Abstract
Coronavirus disease-2019 (COVID-19) is a new types of coronavirus which have turned into a pandemic within a short time. Reverse transcription–polymerase chain reaction (RT-PCR) test is used for the diagnosis of COVID-19 in national healthcare centers. Because the number of PCR test kits is often limited, it is sometimes difficult to diagnose the disease at an early stage. However, X-ray technology is accessible nearly all over the world, and it succeeds in detecting symptoms of COVID-19 more successfully. Another disease which affects people’s lives to a great extent is colorectal cancer. Tissue microarray (TMA) is a technological method which is widely used for its high performance in the analysis of colorectal cancer. Computer-assisted approaches which can classify colorectal cancer in TMA images are also needed. In this respect, the present study proposes a convolutional neural network (CNN) classification approach with optimized parameters using gradient-based optimizer (GBO) algorithm. Thanks to the proposed approach, COVID-19, normal, and viral pneumonia in various chest X-ray images can be classified accurately. Additionally, other types such as epithelial and stromal regions in epidermal growth factor receptor (EFGR) colon in TMAs can also be classified. The proposed approach was called COVID-CCD-Net. AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet architectures were used in COVID-CCD-Net, and the hyperparameters of this architecture was optimized for the proposed approach. Two different medical image classification datasets, namely, COVID-19 and Epistroma, were used in the present study. The experimental findings demonstrated that proposed approach increased the classification performance of the non-optimized CNN architectures significantly and displayed a very high classification performance even in very low value of epoch.
Graphical abstract
Keywords: Colon cancer diagnosis, Convolutional neural network (CNN), COVID-19, Gradient-based optimizer (GBO), Hyperparameter optimization
Introduction
COVID-19 broke out in the world in early December 2019 and rapidly turned into a pandemic. According to the World Health Organization (WHO) data, 227,940,972 people have been infected, while 4,682,899 people have been killed by the disease around the world until today [1]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus which has caused COVID-19 pandemic [2]. Common symptoms of COVID-19 pandemic can be listed as fever, muscle pain, dry cough, head ache, a sore throat ve chest pain [3, 4]. Due to these sypmtoms, COVID-19 has been accepted as a respiratory tract disease. It may take these symptoms 2 to 14 days to appear in a person who has been infected with the virus [5]. Despite recent attempts at finding a treatment method, such as a drug or vaccine, against the disease, no viable solutions to COVID-19 have been found yet. Various medical imaging techniques such as X-ray and computed tomography (CT) can be considered as important tools in the diagnosis of COVID-19 cases [6, 7]. Coronavirus usually causes lung infections. Therefore, chest X-ray and CT images are widely used by physicians and radiologists for an accurate and quick diagnosis in the patients infected with the virus.
Polymerase chain reaction (PCR) test method is widely used for the diagnosis of COVID-19. However, the test is not always accessible at all healthcare points. It must be also noted that compared to PCR tests, X-ray and CT-based imaging techniques are usually more reliable and accessible. When CT and X-ray methods are compared, X-ray machines are preferred more by radiologists and physicians because of their accessibility nearly in every location including remote rural areas, cost-effectiveness, and their capacity to perform imaging in a fairly short period of time [5]. However, it is also time-consuming for physicians and radiologists to evaluate the patients’ X-ray images. Furthermore, it also runs the risk of inaccurate diagnosis because the detection of infected areas in an image requires technical know-how and medical experience. Therefore, an accurate and quick computer-assisted diagnosis system is needed for COVID-19 cases. The following literature review indicated that deep learning (DL) algorithms were used in order to diagnose COVID-19 in X-ray images succesfully [5, 8–12].
Introduced by Kononen [13] in 1998, tissue microarray (TMA) is an innovative and high-performance technique used for the analysis of multiple tissue samples. It is a high-end technology with a remarkable performance and has been used in the analysis of molecular identifiers recently. There is sufficient evidence to claim that epidermal growth factor receptor (EGFR) plays an important role in tumor development [14]. In parallel with this, it was also observed that EGFR played an important role in the initation and progress of colorectal cancer [15].
The present study proposes a convolutional neural network (CNN) classification approach with optimized hyperparameters using gradient-based optimizer (GBO) algorithm [16]. CNN is the most widely used DL model. The proposed approach was used to classify COVID-19, normal, and viral pneumonia. In addition, it can be also used to classify other types such as epithelial and stromal regions in EFGR-colon in digitized tumor TMAs.
Real-world applications in many different fields such as medicine, agriculture, and engineering can be approached as an optimization problem. To this day, numerous optimization approaches have been developed in order to solve real-world problems in an effective way. However, high-performance optimization approaches are needed due to the fact that the difficulty of these optimization problems is increasing day by day. In this respect, metaheuristic algorithms (MAs), which are known as global optimization techniques, have been widely used to solve challenging optimization problems [17–22].
Artificial neural network (ANN) is an important machine learning approach inspired by the neural system in human mind. It involves an input layer, hidden layer, and output layer, and aims to adjust optimal values in relation with the weight of each neuron in ANN following a training process [23]. The performance of an ANN structure is heavily affected by the number and variety of training data. If an insufficient number of data is used in the training process, the performance of ANN is very likely to decrease.
Various changes have been so far applied to ANN structure to design feedback and multi-layer model structures, which paved the way for the solution of non-linear problems. With the advent of multi-layer neural network models, the number of layers in an ANN structure has also increased and led to the development of CNN, which is a high-performance version of ANN models. Introduced during the 1990s, CNN was not preferred due to computer hardware incapacity in this period [23]. However, thanks to the technological developments in computer hardware and graphical processing unit (GPU) in the following years, CNN performances have also increased remarkably in recent years, and it became one of the most widely used machine learning approaches in various fields such as health, transportation, security, stock exchange, and law.
Various CNN architectures have been so far proposed in the existing literature, as manifested by several examples such as MobileNet-V2, ShuffleNet, GoogleNet, VGG-16, VGG-19, and AlexNet. In these CNN architectures, hyperparameters such as learning rate, solver, L2 regularization, gradient threshold method, and gradient threshold are known to affect CNN performance directly. Therefore, it is not surprising that various studies in the existing literature attempted to offer solutions to the optimization of these hyperparameters.
The present study benefited from AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet architectures for the proposed approach, i.e., a COVID-19 and colon cancer diagnosis system with optimized hyperparameters using GBO. In order to optimize hyperparameters such as learning rate, solver, L2 regularization, gradient threshold method, and gradient threshold in these architectures, GBO algorithm proposed by Ahmadianfar et al. [16] was used in the present study. Inspired by Newton’s method, GBO is one of the most recent metaheuristic optimization approaches. The present study aims to optimize hyperparameters in AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet and increase its classification performance.
The main contributions of the present study can be summarized as follows:
The present study proposes a high-performance approach which can classify both COVID-19 and colon cancer in TMAs. No approach which can classify both diseases has been so far proposed in the current literature.
The proposed COVID-CCD-Net approach benefits from GBO [16] algorithm proposed in 2020 in order to optimize hyperparameters in AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet.
The present study aims to obtain a high level of accuracy with a low value of epoch in AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet architectures in the proposed COVID-CCD-Net approach. On the other hand, the non-optimized CNN methods obtained a much lower level of accuracy with the same value of epoch.
The organization of the present study is as follows: “Section 2” describes the related works. “Section 3” presents gradient-based optimizer and convolutional neural networks. “Section 4” describes the proposed COVID-CCD-Net approach. “Section 5” presents experiments and results, and Section 6 concludes the study.
Related works
Hyperparameter optimization
In order to optimize hyperparameters in CNN, various approaches such as adaptive gradient optimizer [24], Adam optimizer [25], Bayesian optimization [26], equilibrium optimization [27], evolutionary algorithm [28], genetic algorithm [29], grid search [30], particle swarm optimization [31, 32], random search [30, 33], simulating annealing [33], and tree-of-parzen estimators [33], whale optimization algorithm [34], and weighted random search [35] have been so far proposed. random search, simulating annealing, and tree-of-parzen estimators.
