Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing an outbreak of coronavirus disease 2019 (COVID-19), is a major threat to public health worldwide. Previous studies have shown that the spike protein of SARS-CoV-2 determines viral infectivity and major antigenicity. However, the spike protein has been undergoing various mutations, which bring a great challenge to the prevention and treatment of COVID-19. Here we present the MutCov, a pipeline for evaluating the effect of mutations in spike protein on infectivity and antigenicity of SARS-CoV-2 by calculating the binding free energy between spike protein and angiotensin-converting enzyme 2 (ACE2) or neutralizing monoclonal antibody (mAb). The predicted infectivity and antigenicity were highly consistent with biologically experimental results, and demonstrated that the MutCov achieved good prediction performance. In conclusion, the MutCov is of high importance for systematically evaluating the effect of novel mutations and improving the prevention and treatment of COVID-19. The source code and installation instruction of MutCov are freely available at http://jianglab.org.cn/MutCov.
Keywords: SARS-CoV-2, Spike protein, Mutation, Infectivity, Antigenicity
1. Introduction
The outbreak of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), has become a pandemic disease globally [[1], [2], [3], [4]]. As of 20 March 2022, over 468 million confirmed cases and over 6 million deaths of COVID-19 have been reported globally according to the World Health Organization [5]. COVID-19 has varied manifestations such as mild infection, pneumonia, lung failure, and even death. It has become a major threat to public health [6].
The genome of SARS-CoV-2 encodes spike (S), envelope (E), membrane (M), and nucleocapsid (N) structural proteins. Among those structural proteins, the surface S protein mediates the process of coronavirus entering into host cells [7], it must be cleaved by the protein convertase furin at the S1/S2 site and the transmembrane serine protease 2 (TMPRSS2) at the S2’ site into S1 and S2 subunits [8,9]. S1 is responsible for binding to the host cell receptor (including ACE2 [10,11], neuropilin-1 [12], and CD-147 [13]) or cathepsin L/B in the endosome pathway, S2 for the fusion of the viral and cellular membranes [7,[14], [15], [16], [17]]. There is a hypothesis that SARS-CoV-2 viral particles may use an alternative strategy for entering cells called antibody-dependent enhancement of infection (ADE) [[18], [19], [20], [21], [22]].
To prevent further dissemination of COVID-19, several promising neutralizing monoclonal antibodies (mAbs) have been developed according to the initial sequence of SARS-CoV-2 [[23], [24], [25], [26], [27], [28]], which have been proved to inhibit viral replication and alleviate severe clinical symptoms effectively [23,29]. Notably, S protein is the major antigen capable of inducing protective immune responses, mAbs inhibit the viral entry into host cells by blocking the binding of S protein to its receptor [[30], [31], [32], [33]].
RNA viruses have much higher mutation rate than DNA viruses due to the error-prone nature of the RNA-dependent RNA polymerase [34]. SARS-CoV-2 has accumulated a considerable amount of mutations in spike protein during the spread of COVID-19 [[35], [36], [37], [38]]. The great majority of those mutations are either ineffective or detrimental to virus function and then removed by natural selection [39]. However, some mutations in the S protein RBD region have been reported to significantly alter viral infectivity and antigenicity [[40], [41], [42], [43]], which bring challenges to the prevention and mAbs development of COVID-19. For example, the N439K in S protein has been proved to emerge independently in multiple SARS-CoV-2 lineages, it both increases S protein affinity for hACE2 and confers virus resistance against several neutralizing antibodies [44]. In addition, there are still many mutations (including K417 N, Y453F, A475V, L452R, V483A, F490L et al.) that are reported to show resistance to some neutralizing antibodies [40,45]. Thus it is crucial to closely monitor the evolution of SARS-CoV-2 and timely investigate the effect of those mutations on viral infectivity and antigenicity [46,47].
To address these questions, we present MutCov, a user-friendly pipeline for evaluating the effect of mutations in spike protein RBD region on SARS-CoV-2 infectivity and antigenicity by calculating the binding free energy between S protein and ACE2 receptor or mAb. MutCov is freely available as Docker images to simplify installation and analysis (http://jianglab.org.cn/MutCov).
2. Methods
2.1. Homology modeling
The build-in three-dimensional structures of wild-type RBD-receptor complexes were downloaded from the Protein Data Bank database [48]. MutCov removes the water and small molecules using PyMOL(The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.), then the amino acid sequence of mutant S protein is constituted according to the amino acid change (user-defined) and wild-type S protein. Finally MODELLER [49] is used for homology modeling of mutant S protein based on the amino acid sequence of mutant S protein and the three-dimensional structures of the wild-type S protein [50]. At the same time, the DOPE and GA341 methods offered by MODELLER are used to assess the reliability of the generated model [51].
