Abstract
Recent studies have shown that passive sampling is a promising tool for SARS-CoV-2 detection for wastewater-based epidemiology (WBE) application. We have previously developed passive sampling of viruses using polymer membranes in seawater. Even though SARS-CoV-2 was not detected yet in seawater, passive sampling could be optimized for future application in coastal areas close to wastewater treatment plant (WWTP). The aim of this study was to optimize passive sampling of SARS-CoV-2 in sewage and seawater by selecting a suitable membrane, to determine whether the quantities of virus increase over time, and then to determine if passive sampling and traditional sampling are correlated when conducted in a wastewater treatment plant. Nylon and Zetapor allowed the detection of heat inactivated SARS-CoV-2 and of the Porcine Epidemic Diarrhea Virus (PEDV), a coronavirus surrogate, in wastewater and seawater spiked with these 2 viruses, showing an increase in detection between 4 h and 24 h of immersion and significantly higher recoveries of both viruses with nylon in seawater (15%) compared to wastewater (4%). On wastewater samples, both membranes detected the virus, the recovery rate was of about 3% for freshly collected samples, and no significant difference was found between SARS-CoV-2 genome concentration on Zetapor and that in water. In sewage spiked seawater, similar concentrations of genome were found on both membranes, with a mean recovery rate of 16% and 11% respectively for nylon and Zetapor. A 3-weeks monitoring with passive sampler allowed the detection of viruses in the influent of a WWTP with a frequency of 100% and 76% for SARS-CoV-2 and norovirus GII respectively. Passive and traditional sampling gave the same evolution of the SARS-CoV-2 concentration over time. All these results confirmed the interest of passive sampling for virus detection and its potential application for monitoring in the wastewater system for targeted public health actions.
Keywords: Passive samplers, Wastewater- seawater, SARS-CoV-2, Norovirus
Graphical abstract
1. Introduction
The SARS-CoV-2 virus responsible for outbreaks of severe acute respiratory disease COVID-19 in Wuhan, China, in late 2019 and causing the ongoing COVID-19 pandemic, is a single stranded RNA virus with a 30 kb-genome. Infection with SARS-CoV-2 is accompanied by the excretion of the virus, particularly in nasal fluids, but also in stool of infected persons (Weiss et al., 2020). Excretion of virus occurred independently of the presence or severity of symptoms, (Gu et al., 2020) and the high rate of asymptomatic infected individuals has led to alternative approaches, such as wastewater-based epidemiology (WBE) to improve the estimation of the infection spread (Ahmed et al., 2020; Buonerba et al., 2021). SARS-CoV-2 genome has been detected in wastewater worldwide, and WBE has been suggested as a tool to determine the extent of the viral circulation in cities and as an early warning for emergence of SARS-CoV-2 circulation in communities (Cluzel et al., 2022; Kitajima et al., 2020). Although the transmission route of the virus is inhalation via person-to-person aerosol or droplet transmission, the issue of transmission through wastewater and seawater aerosol was a concern at the beginning of the pandemic as the transmission of SARS-CoV-1 by aerosols from wastewater was suspected in one outbreak (McKinney et al., 2006). Even though its persistence in terms of infectious particles in aquatic environments, as an enveloped virus, is expected to be short, SARS-CoV-2 genome has been detected in river water due to overflow from sewage plants during rainy periods and in low sanitation country (Fongaro et al., 2021; La Rosa et al., 2020; Rimoldi et al., 2020). Considering the example of human enteric viruses, and more precisely norovirus (NoV), which have long contaminated coastal seawater and oysters through wastewater spill-over, we have adapted methods to investigate the possible presence of SARS-CoV-2 in coastal area in order to the study its fate in marine coastal environment (Desdouits et al., 2021).
The sampling method has a great importance when performing viral or bacteriological monitoring of water. Conventionally, samples for WBE are 24-h composite sampling. In the coastal environment, grab sampling of large volumes of water is reported (Haramoto et al., 2018; Hata et al., 2017). These sampling methods, widely applied on easily accessible sites, only gives a snapshot of the microorganisms present at the time of sampling, which may lead to false estimation of the contamination, especially for human pathogens shed sporadically or pathogens that are not persistent in the environment. Therefore, the ongoing development of passive samplers is of great importance to improve the detection of such human pathogens.
Passive sampling involves the deployment in the waterbody of devices containing membranes that adsorb microorganisms from the water column, avoiding handling and concentration of large volumes of water (Sikorski and Levine, 2020). This method of sampling, based on passive sampling of chemical contaminants and on the original method developed by Moore in the 1950s, performs a continuous and direct extraction of microbial contaminants from the water column, and provides a time-integrated sample of contamination, making it possible to integrate peaks of contamination as well as lower levels (Booij et al., 2016). Passive sampling has so far been little applied to the microbiological analysis of water. Examples include the detection of poliovirus in wastewater or of NoV in continental waters with gauze as the adsorption membrane (Fattal and Katzenelson, 1976; Tian et al., 2017). The Moore swab method has been proved efficient for the detection of Salmonella in wastewater and surface waters (Sikorski and Levine, 2020). Recently, we used passive sampling for the detection of human viruses in coastal waters, NoV and sapovirus, and a virus pathogenic for oyster, the ostreid herpes virus OsHV-1 (Vincent-Hubert et al., 2021). Currently, based on our data, passive sampling seems to be a more qualitative than quantitative approach (Vincent-Hubert et al., 2021). Indeed, the volume of water passing through the membrane during the immersion time is unknown precluding the normalisation of viral concentrations measured on the membranes to the volume of water.
Recently passive sampling has been used to detect SARS-CoV-2 in wastewater from populations with low prevalence of COVID-19 infections demonstrating their ability as a detection tool (Bivins et al., 2021; Hayes et al., 2021; Liu et al., 2020; Schang et al., 2021). These studies have provided a proof of concept that passive samplers can detect the SARS-CoV-2 in wastewater at various scales, ranging from WWTP to building (Bivins et al., 2021; Schang et al., 2021). However, questions remain before they can be used as an early warning tool, concerning the sensitivity of the method and the correlation between passive sampling and traditional sampling. According to published studies, different materials, from natural cotton to synthetic polymer membrane, were used for passive sampling, which probably influenced the recovery rate of the viruses. The nature and charge of the membrane is known to select or to be more suitable for one type of virus, as observed with Zetapor more sensitive than nylon to the enveloped OsHV-1 virus (Vincent-Hubert et al., 2021).
The aim of this study was first to optimize passive sampling of SARS-CoV-2 in wastewater and seawater by selecting a suitable membrane, to test different immersion times to determine whether the quantities of virus increase over time, and then to determine whether passive and traditional sampling are correlated. An application of passive sampling was performed in WWTP to compare the efficiency of both sampling methods in real condition during a 3-week monitoring. For this study, we used as a control NoV, a non-enveloped human enteric virus, known to persist in the environment.