In addition to its comprehensivess as a searching algorithm, grid search aims to identify the most optimal values for hyperparameters through a manually specified subset of hyperparameter space [36]. However, since the grid of configurations grows exponentially depending on the number of hyperparameters during the hyperparameter optimization process, the algorithm is not often useful for the optimization of deep neural networks [36]. During the hyperparameter optimization in CNN, it may take a few hours or a whole day to evaluate a hyperparameter selection, which causes serious computational problems. Similar to grid search algorithm, random search algorithm too encounters various disadvantages in sampling a sufficient number of points to be evaluated [37].
Bayesian optimization has been a popular technique for hyperparameter optimization recently [38]. One of the main advantages in Bayesian optimization–based neural network optimization is that it does not require running neural network completely. On the other hand, its complexity and high-dimensional hyperparameter space makes Bayesian optimization an impractical and expensive approach for hyperparameter optimization [36].
One of the biggest disadvantages of genetic algorithm is that it usually becomes stuck in a local optimal value and, as a result, results in yielding early convergence and non-optimal solutions [39]. Therefore, hyperparameter optimization techniques which benefit from genetic algorithm–based approaches are also likely to be problematic.
Lima [33] compared various hyperparameter optimization algorithms such as random search, simulating annealing, and tree-of-parzen estimators in order to find the most effective CNN architecture in the classification of benign and malignant small pulmonary nodules. Kumar and Hati [24] proposed the adaptive gradient optimizer–based deep convolutional neural network (ADG-dCNN) approach for bearing and rotor faults detection in squirrel cage induction motor. Ilievski et al. [40] used radial basis function (RBF) as a surrogate of hyperparameter optimization in order to reduce the complexity of original network. Talathi [41] proposed a simple sequential model based optimization algorithm in order to optimize hyperparameters in deep CNN architectures.
Rattanavorragant and Jewajinda proposed an approach using an island-based genetic algorithm in order to optimize hyperparameters in DNN automatically [42]. This approach involves two steps: hyperparameter search and a detailed DNN training. Navaneeth and Suchetha proposed the optimized one-dimensional CNN with support vector machine (1-D CNN-SVM) approach in order to diagnose chronic kidney diseases using PSO algorithm [43].
Compared to the literature review above, the main contribution of the present study is that the proposed COVID-CCD-Net approach can detect two important diseases: COVID-19 and colon cancer in TMAs. In addition, the proposed approach benefits from GBO, which is a metaheuristic approach, for the optimization of CNN models to overcome various problems mentioned in the existing literature.
Deep learning approaches for COVID-19
In recent times, many studies focusing on the diagnosis of COVID-19 using CNN have been published [44–50]. The literature review indicates that some of these studies [45–47] focused on the diagnosis of COVID-19 in non-COVID cases. On the other hand, there are also studies which classified cases into three groups as COVID, normal, and pneumonia [48–50]. Within the framework of the present study, the proposed COVID-CCD-Net approach classifies chest X-ray images into three different groups as COVID, normal, and pneumonia.
Shi et al. [51] performed a detailed literature review regarding the state-of-the-art computer-assisted methods for the diagnosis of COVID-19 in X-ray and CT scans. Castiglioni et al. [52] benefited from two chest X-ray datasets containing 250 COVID-19 and 250 non-COVID cases in order to perform training, validation, and testing processes for Resnet-50.
Hemdan et al. [53] proposed a deep learning–based approach called COVIDX-Net in order to diagnose COVID-19 in chest X-ray images automatically. This study involved seven different deep architectures, namely MobileNetV2, VGG19, InceptionV3, DenseNet201, InceptionResNetV2, ResNetV2, and Xception. Khan et al. [54] proposed a CNN-based approach called CoroNet in order to diagnose COVID-19 using X-ray and CT scans based on Xception architecture. The experimental studies demonstrated that the proposed model yielded an overall accuracy rate of 89.6% in four different classes (COVID vs. pneumonia bacterial vs. pneumonia viral vs. normal) and an overall accuracy rate of 95% in three different classes (normal vs. COVID vs. pneumonia).
The proposed COVID-CCD-Net approach differs from other studies on the detection of COVID-19 using CNN models in that it improves classification performance by optimizing hyperparameters of CNN models thanks to GBO approach.
Computer-aided colon cancer detection approaches
As can be seen in various studies in the existing literature, the number of studies dealing with automatic diagnosis of colon cancer in TMAs is limited. Nguyen et al. [55] analyzed different ensemble approaches for colorectal tissue classification using highly efficient TMAs and proposed an ensemble deep learning–based approach with two different neural network architectures called VGG16 and CapsNet. Thanks to this approach, they classified colorectal tissues in highly efficient TMAs into three different categories, namely tumor, normal, and stroma/others.
Xu et al. [56] proposed a deep CNN approach in order to perform the segmentation and classification of epithelial ve stromal regions in TMAs. This study benefited from two different datasets containing breast and colorectal cancer images. Finally, Linder et al. [57] proposed an approach for an automatic detection of epithelial ve stromal regions in colorectal cancer TMAs thanks to texture features and a SVM classifier.
The proposed COVID-CCD-Net approach is superior to other studies on the detection of colon cancer in TMAs using CNN models in that it optimizes the hyperparameters of CNN models, which significantly increases the detection accuracy rates of colon cancer. The effective performance of CNN in image classification contributes to the present study to a higher extent compared to other studies using other approachs for the classification of colon cancer in TMAs in the existing literature.
Theoretical background
Gradient-based optimizer
Inspired by gradient-based Newton’s method, GBO was proposed by Ahmadianfar et al. [16] as one of the most recent metaheuristic algorithms. This algorithm is based on two main operators: gradient search rule (GSR) and local escaping operator (LEO). Main steps of GBO are described below.
Initialization process
In GBO, each member of the population is called a “vector” and, as seen in Eq. 1, the population consists of N number of vectors in a D-dimension search space.
| 1 |
As shown in Eq. 2, each vector in the initial population is created by assigning random values within the boundaries of search space.
| 2 |
Here, Xmin and Xmax are lower and upper boundaries in the search space, respectively, while rand(0,1) is a random number in a range of [0,1].
Gradient search rule
GSR operator is used in GBO in order to increase exploration ability, eliminate local minimum, and accelerate the convergence rate. Thus, optimal solutions can be obtained within the search space [16].
The position of a vector in the next iteration (xnm+1) is calculated using Eqs. 3 and 4 with: X1nm, X2nm, and xnm, which denotes the current position of the vector.
| 3 |
| 4 |
ra and rb are random numbers in a range of [0, 1]. X1nm and X2nm in this equation are shown in the following equations:
| 5 |
| 6 |
Here, xnm and xbest are the current position and the best vector in the population, respectively. GSR denotes the gradient search rule, while DM represents the direction of movement. GSR enables GBO to assign randomly, improve its exploration ability and eliminate local minimals. GSR can be calculated as shown in the following equations [16]:
| 7 |
| 8 |
| 9 |
| 10 |
Here, rand(1:N) is an N-dimensional random number, r1, r2, r3, and r4 denote random integer numbers selected from a range of [1, N], and, finaly, step represents the step size.
DM shown in Eq. 11 helps the current position of the vector (xn) move along the direction of xbest - xn and thus provides local searching in order to improve convergence speed of GBO [16].
| 11 |
Global exploration and local exploitation must be balanced in an algorithm in order to find solutions closer to a global optimal value. p1 and p2 parameters in Eqs. 4, 7, and 11 are used to balance exploration and exploitation in GBO [16]. These parameters are calculated using the following equations:
| 12 |
| 13 |
| 14 |
Here, βmin and βmax are 0.2 and 1.2, respectively, and m denotes the current number of iteration. M represents the maximum number of iteration.
Local escaping operator
LEO is used to improve efficiency of GBO. It can change the position of xnm+1 vector significantly. Thanks to LEO, XLEOm, which is a new vector, is created as shown in Eqs. 15 and 16, and assigned to xnm+1 vector, as shown in Eq. 17.
| 15 |
| 16 |
| 17 |
Here, f1 and f2 are random numbers generated in a range of [−1, 1], and u1, u2, and u3 are three randomly generated and different numbers, while xkm is a newly generated vector. u1, u2, u3, and xkm are defined as shown in the following equations:
| 18 |
| 19 |
| 20 |
| 21 |
Here, rand, μ1, and μ2 are random numbers in a range of [0, 1], xrand denotes a randomly generated new vector, and xpm is a vector randomly selected from the population [16]. Flowchart of the GBO is shown in Fig. 1.
