Abstract
The B.1.617.2 Delta variant is considered to be the most infectious of all SARS-CoV2 variants. Here, an attempt has been made through in-silico screening of 55 bioactive compounds from two selected plants, Saussurea costus and Saussurea involucrata as potential inhibitors of two viral proteases, main protease Mpro (PDB ID:6LU7) and the RBD of SGP of Sars-CoV-2 B1.617.2 Delta variant (PDB ID:7ORB) where the binding energy, molecular interactions, ADMET/Tox, chemical descriptors and Quantum-Chemical Calculations were explored. Molecular docking results demonstrated that the three top docked compounds formed relatively stable complexes within the active site and displayed remarkable binding energy in the order of Tangshenoside III, Rutin and Hesperidin (-9.35, -9.14 and -8.57 kcal/mol, respectively) with Mpro and Rutin, Tangshenoside III and Hesperidin (-9.07, -7.71 and -7.57 kcal/mol) with RBD of SGP. These compounds are non-Mutagen and non-carcinogen. Therefore, according to the Lipinski's Rule of Five they exhibited three violations concerning hydrogen acceptor, donor and molecular weight. However, based on the Quantum-Chemical Calculations results the selected ligands have effective reactivity, as they showed lower band gaps. The difference of the ELUMO and EHOMO was low, ranging from 0.0639 to 0.0978 a.u, implying the strong affinity of these inhibitors towards the target proteins. Among the three inhibitors, Rutin exhibited higher reactivity against two viral proteases, main protease (Mpro) and the Sars-CoV-2 B1.617.2, as the band energy gap was lowest among all the three phytochemicals, 0.0639 a.u This could indicate that Rutincan be potential anti-viral drug candidates against the existing SARS-CoV-2, the B.1.617.2 Delta variant.
Keywords: SARS-CoV-2 Delta, Mpro, Spike, Saussurea costus, Saussurea involucrate Molecular docking
1. Introduction
At the end of year 2019 a novel Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) with human-to-human transmission, was identified [1]. COVID-19 outbreak was first identified in the city of Wuhan, China in December 2019 as an epidemic; afterward it quickly propagated in Chinese territory and outside. World Health Organization declared it, two months later, as a pandemic on March 11, 2020 [2]. SARS-CoV-2 spread rapidly, causing the disease known as COVID-19, has since infected over 198 million people, leading to at least 4.2 million deaths worldwide of August 1st, 2021 [3]. The Alpha Variant of Concern (VOC) (formerly known as the Kent VOC, B.1.1.7, or S gene negative) had been the dominant strain previously, but it has rapidly been replaced on May 19, 2021, by the Delta VOC formerly known as the Indian Delta VOC or B 1.617.2 [4].
The most promising targets so far identified have been the spike protein, RNA-dependent RNA polymerase (RdRp), and the papain-like protease 3CLpro, also known as main protease (Mpro) [5,6]. It is essential for processing the polyprotein that led to the proteolytic activation of the viral functional proteins [7]. Since the main protease (Mpro) is required for viral gene expression and replication, it is a promising target for antagonists to treat novel coronavirus disease.
SARS CoV-2 utilizes the receptor-binding domain (RBD) of trimeric spike glycoprotein (SGP) to gain quick entry into bronchial epithelial cells by binding with a human receptor, Angiotensin-Converting Enzyme-II (ACE-II) on the host cells that located on many body organs [8,9]. It is well reported that the utilization of ACE-II receptor blockers exhibit adverse effect such as hyperkalemia and angioedema [10]. The efficacy of currently licensed COVID-19 vaccines against B.1.617.2 is unknown [4]. Currently, several inhibitors are proposed by researchers across the globe as potential drug targets for the treatment of this disease but they still lack optimum specificity and efficacy; and to date, no highly effective therapy for treating coronavirus infections has been made available, although it possesses 12 mutations in its spike protein relative to the wild type SARS-CoV-2 first detected in Wuhan, China. Thus, blocking of the RBD of SGP, which interacts with amino acid residues of ACE-II could be an attractive target domain for the treatment of the development of new antiviral drugs anti-B.1.617.2 infection and prevention of the risk of multi-organ failure.
