Abstract
Background
Evidence indicates that the dysregulation of extracellular matrix (ECM) components can lead to cardiovascular diseases. The Talin‐1 (TLN1) gene is a major component of the ECM, and it mediates integrin adhesion to the ECM. In this study, we aimed to determine microRNAs (miRs) that regulate the expression of TLN1 and determine expression alterations in TLN1 and its targeting miRs in coronary artery disease (CAD).
Methods
Data sets of CAD and normal samples of blood exosomes were downloaded, and TLN1 was chosen as one of the genes with differential expressions in an in silico analysis. Next, miR‐182‐5p and miR‐9‐5p, which have a binding site on 3´‐UTR of TLN1, were selected using bioinformatics tools. Then, the miR target site was cloned in the psiCHECK‐2 vector, and direct interaction between the miR target site and the TLN1 3′‐UTR putative target site was investigated by luciferase assay. The expression of miR‐182‐5p, miR‐9‐5p, and TLN1 in the serum samples of CAD and non‐CAD individuals was assessed via a real‐time quantitative polymerase chain reaction.
Results
Our data revealed that miR‐182‐5p directly regulated the expression of TLN1. Moreover, miR‐182‐5p and miR‐9‐5p were significantly upregulated in the CAD group. Hence, both bioinformatics and experimental analyses determined the downregulated expression of TLN1 in the CAD samples.
Conclusions
Our findings demonstrated that miR‐182‐5p and miR‐9‐5p could play significant roles in TLN1 regulation and participate in CAD development by targeting TLN1. These findings introduce novel biomarkers with a potential role in CAD pathogenesis.
Keywords: coronary artery disease, miR‐182‐5p, miR‐9‐5p, TLN1 gene
Data sets of CAD and normal samples were downloaded, and TLN1 was chosen as one of the genes with differential expressions. miR‐182‐5p and miR‐9‐5p, which have a binding site on 3´‐UTR of TLN1, were selected by bioinformatics tools. miR‐182‐5p was cloned into the downstream of the green fluorescent protein (GFP) gene of the pEGFP‐N1 vector and was overexpressed in the HEK293T cell line. Then, the miRNA target site was cloned in the psiCHECK‐2 vector, and direct interaction between the miRNA target site and the TLN1 3′‐UTR putative target site was investigated by luciferase assay. A real‐time quantitative polymerase chain reaction was done to assess the expression of miR‐182‐5p, miR‐9‐5p, and TLN1 in the serum samples of CAD and non‐CAD individuals. So, miR‐182‐5p and miR‐9‐5p could play important roles in TLN1 regulation and participate in CAD development by targeting TLN1.

1. INTRODUCTION
Coronary artery disease (CAD) is the most common type of heart disease, with about 18.2 million adult sufferers and more than 3,65,000 deaths in 2017. 1 Considerable efforts to treat CAD have hitherto failed to curb its increasing mortality rate, 2 underscoring the significance of discovering specific and sensitive biomarkers to detect the disease early. 3 , 4 Atherosclerotic plaques constitute a major clinical manifestation of CAD and affect the large arteries. The significant stages involved in atherosclerotic plaque development are normal endothelium function loss due to lipids and inflammatory factors in arteries and the proliferation of smooth muscle cells. 4 , 5 In this disease, immune mechanisms react with metabolic factors and result in arterial damage. The production of inflammatory molecules, proteolytic enzymes, coagulation factors, and a variety of molecules can cause extracellular matrix (ECM) remodeling, plaque rupture, and increased thrombosis. 5 , 6 , 7 In the cardiovascular system, the ECM plays essential roles such as structural integrity preservation, cell adhesion, cell‐cell contacts, and remodeling in inflammation. In addition, the ECM is involved in the pathogenesis of atherosclerosis and heart failure. 8 Talin is a large dimeric protein (270 kDa) that binds to integrin and cytoskeletal actin and creates an essential connection between intra‐ and extracellular spaces. Therefore, it is essential for the attachment of cells to the ECM and the preservation of tissue architecture. 9 , 10 , 11 The expression of talin is reduced in atherosclerotic plaques, 12 while the expression of the talin‐1 (TLN1) gene in patients with CAD is upregulated. 13
MicroRNAs (miRs) are noncoding, small RNAs of about 22 nucleotides that regulate gene expression at the post‐transcriptional level. 14 Several studies have addressed the role of miRs as new biomarkers for CAD. These miRs include miR‐1, miR‐133a, and miR‐499 in patients with acute myocardial infarction and miR‐17, miR‐92a, miR‐126, miR‐133, miR‐140, miR‐145, miR‐155, and miR‐208a in patients with CAD. 15 , 16
A review of the key components of the ECM in CAD development can provide diagnostic and prognostic clues. 17 The regulation of the TLN1 gene could be one of the fascinating aspects of the molecular pathogenesis of CAD. Indeed, a better understanding of the TLN1 gene and its regulating miRs will help us prevent and treat the disease better. In this study, we sought to determine the expression pattern of the TLN1 gene and its candidate regulatory miRs in CAD and non‐CAD patients. In addition, we assessed the suitability of these miRs as novel biomarkers for CAD.