Fig. 1.

Flowchart of the GBO
Convolutional neural networks
Convolutional neural networks (CNN) is a special type of neural network inspired by the biological model of animal visual cortex [58, 59]. They are particularly used in the field of image and sound processing due to their main advantage: the extraction of automatic and adaptive features during a training process [60]. In CNNs, the variable of the network structure (kernel size, stride, padding, etc.) and the network trained (learning rate, momentum, optimization strategies, batch size etc.) are known as hyperparameters [29], which must be adjusted accurately for a more effective CNN performance.
In the present study, learning rate, solver, L2 regularization, gradient threshold method, and gradient threshold value, which are among network trained hyperparameters of AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet, were optimized using GBO algorithm. Learning rate, which is also known as step size, is decisive in terms of updating weights [61, 62]. Solver, on the other hand, represents the optimization method to be used such as Adam, Sgdm, or Rmsprop [63]. The L2 regularization, which is also called weight decay, is a simple regularization method that scales weights down in proportion to their current size [64, 65]. Gradient threshold method and gradient threshold value are parameters related to gradient clipping. If the gradient increases exponentially in magnitude, it means that the training is unstable and can diverge within a few iterations. Gradient clipping helps avoid the exploding gradient problem. If the gradient exceeds the value of gradient threshold, then the gradient is clipped according to gradient threshold method [66, 67].
Input image size in AlexNet architecture, developed by Krizhevsky et al. [68], is 227×227. It consists of 5 convolution and 3 fully connected layers, thus reaching a depth of 8 layers. DarkNet-19 has a depth of 19 layers and its input image size is 256×256 [69]. Introduced by Szegedy et al. [70], Inception-v3 model has a depth of 48 layers with an input image size of 299×299. ResNet-18, which has a depth of 18 layers and an input image size of 224×224, was developed by He et al. [71]. Zhang et al. [72] proposed ShuffleNet model with a depth of 50 layers and an input image size of 224×224. Finally, MobileNet, which was proposed by Sandler et al. [73], has a depth of 53 layers and an input image size of 224×224.
Hyperparameter optimization of CNN models using gradient-based optimizer
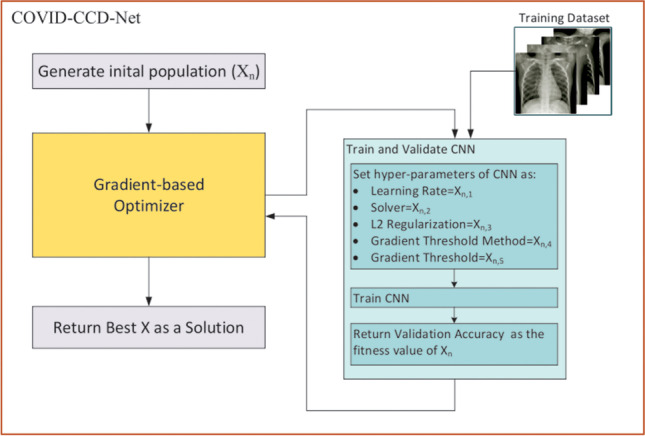
In the present study, hyperparameters of AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet CNN models such as learning rate, solver, L2 regularization, gradient threshold method, and gradient threshold value were optimized using GBO algorithm in order to classify COVID-19, normal, and viral pneumonia in chest X-ray images. In addition, other types such as epithelial and stromal regions in epidermal growth factor receptor (EFGR) colon in TMAs can also be classified. The proposed approach is called COVID-CCD-Net, as shown in the flowchart in Fig. 2.
Fig. 2.
Flowchart of COVID-CCD-Net
In the proposed COVID-CCD-Net approach, initial parameters of GBO such as ε, the number of population and maximum number of iteration are adjusted. Then, an initial population is created by using vectors with randomly assigned values. Each vector consists of 5 dimensions which represent learning rate, solver, L2 regularization, gradient threshold method, and gradient threshold parameters of CNN models. Lower boundary (LB) and upper boundary (UB) values of these parameters are given in Table 1. Learning rate, L2 regularization, and gradient threshold are real values which are randomly generated between LB and UB values. If the solver value is 1, 2, or 3, “sgdm,” “adam,” and “rmsprop” optimization method is selected, respectively. If the gradient threshold method value is 1, 2, or 3, “l2norm,” “global-l2norm,” and “absolute-value” method is selected, respectively. In parallel with these boundaries, each vector in the initial population is generated using the formula in Eq. 22:
| 22 |
Table 1.
Hyperparameters to be optimized and their ranges
| Parameter | LB | UB |
|---|---|---|
| Learning rate | 0.00001 | 0.01 |
| Solver | 1 | 3 |
| L2 regularization | 0.00001 | 0.01 |
| Gradient threshold method | 1 | 3 |
| Gradient threshold | 0.1 | 10 |
The following steps are taken in order to calculate the fitness value of each vector: Firstly, Xn vector whose fitness value will be calculated is sent to CNN model and the values of Xn vector are assigned to learning rate, solver, L2 regularization, gradient threshold method, and gradient threshold parameters of CNN model. Later, CNN model is trained using the training dataset. Following the training processes, validation accuracy value obtained from the training is sent back to GBO and assigned as the fitness value of Xn vector.
As shown in Fig. 2, each step of the algorithm is iterated until it reaches a maximum number of iterations. At the end, the vector with the most optimal fitness value is accepted as the solution of the problem.
Experiments and results
The present study proposes the COVID-CCD-Net approach in which learning rate, solver, L2 regularization, gradient threshold method, and gradient threshold parameters of AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet were optimized using GBO. The classification performance of the proposed approach was tested using two different medical image classification datasets. Additionally, the results of this test were compared with those obtained from non-optimized AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet CNN models. In addition, Quasi-Newton (Q-N) algorithm [74], one of the most fundamental optimization methods, was also used to optimize the hyperparameters of CNN models and compared with the proposed COVID-CCD-Net approach. The following sub-sections describe medical image classification datasets, experiment setup, and present comparative experimental findings.
Medical image classification datasets
COVID-19 [75, 76] and Epistroma [77] datasets were selected for the experimental studies. COVID-19 dataset consists of three classes, namely “Covid-19,” “Normal,” and “Viral Pneumonia,” with a total of 3829 images. Epistroma dataset, on the other hand, consists of two classes, namely “epithelium” and “stroma,” with a total of 1376 images. In both datasets, 80% and 20% images were used for training and testing processes, respectively and we have performed 5-fold cross-validation. Ten percent of the training data in each data set was also used for validation. Samples images from both datasets are shown in Fig. 3.
Fig. 3.
Sample images from datasets
Experimental setup
All experimental studies were carried out on MATLAB R2020a platform. The number of vectors in GBO population and the maximum number of iterations were selected as 10 in the proposed COVID-CCD-Net approach. In other words, the fitness function is called 100 times. Q-N algorithm performs search starting at a single point instead of a population-based search. For a healthier comparison with the proposed approach, the number of maximum iterations was selected as 100 in Q-N algorithm to call the fitness function 100 times. In addition, default MATLAB values for solver, L2 regularization, gradient threshold method, and gradient threshold parameters were selected as “sgdm,” “0.0001,” “l2norm,” and “Inf,” respectively for non-optimized CNN models. Values of epoch for all CNN models were selected as 2 for COVID-19 dataset as 5 for Epistroma dataset. Mini batch size was set to 25. Twenty independent experimental studies were conducted on these datasets for all CNN models, and the obtained mean accuracy, maximum accuracy, F1-score, and standard deviation values were compared to measure the performances of all models.
