Abstract
Background
Some streptococci strains identified as Streptococcus pneumoniae (S. pneumoniae) by routine clinical methods exhibiting negative Quellung reaction results may belong to other species of viridans group streptococci or non‐typeable S. pneumoniae. The purpose of this study was to investigate the identification and molecular characteristics of S. pneumoniae with negative Quellung reaction results.
Methods
One hundred and five isolates identified as S. pneumoniae using routine microbiological methods with negative Quellung reaction results were included. Multilocus sequence analysis (MLSA) was used as a gold standard in species identification, and the capacity of matrix‐assisted laser desorption ionization‐time of flight mass spectrometry (MALDI‐TOF MS) in identification was evaluated. Capsular genes and sequence types of S. pneumoniae isolates were determined by sequential multiplex PCR and multilocus sequence typing. Antimicrobial susceptibility patterns were determined via broth microdilution with a commercialized 96‐well plate.
Results
Among the isolates, 81 were identified as S. pneumoniae and 24 were S. pseudopneumoniae by MLSA. MALDI‐TOF MS misidentified six S. pneumoniae isolates as S. pseudopneumoniae and nine S. pseudopneumoniae isolates as S. pneumoniae or S. mitis/S. oralis. Thirty‐one sequence types (STs) were detected for these 81 S. pneumoniae isolates, and the dominant ST was ST‐bj12 (16, 19.8%). The non‐susceptibility rates of S. pseudopneumoniae were comparable to those of NESp strains.
Conclusions
Some S. pneumoniae isolates identified by routine methods were S. pseudopneumoniae. Most NESp strains have a different genetic background compared with capsulated S. pneumoniae strains. The resistance patterns of S. pseudopneumoniae against common antibiotics were comparable to those of NESp.
Keywords: antimicrobial susceptibility, identification, MLSA, MLST, Streptococcus pneumoniae
Isolates identified as Streptococcus pneumoniae by routine tests were examined by multilocus sequence analysis, multilocus sequence typing, Vitek MS, and sequential multiplex PCR. Some Streptococcus pneumoniae isolates identified by routine methods could be Streptococcus pseudopneumoniae. Most non‐encapsulated Streptococcus pneumoniae strains have a different genetic background with capsulated Streptococcus pneumoniae strains.
1. INTRODUCTION
The routine methods for identifying Streptococcus pneumoniae (S. pneumoniae) in most clinical laboratories are mainly based on phenotypic characteristics, including morphology and biochemical reactions, such as optochin susceptibility test and bile solubility test. 1 However, the bile solubility test is often neglected in daily laboratory analysis. 2 Optochin‐susceptible non‐pneumococcal alpha‐hemolytic streptococci have been reported 3 ; thus, it is not reliable to identify S. pneumoniae using optochin susceptibility test results alone.
The Quellung reaction is the gold standard for serotyping of S. pneumoniae. 4 In previous studies, we noticed that some isolates identified as S. pneumoniae using routine methods displayed negative results in the Quellung reaction test, and such isolates were called non‐typeable S. pneumoniae (NTSp). Most NTSp strains lack a capsule, that is, they are non‐encapsulated S. pneumoniae (NESp). 5 , 6 It is important to make clear whether S. pneumoniae identified by routine methods with negative Quellung reaction result is true S. pneumoniae or other alpha‐hemolytic streptococci.
Some molecular methods targeting at specific genes such as 16S rRNA and lytA (encoding autolysin) have proven ineffective in differentiating S. pneumoniae from other viridans group streptococci (VGS), especially S. mitis. 4 , 7 Multilocus sequence analysis (MLSA) eliminates the potential problems resulting from homologous recombination, and it has been applied in the identification of several bacterial species that are difficult to identify using other methods. 8 MLSA for VGS, developed by Bishop et al., could accurately differentiate VGS at the species level. 9 Due to its rapid, simple, cost‐effective, and high‐throughput performance, matrix‐assisted laser desorption ionization‐time of flight mass spectrometry (MALDI‐TOF MS) has been widely used in clinical laboratories. 10 , 11 In addition, multilocus sequence typing (MLST) and whole‐genome sequencing (WGS) are reliable in the identification of VGS. 4 Hence, MLSA was used to accurately differentiated S. pneumoniae and to evaluate the ability of MALDI‐TOF MS.
Recently, NTSp isolates were categorized into Group I and Group II (null capsule clade1 [NCC1] and NCC2) based on the cps, aliB, and pspK genes. 12 To date, little is known about the genetic background or antibiotic resistance characteristics of NTSp strains.
In this study, 105 clinical isolates identified as S. pneumoniae using routine microbiological methods with negative Quellung reaction results were included. MLSA was used as a gold standard in species identification, and the capacity of MALDI‐TOF MS for identification was evaluated. Molecular epidemiological characteristics and antibiotic susceptibility patterns of these isolates were detected.
