Summary
Background
SARS-CoV-2 Omicron variant is rampantly spreading across the globe. We assessed the pathogenicity and immune response generated by BA.1.1 sub-lineage of SARS-CoV-2 [Omicron (R346K) variant] in 5 to 6-week old Syrian hamsters and compared the observations with that of Delta variant infection.
Methods
Virus shedding, organ viral load, lung disease and immune response generated in hamsters were sequentially assessed.
Findings
The disease characteristics of the Omicron (R346K) variant were found to be similar to that of the Delta variant infection in hamsters like viral replication in the respiratory tract and interstitial pneumonia. The Omicron (R346K) infected hamsters demonstrated lesser body weight reduction and viral RNA load in the throat swab and nasal wash samples in comparison to the Delta variant infection. The viral load in the lungs and nasal turbinate samples and the lung disease severity of the Omicron (R346K) infected hamsters were found comparable with that of the Delta variant infected hamsters. Neutralizing antibody response against Omicron (R346K) variant was detected from day 5 and the cross-neutralization titre of the sera against other variants showed severe reduction ie., 7 fold reduction against Alpha and no titers against B.1, Beta and Delta.
Interpretation
This preliminary data shows that Omicron (R346K) variant infection can produce moderate to severe lung disease similar to that of the Delta variant and the neutralizing antibodies produced in response to Omicron (R346K) variant infection shows poor neutralizing ability against other co-circulating SARS-CoV-2 variants like Delta which necessitates caution as it may lead to increased cases of reinfection.
Funding
This study was supported by Indian Council of Medical Research as an intramural grant (COVID-19) to ICMR-National Institute of Virology, Pune.
Keywords: SARS-CoV-2, Omicron (R346K), BA.1.1, R346K, Syrian hamsters, Pathogenicity, Neutralization
Research in context.
Evidence before this study
SARS-CoV-2 Omicron variant is spreading worldwide rampantly. Preliminary evidences have shown that this Variant of Concern (VOC) has high transmissibility, lesser risk of hospitalization and immune escape property. The variant is further evolving with many descendant lineages and sublineages. Syrian hamster (Mesocricetus auratus) is a valuable model to understand SARS-CoV-2 disease characteristics and has been used widely for characterizing SARS-CoV-2 variants as well as in preclinical studies. The preliminary pathogenicity studies in rodent models have shown limited infectivity or lower pathogenicity of the Omicron variant in comparison to the other SARS-CoV-2 variants. Of the spike gene mutations in the Omicron variant, the substitution R346K is being monitored by the World Health Organization.
Added value of this study
Added value of this study: We analyzed the pathogenesis, disease severity and immune response generated in Syrian hamsters by Omicron (R346K) variant and compared the same with the Delta variant in hamsters. We also studied the neutralization potential of Omicron (R346K) infected hamster sera with other Variant of Concerns like Alpha, Beta and Delta.
Implications of all the available evidence
Implications of all the available evidence: Our findings shows that Omicron variant with R364K mutation replicates to high levels in respiratory tract and is capable of inducing lung disease in hamsters. We found no evidence of prolonged virus shedding in hamsters. The model can be used to assess the efficacy of countermeasures against Omicron variant. Neutralizing antibody response generated against the Omicron (R346K) variant was found poor in neutralizing the earlier as well as currently circulating SARS-CoV-2 VOCs. The findings suggest that the pathogenicity of the circulating Omicron variant lineages could differ.
Alt-text: Unlabelled box
Introduction
The emergence of SARS-CoV-2 Variants of Concerns (VOCs) has caused the most severe crisis and challenge to the public health worldwide. The situation has worsened yet again with the recent emergence of highly mutated and super spreading, Omicron variant (B.1.1.529) in the month of November 2021.1 As of March 22nd 2022, the variant has been reported from 164 countries.1 Increased incidences of COVID-19 cases were reported from countries where Omicron variant has become dominant.1,2 The variant constitutes for more than 99% of the SARS-CoV-2 sequences deposited in the Global Initiative on Sharing Avian Influenza Data (GISAID) database as on the third week of March 2022.1 Millions of people are getting infected with Omicron daily, hence there is an urgent need to deduce the properties of this VOC.
Omicron variant possess around 26 to 32 mutations in the spike protein including mutations associated with increased receptor binding and immune escape property.3 The major descendant lineages identified for the Omicron variant includes BA.1, BA.1.1, BA.2 and BA.3 of which BA.1 and BA.1.1 together contributes to the maximum cases reported around globe.1,4,5 These lineages has been further subdivided into multiple sublineages.4 BA.2 lineage is becoming a dominant lineage across the world now.1 Apart from this some recombinant variants of Delta and Omicron variant are also being reported and has been given pango lineage designations recently as XD, XE and XF.1 The preliminary data available on the phenotypic characteristics of the Omicron variant shows increased transmissibility, risk of reinfection, reduced disease severity and need for hospitalization.1 More data needs to be assessed accounting to the population immunity and other co-morbid conditions to understand the severity of Omicron infection in humans. The studies on the humoral antibody responses have reported a reduction in the neutralizing antibody titers to the Omicron variant in comparison to the wild type SARS-CoV-2 in infected or vaccinated individuals.1,6,7 Apart from the unique mutations identified in the spike protein of this VOC, an additional amino acid change R346K in the variant is being monitored.5,8 This variant with R346K mutation has been now designated as BA.1.1 sublineage of Omicron and has been reported from at least 151 countries.4
Multiple animal models have been characterized for SARS-CoV-2 and have been used in preclinical studies and other allied research.9 Syrian hamsters seems to be an appropriate model for SARS-CoV-2 in terms of ACE-2 binding and also the effective replication of the virus in respiratory tract producing pneumonia.10,11 The model have been effectively used for the studies on the vaccines, antivirals, pathogenesis, transmissibility, virus shedding and disease severity associated with SARS-CoV-2 VOC's.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 Recently, few studies have been published on the pathogenesis aspect of Omicron variant in mice and Syrian hamsters in comparison with other VOC's demonstrating decreased severity of infection.23, 24, 25, 26, 27, 28, 29 Abdelnabi et al. reported limited lung infectivity with no evidence of pneumonia by the Omicron variant in hamsters in contrast to other studies which showed lower viral loads and less severe pneumonia in hamster model in comparison to ancestral variant or other VOCs.23,24,26, 27, 28 The studies in the wild type and hACE2 transgenic mice models also showed milder disease by the Omicron variant.28,29
After multiple failed attempts of isolating the Omicron variant in cell lines reported to be susceptible for SARS-CoV-2, we attempted virus isolation using the Syrian hamster model. The virus grew to high titres in the nasal turbinates’ of the model.30 The deep sequencing of the specimens revealed only 0.0066% difference in the hamster passage sequence with that of the clinical sample with an additional Q19E substitution in the M gene. Apart from the signature mutations of the Omicron variant, the isolate also possessed the R346K mutation. All the published animal studies till date has used the Omicron variant of BA.1 sub lineage without the R346K mutation. Here, we report the characterization of Omicron variant of BA.1.1 sub lineage [Omicron (R346K) variant] in Syrian hamster model for its pathogenicity, virus shedding pattern and immune response.
