Abstract
A mutant of Mycobacterium smegmatis has been isolated that is simultaneously resistant to both d-cycloserine (d-CS) and vancomycin. Genetic complementation with a PBP4 homolog restores sensitivity to both drugs. Resistance to d-CS and vancomycin in this mutant is most likely due to a novel mechanism involving peptidoglycan assembly at the cell surface.
The emergence of antibiotic resistance is a growing problem among pathogenic bacteria. Of great concern is the increased prevalence of multidrug-resistant strains of Mycobacterium tuberculosis (4). To complicate this issue further, mycobacteria are intrinsically resistant to many drugs normally active on other gram-positive microorganisms, due to their distinctive lipid barriers composed of bound mycolic acid and free lipids, which render them impermeable to many substances (3).
To gain greater insight into resistance mechanisms utilized by mycobacteria, we sought to create a laboratory mutant of Mycobacterium smegmatis that is resistant to the peptidoglycan biosynthesis inhibitor d-cycloserine (d-CS). To that end, a colony of wild-type M. smegmatis mc2155 was cultured in minimal medium with increasing concentrations of d-CS ranging from 10 to 150 μg/ml. Aliquots of cells from each culture were plated onto 102 μg of d-CS per ml, the concentration of d-CS that consistently kills wild-type cells. One of the mutants isolated from this selection was chosen for further characterization.
The mutant was evaluated for resistance to other drugs affecting peptidoglycan biosynthesis, including fosfomycin and vancomycin. From these studies, it was determined that the mutant is resistant to low levels of vancomycin. Because the mutant displayed resistance to both d-CS and vancomycin, it was named Cvr-1 for d-cycloserine and vancomycin resistance. The d-CS MIC and minimum bactericidal concentration for wild-type M. smegmatis are both 105 μg/ml, and those for Cvr-1 are 140 and 145 μg/ml, respectively. The vancomycin MIC and minimum bactericidal concentration for wild-type M. smegmatis are 40 and 45 μg/ml, respectively, and those for Cvr-1 are 65 and 70 μg/ml, respectively.
Resistance to both d-CS and vancomycin commonly maps to the d-alanine–d-alanine ligase (encoded by the gene ddlA) in many pathogenic bacterial species. The d-alanine–d-alanine ligase is responsible for catalyzing amide bond formation between two d-alanine molecules (16). The d-alanine–d-alanine dipeptides created by the ligase are then added onto the C termini of UDP-N-acetyl-muramic tripeptides to form peptidoglycan monomers that are eventually added to the peptidoglycan (16). David performed genetic fluctuation analyses on d-CS-resistant mutants of M. tuberculosis and concluded that drug resistance is primarily due to mutations in the d-alanine–d-alanine ligase gene (7). d-CS resistance mechanisms common to the ligase include changes in the specificity of the enzyme such that it is no longer able to recognize and interact with the drug.
Resistance to vancomycin in other bacteria results from the replacement of the terminal d-alanine–d-alanine of the peptidoglycan pentapeptide precursor with either a d-alanine–d-lactate depsipeptide or a d-alanine–d-serine dipeptide. These molecules are produced by resistance plasmid-encoded homologs of d-alanine–d-alanine ligases with altered specificities, best described for vancomycin-resistant enterococcal species (2, 13, 15). Vancomycin has a lower affinity for these modified cell wall precursors and is therefore unable to interact with them to inhibit peptidoglycan assembly at the cell surface.
Because resistance to both d-CS and vancomycin commonly maps to the d-alanine–d-alanine ligase gene, we sought to discover whether the defect in Cvr-1 leading to its cross-resistant phenotype was due to altered ligase activity. We sequenced the ddlA gene from Cvr-1 and compared the sequence to that of the wild-type parent, M. smegmatis mc2155. Sequencing analyses demonstrated that there was no mutation in the d-alanine–d-alanine ligase gene or its promoter in Cvr-1 (data not shown).
