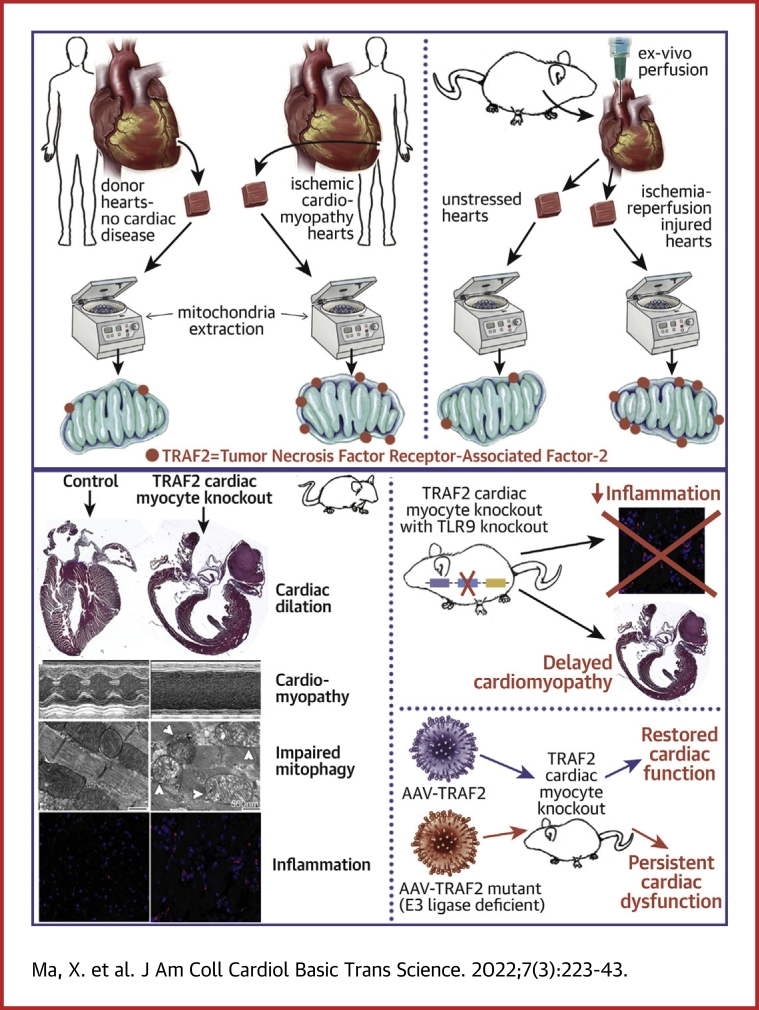
Visual Abstract
Key Words: cell death, inflammation, mitophagy, TRAF2
Abbreviations and Acronyms: AAV9, adeno-associated virus serotype 9; cTnT, cardiac troponin T; ER, endoplasmic reticulum; FS, fractional shortening; GFP, green fluorescent protein; IP, intraperitoneal; LV, left ventricular; MAM, mitochondria-associated membranes; MCM, MerCreMer; MEF, murine embryonic fibroblast; PINK1, PTEN-induced kinase 1; RFP, red fluorescent protein; TLR9, toll-like receptor 9; TRAF2, tumor necrosis factor receptor-associated factor-2; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling
Highlights
-
•
TRAF2, an innate immunity adaptor protein, localizes to the mitochondria in cardiac myocytes in human and mouse hearts during physiology, with increased mitochondrial localization under pathologic stress.
-
•
TRAF2 is essential for physiological mitophagy in cardiac myocytes, with a critical requirement for its E3 ligase domain in this role.
-
•
TRAF2 suppresses expression of TLR9, a sensor for leaked mitochondrial DNA, to suppress sterile inflammation in the myocardium.
-
•
Loss of physiological TRAF2-mediated mitophagy in cardiac myocytes triggers myocardial inflammation and cardiac myocyte cell death. Interruption of TLR9-mediated mitochondrial DNA sensing rescues inflammation, but persistence of damaged mitochondria leads to cell death with aging, pointing to a critical role for physiological cardiac myocyte mitophagy in maintaining myocardial homeostasis.
Summary
Mitochondria are essential for cardiac myocyte function, but damaged mitochondria trigger cardiac myocyte death. Although mitophagy, a lysosomal degradative pathway to remove damaged mitochondria, is robustly active in cardiac myocytes in the unstressed heart, its mechanisms and physiological role remain poorly defined. We discovered a critical role for TRAF2, an innate immunity effector protein with E3 ubiquitin ligase activity, in facilitating physiological cardiac myocyte mitophagy in the adult heart, to prevent inflammation and cell death, and maintain myocardial homeostasis.
Mitochondrial permeabilization results in activation of cell death signaling and is a major pathway for cardiac myocyte death under stress.1 Damaged mitochondria also leak mitochondrial DNA, which is a potent trigger for immune activation due to its physical features that mimic bacterial DNA.1 Indeed, mitochondrial damage resulting in sterile myocardial inflammation and cardiac myocyte death is implicated in causing cardiomyopathy under pressure overload stress2,3 and with ischemia-reperfusion injury,4,5 whereby targeting cell death as well as deleterious myocardial inflammation are regarded as key therapeutic targets to prevent and treat heart failure.1,6 Mitophagy is a lysosomal process to sequester damaged mitochondria (or parts thereof) to facilitate their removal via lysosomal degradation, and serves as a defense mechanism against mitochondrial DNA sensing to drive inflammation and cell death.2,7
Mitophagy plays a critical role in the myocardium during perinatal cardiac growth and development,8 with aging,9,10 with exhaustive exercise,7 and in pathologic states, such as after injury.2,9, 10, 11, 12, 13, 14, 15 Despite these discoveries, its role in physiological function of the adult heart remains unknown. Moreover, although studies with contemporary reporters demonstrate robust rates of cardiac myocyte mitophagy in the unstressed myocardium,16,17 ablation of canonical mitophagy mediators such as PTEN-induced kinase 1 (PINK1) and PARKIN18 does not alter mitophagy in young adult mouse hearts.12,15,19, 20, 21
Prior studies indicate that lysosome function is critical for preventing myocardial inflammation under stress, as ablation of lysosomal DNase II in cardiac myocytes, which degrades mitochondrial DNA, exacerbates myocardial inflammation to accelerate development of cardiomyopathy with pressure overload.2 Whether physiological mitophagy suppresses myocardial inflammatory signaling in the unstressed heart remains unknown. It is also noteworthy that all currently defined mitophagy pathways are intrinsic to damaged mitochondria and recognize mitochondrial damage or dysfunction to facilitate lysosomal removal of damaged mitochondria via mitophagy or other mechanisms,22 whereby participation of innate immune mechanisms in facilitating mitophagy remains a missing piece of this conceptual framework. In this paper, we reveal evidence that tumor necrosis factor receptor-associated factor-2 (TRAF2), an innate immunity effector, acts as a first line of defense against cardiac myocyte cell death and sterile myocardial inflammation by facilitating mitophagy in the adult mouse heart.
Our data demonstrate that a fraction of TRAF2 is associated with mitochondria in homeostasis, and its recruitment to the mitochondria is up-regulated under conditions of mitochondrial damage with ischemia-reperfusion injury and in cardiomyopathic failing human hearts. Inducible ablation of TRAF2 in adult cardiac myocytes impairs mitophagy and provokes accumulation of damaged mitochondria, primarily based upon its role as an E3 ubiquitin ligase. Moreover, inducible TRAF2 ablation up-regulates toll-like receptor 9 (TLR9) expression to drive myocardial dysfunction and knockout of TLR9 delays the onset of cardiomyopathy by curtailing inflammation, but does not prevent eventual cardiomyopathic decompensation due to presence of damaged mitochondria. These studies highlight a central role for the innate immunity pathway as a mitochondria-extrinsic mechanism in sensing mitochondrial damage to effect mitophagy and maintain homeostasis.
Methods
Reagents and samples
We crossed mice homozygous for floxed Traf2 alleles (Traf2 fl/fl)23 (which permits Cre-mediated excision of exon 3 and introduces a frame shift resulting in loss of TRAF2 protein; generously provided by Dr. Robert Brink, Garvan Institute of Medical Research, Australia) with mice carrying the Myh6 promoter driven Mer-Cre-Mer (MCM) transgene (generous gift from Jeffery D. Molkentin, PhD, Cincinnati Children’s Hospital, Cincinnati, Ohio).24 Mice were injected with tamoxifen (Millipore Sigma #T5648, dissolved in ethanol and sunflower oil #S5007 to make a stock solution of 10 mg/mL) at the doses indicated (intraperitoneal [IP]); or fed with tamoxifen chow (Envigo, cat#TD.130857), as described for individual experiments. Tlr9-null mice were generously provided by Drs. Shizou Akira, Laboratory of Host Defense, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan. MitoQC reporter strain was previously described17 and generously provided by Dr. Ian Ganley at University of Dundee, UK. Park2-null (Strain number 006582), Pink1-null (stock number 017946), and the autophagic reporter CAG-RFP-EGFP-LC3 (strain number 027139) mice were purchased from The Jackson Laboratory. All mice were maintained on a C57BL/6J background. Mice of both sexes were studied. No significant differences were observed between sexes for the primary phenotype, thus data for both sexes were combined for presentation. Mouse studies were randomized and observers blinded. All animal studies were approved by the institutional animal care and use committee at Washington University School of Medicine. Studies on human tissue were performed under an exemption by the institutional review board at Washington University School of Medicine because only deidentified human samples were used. Human heart tissue was obtained from Kenneth Margulies, MD, at the University of Pennsylvania, Philadelphia (Supplemental Table 1). Hearts were categorized as either nonfailing (no history of heart failure, obtained from nonfailing brain-dead donors) or ischemic cardiomyopathy (obtained at time of orthotopic heart transplantation). After in situ cold cardioplegia, all hearts were placed on wet ice at 4°C in Krebs-Henseleit buffer. Transmural left ventricular (LV) samples were obtained from the LV free wall with epicardial fat excluded. The mitoKeima construct was generously provided by Dr. Atsushi Miyawaki, RIKEN brain science institute in Saitama, Japan.
Echocardiography and myocardial characterization
Two-dimensional–directed M-mode echocardiography was performed as we have previously described.25 Histologic assessment for myocyte registration was performed with hematoxylin and eosin staining, and myocardial fibrosis was assessed with Masson’s trichrome staining, as we have previously described.25 Quantitative assessment of fibrosis was performed with Picrosirius Red staining, and images were assessed for area of the red signal as a fraction of total tissue area by Visiomorph software (Visiopharm). Transmission electron microscopy was performed in cardiac tissue as previously described.25
Studies with adeno-associated viral vectors
Adeno-associated virus serotype 9 [AAV9] particles coding for TRAF2 or its Rm mutant constructs (which we have described previously26) driven by the cardiac troponin T (cTnT) promoter for conferring cardiac myocyte selective expression,27 were generated by the Hope Center viral vectors core; using AAV backbone constructs generously provided by Dr. Brent French at University of Virginia, Charlottesville.
Ex vivo cardiac ischemia-reperfusion injury
Hearts were isolated and perfused as previously described.28 After a 20-minute stabilization period, mouse hearts were subjected to no-flow ischemia (time [t] = 0 minutes) for 30 minutes followed by reperfusion for 60 minutes (total t = 90 minutes).