Methods
Virus
SARS-CoV-2 Omicron (R346K) variant (GISAID accession no: EPI_ISL_8542938) belonging to the BA.1.1 sub lineage isolated from nasopharyngeal swab of a COVID-19 patient by inoculating into Syrian hamster was used for the study.30 The virus titer was found to be 2·13×105 TCID50/ml on titration in Vero (ATCC® CCL-81™) cells (ATCC, USA) as per the Reed and Muench method. On deep sequencing following amino acid changes were found in the isolate.(NSP5_P132H,Spike_T95I,Spike_K417N,Spike_S373P,Spike_Q493R,Spike_N969K, Spike_H655Y,Spike_N856K,N_R203K,Spike_S371L,NSP3_A1892T,Spike_Q954H,Spike_G339D,N_P13L,Spike_N501Y,NSP14_I42V,Spike_P681H,M_Q19E,Spike_N440K,NSP4_T492I,Spike_S375F,Spike_Q498R,Spike_G446S,Spike_N679K,Spike_N764K,Spike_S477N,Spike_Y505H,NSP3_K38R,Spike_R346K,NSP6_I189V,Spike_T547K,M_D3G,Spike_D796Y,Spike_E484A,N_G204R,Spike_T478K,E_T9I,Spike_L981F,M_A63T,NSP12_P323L,Spike_D614G,Spike_G496S).30 Delta variant (GISAID accession no: EPI_ISL_2400521) isolated similarly as mentioned above with a titer 1 × 105 TCID50/ml was used as comparator virus strain for the study.
Hamster experiments
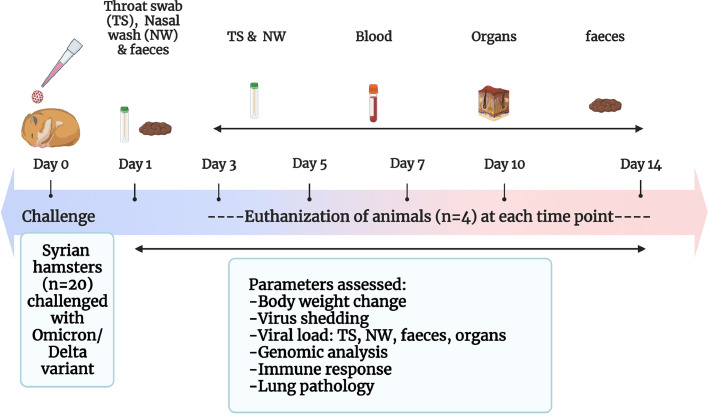
The experiment was approved by the Institutional Animal Ethics Committee of ICMR-National Institute of Virology (ICMR-NIV), Pune under the approval no. NIV/IAEC/2021/MCL/01. The guidelines of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Government of India were followed during the experiment. Forty-four Syrian hamsters (either sex) of 5, 6-week age procured from the laboratory animal facility of ICMR-NIV, Pune licensed by CPCSEA were used for the study. Individually ventilated cage system was used for animal housing in the maximum containment facility with ad libitum food and water and animals were randomly allocated to three groups after a week period of acclimatization. The caretakers and technicians were blinded to the group allocation. Baseline body weights were measured before the experiment. Hamsters were anaesthetized using isoflurane during intranasal virus inoculation and collection of samples (nasal wash and throat swab). Hundred microlitre of a virus [Omicron (R346K) variant] dose of 2·13 × 104 TCID50 was used to infect the twenty hamsters intranasally. Similarly, another group of twenty hamsters were intranasally infected with 1 × 104 TCID50 of the Delta variant. A group of age matched hamsters (n = 4) inoculated with 0.1 ml sterile media were kept till 14 days as uninfected control for the study. The hamsters were monitored for 14 days for body weight changes. Throat swab (TS) samples (n = 8) with thin nylon flocked swabs (Cat # PW1172, Himedia, Mumbai), nasal wash (NW) samples (n = 8) with sterile phosphate buffered saline (PBS) and freshly voided feces samples (n = 8) were collected in 1 ml viral transport media (Cat # AL167, Himedia, Mumbai,) on days 1,3,5,7 and 10 from the same eight hamsters of both the virus infected groups. On the 14th day, samples were collected from the remaining 4 hamsters of both the groups. Four hamsters/group were euthanized on days 3, 5, 7, 10- and 14-days post infection (DPI) using overdose of isoflurane and the control group was sacrificed on 14 DPI. Organ samples collected during the necropsy were weighed and homogenized using a tissue homogenizer (Qiagen, Germany) and were used for viral load estimation. Lung tissues were examined for their gross changes and fixed in 10% neutral buffered formalin for histopathological evaluation. The experimental design and the outcome measures are summarized in the Figure 1. ARRIVE guideline checklist has been attached as supplementary file.
Figure 1.
Study design. (a) Summary of the study.
Viral load estimation
RNA extraction was performed using the MagMAX™ viral/pathogen nucleic acid isolation kit (Cat # A48310, ThermoScientific, USA) as per the manufacturer's instructions. RT-qPCR was performed further using published primers for E gene and N gene for the viral RNA and sub genomic RNA (sgRNA), respectively.31,32 The lungs, nasal turbinates and NW samples were titrated for the live virus in Vero (ATCC® CCL-81™) cells (ATCC, USA) to determine the presence of replication competent virus as described earlier.18 For titration, 10 fold dilutions of samples were added onto cell monolayer in a 24 well plate and were incubated for one hour. After removal of the media, the cell monolayer was washed with phosphate buffered saline (PBS). The cells were further incubated with maintenance cell culture media containing serum (Cat # 11095080, Gibco, ThermoFisher Scientific, USA) in a CO2 incubator. The cells were observed daily for any cytopathic effects (CPE). On observation of CPE, the supernatant from the wells were tested by RT-qPCR for confirmation.
Anti-SARS-CoV-2 IgG ELISA
The hamster serum samples were tested for antibodies using anti-SARS-CoV-2 hamster IgG ELISA33 and S1-Receptor Binding Domain (RBD) ELISA. For S1-RBD ELISA, 96-well polystyrene microtitre plates (Nunc, Germany) coated with 1·5 µg/well concentration of S1-RBD protein (Cat # FS001, Labcare, India) was used. Five percent skimmed milk (Cat #. 232100, Difco, Thermo Fisher Scientific, USA) in 1 X PBS with 0·1% Tween-20 (Cat # P1379, Sigma-Aldrich, USA) was used for blocking the wells. The plates were washed thrice after blocking for 2 h at 37 °C. The serum samples were added to the plates and were further incubated for an hour. After washing the plates, 100 μl of anti-hamster IgG horseradish peroxidase (Cat # 31,115, Thermo Fisher Scientific, USA) was added to each well and incubated for 30 min at 37 °C. 3, 3′, 5, 5′-tetramethylbenzidine (TMB) (Cat # T444, Sigma Aldrich, USA) was used as substrate and the reaction was terminated using 1 N sulphuric acid. The optical density (OD) was measured using an ELISA reader at 450 nm and the cut off was set at twice the OD value of the negative control.