2. MATERIALS AND METHODS
2.1. Bacterial strains
The strains were isolated from children's respiratory tract specimens collected during daily work in the Microbiology Laboratory of Beijing Children's Hospital from 2013 to 2019. These isolates were identified as S. pneumoniae using a standard optochin susceptibility test, but showed negative results when they were serotyped via Quellung reaction test.
2.2. Optochin susceptibility and bile solubility tests
A disk (DD0001B 5 μg of optochin, Oxoid Ltd) was placed on a blood agar plate with the inoculated isolate. Then, the plate was incubated in 37℃ for 24 h. An inhibition zone diameter of ≥14 mm denoted a positive result.
The bile solubility assay was performed according to the standard procedures described by Arbique et al. 13 Visible clearing of bacterial suspensions in the deoxycholate [DZ0240 sodium deoxycholate (10%, pH 7), Leagene Biotechnology] was interpreted as a positive result.
In both experiments, strains with known positive and negative reactions kept in our laboratory were used as positive control and negative control strains, respectively.
2.3. Quellung reaction test
Omni serum (SSI), which contains antibodies to all known pneumococcal serotypes, was used to do the Quellung reaction test for all the isolates. The capsule became visible because of an in situ immunoprecipitation leading to changes in its refractive index, as well as agglutination of the bacterium. 14 Negative Quellung reaction did not have these features.
2.4. MLSA for VGS
Bacterial DNA was extracted using a DNA extraction kit (SBS Genetic Co. Ltd) according to the manufacturer's protocol. The seven housekeeping genes map, pfl, ppaC, pyk, rpoB, soda, and tuf were amplified as previously described. 15 Polymerase chain reaction (PCR) products were sequenced at Tianyihuiyuan Biotechnology Company. The identification of the isolates was performed following the published procedure. 9 Briefly, the sequences of the seven genes were aligned, trimmed, edited, and concatenated using the MEGA 4.0 software (http://www.megasoftware.net). Phylogenetic analysis of the concatenated sequences (map–pfl–ppaC–pyk–rpoB–soda–tuf), including isolates in this study and those reported in the MLSA database (download from http://www.emlsa.net/), was conducted using MEGA 4.0 and the maximum likelihood method with 1000 bootstrap replicates to build a neighbor‐joining phylogenetic tree.
2.5. MALDI‐TOF MS
All isolates were tested using the Vitek MS system (bioMérieux). Plate preparation, mass spectrum generation, and processing were performed using MYLA software affiliated with IVD version 3.0.
2.6. Sequential multiplex PCR for molecular serotyping
The molecular serotypes of S. pneumoniae isolates were determined with multiplex PCR. Twenty‐eight previously described oligonucleotide primers 15 were divided into seven groups and used to detect the serotypes. S. pneumoniae American Type Culture Collection strain 49619 (ATCC49619) was used as the quality‐control strain and included in each set of tests.
2.7. MLST for S. pneumoniae
The housekeeping genes aroE, gdh, gki, recP, spi, xpt, and ddl were amplified via PCR. 16 Each sequence of the seven loci was compared with those of all known alleles at the loci and with the sequence types (STs) in the database of the MLST website (http://spneumoniae.mlst.net). eBURST v3 software (http://spneumoniae.mlst.net/eburst/) was used to determine the relationships between isolates and to assign a clonal complex (CC) based on the stringent group definition of six of seven shared alleles.
2.8. NTSp grouping by PCR
NTSp can be divided into several groups using the cpsA, pspK, aliB‐like ORF1, and aliB‐like ORF2 genes. 12 Among these genes, cpsA is a conserved pneumococcal capsular polysaccharide gene, and aliB‐like ORF1 and aliB‐like ORF2 are frequently present in the capsular region of non‐capsulated pneumococci. PCR were performed following the protocol described in previous reports. 15 , 17 , 18
2.9. Antimicrobial susceptibility test
Susceptibility patterns to penicillin, amoxicillin/clavulanic acid, ceftriaxone, cefotaxime, cefuroxime, meropenem, erythromycin, azithromycin, chloramphenicol, trimethoprim/sulfamethoxazole, tetracycline, vancomycin, levofloxacin, and moxifloxacin were tested for all isolates. Minimum inhibitory concentrations (MICs) were determined via broth microdilution method with a commercialized 96‐well plate (B8283A, STP6F, Trek Diagnostic Systems Ltd), and the result was read by the Sensititre™ ARIS™ 2X ID/AST system (Thermo Fisher Scientific Inc.). The susceptibility rate was interpreted in accordance with the Clinical and Laboratory Standards Institute 2019 guidelines. 19 Quality control was performed using S. pneumoniae ATCC 49619.