Adult cardiac myocyte isolation
Adult mouse hearts were subjected to enzymatic digestion to isolate adult cardiac myocytes, as described.29 The remaining cellular fraction was isolated as the nonmyocyte fraction. The cells were subsequently homogenized in RIPA buffer and subjected to SDS-PAGE gel electrophoresis followed by immunoblotting as we have previously described.26
Preparation of murine embryonic fibroblasts
Traf2 floxed or Pink1 floxed mice were subjected to timed mating and murine embryonic fibroblasts were prepared from embryonic day 14 pups following standard protocols.
Biochemical subcellular fractionation
Mitochondria-enriched and cytosolic fractions were prepared from hearts and cells following the protocols we have previously described.26 Biochemical separation of mitochondria-rich fraction into mitochondria stripped of other organelles, mitochondria-associated membranes (MAM), and endoplasmic reticulum (ER) was performed with Percoll density centrifugation following published protocols.30
In situ hybridization with RNAscope
All procedures for Tlr9 transcript detection with fluorescence in situ hybridization were performed using the RNAscope Multiplex Fluorescent Reagent kit V2 (Cat. No. 323100, Advanced Cell Diagnostics) according to the manufacturer's instructions. RNAscope Probe-Mm-Tlr9, Mouse (Cat. No. 468281, Advanced Cell Diagnostics) and Opal 690 Dye (Cat. No. FP1497001KT, Akoya BioSciences) were used according to the manufacturer's instructions for fluorescence in situ hybridization. Desmin staining was performed subsequently to the horseradish peroxidase blocker step of fluorescence in situ hybridization. Slides were rinsed and covered with 1:50 dilution of goat polyclonal anti-desmin antibody (Cat. No. SC-7559, Santa Cruz Biotechnology) overnight at 4°C. The next morning, slides were rinsed and covered with 1:200 dilution of Alexa Fluor 488 donkey anti-goat secondary antibody (Cat. No. A-11055, Invitrogen) for 45 min at room temperature. Slides were rinsed and mounted, stained with DAPI using Antifade Mounting Medium with DAPI (Cat. No. H-1200, Vector Labs). Slides were stored at 4°C in the dark until image analysis. Images were captured on a Zeiss LSM 700 confocal microscope.
Immunofluorescence Analysis
We followed the protocol we have previously described.25 Paraffin-embedded heart sections (10-μm thick) were subjected to heat-induced epitope retrieval, followed by blocking, and then incubated overnight with primary antibody. After serial washes, samples were stained with Alexa Fluor 594 (Invitrogen) and mounted with fluorescent DAPI mounting medium (Vector Labs, H-1200). Confocal imaging was performed on a Zeiss confocal LSM-700 laser scanning confocal microscope using 639 Zeiss Plan-Neofluar 40/1.3 and 63/1.4 oil immersion objectives, and images were acquired using Zen 2010 software. CD68 (Biolegend, 137001), TLR9 (Novus Biologicals, NBP2-24729), PicoGreen from Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific, P7589). For CD68 staining, we used MaxBlock Autofluorescence Reducing Reagent Kit (MaxVision Biosciences, cat# MB-M) to block the high autofluorescence background.
TUNEL staining
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) was performed on formalin-fixed and paraffin-embedded tissue sections by using the Dead-End fluorometric TUNEL system (Promega, G3250) following the manufacturer's directions, as previously described.25
Assessment of autophagic flux
We performed autophagic flux analyses by immunofluorescence examination of frozen myocardial tissue from mice with expression of the red fluorescent protein (RFP)–green fluorescent protein (GFP)-LC3 reporters, as described.31 Only puncta associated with cells identified as myocytes based upon visualization of boxcar-shaped nuclei were counted.
Assessment of mitophagy
Mice transduced with AAV9 particles coding for CMV promoter–driven mitoKeima were generated by the Hope Center viral vectors core at Washington University at a dose of 3.5 × 1011 viral particles per mouse. Mitophagy was assessed with expression of mitoKeima as previously described.16,32 Briefly, live imaging of freshly harvested cardiac slices were performed with a Zeiss LSM5 Pascal confocal microscope. Two sequential excitations were performed at wavelengths of 458 and 543 nm, followed by imaging with a 560-nm longpass filter for assessment of the mitophagy signal. Laser power was individualized for each condition to optimize Keima imaging. Murine embryonic fibroblasts (MEFs) were transfected with pcDNA3.1 encoding for mitoKeima construct using GenJet transfection reagent (Sinagen) following the manufacturer’s protocols, and mitophagy was assessed with sequential excitation at wavelengths at 458 and 561 nm, followed by emission at 580 nm using a Nikon A1R confocal system at the Washington University Center for Cellular Imaging. Adenoviruses expressing Cre and rat PARKIN were generated as previously described.26 The images were quantitated using ImageJ software (NIH). Mitophagy was also assessed after injection of AAV9-cTnT-TRAF2 or AAV9-cTnT-null viral particles (with 3.5 × 1011 viral particles per mouse) in young adult mitoQC mice. Mitophagy index was calculated as a ratio of the average intensity of the red/green signal using ImageJ software.
Quantitative polymerase chain reaction analyses
Assessment of transcript abundance was performed as previously described25 using SYBR Green with primers reported in Supplemental Table 2. Assessment of Pink1 transcript expression was performed using TaqMan gene expression assays (Applied Biosystems) for mouse Pink1 (Mm00550827_m1).
Immunoblotting
Immunoblotting was performed as previously described.25 Specific antibodies employed are as follows: LC3 (Novus Biologicals, NB100-2220); p62 (Abcam, ab56416); TRAF2 (Abcam, ab126758); COX IV (Abcam, ab14744); TOMM20 (Sigma, WH0009804M1); VDAC (Cell Signaling Technology, 4661S); PARKIN (Abcam, ab15954); FACL4 (Abcam, ab155282); calreticulin Antibody #2891; VAPB (Thermo Fisher Scientific, A302-894A); IRE1α (14C10) (Cell Signaling Technology, 3294S); GAPDH (Abcam, ab22555); TLR9 (Novus Biologicals, NBP2-24729); actin (Sigma, A2066); PINK1 (MRC PPU products and reagents, S774C [DU17570] and S086D [DU34559]); and α-sarcomeric actin (Abcam, ab52219).
Enzyme-linked immunosorbent assays
Enzyme-linked immunosorbent assays were performed on sera collected from mice at terminal bleed with the following kits per the manufacturers’ instructions: mouse IL-1β/IL-1F2 (R&D Systems, MLB00C), mouse TNF-alpha (R&D Systems, MTA00B), mouse IL-6 (R&D Systems, M6000B), and HMGB1 (Chondrex, 6010).
Immunogold detection
For immunolocalization, cells were fixed in 4% paraformaldehyde/0.05% glutaraldehyde (Polysciences) in 100 mmol/L PIPES/0.5 mmol/L MgCl2, pH 7.2 for 1 hour at 4°C. Samples were then embedded in 10% gelatin and infiltrated overnight with 2.3 mol/L sucrose/20% polyvinyl pyrrolidone in PIPES/MgCl2 at 4°C. Samples were trimmed, frozen in liquid nitrogen, and then sectioned with a Leica Ultracut UCT7 cryo-ultramicrotome (Leica Microsystems). Ultrathin sections of 50 nm were blocked with 5% FBS/5% NGS for 30 min and subsequently incubated with rabbit anti-GFP (Life Technologies, 1:200) for 1 hour at room temperature. Following washes in block buffer, sections were incubated with 18-nm colloidal gold conjugated goat anti-rabbit IgG+IgM (Jackson ImmunoResearch Laboratories, 1:30) for 1 hour. Sections were stained with 0.3% uranyl acetate/2% methyl cellulose and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA) equipped with an AMT 8 megapixel digital camera and AMT Image Capture Engine V602 software (Advanced Microscopy Techniques). All labeling experiments were conducted in parallel with control sections omitting the primary antibody.
Statistical analysis
Data are presented as the mean ± SEM. All measurements were obtained on distinct biological replicates. Statistics were performed in Prism Version 8.0.2 (GraphPad Software). Data were tested for assumptions of normality with Shapiro-Wilk normality test. Statistical significance of differences was calculated via unpaired 2-tailed Student’s t-test for 2 group comparisons, or 1-way or 2-way analysis of variance for assessing differences across multiple groups followed by post hoc testing (Tukey’s) to evaluate pairwise differences. A nonparametric test was employed if data were not normally distributed. Graphs containing error bars show the mean ± SEM, with a P value <0.05 considered statistically significant. The datasets from the current study are available from the corresponding author on reasonable request.
Results
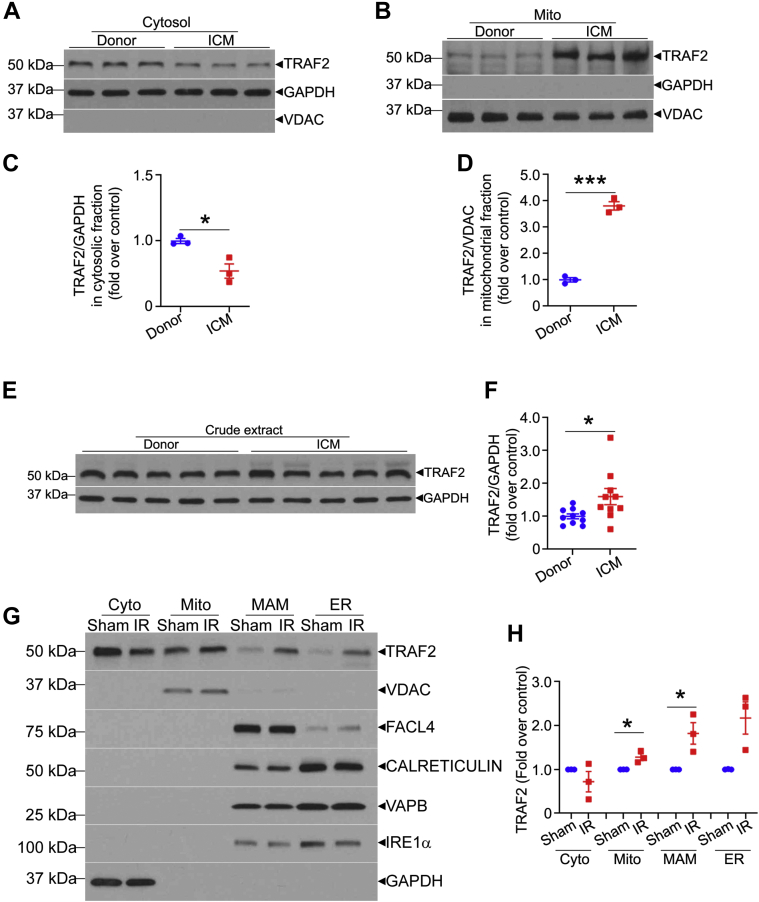
Our published studies in isolated cardiomyocytes indicate that TRAF2, an innate immune adaptor, localizes to mitochondria.26 To determine whether this occurs in human hearts, we obtained heart tissue from donors rejected for transplantation, as well as from patients with ischemic cardiomyopathy, and examined TRAF2 expression following biochemical fractionation into a mitochondria-enriched fraction, and cytosol (Figures 1A to 1D). TRAF2 protein was detected in the mitochondrial fraction, where it was dramatically up-regulated in hearts with ischemic cardiomyopathy and end-stage heart failure (Figures 1B and 1D). This was accompanied by an increase in overall TRAF2 levels (Figures 1E and 1F), as well as a relative decline in cytosolic levels (Figures 1A and 1C), suggesting that TRAF2 translocates to the mitochondria upon injury.