Neutralization test
The neutralization assay was performed for the serum collected on each time point (3, 5, 7, 10 and 14 DPI). A commercially available SARS-CoV-2 surrogate virus neutralizaton test (sVNT) kit (Cat # L00847-A, Genscript, Piscataway, United States) for the Omicron variant based on antibody mediated blockage of the ACE-2 receptor- RBD interaction was used to assess the neutralization of the Omicron (R346K) variant infected hamster sera samples. The assay results were expressed as percent inhibition with a cut off ≥ 30%.
The live virus neutralization assay was performed against B.1 variant (GISAID accession no: EPI_ISL_420546), Alpha (GISAID accession no: EPI_ISL_825086), Beta (GISAID accession no: EPI_ISL_2014131), Delta (GISAID accession no: EPI_ISL_2400521) and Omicron (R346K) VOCs of SARS-CoV-2 as described earlier.34
Histopathology and immunohistochemistry
Lungs samples fixed in formalin were processed for histopathology by hematoxylin and eosin staining.35 Coded tissue samples were blindly scored by a pathologist. Each section was scored for vascular changes, bronchiolar changes, inflammatory cell infiltration, emphysema, oedema, hyaline changes and alveolar pneumocyte hyperplasia.18 Each lesion was given a score of 0 to 1 and the cumulative score was plotted.
For immunohistochemical evaluation, duplicate sections were used as that of histopathology. Lung tissue sections were rehydrated and 0.3% hydrogen peroxide in methanol was used for antigen retrieval. Polyclonal anti-SARS-CoV-2 mouse serum (generated in house using inactivated SARS-CoV-2 antigen in mice) was used as the primary antibody. The antibody was validated for its reactivity and specificity using plaque reduction neutralization test. The sections were incubated with primary antibody (1: 400 dilution) for 60 min and washed with PBS. Goat anti-mouse horse radish peroxidase antibody (Cat # P0447, Agilent Dako, USA) and 3, 3′-diaminobenzidine (DAB) tetrahydrochloride substrate (Cat # D4293, Sigma Aldrich, USA) were used as secondary antibody and for detection, respectively.
Next generation sequencing
Viral RNA was extracted with QIAmp Viral RNA extraction kit (Cat # 52962, Qiagen, Germany). Qubit® 2·0 Fluorometer (Invitrogen, USA) was used for quantifying the RNA with the Qubit RNA High Sensitivity (HS) kit (Cat # 333898, Thermofisher, USA). The RNA library was prepared using the TruSeq Stranded mRNA LT Library preparation kit (Cat # 20020595, Illumina, USA). The library was quantified, normalized (1·8 picomolar) and loaded on to the Illumina Miniseq Next generation sequencing platform. The reads generated were mapped along with the SARS-CoV-2 Wuhan isolate (Accession No.: NC_045512·2) using the reference-based assembly method of the CLC Genomics Workbench v.20. The nucleotide variations were identified using the basic variant tool as implemented in the CLC Genomics Workbench v.20.
Statistics
The experimental data was analyzed using Graph pad Prism version 9·2·0 software. Non parametric two tailed Mann Whitney tests were used for the body weight, viral load and histopathology score comparison of the infected with the control animals or between the infected groups and Kruskal Wallis tests with Dunn's multiple comparison test was used for the comparison of neutralization data. A p-value less than 0·05 were considered as statistically significant.
Role of funding source
The study sponsor has no role in study design, analysis, interpretation of data, in the writing of the report and in the decision to submit the paper for publication.
Results
Omicron (R346K) infection reduced body weight gain in hamsters
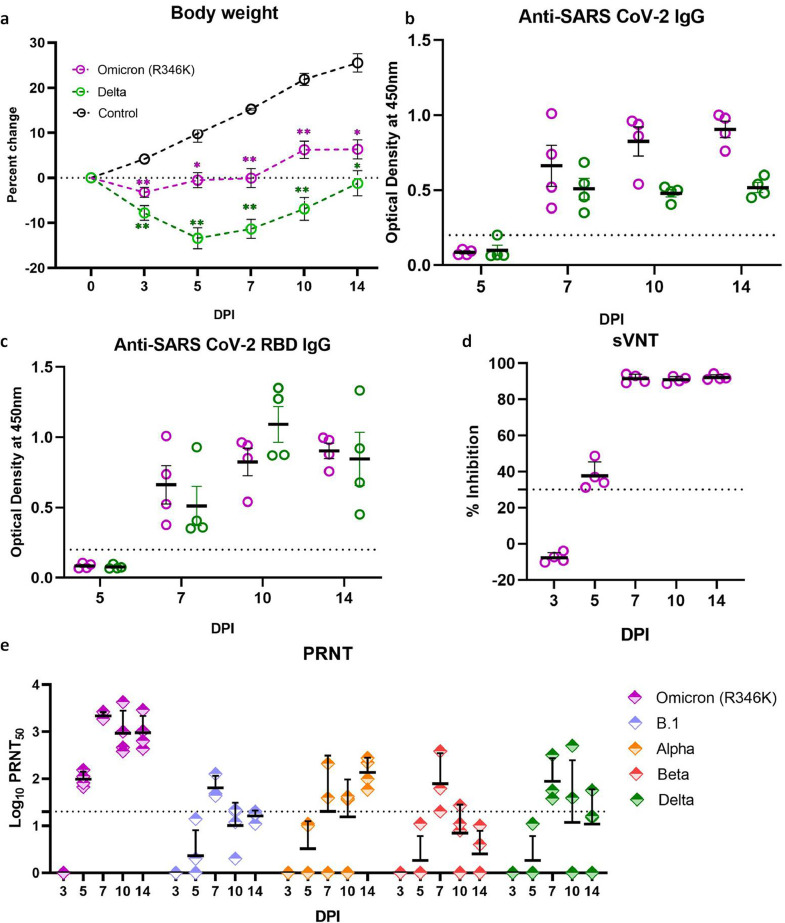
The body weight loss observed in hamsters infected with the Omicron (R346K) variant was lesser than the Delta variant infected hamsters (Figure 2a). Percent mean body weight difference of -3·2%, -0·53% and - 0·08% were observed on day 3, 5 and 7 DPI in the Omicron (R346K) infected animals whereas control group showed an increase of 4·17%, 9·8% and 15·25% on day 3, 5 and 7 DPI, respectively. In case of the Delta variant infected animals, mean weight loss of -7·79%, -13·39% and -11·34% were observed on 3, 5 and 7 DPI, respectively. Thereafter an increase in the mean body weight was seen in the Omicron (R346K) and the Delta variant infected hamsters which was significantly lesser in comparison to the animals from the uninfected control group.
Figure 2.