2.2. Docking
The homology modeling module has got the three-dimensional structure of mutant S protein, the docking module further constructs the structures of mutant RBD-receptor complexes. Since the wild-type and mutant S proteins have similar three-dimensional structures, MutCov aligns the mutant S protein to wild-type S protein in RBD-receptor complexes using PyMOL. Then the wild-type S protein is deleted from the complexes [[52], [53], [54], [55]].
2.3. Molecular dynamics simulations
For wild-type and mutant RBD-receptor complexes, MutCov performs the molecular dynamics simulations with the CHARMM36 [56] force field and TIP3P [57] water model using GROMACS software (version 5.1.4) [58], respectively. In the preparation stage, the complexes are solvated in a rectangular water box with 10 Å distance from box boundaries, then the Na+ and CL− ions are added to neutralize the whole system and achieve a final salt concentration of 0.15 M [[59], [60], [61]].
Subsequently, the MutCov performs 50,000-step energy minimization with all atoms unrestrained. Then 1ns NVT equilibration under constant volume conditions and 1ns NPT equilibration at 1 bar pressure and 310K temperature are performed [62]. Finally, MutCov performs 5ns (MutCov default value) production MD simulations with a 2fs integration step. The Particle Mesh Ewald [63] method is used to compute long-range electrostatic interactions, and the SHAKE [64] method is used to constrain the covalent bonds involving hydrogen atoms [65].
2.4. The analysis of molecular dynamics trajectory
To evaluate the structure change of complexes, the binding free energy, binding free energy contribution of each residue, root mean square deviation (RMSD), root mean square fluctuation (RMSF), solvent accessible surface area (SASA), radius of gyration (RG), and hydrogen bonds (HB) analyses are performed based on the molecular dynamics trajectories in binding free energy calculation module.
The RMSD, RMSF, SASA, RG, and HB are analyzed using GROMACS built-in tools. The RMSD describes the molecule's overall discrepancy with respect to reference at each time of molecular dynamics simulations. The RMSF describes the variation of each atom in RBD-receptor complexes over the whole trajectory. SASA describes the surface area of the RBD-receptor complexes that is accessible to solvent at each time of molecular dynamics simulations. RG describes the radius of gyration of RBD-receptor complexes. HB is defined as the number of hydrogen bonds only between the S protein RBD region and ACE2 receptor or mAb [66].
The binding free energy describes the change in the free energy associated with a binding process [67]. In other words, the binding of RBD-receptor occurs spontaneously only when the binding free energy is negative, and the decrease of binding free energy can promote the RBD-receptor binding [68]. Since the complexes are in an unstable state during the early MD production phase, the binding free energy is calculated according to the last 20% of MD trajectories. For infectivity evaluation, the binding free energy is calculated between the RBD region of S protein and ACE2 protein. Similarly, the binding free energy is calculated between the RBD region and double chains of neutralizing antibody respectively during antigenicity evaluation. The g_mmpbsa tool [69] is used to calculate the binding free energy, which is composed of van der waal energy, electrostatic energy, polar solvation energy, and SASA energy. To evaluate the effect of mutation on virus infectivity and antigenicity, the p-value for the difference between wild-type and mutant binding free energy is calculated with the Wilcoxon rank-sum statistic.
3. Results
3.1. Overview of the MutCov pipeline
MutCov takes the amino acid change in the S protein as input to predict the infectivity and antigenicity changes of mutant SARS-CoV-2 through four analytical modules based on state-of-the-art computational tools. Both single mutation, co-mutation (a virus strain contains multiple mutations), and multi-mutation (multiple virus strains contain multiple mutations) are accepted, and the mutation type can be substitution, insertion, and deletion. In addition to common built-in mAbs, users can evaluate the effect of mutations on the neutralization activity of customized mAbs as well.
MutCov is comprised of the following four modules outlined in Fig. 1 : (1) homology modeling of mutant S protein, (2) mutant S protein docking to ACE2 (in infectivity evaluation) or mAb (in antigenicity evaluation), (3) molecular dynamics (MD) simulations, and (4) binding free energy calculations.
Fig. 1.
Schematic overview of MutCov algorithm. The MutCov algorithm consists of homology modeling, docking, molecular dynamics simulation, and binding free energy calculation modules. The orange box is input, the green box is a module, the blue box is data, and the grey box is output.
