~80% SA-β-Gal–positive cells compared to 10% SA-β-Gal positive in the untreated group (Fig. 1D, right). Second, cell proliferation analysis by measuring 5-bromo-2′-deoxyuridine (BrdU) incorporation showed that cells treated with 50 μM etoposide incorporated significantly less BrdU than untreated cells (Fig. 1E). Third, the expression levels of senescence markers p16 mRNA and IL6 mRNA, monitored by reverse transcription followed by quantitative polymerase chain reaction (RT-qPCR) analysis, were markedly elevated by 10 days of 50 μM etoposide treatment (Fig. 1F).
Given that by 10 days of treatment 50 μM etoposide induced senescence, while 200 μM etoposide triggered cell death, we explored the signaling status of cells in each group just 48 hours into the treatment, when cells were still healthy but death-or-senescence decisions were being made. Phosphoprotein array analysis was used to evaluate the phosphorylation levels of approximately 600 residues across the human kinome to find the most significant changes (table S1), and those above 1.25-fold (40 proteins) and below 0.8-fold (80 proteins) (table S2) were included in the final dataset. Interaction pathways involving the differentially phosphorylated proteins in cells committed to senescence (50 μM) and to apoptosis (200 μM) were studied using the STRING software (Materials and Methods), which enabled the identification of hub proteins affecting many of the differentially phosphorylated proteins. The highest-ranked protein between the two conditions was the kinase SRC (Fig. 1G). SRC is mainly implicated in transducing signals from the plasma membrane through many phosphorylation cascades (12) and was previously described as a prosurvival kinase in cancer, in keeping with a potential role in the survival of etoposide-treated IMR-90 fibroblasts. Treatment of IMR-90 cells with 50 μM etoposide increased SRC phosphorylation at tyrosine (Y) 419, while 200 μM etoposide reduced this phosphorylation event, as found by phosphoarray analysis (Fig. 1H) and confirmed by Western blot analysis (Fig. 1I). Thus, SRC activation appeared linked to the cell fate decision to undergo senescence rather than apoptosis after DNA damage by etoposide.
Genetic modulation supports SRC involvement in promoting cell survival upon DNA damage
Having found SRC and SRC-related proteins at the decision point between senescence and apoptosis, we next asked whether modulating SRC function might affect the outcome of cells responding to DNA damage. First, we reduced SRC levels by silencing using SRC-directed small interfering RNA (siSRC); siCtrl was included in control silencing groups. After confirming the efficiency of silencing SRC protein and SRC mRNA by Western blot and RT-qPCR analyses, respectively (Fig. 1J), we examined the effect of SRC silencing on senescence induced by etoposide (50 μM). Cell viability was markedly reduced when SRC was silenced in cells treated with 50 μM etoposide for 48 hours compared to control cells (siCtrl), where cell survival was unchanged (Fig. 1K), suggesting that SRC contributes to the survival of cells undergoing etoposide-triggered senescence. To examine whether SRC silencing specifically directed senescence-committed cells toward apoptosis, we sought to prevent apoptosis by treating cells with Z-VAD-FMK. As shown in Fig. 1L, the death of SRC-silenced cells treated with 50 μM etoposide was fully rescued by adding Z-VAD-FMK, indicating that SRC silencing in this paradigm sensitized cells to apoptotic death. Furthermore, analysis of caspase 3/7 activity in etoposide-treated cells (50 μM) by luminescence analysis (Materials and Methods) revealed that SRC silencing increased caspase activity (Fig. 1M) and that the increased caspase activity was fully rescued by addition of Z-VAD-FMK. Last, as anticipated, Z-VAD-FMK treatment did not affect the viability or the caspase activity of untreated cells (Fig. 1N). In short, SRC contributed to maintaining a senescence phenotype and avoiding apoptosis in response to etoposide-induced damage.
We then asked whether the effect of SRC in promoting senescent cell survival might be linked to its kinase activity. We generated viral vectors that expressed a constitutively active (CA) form of human SRC (Y530F), hereafter “SRC(CA),” where a substitution of tyrosine 530 to phenylalanine (Y530F) produces constitutive phosphorylation of SRC (13). We also generated a dominant-negative (DN) mutant construct of human SRC featuring K298M/Y530F mutations, hereafter “SRC(DN).” These viral vectors also have a C-terminal myc tag, a doxycycline-inducible promoter, and an internal ribosomal entry site (IRES)–driven red fluorescent protein (RFP) reporter to track their expression in cells. After transducing cells with the lentiviral particles (Fig. 1O), we tested whether expression of SRC(CA) and SRC(DN) influenced the proportion of apoptotic cells in etoposide-treated cells. As shown, expression of SRC(CA) significantly increased the survival of cells treated with 200 μM etoposide, while expression of SRC(DN) reduced the viability of cells treated with 50 μM etoposide (Fig. 1P). No changes were observed in proliferating cells when the SRC mutants were overexpressed (Fig. 1Q). Transduction efficiency was monitored by immunoblot analysis of the myc tag (Fig. 1, R and S) and by tracking RFP fluorescence at 72 hours (fig. S1C). In addition, silencing the SRC-inhibitory kinase CSK (13) recapitulated the apoptosis-rescuing effects of SRC(CA) overexpression (fig. S1, B to D). Together, these data support the notion that SRC activity elicits a prosurvival influence in cells responding to senescence-inducing genotoxic stress.
Pharmacologic inhibition of SRC activity orients cells toward apoptosis instead of senescence
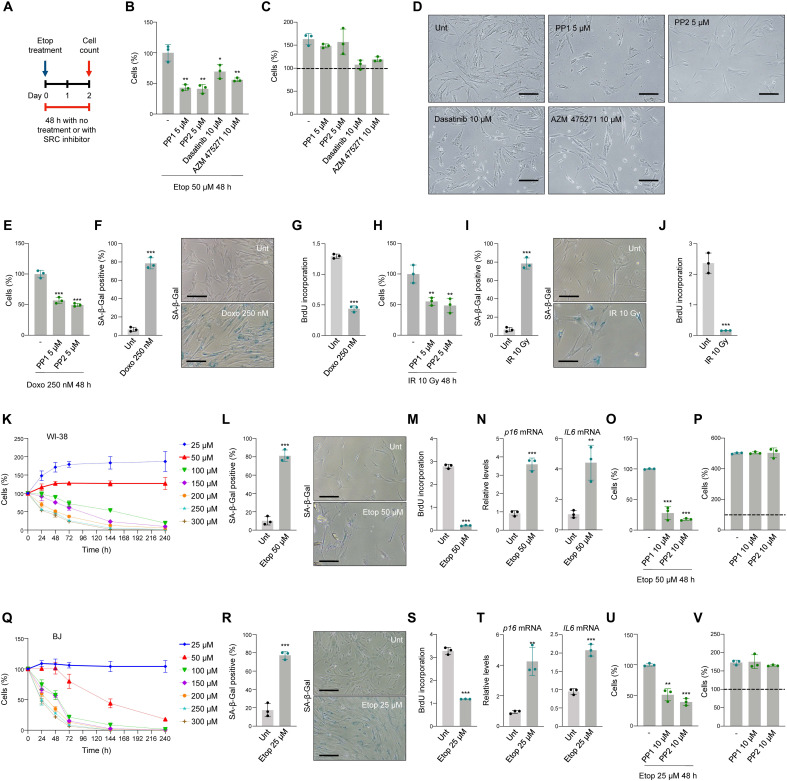
We tested whether commercially available SRC inhibitors could recapitulate the effects seen after silencing SRC. We treated cells simultaneously with 50 μM etoposide and SRC inhibitors PP1, PP2, dasatinib, or AZM 475271 (Fig. 2A). All SRC inhibitors reduced the viability of cells treated with 50 μM etoposide, similar to what was seen after silencing SRC (Fig. 2, B to D); in the absence of etoposide, none of the inhibitors reduced cell viability significantly by 48 hours, compared to untreated controls, although both dasatinib and AZM 475271 reduced proliferation modestly in undamaged cells (Fig. 2C). These results agree with the known function of SRC as a transducer of growth signals from activated RTKs (8).
Fig. 2. Inhibition of the SRC activity orients cells toward apoptosis instead of senescence.
(A) Schedule of SRC inhibitor and etoposide treatments. (B and C) IMR-90 cell viability was measured by direct cell counting 48 hours after treatment with SRC inhibitors and 50 μM etoposide (B) or no etoposide (C). Dashed line, cells present at t = 0 (C). (D) Representative IMR-90 cell micrographs 48 hours after treatment with 50 μM etoposide and SRC inhibitors. Scale bars, 100 μm. (E to G) Cell viability, measured as in (B), 48 hours after treating IMR-90 cells with doxorubicin (Doxo) (250 nM) and SRC inhibitors (E); senescence was assessed by SA-β-Gal staining (F) and BrdU incorporation (G) assays. (H to J) IMR-90 cell viability 48 hours after treatment with IR (10 Gy) and SRC inhibitors (H). Senescence was assessed by SA-β-Gal staining (I) and BrdU incorporation (J) assays. (K to N) Percent live human WI-38 fibroblasts (direct counts) after treatment with different etoposide concentrations (K); senescence at day 10 was assessed by SA-β-Gal staining (L), BrdU incorporation (M), and RT-qPCR analysis of the levels of p16 and IL6 mRNAs, normalized to ACTB mRNA levels (N). (O and P) Viability (direct counts) of WI-38 fibroblasts treated with 50 μM etoposide (O) or untreated (P) and treated with SRC inhibitors; dashed line, cell numbers at t = 0. (Q to T) Percent live human BJ fibroblasts (direct counts) after treatment with different etoposide concentrations (Q); senescence at day 10 was assessed by SA-β-Gal staining (R), BrdU incorporation (S), and RT-qPCR analysis of the levels of p16 and IL6 mRNAs, normalized to ACTB mRNA levels (T). (U and V) Viability (direct counts) of BJ fibroblasts treated with 50 μM etoposide (U) or untreated (V) and treated with SRC inhibitors; dashed line, cell numbers at t = 0. In (B), (C), and (E) to (V), significance was assessed by two-tailed Student’s t test. Graphs represent means ± SD (n = 3 experiments); significance (*P < 0.05, **P < 0.01, ***P < 0.001) was determined using two-tailed Student’s t test.