Percent body weight change and humoral immune response in hamsters post infection. (a) The body weight changes in hamsters after SARS-CoV-2 Omicron (R346K) and Delta variant infection in comparison to that of age matched control animals. [n = 8 on 3, 5, 7 and 10 DPI and n = 4 on 14 DPI for Omicron (R346K) and Delta variant infected group, uninfected control (n = 4), Mann Whitney test]. Mean value ± SEM is represented in the graph. * represents p < 0·05 and ** represents p < 0·001. (b) Anti-SARS-CoV-2 inactivated antigen IgG response and (c) anti-SARS-CoV-2 RBD antibodies in hamsters post infection by ELISA. Mean value ± SEM is represented in the graph. (d) Percentage of inhibition in surrogate virus neutralization assay. Mean value ± SEM is represented in the graph. (e) Neutralizing antibody levels in Omicron (R346K) infected hamsters on 3,5,7,10 and 14 DPI against Omicron (R346K), B.1, Alpha, Beta and Delta variants. Geometric mean± SD is represented in the graph. The dotted line represents the cut-off for the assay.
Immune response in hamsters after infection
The IgG antibodies could be detected from 7 DPI in both the Omicron (R346K) and the Delta variant infected hamsters (Figure 2b,c). The neutralizing antibodies against Omicron (R346K) variant was detected in the infected hamsters from 5 DPI by live as well as surrogate virus neutralization assay (Figure 2d, e). In the live virus neutralization assay, geometric mean titre (GMT) of 98·30, 2170·40, 932·62 and 953 was observed on 5,7,10 and 14 DPI, respectively (Figure 2e). The neutralizing ability of the antibodies against other variants was found to be substantially reduced. Against Alpha variant, GMT was 136·6 on 14 DPI and no neutralization was observed against the B.1, Beta (p = 0·0137, n = 4, Dunns multiple comparison test) and Delta variants (except one hamster sera) (Figure 2e).
Virus shedding following Omicron (R346K) infection in Syrian hamsters
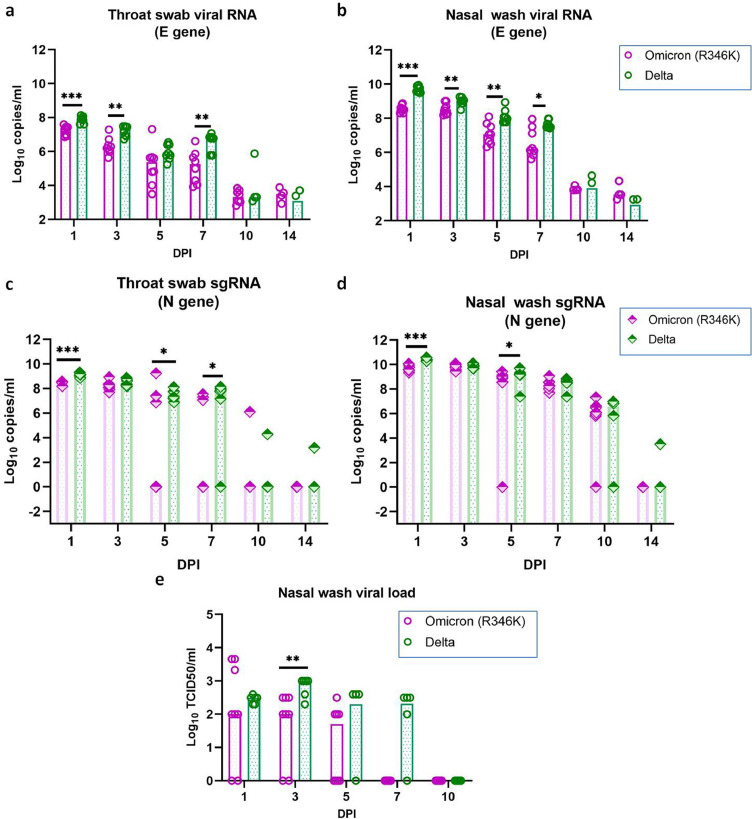
Viral RNA was detected in the NW, TS and faeces samples of Omicron (R346K) infected hamsters and the period of detection varied among samples (Figure 3a, b, Supplementary Figure 1a). The NW and TS samples showed the presence of viral RNA till 14 DPI in all hamsters and till 5 DPI in faeces samples. The highest viral RNA load was observed on 1 and 3 DPI in the TS and NW samples, respectively, which showed decrease in the genome copies on further days. The comparison of the viral RNA in the TS and NW with the Delta variant infected hamsters showed significantly lower viral RNA load during first week post infection in the Omicron (R346K) infected hamsters.
Figure 3.
Viral shedding in hamsters after the SARS-CoV-2 Omicron (R346K) and Delta variant infection. (a) SARS-CoV-2 Omicron (R346K) viral RNA load in a) Throat swab (n = 8, p = 0·0002 on 1DPI, p = 0·0019 on 3DPI, p = 0·0042 on 7DPI, Mann Whitney test), (b) Nasal wash (n = 8, p = 0·0002 on 1DPI, p = 0·0092 on 3DPI, p = 0·0044 on 5DPI, p = 0·019 on 7DPI, Mann-Whitney test) and viral sgRNA load in (c) Throat swab (n = 8, p = 0·0007 on 1DPI, p = 0·049 on 5DPI, p = 0·02 on 7DPI, Mann-Whitney test) and (d) Nasal wash (n = 8, p = 0·0002 on 1DPI, p = 0·0462 on 5DPI, Mann-Whitney test) in hamsters on 1,3,5,7,10 and 14 DPI post Omicron (R346K) infection. (e) Viral load in the nasal wash samples of hamsters, n = 8, p = 0·0037 on 5DPI, Mann-Whitney test. The bars represent the median and *,** and *** represents p < 0·05, p < 0·001 and p < 0·0001, respectively.
Subgenomic RNA detection window was further lower for all samples ie., till 10, 7 and 5 DPI in NW,TS and faeces samples, respectively in the Omicron (R346K) infected hamsters (Figure 3c, d, Supplementary Figure 1b). The sgRNA load was also significantly lower in the NW and TS samples of Omicron (R346K) infected hamsters during the first week. But this difference was not observed on live virus titration, of the NW sample except on 3 DPI. The NW samples of the hamsters infected with the Omicron (R346K) variant had a median titre of 102 TCID50/ml on 1 and 3 DPI and 50 TCID50/ml on 5 DPI (Figure 3e). In case of the Delta variant infected hamsters, the titre was slightly higher with a median TCID50/ml of 3·16 × 102 on 1 DPI, 103 on 3DPI, 1·9 × 102 on 5DPI and 2·08 102 on 7DPI.