2. MATERIALS AND METHODS
2.1. Bioinformatics analysis in the CAD samples
All data sets were downloaded from the NCBI Gene Expression Omnibus (GEO). CAD and normal samples were obtained from GSE99985 and GSE100206, respectively. These data are the transcriptome profiling of RNAs in blood exosomes, and plasma and sera have been used to extract exosomal RNAs in these data sets. Adapter sequences were removed by Trimmomatic, version 0.36, and sequence reads were aligned to the human genome (hg38) with HISAT2 2.1.0. Ref‐seq (hg38), obtained from the University of California at Santa Cruz (UCSC) databases, was chosen as an annotation reference for RNA analyses. The read counts of each transcript were considered by HTSeq‐0.9.1. In this study, 6 CAD patients and 6 non‐CAD samples were analyzed to identify differentially expressed genes. Statistically, significant differentially expressed genes were identified using DEseq2 software in the R program with a log2FoldChange≥1 and an adjusted p value <0.01. The Enrichr database was used to perform pathway analysis with a view to acquiring important pathways among the CAD and non‐CAD samples. In the differentially expressed genes, TLN1 was detected as one of the significant genes in the CAD patients. Additionally, our survey on the Genotype‐Tissue Expression (GTEx) portal revealed that this gene had the highest expression in arteries. The Human miRNA Tissue ATLAS database was drawn upon to investigate miR expression information in different tissues.
2.2. Predicting TLN1‐targeting miRs by bioinformatics tools
For the prediction of miRs that targeted TLN1, the following bioinformatics tools were employed: TargetScan (http://www.targetscan.org/vert_71/), miRmap (http://mirmap.ezlab.org/), miRWalk (http://zmf.umm.uni‐heidelberg.de/apps/zmf/mirwalk2/), PicTar2 (https://pictar.mdc‐berlin.de/), MiRcode (http://www.mircode.org/), and RNAhybrid (http://bibiserv.cebitec.uni‐bielefeld.de/rnahybrid). Because of their high scores in all the tools, miR‐9‐5p and miR‐182‐5p were chosen, and experimental associations between these miRs and CAD and TLN1 regulation were analyzed.
2.3. Gene cloning and overexpression of miRs
The ability of miR‐9‐5p to regulate TLN1 was confirmed by Tang et al. 18 The miR‐182‐5p gene was polymerase chain reaction (PCR)‐amplified and cloned into the downstream of the green fluorescent protein (GFP) gene of the pEGFP‐N1 vector (Clontech, Japan). This miR was overexpressed in the HEK293T cell line. The HEK293T cells were cultured in DMEM‐F12 (Invitrogen, USA), containing 10% FBS, 100 U/ml of penicillin, and 100 µg/ml of streptomycin. The cultured cells were lysed using RiboEx (GeneAll, South Korea), and RNAs were extracted based on the guanidinium‐thiocyanate phenol‐chloroform separation protocol. The mRNA expression level of TLN1 was measured 24 and 48 hours post‐transfection by reverse transcription‐quantitative real‐time polymerase chain reaction (RT‐qPCR). RT‐qPCR was performed using the BIOFACTTM 2X RT‐qPCR master mix (for SYBR Green I; BIOFACT, South Korea). RT‐qPCR was conducted with 20 μl of the reaction mixture, composed of 2 μl of cDNA, 10 μl of SYBR Green I Master, 0.5 μl of forward and reverse primers for miRs and the TLN1 gene, and 6 μl of nuclease‐free water. Reactions were incubated at 95°C for 15 minutes, followed by 40 cycles of 95°C for 15 seconds and 62°C for 20 seconds. Melt curve analysis was performed to validate the specific generation of the expected PCR product.
2.4. Cloning of the 3′‐UTR and 3′‐UTR mutated form of TLN1
Human genomic DNA extracted from a blood sample was used to clone the 3′‐UTR of TLN1 with specific primers and cloned into the psiCHECK‐2 vector (Promega, USA) downstream of the Renilla gene. As controls, constructs, where the whole binding site of nominated miRs was eliminated, were built using SOEing PCR. (For primers sequences please see Table 1).
TABLE 1.
Sequences of the primers utilized in the study
| Primer name | Sequence |
|---|---|
| miR‐182‐5p |
Forward: GGCGGTTTGGCAATGGTAGA Reverse: AGTGCAGGGTCCGAGGTA Stem: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTGTG |
| miR‐9‐5p |
Forward: CGGCGGTCTTTGGTTATCTAGC Reverse: AGTGCAGGGTCCGAGGTA Stem: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCATAC |
| TLN1 |
Forward: TACAGTCAGCCAAGGAGGTG Reverse: AGGAATGCTGGAGAACTCAGG |
| UTR TLN1 |
Forward: CCCTCGAGGGAGAGCTTCGAGATGAGCACT Reverse: ATTTGCGGCCGCGGCTCAGACACTTACAGGC |
| Over miR‐182‐5p |
Forward: CCCTCGAGGGGGACCTTGTCGCAGTTGC Reverse: CGGAATTCCGACCTGCCCTCTGCCACTAC |
| pEGFPN1vector |
Forward: TGTCGTAACAACTCCGCCC Reverse: TGAACAGCTCCTCGCCCTT |
| psiCHECK‐2 vector |
Forward: GAGGACGCTCCAGATGAAATG Reverse: CTCACACAAAAAACCAACACACAG |
| Mutant UTR |
Forward: GCCACTACCAAAGCCTTCTCCAGTCCCGCAGTACATC Reverse: GATGTACTGCGGGACTGGAGAAGGCTTTGGTAGTGGC |
2.5. HEK293T transfection and luciferase assay
Three hundred nanograms of miR‐expressing vectors and 150 ng of wild‐type or mutated 3′‐UTR constructs were co‐transfected in HEK239T (cultured in 48‐well plates) using lipofectamine 2000 (Invitrogen, USA). Additionally, psiCHECK‐2 and pEGFPN1 mock vectors were transfected as controls for luciferase assay and transfection, respectively. Forty‐eight hours after HEK293T transfection, luciferase reporter assay was performed using the Dual‐Luciferase Reporter Assay System (Promega, USA) with a luminometer (Titertek‐Berthold, Germany) according to the manufacturer's protocol. Transfection efficiency was monitored by fluorescence microscopy (Nikon TE2000S, Japan).