Figure 1.
TRAF2 Localizes to the Mitochondria in Human and Mouse Hearts
(A and B) Immunoblots depicting TRAF2 expression in hearts from individuals evaluated as donors for cardiac transplantation, without evidence for cardiomyopathy (donor) or patients with end-stage ischemic cardiomyopathy (ICM) undergoing cardiac transplantation. The hearts were subjected to biochemical fractionation into mitochondria-enriched (B) and cytosolic (A) fractions, shown by segregation of VDAC, a mitochondrial protein and GAPDH, a cytosolic protein, respectively. (C and D) Quantitative assessment of TRAF2 expression in the respective biochemical fractions are shown in A and B. (E and F) Representative immunoblot and quantitation are shown depicting total TRAF2 expression in crude extracts from human hearts from patients with ischemic cardiomyopathy (ICM) or donors as control. (G and H) Representative immunoblot is shown depicting TRAF2 expression in hearts from male C57BL/6J mice (8 weeks old) subjected to ex vivo ischemia-reperfusion injury (IR) or sham (S) treatment as control. Expression of VDAC, FACL4, calreticulin, VAPB, and IRE1α is shown to evaluate cosegregation with mitochondria (mito, with VDAC), mitochondria-associated membranes (MAM, with FACL4), endoplasmic reticulum (ER, with calreticulin, VAPB, and IRE1α), and cytosol (cyto, with GAPDH). ∗P < 0.05 and ∗∗∗P < 0.001, by t-test. No statistically significant differences were observed between the groups for TRAF2 abundance in the cyto fraction (by t-test) or ER (by Mann-Whitney test).
To examine the subcellular localization of TRAF2 in the mouse heart, we subjected wild-type mouse hearts to ex vivo ischemia-reperfusion injury or sham treatment and performed subcellular fractionation to further isolate the mitochondrial biochemical fraction into pure mitochondria, MAM and ER.30 Our data (Figures 1G and 1H) demonstrate that TRAF2 is predominantly located in the pure mitochondrial fraction in the sham (unstressed) heart, with a lesser degree of localization to the MAM and ER. Ex vivo ischemia-reperfusion injury increased TRAF2 localization to mitochondria and the MAM (Figures 1G and 1H). Indeed, TRAF2 was also detected by immunogold analysis and transmission electron microscopy on mitochondrial membranes and MAM in HEK293 cells, with increased localization to these structures as well as to mitochondria in autophagosomes after hypoxia-reoxygenation injury (Supplemental Figure S1). These observations suggest that the stress-induced increase in TRAF2 localization to mitochondria may play a functional role in its cytoprotective effects, as suggested by prior studies showing that TRAF2 is both necessary33 and sufficient28,34,35 to enhance cardiomyocyte survival after ischemia-reperfusion injury.
To examine the mechanisms for recruitment of TRAF2 to the mitochondria, we evaluated whether its mitochondrial localization depends upon PINK1. Indeed, as an executor of the canonical mitophagy pathway via macro-autophagy, PINK1, a serine-threonine kinase, is stabilized upon inner membrane depolarization following mitochondrial damage and recruits the E3 ligase PARKIN.36 PINK1 was not essential for recruitment of TRAF2 to the mitochondrial fraction under normoxic conditions, but loss of PINK1 markedly reduced TRAF2 translocation to the mitochondrial fraction upon hypoxia-reoxygenation injury (Supplemental Figure S2) consistent with a lack of role for PINK1 in executing basal mitophagy in the unstressed heart.19 Taken together, these data suggest a role for TRAF2 in homeostatic mitochondrial quality control in cardiac myocytes, in vivo.
Loss of myocardial TRAF2 impairs physiological mitophagy in the mouse heart
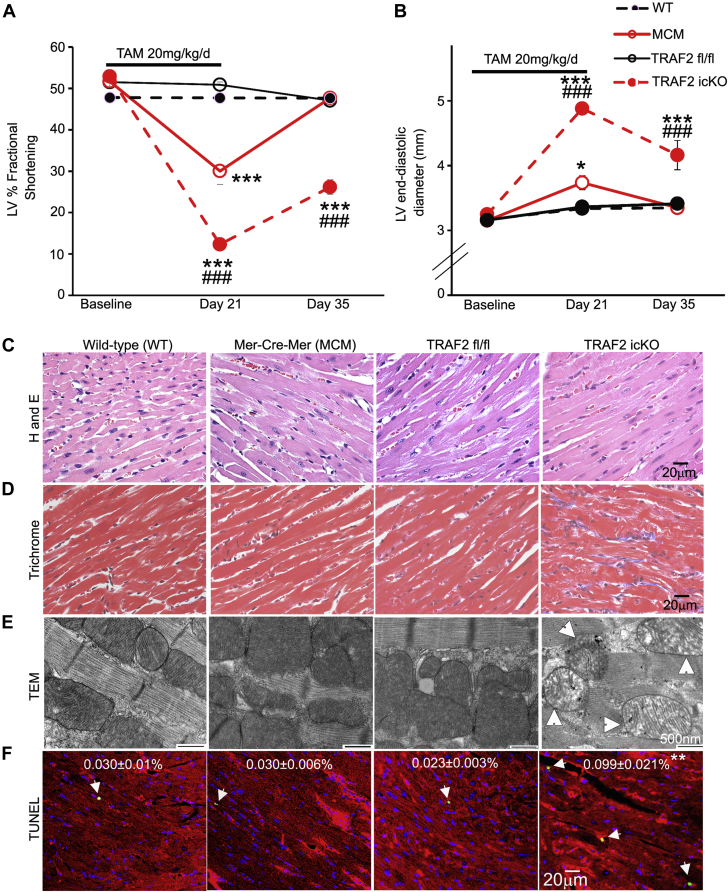
Prior studies have determined that perinatal cardiomyocyte-specific ablation of TRAF2 in the mouse heart provokes necroptosis of cardiac myocytes with systemic inflammation and cardiomyopathy.33 To isolate the effects of TRAF2 on cardiac myocyte homeostasis from developmental signaling, we examined the consequences of loss of TRAF2 in cardiomyocytes in the adult heart. Mice homozygous for floxed Traf2 alleles carrying the Myh6-promoter–driven MCM were injected with tamoxifen (30 mg/kg/d) for 3 consecutive days to generate inducible cardiac-myocyte specific TRAF2 knockout mice (depicted as TRAF2-icKO). Similarly treated Traf2 fl/fl mice (depicted as TRAF2 fl/fl) were studied as control mice. TRAF2-icKO mice manifest symptoms of heart failure with reduced physical activity, hunched posture, labored breathing, and ruffled coat within 5 days of initiating tamoxifen injections. Echocardiographic examination demonstrated marked LV cavity dilation and reduced contractile performance (% fractional shortening [%FS]) (Supplemental Figure S3A). On necropsy, the hearts were enlarged with dilation of the LV cavity (Supplemental Figures S3B and S3C) and significantly increased heart weight (heart weight/tibial length ratio: 7.27 ± 0.5 in TRAF2-icKO mg/mm vs 5.66 ± 0.15 mg/mm in Traf2 fl/fl, n = 3 per group; P = 0.036). Immunoblot analysis demonstrated a 66% reduction in myocardial TRAF2 protein in TRAF2-icKO mice (Supplemental Figures S4A and S4B). Although these data suggest that loss of TRAF2 results in cardiomyopathy, a decline in systolic performance was also detected in MCM transgenic mice as compared with wild-type alleles at the Traf2 locus after tamoxifen injection (%FS: 30% ± 4% vs 51% ± 4% in wild-type control mice; P = 0.016 by t-test. n = 3/group).
Because this signal of Cre-mediated toxicity has been described previously,37,38 we employed 2 other tamoxifen regimens that have been reported to circumvent Cre-mediated toxicity in the heart.38,39 A 3-week regimen of IP tamoxifen injections (20 mg/kg/d for 5 days per week) resulted in a 70% reduction of in whole-heart TRAF2 levels (Supplemental Figures S4C and S4D). As expected, isolation of adult cardiomyocytes from nonmyocytes demonstrated near complete loss of TRAF2 protein in the cardiomyocyte fraction (Supplemental Figures S4E and S4F) in TRAF2-icKO mice. TRAF2 protein expression was not altered by MCM-mediated TRAF2 ablation in noncardiomyocytes (Supplemental Figures S4E and S4F). We observed increased mortality among TRAF2-icKO mice (5/13 vs 0/10 in Traf2 fl/fl control mice; P = 0.046 by Fisher’s exact test) with signs of heart failure such as reduced activity, labored breathing, and generalized swelling before death. None of the mice carrying the MCM transgene or Traf2 fl/fl mice demonstrated signs of heart failure or died during the study. Serial echocardiography was performed at baseline, day 21 (after the last tamoxifen dose) and 2 weeks after the last dose was administered. We observed a decline in LV contractile function as shown by %FS (Figure 2A) with LV dilation (Figure 2B) in TRAF2-icKO mice as compared with Traf2 fl/fl control mice, that persisted at follow-up. We also observed a transient decline in %FS and mild LV dilation in transgenic MCM control mice, which was completely reversed at day 35 (Figures 2A and 2B), demonstrating evidence for transient, but reversible, Cre-mediated toxicity with this protocol. Surviving TRAF2-icKO mice demonstrated increased fibrosis at day 35 (Figures 2C and 2D, Supplemental Figure S5), and ultrastructural examination revealed abnormal swollen mitochondria with cristal rarefication (Figure 2E). This was accompanied by an ∼4-fold increase in TUNEL-positive cardiac myocytes as compared with Traf2 fl/fl control mice (Figure 2F). Importantly, the myocardium in MCM transgenic mice was histologically indistinguishable from wild-type mice without an increase in TUNEL-positive cardiomyocytes (Figures 2C to 2F). Another regimen of a single dose of tamoxifen (40 mg/kg) followed by echocardiography 2 weeks later induced only a modest TRAF2 protein knockdown in the myocardium (Supplemental Figures S4G and S4H) and resulted in a mild cardiomyopathy with mildly dilated LV (3.6 ± 0.1 mm in TRAF2-icKO vs 3.3 ± 0.1 mm in Traf2 fl/fl; P = 0.017, n = 4/group) with mild systolic dysfunction (%FS 41 ± 1 in TRAF2-icKO vs 50 ± 1 in Traf2 fl/fl; P < 0.001). At this dose, we did not observe any cardiac structural or functional abnormalities in MCM transgenic mice as compared with wild-type control mice.
Figure 2.