Viral load in the organs
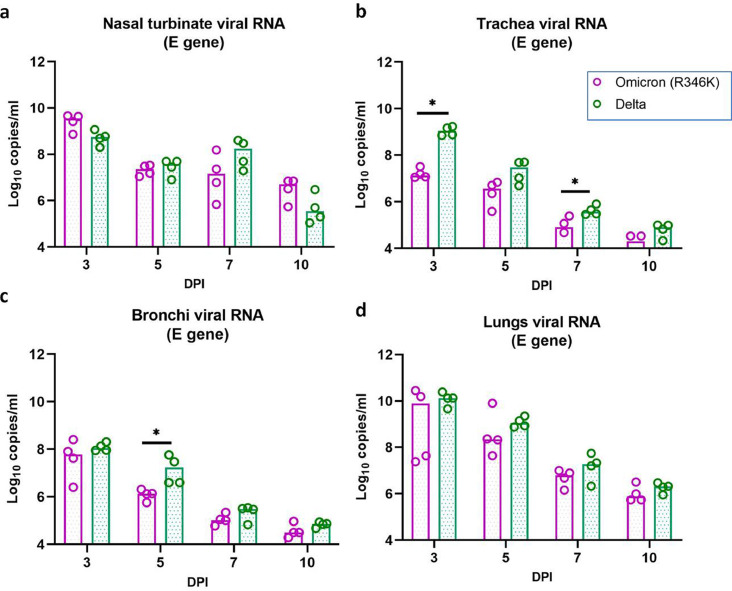
Among the organs tested for viral RNA and sgRNA in the Omicron (R346K) infected hamsters, respiratory organs (nasal turbinates, trachea, bronchi and lungs) showed consistent detection on all the sampled days (Figs. 4, 5a-d). The highest viral RNA load was found on 3 DPI in the respiratory organs. The viral RNA levels showed reduction on further days. Unlike 2 log reduction of viral RNA copies in the nasal turbinate samples on 5 DPI, lung samples showed lesser decrease. Lungs samples showed a median of 7·8 × 109, 2·2 × 108, 6·2 × 106 viral RNA copies/ml on 3, 5 and 7 DPI. All the other organs (brain, liver, heart, spleen intestine and kidney) were negative except that of one hamster in the Omicron (R346K) infected group on 3 DPI which showed viral RNA and sgRNA detection in liver, small intestine and serum samples.
Figure 4.
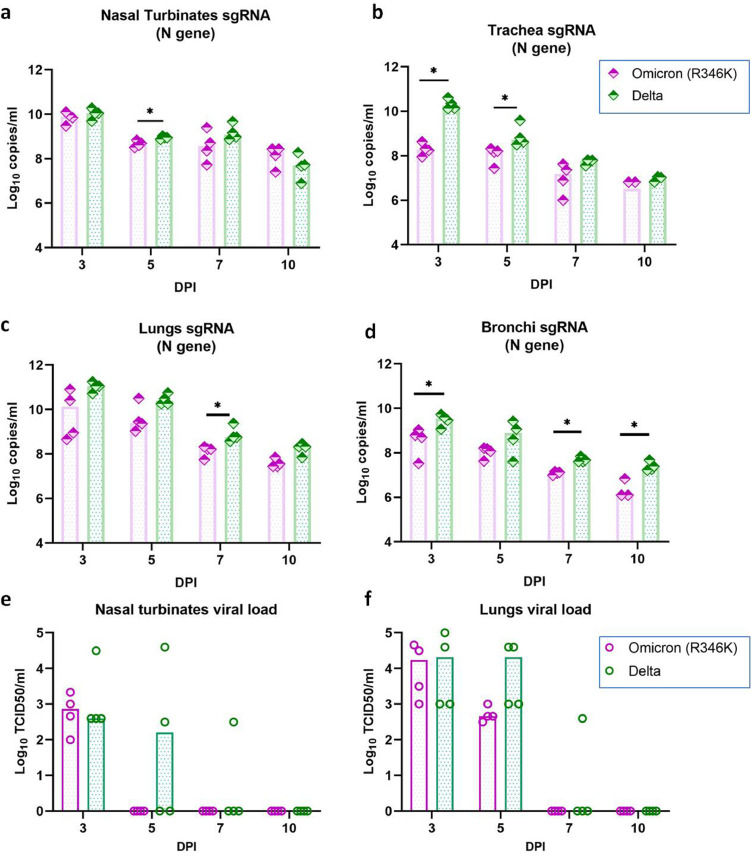
Viral RNA load in organs of hamsters after the SARS-CoV-2 Omicron (R346K) and Delta variant infection. The viral RNA load in (a) Nasal turbinates (b) trachea (n = 4, p = 0·0286 on 3DPI, p = 0·0286 on 7DPI, Mann-Whitney test) (c) Bronchi (n = 4, p = 0·0286 on 5DPI, Mann-Whitney test) and (d) Lungs estimated by E gene qRT-PCR. The bars represent the median and *,** and *** represents p < 0·05, p < 0·001 and p < 0·0001, respectively.
Figure 5.
Viral load in organs of hamsters after the SARS-CoV-2 Omicron (R346K) and Delta variant infection. Viral sgRNA load in (a) Nasal turbinates (n = 4, p = 0·0286 on 5DPI, Mann-Whitney test) (b) Trachea (n = 4, p = 0·0286 on 3DPI, p = 0·0286 on 5DPI, Mann-Whitney test) (c) Lungs (n = 4, p = 0·0286 on 7DPI, Mann-Whitney test) and (d) Bronchi (n = 4, p = 0·0286 on 3DPI, p = 0·0286 on 5DPI, p = 0·0286 on 7DPI, Mann-Whitney test) estimated by N gene qRT-PCR. Viral load (TCID50) in (e) Nasal turbinates and (f) Lung samples of hamsters. The bars represent the median and *,** and *** represents p < 0·05, p < 0·001 and p < 0·0001, respectively.
On comparison with the Delta variant infected hamsters, comparable viral RNA loads were observed in the lungs and nasal turbinates of the Omicron (R346K) infected hamsters. But the viral RNA load in the trachea on 3 and 5 DPI and bronchi samples on 5DPI were lower. The sgRNA levels in the Omicron (R346K) variant infected hamsters were also lower in the trachea on 3 and 5 DPI, in the nasal turbinates on 5 DPI, in the lungs on 7 DPI and in the bronchi on 3,7 and 10 DPI. The liver and intestine samples of 3 out of 4 Delta variant infected hamsters sacrificed on 3DPI were positive for viral RNA and all the other organs were found negative for viral RNA/sgRNA on 3DPI as well as on subsequent days.
On live virus titration, the titres of lungs and nasal turbinate samples did not show any statistically significant difference in both the Delta and Omicron (R346K) infected hamsters. Virus clearance from nasal turbinates in case of the Omicron (R346K) infection was found earlier by day 5 compared to Delta infection. No viral titre could be detected in the lungs and nasal turbinate samples on further days (Figure 5e, f).