Experimental results
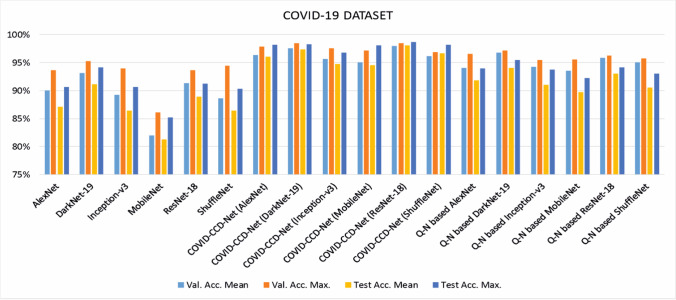
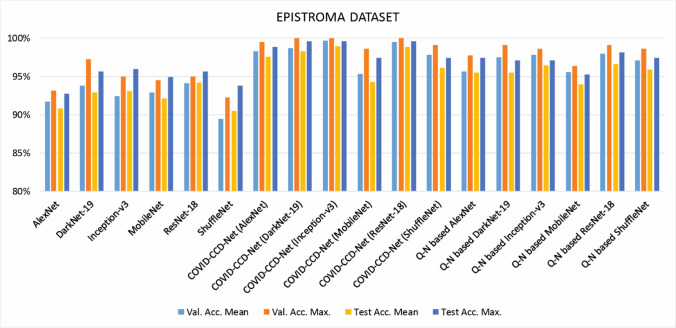
Mean accuracy, maximum accuracy, F1-score, and standard deviation values obtained from 20 different independent studies on COVID-19 and Epistroma datasets are given in Tables 2 and 5, respectively. The findings were also shown in bar charts in Figs. 4 and 5 to give a clearer picture of the overall findings.
Table 2.
Accuracy, F1-score and Std. dev. results of validation and test for the COVID-19 dataset
| Validation | Test | |||||||
|---|---|---|---|---|---|---|---|---|
| Acc. (mean) | Acc. (max) |
F1-score | Std. dev. | Acc. (mean) | Acc. (max) |
F1-score | Std. dev. | |
| AlexNet | 90.065 | 93.638 | 90.254 | 2.627 | 87.089 | 90.601 | 87.305 | 2.590 |
| DarkNet-19 | 93.189 | 95.269 | 93.292 | 1.833 | 91.149 | 94.125 | 91.309 | 1.526 |
| Inception-v3 | 89.233 | 93.964 | 89.291 | 3.610 | 86.436 | 90.601 | 86.631 | 3.171 |
| MobileNet | 82.007 | 86.134 | 81.716 | 3.052 | 81.312 | 85.248 | 81.333 | 2.470 |
| ResNet-18 | 91.313 | 93.638 | 91.418 | 1.460 | 88.962 | 91.253 | 89.115 | 1.793 |
| ShuffleNet | 88.679 | 94.454 | 88.665 | 4.035 | 86.449 | 90.339 | 86.535 | 3.075 |
| COVID-CCD-Net (AlexNet) | 96.354 | 97.879 | 96.460 | 1.166 | 96.044 | 98.172 | 96.138 | 0.927 |
| COVID-CCD-Net (DarkNet-19) | 97.553 | 98.532 | 97.654 | 0.631 | 97.369 | 98.303 | 97.458 | 0.496 |
| COVID-CCD-Net (Inception-v3) | 95.718 | 97.553 | 95.830 | 1.139 | 94.791 | 96.736 | 94.900 | 1.736 |
| COVID-CCD-Net (MobileNet) | 95.057 | 97.227 | 95.179 | 1.502 | 94.608 | 98.042 | 94.718 | 1.816 |
| COVID-CCD-Net (ResNet-18) | 97.977 | 98.532 | 98.063 | 0.301 | 98.107 | 98.695 | 98.158 | 0.353 |
| COVID-CCD-Net (ShuffleNet) | 96.223 | 96.900 | 96.312 | 0.424 | 96.730 | 98.172 | 96.805 | 0.623 |
| Quasi-Newton-based AlexNet | 94.043 | 96.574 | 94.205 | 4.123 | 91.879 | 93.994 | 92.099 | 3.038 |
| Quasi-Newton-based DarkNet-19 | 96.807 | 97.226 | 96.914 | 0.373 | 94.072 | 95.430 | 94.225 | 1.399 |
| Quasi-Newton-based Inception-v3 | 94.266 | 95.432 | 94.331 | 0.868 | 91.051 | 93.733 | 91.179 | 2.554 |
| Quasi-Newton-based MobileNet | 93.541 | 95.595 | 93.653 | 1.723 | 89.713 | 92.298 | 89.925 | 1.990 |
| Quasi-Newton-based ResNet-18 | 95.834 | 96.248 | 95.916 | 0.511 | 93.019 | 94.125 | 93.124 | 0.869 |
| Quasi-Newton-based ShuffleNet | 95.050 | 95.759 | 95.142 | 1.055 | 90.551 | 93.081 | 90.731 | 2.336 |
Table 5.
Accuracy, F1-score, and Std. dev. result of validation and test for the Epistroma dataset
| Validation | Test | |||||||
|---|---|---|---|---|---|---|---|---|
| Acc. (mean) | Acc. (max) |
F1-score | Std. dev. | Acc. (mean) | Acc. (max) |
F1-score | Std. dev. | |
| AlexNet | 91.750 | 93.182 | 91.615 | 0.924 | 90.800 | 92.727 | 90.691 | 1.144 |
| DarkNet-19 | 93.773 | 97.273 | 93.608 | 2.017 | 92.909 | 95.636 | 92.714 | 2.518 |
| Inception-v3 | 92.477 | 95 | 92.264 | 1.255 | 93.073 | 96 | 92.870 | 1.481 |
| MobileNet | 92.886 | 94.545 | 92.727 | 1.066 | 92.145 | 94.909 | 92.009 | 1.641 |
| ResNet-18 | 94.091 | 95 | 93.950 | 0.692 | 94.218 | 95.636 | 94.097 | 1.048 |
| ShuffleNet | 89.454 | 92.273 | 89.149 | 2.133 | 90.491 | 93.818 | 90.270 | 2.105 |
| COVID-CCD-Net (AlexNet) | 98.341 | 99.545 | 98.278 | 0.711 | 97.618 | 98.909 | 97.542 | 0.683 |
| COVID-CCD-Net (DarkNet-19) | 98.727 | 100 | 98.676 | 0.996 | 98.273 | 99.636 | 98.211 | 1.538 |
| COVID-CCD-Net (Inception-v3) | 99.705 | 100 | 99.692 | 0.424 | 98.964 | 99.636 | 98.924 | 0.476 |
| COVID-CCD-Net (MobileNet) | 95.318 | 98.636 | 95.161 | 3.561 | 94.255 | 97.455 | 94.096 | 3.752 |
| COVID-CCD-Net (ResNet-18) | 99.545 | 100 | 99.526 | 0.330 | 98.836 | 99.636 | 98.793 | 0.481 |
| COVID-CCD-Net (ShuffleNet) | 97.818 | 99.091 | 97.736 | 0.880 | 96.164 | 97.455 | 96.046 | 0.955 |
| Quasi-Newton based AlexNet | 95.636 | 97.727 | 95.505 | 2.286 | 95.491 | 97.455 | 95.362 | 2.337 |
| Quasi-Newton based DarkNet-19 | 97.545 | 99.091 | 97.461 | 2.073 | 95.491 | 97.091 | 95.333 | 1.326 |
| Quasi-Newton based Inception-v3 | 97.818 | 98.636 | 97.739 | 0.985 | 96.436 | 97.091 | 96.310 | 0.598 |
| Quasi-Newton based MobileNet | 95.545 | 96.364 | 95.407 | 1.132 | 93.964 | 95.273 | 93.802 | 0.913 |
| Quasi-Newton based ResNet-18 | 98.001 | 99.091 | 97.929 | 1.944 | 96.655 | 98.182 | 96.541 | 1.530 |
| Quasi-Newton based ShuffleNet | 97.091 | 98.636 | 96.991 | 1.997 | 95.927 | 97.455 | 95.805 | 2.063 |
Fig. 4.
Bar charts for the COVID-19 dataset
Fig. 5.