TRAF2 Ablation in Adult Cardiac Myocytes Induces Cardiomyopathy With Damaged Mitochondria and Cell Death
(A and B) Left ventricular (LV) endocardial fractional shortening (%FS (A) and end-diastolic diameter (B) in wild-type, Traf2 floxed (TRAF2 fl/fl), MerCreMer (MCM) mice, and mice homozygous for Traf2 floxed alleles carrying the MerCreMer transgene (to generate TRAF2-icKO), 14 days after tamoxifen treatment (20 mg/kg/d, IP 5 days per week for 3 weeks). n = 9 to 10 mice per group. ∗P < 0.05 and ∗∗∗P < 0.001 for comparison versus wild-type group; ###P < 0.001 for comparison versus Traf2 fl/fl group by post hoc test after 2-way ANOVA. (C and D) Representative images with hematoxylin and eosin (H and E) staining (C), and trichrome staining (D) to evaluate myocardial structure and fibrosis, respectively, in mice as treated in A and B. (E) Representative transmission electron microscopy images to evaluate myocardial ultrastructure in mice as in A. Arrows indicate swollen mitochondria with cristal rarefication. (F) Representative TUNEL-stained images to evaluate cardiac myocyte TUNEL positivity (green) in mice as in A. DAPI staining identifies nuclei. Staining for α-sarcomeric actin was performed to evaluate TUNEL staining in cardiac myocyte nuclei. %TUNEL-positive nuclei/total cardiac myocyte nuclei are depicted. n = 4 to 6/group. ∗∗P < 0.01 versus Traf2 fl/fl by post hoc test after 1-way ANOVA. ANOVA = analysis of variance; TAM = tamoxifen; TEM = transmission electron microscopy; TUNEL = terminal deoxynucleotidyl transferase dUTP nick end labeling; WT = wild type.
Overall, we observed a strong correlation between the degree of TRAF2 protein knockdown in the myocardium in the TRAF2-icKO mice and worsening fractional shortening and LV dilation (Supplemental Figures S4I and S4J) with different tamoxifen regimens. We also set up a model of inducible cardiac myocyte TRAF2 ablation by feeding tamoxifen chow. Mice treated with tamoxifen chow for 7 days demonstrated a similar degree of TRAF2 deletion as the 21-day injection regimen (Supplemental Figures S6A and S6B) and with comparable LV dilation and systolic dysfunction as compared with floxed control mice (data not shown). Taken together, these data indicate that the loss of TRAF2 in cardiac myocytes provokes cardiomyocyte death in unstressed hearts with spontaneous development of cardiomyopathy.
Prior studies with perinatal ablation of TRAF2 in cardiomyocytes (with Myh6-Cre) reported cardiac myocyte necrosis with systemic inflammation.33 Accordingly, we assessed for circulating markers noted to be elevated with TRAF2 ablation in the prior work. Our data demonstrate that circulating HMGB1 levels, a marker of necrotic cell death (Supplemental Figure S7A), or circulating cytokines (IL-1β, TNF, and IL-6) (Supplemental Figures S7B to S7D) are not elevated in TRAF2-icKO mice as compared with Traf2 fl/fl control mice, pointing to a different mechanism for the observed cardiomyopathy in mice with adult onset TRAF2 ablation.
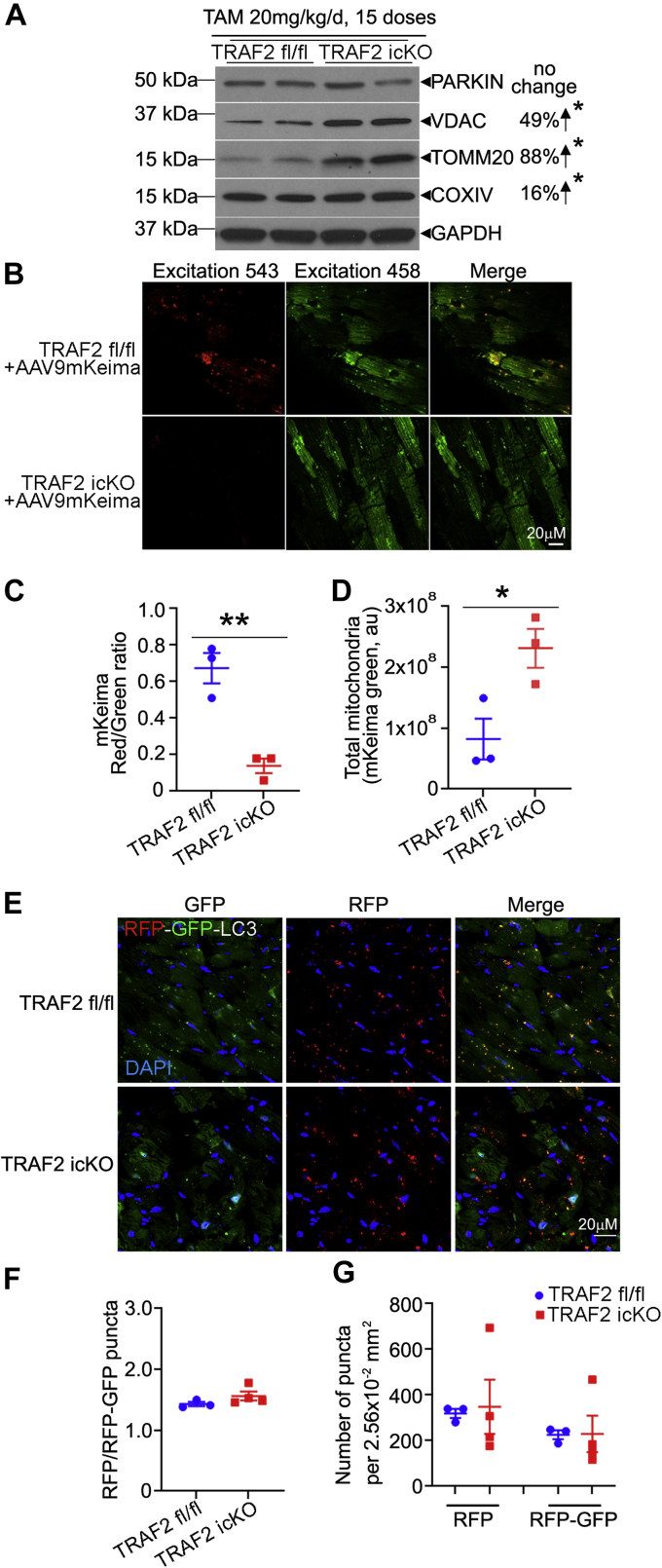
We observed that mitochondrial proteins accumulate upon TRAF2 ablation in the TRAF2-icKO mice (Figure 3A, Supplemental Figures S6B to S6F), pointing to impaired removal of damaged mitochondria with TRAF2 deficiency (as seen with the TEM analysis in Figure 2E). Indeed, examination of basal mitophagy with AAV9-mediated transduction of mKeima reporter32 revealed marked suppression of cardiac myocyte mitophagy in the TRAF2-icKO mice (Figures 3B to 3D). To determine whether the mitophagy deficit was secondary to defects in macro-autophagy, we examined flux in TRAF2-icKO mice expressing a dual fluorescent LC3-based flux reporter. The prevalence of autolysosomes relative to total autophagic structures, as well as the abundance of autolysosomes and autophagosomes, was not reduced in TRAF2-icKO as compared with Traf2 fl/fl mice (Figures 3E to 3G). Moreover, the abundance of autophagosome-bound LC3-II and p62, an adaptor protein that binds to autophagic cargo to sequester them within autophagosomes, was not decreased (Supplemental Figures S8A to S8C); and transcripts for Map1lc3b (that encodes for LC3 protein), Sqstm1 (which encodes for p62) and Becn1 (which encodes for BECLIN-1, a protein essential for autophagosome formation) were not reduced in TRAF2-icKO hearts (Supplemental Figures S8D to S8F). These data indicate that the observed defect with TRAF2 ablation is with organelle-specific autophagy, that is, mitophagy, without impairment of macro-autophagy in TRAF2-icKO hearts.
Figure 3.
TRAF2 Ablation in Adult Cardiac Myocytes Impairs Homeostatic Mitophagy
(A) Representative immunoblot is shown depicting expression of PARKIN and mitochondrial proteins (VDAC, TOMM20, COXIV) in cardiac extracts from mice at day 35 after inducible adult-onset ablation of TRAF2 (TRAF2-icKO), 14 days after tamoxifen (TAM) treatment (20 mg/kg/d, intraperitoneal for 5 days per week for 3 weeks). n = 4/group. Protein levels were normalized to GAPDH. Upward arrow points to up-regulation in abundance of indicated protein indicated as a % change. ∗P < 0.05 for TRAF2-icKO versus Traf2 fl/fl groups by t-test; and. (B to D) Representative images (B) from hearts transduced with AAV9-mitoKeima in mice as in A; with the ratio of quantitation of mitoKeima emission in the red channel over green as an index of mitophagy (C). Total mitochondria were assessed by abundance of the green signal (arbitrary units, au) per unit area of transduced myocytes (D). ∗P < 0.05 and ∗∗P < 0.01 by t-test. (E-G) Representative images (E) for assessment of flux through macroautophagy in TRAF2-icKO versus Traf2 fl/fl mice carrying the RFP-GFP-LC3 reporter transgene, with quantitative assessment of the ratio of autolysosomes (punctate RFP signal) to autophagosomes (punctate RFP+GFP signal, in F); and total autophagosomes (RFP+GFP puncta) and autolysosomes (RFP puncta) per unit myocardial area in G. No statistically significant differences were observed between TRAF2-icKO versus Traf2 fl/fl mice by t-test in F and G. GFP = green fluorescent protein; RFP = red fluorescent protein.