To test whether the observed effects of SRC were seen with other DNA-damaging agents capable of triggering senescence, we studied the impact of doxorubicin (250 nM) and ionizing radiation (IR; 10 Gy) (14, 15). Simultaneous treatment with PP1 and PP2 for 48 hours reduced cell viability relative to doxorubicin treatment alone (Fig. 2E) at a concentration of doxorubicin that caused senescence by day 10, as measured by SA-β-Gal staining and BrdU incorporation analyses (Fig. 2, F and G). Similarly with IR, joint treatment with PP1 or PP2 reduced the cell viability relative to IR treatment alone by 48 hours (Fig. 2H) at doses of IR that triggered senescence by day 10, as assessed by SA-β-Gal staining and BrdU incorporation analyses (Fig. 2, I and J). In summary, SRC inhibition reduces cell viability after exposure to different DNA-damaging agents.
To further study whether the effect of SRC inhibition on survival was extended to other cell systems, we tested human diploid WI-38 and BJ fibroblasts, which differ from IMR-90 fibroblasts but are commonly used to study cell senescence. Analysis of the thresholds of etoposide that induced senescence or apoptosis revealed that, like IMR-90 fibroblasts, WI-38 fibroblasts also underwent senescence with 50 μM etoposide (Fig. 2K); this phenotype was confirmed by increased SA-β-Gal staining, decreased BrdU incorporation, and increased levels of p16 and IL6 mRNAs (Fig. 2, L to N). Further treatment with PP1 and PP2, two SRC inhibitors with strong effect in IMR-90 cells, significantly sensitized WI-38 cells to apoptotic cell death (Fig. 2O). This sensitization was specific for etoposide-treated cells, as untreated, proliferating WI-38 cells showed neither cell death nor decreased proliferation when treated with PP1 or PP2 (Fig. 2P). BJ fibroblasts were substantially more sensitive to DNA damage caused by etoposide (Fig. 2Q), and thus, we used 25 μM etoposide to induce senescence, which was confirmed by SA-β-Gal staining, reduced BrdU incorporation, and increased levels of p16 and IL6 mRNAs (Fig. 2, R to T). Treating senescence-committed BJ cells with PP1 and PP2 also triggered a marked increase in cell death, similar to the findings in IMR-90 and WI-38 fibroblasts (Fig. 2U), while untreated BJ cells were unaffected by SRC inhibitors (Fig. 2V). Collectively, these results indicate that early pharmacological inhibition of SRC could potentially offer a strategy to redirect cells with sublethal DNA damage from senescence to apoptosis.
Essential factors drive dichotomous cell fate determination downstream of SRC signaling
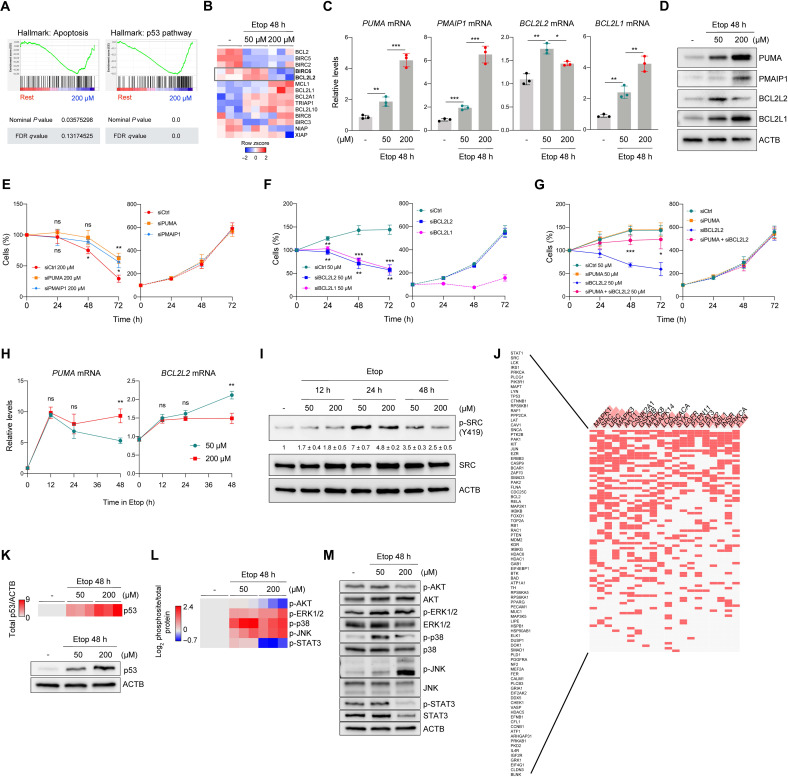
The fate of damaged cells is determined by a tightly regulated balance between proapoptotic and prosurvival proteins (11). To gain further insight into the mechanisms underlying the cell fate determination during early senescence, we carried out microarray analysis to compare the transcriptomic profiles of cells treated with 50 and 200 μM etoposide (GSE169037). Gene Set Enrichment Analysis (GSEA) associated with 200 μM etoposide treatment identified both apoptotic (Hallmark: Apoptosis) and p53 (Hallmark: p53 pathway) transcriptomic programs (Fig. 3A), in keeping with the role of p53 in apoptotic cell death. We hypothesized that the most prominent mRNAs encoding antiapoptotic proteins should increase in senescence-committed cells but not in proapoptotic conditions (Fig. 3B), as they might be key to triggering cell death when damage is excessive. Only 2 of the 14 mRNAs (box), encoding prosurvival proteins BIRC6 and BCL2L2, displayed that trend; BCL2L2 is an antiapoptotic, senescence-associated protein of the BCL2 prosurvival protein family usually targeted by senolytic drugs (9).
Fig. 3. Essential factors for cell fate determination are regulated by signaling through SRC.
(A and B) GSEA plots (A) and heatmap (B) depicting microarray analysis of mRNAs encoding proapoptotic (A) or prosurvival (B) proteins in untreated or etoposide-treated IMR-90 fibroblasts; data in (B) are row z score of log2-normalized transcript values (n = 3). (C) Validation of the top microarray targets PUMA, PMAIP1, BCL2L2, and BCL2L1 mRNAs by RT-qPCR analysis (see also fig. S2A). (D) Western blot analysis of proteins expressed in cells treated as in (A) to (C). (E) Viability (direct counts) of cells transfected with the siRNAs shown, then treated with 200 μM etoposide (left) or left untreated (right). Significant differences are indicated above (siPUMA versus siCtrl) and below (siPMAIP1 versus siCtrl) the curves. (F) Viability (direct counts) of cells that were transfected with the siRNAs shown, either treated with 50 μM etoposide (left) or left untreated (right). Significant differences are indicated above (siBCL2L1 versus siCtrl) and below (siBCL2L2 versus siCtrl) the curves. (G) Viability (direct counts) of cells transfected with the siRNAs shown, then treated with 50 μM etoposide (left) or left untreated (right). (H) RT-qPCR analysis of PUMA and BCL2L2 mRNAs (normalized to ACTB mRNA) after etoposide treatment for different times. (I) Western blot analysis of the proteins shown; means ± SD from densitometric quantification of phosphorylated SRC(Y419) (n = 3) is shown. (J) Enrichr analysis of terms (columns, hub proteins) and input proteins (rows); filled cells designate row proteins associated with hub proteins (Materials and Methods). (K) Heatmap representation (top) of quantified densitometry signals from p53 by Western blot analysis (bottom) in IMR-90 cells treated as indicated. (L and M) Heatmap representation (L) of the quantified densitometry signals on Western blots (M) detecting phosphorylated protein relative to corresponding total protein. Data represent log2 of the normalized values. Graphs (C) and (E) to (I) represent means ± SD (n = 3 experiments); significance (*P < 0.05, **P < 0.01, ***P < 0.001) was determined using two-tailed Student’s t test.
RT-qPCR analysis confirmed the expression profiles of mRNAs encoding proapoptotic proteins (BBC3/PUMA, PMAIP1, BAX, and APAF1 mRNAs) and prosurvival proteins (BCL2L2, BIRC6, and BCL2L1 mRNAs); only the levels of PUMA and PMAIP1 mRNAs increased robustly in cells treated with both 50 and 200 μM etoposide, while BCL2L2 mRNA increased selectively by 50 μM, but not by 200 μM, etoposide (Fig. 3C and fig. S2A). Western blot analysis revealed that the levels of PUMA, PMAIP1, BCL2L2 (BCL-W), and BCL2L1 (BCL-XL) reflected the changes in mRNA levels (Fig. 3D).
To directly test whether PUMA or PMAIP1 reduced IMR-90 cell survival, we silenced them using specific small interfering RNAs (siRNAs) and challenged cells with 200 μM etoposide. We found that silencing either PUMA mRNA or PMAIP1 mRNA partially but significantly reduced cell death caused by etoposide (Fig. 3E, left). Silencing these genes did not affect survival of proliferating cells (Fig. 3E, right). Both BCL2L2 and BCL2L1 contributed to maintaining cell viability during senescence, as silencing them using dedicated siRNAs and challenging them with 50 μM etoposide resulted in enhanced cell death (Fig. 3F, left). In a complementary experiment, treatment with ABT-737 (a senolytic agent that inhibits BCL2L1 and BCL2L2) diverted senescent cells toward apoptosis (fig. S2B). In proliferating cells, simply silencing BCL2L1 mRNA reduced cell viability (Fig. 3F, right); since silencing BCL2L2 mRNA did not have this effect, we propose that BCL2L2 more specifically protects senescence-committed cells. We thus focused on PUMA and BCL2L2 as representative factors in the survival-versus-apoptosis decision made in early senescence. To test their direct impact on this decision, we studied whether cell death caused by BCL2L2 silencing might be rescued by simultaneously depleting PUMA. Notably, while BCL2L2 silencing decreased viability in cells treated with 50 μM etoposide, simultaneous silencing of PUMA partially prevented this reduction (Fig. 3G, left). Silencing PUMA alone had no effect on survival (Fig. 3G, left), and these silencing interventions did not affect proliferating cells (Fig. 3G, right); the silencing interventions were confirmed by RT-qPCR analysis (fig. S2C). These results suggest that proapoptotic PUMA and antiapoptotic BCL2L2 proteins directly contribute to the survival-versus-apoptosis program implemented by SRC early following etoposide treatment. Further analysis of senescence-associated markers p21 and p16 mRNAs revealed that they increased in the presence of 50 and 200 μM etoposide (fig. S2D) and that silencing p21 or p16 mRNAs increased apoptosis after 50 μM etoposide (fig. S2, E to G). These results are in line with previous findings suggesting that disrupting the growth inhibitory program in senescence can increase apoptosis (16–18).