It is well known that, over the past decades, medicinal plants have been attracting attention in the search for bioactive compounds to develop new drugs and healthy foods among which polyphenols, lipids, polysaccahrides, sterols, terpenes. Furthermore, various plant extracts were reported to have high antioxidant, anti-cancer activity and to influence anti-inflammatory responses. However, plant based medicines are widely used due to their wide acceptance and therapeutic effects with minimal side effects [11,12]. Hence, at such a time, herbal medicine have shown experimentally promising results in SARS CoV-2 inhibition. Recently, various studies have highlighted the implication of a variety of phytoconstituents which has the potential to be developed as antiviral agents for SARS-CoV-2 [13], [14], [15].
The genus Saussurea DC. of the flowering plant family Asteraceae comprises about 300 species in the world. A family that is rich with well-known medicinal plants of highly economic importance. Many of which have been intensively studied for their phytochemical constituents and medicinal applications, including S. lappa. [16] and S. involucrata. [17]. Traditional clinical applications were considered guide for bioactivity evaluations, and many correlations were found between the pharmacological actions in traditional records of the herb and bioactivities confirmed in modern studies [18]. S. lappa. and S. involucrata are traditionally used to treat a wide spectrum of disorders; its pharmacological evidence is grounded by long history of usage and promising experimental results.
Saussurea costus (Falc.) Lipschitz, a perennial herbaceous plant, one of the best-known species within this genus, is commonly known as costus in English. It has several synonyms, such as Aucklandia costus Falc., Aplotaxis lappa Decne., Saussurea costus (Falc,) Sch.-Bip., Aucklandia lappa Decne, Saussurea lappa (Decne.) C. B. Clarke., Theodorea costus O. Kuntz [19]. Several workers have reported on the different biological activities of Saussurea costus in various in vitro and in vivo test models. Pharmaceutical effects investigations of extracts from Saussurea costus roots have been found to exhibit anti-inflammatory [20], hepatoprotective [21], anti-ulcer [22], anticancer [23], would-healing [24], antifungal and antimicrobial [25], anticonvulsant [26], larvicidal [27] and protein tyrosine phosphatase 1B inhibitory [28] activities.
Another Saussurea species, S. involucrata Karel. & Kir. with large translucent cream-colored bracts concealing the huge inflorescence, are named ‘glasshouse plant’ [29]. According to “The Plant List”, S. involucrata Matsum. & Koidz is the only accepted name for the herb, with asynonym “S. involucrata (Kar. & Kir.) Sch. Bip.” [30].
More recent studies have shown that this plant have anti-inflammatory, analgesic [31]. Immunomodulatory [32], antioxidative [33], antihypoxic and neuroprotective [34], cardiovascular diseases [35], allergies and asthma [36].
The research and the development of a drug molecule “drug discovery” is often an extremely long, tedious and very expensive process at high risk of failure. For each new drug, the time elapsing between the first step, during which a relevant biological target in a given disease process is identified, until a drug is placed on the market is estimated at an average of 12 to 14 years [37]. An alternative approach, such as virtual screening or rational drug design, are now routinely used to guide drug discovery. The use of these in silico techniques could improve the efficiency and save the cost of drug discovery [38].Virtual screening approaches based on the structure of the target, more specifically the molecular docking, predict possible modes of interaction between a ligand and the therapeutic target and provide a way of studying interactions at the molecular level and are, therefore, an indication of the biological activity of new molecules taking into account only structural criteria.
In this context, we conducted computational screening study on a library of 55 isolated molecules from Saussurea costus and Saussurea involucrata, investigated the ADMET/tox studies, the DFT calculations and the binding affinity, of these compounds with the key target RBD of SGP SARS-CoV-2 B1.617.2 and the main protease (Mpro) through molecular docking analysis.
2. Materials and methods
2.1. Molecular modeling platform
Molecular docking and ADMET/Tox studies were performed using Molecular Operating Environment (MOE) version 2019.10 molecular modeling software and Discovery Studio (DS) 2.0 Software (Studio 2.5, Accelrys, Co. Ltd., San Diego, CA, USA), respectively. DFT calculations were performed by Gaussian 09 suite of program at 6–31 G (d,p) basis set.
2.2. Library preparation
A total of 55 compounds were included in the screening (Table 1 ), encoded by S. lappa. and S. involucrate medicinal plants, after a profound literature review. Structure Data Format of these bioactive phytoconstituents were retrieved from the PubChem database (www.pubchem.ncbi.nlm.nih.gov) alongside the reference inhibitors (N3 and Hydroxychloroquine for Mpro and Spike glycoprotein, respectively).