2. Materials and methods
2.1. Virus stocks
Porcine Epidemic Diarrhea Virus (PEDV) strain CV777 (kindly provided by Dr. Y. Blanchard, ANSES, Ploufragan) was produced in vero-E6 cells as described previously (Bigault et al., 2020). The heat inactivated SARS-CoV-2 (kindly provided by Dr. C. Bressolette-Bodin Nantes Université, Centre de Recherche en Transplantation et Immunologie, UMR 1064, ITUN, Nantes, France), was inactivated 15 s at 60 °C, and its inactivation was verified by TCID50 assay.
2.2. Wastewater samples
Wastewater samples were collected in the western region of France from two large wastewater treatment plants (WWTP) located in a big city (A: 600,000 and B: 180,000 Person equivalent (P.E.)) from June 2020 to April 2021 and from one WWTP located in a smaller city (40,000 P.E.) from November to December 2020. These samples were raw sewage 24-h samples collected from the influent WWTP. Samples were carefully homogenized, distributed in a 1 L polyethylene bottle, transported to the laboratory at 4 °C and analysed the same day for the detection of SARS-CoV-2 and NoV.
2.3. Preparation of passive samplers
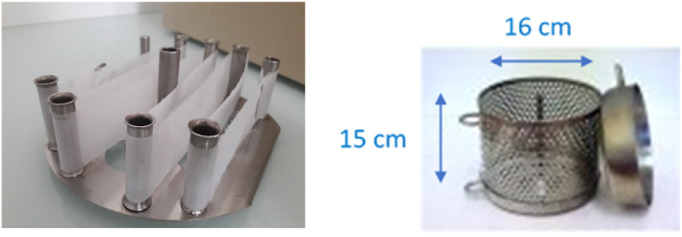
Two types of membranes, Zetapor and nylon, were used as passive samplers based on the data of our previous study (Vincent-Hubert et al., 2017). Zetapor filter (0.45 μm), an electropositive, charge-modified nylon filter, was purchased from 3 M (Alisman filtration, Sainte-Luce sur Loire, France) and nylon nets (thickness of 100 μm) from Mougel (Nantes, France). For laboratory exposures, the membrane surfaces were 32 cm2 for all experiments: nylon membrane (4 cm × 8 cm) was cut from a roll and for Zetapor, 2 discs of 4.5 cm diameter were used. For field experiments, only nylon was used, a piece of 3 × 92 cm of nylon was cut from a roll, directly attached on a “spider” carrier and placed inside the small deployment canister as shown on Fig. 1 . This device, SPMD canister, is the same as the one used for passive sampling of organic contaminants (E&H, Czech Republic).
Fig. 1.
Device used for field deployment
The spider carrier for nylon membrane (surface: 3 × 92 cm) (left) was place inside the small deployment canister (right), also named SPMD canister.
2.4. Exposure of the membranes in contaminated water samples
Three types of mixtures for membranes exposure were prepared to determine if the SARS-CoV-2 genome can be detected with the nylon and Zetapor membranes as passive sampling material:
1-Spiked water samples: Two types of water (seawater, wastewater) scored negative in SARS-CoV-2 and PEDV were spiked with heat-inactivated SARS-CoV-2 and PEDV, at a concentration of respectively 4.3.107 genome copies and 3.3.108 genome copies in 250 ml of water.
2-Seawater artificially contaminated with wastewater: 200 ml of clean seawater were mixed with 50 ml of raw wastewater scored positive for SARS-CoV-2. SARS-CoV-2 concentration was 103 to 104 genome copies per 250 ml. Seawater parameters were: salinity = 33.9 g/l, turbidity = 2.7 NTU and pH = 8.7.
3-Influent WWTP samples: 250 ml of frozen or of freshly collected raw wastewaters were used, all samples being scored positive for SARS-CoV-2 (Table 2).
Table 2.
Concentrations of SARS-CoV-2 measured with traditional method and passive sampling.
| WWTP | Collection date | Wastewater (Log gc /100 ml) | Zetapor (Log gc/32 cm2) |
Nylon (Log gc/32 cm2) |
||
|---|---|---|---|---|---|---|
| 4 h | 24 h | 4 h | 24 h | |||
| A | 6/9/2020 | 3.5 | ND | ND | ND | ND |
| B | 6/2/2020 | 3.4 | ND | ND | ND | ND |
| B | 6/9/2020 | 3.5 | ND | ND | ND | ND |
| A | 8/4/2020 | 3.2 | 3.2 | ND | 3.2 | 2.4 |
| B | 8/4/2020 | 3.3 | 2.2 | 2.3 | ND | 2.2 |
| B | 8/25/2020 | 3.5 | 1.1 | ND | 1.3 | 1.9 |
| B | 9/22/2020 | 4.2 | 2.7 | 3.0 | ND | 3.5 |
| A | 10/20/2020 | 4.3 | 3.2 | 2.5 | ND | 3.2 |
| A | 11/4/2020 | 4.5 | 2.6 | 3.7 | 2.6 | 3.6 |
| A | 11/9/2020 | 3.3 | ND | 3.7 | 3.0 | 3.7 |
| A | 11/17/2020 | 4.1 | 2.4 | 3.5 | 3.4 | 3.4 |
| A | 11/24/2020 | 3.7 | ND | 2.5 | ND | 3.6 |
| B | 11/24/2020 | 4.2 | 1.8 | ND | ND | 2.6 |
| Ba | 3/31/2121 | 3.7 | 2.5 | 2.9 | 2.5 | 2.8 |
| Aa | 3/31/2121 | 3.9 | 2.5 | 2.8 | 2.4 | 2.4 |
| Ba | 4/6/2121 | 4.3 | 3.0 | 3.1 | 2.25 | 2.7 |
| Aa | 4/6/2021 | 4.4 | 3.1 | 3.1 | 1.0 | 2.2 |
| Ba | 4/13/2021 | 4.3 | 3.4 | 3.2 | 3.5 | 3.4 |
| Aa | 4/13/2021 | 4.02 | 3.1 | 3.0 | 2.9 | 2.7 |
Wastewater samples were collected from two WWTP (sites A & B), from June 2020 to April 2021. Nylon and Zetapor membranes were immersed for 4 h or 24 h either in thawed or freshly collected samples. ND = not detected (No Ct).
Freshly collected samples.
The conditions of exposure were identical for the three mixtures tested: membranes of nylon and Zetapor were immersed in a beaker containing 250 ml of mixture, for 4 h and 24 h at room temperature (20 ± 2 °C) with continuous stirring. At the end of the exposure period, the membranes were rinsed in sterile water for 30 s to eliminate non-adsorbed particles and stored at −20 °C. For each experiment, a fraction of the viral inoculum and 1 ml of the exposure mixture was sampled at the beginning were titrated in parallel by qRT-PCR to calculate the titer of the inoculum and of the exposure mixture. Two or three independent experiments were carried out for each condition tested. For seawater spiked with wastewater, the concentration of SARS-CoV-2 was also measured at the end of exposure to evaluate the potential degradation of the virus after 24 h in exposure mixture.