Lung pathological changes in hamsters after infection
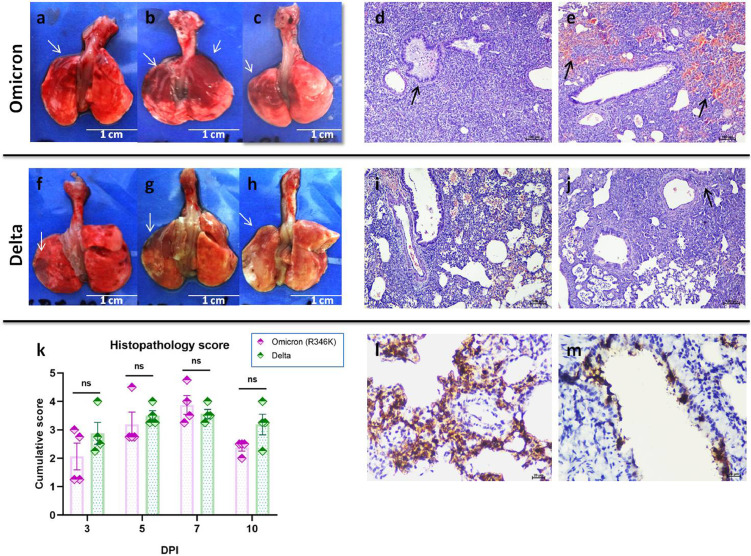
Gross changes like congestion and haemorrhages were visible in the lungs of hamsters from 3 DPI which became pronounced on 5 and 7 DPI in both the Omicron (R346K) and Delta variant infected hamsters (Figure 6a–c, f–h). Histopathologically early changes observed were congestion and bronchial epithelial necrosis with exudates on 3 DPI which progressed to diffuse alveolar damage on 5 and 7 DPI. Emphysema, haemorrhages, hyperplasia of alveolar epithelial cells, oedema and mononuclear cell infiltration were also observed in the alveolar parenchyma. Peribronchial and perivascular inflammatory cell infiltration was found during 7 DPI. The histopathological changes observed were similar in both the Omicron (R346K) and Delta variant infected hamsters and the cumulative histopathological score were comparable in both groups (Figure 6d, e, i–k). By 10 DPI, the severity of lesions observed were found to be reduced in the Omicron (R346K) infected group, whereas in the Delta variant group lesions were still prominent. The lungs sections from the mock infected control animals were found showing normal histology whereas in case of the Omicron (R346K)/Delta variant infected animals reparatory changes were prominent on 14 DPI (Supplementary Figure 2). Viral antigen could be detected in the bronchial epithelium, alveolar epithelial cells and macrophages. The immunostaining was prominent in the bronchiolar epithelial cells on 3 DPI and in both bronchiolar and alveolar on 5 and 7 DPI. On further days only faint or focal immunostaining was observed in the alveolar epithelial cells (Figure 6l, m).
Figure 6.
Pathological changes in hamsters after SARS-CoV-2 infection. Lungs of hamsters infected with the Omicron (R346K) variant (a–c) showing varying degree of consolidation and haemorrhages, scale bar = 1 cm. (d) Lungs sections showing bronchioles filled with exudates (arrow) and denuded epithelial cells as well as inflammatory cells, diffuse consolidation and alveolar septal thickening in the parenchyma as well as peribronchial and perivascular mononuclear cell infiltration post Omicron (R346K) infection, H& E, scale bar = 100 µm. (e) Lung section showing diffuse alveolar haemorrhages (arrows), peribronchial and perivascular mononuclear cell infiltration and bronchiolitis, post Omicron (R346K) infection, H& E, scale bar = 100 µm. Lungs of hamsters infected with the Delta variant (f–h) showing varying degree of consolidation and haemorrhages (arrows), scale bar = 1 cm. Lung sections from Delta variant infected hamsters showing (i) diffuse alveolar septal thickening, alveolar haemorrhages (j) diffuse alveolar septal thickening and peribronchial mononuclear cell infiltration (arrow), H& E, scale bar = 100 µm. (k) Cumulative histopathological score of lung lesions observed on 3, 5, 7 and 10 DPI in hamsters, which was scored out of 8, (n = 4, p= non-significant (ns), Mann-Whitney test). (l) Alveolar epithelial cells showing intense immunostaining on 5 DPI by immunohistochemistry, DAB, scale bar = 20 µm. (m) Bronchiolar epithelial cells showing intense immunostaining on 5 DPI by immunohistochemistry, DAB, scale bar = 20 µm.
SARS-CoV-2 variant analysis
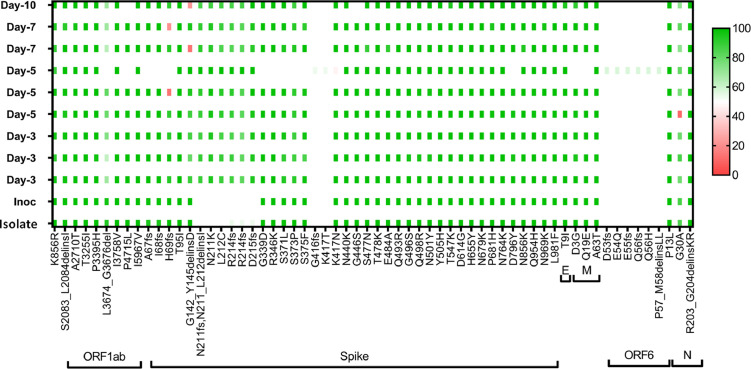
We performed the variant analysis of the Omicron (R346K) virus isolate, inoculum as well as the Omicron (R346K) infected lung samples collected on 3, 5, 7 and 10 DPI. The analysis showed no additional mutations in the genome other than that of the virus isolate (Figure 7).
Figure 7.
SARS-CoV-2 variant analysis. The plot showing the amino acid substitutions in the virus inoculum and lung samples post infection on 3, 5, 7 and 10 DPI.
Discussion
The Omicron variant is spreading worldwide at an alarming rate and the researchers across the globe are putting their best efforts to understand the characteristics of the virus. The Omicron variant we used in this study had an R346K substitution unlike other studies available in public domain. Though specific role of this mutation is not well studied but the substitution is being monitored.
The disease characteristics of the Omicron (R346K) variant observed here are similar to those reported for other SARS-CoV-2 variants like Alpha, Beta, Kappa and Delta in hamsters by earlier studies except that of the body weight loss.12,21,36 Our finding corroborated with the few recent studies which also reported the limited or negligible body weight loss in hamsters post infection with Omicron variants of B.1.1.529/ BA.1 lineage.24,27, 28, 29 These findings suggest that the pathogenicity of circulating Omicron lineages can differ. A recent study on the BA.2 infection in hamster model demonstrated higher pathogenicity in comparison to the BA.1 variant.37 The age and virus dose can affect the body weight loss and disease severity in hamsters post SARS-CoV-2 infection.10,20 We have used 5–6 week old hamsters and other published studies on Omicron infection in Syrian hamsters have also used animals of the age range of 4–8 weeks except Ryan et al., have used 20–22 week animals.23, 24, 25, 26, 27, 28 Even high Omicron virus doses could not result in body weight loss in hamsters.26 In the present study we have used only a single dose for virus infection.