2.10. Data analysis
The phylogenetic tree was built and analyzed using MEGA 4.0 software. Antimicrobial susceptibility and MLST data were analyzed using the WHONET 5.6 software as recommended by the World Health Organization.
3. RESULTS
3.1. Identification by MLSA
We observed that most of the isolates displayed small, shiny, smooth, and domed colonies without depressed centers as typical S. pneumoniae strains.
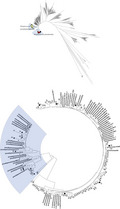
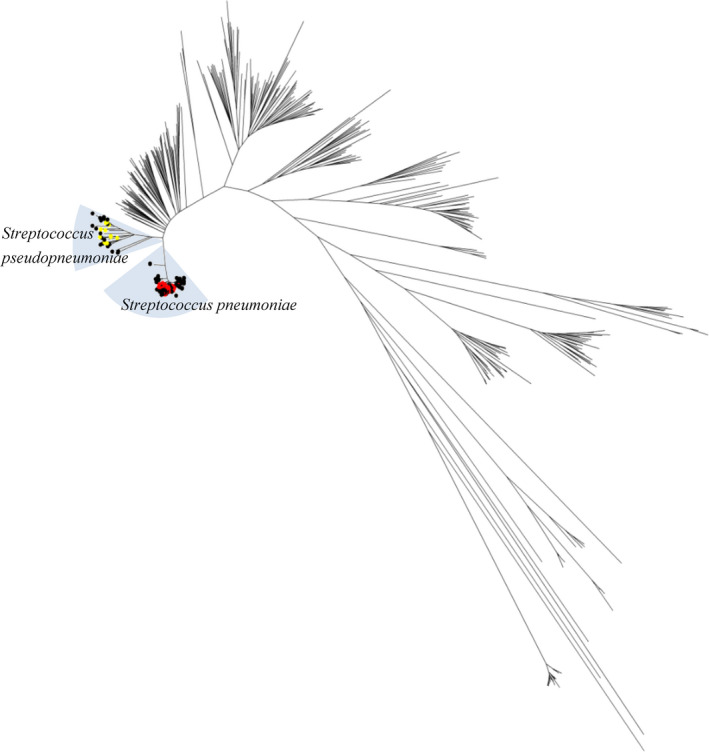
The 105 strains in this study and strains in the MLSA database (http://www.emlsa.net/) were analyzed together in the phylogenetic tree, and the results were presented in Figure 1 and Figure 2. As shown in Figure 1, the 105 isolates in the present study were significantly divided into S. pneumoniae (n = 81) branch and S. pseudopneumoniae (n = 24) branch. No other VGS were detected.
FIGURE 1.
Genetic relationships of the 105 isolates determined by multilocus sequence analysis (MLSA). The symbols indicate the following: ● isolates obtained in this study;  Streptococcus pneumoniae strains from the MLSA database;
Streptococcus pneumoniae strains from the MLSA database;  S. pseudopneumoniae strains from the MLSA database
S. pseudopneumoniae strains from the MLSA database
FIGURE 2.
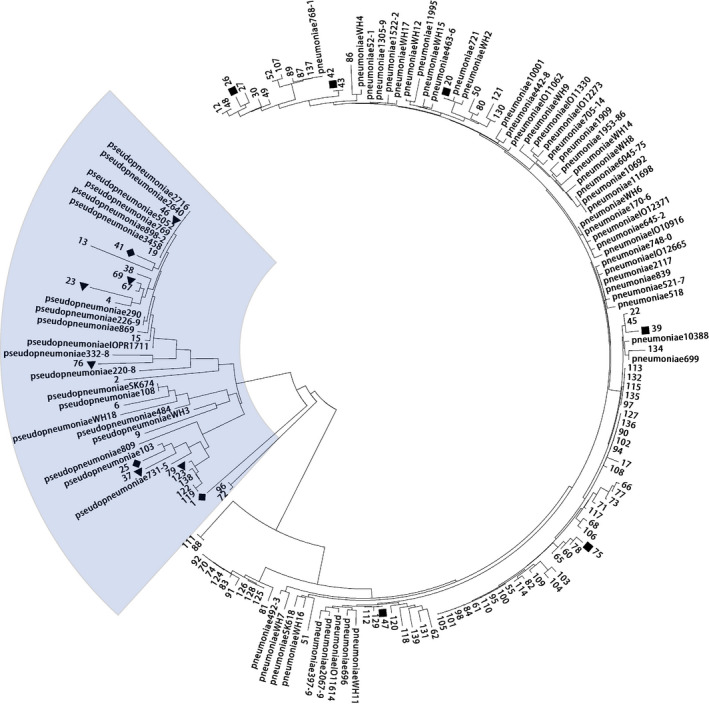
Phylogenetic tree of 105 isolates in this study with Streptococcus pneumoniae and S. pseudopneumoniae strains from the multilocus sequence analysis (MLSA) database. ■ S. pneumoniae isolates that were wrongly identified as S. pseudopneumoniae by Vitek MS. ▲ S. pseudopneumoniae isolates that were wrongly identified as S. pneumoniae by Vitek MS. ◆ S. pseudopneumoniae isolates that were wrongly identified as S. mitis/S. oralis by Vitek MS
3.2. Other identification methods
The results of MLSA analysis were used as a standard to evaluate the other identification methods. The optochin susceptibility test showed that the inhibition zone diameters caused by optochin ranged from 16 to 25 mm for the 81 S. pneumoniae isolates (median, 19 mm), whereas the diameters ranged from 14 to 22 mm for the 24 S. pseudopneumoniae isolates (median, 15 mm).