2.5. Field study on a waste water treatment plant: Passive sampling and traditional sampling
For field study, passive sampling using nylon and traditional sampling (24 h composite samples) were performed in parallel on a WWTP located in a small city southern Brittany (40,000 P.E.). A daily monitoring was performed for 3 weeks from November to December 2020. The devices were immersed for 24 h in the flow of the influent ensuring that the membrane was constantly immersed. At the end of exposure, nylon was rinsed in sterile water to remove large particles and stored at −20 °C.
2.6. Nucleic acids extraction
Raw wastewater samples were homogenized and 11 mL were ultracentrifugated as described previously (Wurtzer et al., 2020). Viral pellets were resuspended in 200 μL of PBS 1×. The membranes of zetapor and nylon were immersed directly in 8 ml of lysis solution to which 4 ml of 1× PBS were added. For virus titration, RNA was extracted from 10 μl of virus stock solution and from 1 ml of exposure mixture. All nucleic acids were extracted using a NucliSENS extraction kit (bioMérieux, Lyon France) as previously described (Vincent-Hubert et al., 2021), eluted with 100 μl of nuclease-free water and kept frozen at −20 °C until purification. Nucleic acids extracted from membranes exposed in the laboratory were then purified using a Qiagen kit (RNA MinElute Clean up, Qiagen, France) to eliminate potential PCR inhibitors, eluted with 100 μl of nuclease-free water (Qiagen, France) and kept frozen at −20 °C. As partial inhibition of qRT-PCR was found with wastewater samples from field study, we used a One Step PCR Inhibitor removal kit (Zymo Research Kit, USA) for all these samples. Inhibited samples represented 25% and 37% of membranes exposed respectively in seawater and wastewater.
2.7. Detection of viral genomes by one-step quantitative RT-PCR
qRT-PCR was performed using an UltraSense One-Step quantitative RT-PCR system (Invitrogen) on an MX3000 (Stratagene, Massy, France). For SARS-CoV-2, one set of primers and probe IP4, targeting the polymerase gene was used (Etievant et al., 2020), with cycling condition as described previously (Desdouits et al., 2021). For NoV genogroup I (GI) and II (GII), qRT-PCR were carried out as described previously (Le Guyader et al., 2009). For PEDV, previously described primers (Bigault et al., 2020) and probe (Kim et al., 2007) were used based on the same cycling conditions as NoV GII. All samples were analysed in duplicate using 5 μl of undiluted or tenfold-diluted nucleic acid extracts. A negative amplification control (sterile water) was included in each amplification series. Inhibition of RT-PCR reaction was controlled by comparing the Ct values of pure and tenfold-diluted nucleic acid extracts. For quantification, duplicate 5-points standard curves were made with PEDV in-vitro transcript T171 (Bigault et al., 2020) and SARS-CoV-2 RNA transcript (CNR des virus respiratoires, Pasteur Institute, Paris), and the synthetic plasmids containing nucleotides 146–6935 of the GI.1 Norwalk virus (Genbank M87661) or nucleotides 4191–5863 of the GII.4 Houston virus (Genbank EU310927). Only samples that yielded a Ct value of less than 39 were included in the quantitative analysis. For their quantification, duplicates of undiluted and 1/10 -diluted extracts were used. Samples presenting a difference between Ct pure and Ct diluted corrected by the slope (ΔCt) < 1 were quantified using mean Ct pure values. For samples presenting a ΔCt > 1, the Ct value used for quantification was the Ct value obtained for the tenfold-diluted sample corrected using the slope of the standard curve.
2.8. Calculation of viral recovery on membranes
The recovery rate was calculated to compare the performance of each membrane. The formula used was:
where gc M = copies of viral genome measured with qRT-PCR in the membrane nucleic acid extract and gc EM = copies of viral genome added in the exposure media (virus inoculum or genome copies in sewage).
2.9. Statistical analysis
The viral RNA copy numbers were log10-transformed before statistical analysis. To determine the effects of factors (type of membrane, duration of exposure, type of waters, WWTP site) on the concentration of SARS-CoV-2 genome on membrane, ANOVA or Kruskal–Wallis test were performed. Multiple comparison tests were then performed: Tukey HSD test was used for pairwise comparisons or Dunn's test, respectively following ANOVA and Kruskal-Wallis test. The efficiency of passive sampling and traditional sampling was compared with Kruskal-Wallis test. Three levels were considered significant: p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***). All statistical analysis and data plotting were performed with R Studio v 3.6.
3. Results
3.1. Detection of SARS-CoV-2 and PEDV genomes on membranes immersed in spiked waters
In order to confirm that SARS-CoV-2 genome could be detected with passive sampling using Zetapor and nylon, we first tested the adsorption capacities of a SARS-CoV-2 surrogate, the PEDV, and of heat-inactivated SARS-CoV-2. Membranes were immersed in seawater and wastewater spiked with these two viruses for 4 h and 24 h.
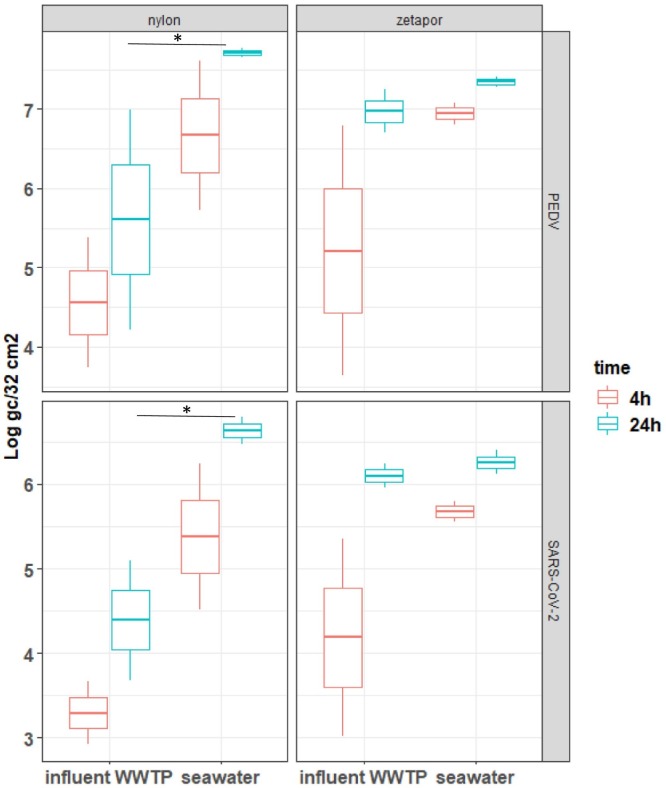
After 4 h of exposure, viruses were detected on both membranes and quantification showed high concentrations ranging from 3 to 7.7 Log10 genome copies/32 cm2 membrane with a high variability (Fig. 2 ). For PEDV and SARS-CoV-2, the concentrations of genome per membrane increased between 4 h and 24 h whatever the type of membrane and of water. A higher but not significant concentration of both viruses were measured on Zetapor in wastewater compared with nylon (Fig. 2). A significant effect of water was observed with nylon membrane immersed 24 h for both viruses, with higher concentrations when membranes were immersed in seawater compared with wastewater (p < 0.05).