Further assessment of the expression levels of PUMA and BCL2L2 mRNAs including earlier time points revealed that their abundance at 50 and 200 μM etoposide treatments diverged at 48 hours (Fig. 3H), as cell fates began to be apparent. SRC phosphorylation remained higher in the 50 μM than in the 200 μM treatment groups, also starting to diverge after 24 hours of etoposide treatment (Fig. 3I), suggesting a potential coordination between SRC activation and the expression of cell fate proteins.
The phosphoarray data (Fig. 1H) were used to identify signaling pathways altered between senescence- and apoptosis-triggering conditions. By including all proteins differentially phosphorylated in both conditions to find central hub signaling proteins [PPI Hub Proteins app within the Enrichr platform; (19)], the highest-ranking hub proteins aside from SRC family members were key signaling elements downstream of SRC, including MAPK1 [extracellular signal–regulated kinase 2 (ERK2)], MAPK3 (ERK1), AKT1 (AKT), MAPK8 [c-Jun N-terminal kinase 1 (JNK1)], MAPK14 (p38-α), and STAT3 (Fig. 3J). In addition, p53, also regulated by SRC, was one of the top proteins showing increased phosphorylation of S315, a residue important for its activation (table S1). GSEA further suggested a strong association of p53 activation with 200 μM etoposide treatment, which was also confirmed by increased p53 protein levels (Fig. 3K) and increased levels of PUMA and PMAIP1 mRNAs (Fig. 3C) in this treatment group. Etoposide treatment strongly increased the phosphorylation of activating residues in the MAPKs (ERK1/2, p38, and JNK) and increased more modestly STAT3 phosphorylation with 50 μM etoposide only (Fig. 3, L and M). Both ERK1/2 and JNK demonstrated stronger increases of phosphorylation by 200 μM compared to 50 μM etoposide (Fig. 3, L and M), while p38 showed the opposite pattern. Although MAPKs play differing roles in regulating the apoptosis-survival balance (10), p38 was recently identified as a prosurvival signaling kinase in senescence (20), was found to increase BCL2L2 mRNA levels (21), and was the only MAPK showing higher activation in senescence-committed cells in our experiments (Fig. 3, L and M). Phosphorylation of another prosurvival protein, AKT, was not observed in the 50 μM etoposide group and was reduced with 200 μM etoposide (9). Last, STAT3 phosphorylation at Y705 increased modestly in senescence-committed and declined in apoptosis-committed IMR-90 cells. In summary, coordinated signaling through SRC, p53, and MAPKs, with ensuing gene expression programs, appears important for translating the cellular responses to DNA damage into cell fate determination.
p53 repression and p38 activation by SRC promote prosurvival responses in early senescence
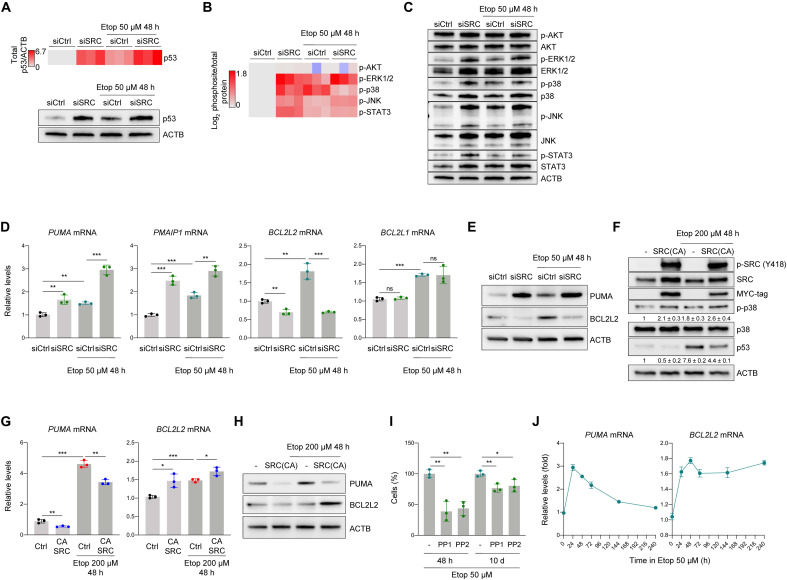
To further evaluate the role of SRC in the differential response to etoposide, we silenced SRC using siRNAs. By 48 hours of treatment with 50 μM etoposide, SRC silencing further elevated p53 levels, suggesting that SRC might prevent an excessive rise in p53 levels (Fig. 4A). Moreover, silencing SRC decreased p38 phosphorylation, but increased phosphorylation of ERK1/2, JNK, and STAT3 to different degrees (Fig. 4, B and C), suggesting that SRC influences these signaling cascades in senescent cells. SRC silencing did not affect AKT status, indicating that these two kinases are not closely related in early senescence. Thus, SRC silencing in cells treated with 50 μM etoposide increased p53 levels and diminished p38 activation, mirroring the proapoptotic pattern observed in cells treated with 200 μM etoposide.
Fig. 4. SRC-mediated p53 repression and p38 activation promotes prosurvival responses in early senescence.
(A) Heatmap representation of p53 levels (top) quantified by densitometry from signals on Western blots (bottom, representative panel) in IMR-90 cells transfected with siCtrl or siSRC siRNAs, either untreated or treated with 50 μM etoposide for 48 hours. (B) Heatmap representation of densitometry quantification of signals on Western blots detecting phosphorylated residues, normalized to protein levels assessed in cells transfected with siCtrl or siSRC siRNAs that were either left untreated or treated with 50 μM etoposide for 48 hours. (C) Representative Western blot analysis of the data shown in (B). (D) Quantification of PUMA, PMAIP1, BCL2L2, and BCL2L1 mRNAs by RT-qPCR analysis. Data were normalized to the levels of ACTB mRNA. (E) Representative Western blot analysis of the levels of PUMA and BCL2L2 proteins. (F) Levels of phosphorylated SRC (Y419), total SRC, myc tag, phospho-p38 (T180/Y182), p38, and p53 in the different conditions tested. The levels of ACTB were assessed to evaluate loading differences. Values indicate means ± SD from signals obtained in three independent experiments. (G) Quantification of PUMA and BCL2L2 mRNAs by RT-qPCR analysis; data were normalized to the levels of ACTB mRNA. (H) Representative Western blot analysis of the levels of proteins expressed from the transcripts in (F). (I) Cell viability was assessed in IMR-90 fibroblasts treated with 50 μM etoposide and either left without further treatment or treated with PP1 or PP2 for 48 hours or 10 days. (J) The levels of PUMA and BCL2L2 mRNAs were quantified by RT-qPCR analysis in cells treated with 50 μM etoposide for the times shown; data were normalized to the levels of ACTB mRNA. Values shown in (D), (G), (I), and (J) are means ± SD; significance (*P < 0.05, **P < 0.01, ***P < 0.001) was determined using two-tailed Student’s t test.
When SRC was silenced in cells exposed to 50 μM etoposide, the levels of PUMA and PMAIP1 mRNAs increased markedly, while the levels of BCL2L2 mRNA decreased and the levels of BCL2L1 mRNA remained unaffected (Fig. 4D). SRC silencing affected PUMA and BCL2L2 protein levels similarly at the protein level (Fig. 4E). Although it was not clear whether classical senescence factors p21 or p16 contributed to a survival program in this context (fig. S2H), the fact that silencing SRC increased PUMA and reduced BCL2L2 suggests that SRC helps to implement a prosurvival program in response to 50 μM etoposide.
To further test this hypothesis, we treated cells with apoptosis-inducing etoposide (200 μM) and attempted to diminish cell death by ectopically expressing SRC(CA). Moderate overexpression of SRC was confirmed by measuring the levels of endogenous and myc-tagged SRC by Western blot analysis (Fig. 4F). SRC(CA) increased p38 phosphorylation and reduced p53 levels as determined by Western blot analysis, resembling the patterns observed after treatment with 50 μM etoposide. Analysis of the levels of PUMA and BCL2L2 mRNAs also revealed prosurvival trends if SRC(CA) was expressed, despite treatment with 200 μM etoposide: PUMA mRNA levels declined and BCL2L2 mRNA levels increased (Fig. 4G), and protein levels reflected these trends (Fig. 4H). In conclusion, SRC silencing reversed the prosurvival phenotype after 50 μM etoposide treatment, while SRC(CA) expression partly rescued the proapoptotic response following 200 μM etoposide treatment.
Last, we explored whether SRC inhibitors could remove already senescent cells. Analysis of the impact of PP1 and PP2 (5 μM, 48 hours) revealed that SRC inhibitors more potently reduced the viability of early-stage (48 hours in 50 μM etoposide) than late-stage (10 days in 50 μM etoposide) senescent cells (Fig. 4I). PUMA mRNA levels peaked after 24 hours, while BCL2L2 mRNA levels gradually increased and were maintained elevated into late senescence (Fig. 4J). These observations further support the view that the influence of SRC on the survival program occurs early after the onset of damage. Taking these results together, we propose that SRC contributes to a senescent, prosurvival phenotype by maintaining reduced p53 activity and elevated p38 activity.