All 81 S. pneumoniae isolates exhibited positive results in the bile solubility test. For the 24 S. pseudopneumoniae strains, 19 showed negative results, and the other 5 showed positive results (Table 1).
TABLE 1.
Results of the bile solubility test and Vitek MS in this study
| Species | Bile solubility test | Vitek MS | |||
|---|---|---|---|---|---|
| Positive | Negative | S. pneumoniae | S. pseudopneumoniae | S. mitis/S. oralis | |
| S. pneumoniae (n = 81) | 81 | 0 | 75 | 6 | 0 |
| S. pseudopneumoniae (n = 24) | 5 | 19 | 6 | 15 | 3 |
Vitek MS could not accurately identify the species for 9 of 24 S. pseudopneumoniae isolates and 6 of 81 S. pneumoniae isolates (Table 1). The sensitivity and specificity of Vitek MS in the identification of S. pneumoniae were 91.7% and 71.4%, respectively. Besides, the isolates mistakenly identified by Vitek MS were distantly separated from each other and located in different sub‐branches in the phylogenetic tree, as presented in Figure 2.
3.3. Multiplex PCR for molecular serotyping and NTSp grouping
None of the isolates could be serotyped by the molecular method. The cpsA gene, which served as the internal control, could not be detected in any of the 105 isolates, although it was identified in the ATCC49619 strain, indicating that all isolates belonged to Group II.
Different NTSp types of these isolates were further identified. Seventeen isolates carrying both aliB‐like ORF1 and aliB‐like ORF2 genes were detected, and thus, the isolates were designated as NCC2. Forty‐six isolates carried the pspK gene, and they were designated as NCC1.
3.4. MLST
As shown in Figure 3, thirty‐one STs belonging to four CCs were detected in the 81 S. pneumoniae isolates, 20 of which were newly identified (named ST‐bj01 to ST‐bj20) with known alleles. CC‐bj12 was the most common CC, being identified in 32.1% of the isolates (26/81). The other three CCs were CC448 (12.3%, 10/81), CC2218 (6.2%, 5/81), and CC10236 (3.7%, 3/81).
FIGURE 3.

Population snapshot of the eBURST analysis for 81 Streptococcus pneumoniae isolates. The numbers in parentheses were the corresponding quantities of isolates for different sequence types (STs). STs without parentheses were linked to only one isolate
Housekeeping genes of the 24 S. pseudopneumoniae isolates were also amplified. No ST could be defined for any of the S. pseudopneumoniae isolates because at least one sequence of the seven loci did not match the known alleles in the database.
3.5. Antimicrobial susceptibility test
The antibiotic susceptibility patterns of the present 105 isolates against 14 antimicrobials are presented in Table 2. All of the isolates were susceptible to vancomycin, levofloxacin, and moxifloxacin. The non‐susceptibility rates (intermediate and resistant) to amoxicillin/clavulanic acid were 8.6% and 8.4%, respectively, for S. pneumonia and S. pseudopneumoniae. The resistance rate of S. pseudopneumoniae to erythromycin and azithromycin was 95.8%. The resistance rate of S. pneumoniae to erythromycin was 74.1% and to azithromycin was 66.7%. The MIC distributions of S. pseudopneumoniae and S. pneumonia isolates were similar.
TABLE 2.