Fig. 2.
SARS-CoV-2 and PEDV genomes concentrations on membranes immersed in spiked wastewater and seawater
Membranes of nylon or zetapor were immersed for 4 h or 24 h in wastewater or seawater spiked with heat-inactivated SARS-CoV-2 or with PEDV. Boxplots show the minimum, 25th percentile, median, 75th percentile, and maximum concentration/32 cm2 membrane. Kruskal-Wallis test followed by a Dunn's multiple comparisons test: *p < 0.05, ** p < 0.01, ***p < 0.001.
Concerning the recovery rate, a significant effect of the type of water was observed at 24 h with nylon membrane: the recovery rate of PEDV and SARS-CoV-2 with nylon being higher in seawater than in influent WWTP, respectively p < 0.01 and p < 0.05 (Table 1 ). The highest recovery rates were obtained with nylon membranes immersed 24 in seawater, with 15.9 ± 2.4% and 15.4 ± 5.6% for PEDV and SARS-CoV-2 respectively, which corresponds for SARS-CoV-2 to a 38 times higher recovery rate in seawater compared to wastewater (Table 1). For wastewater, the highest mean recovery rate of both viruses was around 4% except for nylon (0.04%) (Table 1).
Table 1.
Recovery rate (%) of viruses on membranes immersed in spiked wastewater and seawater.
| PEDV |
SARS-CoV-2 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Nylon |
zetapor |
Nylon |
zetapor |
|||||
| 4 h | 24 h | 4 h | 24 h | 4 h | 24 h | 4 h | 24 h | |
| Influent WWTP | 0.09 ± 0.1 | 3.78 ± 2.6 | 2.4 ± 1.6 | 4.04 ± 4 | 0.03 ± 0.02 | 0.4 ± 0.6 | 1.69 ± 1.19 | 4.5 ± 3.03 |
| Seawater | 7.5 ± 5.5 | 15.9 ± 2.4** | 3.1 ± 2.2 | 6.8 ± 0.8 | 6.5 ± 4.1 | 15.4 ± 5.6* | 1.7 ± 1.2 | 6.6 ± 4.5 |
Recovery rates of viruses with membranes exposed for 4 h and 24 h in waters (influent WWTP or seawater) spiked with PEDV and heat-inactivated SARS-CoV-2. Mean value ±SD. Comparisons were made for seawater/influent WWTP with Kruskal-Wallis test: *p < 0.05; ** p < 0.01.
3.2. Detection of SARS-CoV-2 in wastewater samples
3.2.1. Detection and quantification of SARS-CoV-2 with passive sampling
To further explore the performance of each membrane in recovering SARS-CoV-2, the experiment was carried out with previously tested, SARS-CoV-2 positive wastewater samples. The first series of tests was performed on 13 samples stored at −20 °C since June 2020 to November 2021 and the second series of tests was performed on 6 freshly collected wastewater samples (Table 2 ).
Ten of 13 membrane samples were positive for SARS-CoV-2 whatever the type of membrane and the immersion time with Ct values ranging from 32.2 to 36.8 (Table 2). The concentration of SARS-CoV-2 was between 1.1 and 3.7 Log10/32 cm2 depending on the time of immersion and membrane (Table 2). For three samples, that are the oldest stored samples, the absence of SARS-CoV-2 detection with membrane could be explain by a decrease of virus concentration due to a degradation of the virus caused by freezing. We therefore remeasured the viral concentration in these samples and found a decreased concentration of 1 to 2.5 Log10, which could explain why the virus genome was not detected with membranes. For this reason, the recovery rate was not calculated on this set of data as there is an uncertainty on the concentration value due to the conservation.
As SARS-CoV-2 is known to be damaged by freezing, membranes were immersed in freshly collected wastewater samples. SARS-CoV-2 was detected and quantified after 4 h and 24 h immersion in all samples with both membranes (Table 2). The recovery was calculated on this series only and we found similar recovery rate of: 2.5 ± 1.7% and 3.1 ± 1.4% after 4 h immersion, and 2.4 ± 1.2% and 3.5 ± 1.5% after 24 h immersion, respectively for nylon and Zetapor.
3.2.2. Analysis of the effect of WWTP sampling site, membrane, exposure time and sampling methods
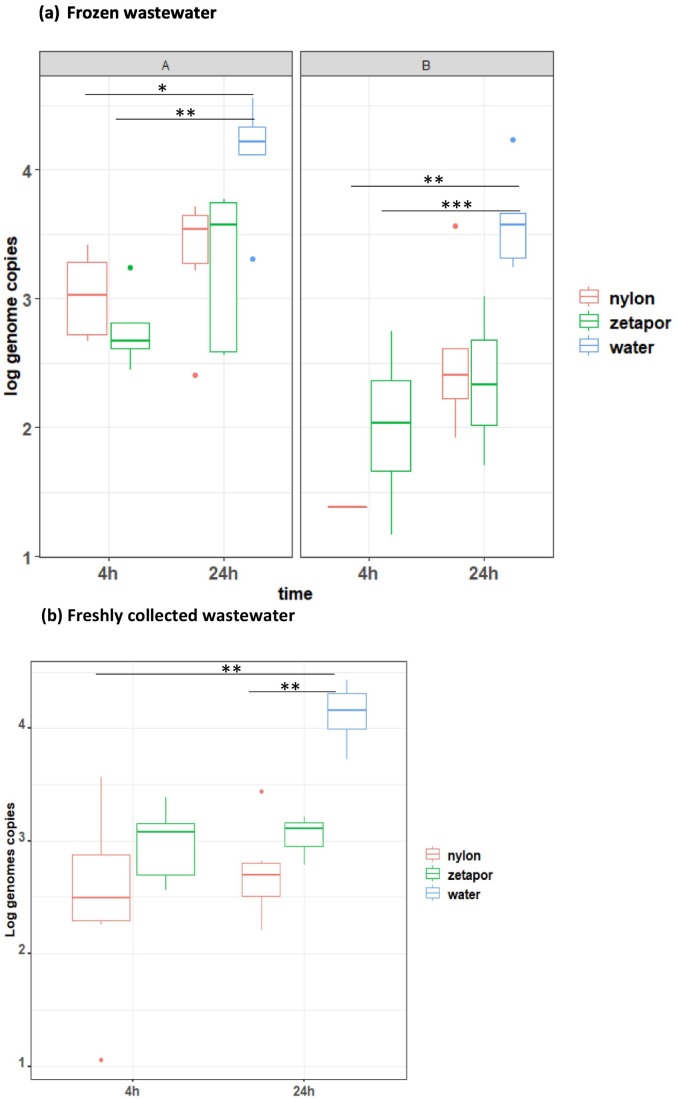
The potential influence of membrane performance and the effect of exposure time were analysed separately for the two complete datasets (“frozen samples” dataset and “freshly collected samples” dataset). No significant effect of membrane and time were found for wastewater samples while a significant effect of the WWTP sampling site was found with nylon membrane after 24 h immersion with the frozen sample data set only (p < 0.05) (Fig. 3A - B).