2.6. Patients and serum sample collection
Sixty patients with chest pain who underwent diagnostic coronary angiography in Rajaie Cardiovascular Medical and Research Center were enrolled from 2019 through 2020 (Code of Ethics: IR.IAU.SRB.REC.1398.006). The patients were divided into 2 groups: 40 patients with CAD and 20 patients without CAD (non‐CAD). The patients in the control group did not have coronary stenosis confirmed by coronary angiography. Whole blood samples (5 ml) were collected before angiography. Serum samples were isolated by centrifugation at 1737 g for 30 minutes at room temperature, and the samples were transferred to RNase/ DNase‐free tubes and stored at −80°C.
2.7. RNA isolation and RT‐qPCR analysis
After the collection of all the samples, the total RNAs of the serum samples were extracted using a plasma/serum RNA purification kit (Norgen Biotek, Canada) and concentrated using the RNA Clean‐Up and Concentration Micro‐Elute Kit (Norgen Biotek, Canada) in accordance with the manufacturer's protocol. Next, cDNA was synthesized using the PrimeScript 1st Strand cDNA Synthesis Kit (Takara Bio, Japan). Through stem‐loop primers (Table 1) and random hexamer primers, 1 µg of total RNA was reverse‐transcribed into cDNA. The expression levels of TLN1 and the nominated miRs were determined using specific primers, listed in Table 1, using RT‐qPCR as was mentioned before. The expression levels of miRs and the gene were normalized to 5s and were calculated by the 2−ΔCt method.
2.8. Statistical analysis
The 2−∆Ct method was used for the miR and gene expression analyses of the patients and the Mann–Whitney for statistical tests. A receiver operating characteristic (ROC) curve was created, and the area under the ROC curve (AUC) was calculated to estimate the specificity and sensitivity of CAD prediction. The 2−∆∆Ct method was also utilized for the cloning data analysis of the miRs. Additionally, GraphPad Prism 6 was employed for the data analysis of RT‐qPCR and luciferase assay, in addition to p value calculation. A p value <0.05 was considered statistically significant for all the experiments.
3. RESULTS
3.1. Downregulation of TLN1 in the CAD samples
The RNA‐sequencing analysis revealed 1920 genes with differential expressions between the CAD and non‐CAD samples (Table S1). Among the genes, the TLN1 (NM_006289) gene was significantly downregulated with a log2FoldChange of −2.52. The pathway analysis on the genes with differential expressions demonstrated the role of TLN1 in platelet activation and focal adhesion signaling pathways involved in ECM regulation and probably CAD development. In addition to the significance of this gene in 2 key CAD pathogenesis pathways obtained from bioinformatics studies, its expression pattern in the GTEx database demonstrated a high expression level for TLN1 in normal coronary arteries, which sustain the most damage in CAD (Figure 1A‐B). The expression patterns of miRs in the Human miRNA Tissue ATLAS database demonstrated higher expression levels for miR‐9‐5p in the brain and spinal cord tissues, lower expression levels for miR‐9‐5p in the artery tissues, and higher expression levels for miR‐182‐5p in the vein, epididymidis, spinal cord, and artery tissues (Figure 1C–D). A negative correlation seemed to exist between the miRs and TLN1 in the brain and heart tissues, which was more common for miR‐9‐5p.
FIGURE 1.
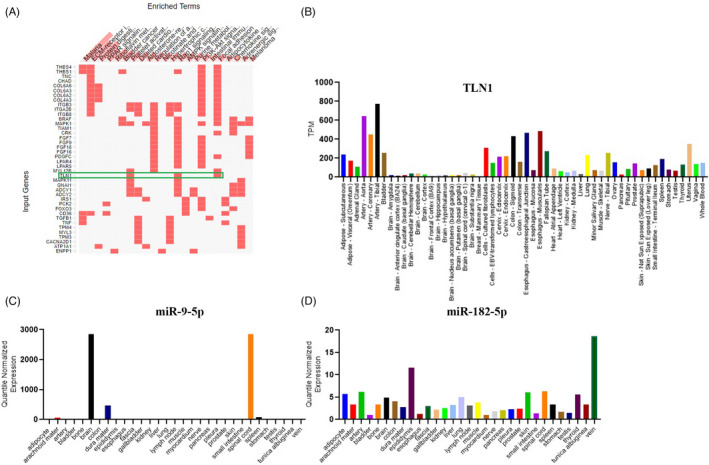
(A) Image illustrates the clustergram of the KEGG pathway for differentially expressed genes. The clustergram shows the role of the talin‐1 (TLN1) gene in platelet activation and focal adhesion pathways. B) The Genotype‐Tissue Expression (GTEx) portal demonstrates that TLN1 has the highest expression in arteries. (C & D) The Human miRNA Tissue ATLAS database shows that miR‐9‐5p has the highest expression in the brain, whereas miR‐182‐5p has the highest expression in vessels
TLN1 was upregulated during cardiac differentiation. (The data are not shown.) Therefore, given the importance of the TLN1 gene in the formation and maintenance of cardiac cells, it was chosen for further analysis.