To confirm the sequencing data, d-alanine–d-alanine ligase activity assays were performed utilizing partially purified d-alanine–d-alanine ligase extracts from either M. smegmatis mc2155 or Cvr-1. This assay was originally used to determine the ligation products of d-alanine–d-alanine ligases present in vancomycin-resistant enterococci (1, 13, 15). The d-alanine–d-alanine ligases from both the wild type and Cvr-1 were able to produce d-alanine–d-alanine dipeptides but not d-alanine–d-lactate depsipeptides (data not shown). The ligases from both strains were also unable to synthesize d-alanine–d-serine dipeptides (data not shown). These results illustrate that the simultaneous d-CS and vancomycin resistance of Cvr-1 is due to a novel mechanism that does not involve the d-alanine–d-alanine ligase.
High-performance liquid chromatography and mass spectrometry were performed on the peptidoglycan precursors from wild-type and mutant M. smegmatis to determine their levels and composition. This analysis demonstrated whether resistance to the two drugs might be due to altered precursor biosynthesis. In order to perform the analysis, the two strains were incubated in the presence of 40 μg of vancomycin per ml to promote the accumulation of the peptidoglycan precursors. The results demonstrated that the composition and the levels of the precursors in the two strains were similar (data not shown), suggesting that a defect in the peptidoglycan precursor biosynthetic pathway of Cvr-1 is unlikely.
Three classes of d-CS-resistant mutants have been described for other bacteria, such as Escherichia coli (17), Bacillus subtilis (8), and the streptococci (14). One of these classes is comprised of mutants defective in transport of d-CS. In E. coli, d-CS is transported by the CycA permease, which also transports d-alanine, glycine, and d-serine. We found no defect in the transport of [14C]d-alanine (of which d-CS is an analog) by the mutant permease suggesting that the mechanism of d-CS resistance in Cvr-1 was not due to a defective CycA permease (data not shown).
The other two classes of d-CS-resistant mutants in bacteria include mutants with alterations in either the d-alanine–d-alanine ligase or the d-alanine racemase (8, 14, 17). As previously mentioned, there is no alteration of ligase activity in Cvr-1. The d-alanine racemase converts l-alanine to d-alanine, which then serves as a substrate for the d-alanine–d-alanine ligase. David (7) speculated that in mycobacteria the inhibition of the alanine racemase by d-CS plays a minor role in the drug's mechanism of action. However, Cáceres et al. (6) demonstrated that d-CS resistance in M. smegmatis can be mediated by overexpression of the gene encoding the alanine racemase (alr), resulting in a 15-fold increase in the specific activity of the enzyme. An increase in the specific activity of the racemase will alter the levels of d-alanine within the cell, resulting in a perturbation of the composition or the levels of the peptidoglycan precursors. Because high-pressure liquid chromatography and mass spectrometry demonstrated that there was no defect in the levels or the composition of the peptidoglycan precursors, it is unlikely that resistance to d-CS in Cvr-1 is due to overexpression of alr.
To determine whether resistance to vancomycin in Cvr-1 results from the inability of vancomycin to interact with d-alanine–d-alanine dipeptides present in the peptidoglycan precursors of the mutant, vancomycin removal assays were performed. The assays are based upon a modification of a technique developed by Payne and Bell to evaluate peptide transport (12). In their assay, uptake is measured by labeling the supernatant of the cells with a fluorescent peptide-labeling chemical to monitor the substrate's removal from the supernatant. The method used here is based upon the fact that the levels of vancomycin in the supernatant can be detected by its absorbance at 282 nm.