Bar charts for the Epistroma dataset
The findings related to COVID-19 dataset demonstrated that in the training process, COVID-CCD-Net (ResNet-18) reached the highest mean validation accuracy, maximum validation accuracy, and F1-score values with 97.977, 98.532, and 98.063, respectively. The second highest values were yielded by COVID-CCD-Net (DarkNet-19) with 97.553, 98.532, and 97.654, while non-optimized MobileNet displayed a lower performance with 82.007, 86.134, and 81.716. In the testing process, COVID-CCD-Net (ResNet-18) classified test images with a mean accuracy rate of 98.107%, followed by Darknet-19 with a mean accuracy rate of 97.369%. MobileNet displayed the lowest performance in terms of training and testing. validation and test accuracy for COVID-19 dataset before and after optimization with COVID-CCD-Net are given in Table 3 and the results demonstrated that COVID-CCD-Net increased the classification performance of the non-optimized CNN models by 6.22–13.29%. The performance was improved when Q-N algorithm was used to optimize the hyperparameters of non-optimized CNN models. However the performance increased between 2.92 and 8.40%, demonstrating that GBO displays a higher performance in the hyperparameter optimization in COVID-19 dataset.
Table 3.
Validation and test accuracy before and after optimization with COVID-CCD-Net on the COVID-19 dataset
| Validation | Test | |||||
|---|---|---|---|---|---|---|
| Before optimization | After optimization | Performance improvement | Before optimization | After optimization | Performance improvement | |
| AlexNet | 90.065 | 96.354 | 6.289 | 87.089 | 96.044 | 8.955 |
| DarkNet-19 | 93.189 | 97.553 | 4.364 | 91.149 | 97.369 | 6.22 |
| Inception-v3 | 89.233 | 95.718 | 6.485 | 86.436 | 94.791 | 8.355 |
| MobileNet | 82.007 | 95.057 | 13.05 | 81.312 | 94.608 | 13.296 |
| ResNet-18 | 91.313 | 97.977 | 6.664 | 88.962 | 98.107 | 9.145 |
| ShuffleNet | 88.679 | 96.223 | 7.544 | 86.449 | 96.73 | 10.281 |
It can understand from the findings related to Epistroma dataset that in the training process, the highest mean accuracy, maximum accuracy, and F1-score values were obtained by COVID-CCD-Net (Inception-v3) with 99.705, 100, and 99.692, respectively. Similarly, COVID-CCD-Net (Inception-v3) also yielded the highest values in the testing process with 98.964, 99.636, and 98.924. It was followed by ResNet-18 with 99.545, 100, and 99.526 for the training and 98.836, 99.636, and 98.793 for the testing process. On the other hand, the lowest performance in the training and testing process was displayed non-optimmized ShuffleNet with 89.454, 92.273, and 89.149 and 90.491, 93.818, and 90.270, respectively. validation and test accuracy for epistroma dataset before and after optimization with COVID-CCD-Net are given in Table 4 and the results demonstrated that COVID-CCD-Net increased the classification performance of the non-optimized CNN models by 2.11–6.81%. The performance was improved when Q-N algorithm was used to optimize the hyperparameters of non-optimized CNN models. It can be seen in Table 5 that the performance increased between 1.81 and 5.43%, demonstrating that GBO displays a higher performance in the hyperparameter optimization in Epistroma dataset.
Table 4.
Validation and test accuracy before and after optimization with COVID-CCD-Net on the Epistroma dataset
| VALIDATION | TEST | |||||
|---|---|---|---|---|---|---|
| Before Optimization | After Optimization | Performance Improvement | Before Optimization | After Optimization | Performance Improvement | |
| AlexNet | 91.75 | 98.341 | 6.591 | 90.8 | 97.618 | 6.818 |
| DarkNet-19 | 93.773 | 98.727 | 4.954 | 92.909 | 98.273 | 5.364 |
| Inception-v3 | 92.477 | 99.705 | 7.228 | 93.073 | 98.964 | 5.891 |
| MobileNet | 92.886 | 95.318 | 2.432 | 92.145 | 94.255 | 2.11 |
| ResNet-18 | 94.091 | 99.545 | 5.454 | 94.218 | 98.836 | 4.618 |
| ShuffleNet | 89.454 | 97.818 | 8.364 | 90.491 | 96.164 | 5.673 |
As shown in Tables 2 and 5, GBO algorithm remarkably improves the performance of non-optimized CNN models in COVID-19 and Epistroma datasets. Additionally, experimental studies indicated that GBO algorithm displayed a higher performance in hyperparameter optimization in both datasets compared to Q-N algorithm.
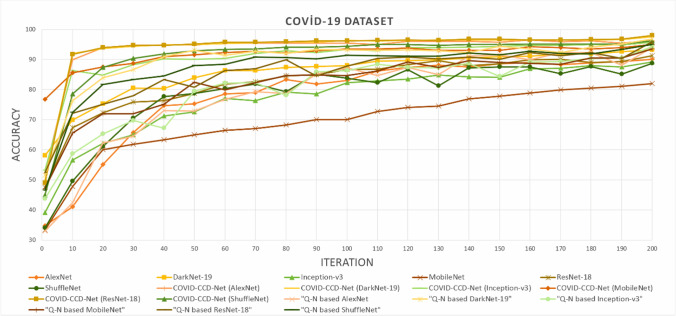
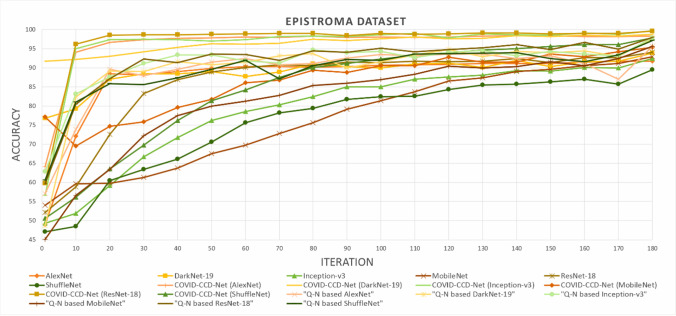
Mean training accuracy curves of all models obtained from COVID-19 dataset are shown in Fig. 6. While COVID-CCD-Net (ResNet-18) displayed a faster convergence, non-optimized MobileNet displayed a slower convergence. Mean training accuracy curves of all models obtained from Epistroma dataset are shown in Fig. 7, COVID-CCD-Net (Inception-v3), COVID-CCD-Net (ResNet-18), and COVID-CCD-Net (DarkNet-19) displayed a fast convergence in the first 20 iterations and a lower convergence in the remaining iterations.
Fig. 6.
Mean training accuracy curves for the COVID-19 dataset
Fig. 7.
Mean training accuracy curves for the Epistroma dataset
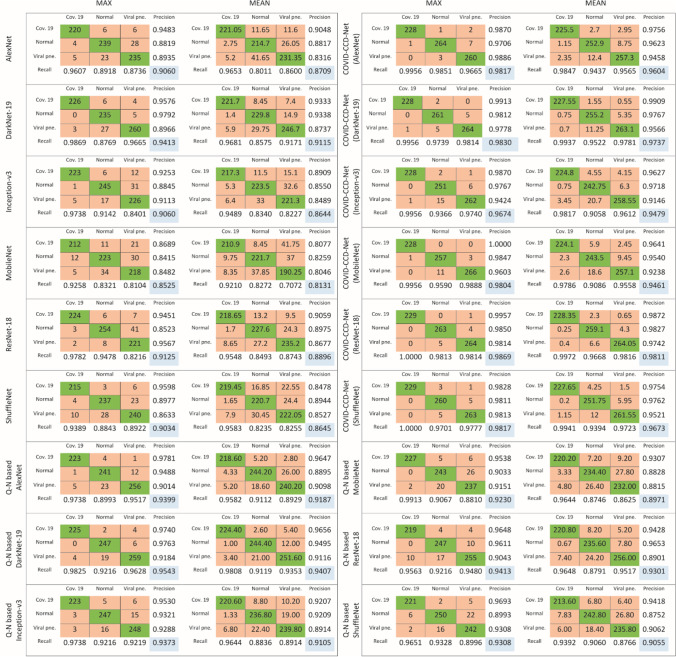
Maximum and mean confusion matrix values of all models obtained from the testing processes for COVID-19 and Epistroma datasets are shown in Fig. 8 and Fig. 9. A confusion matrix is a table which is used to describe the performance of a model by referring to its accuracy rates in each class. Rows and columns in a confusion matrix correspond to the predicted class (output class) and true class (target class), respectively.
Fig. 8.