Table 1.
Informations of the tested library of active phytoconstituents from S. lappa. and S. involucrata.
| no | Compounds | Source | Refs. | PubChem CID | Formula | Type compounds |
|---|---|---|---|---|---|---|
| 1. | 8-cedren-13-ol | S.costus | [27] | 519,545 | C15H24O | Sesquiterpenoid |
| 2. | (7Z,10Z,13Z) 7,10,13-Hexadecatrienal | S.costus | [16] | 556,280 | C16H26O | Trienyl derivative |
| 3. | α-Cyclocostunolide | S.costus | [39] | 385,663,116 | C15H20O2 | Sesquiterpenoid lactones. |
| 4. | α -Curcumene | S.costus | [27] | 92,139 | C15H22 | Sesquiterpenoids |
| 5. | α-Selinene | S.costus | [27] | 10,856,614 | C15H24 | Sesquiterpenoids |
| 6. | β-sitosterol | S.costus | [39] | 348,274,860 | C29H50O | Steroids |
| 7. | Alloisoimperatorin | S. involucrata | [40] | 5,317,436 | C16H14O4 | Coumarins |
| 8. | Apigenin |
S.costus S. involucrata |
[41] [17] |
5,280,443 | C15H10O5 | Flavones, (flavonoïdes) |
| 9. | Arbusculin B | S.costus | [42] | 161,442 | C15H20O2 | Sesquiterpenoid lactones. |
| 10. | Arctigenin | S. involucrata | [43] | 64,981 | C21H24O6 | Dibenzylbutyrolactone lignans |
| 11. | Arctiin | S. involucrata | [41] | 100,528 | C27H34O11 | Lignan glycosides. |
| 12. | Bergapten | S. involucrata | [41] | 2355 | C12H8O4 | Furanocoumarins |
| 13. | Caffeic acid | S.costus | [41] | 689,043 | C9H8O4 | Hydroxycinnamic acid |
| 14. | Catechin | S.costus | [41] | 9064 | C15H14O6 | Flavones, (flavonoïdes) |
| 15. | Chlorogenic acid |
S.costus S. involucrata |
[41] |
1,794,427 | C16H18O9 | Hydroxycinnamic acid |
| 16. | Chrysoeriol 7-O-glucuronide | S. involucrata | [29] | 14,630,703 | C22H20O12 | Flavonoid-7-o-glucuronides |
| 17. | Costunolide | S.costus | [44] | 5,281,437 | C15H20O2 | Germacranolide |
| 18. | Daucosterol | S.costus | [45] | 5,742,590 | C35H60O6 | Steroids-like |
| 19. | Dehydrocostuslactone | S.costus | [39] | 73,174 | C15H18O2 | Guaianolide sesquiterpene lactone |
| 20. | Edultin | S. involucrata | [41] | 5,317,013 | C21H22O7 | Furanocoumarin |
| 21. | elemol | S.costus | [16] | 92,138 | C15H26O | Sesquiterpenoid |
| 22. | Epicatechin gallate | S.costus | [39] | 107,905 | C22H18O10 | Flavanols |
| 23. | Epicatechin | S.costus | [41] | 72,276 | C15H14O6 | Flavanols |
| 24. | Epigallocatechin gallate | S.costus | [41] | 65,064 | C22H18O11 | Flavanols |
| 25. | Eupatoriopicrin | S.costus | [42] | 5,281,461 | C20H26O6 | Sesquiterpenoids. |
| 26. | Gallic acid | S.costus | [41] | 370 | C7H6O5 | Phenolic acids |
| 27. | Hesperidin | S.costus | [41] | 10,621 | C28H34O15 | Flavanone glycoside |
| 28. | Hispidulin | S. involucrata | [46] | 5,281,628 | C16H12O6 | Monomethoxyflavone |
| 29. | hispidulin-7-glucoside | S. involucrata | [46] | 5,318,083 | C22H22O11 | Flavonoid-7-o-glucusides |
| 30. | Hydroxybenzoic acid | S.costus | [41] | 135 | C7H6O3 | phenolic derivative |
| 31. | Isopimpinellin | S. involucrata | [41] | 68,079 | C13H10O5 | 8-methoxypsoralens |
| 32. | Isorhamnetin | S.costus | [41] | 5,281,654 | C16H12O7 | Flavonols |
| 33. | Jaceosidin | S. involucrata | [47] | 5,379,096 | C17H14O7 | 6-o-methylated flavonoids |
| 34. | Kaempferol | S. involucrata | [48] | 5,280,863 | C15H10O6 | Flavonols |
| 35. | Luteolin |
S.costus S. involucrata |
[41] [17] |