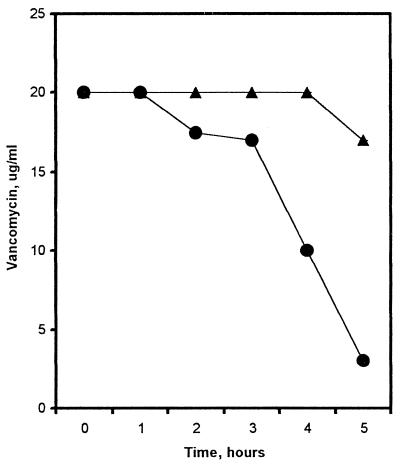
To perform the assay, 20 μg of vancomycin per ml was added to wild-type and mutant cultures. The concentration of vancomycin used was two times below the MIC for wild-type M. smegmatis to prevent killing of the wild-type parent during the assay. The amount of vancomycin remaining in the supernatant was measured over a period of 5 h. To ensure that the vancomycin remained stable over the 5 h, a control with no cells was performed simultaneously (data not shown). Figure 1 shows that after 5 h of incubation, 15% of the vancomycin was removed from the supernatant of Cvr-1. The wild-type parent removed threefold more vancomycin from the supernatant (85%) than the mutant. This result suggests that there is either less substrate (d-alanine–d-alanine of the peptidoglycan precursors) for vancomycin binding on the bacterial cell wall or that the substrate is inaccessible to the drug.
FIG. 1.
Vancomycin removal assay. Removal of vancomycin from the supernatant by M. smegmatis mc2155 (circles) or Cvr-1 (triangles) was monitored at 282 nm at the indicated time points. At each time point, the A282 was converted to micrograms of vancomycin per milliliter remaining in the supernatant. The data are the averages of three experiments performed in duplicate. Error bars are smaller than data points.
One explanation for the reduced removal of vancomycin from the supernatant of the mutant strain is that the strain has less target for binding. To examine this possibility, we compared the levels of incorporation of 14C radioactive label into the peptidoglycan after metabolic labeling of the wild-type and mutant strains, using a protocol developed by Burns-Keliher et al. (5). Following incubation of the wild type and mutant with [14C]d-alanine, the wild-type strain contained 7.05 nmol of 14C label derived from d-alanine/mg (wet weight), whereas the mutant strain contained only 5.45 nmol of 14C label derived from d-alanine/mg (wet weight). As a control, we performed the same experiment with [14C]d-serine because d-serine is not usually a component of peptidoglycan. Following incubation of the wild type and mutant with [14C]d-serine, the wild-type strain contained 1.90 nmol of 14C label derived from d-serine/mg (wet weight), while the mutant strain contained 2.14 nmol of 14C label derived from d-serine/mg (wet weight). The results from this assay suggest that less d-alanine is incorporated into the cell wall of Cvr-1; therefore, less target may be available for vancomycin binding.
One obvious explanation for the simultaneously d-CS and vancomycin-resistant phenotype of Cvr-1 is the presence of two or more unlinked mutations in Cvr-1. To address this issue, we performed conjugative mating assays. Parsons et al. (11) demonstrated that M. smegmatis mc2155 and its derivatives can act as donor strains for the unidirectional conjugal transfer of DNA. The streptomycin-resistant strain MKD8 serves as the acceptor strain for the DNA from M. smegmatis mc2155 or its derivatives.
Upon mating Cvr-1 and MKD8, 4.5 × 104 streptomycin- and d-CS-resistant colonies were isolated, with a transfer frequency (conjugants per donor) of 1.3 × 10−3. To determine whether resistance to d-CS and vancomycin in Cvr-1 could be transferred to the recipient strains simultaneously, the streptomycin- and d-CS-resistant colonies were tested for their resistance to vancomycin. None of the streptomycin- and d-CS-resistant colonies were resistant to vancomycin, indicating that resistance to d-CS is not linked to vancomycin resistance in Cvr-1. This suggests that Cvr-1 contains two or more mutations leading to its simultaneous drug-resistant phenotype.
To gain insight into the nature of the multidrug-resistant phenotype of Cvr-1, we screened for wild-type sequences that would restore to Cvr-1 sensitivity to both d-CS and vancomycin. Using a multicopy M. smegmatis mc2155 library, we screened 600 transformants for resistance to d-CS. One of the d-CS-resistant clones isolated was subcloned into a multicopy vector and tested for its ability to restore vancomycin sensitivity to Cvr-1. This clone, A1-3(10), restored both d-CS and vancomycin sensitivity to Cvr-1, whereas the vector alone did not. Although multicopy suppression is not always successful in identifying actual mutations, the fact that multicopy expression restores both d-CS and vancomycin sensitivity to Cvr-1 can be useful in understanding the mechanism of resistance to both drugs.