Confusion matrices of COVID-19 dataset
Fig. 9.
Confusion matrices of Epistroma dataset
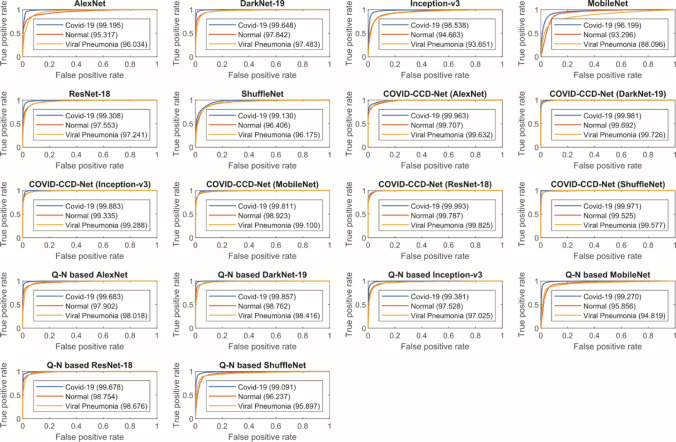
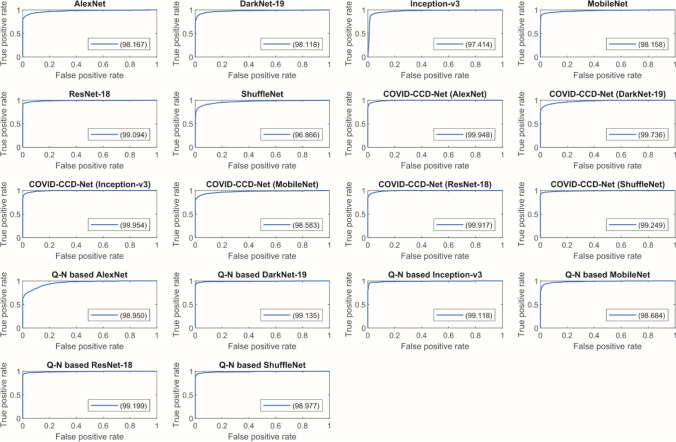
The receiver operating characteristic (ROC) curves of COVID-19 and Epistroma datasets are provided in Fig. 10 and Fig. 11 respectively, which showing the relationship between the false positive rate (FPR) and the true positive rate (TPR). It can be clearly seen, in COVID-19 dataset COVID-CCD-Net (ResNet-18) and in Epistroma dataset COVID-CCD-Net (Inception-v3) have higher true positive rates.
Fig. 10.
ROC curves for COVID-19 dataset
Fig. 11.
ROC curves for Epistroma dataset
Table 6 and Table 7 compare the performance of the COVID-CCD-Net with several state-of-the art methods on COVID-19 and Epistroma datasets. It can be seen obviously; the the COVID-CCD-Net has the highest classification accuracy among the compared methods for both datasets.
Table 6.
Comparison of the results with state-of-the art CNN methods for COVID-19 dataset
| Source | Models/methods | Number of classes | Overall acc (%) |
|---|---|---|---|
| Song et al. [78] | DRE-Net |
2 3 |
86 93 |
| Ozturk et al.[79] | DarkCovidNet |
3 2 |
87.02 98.08 |
| Wang et al. [80] | CNNs, transfer learning | 2 | 89.5 |
| Keidar et al.[81] | Data augmentation, segmentation and CNN | 2 | 90.3 |
| Wang et al. [50] | Customized CNN architectur | 3 | 93.33 |
| Zhang et al.[82] | COVID19XrayNet | 2 | 91.92 |
| Goel et al.[83] | OptCoNet | 3 | 97.78 |
| Proposed | COVID-CCD-Net | 3 | 98.107 |
Table 7.
Comparison of the results with state-of-the art CNN methods for Epistroma dataset
Conclusion
In order to classify Covid-19, normal, and viral pneumonia in chest X-ray images as well as epithelial and stromal regions in TMA images accurately, the present study proposed the COVID-CCD-Net approach with the optimized hyperparameters of AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet CNN models using GBO, which is one of the most recent metaheuristic optimization algorithms. Network-trained parameters of these CNN models such as learning rate, solver, L2 regularization, gradient threshold method, and gradient threshold were optimized and tuned using GBO algorithm. In the GBO, each vector of the population represents a set of CNN’s hyperparameters, and the algorithm searches for the hyperparameter values that help the model display the highest classification performance. Two different medical image classification datasets, i.e., COVID-19 and Epistroma, were used in the experimental study. While GBO hyperparameter optimization improved the performance of non-optimized CNN models in COVID-19 dataset by 6.22% to 13.29%, the contribution of Q-N algorithm did not exceed 2.92% to 8.40%. Similarly, GBO hyperparameter optimization improved the performance of non-optimized CNN models in Epistroma dataset by 2.11% to 6.81%, Q-N algorithm improved it only 1.81% to 4.53%. These results demonstrated that the proposed approach significantly improved the classification performance of AlexNet, DarkNet-19, Inception-v3, MobileNet, ResNet-18, and ShuffleNet CNN models and displayed a better performance compared to non-optimized CNN models. One of the main problems in CNN-based classification approaches is their need for a high number of high-quality images for a succesful classification performance and optimal values for the hyperparameters of CNN architecture. In the present study, a sufficient number of images was used to complete training process for CNN architecture, and the proposed COVID-CCD-Net approach was used to optimize the hyperparameters of CNN architectures to overcome the above-mentioned problems. Future studies will focus on the optimization of different hyperparameters such as filter size, filter number, stride, and padding using various metaheuristic optimization algorithms.
Biographies
Soner Kiziloluk
received the M.Sc. degree (2013) in electrical and electronic engineering at Munzur University and the Ph.D. degree (2017) in Computer Engineering at Fırat University. Currently he is assistant professor at Computer Engineering of Malatya Turgut Ozal University. His research interests include metaheuristic algorithms, optimization and data mining.
Eser Sert
received the M.Sc. degree in 2010 and the Ph.D. degree in 2013, all in Computer Engineering at Trakya University, Turkey. Currently he is associate professor at Computer Engineering of Malatya Turgut Ozal University. His research interests include 3D modeling system, image processing, computer vision, field programmable gate arrays (FPGA) and artificial inteligence.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Soner Kiziloluk, Email: soner.kiziloluk@ozal.edu.tr.
Eser Sert, Email: eser.sert@ozal.edu.tr.