Fig. 3.
Comparison of SARS-CoV-2 concentration measured with traditional method and passive sampling
Membranes of nylon or zetapor were immersed for 4 h or 24 h in wastewater samples collected from two WWTP (sites A & B). (a) frozen wastewater, (b) freshly collected wastewater. Boxplots show the minimum, 25th percentile, median, 75th percentile, and maximum concentration/32 cm2 of membrane or /100 ml of wastewater. Kruskal-Wallis test followed by a Dunn's multiple comparisons test: *p < 0.05, ** p < 0.01, ***p < 0.001.
When comparing passive sampling to traditional water analysis, with the frozen sample dataset, a significant difference was found at 4 h, whatever the membrane, suggesting that 24 h immersion of membranes provides a better estimate of the concentration in wastewater than 4 h immersion (Fig. 3B). With the freshly collected samples, a significant difference was found between the concentrations measured with nylon membrane and with traditional sampling, whatever the duration of immersion, while no significant difference was found between zetapor and traditional sampling, suggesting that the zetapor membrane gives a better estimation of the viral contamination compared to the nylon membrane (Fig. 3B).
3.3. Detection of SARS-CoV-2 in seawater contaminated with wastewater
To determine whether SARS-CoV-2 could be detected with membranes in seawater, seawater has been artificially contaminated with wastewater positive for SARS-CoV-2. The concentration of the virus did not decrease in the contaminated seawater during 24 h (data not shown). SARS-CoV-2 genome was detected and quantified on all membranes, whatever the duration of membrane immersion (Fig. 4 ). Similar concentrations of genome were detected on nylon and Zetapor membranes, and no significant effect of time and membrane was found. The recovery rates of the virus were higher but not significantly with nylon compared with Zetapor, with respectively 13.2 ± 5% and 10.1 ± 4% after 4 h, and 16.5 ± 5% and 11.6 ± 5% after 24 h.
Fig. 4.
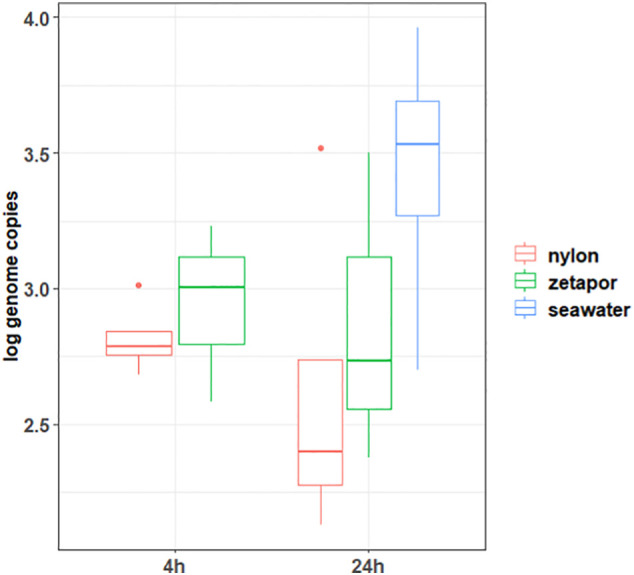
Concentration of SARS-CoV-2 genome on membranes immersed in sewage spiked seawater
Membranes of nylon and zetapor were immersed for 4 h or 24 h in seawater artificially contaminated with wastewater. Boxplots show the minimum, 25th percentile, median, 75th percentile, and maximum concentration/32 cm2 membrane or /100 ml of seawater. n = 5.
Serial dilution of this exposure mixture has been done until 10−2 to determine if the virus concentration on membrane decreases with the virus concentration in the exposure media. The mean concentration of the undiluted medium was 4.2 Log cRNA/250 ml, and in-1 and -2 dilutions, the viral concentrations were respectively 3 Log cRNA/250 ml and 2 Log cRNA/250 ml. The virus was detected on all membranes regardless of the dilution factor, and virus concentration on membrane decreased with the dilution factor. The virus concentrations for zetapor were 3.5 ± 0.1, 3 ± 0.3 and 2.5 ± 0.4 and for nylon they were 2.9 ± 0.7, 2.78 ± 0.5, and 2.26 ± 0.01 respectively for undiluted, −1 and − 2 dilutions. Similar amounts of genome copies were detected on membranes exposed to undiluted and 10-fold dilutions, suggesting that the dilution did not affect significantly the adsorption of SARS-CoV-2 in samples that are 10-fold diluted.
3.4. Detection and quantification of SARS-CoV-2 and NoV on membranes deployed in a WWTP
As nylon and Zetapor tested with freshly collected wastewater samples displayed similar recovery rate during laboratory experiments, nylon has been chosen for deployment in a WWTP as it can easily be used in the selected devices that need to be robust (Fig. 1). In order to determine whether SARS-CoV-2 concentrations measured with passive sampling and composite sampling display the same trend, both sampling methods were used for three weeks in the influent of a WWTP.
SARS-CoV-2 was always detected and quantified on nylon membrane, while NoVs was detected and quantified in 15 of 21 membranes, with 4 samples below LD (Table 3 ), (frequency of detection = 76.2% for NoVs) (Table 3). For the 3-weeks monitoring, the concentrations of SARS-CoV-2 and NoVs were similar ranging from 1.2 to 3.2 Log cRNA/100 cm2 and always lower than the concentrations measured in wastewater (concentration of viruses expressed for 100 ml): for SARS-CoV-2, the mean concentration was 1.9 Log lower, and for NoVs, the mean concentration was 2.4 Log lower (Table 3). Surprisingly, on nylon membrane, NoVs concentrations were low compared to those of SARS-CoV-2 while the opposite was observed in water, with higher NoVs concentrations compared to SARS-CoV-2 (Table 3). No correlation was found between the measured concentrations of SARS-CoV-2 RNA with passive sampling and composite sampling. On the other hand, by using the mean concentration of the values obtained during the three weeks, ANOVA shown that the sampling method significantly influences the measurement of the viral concentration for NoVs (p < 0.001), but not for SARS-CoV-2, suggesting that passive sampling is representative of SARS-CoV-2 contamination in wastewater (Table 3).
Table 3.