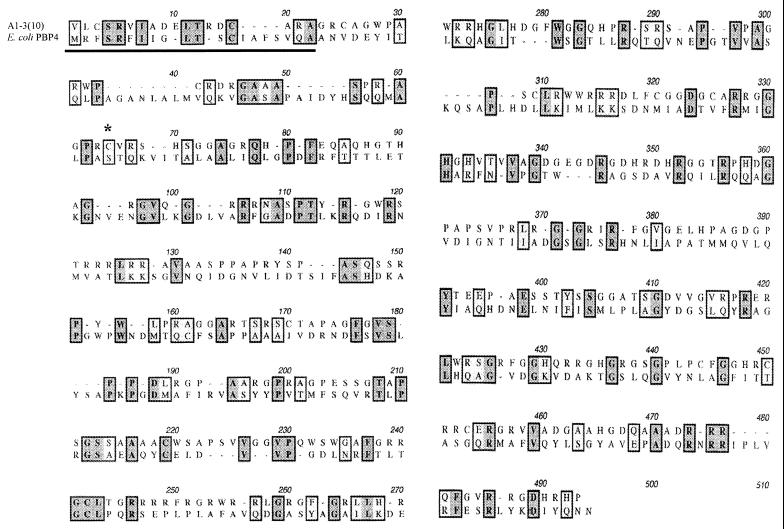
Sequencing of the 1.1-kb PstI insert in A1-3(10) revealed an open reading frame (ORF) with homology (24% identity and 37% similarity) to the E. coli penicillin binding protein 4 (PBP4) gene (10) (Fig. 2). The PBPs are a class of membrane proteins present at the bacterial cell surface which are involved in peptidoglycan chain assembly and turnover (16). Kyte-Doolittle analysis of the putative PBP4 homolog showed that the 17 N-terminal amino acids displayed a hydrophobic pattern (data not shown). The hydrophobicity of the N terminus, along with the presence of several basic amino acids, suggests that the N terminus may act as a signal sequence, targeting the protein to the cell wall. This is a common feature of penicillin binding proteins (9, 10). The critical active-site serine of E. coli PBP4 is replaced with a similar cysteine residue in the A1-3(10) ORF (Fig. 2).
FIG. 2.
Amino acid sequence comparison of the complementing ORF with PBP4 from E. coli. The putative N-terminal signal sequence is underlined, and the active-site residue is indicated by the asterisk (see text). Identical residues are highlighted in darker boxes, whereas similar residues are highlighted in lighter boxes.
In conclusion, a mutant of M. smegmatis resistant to both d-CS and vancomycin has been isolated. Because the d-alanine–d-alanine ligase from Cvr-1 demonstrated wild-type function and because no unusual peptidoglycan precursors were detected, a defect in the peptidoglycan precursor biosynthetic pathway seems unlikely. The vancomycin removal and peptidoglycan labeling assays indicated that there may be less substrate available for vancomycin binding in the cell wall of the mutant. Therefore, the substrate for vancomycin binding in the mutant must be altered during a later step of peptidoglycan biosynthesis, perhaps during cell wall assembly at the cell surface.
Supporting this hypothesis, a multicopy wild-type sequence that complements d-CS and vancomycin resistance in Cvr-1 displayed homology to the gene encoding the PBP4 enzyme from E. coli (10). This protein is involved in cell wall assembly at the cell surface. Further studies of the details of the cell wall assembly reactions in Cvr-1 will elucidate the mechanism behind the interesting phenotypes of this mutant and lead to an increased understanding of mycobacterial cell wall synthesis and architecture.
Acknowledgments
We thank Keiko Taber of Wyeth-Ayerst Research for the mass spectrometric analysis of the precursors. We also thank Robert Donnelly and The Molecular Resource Facility for assistance with the sequencing analysis and Majorie Brandriss for critical reading of the manuscript.