References
- 1.WCOVID-19 Weekly epidemiological update data as received by WHO from national authorities, as of 21 September 2021
- 2.Of the International, C. S. G The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506 [DOI] [PMC free article] [PubMed]
- 4.El Asnaoui K, Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn. 2021;39(10):3615–3626. doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nayak SR, Nayak DR, Sinha U, Arora V, Pachori RB. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed Signal Process Control. 2021;64:102365. doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong D, Tang Z, Wang S, Hui H, Gong L, Lu Y, Xue Z, Liao H, Chen F, Yang F, Jin R, Wang K, Liu Z, Wei J, Mu W, Zhang H, Jiang J, Tian J, Li H (2020) The role of imaging in the detection and management of COVID-19: a review. IEEE Rev Biomed Eng 14:16–29 [DOI] [PubMed]
- 7.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH. Essentials for radiologists on COVID-19: an update—radiology scientific expert panel. Radiology. 2020;296(2):E113–E114. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karakanis S, Leontidis G. Lightweight deep learning models for detecting COVID-19 from chest X-ray images. Comput Biol Med. 2021;130:104181. doi: 10.1016/j.compbiomed.2020.104181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouchicha C, Ammor O, Meknassi M. CVDNet: a novel deep learning architecture for detection of coronavirus (Covid-19) from chest x-ray images. Chaos, Solitons Fractals. 2020;140:110245. doi: 10.1016/j.chaos.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ. Deep-covid: predicting covid-19 from chest x-ray images using deep transfer learning. Med Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussain E, Hasan M, Rahman MA, Lee I, Tamanna T, Parvez MZ. CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos Solitons Fractals. 2021;142:110495. doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta A, Gupta S, Katarya R. InstaCovNet-19: a deep learning classification model for the detection of COVID-19 patients using chest X-ray. Appl Soft Comput. 2021;99:106859. doi: 10.1016/j.asoc.2020.106859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kononen J, Bubendorf L, Kallionimeni A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallionimeni OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 14.Spano JP, Lagorce C, Atlan D, Milano G, Domont J, Benamouzig R, Attar A, Benichou J, Martin A, Morere JF, Raphael M, Llorca FP, Breau JL, Fagard R, Khayat D, Wind P. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 2005;16(1):102–108. doi: 10.1093/annonc/mdi006. [DOI] [PubMed] [Google Scholar]
- 15.Markman B, Javier Ramos F, Capdevila J, Tabernero J. EGFR and KRAS in colorectal cancer. Adv Clin Chem. 2010;51:72. doi: 10.1016/s0065-2423(10)51004-7. [DOI] [PubMed] [Google Scholar]
- 16.Ahmadianfar I, Bozorg-Haddad O, Chu X. Gradient-based optimizer: a new metaheuristic optimization algorithm. Inf Sci. 2020;540:131–159. doi: 10.1016/j.ins.2020.06.037. [DOI] [Google Scholar]
- 17.Yang XS. Nature-inspired metaheuristic algorithms. 2. UK: Luniver Press; 2010. [Google Scholar]
- 18.Beheshti Z, Shamsuddin SMH. A review of population-based meta-heuristic algorithms. Int. J. Adv. Soft Comput. Appl. 2013;5(1):1–35. [Google Scholar]
- 19.Albani RA, Albani VV, Neto AJS. Source characterization of airborne pollutant emissions by hybrid metaheuristic/gradient-based optimization techniques. Environ Pollut. 2020;267:115618. doi: 10.1016/j.envpol.2020.115618. [DOI] [PubMed] [Google Scholar]
- 20.Ramos-Figueroa O, Quiroz-Castellanos M, Mezura-Montes E, Schütze O. Metaheuristics to solve grouping problems: a review and a case study. Swarm Evol Comput. 2020;53:100643. doi: 10.1016/j.swevo.2019.100643. [DOI] [Google Scholar]
- 21.Taramasco C, Crawford B, Soto R, Cortés-Toro EM, Olivares R. A new metaheuristic based on vapor-liquid equilibrium for solving a new patient bed assignment problem. Expert Syst Appl. 2020;158:113506. doi: 10.1016/j.eswa.2020.113506. [DOI] [Google Scholar]
- 22.Latha M, Kavitha G. Combined Metaheuristic Algorithm and Radiomics Strategy for the Analysis of Neuroanatomical Structures in Schizophrenia and Schizoaffective Disorders. IRBM. 2021;42(5):353–368. doi: 10.1016/j.irbm.2020.10.006. [DOI] [Google Scholar]
- 23.Lee WY, Park SM, Sim KB. Optimal hyperparameter tuning of convolutional neural networks based on the parameter-setting-free harmony search algorithm. Optik. 2018;172:359–367. doi: 10.1016/j.ijleo.2018.07.044. [DOI] [Google Scholar]
- 24.Kumar P, Hati AS. Deep convolutional neural network based on adaptive gradient optimizer for fault detection in SCIM. ISA transactions. 2021;111:350–359. doi: 10.1016/j.isatra.2020.10.052. [DOI] [PubMed] [Google Scholar]
- 25.Waheed A, Goyal M, Gupta D, Khanna A, Hassanien AE, Pandey HM. An optimized dense convolutional neural network model for disease recognition and classification in corn leaf. Comput Electron Agric. 2020;175:105456. doi: 10.1016/j.compag.2020.105456. [DOI] [Google Scholar]
- 26.Bergstra JS, Bardenet R, Bengio Y, Kégl B. Advances in neural information processing systems. 2011. Algorithms for hyperparameter optimization; pp. 2546–2554. [Google Scholar]
- 27.Nguyen T, Nguyen G, Nguyen BM. EO-CNN: an enhanced CNN model trained by equilibrium optimization for traffic transportation prediction. Procedia Comput Sci. 2020;176:800–809. doi: 10.1016/j.procs.2020.09.075. [DOI] [Google Scholar]
- 28.Bochinsk E, Senst T, Sikora T (2017) Hyper-parameter optimization for convolutional neural network committees based on evolutionary algorithms. In 2017 IEEE International Conference on Image Processing (ICIP) (pp. 3924-3928). IEEE. 10.1109/ICIP.2017.8297018
- 29.Aszemi NM, Dominic PDD. Hyperparameter optimization in convolutional neural network using genetic algorithms. Int J Adv Comput Sci Appl. 2019;10(6):269–278. [Google Scholar]
- 30.Bergstra J, Bengio Y. Random search for hyper-parameter optimization. J Mach Learn Res. 2012;13:281–305. doi: 10.5555/2188385.2188395. [DOI] [Google Scholar]
- 31.Wang Y, Zhang H, Zhang G. cPSO-CNN: an efficient PSO-based algorithm for fine-tuning hyper-parameters of convolutional neural networks. Swarm Evol Comput. 2019;49:114–123. doi: 10.1016/j.swevo.2019.06.002. [DOI] [Google Scholar]
- 32.Soon FC, Khaw HY, Chuah JH, Kanesan J. Hyper-parameters optimisation of deep CNN architecture for vehicle logo recognition. IET Intell Transp Syst. 2018;12(8):939–946. doi: 10.1049/iet-its.2018.5127. [DOI] [Google Scholar]
- 33.Lima LL, Ferreira Junior JR, Oliveira MC. Toward classifying small lung nodules with hyperparameter optimization of convolutional neural networks. Comput Intell. 2021;37(4):1599–1618. doi: 10.1111/coin.12350. [DOI] [Google Scholar]
- 34.Dixit U, Mishra A, Shukla A, Tiwari R (2019) Texture classification using convolutional neural network optimized with whale optimization algorithm. SN. Appl Sci 1(6). 10.1007/s42452-019-0678-y
- 35.Andonie R, Florea AC (2020) Weighted random search for CNN hyperparameter optimization. arXiv preprint arXiv:2003.13300
- 36.Mahdaddi A, Meshoul S, Belguidoum M. EA-based hyperparameter optimization of hybrid deep learning models for effective drug-target interactions prediction. Expert Syst Appl. 2021;185:115525. doi: 10.1016/j.eswa.2021.115525. [DOI] [Google Scholar]
- 37.Zhang M, Li H, Pan S, Lyu J, Ling S, Su S. Convolutional neural networks-based lung nodule classification: A surrogate-assisted evolutionary algorithm for hyperparameter optimization. IEEE Transactions on Evolutionary Computation. 2021;25(5):869–882. doi: 10.1109/TEVC.2021.3060833. [DOI] [Google Scholar]
- 38.Shahriari B, Swersky K, Wang Z, Adams RP, De Freitas N. Taking the human out of the loop: a review of bayesian optimization. Proc IEEE. 2015;104:148–175. doi: 10.1109/JPROC.2015.2494218. [DOI] [Google Scholar]
- 39.Fatyanosa TN, Aritsugi M. An automatic convolutional neural network optimization using a diversity-guided genetic algorithm. IEEE Access. 2021;9:91410–91426. doi: 10.1109/ACCESS.2021.3091729. [DOI] [Google Scholar]
- 40.Ilievski I, Akhtar T, Feng J, Shoemaker C (2017) Efficient hyperparameter optimization for deep learning algorithms using deterministic rbf surrogates. In Proceedings of the AAAI Conference on Artificial Intelligence. 31(1): 822–829
- 41.Talathi SS (2015) Hyper-parameter optimization of deep convolutional networks for object recognition. In 2015 IEEE International Conference on Image Processing (ICIP) (pp. 3982-3986). IEEE
- 42.Rattanavorragant R, Jewajinda Y (2019) A hyper-parameter optimization for deep neural network using an island-based genetic algorithm. In 2019 16th International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology (ECTI-CON) (pp. 73-76). IEEE