Non‐susceptibility rates and MIC distribution of the Streptococcus pneumoniae and Streptococcus pseudopneumoniae isolates for 14 antimicrobials
| Antibiotics | S. pneumoniae (n = 81) | S. pseudopneumoniae (n = 24) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Non‐susceptibility | MIC (μg/ml) | Non‐susceptibility | MIC (μg/ml) | |||||||
| I% | R% | MIC50 | MIC90 | Range | I% | R% | MIC50 | MIC90 | Range | |
| Penicillin | ||||||||||
| Oral | 56.8 | 13.6 | 0.25 | 2 | ≤0.03–>4 | 54.2 | 16.6 | 0.5 | 2 | ≤0.03–>4 |
| Meningitis‐parenteral | ‐ a | 70.4 | 0.25 | 2 | ≤0.03–>4 | ‐ a | 70.8 | 0.5 | 2 | ≤0.03–>4 |
| Nonmeningitis‐parenteral | 3.7 | 3.7 | 0.25 | 2 | ≤0.03–>4 | 4.2 | 4.2 | 0.5 | 2 | ≤0.03–>4 |
| Amoxicillin/clavulanic acid | 3.7 | 4.9 | ≤2 | ≤2 | ≤2–16 | 4.2 | 4.2 | ≤2 | ≤2 | ≤2–8 |
| Ceftriaxone | ||||||||||
| Meningitis | 7.4 | 9.9 | ≤0.12 | 1 | ≤0.12–>2 | 8.4 | 8.4 | 0.25 | 1 | ≤0.12–>2 |
| Nonmeningitis | 3.7 | 6.2 | ≤0.12 | 1 | ≤0.12–>2 | 0 | 8.4 | 0.25 | 1 | ≤0.12–>2 |
| Cefotaxime | ||||||||||
| Meningitis | 4.9 | 6.2 | ≤0.12 | 1 | ≤0.12–>4 | 8.4 | 8.4 | 0.25 | 1 | ≤0.12–>4 |
| Nonmeningitis | 2.5 | 3.7 | ≤0.12 | 1 | ≤0.12–>4 | 0 | 8.4 | 0.25 | 1 | ≤0.12–>4 |
| Cefuroxime (parenteral) | 8.6 | 19.8 | ≤0.5 | ≥4 | ≤0.5–>4 | 8.4 | 50 | 1 | 4 | ≤0.5–>4 |
| Meropenem | 9.9 | 7.4 | ≤0.25 | 0.5 | ≤0.25–2 | 20.8 | 8.4 | ≤0.25 | 0.5 | ≤0.25–2 |
| Erythromycin | 3.7 | 74.1 | >2 | >2 | ≤0.25–>2 | 0 | 95.8 | >2 | >2 | ≤0.25–>2 |
| Azithromycin | 8.6 | 66.7 | >2 | >2 | ≤0.25–>2 | 0 | 95.8 | >2 | >2 | ≤0.25–>2 |
| Chloramphenicol | ‐ a | 39.5 | 2 | 16 | ≤1–16 | ‐ a | 4.2 | 2 | 4 | ≤1–8 |
| Trimethoprim/sulfamethoxazole | 44.4 | 28.4 | 2 | 4 | ≤0.5–>4 | 8.4 | 70.8 | >4 | >4 | ≤0.5–>4 |
| Tetracycline | 4.9 | 82.8 | >8 | >8 | ≤1–>8 | 0 | 91.6 | >8 | >8 | ≤1–>8 |
| Vancomycin | ‐ a | ‐ a | ≤0.5 | ≤0.5 | ≤0.5 | ‐ a | ‐ a | ≤0.5 | ≤0.5 | ≤0.5 |
| Levofloxacin | 0 | 0 | ≤0.5 | 1 | ≤0.5–1 | 0 | 0 | 1 | 1.5 | ≤0.5–2 |
| Moxifloxacin | 0 | 0 | ≤1 | ≤1 | ≤1 | 0 | 0 | ≤1 | ≤1 | ≤1 |
Abbreviation: MIC, minimum inhibitory concentration.
No breakpoints listed.
4. DISCUSSION
Our present study showed that MLSA could differentiate S. pseudopneumoniae from S. pneumoniae well, while the bile solubility test and MALDI‐TOF MS could not always completely and accurately distinguish S. pseudopneumoniae and S. pneumoniae.
S. pseudopneumoniae was first described by Arbique et al. 13 in 2004. Arbique et al. 13 believed that S. pseudopneumoniae was resistant or intermediate resistant to optochin when conducting the optochin test in CO2 atmosphere, and was negative in the bile solubility test. In 2010, Leegaard et al. 20 reported one S. pseudopneumoniae isolate with positive result in bile solubility test. A Spanish study examining 61 S. pseudopneumoniae strains reported that only 50.8% of the strains exhibited a typical optochin phenotype and 36.1% were bile‐soluble. 21 The findings of the above studies, combined with the results of our present study, suggested that the optochin test or the bile solubility test alone is not enough to identify S. pseudopneumoniae.