The viral RNA shedding and live virus clearance observed here was similar to earlier VOCs studied in hamster model.22,36 Replication competent virus could be detected in NW till 5 DPI. The clearance of virus was observed by 1 week in hamsters. Virus shedding period found in human Omicron cases are also suggesting that infectious period is lesser than 10 days since symptom onset.38
We observed moderate to severe changes in the lungs of hamsters infected with Omicron (R346K). In contrary to our findings, a recent ex vivo study reported that Omicron infectivity of lungs is lower in comparison to the Delta variant.39 Lung pathology observed in the Omicron BA.1 studies in hamsters also reports that lung disease is milder in comparison to the ancestral strains.24, 25, 26,28 The virus dose used in each study differed and the time point used for assessing histopathological disease score also varied. Here we have selected multiple time points to understand the difference in lung disease severity. But we have used only a single virus dose for infection. In comparison to our earlier studies of SARS-CoV-2 variants in Syrian hamster model and also the Delta variant infected group used for comparison, the lung lesions observed with Omicron (R346K) was prominent. We also observed higher viral load in the lungs similar to that of upper respiratory tract in hamsters. The virus titration showed early virus clearance from the nasal turbinates by day 5 and from lungs by day 7. We found no evidence that Omicron (R346K) produce less severe lung disease or more predilection to upper respiratory tract in comparison to lung as reported earlier.23, 24, 25, 26, 27, 28, 29
Neutralizing antibodies could be detected from 5th DPI in Omicron (R346K) infected hamsters, which showed an increase in titre on further days similar to other VOCs.21 The serum samples of hamster infected with Omicron (R346K) showed poor neutralization against other VOCs tested like Alpha, Beta and Delta variants. As Omicron variant harbours around 26–32 mutations in the spike protein RBD and NTD, which are the most antigenic/ immunodominant region of the virus, immune evasion was speculated for the variant.40 It harbours N501Y, K417N, T478K, E484A which are amino acid substitutions linked with immune escape.41 In addition virus isolate used in study also possess R346K mutation which is also present in the Mu variant known for immune escape.42 So far, neutralization studies on convalescent sera of Omicron infected patients and animal models are not available. The studies published on the neutralization potential of the convalescent sera samples of COVID-19 recovered as well as vaccinated individuals showed poor neutralization against the Omicron variant.43,44 Our preliminary data suggests that humoral immune response generated by Omicron (R346K) infection may not confer much protection to other currently circulating variants like Delta. The other studied variants like B.1, Beta and Alpha variants have almost disappeared from circulation globally. This suggests constant monitoring of immunized population for their antibody levels and studies on vaccine efficacy against emerging variants. The antigenicity of the BA.2 variant is also reported to differ from BA.1 and have shown resistance to the BA.1 generated humoral immune response.37
Here, we have characterized an Omicron isolate of BA.1.1 sub lineage with R346K mutation in Syrian hamster model. We observed limited body weight gain, viral replication in the upper and lower respiratory tract and interstitial pneumonia in Syrian hamsters following infection with the Omicron (R346K) variant. The lung disease severity was comparable to that of the Delta variant. Moreover, the neutralizing antibodies generated after Omicron (R346K) infection showed substantial reduction or negligible neutralization against other variants like B.1, Alpha, Beta and Delta. The presented data provides new insight into the neutralization potential and cross protection of Omicron (R346K) variant against other VOCs and suggest that the pathogenicity of the circulating Omicron lineages could differ.
Contributors
PD Yadav conceived and designed the study. S Mohandas performed the animal experiments. A Shete co-ordinated and performed the real time PCR assays, sample processing and testing. G Sapkal and G Deshpande performed the PRNT assays. D Nyayanit performed the statistical analysis. R Jain and A Shete performed the ELISA based experiments. M Kadam and A Kumar performed the daily sampling and sample processing.C Mote performed the histopathological and immunohistochemical processing and examination. P D Yadav, and S Mohandas verified the data, analysed and interpreted the results. S Mohandas and DY Patil wrote the preliminary manuscript draft. All authors read, approved and substantially revised the final version of the manuscript. PD Yadav, S Mohandas and A Shete have accessed and verified the data, and decided to submit the manuscript.
Declaration of interests
The authors declare no competing financial interests.
Acknowledgments
Acknowledgments
This study was supported by Indian Council of Medical Research as an intramural grant (COVID-19) to ICMR-National Institute of Virology, Pune. Authors acknowledge the support of Prof. Priya Abraham, Director, ICMR-NIV, Pune. The authors also acknowledge the support received from the laboratory team of Maximum Containment Facility of ICMR-NIV, Pune ie., Mr Prasad Sarkale, Mr Hitesh Dighe and Mr Rajen Lakra for tissue culture support, Mr Annasaheb Suryawanshi and Mr Kundan Wakchaure for laboratory animal care and sample processing, Mrs Kaumudi Kalele, Ms Jyoti Yemul, Ms Pranita Gawande, Mrs Poonam Bhodke, Mrs Priyanka Waghmare, Mrs Shilpa Ray for sample testing and Dr Abhinendra Kumar, Mrs Triparna Majumdar, Mrs Savita Patil, Mr Yash Joshi and Ms Manisha Dudhmal for support in sequencing and analysis.
Data sharing statement
All the data pertaining to the study are available in the manuscript or in the supplementary materials.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103997.
Appendix. Supplementary materials
References
- 1.World Health Organization. Weekly epidemiological update on COVID-19 - 22 March 2022, edition 84. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19—22-march-2022. Accessed 25 March 2022.
- 2.United Kingdom Health Security Agency. SARS-CoV-2 variants of concern and variants under investigation in England 2021: technical briefing 34, 14 January 2022. https://www.gov.uk/government/publications/investigation-of-sars-cov-2-variants-technical-briefings. Accessed 14 January 2022.
- 3.Venkatakrishnan AJ, Anand P, Lenehan PJ, et al. Omicron variant of SARS-CoV-2 harbors a unique insertion mutation of putative viral or human genomic origin 2021. OSF preprints, 2021. doi: 0.31219/osf.io/f7txy. [Preprint].
- 4.PANGO lineages: Latest Epidemiological lineages of SARS-CoV-2, Lineage list: Lineage Omicron. https://cov-lineages.org/lineage_list.html. Accessed 25 March 2022.
- 5.World Health Organization. Tracking of SARS-CoV-2 Variants 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Accessed 25 March 2022.
- 6.Schmidt F., Muecksch F., Weisblum Y., et al. Plasma neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iketani S., Liu L., Guo Y., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022:1. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Global Initiative on Sharing All Influenza Data (GISAID) database. Tracking of variants. https://www.gisaid.org/. Accessed 25 March 2022.