SRC influence on viability is modulated by signaling through FAK, p38, and p53
We sought to further delineate how SRC activation led to senescence over apoptosis. First, we examined whether p38 inhibition or p53 activation reproduced the effects of SRC on apoptosis. We used AL 8697, a potent and selective inhibitor of p38 (p38i) (22), and Nutlin-3a, a strong inhibitor of the interaction between MDM2 and p53 that renders p53 stable and active (p53a) (23). As shown, in the presence of 50 μM etoposide, treatment with p38i (AL 8697) or p53a (Nutlin-3a) (10 μM each) decreased cell viability compared to 50 μM etoposide alone (Fig. 5A). Each compound sensitized cells to apoptosis significantly less than the SRC inhibitors did (Fig. 5A). Inhibition of the other two MAPKs increased by SRC silencing, ERK1/2 and JNK (using U0126 and SP 600125, respectively, 10 μM each), revealed no change in cell viability in the presence of 50 μM etoposide (fig. S3A), further supporting a prominent role for p38 in maintaining viability during early senescence. As SRC inhibition affects both p53 and p38, we tested the effect of combining p38i and p53a. Both pathways appear important, as there was greater sensitization to apoptosis by 48 hours in 50 μM etoposide by the joint treatment relative to treatments with p38i or p53a alone, and it was comparable to the sensitizing effects of SRC inhibitors PP1 and PP2 (Fig. 5A and fig. S3B).
Fig. 5. SRC-FAK activation influences cell viability by signaling through p38 and p53.
(A) Viable IMR-90 cells (direct counts) following 48 hours of treatment with etoposide (50 μM) plus p38i (AL 8697, 10 μM), p53a (Nutlin-3a, 10 μM), p53a + p38i, PP1 (5 μM), or PP2 (5 μM); see also fig. S3B. (B) RT-qPCR analysis of PUMA and BCL2L2 mRNAs (normalized to ACTB mRNA) in cells treated as in (A). (C) Western blot analysis of the proteins shown (and loading control ACTB). (D) Cell viability (direct counting) after 48-hour treatment as indicated; doxycycline-induced SRC(CA) expression was performed as in Fig. 1R simultaneously with the other treatments. (E) RT-qPCR analysis of PUMA and BCL2L2 mRNAs (normalized to ACTB mRNA) in cells treated as in (D). (F) Analysis of proteins preferentially phosphorylated at 50 μM etoposide ranked CAS-SRC-FAK at the top using CORUM database and Enrichr analysis using data from Fig. 1H. (G) Western blot analysis of p-FAK (activating phosphorylation Y576/Y577), FAK, and loading control ACTB. (H and I) Viability (direct counting) of cells transfected with the siRNAs shown and treated with etoposide (H), and Western blot analysis (I) of the proteins shown (p-p38 is phosphorylated at T180/Y182). (J and K) GSEA (J) and heatmap (K) analyses of the levels of mRNAs encoding EMT and integrin proteins (z scores of the log2-normalized expression values, n = 3). (L and M) GSEA of relevant gene sets in the conditions indicated after expressing SRC(DN) (see also fig. S3E) (L) or SRC(CA) (M). (N) Proposed model (created using BioRender). Senescence-causing, low-level DNA damage (lower half) activates SRC, decreasing p53 function and activating p38 (lowering PUMA, elevating BCL2L2). Apoptosis-causing, high-level DNA damage (top half) does not activate SRC and instead activates p53 and represses p38, in turn increasing PUMA and lowering BCL2L2 levels and leading to apoptosis. Values shown in (A), (B), (D), (E), and (H) are means ± SD (n ≥ 3); significance (*P < 0.05, **P < 0.01, ***P < 0.001) was determined by using two-tailed Student’s t test.
We then assessed the impact of p53 and p38 on the production of PUMA and BCL2L2. In line with earlier findings (10), p38i reduced the abundance of BCL2L2 mRNA, but not PUMA mRNA (Fig. 5B), while p53a increased the levels of PUMA mRNA, a transcriptional target of p53, but not the levels of BCL2L2 mRNA. The encoded proteins generally showed similar trends, although BCL2L2 levels decreased by p53a treatment when compared to etoposide treatment only (Fig. 5C). As expected, p38 inhibition prevented the phosphorylation of p38 target HSP27, and Nutlin-3a elevated total p53 levels. These results suggest that p53 activation during early senescence directly increases PUMA mRNA levels, while p38 activity augments BCL2L2 mRNA levels. Combined p38i + p53a treatment further increased the levels of PUMA mRNA, encoding the proapoptotic protein PUMA, and prevented the increase in BCL2L2 mRNA elicited by 50 μM etoposide only, thus suppressing a rise in the prosurvival protein BCL2L2. These results illustrate the change in gene expression programs driven by p53 and p38 as effectors of SRC-driven responses. Our findings further support the notion that SRC inhibition, rather than p53 activation or p38 inhibition alone, effectively orchestrates the shifting of the cell response to damage from senescence to apoptosis.
To further examine a model whereby SRC mounts a prosurvival program, we expressed SRC(CA) and found that it increased cell viability of cells treated with 200 μM etoposide, but this protection was significantly reversed by combined treatment with p38i + p53a (Fig. 5D). Accordingly, in the presence of SRC(CA), both the decrease in PUMA mRNA levels and the rise in BCL2L2 mRNA levels were reversed by the p38i + p53a combination (Fig. 5E). These results indicate that the PUMA and BCL2L2 mRNAs were independently regulated by p53 and p38, respectively, and further support the more substantial effect of inhibiting SRC instead of individually activating p53 or inhibiting p38.
To investigate the possible mechanism by which SRC can modulate p38 and p53 simultaneously, we used the Enrichr platform to identify pathways activated by 50 μM etoposide. The plot in Fig. 5F represents the significance of each gene set from the CORUM database versus its odds ratio; each point represents a gene set, the x axis indicates the odds ratio for the gene set, while the y axis refers to the −log(P value) of the gene set. The top-ranked complex was the CAS-SRC-FAK complex. The SRC-FAK interaction is widely known to control processes including the balance between apoptosis and survival (24). Analysis of the influence of SRC on FAK activation revealed that SRC silencing markedly reduced FAK (activating) phosphorylation at tyrosines 576 and 577 after 48 hours with 50 μM etoposide, while SRC(CA) ectopic expression markedly increased FAK phosphorylation in the presence of apoptosis-causing 200 μM etoposide (Fig. 5G). FAK silencing also reduced cell viability, prevented p38 phosphorylation, and elevated p53 when exposed to 50 μM etoposide for 48 hours, recapitulating the effects observed after SRC inhibition or silencing (Fig. 5, H and I). These results suggest that SRC modulation of p53 and p38 potently relies on its impact on FAK, as previously described in other systems (25).
In light of these observations, we reexamined the transcriptomic data and discovered that the senescence-committed cells appeared to mount an EMT-like transcriptomic program (Fig. 5J and fig. S3E, left). The EMT phenotype usually includes integrins and integrin-dependent signaling (often involving SRC-FAK) in many contexts (26), integrin activation in turn reinforces the EMT phenotype (27), and FAK signaling by integrins was recently reported in senescent cells (28). Most integrin ligands are extracellular matrix (ECM) proteins that participate in profibrotic stages during repair processes related to cellular senescence (29–31). Moreover, 50 μM etoposide increased expression of most integrins (except for ITGA5-7) by 48 hours (Fig. 5K). This finding supports a model whereby integrins, which are upstream activators of SRC-FAK, might induce this signaling complex in senescent cells. Therefore, we tested two different compounds that interfere with integrin function: SB273005, a pharmacological inhibitor of αvβ3, and RGDS peptide, which inhibits several integrin heterodimers (32). Both SRC phosphorylation and viability of IMR-90 cells were reduced by 48 hours in 50 μM etoposide (fig. S3, C and D), suggesting that SRC phosphorylation in our system depends on integrin binding to its ligands.
Transcriptomic analysis of cells expressing SRC(DN) after etoposide treatment revealed that SRC function is required for complete EMT and integrin-dependent transcriptomic programs in 50 μM etoposide–treated cells (Fig. 5L and fig. S3E, right). Moreover, ectopic SRC(CA) expression in 200 μM etoposide–treated cells restored this integrin-initiated program to some extent in apoptosis-committed cells (Fig. 5M). Sublethal damage activates SRC and FAK through integrins, leading to inhibition of p53 and activation of p38. The establishment of an EMT-like phenotype reinforces a SRC-dependent suppression of apoptosis and implementation of a survival gene expression program with low PUMA and high BCL2L2 levels that leads to senescence. Lethal damage only minimally activated SRC, in turn enabling p53 activation and p38 repression and mounting a gene expression program (high PUMA, low BCL2L2) that leads to apoptosis (Fig. 5N). In summary, SRC signaling appears to both respond to and enhance a prosurvival EMT/integrin program in cells progressing toward senescence.