3.2. Bioinformatics prediction of TLN1‐targeting miRs
Several target prediction databases, including miRWalk, miRmap, PicTar2, RNAhybrid, TargetScan, and MiRcode, were utilized to find candidate miRs capable of targeting the 3′‐UTR of TLN1. The analysis predicted 978 miRs that could target the 3´‐UTR of the TLN1 gene. Among these miRs, miR‐182‐5p and miR‐9‐5p were chosen based on their more frequent appearances in all the mentioned databases for further analysis. Both miRs had 1 target site on the TLN1 3′‐UTR (Figure 2A–B).
FIGURE 2.

Image depicts the predicted miR‐182‐5p and miR‐9‐5p target sites on the talin‐1 (TLN1) transcript. (A) Exon, intron, and miR target sites are shown on the 3′ ‐UTR of the TLN1 sequence. (B) The target sites of the miRs and their nucleotide hybridization positions are illustrated
3.3. Direct regulation of TLN1 expression by miR‐182‐5p
Luciferase assay was used to investigate the interaction between miR‐182‐5p and the 3′‐UTR of TLN1. A miR‐overexpressing plasmid, which contained the precursor of the miRs, and a psiCHECK‐2 plasmid, which carried the TLN1 3′‐UTR in the wild‐type and mutant status downstream of a luciferase‐coding sequence, were constructed. Forty‐eight hours after the co‐transfection of the miR‐overexpressing and psiCHECK‐2 plasmids, a reduction was detected in relative luciferase activity (p = 0.008) in the cells co‐transfected with a vector containing a luciferase‐coding sequence upstream of the wild‐type 3′‐UTR of TLN1. In contrast, a mutated form of the TLN1 3′‐UTR, in which the binding site of miR‐182‐5p was deleted, failed to exert the same effect on the transfected cells (p = 0.39) (Figure 3A–B).
FIGURE 3.
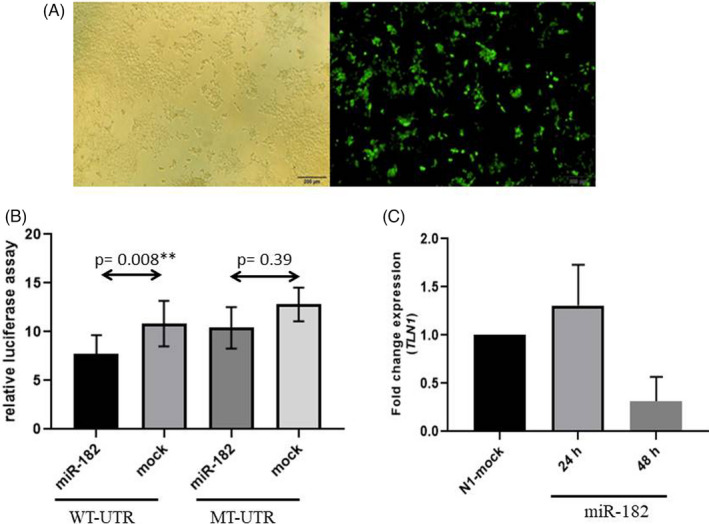
Image presents the luciferase assay results. (A) HEK293T transfection is illustrated herein. The green fluorescent protein (GFP) signal shows that the pEGFP‐N1 vector is functional. (B) Relative luciferase activity is decreased in wild‐type 3′ ‐UTR (WT‐UTR) (p = 0.008) after miR‐182‐5p overexpression, but no significant alterations are observed in relative luciferase activity when the miR‐182‐5p target site is deleted from 3′ ‐UTR (MT‐UTR) (p = 0.39). (C) The miR fold change in the talin‐1 (TLN1) gene is shown herein after miR‐182‐5p overexpression in the HEK293T cell line. There is a decline in the TLN1 miR level after the overexpression in the miR 48 hours after transfection. The GAPDH gene was used as an internal control for TLN1 expression normalization. The bars represent the mean ± SEM
Next, the expression of TLN1 at the miR level was assessed 24 and 48 hours after the transfection of the Hek293T cells with the miR‐overexpressing vector. The results demonstrated significant downregulation in TLN1 expression 48 hours following transfection, whereas no significant change was illustrated in the expression of TLN1 at the miR level 24 hours after transfection (Figure 3C).
Thus, miR‐182‐5p can regulate the expression of the TLN1 gene by interacting with its 3′‐UTR.
3.4. Reciprocal patterns of expression alterations between miR‐182‐5p, miR‐9‐5p, and TLN1 in the CAD group
In the next stage, the expression patterns of miR‐182‐5p, miR‐9‐5p, and TLN1 were investigated in the serum samples of the CAD group (n = 40) and the non‐CAD group (n = 20). The clinicopathological characteristics of the patients are presented in Table 2.
TABLE 2.