TRAF2 ablation results in mitochondrial DNA release and cardiac inflammation via TLR9
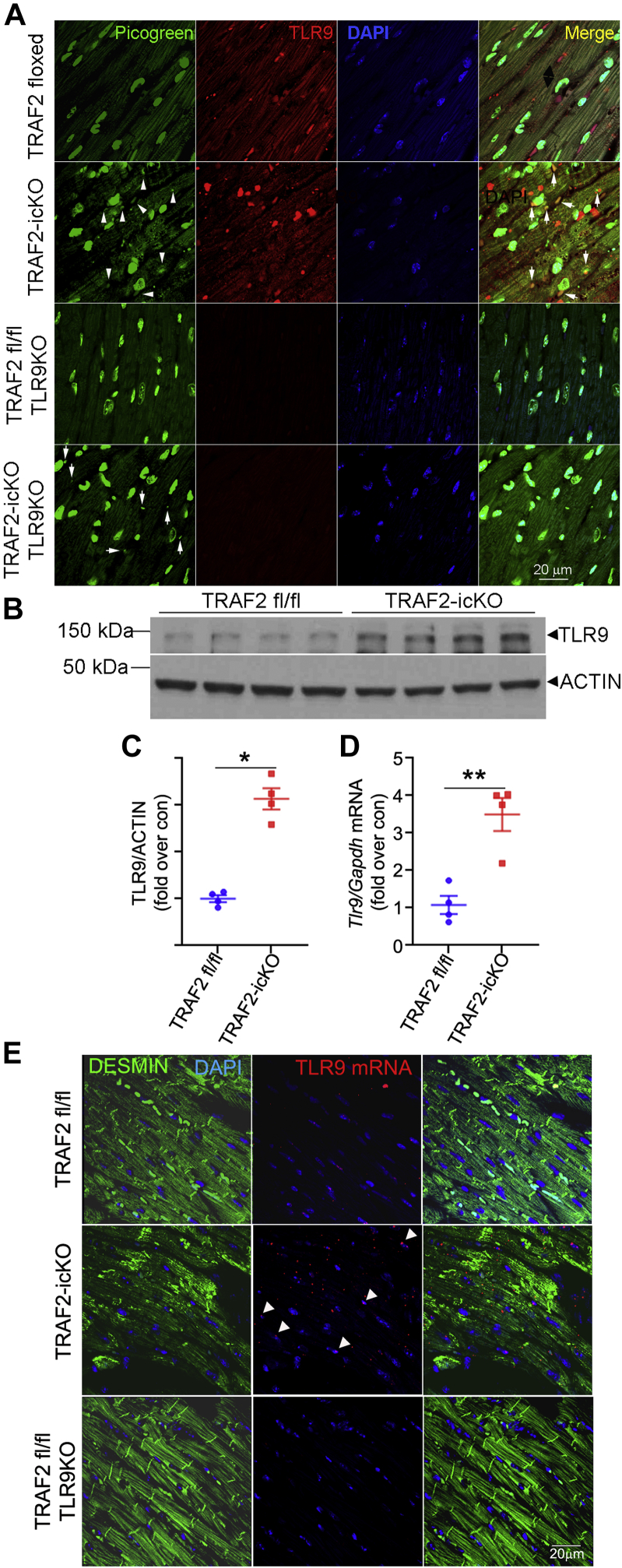
Impaired mitophagy results in mitochondrial DNA leak from damaged mitochondria, and TLR9-mediated mitochondrial DNA sensing has previously been shown to drive myocardial pathology under pressure overload stress.2,4 TRAF2-icKO myocardium demonstrated increased co-localization of DNA detected by PicoGreen with TLR9 in cardiac myocytes (Figure 4A). Although PicoGreen is not specific for mitochondrial origin of the visualized DNA,2 its cytosolic staining in the context of damaged mitochondria suggests release of mitochondrial DNA. Furthermore, TRAF2 ablation resulted in transcriptional up-regulation of TLR9 expression (Figures 4B to 4D), which was localized to cardiac myocytes (Figure 4E) per in situ hybridization to detect Tlr9 transcript expression.
Figure 4.
TRAF2 Ablation in Adult Cardiac Myocytes Up-Regulates TLR9 and Mitochondrial DNA Sensing
(A) Representative images demonstrating colocalization of PicoGreen with TLR9 in myocardial sections from mice with adult-onset inducible TRAF2 ablation (TRAF2-icKO) and Traf2 floxed control mice, without and with concomitant germline TLR9 ablation. Nuclei are stained blue (DAPI). Arrowheads demonstrate colocalization of PicoGreen with TLR9. Arrows point to cytosolic detection of DNA by PicoGreen in TRAF2-icKO-TLR9KO myocardium. (B and C) Immunoblot (B) and quantitation (C) depicting TLR9 expression in cardiac extracts from TRAF2-icKO and Traf2 floxed control mice modeled as in A. ∗P < 0.05 and ∗∗P < 0.01 by t-test. (D)Tlr9 transcript expression in cardiac extracts from TRAF2-icKO and Traf2 floxed control mice as in A. The P value depicted is by t-test. (E) In situ hybridization to detect Tlr9 transcript (red) and its colocalization with DESMIN (green) to identify cardiac myocytes. Arrowheads point to TLR9 transcripts in cardiac myocytes. Representative section from Traf2 fl/fl Tlr9-null mouse is shown as a control.
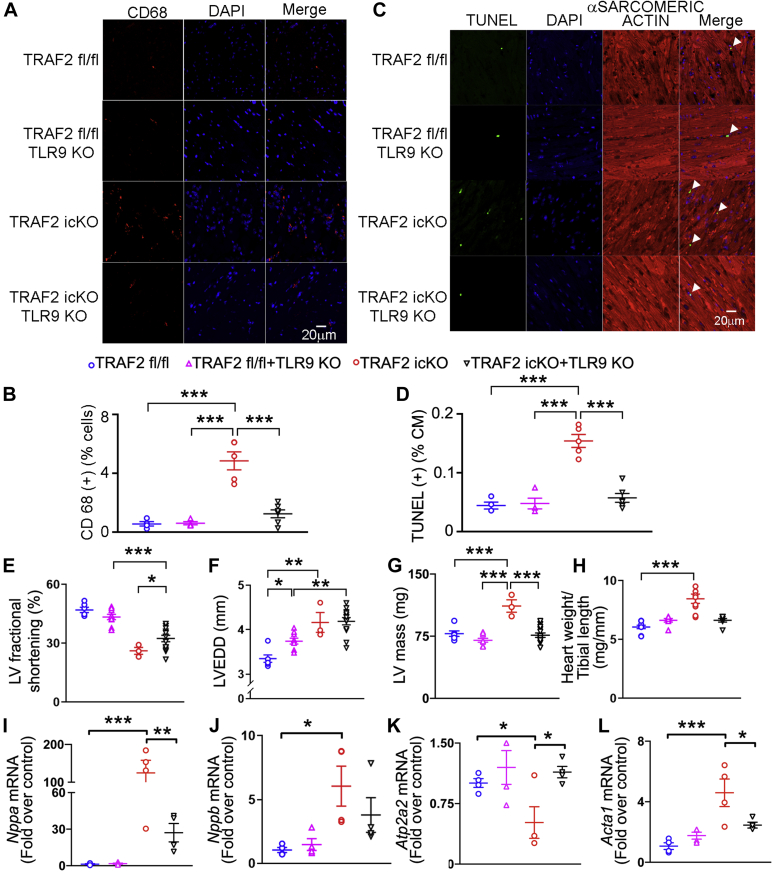
To determine whether increased TLR9 activation led to increased inflammation, we assessed the prevalence of CD68+ macrophages. CD68+ cells were increased in TRAF2-icKO myocardium (Figures 5A and 5B), indicating increased inflammatory cell infiltrate. Concomitant ablation of TLR9 in TRAF2-icKO mice (Supplemental Figures S9A to S9C) markedly reduced cardiac CD68+ cells (Figures 5A and 5B) and TUNEL+ cardiomyocytes (Figure 5C and 5D) to levels observed in control mice. These data indicate that loss of TRAF2 sensitizes the myocardium to proinflammatory signaling driven by TLR9 sensing of leaked mitochondrial DNA.
Figure 5.
TLR9 Ablation Rescues Inflammatory Cell Infiltration and Cell Death in Mice With Inducible TRAF2 Ablation in Adult Cardiac Myocytes, in the Short Term
(A) Representative images depicting CD68+ cells in myocardial section from mice with adult-onset inducible TRAF2 ablation (TRAF2-icKO) and Traf2 floxed control mice, without and with concomitant germline TLR9 ablation, 2 weeks after TRAF2 ablation. Nuclei are stained blue (DAPI). (B) Quantitation of CD68+ cells is shown as % of all DAPI-stained nuclei in mice as in A. P values are by post hoc test after 1-way ANOVA. (C and D) Representative TUNEL-stained images (C) with quantitative assessment (D) of cardiac myocyte TUNEL positivity (green; %TUNEL-positive nuclei/total cardiac myocyte [CM] nuclei) in mice as in A. DAPI staining identifies nuclei. Staining for α-sarcomeric actin was performed to evaluate TUNEL staining in cardiac myocyte nuclei. P values are by post hoc test after 1-way ANOVA. (E to H) Left ventricular endocardial fractional shortening (%FS, E), end-diastolic diameter (LVEDD, in millimeters, F), LV mass (in milligrams, G), and heart weight normalized to tibial length (in milligrams per millimeter, H) in mice modeled as in A. P values are by post hoc test after 1-way ANOVA. (I-L) Expression of Nppa(I), Nppb(J), Atp2a2(K), and Acta1(L) transcripts in mice modeled as in A. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 by post hoc test after 1-way ANOVA for all panels except by Kruskal-Wallis test for H and J. Abbreviations as in Figure 2.
Concomitant TLR9 ablation also attenuated the decline in systolic function (Figure 5E) and left ventricular hypertrophy (Figures 5G and 5H) without significantly affecting left ventricular dimensions (Figure 5F) in TRAF2-icKO mice. This was accompanied by reduced myocardial fibrosis (Supplemental Figures S9D-S9F). Assessment of the fetal gene expression program as a marker for pathologic hypertrophy showed reductions in Nppa (encoding for ANP), Nppb (encoding for BNP), Acta1 (encoding for skeletal α-actin), and restoration of Atp2a2 (encoding for SERCA2a) transcripts (Figures 5I to 5L), without an effect on Myh6 and Myh7 expression (Supplemental Figures S9G and S9H) in TRAF2-icKO mice with concomitant TLR9 expression versus those that were wild type at the Tlr9 locus. Taken together, these data suggest that TLR9-mediated inflammatory cell infiltration in the myocardium drives cell death and myocardial pathology in TRAF2-icKO mice without a contribution from systemic inflammation driven by necroptotic cell death that was observed with perinatal TRAF2 ablation.33
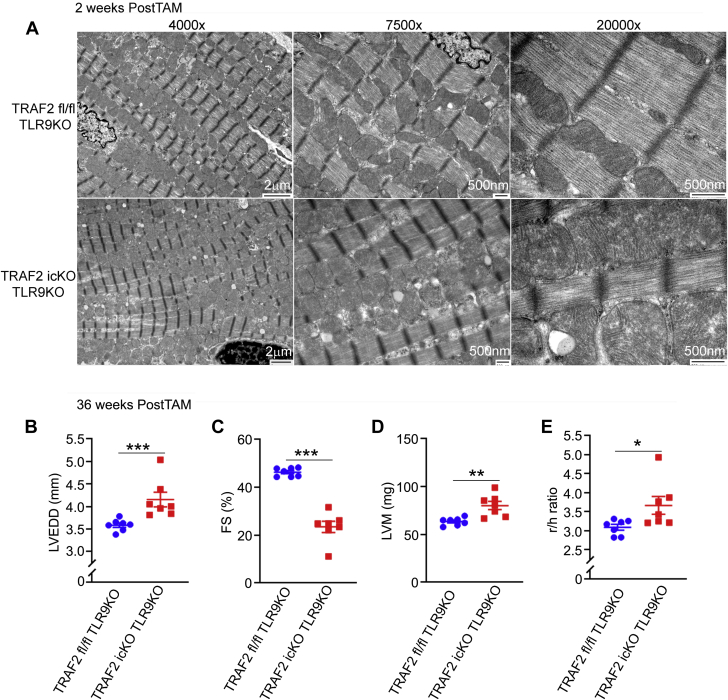
Evaluation of mitochondrial ultrastructure revealed that concomitant ablation of TLR9 in TRAF2-icKO mice did not rescue the mitochondrial defects observed with TRAF2 ablation in cardiac myocytes (Figures 2E and 6A). Damaged mitochondria trigger cell death signaling,1 which prompted us to follow these animals over a longer time period to determine whether impaired mitophagy would result in cardiomyopathy independent of TLR9-mediated inflammation. Re-examination of these animals at 36 weeks after tamoxifen treatment (to induce TRAF2 ablation) demonstrated LV dilation (Figure 6B), LV systolic dysfunction (Figure 6C), increased LV mass (Figure 6D), and an increased ratio of cavity size to wall thickness (Figure 6E) in TRAF2-icKO-TLR9–null mice (as compared with control mice), indicating that persistence of damaged mitochondria is sufficient to induce cardiomyopathy independent of sustained inflammatory signaling.