Concentration of SARS-CoV-2 and NoV GII in WWTP influent measured with passive sampling and composite sampling.
| SARS-CoV-2 |
NoV GII |
|||
|---|---|---|---|---|
| Date of sampling | Passive sampling |
Composite sampling |
Passive sampling |
Composite sampling |
| (Log gc /100cm2) | (Log gc /100 ml) | (Log gc /100cm2) | (Log gc /100 ml) | |
| 24-Nov-2020 | 1.7 | 4.1 | 1.2 | 3.2 |
| 25-Nov-2020 | 1.6 | 3.8 | 2.7 | 4.7 |
| 26-Nov-2020 | 1.8 | 4.4 | 2.8 | 4.6 |
| 27-Nov-2020 | 2.2 | 4.3 | 2.0 | 5.1 |
| 28-Nov-2020 | 2.2 | 4.0 | <LD | 5.2 |
| 29-Nov-2020 | 1.5 | 4.1 | 1.9 | 4.7 |
| 30-Nov-2020 | 1.4 | 4.1 | <LD | 4.6 |
| 1-Dec-2020 | 1.4 | 4.2 | <LD | 4.5 |
| 2-Dec-2020 | 1.6 | 3.8 | 2.3 | 4.5 |
| 3-Dec-2020 | 1.4 | 4.1 | 1.7 | 4.9 |
| 4-Dec-2020 | 2.4 | 3.9 | ND | 5.0 |
| 5-Dec-2020 | 2.8 | 4.0 | ND | 4.8 |
| 6-Dec-2020 | 1.8 | 3.9 | ND | 4.3 |
| 7-Dec-2020 | 2.4 | 3.7 | ND | 4.6 |
| 8-Dec-2020 | 2.4 | 3.9 | ND | 4.7 |
| 9-Dec-2020 | 2.3 | 4.2 | ND | 4.5 |
| 10-Dec-2020 | 2.6 | 4.1 | 1.9 | 4.1 |
| 11-Dec-2020 | 2.8 | 4.0 | 2.7 | 4.7 |
| 12-Dec-2020 | 3.0 | 4.0 | 1.8 | 4.1 |
| 13-Dec-2020 | 2.4 | 3.7 | <LD | 3.9 |
| 14-Dec-2020 | 3.2 | 4.1 | 1.4 | 4.0 |
| mean ± SD | 2.1 ± 0.5 | 4 ± 0.2 | 2.03 ± 0.5*** | 4.51 ± 0.4 |
| Frequency of detection (%) | 100 | 100 | 76.2 | 100 |
Nylon membrane (100 cm2) were immersed in the influent of a WWTP during 24 h from the 24/11/2020 to 14/12/2020; composite sampling: 24 h composite sample of raw wastewater. LD: limit of detection; ND: not determined. ANOVA was followed by Tukey's multiple comparisons test: ***p < 0.001.
4. Discussion
This paper presented the optimization of SARS-CoV-2 passive sampling with two synthetic membranes that we have previously selected for the passive sampling of other microorganisms, such as NoVs, sapovirus, the ostreid herpes virus OsHV-1 and a marine bacteria Vibrio spp. (Vincent-Hubert et al., 2021). The conditions tested in this study allowed a rapid detection and quantification of the SARS-CoV-2 genome in wastewater and seawater samples during laboratory experiments, with both membranes, confirming that SARS-CoV-2 can be detected with passive samplers in wastewater samples as published recently with other types of membranes (Bivins et al., 2021; Hayes et al., 2021; Liu et al., 2020; Schang et al., 2021). To our knowledge this is the first published study that evaluate the possible use of passive samplers in seawater for SARS-CoV-2 detection and in wastewater influent for NoV detection while it was detected before in continental waters and estuary (Tian et al., 2017; Vincent-Hubert et al., 2021).
Based on our data, we found that the recovery rate of coronaviruses, whether it is PEDV, inactivated or native SARS-CoV-2 were similar, indicating that PEDV is a good surrogate and that heat inactivation does not appear to alter the adsorption of the virus to the membrane. However, the recovery rates of all viruses seem to be dependent on matrices, as it was always much higher in seawater than in wastewater. These matrices are known to be highly contrasted and differ in pH, concentration of inorganic salts, chemical contaminants, particulate and dissolved organic matters and PCR inhibitors, all parameters that can modulate either the persistence of the virus in seawater, its adsorption onto the membranes or its detection by PCR. Indeed, as we have already observed with norovirus in serial dilution experiments of the exposure medium, the adsorption of SARS-CoV-2 on the membrane did not decrease proportionally, suggesting that the characteristics of the exposure medium are determinant (Vincent-Hubert et al., 2017).The persistence of the virus studied in laboratory under controlled conditions, the times for 90% reduction (T 90) of viable SARS-CoV-2 in wastewater and seawater were respectively 1.7 days and 1.1 days at room temperature (Ahmed et al., 2020; Bivins et al., 2020; Sala-Comorera et al., 2021). SARS-CoV-2 RNA concentrations are higher in settled solids than in the liquid components of wastewater as shown in many recent studies suggesting that the virus is preferentially adsorbed onto particles (Graham et al., 2021; Li et al., 2021) and confirming the initial observations of (Ye et al., 2016) on the partitioning of model enveloped viruses in raw wastewater samples. However, we still do not know whether the genome or the viral particle is really adsorbed on membranes, which can also explain the recovery rate. Indeed, in contrast to the rapid inactivation of infectious SARS-CoV-2 in water, viral RNA was relatively stable in seawater at 20 °C (> 20 days) (Sala-Comorera et al., 2021) and a T 90 values of 3.3 days and 26.2 days were found in wastewater at respectively high and low titers (Bivins et al., 2020).
Many parameters may influence the detection of virus with passive samplers, such as the type of membrane and exposure time as well as the efficiency of the method used for virus/nucleic acids recovery. The choice of the membrane tested in this study was based on previous observations demonstrating that an enveloped virus, the ostreid herpes virus OsHV-1, was more frequently detected with Zetapor membrane compared with nylon and LDPE (Vincent-Hubert et al., 2021). We show here that Zetapor and nylon displayed a similar recovery rate of SARS-CoV-2 from frozen wastewater samples, and that Zetapor seems to be more efficient than nylon with freshly collected wastewater samples. It is difficult to compare our data with published data as we did not use the same membranes neither the same method for virus recovery. First published papers reported the use of cotton gauze, cotton cheesecloth, cellulose sponges and electronegative filters. According to Schang et al. (2021), a higher proportion of electronegative membrane was positive compared with gauze and cotton puds and in the same way, Hayes et al. (2021) showed that the highest virus recovery was achieved using both cheesecloth and electronegative filters compare with gauze and sponge.
Another factor that can increase the SARS-CoV-2 detection is the method used for virus particle recovery and the addition of a nucleic acid purification step. According to published studies, elution of viral particles is used with gauze (Bivins et al., 2021; Liu et al., 2020; Schang et al., 2021), whereas it is rarely used with other membranes (Hayes et al., 2021). Elution of virus particle was not used in our study with nylon and Zetapor, as it was not necessary for NoV recovery, neither with cotton and electronegative membrane (Schang et al., 2021; Vincent-Hubert et al., 2021). With cheesecloth and electronegative filter, elution seemed to increase the recovery rate compared to direct lysis of virus particle (Hayes et al., 2021). Concerning the purification of nucleic acids, in these matrices, and particularly in sewage characterized by high concentrations of particulate matters, it is useful to include a final purification step to remove PCR inhibitors as they can ultimately decrease the efficiency of detection as we did, or to dilute the RNA to decrease the inhibition (Hayes et al., 2021).