Patient Characteristics (p < 0.001)
| Variable | CAD (n = 40) | No_CAD (n = 20) | p value |
|---|---|---|---|
| Age (years), mean ± SD | 42.36 ± 5.2 | 42.9 ± 4.6 | 0.72 |
| Male/Female, (n / n) | (19,21) | (4,16) | 0.051 |
| Smoking, (n, %) | (19, 48.7%) | (4, 21.1%) | 0.051 |
| Systolic blood pressure (mmHg), mean ± SD | 128.3 ± 17.8 | 127.3 ± 21.2 | 0.54 |
| Diastolic blood pressure (mmHg), mean ± SD | 77.9 ± 7 | 74.9 ± 17.3 | 0.86 |
| Total cholesterol (mg/dl), mean ± SD | 143.7 ± 43.2 | 130.4 ± 22.2 | 0.151 |
| HDL (mg/dl), mean ± SD | 37.3 ± 12.2 | 37.7 ± 8.5 | 0.56 |
| LDL (mg/dl), mean ± SD | 80.7 ± 33.7 | 72.4 ± 18.1 | 0.507 |
| Triglyceride(mg/dl), mean ± SD | 161.2 ± 70.4 | 105.3 ± 43.4 | 0.005 |
| Glucose (mg/dl), mean ± SD | 139.8 ± 60.9 | 100.2 ± 17.2 | 0.001 |
| History of stroke (n, %) | (12, 30.8%) | (2, 10%) | 0.109 |
The RT‐qPCR results demonstrated that the expression levels of miR‐182‐5p (p = 0.005) and miR‐9‐5p (p = 0.0198) were considerably higher in the CAD group than in the non‐CAD group, while the miR expression level of TLN1 was significantly decreased (p = 0.042) in the CAD group by comparison with the non‐CAD group (Figure 4A‐C).
FIGURE 4.
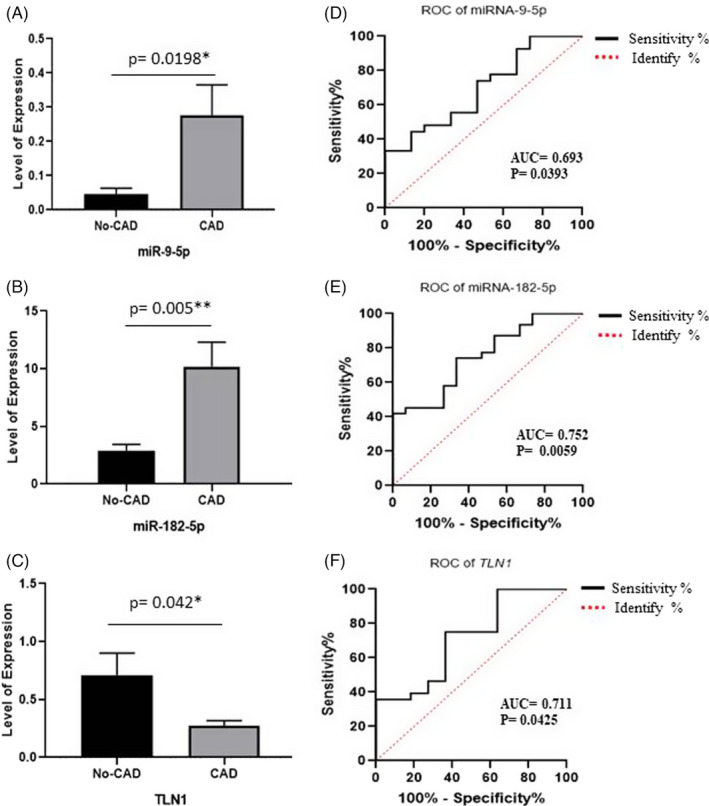
Image depicts the validation of miR‐182‐5p, miR‐9‐5p, and the talin‐1 (TLN1) gene between the 2 study groups: patients with coronary artery disease (CAD) and subjects without CAD. The expression levels of miR‐182‐5p (A), miR‐9‐5p (B), and TLN1 (C) by reverse transcription‐quantitative real‐time polymerase chain reaction are compared between the CAD and non‐CAD groups. Also presented is the receiver operating characteristic (ROC) curve analysis of miR‐182‐5p, miR‐9‐5p, and TLN1, acting as potential biomarkers for CAD. (D) The ROC curve analysis of miR‐182‐5p, (E) the ROC curve analysis of miR‐9‐5p, and (F) the ROC curve analysis of TLN1 are depicted herein. The bars represent the mean ± SEM
For the evaluation of the suitability of the differentially expressed miR‐182‐5p and miR‐9‐5p as diagnostic markers for CAD, ROC curve analysis was performed. AUC was 0.752 (95% CI, 0.608 to 0.897; p = 0.0059) for miR‐182‐5p, with a sensitivity of 74.19% and a specificity of 66.67% (cutoff ≥3.325). In addition, AUC was 0.693 (95% CI, 0.530 to 0.857; p = 0.0393) for miR‐9‐5p, with a sensitivity of 74.07% and a specificity of 53.33% (cutoff ≥0.023). Thus, elevated expression levels of miR‐182‐5p and miR‐9‐5p may act as diagnostic biomarkers for CAD. Furthermore, AUC was 0.711 (95% CI, 0.528 to 0.893; p = 0.0425) for TLN1, with a sensitivity of 75% and a specificity of 63.64% (cutoff ≤0.4218) (Figure 4D‐F).