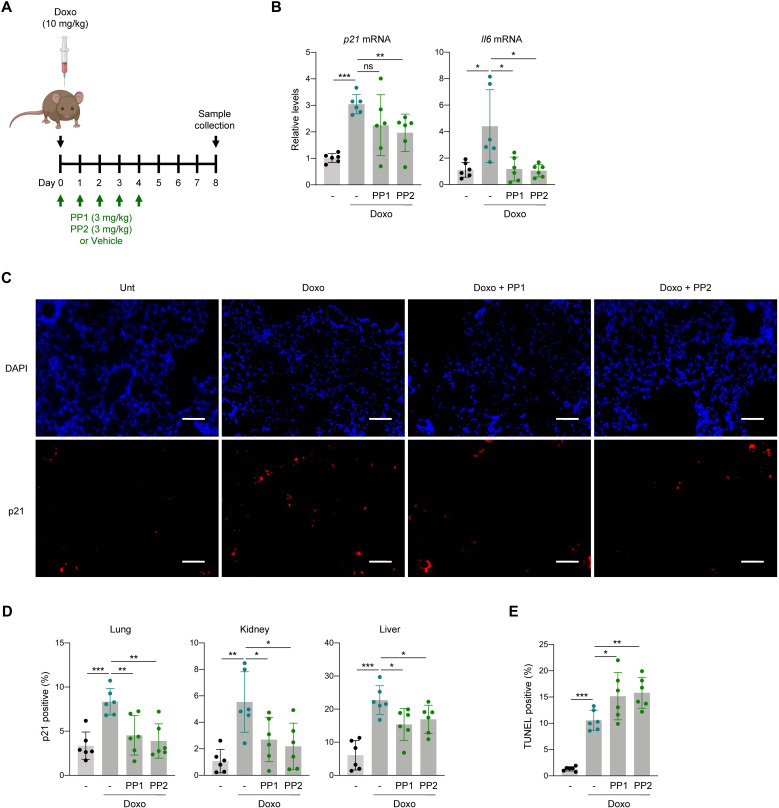
Inhibiting SRC in mice prevents the accumulation of senescent cells after treatment with doxorubicin
To evaluate the physiologic relevance of our findings, we tested the effectiveness of SRC inhibition in senescence versus apoptosis fates in an established in vivo mouse model of senescence (14), triggered by a single intraperitoneal injection of doxorubicin (Fig. 6A). Mice were either left untreated or treated with a single dose of doxorubicin (10 mg/kg) and then given either vehicle [10% dimethyl sulfoxide (DMSO) in phosphate-buffered saline (PBS)], PP1, or PP2 consecutively for 5 days (Fig. 6A); 8 days after doxorubicin administration, mouse tissues were harvested for analysis. In the lung, RT-qPCR analysis revealed increased levels of p21 and Il6 mRNAs, two senescence-associated transcripts known to be elevated in this model (Fig. 6B) (14, 33). Treatment with either SRC inhibitor reduced the increases in p21 and Il6 mRNAs seen in the doxorubicin-treated mice, bringing them to near control levels. Next, we monitored the accumulation of senescent cells by measuring p21-positive cells in lung sections. As shown by immunofluorescence microscopy and counting p21-positive cells, doxorubicin treatment significantly elevated the percentage of p21-positive cells in the lungs (Fig. 6, C and D), but PP1 and PP2 treatments reversed this rise.
Fig. 6. SRC inhibition prevents accumulation of senescent cells in doxorubicin-induced senescence in mice.
(A) Overview of the schedule of experiments performed in mice. (B) Measurement of the levels of p21 and Il6 mRNAs by RT-qPCR analysis in lungs treated as in (A); data were normalized to Actb mRNA levels. (C) Representative micrographs of p21 immunofluorescent staining of the experimental groups indicated. Scale bar, 50 μm. (D) Quantification of p21-positive cells in each tissue assayed; see also fig. S4 (A and B). (E) Percentages of TUNEL-positive cells in lungs from the indicated experimental groups. See also fig. S4C. Graph in (B) shows the mean values ± SD of n = 3 independent experiments. Graphs in (B), (D), and (E) display the means (bars) and each individual value as a dot ± SD of n = 6 mice; significance (*P < 0.05, **P < 0.01, ***P < 0.001) was determined using two-tailed Student’s t test.
We expanded this analysis to other tissues where doxorubicin treatment significantly induces p21 expression, such as kidney and liver (33); here, too, p21-positive cells accumulated by 8 days after doxorubicin exposure, but treatment with PP1 or PP2 prevented this accumulation (Fig. 6D and fig. S4, A and B). Last, to test whether SRC inhibition following doxorubicin damage in vivo favored apoptosis instead of senescence, we performed terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) assays (Materials and Methods) on lung sections. Notably, TUNEL-positive cells were more abundant in mice that received doxorubicin than in untreated mice (Fig. 6E and fig. S4C), but treatment with PP1 or PP2 led to an even higher proportion of TUNEL-positive cells within the lungs. These data suggest that SRC inhibition, favoring apoptotic cell death following DNA damage, also prevents senescence in vivo.
DISCUSSION
There is intense interest in identifying strategies to eliminate senescent cells to improve health span and disease outcomes (4). Given that senescent cells are selectively resistant to apoptotic cell death, elucidating the mechanisms that engender protection to senescent cells has become an area of immense effort. While our knowledge of senolysis has expanded in recent years, much remains to be elucidated, particularly at the molecular level (9). Here, we describe the identification of a mechanism whereby cells responding to DNA damage levels capable of triggering senescence can instead be redirected to undergo apoptosis. Following DNA damage, early changes in expressed proteins are known to drive cell fate by shifting the balance between proapoptotic and prosurvival programs; these programs are largely controlled by proteins in the BCL2 family (11). In this regard, prosurvival members of the BCL2 family (e.g., BCL2, BCL2L2, and BCL2L1) promote senescence, while proapoptotic members of the BCL2 family (e.g., PUMA and PMAIP1) enhance cell death (6, 9). Delineating the different signaling pathways is complex, given that in many cases the upstream regulators are shared. For example, a major effector of both apoptosis and senescence, p53, transcriptionally induces expression of p21, which can protect against apoptosis, and also induces the production of proapoptotic proteins like PUMA, PMAIP1, BAX, or APAF1 (34). Similarly, another key regulator, p38, can promote the expression of both prosurvival proteins such as BCL2L2 (21). Here, we found that SRC is an upstream regulator of this balance between senescence and apoptosis. In response to low etoposide treatment, activation of SRC simultaneously decreased p53 signaling and increased p38 signaling. As a result, silencing or inhibiting SRC led to increased production of PUMA (and other p53-dependent proapoptotic proteins) and reduced BCL2L2, tilting the response from senescence/survival to apoptosis.
SRC as a prosurvival factor in senescence
Traditionally considered a strong prosurvival protein, SRC belongs to a family of non-RTKs that are usually activated in cancer and other stress conditions. Although we focused on genotoxic stress, SRC can also be activated by RTKs, integrins, and oxidative stress conditions (35). Despite its prominent role in stress response programs, SRC was not previously linked to the balance between prosurvival and proapoptotic pathways that characterize the senescence response. Our results further support the view that SRC activation following sublethal DNA damage promotes cell survival and that this function relies on the coordination of p53 and MAPK signaling at an early point that defines the ensuing stress response. Our results complement earlier reports that signaling through integrins (α6β4), SRC, and AKT conferred survival of cancer cells in response to IR (36). They also provide molecular context to an earlier study identifying SRC as a suppressor of apoptosis triggered by staurosporine in cells already rendered senescent (37) and to a recent demonstration that silencing FAK prevents senescence in lungs (28), supporting the involvement of integrins and the EMT program to complement SRC function enabling senescence. Our findings further implicate SRC function in several processes linking senescence to tissue repair and development (17, 18, 29). Namely, SRC could participate in the profibrotic EMT-like response of early senescent cells in tissues both by becoming activated by integrin ligands (such as ECM proteins present at injury sites) and by triggering a protective program of senescence implementation over apoptosis. At the same time, SRC inhibition could be exploited to eliminate senescent cells when their presence exacerbates the decline of organ function (30, 38).
Inhibition of p53 by SRC
In the proapoptotic arm of our study, 200 μM etoposide increased the expression of several p53-dependent proteins. Although the influence of p53 on lethal stress signals is quite complex, there is ample support for the notion that p53 levels dictate the decision between cell senescence and apoptosis (39). We found that FAK cooperated with SRC to prevent apoptosis and favor senescence following low-level DNA damage. FAK and p53 are mutually suppressive, as p53 inhibits the transcription of FAK mRNA and FAK protein in turn represses the transcriptional activity of p53 (25, 40); in addition, signaling through integrins prevents p53 induction in several cell models (27). In keeping with the inhibition of p53, our phosphoprotein array data revealed that one of the top phosphorylated residues of p53 in senescence-committed cells was S315, which promotes p53 proteasomal degradation by enhancing its interaction with MDM2 (41). Phosphorylation of p53 at S315 may be mediated by AURKA (Aurora kinase A), a direct SRC substrate, or by NEK2 (41, 42). Furthermore, SRC may also decrease p53 levels directly in other ways, including by phosphorylating HIPK2, which is then unable to bind p53, and this leads to decreased p53 activity (43). In summary, activation of SRC and FAK decreases p53 function, which is essential for favoring senescence over apoptosis, through many paths.
Activation of p38 by SRC
A key mechanism whereby SRC and FAK promote cell survival is by activating p38. In keeping with evidence that the two kinases induce p38 signaling, silencing of either SRC or FAK reduced p38 phosphorylation after etoposide treatment. FAK signaling activates p38 in many systems (44, 45), and SRC can activate p38 through different mechanisms (8, 46). We found that p38 increased the levels of BCL2L2 mRNA, which are tightly regulated by transcription factors including MEF2, ETS1/2, C/EBP, and NF-κB (47). Despite its complex influence on survival and apoptosis, p38 has been found to mainly promote the viability of nontransformed cells (48). In senescence, p38 likely favors cell survival by promoting autophagy (20), and perhaps by activating pathways such as those that increase NF-κB activity (49) or augment p21 expression levels (16). Last, active p38 can prevent cell death by inhibiting signaling through JNK (50), a proapoptotic MAPK.
Targeting SRC in senolytic strategies
Our results point to SRC as a promising target to prevent the accumulation of senescent cells. Although SRC is widely expressed and active in homeostatic conditions, SRC activity increases in response to stress conditions (35). SRC hyperactivation is common in cancer cells, and therefore, strategies directed at SRC have been studied extensively in cancer treatment (51). Moreover, dasatinib, an inhibitor of SRC and other kinases, was included in some of the first cocktails (quercetin + dasatinib) reported to selectively eliminate senescent cells (52). Dasatinib was less effective than SRC inhibitors PP1 or PP2 in sensitizing cells to apoptosis, suggesting that current senolytics might be optimized for more efficient reduction of senescent cell viability. It is important to point out that SRC inhibition by PP1 or PP2 was more effective in reducing viability at early rather than late phases of senescence, although we did not directly compare this effect with dasatinib or other established senolytics. In summary, although SRC-specific inhibitors effectively reduced viability during early senescence, more evidence is required to determine their efficacy in modulating late-stage senescence. Similarly, the usefulness of targeting EMT components, including integrins and TGFB1, in early or late senescence remains to be explored (26, 53, 54).