In recent years, MALDI‐TOF MS has shown a promising application prospect in the identification of species within the mitis group. 22 In the present study, we found that MALDI‐TOF MS could not always distinguish S. pneumoniae from S. pseudopneumoniae accurately. In accordance with our finding, Van Prehn et al. 23 reported one S. pneumoniae isolates mistakenly identified as S. mitis/S. oralis group using the Vitek MS platform. A recent study reported that the Vitek MS only identified 2 of 17 S. pseudopneumoniae isolates. 22 The discrepancy may be due to different databases or software versions used for MALDI‐TOF MS analysis. Improvement of database entries with multiple spectra of well‐characterized species has yielded high identification rates for Mycobacterium spp. 24 and Helicobacter pylori. 25
MLSA, a method in prokaryotic taxonomy considering the internal fragments of several genes, 26 is increasingly applied to obtain a higher resolution between species within a genus. The phylogenetic trees of MLSA were based on the concatenated aligned gene sequences and can reflect the accurate relationship of bacterial taxa. 27 Now, MLSA has been successfully used in the identification of S. anginosus, 28 Pseudomonas spp., 29 and other species. 30 , 31 MLSA has also been validated as a tool for the identification of reliable species among VGS, 32 and thus, it served as the standard in the present study. Most of the isolates in this study were identified as S. pneumoniae by MLSA. Combined with the negative Quellung reaction result and molecular biological characteristics, these strains were determined as NTSp. Our sequential multiplex PCR results showed that these 81 S. pneumoniae isolates did not carry cpsA gene fragment. It is important to realize that the regulatory and processing genes cpsABCD (also known as wzg, wzh, wzd, and wze) are conserved with high sequence identity in all S. pneumoniae isolates for the generation of capsular polysaccharides (CPSs). 33 They are common to all pneumococcal serotypes, excluding serotypes 3 and 37, and deletion of any of these genes will affect the production of CPSs. 34 Thus, we considered that the NTSp isolates in our present study may lack the capsule and could be designated as NESp.
NTSp can be divided into two groups based on the capsular genes. Group I strains carry the cps locus sequences of conventional capsule types but with disrupted ability to produce CPSs. Group II strains, lacking all of the genes usually found in the cps sequences in encapsulated S. pneumoniae isolates, 35 can be further divided into different cps types (NCC1, NCC2, and NCC3). NCC1 strains carry the pspK gene, which encodes a novel pneumococcal surface protein with several features playing a role in cell adhesion and enhanced colonization. NCC2 strains carry both aliB‐like ORF1 and aliB‐like ORF2 genes, which are predicted to encode lipoproteins. NCC3 strains carry aliB‐like ORF2 but not aliB‐like ORF1, and they were revealed to be not pneumococci. 12 All of the 81 S. pneumoniae isolates in our present study were categorized into Group II. Of these isolates, 17 isolates were classified as NCC2, and 46 isolates were classified as NCC1. The remaining 18 isolates failed to generate any production of PspK, aliB‐like ORF1, or aliB‐like ORF2. A recent study described a Group II isolate with only transposable elements in its cps locus and proposed that NCC4 be named for this isolate. 36 The 18 strains in this study may be belonged to NCC4.
The common STs of NTSp found in the present study were different from those described in a similar study from Taiwan, which reported that the three dominant STs were ST1106 (19, 48.7%), ST7494 (6, 15.4%), and ST7502 (2, 5.1%) among 39 NTSp. 37 A study from South Korea reported that the three dominant STs among 26 NTSp strains were ST271 (3, 11%), ST320 (6, 23%), and ST1464 (6, 23%). 38 The variation of ST prevalence among different reports was not only related to the region, but also indicated that the homology of NTSp strains was very different.
Antimicrobial susceptibility data on S. pseudopneumoniae are relatively limited, and there are no judgment criteria in the Clinical and Laboratory Standards Institute guidelines. 19 NTSp and S. pseudopneumoniae in the present study showed similar non‐susceptibility rates to penicillin, amoxicillin/clavulanic acid, ceftriaxone, and cefotaxime as S. pneumoniae in our previous study. 39
There were also some limitations in our study. First, the strains were not collected continuously over time, and there may be differences in molecular epidemiological characteristics and antibiotic susceptibility patterns of strains from different ages. Second, some clinical information on these strains is missing. However, this study is the first to explore the molecular epidemiology and antibiotic susceptibility characteristics of Streptococcus pneumoniae strains with negative Quellung reaction test results from children in China. In future study, we will continuously collect more strains and include clinical isolation information of strains as much as possible, so as to fully understand the characteristics of this group of strains.
5. CONCLUSION
Commonly used methods, including MALDI‐TOF MS, could not always accurately differentiate S. pneumoniae from S. pseudopneumoniae. Most NESp isolates had heterologous genetics as encapsulated S. pneumoniae, and the NCC1 and NCC2 types of NESp were first reported in mainland China in this study. S. pneumoniae and S. pseudopneumoniae have similar antibiotic susceptibility patterns.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
All authors contributed to the study conception and design. Fang Dong, Qingying Meng, Lin Yuan, and Changhui Chen collected the strains. Ju Jia, Fang Dong, and Qingying Meng performed the identification tests. Wei Shi and Lin yuan performed the antibiotic susceptibility test. Ju Jia, Wei Shi, and Kaihu Yao collected data and drafted the article.