4. DISCUSSION
CAD is a major cause of death worldwide as is attested to by its rapidly increasing prevalence. 1 The ECM not only is involved in controlling numerous functions of vessels and the heart but also preserves the structural integrity of the heart and the vascular network and promotes post‐vascular injury remodeling. 19 Many genes are associated with cellular mechanical stability and CAD progression. The TLN1 gene is among the major components of the ECM. TLN1 expression is crucial to tissue remodeling and the maintenance of tissue integrity. 12 , 20 , 21 , 22 Moreover, reduced TLN1 expression levels could compromise ECM constituents, cause endothelial injury, and trigger an inflammatory response to CAD. 12 , 23
In both normal and pathologic states, miRs have been firmly established as significant molecular components of the regulatory network in cells, and their expression alterations have been demonstrated in CAD, as well as cancer and other diseases. 24 Accordingly, alterations in the expression levels of miRs are utilized as valuable biomarkers for the diagnosis, prognosis, and further treatment of diseases. Our results introduce miR‐182‐5p as a natural regulator of TLN1. Based on our bioinformatics and in vitro functional analysis, TLN1 expression is directly targeted by miR‐182‐5p. Notably, during our research experiment, Tang et al 18 reported that miR‐9‐5p, one of our candidate miRs, could target the TLN1 3′‐UTR. Consequently, we eliminated miR‐9‐5p from the miR‐target interaction experiments.
Previous studies have demonstrated that miR‐330‐5p and miR‐124 regulate the expression of TLN1. Overexpressed levels of miR‐330‐5p have been confirmed in unstable plaques, leading to significant downregulation in TLN1 expression. This finding suggests that miR‐330‐5p may play a role in TLN1 downregulation in atherosclerotic plaques. 25 Zhang et al 26 showed that miR‐124 impaired the adhesion, migration, and invasion of prostate cancer cells by downregulating TLN1.
Research has shown that miR‐182‐5p targets the NRF‐1 and CLAPIN1 genes, which are associated with a reduction in reperfusion injury‐induced myocardial ischemia and the protection of cardiomyocytes against hypoxia‐induced injury. 27 , 28 Furthermore, miR‐182/FoxF2 may play a role in colorectal cancer by regulating ECM remodeling. A significant decline in the histological scores of inflammation and tissue destruction was confirmed in a murine model of arthritis after the inhibition of miR‐182‐5p. 29 , 30 Our results chime in with those reported by previous studies insofar as miR‐182‐5p plays a role in CAD progression by directly regulating a principal component of the ECM, the TLN1 gene.
We also sought to determine the potential expression alterations of TLN1 and its targeting miRs, namely miR‐182‐5p and miR‐9‐5p, in patients with CAD by comparison with a non‐CAD group. Our results exhibited that miR‐182‐5p and miR‐9‐5p were significantly upregulated in the CAD group, whereas the expression of the TLN1 gene was downregulated in the non‐CAD group. Our findings are concordant with some other studies. Taurino et al 31 drew upon the miR microarray approach and identified upregulation in miR‐140‐3p and miR‐182 among patients with CAD. Zhu et al 32 showed that miR‐182‐5p was upregulated in the plasma of patients with unprotected left main CAD. In patients suffering from chronic heart failure, Cakmak et al 33 showed that miR‐182 had a higher prognostic value for cardiovascular mortality and that miR‐182 might serve as a biomarker for atherosclerosis progression.
The association between miR‐9‐5p and the talin‐1/FAK/Akt pathway has already been reported.18 Previous investigations have revealed significantly higher miR‐9 levels in patients with type II diabetes mellitus, the main risk factor for CAD development. 34 , 35 Bazzoni et al 36 highlighted the role of miR‐9 as a proinflammatory signal regulator in monocytes and neutrophils. Further, miR‐9‐5p can affect some matrix metalloproteinases (MMPs) involved in ECM degradation, as an important step in CAD progression. 37 , 38 Xiao et al 39 showed that miR‐9‐5p suppression could prevent cardiac remodeling after acute myocardial infarction.
Downregulation in the expression level of the TLN1 gene has been previously reported in atherosclerotic plaques and aortic dissection. 12 , 40
A proteomic study also demonstrated the downregulation of TLN1 in atherosclerotic coronary media. 41 However, Aoyama et al 13 reported that plasma TLN1 levels in patients with CAD were high and associated with the presence and severity of CAD. In the present study, TLN1 was downregulated in patients with CAD, which is in line with the aforementioned investigations, which confirmed reduced TLN1 expression in CAD.
The observed upregulation of miR‐182‐5p and miR‐9‐5p in CAD samples is consistent with the downregulation of TLN1 as their validated target. Given that an ideal biomarker should have high sensitivity and specificity, 42 our data suggest that overexpressed miR‐182‐5p and miR‐9‐5p and downregulated TLN1 are suitable biomarkers for CAD. Notably, downregulation in TLN1 expression may be only one of the initial triggers of CAD that can affect the connection of endothelial cells to each other and the ECM. Thus, not only does it allow the progression of inflammatory mediators and plaque formation but also it damages vessel integrity, prompting platelet aggregation. 43 , 44
The findings of the present study demonstrated that miR‐182‐5p and miR‐9‐5p could play significant roles in TLN1 regulation, which is crucial to ECM integrity. Plaque formation disturbs the balance between ECM formation and degradation, possibly leading to cardiovascular events. Given the importance of TLN1 regulation in CAD development, these miRs could be considered diagnostic biomarkers and future therapeutic targets in CAD research. Our data indicated that upregulated miR‐182‐5p and miR‐9‐5p and downregulated TLN1 could be potential biomarkers for CAD since they reach the score needed for a reliable biomarker. Nonetheless, further investigations are warranted to clarify the possible effects of miR‐182‐5p and miR‐9‐5p manipulation on CAD pathogenesis pathways.