Senescence can be beneficial in some contexts, but there are many scenarios in which senescence is strongly detrimental. Chemotherapy-induced senescence of nonmalignant cells is an example of cellular senescence that has proven to be quite harmful (14). Another example of deleterious senescence is the systemic reaction to the transplantation of organs (55). Preventing the accumulation and deleterious impact of senescent cells could be alleviated by SRC inhibitors, potentially improving therapeutic outcomes such as renal transplantation (56). Moreover, senescence of liver stellate cells from young mice prevents fibrosis, while their accumulation with age promotes steatosis (57, 58). An increased proportion of senescent hepatocytes drives liver fibrosis, while prevention of senescence within the liver of adult mice enhances liver regeneration after partial hepatectomy or acetaminophen poisoning (30, 38, 59). It remains to be seen whether senescent cells improve or worsen the liver response to injury, although treatment with SRC inhibitors improved liver injury outcomes in senescence-related processes such as fibrosis, steatosis, and regeneration (60–62). In this regard, the finding that SRC inhibitors prevented the accumulation of senescent cells in mouse lung, liver, and kidney after treatment with doxorubicin (Fig. 6), while it increased apoptosis in this model, suggests that interventions directed at SRC deserve strong consideration as part of efforts to exploit vulnerabilities in senescent cells for therapeutic benefit.
MATERIALS AND METHODS
Cell culture, treatment, and detection of SA-β-Gal
Human IMR-90 [American Type Culture Collection (ATCC)], WI-38 (Coriell Institute), and BJ fibroblasts (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco), penicillin-streptomycin (Gibco), sodium pyruvate (Gibco), and nonessential amino acids (Gibco) in a 5% CO2 incubator. Cells were maintained at low population doubling levels (PDLs) ranging between PDL15 and PDL25. Every control condition included equivalent concentrations of vehicle (DMSO). Doxycycline (1 μg/ml) was refreshed every 24 hours. Etoposide and doxorubicin were added to the media as indicated every 3 days. The remaining drugs used were refreshed every 48 hours. The drugs used are listed (table S3). For senescence triggered by IR, cells were exposed to 10 Gy and assayed at the indicated times afterward.
Cells were transfected with RNAiMAX (Invitrogen) following the manufacturer’s instructions. Using RNAiMAX, 50% confluent cells were transfected with ON-TARGETplus SMARTPool (Dharmacon) nontargeting siCtrl (catalog ID: D-001810-10-05), siSRC (catalog ID: L-003175-00-0005), siFAK (catalog ID: L-003164-00-0005), siBCL2L2 (catalog ID: L-004384-00-0005), siBCL2L1 (catalog ID: L-003458-00-0005), siPUMA/BBC3 (catalog ID: L-004380-00-0005), sip21 (catalog ID: L-003471-00-0005), sip16 (catalog ID: L-011007-00-0005), siCSK (catalog ID: L-003110-00-0005), and siPMAIP1 (catalog ID: L-005275-00-0005) siRNAs at a final concentration of 25 nM. Forty-eight hours later, additional treatments were initiated as indicated. Viability was assessed by cell counting and represented as the percentage of remaining cells compared to the number of cells present at the beginning of the experiment. SA-β-Gal activity was evaluated following the manufacturer’s instructions (Cell Signaling Technology). All cell counts were performed manually by using ImageJ and performed in at least three independent replicates. From each replicate, three fields were counted.
Lentiviral vectors
Lentiviral constructs were commercially produced by Cellecta. Either SRC(CA) (human SRC Y530F) or SRC(DN) (human SRC K298M/Y530F) was cloned into Cellecta-pTMONRB-TRE-MCS-I2-TagRFP-EFS-rtTA-2A-Blast lentiviral plasmid with a C-terminal myc tag. The mutations generated were based on extensive literature on SRC structure and function (13). Lentiviral constructs were also packaged by Cellecta and transduced at 3 MOI (multiplicity of infection) with polybrene (1 μg/ml; Millipore).
RT-qPCR analysis
Samples (tissues or cells) were lysed in Tri-Reagent (Invitrogen). The extracted aqueous phase was processed with QIAcube (Qiagen) to purify total RNA, which was then subjected to RT to create complementary DNA (cDNA) using Maxima reverse transcriptase (Thermo Fisher Scientific) and random hexamers. Following RT, qPCR analyses were performed using SYBR Green mix (Kapa Biosystems). Relative expression was determined by the 2-ΔΔCt method and normalized to ACTB mRNA levels. The primers used were as follows, each forward and reverse, respectively:
GCACAGAGCCTCGCCTT and GTTGTCGACGACGAGCG for human ACTB mRNA,
GTTACGGTCGGAGGCCG and GTGAGAGTGGCGGGGTC for human p16 mRNA,
AGTCAGTTCCTTGTGGAGCC and CATGGGTTCTGACGGACAT for human p21 mRNA,
AGTGAGGAACAAGCCAGAGC and GTCAGGGGTGGTTATTGCAT for human IL6 mRNA,
GATGGTGGCCTACCTGGAGA and AGAGCTGTGAACTCCGCCCA for human BCL2L2 mRNA,
CCCGAGAGGTCTTTTTCCGAG and CCAGCCCATGATGGTTCTGAT for human BAX mRNA,
AAGGTGGAGTACCACAGAGG and TCCATGTATGGTGACCCATCC for human APAF1 mRNA,
GAGCTGGTGGTTGACTTTCTC and TCCATCTCCGATTCAGTCCCT for human BCL2L1 mRNA,
GAGCGGCTCCAGATTGTCAA and CTGGGGATGTAGCCTGTCTGT for human SRC mRNA,
CAGCAGCTCTTATCAGCATGT and AACTGTGGCCCACTTAGCAAC for human BIRC6 mRNA,
ACCAAGCCGGATTTGCGATT and ACTTGCACTTGTTCCTCGTGG for human PMAIP1 mRNA,
GAGCGGCTCCAGATTGTCAA and CTGGGGATGTAGCCTGTCTGT for human SRC mRNA,
AGGACCCCAACTGGTACAAAG and CGTGGAACCAAGGCATGAG for human CSK mRNA,
GACCTCAACGCACAGTACGAG and AGGAGTCCCATGATGAGATTGT for human BBC3/PUMA mRNA, TTCTTTGCAGCTCCTTCGTT and ATGGAGGGGAATACAGCCC for mouse Actb mRNA,
TTGCCAGCAGAATAAAAGGTG and TTTGCTCCTGTGCGGAAC for mouse Cdkn1a mRNA, and ACCAGAGGAAATTTTCAATAGGC and TGATGCACTTGCAGAAAACA for mouse Il6 mRNA.
Western blot analysis
Protein extracts were obtained by lysing cells with a denaturing buffer containing 2% SDS (Sigma-Aldrich) in 50 mM Hepes. After boiling and sonication, whole-cell protein extracts were size-fractionated through polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% nonfat dry milk and immunoblotted. Specific primary antibodies recognized total p38 MAPK (Cell Signaling Technology, ref. 9212S), phosphorylated p38 MAPK (T180/Y182) (BioLegend, ref. 903501), total SRC (Cell Signaling Technology, ref. 2108S), phosphorylated SRC (Y416) (Cell Signaling Technology, 6943S), total FAK (D5O7U) XP (Cell Signaling Technology, 71433S), phosphorylated FAK (Y576/577) (Cell Signaling Technology, 3281S), total SAPK/JNK antibody (Cell Signaling Technology, 9252S), phosphorylated SAPK/JNK (T183/Y185) (81E11) (Cell Signaling Technology, 4668S), total ERK1/2 (Cell Signaling Technology, 4695S), phosphorylated ERK1/2 (T202/Y204) (BioLegend, 675502), total AKT (pan) (40D4) (Cell Signaling Technology, 2920S), phosphorylated AKT (Ser473) (Cell Signaling Technology, 4060S), total STAT3 (124H6) (Cell Signaling Technology, 9139T), phosphorylated STAT3 (Y705) (D3A7) XP (Cell Signaling Technology, 9145S), p53 (DO-1) (Cell Signaling 9139, 18032S), ACTB (β-actin C4) (Santa Cruz Biotechnology, sc-47778), cleaved CASP3 (caspase-3) (Asp175) (Cell Signaling Technology, 9661S), PUMA/BBC3 (Cell Signaling Technology, 98672S), NOXA/PMAIP1 (Cell Signaling Technology, 4766S), BCL2L1/BCL-xL (Cell Signaling Technology, 2764S), and BCL2L2/BCL-w (Cell Signaling Technology, 2724S). After incubation with the appropriate secondary antibodies conjugated with horseradish peroxidase (Jackson ImmunoResearch), the chemiluminescent signals were detected by using the ChemiDoc system (Bio-Rad).
BrdU incorporation
Forty thousand cells were seeded in 24-well plates and incubated with BrdU diluted in DMEM with 10% FBS for 24 hours. BrdU incorporation was measured following the manufacturer’s protocol (Cell Signaling Technology). Briefly, cells were fixed and denatured before the addition of BrdU mouse monoclonal antibody, and BrdU was detected using a GloMax plate reader (Promega).
Caspase 3/7 activity
Apoptosis was monitored by measuring caspase 3/7 activity using the Caspase-Glo 3/7 Assay System (Promega). Briefly, Caspase-Glo 3/7 solution at 25°C was added directly to each well. The plate was then shaken vigorously for 30 s and incubated at 25°C in the dark for 30 to 180 min. Luminescence was then measured using a GloMax plate reader (Promega).
Phosphoarray analysis
The Phospho Explorer Antibody Array (Full Moon Biosystems) platform was used to assess the phosphorylation status of protein samples in different conditions tested (n = 2 samples per condition). Cell lysates were prepared using the Protein Extraction Kit, and lysates were biotinylated and incubated on the array slides for 2 hours at 25°C. After washes following the manufacturer’s protocol, the array slides were incubated with Cy3-streptavidin for 45 min at 25°C. The arrays were sent to the manufacturer for signal extraction. The data were normalized to the median signal intensity of all signals on the slide. Fold changes among samples were calculated (the complete dataset is in table S1), and those changes above 1.25-fold and below 0.8-fold (table S2) were included in the final dataset. The STRING platform was used to identify and count the number of interactions of each of the proteins differentially phosphorylated. Enrichr analysis was performed by introducing the list of proteins whose phosphorylation was significantly changed between conditions. In the clustergram shown in Fig. 3J, the enriched terms (in this case, the hub proteins) are the columns, input proteins are the rows, and the filled cells in the matrix designate if a protein is associated with a hub protein. Columns are rank-ordered by the P value score obtained for each hub protein. In Fig. 5F, a volcano plot shows the significance of each gene set from the CORUM database versus its odds ratio. Each point represents a gene set; the x axis indicates the odds ratio (0, infinity) for the gene set, while the y axis indicates the −log(P value) of the gene set.