- 43.Navaneeth B, Suchetha M. PSO optimized 1-D CNN-SVM architecture for real-time detection and classification applications. Comput Biol Med. 2019;108:85–92. doi: 10.1016/j.compbiomed.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 44.Elkorany AS, Elsharkawy ZF (2021) COVIDetection-Net: A tailored COVID-19 detection from chest radiography images using deep learning. Optik 231 [DOI] [PMC free article] [PubMed]
- 45.Brunese L, Mercaldo F, Reginelli A, Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput Methods Prog Biomed. 2020;196:105608. doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panwar H, Gupta PK, Siddiqui MK, Morales-Menendez R, Singh V. Application of deep learning for fast detection of COVID-19 in X-rays using nCOVnet. Chaos Solitons Fractals. 2020;138:109944. doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh D, Kumar V, Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur J Clin Microbiol Infect Dis. 2020;39(7):1379–1389. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makris A, Kontopoulos I, Tserpes K (2020) COVID-19 detection from chest X-ray images using deep learning and convolutional neural networks. In 11th Hellenic Conference on Artificial Intelligence (pp. 60-66)
- 50.Wang L, Lin ZQ, Wong A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, He K, Shi Y, Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19. IEEE Rev Biomed Eng. 2020;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 52.Castiglioni I, Ippolito D, Interlenghi M, Monti CB, Salvatore C, Schiaffino S, Polidori A, Gandola D, Messa C, Sardanelli F. Machine learning applied on chest x-ray can aid in the diagnosis of COVID-19: a first experience from Lombardy, Italy. Eur Radiol Exp. 2021;5(1):1–10. doi: 10.1186/s41747-020-00203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hemdan EED, Shouman MA, Karar ME (2020) Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055
- 54.Khan AI, Shah JL, Bhat MM. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Prog Biomed. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen HG, Blank A, Dawson HE, Lugli A, Zlobec I. Classification of colorectal tissue images from high throughput tissue microarrays by ensemble deep learning methods. Sci Rep. 2021;11(1):1–11. doi: 10.1038/s41598-021-81352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu J, Luo X, Wang G, Gilmore H, Madabhushi A. A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing. 2016;191:214–223. doi: 10.1016/j.neucom.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linder N, Konsti J, Turkki R, Rahtu E, Lundin M, Nordling S, Haglund C, Ahonen T, Pietikäinen M, Lundin J. Identification of tumor epithelium and stroma in tissue microarrays using texture analysis. Diagn Pathol. 2012;7(1):1–11. doi: 10.1186/1746-1596-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo T, Dong J, Li H, Gao Y (2017). Simple convolutional neural network on image classification. In 2017 IEEE 2nd International Conference on Big Data Analysis (ICBDA)(pp. 721-724). IEEE
- 59.Nhat-Duc H, Nguyen QL, Tran VD. Automatic recognition of asphalt pavement cracks using metaheuristic optimized edge detection algorithms and convolution neural network. Autom Constr. 2018;94:203–213. doi: 10.1016/j.autcon.2018.07.008. [DOI] [Google Scholar]
- 60.Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9(4):611–629. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoo JH, Yoon HI, Kim HG, Yoon HS, Han SS (2019) Optimization of hyper-parameter for CNN model using genetic algorithm. In 2019 1st International Conference on Electrical, Control and Instrumentation Engineering (ICECIE) (pp. 1-6). IEEE
- 62.Mustafa EM, Elshafey MA, Fouad MM (2019) Accuracy enhancement of a blind image steganalysis approach using dynamic learning rate-based CNN on GPUs. In 2019 10th IEEE International Conference on Intelligent Data Acquisition and Advanced Computing Systems: Technology and Applications (IDAACS) (Vol. 1, pp. 28-33). IEEE
- 63.Jaworska T. International Workshop on Intuitionistic Fuzzy Sets and Generalized Nets. Cham: Springer; 2018. Image segment classification using CNN; pp. 409–425. [Google Scholar]
- 64.Zeng M, Nguyen LT, Yu B, Mengshoel OJ, Zhu J, Wu P, Zhang J (2014) Convolutional neural networks for human activity recognition using mobile sensors. In 6th International Conference on Mobile Computing, Applications and Services (pp. 197-205). IEEE
- 65.Si L, Xiong X, Wang Z, Tan C. A deep convolutional neural network model for intelligent discrimination between coal and rocks in coal mining face. Math Probl Eng. 2020;2020:1–12. doi: 10.1155/2020/2616510. [DOI] [Google Scholar]
- 66.Pascanu R, Mikolov T, Bengio Y (2013) On the difficulty of training recurrent neural networks. In International conference on machine learning (pp. 1310-1318). PMLR.
- 67.Lin Y, Han S, Mao H, Wang Y, Dally WJ (2017) Deep gradient compression: Reducing the communication bandwidth for distributed training. arXiv preprint arXiv:1712.01887
- 68.Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems (pp. 1097–1105)
- 69.Redmon J (2013) Darknet: Open source neural networks in C. http://pjreddie.com/darknet/. 2013–2016. Accessed 12 Aug 2020
- 70.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 2818-2826)
- 71.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770-778)
- 72.Zhang X, Zhou X, Lin M, Sun J (2018) Shufflenet: an extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 6848-6856)
- 73.Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) Mobilenetv2: inverted residuals and linear bottlenecks. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 4510-4520)
- 74.Nocedal J, Wright SJ, editors. Numerical optimization. New York: Springer New York; 1999. [Google Scholar]
- 75.Rahman T, Chowdhury ME, Khandakar A (2020) COVID-19 chest X-ray database. Kaggle Data, v3. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. Accessed 20 Dec 2020
- 76.Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Emadi NA, Reaz MBI, Islam MT. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 77.Webmicroscope. EGFR colon TMA stroma LBP classification (2012) http://fimm.webmicroscope.net/Research/Supplements/epistroma. Accessed 20 Dec 2020
- 78.Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Wang R, Zhao H, Zha Y, Shen J, Chong Y, Yang Y (2021) Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. in IEEE/ACM Transactions on Computational Biology and Bioinformatics. 10.1109/TCBB.2021.3065361 [DOI] [PMC free article] [PubMed]
- 79.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X, Xu B. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) Eur Radiol. 2021;31:6096–6104. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keidar D, Yaron D, Goldstein E, Shachar Y, Blass A, Charbinsky L, Aharony I, Lifshitz L, Lumelsky D, Neeman Z, Mizrachi M, Hajouj M, Eizenbach N, Sela E, Weiss CS, Levin P, Benjaminov O, Bachar GN, Tamir S, et al. COVID-19 classification of X-ray images using deep neural networks. European radiology. 2021;31(12):9654–9663. doi: 10.1007/s00330-021-08050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang R, Guo Z, Sun Y, Lu Q, Xu Z, Yao Z, Duan M, Liu S, Ren Y, Huang L, Zhou F. COVID19XrayNet: a two-step transfer learning model for the COVID-19 detecting problem based on a limited number of chest X-ray images. Interdiscip Sci Comput Life Sci. 2020;12(4):555–565. doi: 10.1007/s12539-020-00393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goel T, Murugan R, Mirjalili S, Chakrabartty DK. OptCoNet: an optimized convolutional neural network for an automatic diagnosis of COVID-19. Appl Intell. 2021;51(3):1351–1366. doi: 10.1007/s10489-020-01904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alinsaif S, Lang J (2020) Histological image classification using deep features and transfer learning. In 2020 17th Conference on Computer and Robot Vision (CRV) (pp. 101-108). IEEE
- 85.Cascianelli S, Bello-Cerezo R, Bianconi F, Fravolini ML, Belal M, Palumbo B, Kather JN. International conference on intelligent interactive multimedia systems and services. Cham: Springer; 2018. Dimensionality reduction strategies for cnn-based classification of histopathological images; pp. 21–30. [Google Scholar]
- 86.Huang Y, Zheng H, Liu C, Ding X, Rohde GK. Epithelium-stroma classification via convolutional neural networks and unsupervised domain adaptation in histopathological images. IEEE J Biomed Health Inform. 2017;21(6):1625–1632. doi: 10.1109/JBHI.2017.2691738. [DOI] [PubMed] [Google Scholar]
- 87.Bianconi F, Bello-Cerezo R, Napoletano P. Improved opponent color local binary patterns: an effective local image descriptor for color texture classification. J Electron Imaging. 2017;27(1):011002. doi: 10.1117/1.JEI.27.1.011002. [DOI] [Google Scholar]