CONFLICT OF INTEREST
The authors hereby declare no conflict of interests.
Supporting information
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Research Deputyship of Rajaie Cardiovascular Medical and Research Center (94111).
Gholipour A, Shakerian F, Zahedmehr A, Irani S, Mowla SJ, Malakootian M. Downregulation of Talin‐1 is associated with the increased expression of miR‐182‐5p and miR‐9‐5p in coronary artery disease. J Clin Lab Anal. 2022;36:e24252. doi: 10.1002/jcla.24252
Contributor Information
Seyed Javad Mowla, Email: sjmowla@yahoo.com.
Mahshid Malakootian, Email: Malakootian@rhc.ac.ir.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in this article, and bioinformatics results from GSE99985 and GSE100206 are available in Table S1.
REFERENCES
- 1. Benjamin EJ, Muntner P, Alonso A, et al. Heart disease and stroke statistics‐2019 update: a report from the American Heart Association. Circulation. 2019;139(10):e56‐e528. [DOI] [PubMed] [Google Scholar]
- 2. Roth GA, Huffman MD, Moran AE, et al. Global and regional patterns in cardiovascular mortality from 1990 to 2013. Circulation. 2015;132(17):1667‐1678. [DOI] [PubMed] [Google Scholar]
- 3. World Health O . The atlas of heart disease and stroke / Judith Mackay and George Mensah; with Shanthi Mendis and Kurt Greenland. World Health Organization; 2004. https://apps.who.int/iris/handle/10665/43007 [Google Scholar]
- 4. Watkins H, Farrall M. Genetic susceptibility to coronary artery disease: from promise to progress. Nat Rev Genet. 2006;7(3):163‐173. [DOI] [PubMed] [Google Scholar]
- 5. Fuster V, Badimon L, Badimon JJ, Chesebro JH. The pathogenesis of coronary artery disease and the acute coronary syndromes (1). N Engl J Med. 1992;326(4):242‐250. [DOI] [PubMed] [Google Scholar]
- 6. Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685‐1695. [DOI] [PubMed] [Google Scholar]
- 7. Rosner D, Stoneman V, Littlewood T, et al. Interferon‐gamma induces Fas trafficking and sensitization to apoptosis in vascular smooth muscle cells via a PI3K‐ and Akt‐dependent mechanism. Am J Pathol. 2006;168(6):2054‐2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Newby AC. Metalloproteinase expression in monocytes and macrophages and its relationship to atherosclerotic plaque instability. Arterioscler Thromb Vasc Biol. 2008;28(12):2108‐2114. [DOI] [PubMed] [Google Scholar]
- 9. Zhang X, Jiang G, Cai Y, Monkley SJ, Critchley DR, Sheetz MP. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat Cell Biol. 2008;10(9):1062‐1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jahed Z, Shams H, Mehrbod M, Mofrad MR. Mechanotransduction pathways linking the extracellular matrix to the nucleus. Int Rev Cell Mol Biol. 2014;310:171‐220. [DOI] [PubMed] [Google Scholar]
- 11. Hákonardóttir GK, López‐Ceballos P, Herrera‐Reyes AD, Das R, Coombs D, Tanentzapf G. In vivo quantitative analysis of Talin turnover in response to force. Mol Biol Cell. 2015;26(22):4149‐4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. von Essen M, Rahikainen R, Oksala N, et al. Talin and vinculin are downregulated in atherosclerotic plaque. Tampere Vascular Study. Atherosclerosis. 2016;255:43‐53. [DOI] [PubMed] [Google Scholar]
- 13. Aoyama M, Kishimoto Y, Saita E, et al. High plasma levels of soluble talin‐1 in patients with coronary artery disease. Dis Markers. 2020;2020:2479830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fichtlscherer S, De Rosa S, Fox H, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107(5):677‐684. [DOI] [PubMed] [Google Scholar]
- 15. Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110(3):483‐495. [DOI] [PubMed] [Google Scholar]
- 16. Takeishi Y. Biomarkers in heart failure. Int Heart J. 2014;55(6):474‐481. [DOI] [PubMed] [Google Scholar]
- 17. Holm Nielsen S, Jonasson L, Kalogeropoulos K, et al. Exploring the role of extracellular matrix proteins to develop biomarkers of plaque vulnerability and outcome. J Intern Med. 2020;287(5):493‐513. [DOI] [PubMed] [Google Scholar]
- 18. Tang H, Yao L, Tao X, et al. miR‐9 functions as a tumor suppressor in ovarian serous carcinoma by targeting TLN1. Int J Mol Med. 2013;32(2):381‐388. [DOI] [PubMed] [Google Scholar]
- 19. Chistiakov DA, Sobenin IA, Orekhov AN. Vascular extracellular matrix in atherosclerosis. Cardiol Rev. 2013;21(6):270‐288. [DOI] [PubMed] [Google Scholar]
- 20. Goult BT, Gingras AR, Bate N, Barsukov IL, Critchley DR, Roberts GC. The domain structure of talin: Residues 1815‐1973 form a five‐helix bundle containing a cryptic vinculin‐binding site. FEBS Lett. 2010;584(11):2237–2241. 10.1016/j.febslet.2010.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goult BT, Zacharchenko T, Bate N, et al. RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J Biol Chem. 2013;288(12):8238‐8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun N, Critchley DR, Paulin D, Li Z, Robson RM. Identification of a repeated domain within mammalian alpha‐synemin that interacts directly with talin. Exp Cell Res. 2008;314(8):1839‐1849. [DOI] [PubMed] [Google Scholar]