The recovery rates of SARS-CoV-2 with passive samplers were evaluated in only one published study using gauze, cheesecloth, sponge and electronegative membranes (Hayes et al., 2021). We found that the recovery rate of the virus is similar with the Zetapor membrane, an electropositive membrane (Hayes et al., 2021). The recovery rate can vary depending on the virus itself, as we have shown with laboratory exposures that it was always lower for NoV (less than 0.1% in undiluted sewage) than for SARS-CoV-2 (around 3% in undiluted sewage), whatever the matrices and the membranes (Vincent-Hubert et al., 2017). This corroborates the field results where it is interesting to note that NoV is less efficiently detected than SARS-CoV-2 while the contrary was observed with traditional sampling, suggesting that NoV is less adsorbed than SARS-CoV-2 on nylon.
When we compared the effectiveness of passive sampling and traditional sampling in term of detection, we showed that both sampling methods display the same pattern of SARS-CoV-2 and NoVs detection during the three-week monitoring, indicating that nylon membrane is appropriate for SARS-CoV-2 detection. However, the concentration of viruses on membranes measured during field experiment were always lower compared with traditional sampling. This can be explained by a desorption of the viral particles due to the intensity of the flow at the entrance of the WWTP, the membrane would then undergo a washout when the flow is important. Another hypothesis is that the membrane selects a fraction of virus. Indeed, as SARS-CoV-2 is preferentially adsorbed on solids, it is possible that solids are not efficiently sampled with membrane, in contrast water analysis include the particulate organic matter which could explain the differences in concentration observed between the two methods. Regarding the difference between the spiked experiments and the experiments in WWTP, in the first case the viral concentration is relatively constant and high and there is no flow, while at the entrance of the WWTP, the viral concentration, the flow intensity and the type of waters vary. Therefore, since the membranes immersed in WWTP are not in permanent contact with virus, their recovery efficiency could be lower compared to the recovery rate measured in spiked experiments.
Regarding the expression of SARS-CoV-2 concentrations on membrane, they can only be reported at the surface and not at the volume of water because we do not know the volume of water flowing over the membranes, which does not allow for a comparison of methods in terms of concentration. For this reason, passive sampling with zetapor and nylon gives qualitative information. If in the future the objective of using passive samplers is to estimate virus concentration, further experiments are needed to investigate the notion of steady state that is very well described for chemical contaminants, and which conditions the immersion time of the samplers and allows the calculation of the concentrations (Booij et al., 2016). Recently, calibrations of various passive samplers have been performed for enteric viruses and gave promising information concerning the sampling kinetic of viruses confirming our previous data (Li et al., 2022; Vincent-Hubert et al., 2017). Indeed, the aim of these studies today is mainly to improve detection over short times rather than to estimate the integrating effect of the membranes. Concerning the immersion time of the membranes, most of the published studies have only tested an immersion of 24 h, and we show here that an immersion of 4 h is sufficient to detect the virus, as already shown by (Bivins et al., 2021) and that its concentration increases with time.
The difference in SARS-CoV-2 concentrations measured with passive sampling and traditional sampling is still an issue and need to be improved. Nevertheless, passive samplers could be used as an early warning tool to alert of the rise of the concentration particularly at the city-scale and neighborhood-scale. For seawater, the approach needs to be validated in field experiment, but results presented here are promising. This is of special interest as seawater sampling is difficult to performed and virus extraction from this matrix is difficult to perform (Desdouits et al., 2021). Moreover, recent development of biophysical model-based scenarios with different virus half-life concluded that SARS-CoV-2 may represents a threat in coastal waters especially in winter (Guo et al., 2021).
5. Conclusion
We have successfully developed passive sampling of SARS-CoV-2 with nylon and Zetapor membrane in sewage. For seawater, passive sampling efficient on a laboratory scale could be applied in case of coastal area contamination. Based on our data analysis, both membranes are suitable however, Zetapor seems to be more efficient for wastewater on freshly collected water sample and a 24 h immersion time was more representative of viral contamination. Even though concentrations measured with both methods were not correlated, the methods have shown the same trend. Both approaches present some interests and may be applied in diverse situations. For sewage, we proposed a qualitative method that gives results within a day which allows a great reactivity. Passive sampling could be an early warning tool if the samplers are placed on the wastewater network, for example when grab sampling is not possible, which would allow for targeted public health actions. This promising approach will be useful for the detection of other viruses or bacteria, being complementary of epidemiological data obtained.
CRediT authorship contribution statement
FVH: Conceptualization; Funding acquisition; Formal analysis; Methodology; Writing original draft, review & editing. CW: Investigation; Writing - reviewing & editing, MD: Conceptualization; Formal analysis; Funding acquisition; Project administration; reviewing & editing. SJ: Investigation; Writing - reviewing & editing. JS: Investigation; Writing - reviewing & editing. PLM: Investigation; Formal analysis; Writing - reviewing & editing. FND: Writing - reviewing & editing. O: Funding acquisition; Supervision; Visualization; Writing - reviewing & editing. FSLG: Conceptualization; Formal analysis; Funding acquisition; Methodology; Project administration; Resources; Supervision; Validation; Visualization; Writing - reviewing & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Céline Bressolette-Bodin (Service de Virology, Centre Hospitalier Universitaire & Université de Nantes, France) and Dr. Yannick Blanchard (ANSES, Génétique Virale et Biosécurité, Ploufragan, France) for helpful discussions and for providing heat-inactivated SARS-CoV-2 and PEDV.
This research project was supported by the Agence Nationale de la Recherche and the Fondation de France (ANR RA-Covid wave 5, n°00109676) and the Obepine network.