Figure 6.
TLR9 Ablation Does Not Rescue Mitochondrial Abnormalities and Does Not Prevent Cardiomyopathy With Inducible Cardiac Myocyte TRAF2 Ablation With Longer-Term Follow-Up
(A) Representative transmission electron microscopy images to evaluate myocardial ultrastructure in mice as with adult-onset inducible TRAF2 ablation (TRAF2-icKO) and Traf2 floxed control mice, without and with concomitant germline TLR9 ablation, 2 weeks after TRAF2 ablation. (B to E) Mice with adult-onset inducible TRAF2 ablation or Traf2 floxed alleles in TLR9-null (knockout [KO]) background (TRAF2-icKO TLR9KO and TRAF2 fl/fl TLR9KO) were followed by echocardiographic evaluation at 36 weeks after tamoxifen (TAM) treatment to induce TRAF2 ablation. Left ventricular end-diastolic diameter (LVEDD, mm (B), endocardial fractional shortening (%FS) (C), mass (LVM, in milligrams (D), and ratio of the chamber diameter to wall thickness (r/h) (E) was assessed as shown. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 by t-test.
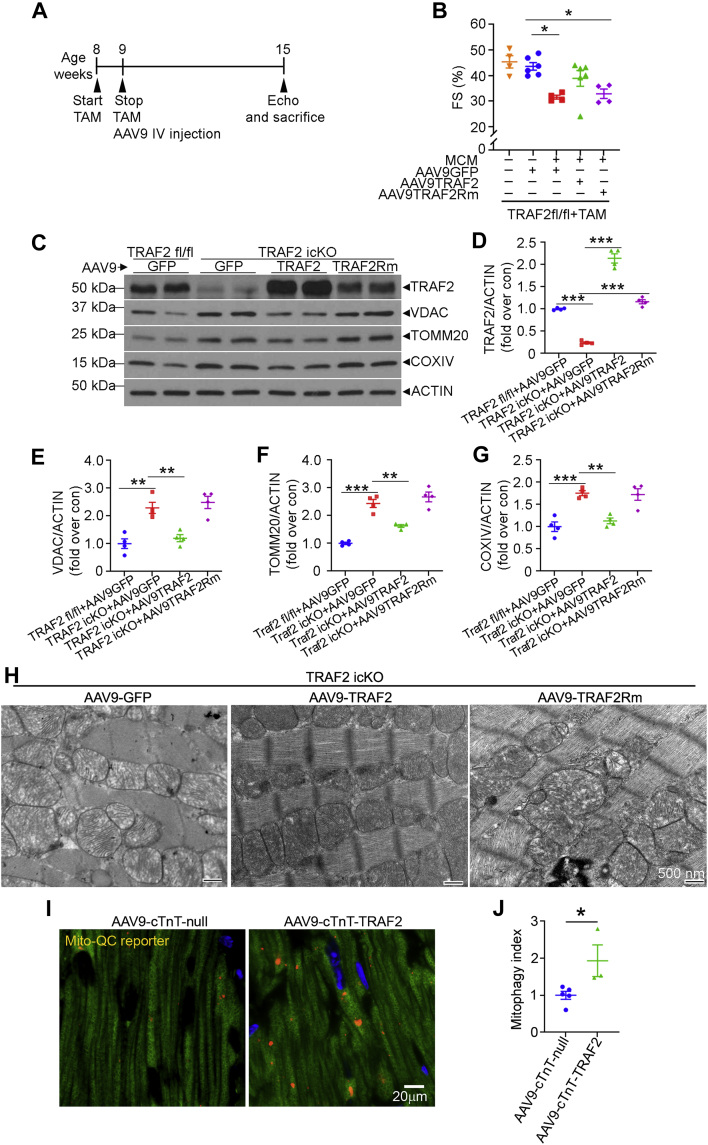
AAV9-mediated restoration of TRAF2, but not its E3 ligase–deficient mutant, rescues cardiomyopathy with cardiomyocyte TRAF2 ablation
To gain a mechanistic understanding of whether TRAF2’s E3 ligase function was critical, as we have observed previously in cardiomyocytes in cell culture,26 we used an AAV9-based approach to restore TRAF2 expression in TRAF2-icKO myocardium. AAV9-mediated expression of TRAF2 targeted to cardiomyocytes with a cTnT promoter restored TRAF2 protein to levels ∼2-fold higher than in Traf2 fl/fl mice treated with a control (AAV9-cTnT-GFP) viral particle (Figures 7C and 7D). AAV9-mediated expression of TRAF2 in TRAF2-icKO mice rescued the decline in systolic function (Figures 7A and 7B) and restored mitochondrial protein levels to those observed in control mice (Figures 7C and 7E to 7G). Mitochondrial morphology was also normalized in TRAF2-icKO mice with AAV9-mediated TRAF2 delivery (Figure 7H). In contrast, AAV9-mediated restoration of TRAF2 with ring domain mutations ablating its E3 ligase activity (ie, TRAF2-Rm mutant, as previously described26,40) (Figure 7C) did not rescue the systolic dysfunction (Figures 7A and 7B), restore abundance of mitochondrial proteins (Figures 7C and 7E to 7G), or rescue the mitochondrial defects (Figure 7H). These data demonstrate that the E3 ligase domain of TRAF2 is required for myocardial homeostasis and mitochondrial quality control, mirroring prior observations in isolated cultured primary cardiomyocyte.26
Figure 7.
Restoration of TRAF2, but Not Its E3 Ligase–Deficient Mutant, Rescues Cardiomyopathy and Mitochondrial Abnormalities With Inducible Cardiac Myocyte TRAF2 Ablation
(A) Schematic depicting experimental strategy for tamoxifen-inducible TRAF2 ablation in mice homozygous for floxed Traf2 alleles carrying the Myh6-MerCreMer transgene, followed by AAV9-cTnT-TRAF2 or TRAF2Rm particles at 5.0 × 1011 viral particles per mouse. (B) Left ventricular endocardial fractional shortening (%FS) in TRAF2icKO mice or TRAF2 floxed mice transduced with AAV9 particles coding for TRAF2 or TRAF2Rm, driven by a cardiac troponin T promoter as shown in A. ∗P < 0.05 by post hoc test after 1-way ANOVA. (C-G) Representative immunoblot (C) with quantitation of TRAF2 expression (D) and abundance of mitochondrial proteins (VDAC, E; TOMM20, F; COXIV, G); all normalized to actin) in cardiac extracts from mice modeled as in A. ∗∗P < 0.01 and ∗∗∗P < 0.001 by post hoc test after 1-way ANOVA. (H) Representative transmission electron microscopy images to evaluate myocardial ultrastructure in mice as in A. Scale bar = 500 nm. (I and J) Representative images (I) are shown demonstrating mitophagy with mitoQC reporter expression with quantitative assessment (J), 4 weeks after injections of AAV9-cTnT-TRAF2 or empty viral particles (AAV9-cTnT-null) in 8-week-old mitoQC reporter mice. ∗P < 0.05 by t-test. Abbreviations as in Figure 2.
We next examined whether TRAF2 overexpression was sufficient to induce mitophagy in cardiac myocytes in the mouse heart. To test this, we employed AAV9-cTnT promoter-driven TRAF2 expression (Figures 7C and 7D) and compared that with expression of empty AAV9-cTnT null viral particles (as control) in hearts from mice expressing the mitoQC mitophagy reporter.17 Our data demonstrate that exogenous TRAF2 is sufficient to induce mitophagy versus the control condition (Figures 7I and 7J).
Parkin expression is not redundant to TRAF2 function; but exogenous Parkin-induced mitophagy can rescue cell death with TRAF2 ablation
In isolated cardiac myocytes, we have previously observed that TRAF2 colocalizes and interacts with PARKIN in the setting of ionophore-induced mitochondrial damage.26 We examined PARKIN levels in TRAF2-icKO mice and did not note them to be significantly altered (Figure 3A). To determine whether endogenous Parkin was playing role in the cardiomyopathy observed with TRAF2 ablation, we generated TRAF2-icKO mice in the Park2 null (Parkin deficient) background. Loss of PARKIN did not worsen LV dysfunction or dilation, or affect histology or fibrosis in TRAF2-icKO mice (Supplemental Figure S10). These data are consistent with a lack of role for Parkin in basal mitophagy in cardiomyocytes in young adult mouse hearts.8,21
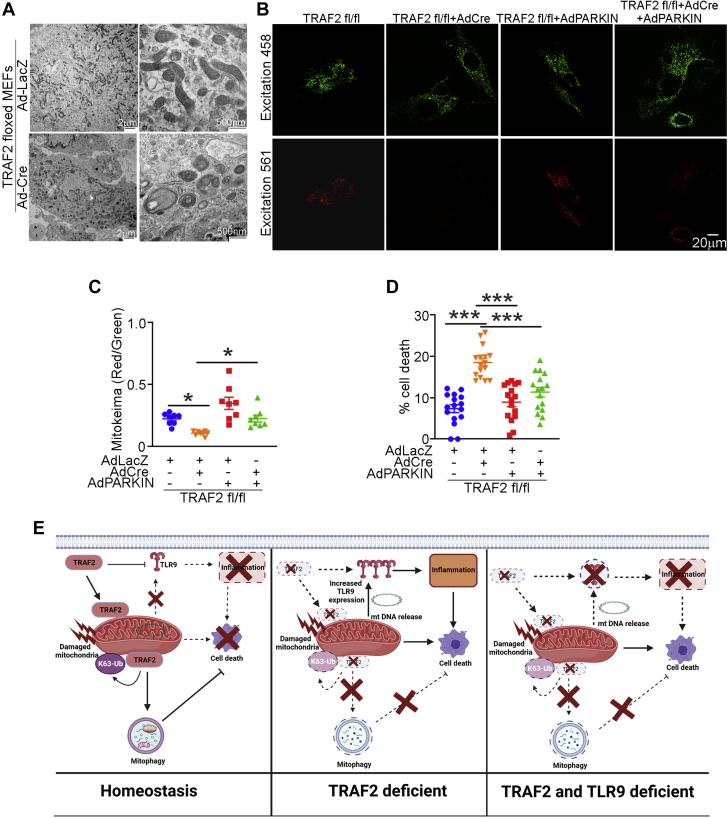
To determine whether the defective mitophagy induced by TRAF2 deficiency was responsible for increased cell death, we studied Traf2 floxed MEFs (Traf2 fl/fl MEFs), where TRAF2 was deleted with adenoviral-Cre expression (Supplemental Figure S11). Loss of TRAF2 in MEFs resulted in accumulation of damaged mitochondria by ultrastructural examination (Figure 8A) with impaired mitophagy assessed with mKeima expression (Figures 8B and 8C), and cell death (Figure 8D). Importantly, ablation of TRAF2 did not alter expression of LC3 or p62 two markers of macroautophagy (Supplemental Figure S11B) mirroring the findings in TRAF2-icKO mice (Figures 3E to 3G) and confirming a specific role for TRAF2 in mitophagy. Adenoviral overexpression of Parkin, as we have previously described,26 rescued mitophagy to control levels and attenuated cell death with TRAF2 ablation (Figures 8B to 8D). Taken together, these data indicate that TRAF2 mediates mitophagy independent of Parkin, and the mitophagy defect is the proximate cause of cell death in the absence of TRAF2.