Transcriptomic analysis
RNA was extracted with RLT lysis buffer (Qiagen) and purified using the QIAcube system (Qiagen). RNA quality and quantity were determined using the Agilent Bioanalyzer RNA6000 Chip (Agilent, Santa Clara, CA). Total RNA (200 ng) was labeled according to the manufacturer’s recommendations using the Quick Amp Labeling Kit (Agilent 5190-2305). Cy3-labeled complementary RNA (cRNA) (600 ng) was hybridized for 17 hours to Agilent SurePrint G3 Human 8x60K V3 microarrays, rinsed, and scanned at 3 μm using an Agilent SureScan scanner. Hybridization intensity data were extracted using Agilent’s Feature Extraction Software. Quantile-normalized data were further used for the subsequent analyses. GSEA analysis was performed as previously described, using non–log2-transformed data (63). The heatmap represents relative differences (log2), displayed by row z score calculations. The complete microarray dataset is provided in GSE169037.
Mice and doxorubicin-induced in vivo senescence
All mouse work, including the import, housing, experimental procedures, and euthanasia, were approved by the Animal Care and Use Committee of the National Institute on Aging (NIA). Ten- to 12-week-old C57BL/6J mice (stock no. 000664) were imported from The Jackson Laboratory (Bar Harbor, ME) and housed in the animal facility in NIA. Mice were provided standard chow ad libitum and maintained under a 12:12-hour light/dark cycle. Systemic cellular senescence was introduced by treating mice with doxorubicin, as described (18). Briefly, a single dose of doxorubicin (10 mg/kg) and/or SRC inhibitor was injected intraperitoneally, and organs (lung, kidney, and liver) were collected.
TUNEL assay and p21 immunofluorescence in tissue sections
To prepare sections, mouse organs were fixed in 4% paraformaldehyde at 4°C for 16 hours, followed by cryopreservation pretreatment in 30% sucrose for additional 16 hours. The organs were embedded in OCT (optimal cutting temperature) compound and frozen at −80°C; 16 hours later, blocks were cut into 10-μm sections by using a Leica cryostat. Slide-mounted sections were dried for 16 hours at 25°C. Immunofluorescent detection of p21 was performed by permeabilizing sections with 0.1% Triton X-100 in PBS for 2 min at 25°C, blocking with 10% goat serum for 1 hour at 25°C, and incubating with anti-p21 antibody (Abcam, [EPR18021], ab188224) in 10% goat serum for 16 hours at 4°C. A fluorescent secondary antibody [Invitrogen, goat anti-rabbit Immunoglobulin G (H+L) superclonal, Alexa Fluor 647] was then incubated in 10% goat serum for 1 hour at 25°C, and slides were mounted with ProLong Diamond Antipode Mountant with 4′,6-diamidino-2-phenylindole (Life Technologies) and coverslips. TUNEL assay (In Situ Cell Death Detection Kit, Fluorescein, Roche) was performed following the manufacturer’s instructions. Signals were collected using a fluorescence microscope (BZ-X Analyzer, Keyence) and analyzed with ImageJ.
Statistical analysis
Data are presented as means ± SD of at least n = 3 independent experiments. Significance was established using two-tailed Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001). All statistical analyses were performed with Prism 9.
Acknowledgments
Funding: This work was funded in its entirety by the National Institute on Aging Intramural Research Program of the National Institutes of Health.
Author contributions: C.A. and M.G. conceived the study; C.A., A.B.H., K.A., and C.-Y.C. designed experiments; C.A., A.B.H., M.R., R.M., E.L., J.L.M., and S.D. performed and analyzed experiments; C.A., A.B.H., and M.G. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The transcriptomic data have been deposited in GSE169037.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.Gorgoulis V., Adams P. D., Alimonti A., Bennett D. C., Bischof O., Bishop C., Campisi J., Collado M., Evangelou K., Ferbeyre G., Gil J., Hara E., Krizhanovsky V., Jurk D., Maier A. B., Narita M., Niedernhofer L., Passos J. F., Robbins P. D., Schmitt C. A., Sedivy J., Vougas K., von Zglinicki T., Zhou D., Serrano M., Demaria M., Cellular senescence: Defining a path forward. Cell 179, 813–827 (2019). [DOI] [PubMed] [Google Scholar]
- 2.McHugh D., Gil J., Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 217, 65–77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Micco R., Krizhanovsky V., Baker D., d’Adda di Fagagna F., Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Childs B. G., Baker D. J., Kirkland J. L., Campisi J., van Deursen J. M., Senescence and apoptosis: Dueling or complementary cell fates? EMBO Rep. 15, 1139–1153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yosef R., Pilpel N., Tokarsky-Amiel R., Biran A., Ovadya Y., Cohen S., Vadai E., Dassa L., Shahar E., Condiotti R., Ben-Porath I., Krizhanovsky V., Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 7, 11190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baar M. P., Brandt R. M. C., Putavet D. A., Klein J. D. D., Derks K. W. J., Bourgeois B. R. M., Stryeck S., Rijksen Y., van Willigenburg H., Feijtel D. A., van der Pluijm I., Essers J., van Cappellen W. A., van Ijcken W. F., Houtsmuller A. B., Pothof J., de Bruin R. W. F., Madl T., Hoeijmakers J. H. J., Campisi J., de Keizer P. L. J., Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169, 132–147.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roskoski R. Jr., Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol. Res. 94, 9–25 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Soto-Gamez A., Quax W. J., Demaria M., Regulation of survival networks in senescent cells: From mechanisms to interventions. J. Mol. Biol. 431, 2629–2643 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Anerillas C., Abdelmohsen K., Gorospe M., Regulation of senescence traits by MAPKs. Geroscience 42, 397–408 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh R., Letai A., Sarosiek K., Regulation of apoptosis in health and disease: The balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 20, 175–193 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anguita E., Villalobo A., Src-family tyrosine kinases and the Ca2+ signal. Biochim. Biophys. Acta Mol. Cell Res. 1864, 915–932 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Cai H., Smith D. A., Memarzadeh S., Lowell C. A., Cooper J. A., Witte O. N., Differential transformation capacity of Src family kinases during the initiation of prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 6579–6584 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demaria M., O’Leary M. N., Chang J., Shao L., Liu S., Alimirah F., Koenig K., Le C., Mitin N., Deal A. M., Alston S., Academia E. C., Kilmarx S., Valdovinos A., Wang B., de Bruin A., Kennedy B. K., Melov S., Zhou D., Sharpless N. E., Muss H., Campisi J., Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 7, 165–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman A. B., Anerillas C., Harris S. C., Munk R., Martindale J. L., Yang X., Mazan-Mamczarz K., Zhang Y., Heckenbach I. J., Scheibye-Knudsen M., De S., Sen P., Abdelmohsen K., Gorospe M., Reduction of lamin B receptor levels by miR-340-5p disrupts chromatin, promotes cell senescence and enhances senolysis. Nucleic Acids Res. 49, 7389–7405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yosef R., Pilpel N., Papismadov N., Gal H., Ovadya Y., Vadai E., Miller S., Porat Z., Ben-Dor S., Krizhanovsky V., p21 maintains senescent cell viability under persistent DNA damage response by restraining JNK and caspase signaling. EMBO J. 36, 2280–2295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munoz-Espin D., Canamero M., Maraver A., Gomez-Lopez G., Contreras J., Murillo-Cuesta S., Rodriguez-Baeza A., Varela-Nieto I., Ruberte J., Collado M., Serrano M., Programmed cell senescence during mammalian embryonic development. Cell 155, 1104–1118 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Storer M., Mas A., Robert-Moreno A., Pecoraro M., Ortells M. C., Di Giacomo V., Yosef R., Pilpel N., Krizhanovsky V., Sharpe J., Keyes W. M., Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 155, 1119–1130 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Xie Z., Bailey A., Kuleshov M. V., Clarke D. J. B., Evangelista J. E., Jenkins S. L., Lachmann A., Wojciechowicz M. L., Kropiwnicki E., Jagodnik K. M., Jeon M., Ma’ayan A., Gene set knowledge discovery with enrichr. Curr. Protoc. 1, e90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slobodnyuk K., Radic N., Ivanova S., Llado A., Trempolec N., Zorzano A., Nebreda A. R., Autophagy-induced senescence is regulated by p38α signaling. Cell Death Dis. 10, 376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danckwardt S., Gantzert A. S., Macher-Goeppinger S., Probst H. C., Gentzel M., Wilm M., Grone H. J., Schirmacher P., Hentze M. W., Kulozik A. E., p38 MAPK controls prothrombin expression by regulated RNA 3′ end processing. Mol. Cell 41, 298–310 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Aiguade J., Balague C., Carranco I., Caturla F., Dominguez M., Eastwood P., Esteve C., Gonzalez J., Lumeras W., Orellana A., Preciado S., Roca R., Vidal L., Vidal B., Novel triazolopyridylbenzamides as potent and selective p38α inhibitors. Bioorg. Med. Chem. Lett. 22, 3431–3436 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Efeyan A., Ortega-Molina A., Velasco-Miguel S., Herranz D., Vassilev L. T., Serrano M., Induction of p53-dependent senescence by the MDM2 antagonist nutlin-3a in mouse cells of fibroblast origin. Cancer Res. 67, 7350–7357 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Zhou J., Yi Q., Tang L., The roles of nuclear focal adhesion kinase (FAK) on Cancer: A focused review. J. Exp. Clin. Cancer Res. 38, 250 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim S. T., Chen X. L., Lim Y., Hanson D. A., Vo T. T., Howerton K., Larocque N., Fisher S. J., Schlaepfer D. D., Ilic D., Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol. Cell 29, 9–22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamouille S., Xu J., Derynck R., Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez D. M., Medici D., Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 7, re8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin E. Y., Park J. H., You S. T., Lee C. S., Won S. Y., Park J. J., Kim H. B., Shim J., Soung N. K., Lee O. J., Schwartz M. A., Kim E. G., Integrin-mediated adhesions in regulation of cellular senescence. Sci. Adv. 6, eaay3909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demaria M., Ohtani N., Youssef S. A., Rodier F., Toussaint W., Mitchell J. R., Laberge R. M., Vijg J., Van Steeg H., Dolle M. E., Hoeijmakers J. H., de Bruin A., Hara E., Campisi J., An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bird T. G., Müller M., Boulter L., Vincent D. F., Ridgway R. A., Lopez-Guadamillas E., Lu W. Y., Jamieson T., Govaere O., Campbell A. D., Ferreira-Gonzalez S., Cole A. M., Hay T., Simpson K. J., Clark W., Hedley A., Clarke M., Gentaz P., Nixon C., Bryce S., Kiourtis C., Sprangers J., Nibbs R. J. B., Van Rooijen N., Bartholin L., McGreal S. R., Apte U., Barry S. T., Iredale J. P., Clarke A. R., Serrano M., Roskams T. A., Sansom O. J., Forbes S. J., TGFβ inhibition restores a regenerative response in acute liver injury by suppressing paracrine senescence. Sci. Transl. Med. 10, eaan1230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues M., Kosaric N., Bonham C. A., Gurtner G. C., Wound healing: A cellular perspective. Physiol. Rev. 99, 665–706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humphries J. D., Byron A., Humphries M. J., Integrin ligands at a glance. J. Cell Sci. 119, 3901–3903 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtani N., Imamura Y., Yamakoshi K., Hirota F., Nakayama R., Kubo Y., Ishimaru N., Takahashi A., Hirao A., Shimizu T., Mann D. J., Saya H., Hayashi Y., Arase S., Matsumoto M., Kazuki N., Hara E., Visualizing the dynamics of p21(Waf1/Cip1) cyclin-dependent kinase inhibitor expression in living animals. Proc. Natl. Acad. Sci. U.S.A. 104, 15034–15039 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer M., Census and evaluation of p53 target genes. Oncogene 36, 3943–3956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun G., Kemble D. J., To C or not to C: Direct and indirect redox regulation of Src protein tyrosine kinase. Cell Cycle 8, 2353–2355 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Jung S. H., Lee M., Park H. A., Lee H. C., Kang D., Hwang H. J., Park C., Yu D. M., Jung Y. R., Hong M. N., Kim Y. N., Park H. J., Ko Y. G., Lee J. S., Integrin α6β4-Src-AKT signaling induces cellular senescence by counteracting apoptosis in irradiated tumor cells and tissues. Cell Death Differ. 26, 245–259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryu S. J., Cho K. A., Oh Y. S., Park S. C., Role of Src-specific phosphorylation site on focal adhesion kinase for senescence-associated apoptosis resistance. Apoptosis 11, 303–313 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Ritschka B., Knauer-Meyer T., Goncalves D. S., Mas A., Plassat J. L., Durik M., Jacobs H., Pedone E., Di Vicino U., Cosma M. P., Keyes W. M., The senotherapeutic drug ABT-737 disrupts aberrant p21 expression to restore liver regeneration in adult mice. Genes Dev. 34, 489–494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hafner A., Bulyk M. L., Jambhekar A., Lahav G., The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 20, 199–210 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Golubovskaya V. M., Cance W. G., FAK and p53 protein interactions. Anticancer Agents Med. Chem. 11, 617–619 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barretta M. L., Spano D., D’Ambrosio C., Cervigni R. I., Scaloni A., Corda D., Colanzi A., Aurora-A recruitment and centrosomal maturation are regulated by a Golgi-activated pool of Src during G2. Nat. Commun. 7, 11727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi B. K., Dayaram T., Parikh N., Wilkins A. D., Nagarajan M., Novikov I. B., Bachman B. J., Jung S. Y., Haas P. J., Labrie J. L., Pickering C. R., Adikesavan A. K., Regenbogen S., Kato L., Lelescu A., Buchovecky C. M., Zhang H., Bao S. H., Boyer S., Weber G., Scott K. L., Chen Y., Spangler S., Donehower L. A., Lichtarge O., Literature-based automated discovery of tumor suppressor p53 phosphorylation and inhibition by NEK2. Proc. Natl. Acad. Sci. U.S.A. 115, 10666–10671 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polonio-Vallon T., Kirkpatrick J., Krijgsveld J., Hofmann T. G., Src kinase modulates the apoptotic p53 pathway by altering HIPK2 localization. Cell Cycle 13, 115–125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aikawa R., Nagai T., Kudoh S., Zou Y., Tanaka M., Tamura M., Akazawa H., Takano H., Nagai R., Komuro I., Integrins play a critical role in mechanical stress-induced p38 MAPK activation. Hypertension 39, 233–238 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Jain M., Dev R., Doddapattar P., Kon S., Dhanesha N., Chauhan A. K., Integrin α9 regulates smooth muscle cell phenotype switching and vascular remodeling. JCI Insight 6, e147134 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim M. J., Byun J. Y., Yun C. H., Park I. C., Lee K. H., Lee S. J., c-Src-p38 mitogen-activated protein kinase signaling is required for Akt activation in response to ionizing radiation. Mol. Cancer Res. 6, 1872–1880 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Hartman M. L., Czyz M., BCL-w: Apoptotic and non-apoptotic role in health and disease. Cell Death Dis. 11, 260 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Igea A., Nebreda A. R., The stress kinase p38α as a target for cancer therapy. Cancer Res. 75, 3997–4002 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Freund A., Patil C. K., Campisi J., p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 30, 1536–1548 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miura H., Kondo Y., Matsuda M., Aoki K., Cell-to-cell heterogeneity in p38-mediated cross-inhibition of JNK causes stochastic cell death. Cell Rep. 24, 2658–2668 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Kim L. C., Song L., Haura E. B., Src kinases as therapeutic targets for cancer. Nat. Rev. Clin. Oncol. 6, 587–595 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Zhu Y., Tchkonia T., Pirtskhalava T., Gower A. C., Ding H., Giorgadze N., Palmer A. K., Ikeno Y., Hubbard G. B., Lenburg M., O’Hara S. P., LaRusso N. F., Miller J. D., Roos C. M., Verzosa G. C., LeBrasseur N. K., Wren J. D., Farr J. N., Khosla S., Stout M. B., McGowan S. J., Fuhrmann-Stroissnigg H., Gurkar A. U., Zhao J., Colangelo D., Dorronsoro A., Ling Y. Y., Barghouthy A. S., Navarro D. C., Sano T., Robbins P. D., Niedernhofer L. J., Kirkland J. L., The Achilles’ heel of senescent cells: From transcriptome to senolytic drugs. Aging Cell 14, 644–658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoare M., Ito Y., Kang T. W., Weekes M. P., Matheson N. J., Patten D. A., Shetty S., Parry A. J., Menon S., Salama R., Antrobus R., Tomimatsu K., Howat W., Lehner P. J., Zender L., Narita M., NOTCH1 mediates a switch between two distinct secretomes during senescence. Nat. Cell Biol. 18, 979–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rapisarda V., Borghesan M., Miguela V., Encheva V., Snijders A. P., Lujambio A., O’Loghlen A., Integrin Beta 3 regulates cellular senescence by activating the TGF-β pathway. Cell Rep. 18, 2480–2493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braun H., Schmidt B. M., Raiss M., Baisantry A., Mircea-Constantin D., Wang S., Gross M. L., Serrano M., Schmitt R., Melk A., Cellular senescence limits regenerative capacity and allograft survival. J. Am. Soc. Nephrol. 23, 1467–1473 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei C., Li L., Menon M. C., Zhang W., Fu J., Kidd B., Keung K. L., Woytovich C., Greene I., Xiao W., Salem F., Yi Z., He J. C., Dudley J. T., Murphy B., Genomic analysis of kidney allograft injury identifies hematopoietic cell kinase as a key driver of renal fibrosis. J. Am. Soc. Nephrol. 28, 1385–1393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krizhanovsky V., Yon M., Dickins R. A., Hearn S., Simon J., Miething C., Yee H., Zender L., Lowe S. W., Senescence of activated stellate cells limits liver fibrosis. Cell 134, 657–667 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogrodnik M., Miwa S., Tchkonia T., Tiniakos D., Wilson C. L., Lahat A., Day C. P., Burt A., Palmer A., Anstee Q. M., Grellscheid S. N., Hoeijmakers J. H. J., Barnhoorn S., Mann D. A., Bird T. G., Vermeij W. P., Kirkland J. L., Passos J. F., von Zglinicki T., Jurk D., Cellular senescence drives age-dependent hepatic steatosis. Nat. Commun. 8, 15691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jurk D., Wilson C., Passos J. F., Oakley F., Correia-Melo C., Greaves L., Saretzki G., Fox C., Lawless C., Anderson R., Hewitt G., Pender S. L., Fullard N., Nelson G., Mann J., van de Sluis B., Mann D. A., von Zglinicki T., Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nat. Commun. 5, 4172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aspinall R. J., Weis S. M., Barnes L., Lutu-Fuga K., Bylund D. J., Pockros P. J., Cheresh D. A., A Src family kinase inhibitor improves survival in experimental acute liver failure associated with elevated cerebral and circulating vascular endothelial growth factor levels. Liver Int. 31, 1222–1230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lakhani H. V., Sharma D., Dodrill M. W., Nawab A., Sharma N., Cottrill C. L., Shapiro J. I., Sodhi K., Phenotypic alteration of hepatocytes in non-alcoholic fatty liver disease. Int. J. Med. Sci. 15, 1591–1599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seo H. Y., Lee S. H., Lee J. H., Kang Y. N., Hwang J. S., Park K. G., Kim M. K., Jang B. K., Src inhibition attenuates liver fibrosis by preventing hepatic stellate cell activation and decreasing connetive tissue growth factor. Cells 9, 558 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1 to S3