The first module takes amino acid change and wild-type structures of S protein binding with its receptors (built-in or user-defined) as input to predict the three-dimensional structures of mutant S protein using MODELLER [49]. To further obtain complete structures of mutant RBD-receptor complexes, the second module performs the molecular docking of mutant S protein to its receptor with PyMOL. The third module adopts the molecular dynamics method to simulate the infection or neutralization process of SARS-CoV-2 using GROMACS [58]. The fourth module performs root mean square deviation (RMSD), root mean square fluctuation (RMSF), solvent accessible surface area (SASA), radius of gyration (RG), and hydrogen bonds (HB) analyses based on molecular dynamics trajectories. Subsequently, the binding free energy of S protein binding to ACE2 (infectivity) or mAbs (antigenicity) in both wild-type and mutant RBD-receptor complexes are calculated to evaluate the effect of mutation on infectivity or antigenicity.
MutCov is available as ready-to-use Docker images containing all the necessary software and dependencies. This allows running the pipeline in an isolated environment, preventing conflicts with other programs in the hosting environment.
3.2. Assessing the power of MutCov for infectivity prediction
To assess the power of MutCov for infectivity prediction, we test MutCov on previous research that verified the effect of mutations on SARS-CoV-2 infectivity and antigenicity by pseudotyped virus experiments [40]. Among them, 14 mutations located in the S protein RBD region are reported to significantly alter SARS-CoV-2 infectivity more than 4-fold, so the power of MutCov for infectivity prediction is assessed utilizing those 14 mutations (Fig. 2 A). The wild-type three-dimensional structure of the S protein RBD region binding to ACE2 (PDB ID: 6M0J) is used for the homology modeling of mutant RBD-ACE2 complexes. Then MutCov pipeline performs the molecular dynamics simulations for both wild-type and mutant complexes with default parameters. Finally, the binding free energy of the S protein binding to ACE2 is calculated, and the effect of mutation on infectivity is inferred by comparing the binding free energy between wild-type and mutant complexes (see the “Methods” section for details).
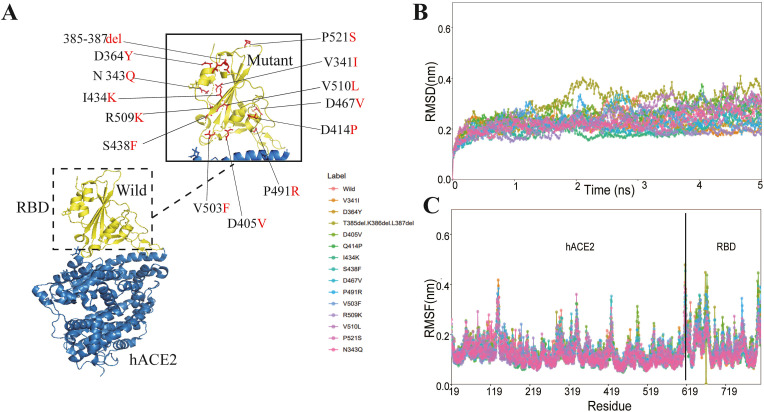
Fig. 2.
The RBD-hACE2 interaction profile of MD simulations. (A)The structure of SARS-CoV-2 RBD-hACE2 (PDB ID: 6M0J). SARS-CoV-2 RBD region is shown in yellow and its interacting hACE2 is colored green. The mutations reported altering the infectivity of SARS-CoV-2 2 are colored red. (B) The RMSD changes of backbone atoms for the wild and mutant RBD-hACE2 complexes during the 5 ns MD trajectories. (C) The RMSF of atoms for the wild and mutant RBD-hACE2 complexes.
To verify the convergence of MD simulations equilibrium, MutCov estimates the root mean square deviations (RMSD) of backbone atoms relative to the corresponding crystal structure, and RMSD results show all the RBD-ACE2 complexes have reached stabilization at 3 ns(Fig. 2B and Supplementary Table S1). The flexibility patterns of residues in the mutant RBD-hACE2 complex display similar fluctuations to the wild-type complex (Fig. 2C). To verify the solvent exposure degree of RBD-ACE2 complexes, MutCov also calculated the solvent accessible surface area (SASA), and all the systems show a relatively stable solvent exposure degree (Supplementary Table S1).