5,280,445 | C15H10O6 | Flavone |
| 36. | Luteolin-7-glucoside | S. involucrata | [17] | 5,280,637 | C21H20O11 | Flavonoid-7-o-glucusides |
| 37. | Malic acid | S.costus | [41] | 525 | C4H6O5 | Alpha‑hydroxy acid |
| 38. | Nepetin | S. involucrata | [49] | 5,317,284 | C16H12O7 | Flavonoid |
| 39. | Nepetin-7-glucoside | S. involucrata | [29] | 12,314,010 | C22H22O12 | Flavonoid-7-o-glucusides |
| 40. | Oroselol | S. involucrata | [41] | 160,600 | C14H12O4 | Coumarins |
| 41. | Osthol | S. involucrata | [41] | 10,228 | C15H16O3 | A derivative of coumarin |
| 42. | Protocatechuic acid |
S.costus S. involucrata |
[41] [50] |
72 | C7H6O4 | Dihydroxybenzoic acid |
| 43. | Quercetin | S. involucrata | [46] | 5,280,343 | C15H10O7 | Flavonols |
| 44. | Quercetin-3-glucoside |
S.costus S. involucrata |
[41] [29] |
25,203,368 | C21H19O12− | Flavonoid-3-o-glycosides |
| 45. | Quercitrin | S. involucrata | [29] | 5,280,459 | C21H20O11 | Flavonoid-3-o-glycosides |
| 46. | quinic acid | S.costus | [41] | 6508 | C7H12O6 | Organic acide |
| 47. | Rutin |
S.costus S. involucrata |
[41] [46] |
5,280,805 | C27H30O16 | Flavonols |
| 48. | Spathulenol | S.costus | [27] | 92,231 | C15H24O | Sesquiterpenoids |
| 49. | Syringin | S. involucrata | [41] | 5,316,860 | C17H24O9 | Phenolic glycoside |
| 50. | Tangshenoside III | S. involucrata | [51] | 11,968,970 | C34H46O17 | Phenylpropanoid |
| 51. | Vaginidiol diacetate | S. involucrata | [41] | 9,997,718 | C18H18O7 | Organic Compounds |
| 52. | Valerenol | S.costus | [16] | 91,699,505 | C15H24O | sesquiterpenoids |
| 53. | Vulgarol B | S.costus | [39] | 91,748,781 | C15H24O | sesquiterpene ketone |
| 54. | Xanthotoxol | S. involucrata | [39] | 65,090 | C11H6O4 | furanocoumarin |
| 55. | Zaluzanin | S.costus | [42] | 12,445,012 | C17H20O4 | Sesquiterpene lactone |
2.3. Molecular docking studies
As molecular targets, three important proteins for SARS-CoV2 infection were obtained from protein data bank of the Research Collaboration for Structural Bioinformatics (RCSB), website (http://www.rcsb.org/pdb) in PDB formats. This including the Main Protease (Mpro) (also called 3C-like protease _ 3CLpro), which represents a pivotal role in the propagation of SARS-CoV–2. The crystal structure of this enzyme was downloaded in complex with its inhibitor N3, (PDB ID: 6LU7; 2.16 A°). The crystalline structure of the L452R mutant receptor binding domain (RBD) of SARS-CoV-2 Spike glycoprotein (SGP) was downloaded in complex with COVOX-75 and COVOX-253 Fabs (PDB code: 7ORB; 2.5 A°). However, the crystalline structure of the Native Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) was downloaded without inhibitor (PDB ID: 1R42; 2.20 A°).
2.3.1. Ligand and protein structures preparation
Retrieval and protein structure preparation of active sites
The three targets proteins, 6LU7, 7ORB and 1R42 used in this study, was protonated where hydrogen atoms were added with their 3D geometry, corrected for any found errors in the connection or type of different atoms, and then energy minimized at the end of the preparation steps. This step followed by site-finder to define and isolate the same binding pocket of the co-crystallized native inhibitor as dummy atoms over helix for the docking step.