Figure 8.
TRAF2 Ablation Induced Cell Death, Which Can Be Rescued by Parkin-Induced Mitophagy
(A) Murine embryonic fibroblasts (MEFs) carrying floxed Traf2 alleles were modeled for TRAF2 ablation with adenoviral Cre (or LacZ as control, each at a multiplicity of infection [MOI] = 100) treatment for 72 hours as in Figure S11 and subjected to transmission electron microscopic (TEM) analysis. Representative TEM images are shown demonstrating mitochondrial abnormalities noted after Traf2 ablation as compared with control mice. (B and C) Representative images (B) and quantitative analyses of mitoKeima emission in the red channel over green as an index of mitophagy (C), in Traf2-null MEFs (modeled as in A) expressing mitoKeima transduced with adenoviral Parkin or LacZ (MOI = 100 for 48 hours) as control. Adenoviral particles coding for LacZ were added to equalize the number of viral particles per treatment. ∗P < 0.05 by post hoc test after 1-way ANOVA. (D) Cell death in Traf2 null MEFs (modeled as in A) and transduced with adenoviral Parkin or LacZ (MOI = 100 for 72 hours) as control. Adenoviral particles coding for LacZ were added to equalize the number of viral particles per treatment. ∗∗∗P < 0.001 by post hoc test after 1-way ANOVA. (E) Schematic depicting role of TRAF2-induced mitophagy in preventing inflammation and cell death in cardiac myocyte homeostasis. TRAF2 localizes to the mitochondria and promotes mitophagy via K63-ubiquitination activity in unstressed cardiac myocytes (left). Ablation of TRAF2 results in mitochondrial DNA leak, and up-regulates TLR9 abundance to provoke inflammation via mitochondria DNA sensing. The resultant inflammation and accumulation of damaged mitochondria provoke cardiac myocyte cell death and cardiomyopathy (middle). Concomitant ablation of TLR9 in the setting of TRAF2 deficiency prevents inflammation and cell death, and rescues left ventricular hypertrophy and function in the short term; but does not prevent accumulation of damaged mitochondria and cell death inexorably resulting in cardiomyopathy during follow up (right).
Discussion
Purported as ancient symbionts, mitochondria act as arbiters of cellular viability and death.1 In this work, we have identified a physiological role for mitophagy to remove damaged mitochondria in cardiomyocytes and distinguished the effects of inflammation from those due to persistent cell death signaling from damaged mitochondria in the development of cardiomyopathy. Our studies reveal a novel regulatory pathway that links 2 key elements of the innate immune system, namely TRAF2 and TLR9 to balance physiological mitophagy and suppress inflammatory signaling for maintaining myocardial homeostasis in the unstressed heart (Figure 8E).
These findings indicate that despite a vital role for mitochondria in energy generation in cardiac myocytes, rapid removal of damaged mitochondria via mitophagy is essential to maintain physiological functioning of the heart. TRAF2-mediated physiological mitophagy is also a critical mechanism to prevent sterile myocardial inflammation, provoked by sensing of mitochondrial DNA leaked from damaged mitochondria. This is particularly relevant because TLR9 signaling has been shown to drive inflammatory signaling in the setting of mitochondrial DNA release, when mitochondria are increasingly damaged, such as with pressure overload–induced stress or with ischemia-reperfusion injury.2,4 Degradation of cytokine mRNAs by Regnase-1 is another mechanism to prevent sterile inflammation in the myocardium in the setting of pressure overload hypertrophy.41 Curiously, although lysosomal DNase2a and Regnase-1 have been shown to be critical for protecting against myocardial inflammation under stress, mice deficient in these genes in cardiac myocytes do not have detectable myocardial inflammation, or cardiac structure or function abnormalities in the unstressed state.2,41 These observations mimic the findings in the myocardium of unstressed adult Pink1- and Park2-null mice7 and strongly support the contention that robust and efficient physiological mitophagy removes damaged mitochondria, to prevent mitochondrial DNA sensing and sterile inflammation, and maintain homeostasis. Our studies point to TRAF2 as the sensor that resides in the mitochondria to effect homeostatic mitophagy upon mitochondrial damage to prevent cardiac myocyte cell death. Indeed, TRAF2-mediated mitophagy prevents mitochondrial DNA release and DNA sensing to drive inflammation, as demonstrated with attenuation of myocardial CD68+ macrophage abundance to levels seen in the unstressed state in TRAF2-icKO mice lacking TLR9. It is important to note that whereas TLR9 ablation rescued cardiac myocyte cell death in the short term by preventing mitochondrial DNA sensing and inflammation, the persistence of damaged mitochondria in the absence of TRAF2-induced mitophagy led to cardiomyopathy over a longer-term follow-up. These data underscore the critical importance for removal of damaged mitochondria to maintain cellular homeostasis.
TRAF2, an adaptor protein, has been ascribed critical roles in innate immunity and inflammatory signaling.42 Our studies demonstrate a novel role for TRAF2 as the first mediator to be uncovered for transducing basal cardiac myocyte mitophagy in young adult mice, distinct from previously described roles for other pathways in stress-induced mitophagy.9, 10, 11, 12, 13, 14, 15 Similarly, TRAF2-mediated physiological mitophagy in young adult myocardium differs from mitophagy during perinatal cardiac growth and development8 or mitophagy induced by exercise, a form of physiological stress.7 Indeed, it is important to note the difference between basal and stress-induced mitophagy. In stress-induced mitophagy, prior studies describe a critical role for PINK1, a mitochondrially targeted serine-threonine kinase, which gets stabilized upon mitochondrial damage and recruits PARKIN,43 an E3 ubiquitin ligase, that ubiquitinates mitochondrial proteins and targets the damaged mitochondria to autophagosomes.44 Our data indicate that TRAF2 localizes to mitochondria independently of PINK1 in the unstressed state (Supplemental Figure S2) and mediates mitophagy independently of endogenous Parkin (Supplemental Figure S10). Moreover, exogenous TRAF2 is sufficient to induce mitophagy in cardiac myocytes (Figures 7I and 7J). Indeed, TRAF2 was uncovered in a screen for PINK1 and PARKIN-independent mitophagy mediators,45 and inflammatory stimuli were observed to induce TRAF2-mediated mitophagy via ubiquitination of orphan nuclear receptor, Nur77,46 independently confirming our prior observations that TRAF2 mediates mitophagy in cardiac myocytes in culture26 and in unstressed hearts, in vivo (current dataset). Consistent with a role for PINK1-mediated mitophagy under stress, we found that PINK1 was essential for recruitment of TRAF2 to mitochondrial fraction upon hypoxia-reoxygenation injury, but did not affect TRAF2’s localization to the mitochondria under unstressed normoxic conditions (Supplemental Figure S2).
Notably, our studies demonstrate induction of cellular inflammation with up-regulated macrophage abundance in the myocardium of mice with inducible adult-onset TRAF2 ablation, without evidence for systemic inflammation. These findings are in contrast to prior studies with perinatal ablation of TRAF2 in cardiac myocytes which demonstrate induction of systemic inflammation with increased circulating TNF levels (along with other cytokines) that transduces TNF receptor 1 (TNFR1)-mediated activation of cardiac myocyte necroptosis.33 Similar observations were made in germline TRAF2-deficient mice47 that demonstrate systemic inflammation, markedly elevated TNF-induced cell death, and early lethality. On the contrary, ablation of TRAF2 in myeloid cells,48 hepatocytes,49 or B cells23 did not induce systemic inflammation, cell death, or mortality in unstressed mice. Taken together, these observations argue for developmental and/or cell-type specific effects of TRAF2 signaling in preventing necroptosis that are not observed in adult cardiac myocytes.
Our findings also confirm an important role for TRAF2’s E3 ubiquitin ligase signaling function in facilitating mitophagy. TRAF2 facilitates K63 ubiquitination50 and K63 poly-ubiquitination has been shown to selectively target proteins for lysosomal degradation.51 We propose that the subcellular localization of TRAF2 mediates differential signaling through its E3 ligase activity. The fraction of TRAF2 that resides in the mitochondria acts as a sensor to effect physiological mitophagy upon mitochondrial damage to prevent mitochondrial DNA leak and cardiac myocyte cell death.
Conclusions
In summary, the data presented herein identify physiological cardiac myocyte mitophagy mediated by TRAF2 as an essential homeostatic phenomenon in the adult myocardium. In doing so, TRAF2 minimizes deleterious intracellular inflammatory signaling, both by preventing the accumulation of depolarized damaged mitochondria, as well as mitigating the impact of released mtDNA by suppressing the TLR9 levels to attenuate mtDNA sensing. A growing body of published reports supports a cardioprotective role for enhanced TRAF2 signaling in the setting of injury,28,34,35 and TRAF2-stimulated mitophagy is likely to play an important role in transducing the observed benefits. Indeed, augmenting TRAF2 signaling to enhance mitochondrial quality control should be explored as a novel therapeutic strategy in cardiomyopathy.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Understanding the mediators of mitochondrial quality control in mitochondria-rich cardiac myocytes and their relevance to myocardial homeostasis is critical for targeting these pathways for clinical benefit. Our studies have uncovered the innate immune system, specifically the TRAF2 protein, as a first responder to mitochondrial damage and an effector of mitophagy, a lysosomal degradative process that removes damaged mitochondria. By facilitating mitophagy in cardiac myocytes, the innate immune response suppresses myocardial inflammation provoked by sensing of leaked mitochondrial DNA and prevents cardiac myocyte cell death triggered by activation of cell death signaling from persistence of damaged mitochondria.
TRANSLATIONAL OUTLOOK: We demonstrate that TRAF2-mediated mitophagy prevents myocardial inflammatory cell infiltration and cardiac myocyte death, which are recognized pathophysiological mechanisms for development of cardiomyopathy and its progression to heart failure. Taken together with prior preclinical reports demonstrating cardioprotective effects of modest activation of TRAF2 signaling under stress, these studies support a translational strategy for enhancing TRAF2-mediated mitophagy to suppress inflammation and prevent cell death to treat heart failure.