The binding free energy of the S protein RBD region binding to ACE2 in wild-type and mutant complexes are shown in Table 1 , and 9 out of 14 infectivity changes predicted by MutCov are consistent with the experiment results. As shown in Supplementary Table S1, the binding free energy changes are mainly focused on electrostatic energy and polar solvation energy. For example, the 385-387del mutant RBD-ACE2 complexes show significantly higher electrostatic energy (−668.850 ± 48.660 kJ/mol) compared to wild-type (−1329.412 ± 58.592 kJ/mol), and the electrostatic energy of V503F R509K, V510L, and P521S mutant RBD-ACE2 complexes also significantly increase by more than 100 kJ/mol. In addition, the polar solvation energy of Q414P and S438F mutant RBD-ACE2 complexes also increase by more than 100 kJ/mol compared with wild-type. Subsequently, MutCov also explores the critical residues involved in the RBD-hACE2 binding by calculating the per-residue decomposition of binding free energy (Supplementary Table S1). The contribution energy of per-residue shows that the LYS386 residue (−140.11 kJ/mol) promotes RBD-ACE2 complexes binding, which is deleted in 385–387del RBD-hACE2 complexes.
Table 1.
The infectivity change of mutant SARS-CoV-2 virus evaluated by MuCov pipeline and pseudotyped virus experiments.
| Mutation | BFEa of wild-type | BFEa of mutation | P-valueb | Infectivity predicted by MutCov | Infectivity verified by Experimentsd |
|---|---|---|---|---|---|
| V341I | −1063.82 | −968.03 | 2.43E-05 | Decreasec | Decrease |
| N343Q | −1063.82 | −954.17 | 1.53E-08 | Decreasec | Decrease |
| D364Y | −1063.82 | −1139.9 | 2.28 E−02 | Increase | Decrease |
| 385-387del | −1063.82 | −580.49 | 9.05E-14 | Decreasec | Decrease |
| D405V | −1063.82 | −1404.58 | 1.21E-13 | Increase | Decrease |
| Q414P | −1063.82 | −952.87 | 2.68E-06 | decreasec | decrease |
| I434K | −1063.82 | −1140.21 | 2.98E-04 | Increase | Decrease |
| S438F | −1063.82 | −1056.02 | 1.65E-03 | Decreasec | Decrease |
| D467V | −1063.82 | −1066.04 | 0.80 | Increase | Decrease |
| P491R | −1063.82 | −1285.05 | 2.61E-11 | Increase | Decrease |
| V503F | −1063.82 | −905.23 | 1.91E-09 | Decreasec | Decrease |
| R509K | −1063.82 | −960.91 | 0.00348688 | Decreasec | Decrease |
| V510L | −1063.82 | −821.38 | 6.63E-14 | Decreasec | Decrease |
| P521S | −1063.82 | −945.07 | 1.61E-09 | Decreasec | Decrease |
The binding free energy (kJ/mol) of RBD region of S protein (wild-type or mutant) binding to ACE2.
The p value of Wilcoxon rank-sum statistic for the binding energy change between wild-type and mutation.
The antigenicity predicted by MutCov show a consistent change with pseudotyped virus experiments.
The infectivity verified by pseudotyped virus experiments.
3.3. Assessing the power of MutCov for antigenicity prediction
Similarly, to assess the power of MutCov for antigenicity prediction, we then test MutCov based on the reported mutations which significantly alter the neutralization activity of mAb [40]. Part of the mutations are excluded from the following analysis because of lacking the three-dimensional structure, so MutCov simulates the effect of the remaining 6 mutations in spike protein on the neutralization activity of their corresponding mAbs including CB6 (PDB ID: 7C01), H014 (PDB ID: 7CAH), and B38 (PDB ID: 7BZ5). MutCov pipeline performs the molecular dynamics simulations for both wild-type and mutant RBD-mAb complexes with the “--Model” parameter to specify the mAb to be used (see the “Methods” section for details).