Jia J, Shi W, Dong F, et al. Identification and molecular epidemiology of routinely determined Streptococcus pneumoniae with negative Quellung reaction results. J Clin Lab Anal. 2022;36:e24293. doi: 10.1002/jcla.24293
Ju Jia and Wei Shi contributed equally to this work.
Funding information
This study was supported by Beijing Natural Science Foundation (L202004) and the Medical Research Project of Chongqing (2016ZDXM041)
DATA AVAILABILITY STATEMENT
The datasets analyzed during the present study are available from the corresponding author Kaihu Yao (email address: jiuhu2655@ sina.com) on reasonable request.
REFERENCES
- 1. Sadowy E, Hryniewicz W. Identification of Streptococcus pneumoniae and other Mitis streptococci: importance of molecular methods. Eur J Clin Microbiol Infect Dis. 2020;39(12):2247‐2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ercibengoa M, Morales M, Alonso M, Ardanuy C, Marimón MJ. Variants of the bile: solubility test to differentiate Streptococcus pneumoniae from other viridans group streptococci. Future Microbiol. 2019;14:949‐955. [DOI] [PubMed] [Google Scholar]
- 3. Balsalobre L, Hernández‐Madrid A, Llull D, et al. Molecular characterization of disease‐associated streptococci of the mitis group that are optochin susceptible. J Clin Microbiol. 2006;44(11):4163‐4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Varghese R, Jayaraman R, Veeraraghavan B. Current challenges in the accurate identification of Streptococcus pneumoniae and its serogroups/serotypes in the vaccine era. J Microbiol Methods. 2017;141:48‐54. [DOI] [PubMed] [Google Scholar]
- 5. Sá‐Leão R, Simões AS, Nunes S, Sousa NG, Frazão N, Lencastre H. Identification, prevalence and population structure of non‐typable Streptococcus pneumoniae in carriage samples isolated from preschoolers attending day‐care centres. Microbiology. 2006;152:367‐376. [DOI] [PubMed] [Google Scholar]
- 6. Hanage WP, Kaijalainen T, Saukkoriipi A, Rickcord JL, Spratt BG. A successful, diverse disease‐associated lineage of nontypeable pneumococci that has lost the capsular biosynthesis locus. J Clin Microbiol. 2006;44:743‐749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jensen A, Scholz CFP, Kilian M. Re‐evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus . Int J Syst Evol Microbiol. 2016;66(11):4803‐4820. [DOI] [PubMed] [Google Scholar]
- 8. Scholz CF, Poulsen K, Kilian M. Novel molecular method for identification of Streptococcus pneumoniae applicable to clinical microbiology and 16S rRNA sequence‐based microbiome studies. J Clin Microbiol. 2012;50:1968‐1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bishop CJ, Aanensen DM, Jordan GE, Kilian M, Hanage WP, Spratt BG. Assigning strains to bacterial species via the internet. BMC Biol. 2009;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bilecen K, Yaman G, Ciftci U, Laleli YR. Performances and reliability of Bruker Microflex LT and VITEK MS MALDI‐TOF mass spectrometry systems for the identification of clinical microorganisms. Biomed Res Int. 2015;2015:516410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fenselau C, Demirev PA. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom Rev. 2001;20:157‐171. [DOI] [PubMed] [Google Scholar]
- 12. Park IH, Kim KH, Andrade AL, Briles DE, McDaniel LS, Nahm MH. Nontypeable pneumococci can be divided into multiple cps types, including one type expressing the novel gene pspK. MBio. 2012;3:e00035‐e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arbique JC, Poyart C, Trieu‐Cuot P, et al. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J Clin Microbiol. 2004;42:4686‐4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lund E. Laboratory diagnosis of Pneumococcus infections. Bull World Health Organ. 1960;23:5‐13. [PMC free article] [PubMed] [Google Scholar]
- 15. Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol. 2006;44:124‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049‐3060. [DOI] [PubMed] [Google Scholar]
- 17. Na IY, Baek JY, Park IH, Kim DH, Song JH, Ko KS. pspK gene prevalence and characterization of non‐typable Streptococcus pneumonia isolates from Asian countries. Microbiology. 2015;161:973‐979. [DOI] [PubMed] [Google Scholar]
- 18. Martin CS, Bradshaw JL, Pipkins HR, McDaniel LS. Pulmonary disease associated with nonencapsulated Streptococcus pneumoniae. Open Forum. Infect Dis. 2018;5:ofy135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clinical and Laboratory Standards Institute (CLSI) . Performance Standards for antimicrobial susceptibility testing. In: Twenty‐ninth Informational Supplement. Clinical and Laboratory Standards Institute; 2019:M100‐S29. [Google Scholar]