Ligands preparation
Structures of the phytoconstituents of Saussurea genus were surveyed and downloaded from the PubChem database in structure data format (SDF). Then, Energy minimizing of these ligands was done under the following conditions: temperature = 300 K, pH = 7. Furthermore, the geometry was performed using the field strengths in the MMFF94x implanted in MOE and Austin model 1 (AM 1) with gradient value of 0.0001 kcal/mol. Then, the compounds were saved in mdb format as a new database.
2.3.2. Docking
The dock tool of Molecular Operating Environment (MOE) 2019.0102 software was used for fitting of the isolated compounds' database into the active site of SARS-CoV-2 and ACE-2 enzymes. Docking site was selected as dummy atoms, alpha triangle as the placement methodology, and London dG as the scoring methodology. The docking process was run followed by evaluation of poses. Poses with the highest energy scores and best ligand-enzyme interactions were selected and recorded.
2.4. Chemical descriptors calculation
Lipinski's Rule of Five [52] was used to investigate phytoconstituents that were selected for this study. Filters like Molecular weight of the ligand (<500 Da), high lipophilicity (LogP<5), number of hydrogen bonds donors (<5), number of hydrogen bond acceptors (<10) and lip druglike were used to carry out the further selection of this compounds. Violation of more than 2 of the above-stated parameters debarred further analysis of a particular molecule. Parameter details were calculated using Molecular Operating Environment (MOE) 2019.0102 software.
2.5. ADMET analysis
A computational study of the top ten docking inhibitor compounds of each protein was carried out for the prediction of pharmacokinetics ADMET (Absorption, Distribution, Metabolism, Excretion and Hepatotoxicity) properties. In this module, eight mathematical models, such as solubility and its level, HIA (human intestinal absorption), ADMET AlogP98, ADMET PSA-2D, BBB penetration level PPB (plasma protein binding), ADMET CYP2D6 and hepatotoxicity were predicted via Discovery Studio (DS) 2.0 Software (Studio 2.5, Accelrys, Co. Ltd., San Diego, CA, USA)
2.6. Toxicity risks assessment
Another method used to determine the therapeutic compatibility of the drug is toxicity prediction by using the USFDA (US FDA, United States Food and Drug Administration) standard toxicity risk predictor software TOPKAT in the Discovery Studio. Toxicity by TOPKAT and prediction, TOPKAT rat female NTP probability and prediction TOPKAT rat male NTP probability and prediction, developmental toxicity potential, aerobic biodegradability, ocular irritancy, skin irritancy and Daphnia EC50.
2.7. Quantum chemical calculations
All Density Functional Theory (DFT) calculations were performed by using the Gaussian 09 program package [53] and the output files were visualized by means of Gauss View 5.0.8 program [54]. Quantum chemical calculations such as optimized geometries and corresponding molecule orbitals (MOs) energies, Mulliken atomic charge and molecular electrostatic potential (MEP) were obtained at B3LYP (Becke's three parameter hybrid model using the Lee-Yang Parr correlation functional) [55,56] level of density functional theory (DFT) with 6–31G(d,p) basis set [57].
3. Results and discussion
3.1. Phytochemical constituents of S. lappa. and S. involucrata
The tested library of active constituents from S. lappa. and S. involucrata comprised phytochemicals that cover major classes of natural products. Phytochemical studies of the two Saussurea plants showed the presence of terpenes (sesquiterpenes and steroids); polyphenols (flavonoids and lignans); aromatic acids (phenolic and hydroxycinnamic acids) and others.
Closely-related plants species often share similar secondary metabolites and bioactivities. The two Saussurea species shows resemblance in bioactivities as well as phytochemical constituents (References against each phytochemical constituents candidate plants have been shown in Table 1).