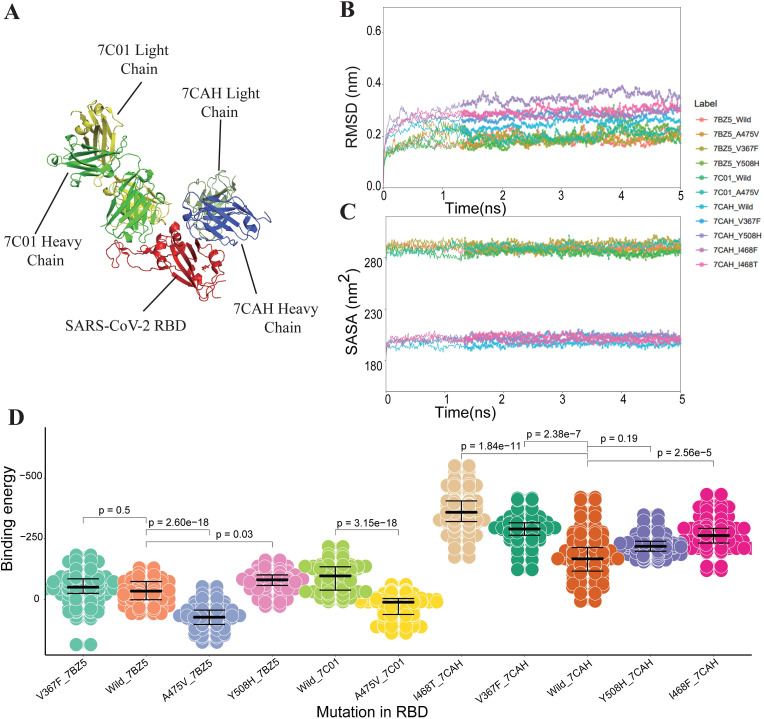
As is shown in the three-dimensional structure of RBD-mAb complexes (Fig. 3 A), the 7CAH and 7C01 mAbs bind to the different RBD regions of S protein, and 7BZ5 mAb show a similar RBD-mAb binding pattern with 7C01. The RMSD results (Fig. 3B) show all the RBD-mAb complexes also reached stabilization at 3 ns. The SASA plot (Fig. 3C) shows the solvent exposure degree of 7CAH RBD-mAb complexes is quite different from that of 7C01 and 7BZ5, which may mainly cause by the different structural patterns.
Fig. 3.
The structural and energetic of both wild and mutant RBD-mAbs interactions. (A) Crystal structures of RDB-7C01/7CAH complexes. The RBD region of S protein is colored red, the heavy and light chains of 7C01 mAbs are represented as light green and yellow respectively, the heavy and light chains of 7CAH mAbs are represented as blue and dark green respectively. (B) The changes in the RMSD values of backbone atoms for the different wild and mutant RBD-mAb complexes during the 5 ns MD trajectories. (C) The residues wise SASA value for wild and mutant RBD-mAbs complexes. (D) The binding free energies for both wild and mutant complexes of RBD-mAb (including 7BZ5, 7C01, and 7CAH), the Wilcoxon rank-sum statistic are conducted to check the statistical significance of binding free energy changes between wild and mutant complexes.
The binding free energy of the RBD region of S protein binding to heavy chains of mAbs are shown in Table 2 and Fig. 3D, and 7 out of 8 antigenicity changes predicted by MutCov are consistent with the experiment results. The binding free energy of the RBD region binding to light chains shows a similar result with heavy chains (Supplementary Table S1). Both the heavy and light chains of 7BZ5 and 7C01 mAbs show higher binding free energy with A475V-mutant S protein than wild-type which indicates A475V is resistant to 7BZ5 and 7C01 mAbs. Oppositely, double chains of 7BZ5 and 7CAH mAbs show lower binding free energy with V367F-mutant S protein which indicates V367F becomes more sensitive to 7BZ5 and 7CAH mAbs. In addition, some mutations in the RBD region, including Y508H, I468F, and I468T, are observed to be more susceptible to the neutralization activity of their corresponding mAbs (Table 2 and Supplementary Table S1).
Table 2.
The antigenicity change of mutant SARS-CoV-2 virus evaluated by MuCov pipeline and pseudotyped virus experiments.
| Antibody | Mutation | BFEa of wild type | BFEa of mutation | P-valueb | Antigenicity predicted by MutCov | Antigenicity verified by experimentsd |
|---|---|---|---|---|---|---|
| 7BZ5 [25] | A475V | −34.81 | 72.34 | 2.60E-18 | Decreasec | Decrease |
| 7BZ5 | V367F | −34.81 | −49.59 | 0.50 | Increasec | Increase |
| 7BZ5 | Y508H | −34.81 | −77.71 | 0.03 | Increasec | Increase |
| 7C01 [26] | A475V | −87.28 | 25.85 | 3.15E-18 | Decreasec | Decrease |
| 7CAH [27] | V367F | −168.99 | −286.56 | 2.38E-07 | Increasec | Increase |
| 7CAH | Y508H | −168.99 | −225.19 | 0.19 | Increase | Decrease |
| 7CAH | I468F | −168.99 | −267.97 | 2.56E-05 | Increasec | Increase |
| 7CAH | I468T | −168.99 | −366.67 | 1.84E-11 | Increasec | Increase |
The binding free energy (kJ/mol) of RBD region of S protein (wild-type or mutant) binding to heavy chain of neutralizing antibody.
The p value of Wilcoxon rank-sum statistic for the binding energy change between wild-type and mutation.
The antigenicity predicted by MutCov show a consistent change with pseudotyped virus experiments.
The antigenicity change verified by pseudotyped virus experiments [40].