Funding Support and Author Disclosures
This study was supported by National Institutes of Health (NIH) grant HL107594 and by the Hope Center Viral Vectors Core at Washington University School of Medicine. Experiments were performed in part through the use of Washington University Center for Cellular Imaging supported by Washington University School of Medicine, The Children’s Discovery Institute of Washington University, and St. Louis Children’s Hospital grants CDI-CORE-2015-505 and CDI-CORE-2019-813; and the Foundation for Barnes-Jewish Hospital grants 3770 and 4642. Dr Rawnsley is supported by NIH grant T32 HL007081. Dr Javaheri is supported by NIH grants K08HL138262 and 1R01HL155344; by the Children's Discovery Institute of Washington University (MC-FR-2020-919) and St. Louis Children's Hospital, as well as the Diabetes Research Center grant P30DK020579; and the Nutrition Obesity Research Center at Washington University grant P30DK056341. Dr Mani was supported by a Seed Grant from the St. Louis VA Medical Center and by a Pilot and Feasibility grant from the Diabetes Research Center at Washington University (NIDDK grant No. P30 DK020579). Dr Abhinav Diwan is supported by NIH grants HL143431 and NS094692, and the Department of Veterans Affairs grant I01BX004235. Dr Mani serves as a member of the Cardiovascular Scientific Advisory Board at Dewpoint Therapeutics. Dr Abhinav Diwan provides consulting services to ERT systems for interpretation of echocardiograms in clinical trials; and serves as a member of the Cardiovascular Scientific Advisory Board at Dewpoint Therapeutics. These interests are not related to and did not influence the current study. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors are grateful to Jeanne M. Nerbonne, PhD, Cardiovascular Division and Center for Cardiovascular Research at Washington University School of Medicine for assistance with adult cardiac myocyte isolation studies. The authors are also grateful for Wandy Beatty, PhD, Department of Molecular Biology at Washington University School of Medicine for assistance with electron microscopy studies, and they wish to thank Joan Avery and Katie Kyle at the Center for Cardiovascular Research at Washington University School of Medicine, for their administrative assistance with conducting the study. Graphical abstract was created with BioRender.com.
Footnotes
Junichi Sadoshima, MD, served as Guest Associate Editor for this paper. Michael Bristow, MD, served as Guest Editor-in-Chief for this paper.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental tables and figures, please see the online version of this paper.
Appendix
References
- 1.Del Re D.P., Amgalan D., Linkermann A., Liu Q., Kitsis R.N. Fundamental mechanisms of regulated cell death and implications for heart disease. Physiol Rev. 2019;99:1765–1817. doi: 10.1152/physrev.00022.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oka T., Hikoso S., Yamaguchi O., et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diwan A., Wansapura J., Syed F.M., Matkovich S.J., Lorenz J.N., Dorn G.W. Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008;117:396–404. doi: 10.1161/CIRCULATIONAHA.107.727073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitazume-Taneike R., Taneike M., Omiya S., et al. Ablation of Toll-like receptor 9 attenuates myocardial ischemia/reperfusion injury in mice. Biochem Biophys Res Comm. 2019;515:442–447. doi: 10.1016/j.bbrc.2019.05.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diwan A., Krenz M., Syed F.M., et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest. 2007;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adamo L., Rocha-Resende C., Prabhu S.D., Mann D.L. Reappraising the role of inflammation in heart failure. Nat Rev Cardiol. 2020;17:269–285. doi: 10.1038/s41569-019-0315-x. [DOI] [PubMed] [Google Scholar]
- 7.Sliter D.A., Martinez J., Hao L., et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561:258–262. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Gong G., Song M., Csordas G., Kelly D.P., Matkovich S.J., Dorn G.W., 2nd Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoshino A., Wang W.J., Wada S., et al. The ADP/ATP translocase drives mitophagy independent of nucleotide exchange. Nature. 2019;575:375–379. doi: 10.1038/s41586-019-1667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubli D.A., Quinsay M.N., Gustafsson A.B. Parkin deficiency results in accumulation of abnormal mitochondria in aging myocytes. Commun Integr Biol. 2013;6 doi: 10.4161/cib.24511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshino A., Mita Y., Okawa Y., et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- 12.Kubli D.A., Zhang X., Lee Y., et al. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong M., Saito T., Zhai P., et al. Mitophagy is essential for maintaining cardiac function during high fat diet-induced diabetic cardiomyopathy. Circ Res. 2019;124:1360–1371. doi: 10.1161/CIRCRESAHA.118.314607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirakabe A., Zhai P., Ikeda Y., et al. Drp1-dependent mitochondrial autophagy plays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure. Circulation. 2016;133:1249–1263. doi: 10.1161/CIRCULATIONAHA.115.020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song M., Gong G., Burelle Y., et al. Interdependence of parkin-mediated mitophagy and mitochondrial fission in adult mouse hearts. Circ Res. 2015;117(4):346–351. doi: 10.1161/CIRCRESAHA.117.306859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun N., Yun J., Liu J., et al. Measuring in vivo mitophagy. Mol Cell. 2015;60:685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McWilliams T.G., Prescott A.R., Allen G.F.G., et al. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. 2016;214:333–345. doi: 10.1083/jcb.201603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McWilliams T.G., Prescott A.R., Montava-Garriga L., et al. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 2018;27:439–449.e5. doi: 10.1016/j.cmet.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodall B.P., Orogo A.M., Najor R.H., et al. Parkin does not prevent accelerated cardiac aging in mitochondrial DNA mutator mice. JCI Insight. 2019;5(10) doi: 10.1172/jci.insight.127713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubli D.A., Cortez M.Q., Moyzis A.G., Najor R.H., Lee Y., Gustafsson A.B. PINK1 is dispensable for mitochondrial recruitment of parkin and activation of mitophagy in cardiac myocytes. PloS One. 2015;10 doi: 10.1371/journal.pone.0130707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng M.Y.W., Wai T., Simonsen A. Quality control of the mitochondrion. Dev Cell. 2021;56:881–905. doi: 10.1016/j.devcel.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Grech A.P., Amesbury M., Chan T., Gardam S., Basten A., Brink R. TRAF2 differentially regulates the canonical and noncanonical pathways of NF-kappaB activation in mature B cells. Immunity. 2004;21:629–642. doi: 10.1016/j.immuni.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Sohal D.S., Nghiem M., Crackower M.A., et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 25.Ma X., Mani K., Liu H., et al. Transcription factor EB activation rescues advanced alphaB-crystallin mutation-induced cardiomyopathy by normalizing desmin localization. J Am Heart Assoc. 2019;8 doi: 10.1161/JAHA.118.010866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang K.C., Ma X., Liu H., et al. Tumor necrosis factor receptor-associated factor 2 mediates mitochondrial autophagy. Circ Heart Fail. 2015;8:175–187. doi: 10.1161/CIRCHEARTFAILURE.114.001635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prasad K.M., Xu Y., Yang Z., Acton S.T., French B.A. Robust cardiomyocyte-specific gene expression following systemic injection of AAV: in vivo gene delivery follows a Poisson distribution. Gene Ther. 2011;18(1):43–52. doi: 10.1038/gt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans S., Tzeng H.P., Veis D.J., et al. TNF receptor-activated factor 2 mediates cardiac protection through noncanonical NF-κB signaling. JCI Insight. 2018;3(3) doi: 10.1172/jci.insight.98278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunet S., Aimond F., Li H., et al. Heterogeneous expression of repolarizing, voltage-gated K+ currents in adult mouse ventricles. J Physiol. 2004;559:103–120. doi: 10.1113/jphysiol.2004.063347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wieckowski M.R., Giorgi C., Lebiedzinska M., Duszynski J., Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nature Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 31.Javaheri A., Bajpai G., Picataggi A., et al. TFEB activation in macrophages attenuates postmyocardial infarction ventricular dysfunction independently of ATG5-mediated autophagy. JCI Insight. 2019;4(21) doi: 10.1172/jci.insight.127312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirakabe A., Fritzky L., Saito T., et al. Evaluating mitochondrial autophagy in the mouse heart. J Mol Cell Cardiol. 2016;92:134–139. doi: 10.1016/j.yjmcc.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Guo X., Yin H., Li L., et al. Cardioprotective role of tumor necrosis factor receptor-associated factor 2 by suppressing apoptosis and necroptosis. Circulation. 2017;136:729–742. doi: 10.1161/CIRCULATIONAHA.116.026240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burchfield J.S., Dong J.W., Sakata Y., et al. The cytoprotective effects of tumor necrosis factor are conveyed through tumor necrosis factor receptor-associated factor 2 in the heart. Circ Heart Fail. 2010;3:157–164. doi: 10.1161/CIRCHEARTFAILURE.109.899732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzeng H.P., Evans S., Gao F., et al. Dysferlin mediates the cytoprotective effects of TRAF2 following myocardial ischemia reperfusion injury. J Am Heart Assoc. 2014;3 doi: 10.1161/JAHA.113.000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazarou M., Sliter D.A., Kane L.A., et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koitabashi N., Bedja D., Zaiman A.L., et al. Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circ Res. 2009;105:12–15. doi: 10.1161/CIRCRESAHA.109.198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lexow J., Poggioli T., Sarathchandra P., Santini M.P., Rosenthal N. Cardiac fibrosis in mice expressing an inducible myocardial-specific Cre driver. Dis Mod Mech. 2013;6(6):1470–1476. doi: 10.1242/dmm.010470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schugar R.C., Moll A.R., Andre d'Avignon D., Weinheimer C.J., Kovacs A., Crawford P.A. Cardiomyocyte-specific deficiency of ketone body metabolism promotes accelerated pathological remodeling. Mol Metab. 2014;3:754–769. doi: 10.1016/j.molmet.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Habelhah H., Takahashi S., Cho S.G., Kadoya T., Watanabe T., Ronai Z. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. EMBO J. 2004;23:322–332. doi: 10.1038/sj.emboj.7600044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Omiya S., Omori Y., Taneike M., et al. Cytokine mRNA degradation in cardiomyocytes restrains sterile inflammation in pressure-overloaded hearts. Circulation. 2020;141:667–677. doi: 10.1161/CIRCULATIONAHA.119.044582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X.D., Sun S.C. Targeting signaling factors for degradation, an emerging mechanism for TRAF functions. Immunol Rev. 2015;266:56–71. doi: 10.1111/imr.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narendra D.P., Jin S.M., Tanaka A., et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moyzis A.G., Sadoshima J., Gustafsson A.B. Mending a broken heart: the role of mitophagy in cardioprotection. Am J Physiol Heart Circ Physiol. 2015;308(3):H183–H192. doi: 10.1152/ajpheart.00708.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zachari M., Gudmundsson S.R., Li Z., et al. Selective autophagy of mitochondria on a ubiquitin-endoplasmic-reticulum platform. Dev Cell. 2020;55:251. doi: 10.1016/j.devcel.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu M., Luo Q., Alitongbieke G., et al. Celastrol-induced Nur77 interaction with TRAF2 alleviates inflammation by promoting mitochondrial ubiquitination and autophagy. Mol Cell. 2017;66:141–153.e6. doi: 10.1016/j.molcel.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeh W.C., Shahinian A., Speiser D., et al. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 48.Jin J., Xiao Y., Hu H., et al. Proinflammatory TLR signalling is regulated by a TRAF2-dependent proteolysis mechanism in macrophages. Nat Commun. 2015;6:5930. doi: 10.1038/ncomms6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z., Sheng L., Shen H., et al. Hepatic TRAF2 regulates glucose metabolism through enhancing glucagon responses. Diabetes. 2012;61:566–573. doi: 10.2337/db11-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarez S.E., Harikumar K.B., Hait N.C., et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nathan J.A., Kim H.T., Ting L., Gygi S.P., Goldberg A.L. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013;32:552–565. doi: 10.1038/emboj.2012.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.