Editor: Warish Ahmed
Contributor Information
Obepine consortium:
I. Bertrand, M. Boni, C. Gantzer, Y. Maday, V. Marechal, J.-M. Mouchel, L. Moulin, and S. Wurtzer
References
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigault L., Brown P., Bernard C., Blanchard Y., Grasland B. Porcine epidemic diarrhea virus: viral RNA detection and quantification using a validated one-step real time RT-PCR. J. Virol. Methods. 2020;283 doi: 10.1016/j.jviromet.2020.113906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(12):937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Lott M., Shaffer M., Wu Z., North D., Lipp E., Bibby K. 2021. Building-level Wastewater Monitoring for COVID-19 Using Tampon Swabs and RT-LAMP for Rapid SARS-Cov-2 RNA Detection. [DOI] [Google Scholar]
- Booij K., Robinson C.D., Burgess R.M., Mayer P., Roberts C.A., Ahrens L., Allan I.J., Brant J., Jones L., Kraus U.R., Larsen M.M., Lepom P., Petersen J., Profrock D., Roose P., Schafer S., Smedes F., Tixier C., Vorkamp K., Whitehouse P. Passive sampling in regulatory chemical monitoring of nonpolar organic compounds in the aquatic environment. Environ. Sci. Technol. 2016;50(1):3–17. doi: 10.1021/acs.est.5b04050. [DOI] [PubMed] [Google Scholar]
- Buonerba A., Corpuz M.V.A., Ballesteros F., Choo K.H., Hasan S.W., Korshin G.V., Belgiorno V., Barcelo D., Naddeo V. Coronavirus in water media: analysis, fate, disinfection and epidemiological applications. J. Hazard. Mater. 2021;415 doi: 10.1016/j.jhazmat.2021.125580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzel N., Courbariaux M., Wang S.Y., Moulin L., Wurtzer S., Bertrand I., Laurent K., Monfort P., Gantzer C., Le Guyader S., Boni M., Mouchel J.M., Marechal V., Nuel G., Maday Y., Obepine C. A nationwide indicator to smooth and normalize heterogeneous SARS-CoV-2 RNA data in wastewater. Environ. Int. 2022;158 doi: 10.1016/j.envint.2021.106998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdouits M., Piquet J.C., Wacrenier C., Le Mennec C., Parnaudeau S., Jousse S., Rocq S., Bigault L., Contrant M., Garry P., Chavanon F., Gabellec R., Lamort L., Lebrun L., Le Gall P., Meteigner C., Schmitt A., Seugnet J.L., Serais O., Peltier C., Bressolette-Bodin C., Blanchard Y., Le Guyader F.S. Can shellfish be used to monitor SARS-CoV-2 in the coastal environment? Sci. Total Environ. 2021;778 doi: 10.1016/j.scitotenv.2021.146270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etievant S., Bal A., Escuret V., Brengel-Pesce K., Bouscambert M., Cheynet V., Generenaz L., Oriol G., Destras G., Billaud G., Josset L., Frobert E., Morfin F., Gaymard A. Performance assessment of SARS-CoV-2 PCR assays developed by WHO referral laboratories. J Clin. Med. 2020;9(6) doi: 10.3390/jcm9061871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattal B., Katzenelson E. Evaluation of gauze pad method to recover viruses from water. Water Res. 1976;10(12):1135–1140. doi: 10.1016/0043-1354(76)90047-6. [DOI] [Google Scholar]
- Fongaro G., Rogovski P., Savi B.P., Cadamuro R.D., Pereira J.V.F., Anna I.H.S., Rodrigues I.H., Souza D.S.M., Saravia E.G.T., Rodriguez-Lazaro D., Lanna M.C.D. SARS-CoV-2 in human sewage and river water from a remote and vulnerable area as a surveillance tool in Brazil. Food Environ. Virol. 2021 doi: 10.1007/s12560-021-09487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Grijalva L.M.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55(1):488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Gu J.Y., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020;158(6):1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Cao Y., Kong X., Kong S., Xu T. The Potential threat of SARS-CoV-2 in coastal waters. Ecotoxicol. Environ. Saf. 2021;220 doi: 10.1016/j.ecoenv.2021.112409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Hata A., Inaba M., Katayama H., Furumai H. Characterization of natural organic substances potentially hindering RT-PCR-based virus detection in large volumes of environmental water. Environ. Sci. Technol. 2017;51(23):13568–13579. doi: 10.1021/acs.est.7b00306. [DOI] [PubMed] [Google Scholar]
- Hayes E.K., Sweeney C.L., Anderson L.E., Li B., Erjavec G.B., Gouthro M.T., Krkosek W.H., Stoddart A.K., Gagnon G.A. A novel passive sampling approach for SARS-CoV-2 in wastewater in a Canadian province with low prevalence of COVID-19. Environ. Sci.Water Res. Technol. 2021 doi: 10.1039/d1ew00207d. [DOI] [Google Scholar]
- Kim S.H., Kim I.J., Pyo H.M., Tark D.S., Song J.Y., Hyun B.H. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol. Methods. 2007;146(1–2):172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guyader F.S., Parnaudeau S., Schaeffer J., Bosch A., Loisy F., Pommepuy M., Atmar R.L. Detection and quantification of noroviruses in shellfish. Appl. Environ. Microbiol. 2009;75(3):618–624. doi: 10.1128/aem.01507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Di D., Saingam P., Jeon M.K., Yan T. Fine-scale temporal dynamics of SARS-CoV-2 RNA abundance in wastewater during a COVID-19 lockdown. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Verhagen R., Ahmed W., Metcalfe S., Thai P., Kaserzon S., Smith W., Schang C., Simpson S., Thomas K., Mueller J., Mccarthy D. In situ calibration of passive samplers for viruses in wastewater. Environ. Sci. Technol. 2022 doi: 10.1021/acsestwater.1c00406. [DOI] [Google Scholar]
- Liu P., Ibaraki M., VanTassell J., Geith K., Cavallo M., Kann R., Moe C. 2020. A Novel COVID-19 Early Warning Tool: Moore Swab Method for Wastewater Surveillance at an Institutional Level. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K.R., Gong Y.Y., Lewis T.G. Environmental transmission of SARS at Amoy Gardens. J. Environ. Health. 2006;68(9):26–30. Retrieved from <Go to ISI>://WOS:000237130600002. [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Comorera L., Reynolds L.J., Martin N.A., O'Sullivan J.J., Meijer W.G., Fletcher N.F. Decay of infectious SARS-CoV-2 and surrogates in aquatic environments. Water Res. 2021;201 doi: 10.1016/j.watres.2021.117090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schang C., Crosbie N.D., Nolan M., Poon R., Wang M., Jex A., John N., Baker L., Scales P., Schmidt J., Thorley B.R., Hill K., Zamyadi A., Tseng C.W., Henry R., Kolotelo P., Langeveld J., Schilperoort R., Shi B.Q., Einsiedel S., Thomas M., Black J., Wilson S., McCarthy D.T. Passive sampling of SARS-CoV-2 for wastewater surveillance. Environ. Sci. Technol. 2021;55(15):10432–10441. doi: 10.1021/acs.est.1c01530. [DOI] [PubMed] [Google Scholar]
- Sikorski M.J., Levine M.M. Reviving the "Moore Swab": a classic environmental surveillance tool involving filtration of flowing surface water and sewage water to recover typhoidal salmonella bacteria. Appl. Environ. Microbiol. 2020;86(13) doi: 10.1128/aem.00060-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P., Yang D., Shan L., Wang D.P., Li Q.Q., Gorski L., Lee B.G., Quinones B., Cooley M.B. Concurrent detection of human norovirus and bacterial pathogens in water samples from an agricultural region in Central California coast. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent-Hubert F., Morga B., Renault T., Le Guyader F.S. Adsorption of norovirus and ostreid herpesvirus type 1 to polymer membranes for the development of passive samplers. J. Appl. Microbiol. 2017;122(4):1039–1047. doi: 10.1111/jam.13394. [DOI] [PubMed] [Google Scholar]
- Vincent-Hubert F., Wacrenier C., Morga B., Lozach S., Quenot E., Mege M., Lecadet C., Gourmelon M., Hervio-Heath D., Le Guyader F.S. Passive samplers, a powerful tool to detect viruses and bacteria in marine coastal areas. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.631174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A., Jellingso M., Sommer M.O.A. Spatial and temporal dynamics of SARS-CoV-2 in COVID-19 patients: a systematic review and meta-analysis. Ebiomedicine. 2020;58 doi: 10.1016/j.ebiom.2020.102916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Eurosurveillance. 2020;25(50):38–44. doi: 10.2807/1560-7917.Es.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y.Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]