4. Discussion
As the SARS-CoV-2 has been undergoing various mutations which bring a great challenge to the prevention and treatment of COVID-19, it is vital to identify the high-risk mutations in the spike protein of SARS-CoV-2. Here, we presented the first computational pipeline to evaluate the infectivity and antigenicity of mutant SARS-CoV-2 on the basis of homology modeling, molecular dynamics simulations, and binding free energy calculations. When MutCov is tested on a public dataset, 9/14 infectivity and 7/8 antigenicity changes predicted by MutCov are consistent with the pseudotyped virus experiments. We expect the MutCov pipeline will provide references for the development of neutralizing antibodies and the prevention of COVID-19.
For the sake of creating a more practical pipeline, the MutCov accepts both substitution, insertion, and deletion of amino acids. As most SARS-CoV-2 virus strains contain multiple mutations, both single mutation, co-mutation, and multi-mutation can be accepted by MutCov. When facing multiple mutations, we recommend the user run the MutCov in a multi-mutation model instead of several single mutation models. In the multi-mutation model, MutCov will only calculate the binding free energy of wild-type for one time, which will save loads of computing resources. In addition, with the emergence of new mAbs, the user also can run MutCov with customized mAbs using the “--type CustomizedAntibody” parameter.
The prediction of each infectivity and antigenicity change takes on average about 14.4 h on a high-performance computing server (Intel Xeon Platinum 8260 CPU @ 2.40 GHz) utilizing 100 threads. Part of the infectivity and antigenicity changes predicted by MutCov are not in agreement with the pseudotyped virus experiments, the conformation of RBD-ACE2 complexes may not reach a stable status during the 5ns production MD simulations. To improve the accuracy and stability of prediction, the time scale of molecular dynamics simulations can be appropriately extended upon the computing resources using the '--MDtime’ parameter.
In the future, using MutCov, it is planned to study the structural determinants of recognition in models of the RBD domain complexes of the spike protein with the ACE2, CD-147, and neuropilin-1 receptors, by a complete enumeration of amino acid residues in the binding interface both in the RBD domain and in the receptor, with the estimation of binding energy and construction of a multidimensional surface of interaction energy. This will make it possible to understand the change in the species specificity of the virus and predict its further spread pathways.
5. Conclusions
In summary, MutCov is a user-friendly pipeline for evaluating the effect of mutations in spike protein on SARS-CoV-2 infectivity and antigenicity. During the testing phase, the predicted infectivity and antigenicity changes are highly consistent with experimental results. The MutCov source code, installation, usages, and output instructions are freely available from http://jianglab.org.cn/MutCov. As SARS-CoV-2 keeps evolving and adapting, MutCov will be of value for the immediate and accurate evaluation of novel mutations, which will improve the prevention of SARS-COV-2 and accelerate the development of therapeutic neutralizing antibodies.
Ethics approval and consent to participate
Not applicable.
Availability and requirements
Project name: MutCov.
Project home page: http://jianglab.org.cn/MutCov.
Operating system(s): Linux/Unix.
Programming language: Python (v2.7 or later).
Other requirements: Docker (version 19.03 or later).
License: GNU GPL.
Any restrictions to use by non-academics: None.
Availability of data and materials
The MutCov source code and installation instructions are freely available from http://jianglab.org.cn/MutCov. The build-in three-dimensional structures of wild-type RBD-receptor complexes were downloaded from the Protein Data Bank database (PDB ID: 6M0J, 6LZG, 6VW1, 7C01,7BWJ,6W41,6XC2,6XC3,6XCN,6WPT,7CAH, and 7BZ5) [48]. The verifications of the effect of mutations are available at https://doi.org/10.1016/j.cell.2020.07.012 [40].