- 9.de Vries R.D., Rockx B., Haagmans B.L., Herfst S., Koopmans M.P., de Swart R.L. Animal models of SARS-CoV-2 transmission. Curr Opin Virol. 2021;50:8–16. doi: 10.1016/j.coviro.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai M., Iwatsuki-Horimoto K., Hatta M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohandas S., Jain R., Yadav P.D., et al. Evaluation of the susceptibility of mice & hamsters to SARS-CoV-2 infection. Indian J Med Res. 2020;151:479–482. doi: 10.4103/ijmr.IJMR_2235_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdelnabi R., Boudewijns R., Foo C.S., et al. Comparing infectivity and virulence of emerging SARS-CoV-2 variants in Syrian hamsters. EBioMedicine. 2021;68 doi: 10.1016/j.ebiom.2021.103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohandas S., Yadav P.D., Nyayanit D., et al. Comparison of SARS-CoV-2 variants of concern 202012/01 (U.K. variant) and D614G variant transmission by different routes in Syrian hamsters. Vector Borne Zoonotic Dis. 2021;21:638–641. doi: 10.1089/vbz.2021.0032. [DOI] [PubMed] [Google Scholar]
- 14.Horiuchi S., Oishi K., Carrau L., et al. Immune memory from SARS-CoV-2 infection in hamsters provides variant-independent protection but still allows virus transmission. Sci Immunol. 2021;6:eabm3131. doi: 10.1126/sciimmunol.abm3131. [DOI] [PubMed] [Google Scholar]
- 15.Tostanoski L.H., Yu J., Mercado N.B., et al. Immunity elicited by natural infection or Ad26.COV2.S vaccination protects hamsters against SARS-CoV-2 variants of concern. Sci Transl Med. 2021;13:eabj3789. doi: 10.1126/scitranslmed.abj3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trimpert J., Adler J.M., Eschke K., et al. Live attenuated virus vaccine protects against SARS-CoV-2 variants of concern B.1.1.7 (Alpha) and B.1.351 (Beta) Sci Adv. 2021;7:eabk0172. doi: 10.1126/sciadv.abk0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mok B.W.Y., Liu H., Deng S., et al. Low dose inocula of SARS-CoV-2 Alpha variant transmits more efficiently than earlier variants in hamsters. Commun Biol. 2021;4:1–8. doi: 10.1038/s42003-021-02640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohandas S., Yadav P.D., Shete-Aich A., et al. Immunogenicity and protective efficacy of BBV152, whole virion inactivated SARS- CoV-2 vaccine candidates in the Syrian hamster model. IScience. 2021;24 doi: 10.1016/j.isci.2021.102054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdelnabi R., Foo C.S., De Jonghe S., Maes P., Weynand B., Neyts J. Molnupiravir inhibits replication of the emerging SARS-CoV-2 variants of concern in a hamster infection model. J Infect Dis. 2021;224:749–753. doi: 10.1093/infdis/jiab361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yinda C.K., Port J.R., Bushmaker T., et al. Prior aerosol infection with lineage A SARS-CoV-2 variant protects hamsters from disease, but not reinfection with B.1.351 SARS-CoV-2 variant. Emerg Microbes Infect. 2021;10:1284–1292. doi: 10.1080/22221751.2021.1943539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohandas S., Yadav P.D., Shete A., et al. SARS-CoV-2 delta variant pathogenesis and host response in Syrian hamsters. Viruses. 2021;13:1773. doi: 10.3390/v13091773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohandas S., Yadav P.D., Shete A., et al. Protective Immunity of the primary SARS-CoV-2 infection reduces disease severity post re-infection with Delta variants in Syrian hamsters. Viruses. 2022;14:596. doi: 10.3390/v14030596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan K.A., Watson R.J., Bewley K.R., et al. Convalescence from prototype SARS-CoV-2 protects Syrian hamsters from disease caused by the Omicron variant. bioRixv. 2021 doi: 10.1101/2021.12.24.474081. [Preprint] [DOI] [Google Scholar]
- 24.Suzuki R., Yamasoba D., Kimura I., et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603:700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shuai H., Chan J.F.W., Hu B., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B1.1.529 Omicron. Nature. 2022;603(7902):693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 26.McMahan K., Giffin V., Tostanoski L.H., et al. Reduced Pathogenicity of the SARS-CoV-2 Omicron Variant in Hamsters. Med N Y. 2022 doi: 10.1016/j.medj.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdelnabi R., Foo C.S., Zhang X., et al. The omicron (B.1.1.529) SARS-CoV-2 variant of concern does not readily infect Syrian hamsters. Antiviral Res. 2022;198 doi: 10.1016/j.antiviral.2022.105253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley EG, Kirby A, Sharma P, et al. SARS-CoV-2 Omicron-B.1.1.529 Variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19. biorixv 2021. 10.1101/2021.12.26.474085.
- 30.Yadav P.D., Gupta N., Potdar V., et al. Isolation and genomic characterization of SARS-CoV-2 omicron variant obtained from human clinical specimens. Viruses. 2022;14:461. doi: 10.3390/v14030461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudhary M.L., Vipat V., Jadhav S., et al. Development of in vitro transcribed RNA as positive control for laboratory diagnosis of SARS-CoV-2 in India. Indian J Med Res. 2020;151:251–254. doi: 10.4103/ijmr.IJMR_671_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perera R.A., Tso E., Tsang O.T., et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis. 2020;26:2701. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shete A., Mohandas S., Jain R., Yadav P.D. A qualitative IgG ELISA for detection of SARS-CoV-2-specific antibodies in Syrian hamster serum samples. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deshpande G.R., Sapkal G.N., Tilekar B.N., et al. Neutralizing antibody responses to SARS-CoV-2 in COVID-19 patients. Indian J Med Res. 2020;152(1–2):82–87. doi: 10.4103/ijmr.IJMR_2382_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Culling C.F.A. 3rd ed. Butterworths; 1974. Handbook of Histopathological and Histochemical Techniques. [Google Scholar]
- 36.Mohandas S., Yadav P.D., Nyayanit D., et al. Comparison of the pathogenicity and virus shedding of SARS CoV-2 VOC 202012/01 and D614G variant in hamster model. bioRixv. 2021 doi: 10.1101/2021.02.25.432136. [Preprint] [DOI] [Google Scholar]
- 37.[Preprint]. Yamasoba D., Kimura I., Nasser H., et al. Virological characteristics of SARS-CoV-2 BA.2 variant. bioRxiv. 2022;2022 doi: 10.1101/2022.02.14.480335. [Preprint] [DOI] [Google Scholar]
- 38.National Institute of Infectious Diseases, Japan. Active epidemiological investigation on SARS-CoV-2 infection caused by Omicron variant (Pango lineage B.1.1.529) in Japan: preliminary report on infectious period 2022, January 5, 2022. https://www.niid.go.jp/niid/en/2019-ncov-e/10884-covid19-66-en.html. Accessed 14 January 2022.
- 39.Li Ka Shing Faculty of Medicine, The University of Hong Kong. HKUMed finds Omicron SARS-CoV-2 can infect faster and better than Delta in human bronchus but with less severe infection in lung, 15 December, 2021. https://www.med.hku.hk/en/news/press/20211215-omicron-sars-cov-2-infection. Accessed 14 January 2022.
- 40.Harvey W.T., Carabelli A.M., Jackson B., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazarevic I., Pravica V., Miljanovic D., Cupic M. Immune evasion of SARS-CoV-2 emerging variants: what have we learnt so far? Viruses. 2021;13:1192. doi: 10.3390/v13071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uriu K., Kimura I., Shirakawa K., et al. Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N Engl J Med. 2021;385:2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rössler A., Riepler L., Bante D., von Laer D., Kimpel J. SARS-CoV-2 omicron variant neutralization in serum from vaccinated and convalescent persons. N Engl J Med. 2022;386:698–700. doi: 10.1056/NEJMc2119236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carreño J.M., Alshammary H., Tcheou J., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2021;602(7898):682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.