- 20. Leegaard TM, Bootsma HJ, Caugant DA, et al. Phenotypic and genomic characterization of pneumococcus‐like streptococci isolated from HIV‐seropositive patients. Microbiology. 2010;156:838‐848. [DOI] [PubMed] [Google Scholar]
- 21. Rolo D, Simões AS, Domenech A, et al. Disease isolates of Streptococcus pseudopneumoniae and non‐typeable S. pneumoniae presumptively identified as atypical S. pneumoniae in Spain. PLoS One. 2013;8:e57047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Angeletti S, Dicuonzo G, Avola A, et al. Viridans Group Streptococci clinical isolates: MALDI‐TOF mass spectrometry versus gene sequence‐based identification. PLoS One. 2015;10:e0120502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prehn J, Veen SQ, Schelfaut JJ, Wessels E. MALDI‐TOF mass spectrometry for differentiation between Streptococcus pneumoniae and Streptococcus pseudopneumoniae . Diagn Microbiol Infect Dis. 2016;85:9‐11. [DOI] [PubMed] [Google Scholar]
- 24. Khéchine AE, Couderc C, Flaudrops C, Raoult D, Drancourt M. Matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry identification of mycobacteria in routine clinical practice. PLoS One. 2011;6:e24720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wieser A, Schneider L, Jung J, Schubert S. MALDI‐TOF MS in microbiological diagnostics‐identification of microorganisms and beyond (mini review). Appl Microbiol Biotechnol. 2012;93:965‐974. [DOI] [PubMed] [Google Scholar]
- 26. Feil EJ, Stackebrandt E, Peer YV, Vandamme P, Thompson FL, Swings J. Opinion: Re‐evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733‐739. [DOI] [PubMed] [Google Scholar]
- 27. Glaeser SP, Kämpfer P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst Appl Microbiol. 2015;38:237‐245. [DOI] [PubMed] [Google Scholar]
- 28. Jensen A, Hoshino T, Kilian M. Taxonomy of the Anginosus group of the genus Streptococcus and description of Streptococcus anginosus subsp. whileyi subsp. nov. and Streptococcus constellatus subsp. viborgensis subsp. nov. Int J Syst Evol Microbiol. 2013;63:2506‐2519. [DOI] [PubMed] [Google Scholar]
- 29. Vásquez‐Ponce F, Higuera‐Llantén S, Pavlov MS, Marshall SH, Olivares‐Pacheco J. Phylogenetic MLSA and phenotypic analysis identification of three probable novel Pseudomonas species isolated on King George Island, South Shetland, Antarctica. Braz J Microbiol. 2018;49:695‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ashford RT, Muchowski J, Koylass M, Scholz HC, Whatmore AM. Application of whole genome sequencing and pan‐family multi‐locus sequence analysis to characterize relationships within the family Brucellaceae. Front Microbiol. 2020;11:1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei M, Xu X, Yang J, et al. MLSA phylogeny and antimicrobial susceptibility of clinical Nocardia isolates: a multicenter retrospective study in China. BMC Microbiol. 2021;21(1):342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wen SCH, Anderson T, Murdoch D. Streptococcus pseudopneumoniae. Clin Microbiol Newslett. 2014;36:65‐71. [Google Scholar]
- 33. Bentley SD, Aanensen DM, Mavroidi A, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paton JC, Trappetti C. Streptococcus pneumoniae Capsular Polysaccharide. Microbiol Spectr. 2019;7(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hathaway LJ, Stutzmann Meier P, Battig P, Aebi S, Muhlemann K. A homologue of aliB is found in the capsule region of nonencapsulated Streptococcus pneumoniae . J Bacteriol. 2004;186:3721‐3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park IH, Geno KA, Sherwood LK, Nahm MH, Beall B. Population‐based analysis of invasive nontypeable pneumococci reveals that most have defective capsule synthesis genes. PLoS One. 2014;9:e97825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen HH, Hsu MH, Wu TL, et al. Non‐typeable Streptococcus pneumoniae infection in a medical center in Taiwan after wide use of pneumococcal conjugate vaccine. J Microbiol Immunol Infect. 2020;53:94–98.36. [DOI] [PubMed] [Google Scholar]
- 38. Cheong HJ, Song JY, Choi MJ, et al. Clinical and microbiological characterization of serotype 6D pneumococcal infections in South Korea. J Infect Chemother. 2016;22:515‐520. [DOI] [PubMed] [Google Scholar]
- 39. Lyu S, Yao KH, Dong F, et al. Vaccine serotypes of Streptococcus pneumoniae with high‐level antibiotic resistance isolated more frequently seven years after the licensure of PCV7 in Beijing. Pediatr Infect Dis J. 2016;35:316‐321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the present study are available from the corresponding author Kaihu Yao (email address: jiuhu2655@ sina.com) on reasonable request.