Funding
This work has been supported by the National Natural Science Foundation of China (No. 62032007 to Q.J.), and the Emergency Research Project for COVID-19 of Harbin Institute of Technology (No. 2020–001 to Q.J.). The funding body did not participate in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Authors' contributions
A.A. and Q.J. conceived the project. W.Z. A.A. and C.X. collected mutation data and the three-dimensional structure of complexes. W.Z, C.X, M.L., P.W., Z.X., G.X., X.J., Y.H., Y.L., and H.N. contributed to the development and testing of pipeline software. G.X. and M.L. contributed to the website shown. Q.J., A.A., W.Z., and C.X. wrote the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
None Declared.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105509.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.C., et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., et al. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382:727–733. doi: 10.1056/NEJMoa2001017. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S., et al. The miRNA: a small but powerful RNA for COVID-19. Briefings Bioinf. 2021;22:1137–1149. doi: 10.1093/bib/bbab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Weekly epidemiological update on COVID-19 - 22 March 2022. 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---22-march-2022 Available from:
- 6.Ren X., et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913. doi: 10.1016/j.cell.2021.01.053. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tortorici M.A., Veesler D. 2019. Structural Insights into Coronavirus Entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bestle D., et al. vol. 3. Life Sci Alliance; 2020. (TMPRSS2 and Furin Are Both Essential for Proteolytic Activation of SARS-CoV-2 in Human Airway Cells). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guadalupe Benitez-Cardoza C., Luis Vique-Sanchez J. Identifying compounds that prevent the binding of the SARS-CoV-2 S-protein to ACE2. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantuti-Castelvetri L., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bittmann S. COVID-19: ACE-2 receptor, TMPRSS2, cathepsin-L/B and CD-147 receptor. J. Regenerat. Biol. Med. 2020 doi: 10.37191/Mapsci-2582-385X-2(3)-031. [DOI] [Google Scholar]
- 14.Zhao W., et al. Identification of nut protein-derived peptides against SARS-CoV-2 spike protein and main protease. Comput. Biol. Med. 2021;138 doi: 10.1016/j.compbiomed.2021.104937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suleman M., et al. Bioinformatics analysis of the differences in the binding profile of the wild-type and mutants of the SARS-CoV-2 spike protein variants with the ACE2 receptor. Comput. Biol. Med. 2021;138 doi: 10.1016/j.compbiomed.2021.104936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakib M.M.H., et al. Computational screening of 645 antiviral peptides against the receptor-binding domain of the spike protein in SARS-CoV-2. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S., et al. RNA-RNA interactions between SARS-CoV-2 and host benefit viral development and evolution during COVID-19 infection. Briefings Bioinf. 2021;23:bbab397. doi: 10.1093/bib/bbab397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nechipurenko Y.D., Anashkina A.A., Matveeva O.V. Change of antigenic determinants of SARS-CoV-2 virus S-protein as a possible cause of antibody-dependent enhancement of virus infection and cytokine storm. Biophysics. 2020;65:703–709. doi: 10.1134/S0006350920040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., et al. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect. Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaichuk T.A., et al. The challenges of vaccine development against betacoronaviruses: antibody dependent enhancement and sendai virus as a possible vaccine vector. Mol. Biol. 2020:1–15. doi: 10.1134/S0026893320060151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F., et al. 2020. Single Cell RNA and Immune Repertoire Profiling of COVID-19 Patients Reveal Novel Neutralizing Antibody, Protein Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi R., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 27.Lv Z., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin X., et al. Global characterization of B cell receptor repertoire in COVID-19 patients by single-cell V(D)J sequencing. Briefings Bioinf. 2021:6. doi: 10.1093/bib/bbab192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Addetia A., et al. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P., et al. Identification of potential vaccine targets for COVID-19 by combining single-cell and bulk TCR sequencing. Clin. Transl. Med. 2021;11:e430. doi: 10.1002/ctm2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P., et al. Comprehensive analysis of TCR repertoire in COVID-19 using single cell sequencing. Genomics. 2021;113:456–462. doi: 10.1016/j.ygeno.2020.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., et al. Rational optimization of a human neutralizing antibody of SARS-CoV-2. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheikh J.A., et al. Emerging genetic diversity among clinical isolates of SARS-CoV-2: lessons for today. Infect. Genet. Evol. 2020;84 doi: 10.1016/j.meegid.2020.104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Dorp L., et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zelenova M., et al. Analysis of 329,942 SARS-CoV-2 records retrieved from GISAID database. Comput. Biol. Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104981. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Dey T., et al. Identification and computational analysis of mutations in SARS-CoV-2. Comput. Biol. Med. 2021;129 doi: 10.1016/j.compbiomed.2020.104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grubaugh N.D., Petrone M.E., Holmes E.C. We shouldn't worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020;5:529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korber B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W., et al. Impact of mutations in SARS-COV-2 spike on viral infectivity and antigenicity. Briefings Bioinf. 2022;23 doi: 10.1093/bib/bbab375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou W., et al. N439K variant in spike protein alter the infection efficiency and antigenicity of SARS-CoV-2 based on molecular dynamics simulation. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.697035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson E.C., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187. doi: 10.1016/j.cell.2021.01.037. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starr T.N., et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mujwar S. Computational repurposing of tamibarotene against triple mutant variant of SARS-CoV-2. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan A., et al. Immunogenomics guided design of immunomodulatory multi-epitope subunit vaccine against the SARS-CoV-2 new variants, and its validation through in silico cloning and immune simulation. Comput. Biol. Med. 2021;133 doi: 10.1016/j.compbiomed.2021.104420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berman H.M., et al. The protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marti-Renom M.A., et al. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 50.Abbass J., Nebel J.C. Rosetta and the journey to predict proteins' structures, 20 years on. Curr. Bioinf. 2020;15:611–626. doi: 10.2174/1574893615999200504103643. [DOI] [Google Scholar]
- 51.Smolarczyk T., Roterman-Konieczna I., Stapor K. Protein secondary structure prediction: a review of progress and directions. Curr. Bioinf. 2020;15:90–107. doi: 10.2174/1574893614666191017104639. [DOI] [Google Scholar]
- 52.Gupta M., Sharma R., Kumar A. Docking techniques in toxicology: an overview. Curr. Bioinf. 2020;15:600–610. doi: 10.2174/1574893614666191003125540. [DOI] [Google Scholar]
- 53.Jiang M., et al. Function analysis of human protein interactions based on a novel minimal loop algorithm. Curr. Bioinf. 2019;14:164–173. doi: 10.2174/1574893613666180906103946. [DOI] [Google Scholar]
- 54.Zhu H., Du X., Yao Y. ConvsPPIS: identifying protein-protein interaction sites by an ensemble convolutional neural network with feature graph. Curr. Bioinf. 2020;15:368–378. doi: 10.2174/1574893614666191105155713. [DOI] [Google Scholar]
- 55.Jakhar R., et al. Relevance of molecular docking studies in drug designing. Curr. Bioinf. 2020;15:270–278. doi: 10.2174/1574893615666191219094216. [DOI] [Google Scholar]
- 56.Best R.B., et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theor. Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price D.J., Brooks C.L. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 58.Pronk S., et al. Gromacs 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skariyachan S., et al. Structural insights on the interaction potential of natural leads against major protein targets of SARS-CoV-2: molecular modelling, docking and dynamic simulation studies. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu T., et al. Exploring the binding mechanism of metabotropic glutamate receptor 5 negative allosteric modulators in clinical trials by molecular dynamics simulations. ACS Chem. Neurosci. 2018;9:1492–1502. doi: 10.1021/acschemneuro.8b00059. [DOI] [PubMed] [Google Scholar]
- 61.Xue W., et al. Computational identification of the binding mechanism of a triple reuptake inhibitor amitifadine for the treatment of major depressive disorder. Phys. Chem. Chem. Phys. 2018;20:6606–6616. doi: 10.1039/c7cp07869b. [DOI] [PubMed] [Google Scholar]
- 62.Wang J., et al. Molecular simulation of SARS-CoV-2 spike protein binding to pangolin ACE2 or human ACE2 natural variants reveals altered susceptibility to infection. J. Gen. Virol. 2020 doi: 10.1099/jgv.0.001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H., Gao X., Fang J. Multiple staggered Mesh Ewald: boosting the accuracy of the smooth particle Mesh Ewald method. J. Chem. Theor. Comput. 2016;12:5596–5608. doi: 10.1021/acs.jctc.6b00701. [DOI] [PubMed] [Google Scholar]
- 64.Kräutler V., Gunsteren W.F.V., Hünenberger P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2015;22:501–508. doi: 10.1002/1096-987X(20010415)22:5<501::AID-JCC1021>3.0.CO;2-V. [DOI] [Google Scholar]
- 65.Zhang Y., et al. Review of the applications of deep learning in bioinformatics. Curr. Bioinf. 2020;15:898–911. doi: 10.2174/1574893615999200711165743. [DOI] [Google Scholar]
- 66.Huang H., Gong X. A review of protein inter-residue distance prediction. Curr. Bioinf. 2020;15:821–830. doi: 10.2174/1574893615999200425230056. [DOI] [Google Scholar]
- 67.Wan S., et al. Rapid, accurate, precise and reproducible ligand-protein binding free energy prediction. Interface Focus. 2020;10 doi: 10.1098/rsfs.2020.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Du X., et al. Insights into protein-ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumari R., et al. g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MutCov source code and installation instructions are freely available from http://jianglab.org.cn/MutCov. The build-in three-dimensional structures of wild-type RBD-receptor complexes were downloaded from the Protein Data Bank database (PDB ID: 6M0J, 6LZG, 6VW1, 7C01,7BWJ,6W41,6XC2,6XC3,6XCN,6WPT,7CAH, and 7BZ5) [48]. The verifications of the effect of mutations are available at https://doi.org/10.1016/j.cell.2020.07.012 [40].