This work was supported in part by the UMDNJ Graduate School of Biomedical Sciences, the New Jersey Medical School National Tuberculosis Center, and Public Health Service grant R29AI34436 to N.D.C.
REFERENCES
- 1.Arthur M, Molinas C, Bugg T D H, Wright G D, Walsh C T, Courvalin P. Evidence for in vivo incorporation of d-lactate into peptidoglycan precursors of vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1992;36:867–869. doi: 10.1128/aac.36.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/aac.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry C E, III, Mdluli K. Drug sensitivity and environmental adaptation of mycobacterial cell wall components. Trends Microbiol. 1996;4:275–281. doi: 10.1016/0966-842x(96)10031-7. [DOI] [PubMed] [Google Scholar]
- 4.Bloom B R, Murray C J. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 5.Burns-Keliher L L, Portteus A, Curtiss R., III Specific detection of Salmonella typhimurium proteins synthesized intracellularly. J Bacteriol. 1997;179:3604–3612. doi: 10.1128/jb.179.11.3604-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cáceres N E, Harris N B, Wellehan J F, Feng Z, Kapur V, Barletta R G. Overexpression of the d-alanine racemase gene confers resistance to d-cycloserine in Mycobacterium smegmatis. J Bacteriol. 1997;179:5046–5055. doi: 10.1128/jb.179.16.5046-5055.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David H L. Resistance to d-cycloserine in the tubercle bacilli: mutation rate and transport of alanine in parental cells and drug-resistant mutants. Appl Microbiol. 1971;21:888–892. doi: 10.1128/am.21.5.888-892.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallori E, Fani R. Characterization of d-cycloserine resistant mutants in Bacillus subtilis. Microbiologica. 1983;6:19–26. [PubMed] [Google Scholar]
- 9.Granier B, Deuz C, Lepage S, Englebert S, Dusart J, Dideberg O, Van Beeumen J, Frere J M, Ghuysen J M. Primary and predicted secondary structures of the Actinomadura R39 extracellular dd-peptidase, a penicillin-binding protein (PBP) related to the Escherichia coli PBP4. Biochem J. 1992;282:781–788. doi: 10.1042/bj2820781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korat B, Mottl H, Keck W. Penicillin-binding protein 4 of Escherichia coli: molecular cloning of the dacB gene, controlled overexpression, and alterations in murein composition. Mol Microbiol. 1991;5:675–684. doi: 10.1111/j.1365-2958.1991.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 11.Parsons L M, Jankowski C S, Derbyshire K M. Conjugal transfer of chromosomal DNA in Mycobacterium smegmatis. Mol Microbiol. 1998;28:571–582. doi: 10.1046/j.1365-2958.1998.00818.x. [DOI] [PubMed] [Google Scholar]
- 12.Payne J W, Bell G. Direct determination of the properties of peptide transport systems in Escherichia coli, using a fluorescent-labeling procedure. J Bacteriol. 1979;137:447–455. doi: 10.1128/jb.137.1.447-455.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perichon B, Reynolds P, Courvalin P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother. 1997;41:2016–2018. doi: 10.1128/aac.41.9.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reitz R H, Slade H D, Neuhaus F C. The biochemical mechanisms of resistance by streptococci to the antibiotics d-cycloserine and o-carbamyl-d-serine. Biochemistry. 1967;6:2561–2570. doi: 10.1021/bi00860a038. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds P E, Snaith H A, Maguire A J, Dutka-Malen S, Courvalin P. Analysis of peptidoglycan precursors in vancomycin-resistant Enterococcus gallinarum BM4174. Biochem J. 1994;301:5–8. doi: 10.1042/bj3010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1025–1034. [Google Scholar]
- 17.Wargel R J, Shadur C A, Neuhaus F C. Mechanism of d-cycloserine action: transport systems for d-alanine, d-cycloserine, l-alanine, and glycine. J Bacteriol. 1970;103:778–788. doi: 10.1128/jb.103.3.778-788.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]