- 23. Davies PF. Hemodynamic shear stress and the endothelium in cardiovascular pathophysiology. Nat Clin Pract Cardiovasc Med. 2009;6(1):16‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arabian M, Mirzadeh Azad F, Maleki M, Malakootian M. Insights into role of microRNAs in cardiac development, cardiac diseases, and developing novel therapies. Iran J Basic Med Sci. 2020;23(8):961‐969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wei X, Sun Y, Han T, et al. Upregulation of miR‐330‐5p is associated with carotid plaque's stability by targeting Talin‐1 in symptomatic carotid stenosis patients. BMC Cardiovasc Disord. 2019;19(1):149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang W, Mao YQ, Wang H, Yin WJ, Zhu SX, Wang WC. MiR‐124 suppresses cell motility and adhesion by targeting talin 1 in prostate cancer cells. Cancer Cell Int. 2015;15:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhai C, Tang G, Peng L, et al. Inhibition of microRNA‐1 attenuates hypoxia/re‐oxygenation‐induced apoptosis of cardiomyocytes by directly targeting Bcl‐2 but not GADD45Beta. Am J Transl Res. 2015;7(10):1952‐1962. [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y, Fang J, Ma H. Inhibition of miR‐182‐5p protects cardiomyocytes from hypoxia‐induced apoptosis by targeting CIAPIN1. Biochem Cell Biol. 2018;96(5):646‐654. [DOI] [PubMed] [Google Scholar]
- 29. Zhang Y, Wang X, Wang Z, Tang H, Fan H, Guo Q. miR‐182 promotes cell growth and invasion by targeting forkhead box F2 transcription factor in colorectal cancer. Oncol Rep. 2015;33(5):2592‐2598. [DOI] [PubMed] [Google Scholar]
- 30. Stittrich AB, Haftmann C, Sgouroudis E, et al. The microRNA miR‐182 is induced by IL‐2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11(11):1057‐1062. [DOI] [PubMed] [Google Scholar]
- 31. Taurino C, Miller WH, McBride MW, et al. Gene expression profiling in whole blood of patients with coronary artery disease. Clin Sci (Lond). 2010;119(8):335‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu L, Chen T, Ye W, et al. Circulating miR‐182‐5p and miR‐5187‐5p as biomarkers for the diagnosis of unprotected left main coronary artery disease. J Thorac Dis. 2019;11(5):1799‐1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cakmak HA, Coskunpinar E, Ikitimur B, et al. The prognostic value of circulating microRNAs in heart failure: preliminary results from a genome‐wide expression study. J Cardiovasc Med (Hagerstown). 2015;16(6):431‐437. [DOI] [PubMed] [Google Scholar]
- 34. Herder C, Karakas M, Koenig W. Biomarkers for the prediction of type 2 diabetes and cardiovascular disease. Clin Pharmacol Ther. 2011;90(1):52‐66. [DOI] [PubMed] [Google Scholar]
- 35. Motawae TM, Ismail MF, Shabayek MI, Seleem MM. MicroRNAs 9 and 370 Association with Biochemical Markers in T2D and CAD Complication of T2D. PLoS One. 2015;10(5):e0126957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bazzoni F, Rossato M, Fabbri M, et al. Induction and regulatory function of miR‐9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. 2009;106(13):5282‐5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones SW, Watkins G, Le Good N, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF‐alpha and MMP13. Osteoarthritis Cartilage. 2009;17(4):464‐472. [DOI] [PubMed] [Google Scholar]
- 38. Zhang H, Qi M, Li S, et al. microRNA‐9 targets matrix metalloproteinase 14 to inhibit invasion, metastasis, and angiogenesis of neuroblastoma cells. Mol Cancer Ther. 2012;11(7):1454‐1466. [DOI] [PubMed] [Google Scholar]
- 39. Xiao Y, Zhang Y, Chen Y, et al. Inhibition of MicroRNA‐9‐5p protects against cardiac remodeling following myocardial infarction in mice. Hum Gene Ther. 2019;30(3):286‐301. [DOI] [PubMed] [Google Scholar]
- 40. Wei X, Sun Y, Wu Y, et al. Downregulation of Talin‐1 expression associates with increased proliferation and migration of vascular smooth muscle cells in aortic dissection. BMC Cardiovasc Disord. 2017;17(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de la Cuesta F, Zubiri I, Maroto AS, et al. Deregulation of smooth muscle cell cytoskeleton within the human atherosclerotic coronary media layer. J Proteomics. 2013;82:155‐165. [DOI] [PubMed] [Google Scholar]
- 42. Hartwell L, Mankoff D, Paulovich A, Ramsey S, Swisher E. Cancer biomarkers: a systems approach. Nat Biotechnol. 2006;24(8):905‐908. [DOI] [PubMed] [Google Scholar]
- 43. Ruggeri ZM. Platelets in atherothrombosis. Nat Med. 2002;8(11):1227‐1234. [DOI] [PubMed] [Google Scholar]
- 44. Nieswandt B, Watson SP. Platelet‐collagen interaction: is GPVI the central receptor? Blood. 2003;102(2):449‐461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
The data that support the findings of this study are available in this article, and bioinformatics results from GSE99985 and GSE100206 are available in Table S1.