Saussurea is a medicinal plant of immense medicinal importance having a variety of compounds and diversity in the pharmacological spectrum. It is used to treat fever, headache, cough, bronchial asthma [58], diarrhea [59], cholera [60], tuberculosis, vomiting, dyspepsia [58,59], gastric ulcer [59], abdominal pain [58,59]. Different studies reported that Saussuria active compounds have antiviral activity including SARS-CoV-2. Prawiro et al. searched to make evidence-based medicine that Honey, Saussuria coctus and Nigella may cure the Covid-19. They divided mice into two groups, the first group as control received PBS as a placebo. Then the second group, received Honey, Saussuria coctus and Nigella sativa. After administration regiments a long three weeks, they sacrificed the mice and evaluated the immune responses markers that are Th2, Th17 for cellular, and NK cells, TGF-β, IL-17A, sIgA, IL-4, IL4, B-def, and IgG for humoral. As results, they reported a statistically deference in the cellular immune responses markers, Th2 and Th17, between control with treatment group (p = 0.05) and a deference in the markers of humoral immunity (p=>0.05). However, the markers of humoral immunity (IL-17A) have no statistical difference [61]. Another study evaluated the crude extract prepared from Saussuria coctus for its antiviral activity against Hepatitis B Virus (HBV). Two compounds Costunolide and Dehdrocostuslactone suppressed the expression of Hepatitis B surface antigen (HBsAg) in a dose dependent manner with an IC50 value of 1.0 and 2.0 mM in Human hepatoma Hep3B cells. Significant suppression was also observed in human hepatoma cell line HepA2 derived from the HepG2 cells [62]. These findings suggest that Saussuria coctus may have the potential to develop as specific anti-virus drugs such as anti- SARS-CoV 2.
3.2. Docking studies
Plants are sources of phytoconstituents which has the potential to be developed as antiviral agents for SARS-CoV-2, as has been reported by previous studies [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [52], [53], [54], [55], [56], [57], [39], [40], [41], [42], [43], [44], [45], [46], [63], [47], [48], [49], [50], [51], [58], [59], [60], [61], [62], [64]. Since ligand binding to a protein of interest is the first step in drug discovery, molecular docking is widely used to predict and identify ligands that fit into the binding pocket of a protein of interest [65]. Molecular docking is not only cost effective but also time effective approach to identify a promising therapeutics agent of COVID-19.
3.2.1. Evaluation test of the program used
Several methods are used to evaluate the performance of the different programs of docking for each application. The ability of an algorithm to find the correct location of the ligand in relation to its receptor is usually determined by means of the deviation root mean-square-deviation (RMSD) of the model designed by the software against the crystal structure. The RMSD test was performed on complexes formed between the SARS-CoV-2 receptors and different ligands. The obtained molecular docking results were considered valid since redocking the orginal ligands (N3). The accepted value is a maximum difference of 2Å beyond which the prediction is considered as inadequate [66]. The obtained molecular docking results were considered valid since redocking the crystalized ligands (N3 and Hydroxychloroquine) furnished great ligand superposition with root-mean standard deviation (RMSD) below 2 Å. Scores, RMSD_refine values, and diverse interactions of the best top scoring ligands with the amino acids of the Mpro, RBD of SGP and ACE-2 pockets are described in Table 2 .
Table 2.
Molecular docking results and interactions of the top scoring compounds of Saussurea species with SARS-CoV-2 proteins and ACE-2.
| Protein | ligand | Binding energy Score (Kcal/mol) | RMSD refine |
Bonds between atoms of compounds and residues of active site |
|||
|---|---|---|---|---|---|---|---|
| Atom of ligand |
Involved Receptor residues |
Bond type |
Bond length (A°) |
||||
| Tangshenoside III | −9.3479 | 1.6416 | O6 O11 C49 C39 |
Thr 24 Asn 142 Met 49 His 41 |
H-donor H-donor H-donor H-Pi |
2.90 3.06 3.94 4.35 |
|
| Rutin | −9.1420 | 1.2988 | O4 O8 O9 O12 O4 O7 O10 6-ring |
Cys 145 Cys 145 Thr 26 Cys 145 Gly 143 His 163 Thr 26 Gln 189 |
H-donor H-donor H-donor H-donor H-acceptor H-acceptor H-acceptor Pi-H |
4.08 3.55 2.89 3.71 2.77 3.23 3.02 3.67 |
|
| Hesperidin | −8.5672 | 1.6829 | O7 O8 O10 O13 O12 6-ring 6-ring |
Asn 142 Met 165 Thr 190 Cys 145 Cys 145 Thr 25 Asn 142 |
H-donor H-donor H-donor H-donor H-acceptor Pi-H Pi-H |
3.04 3.61 2.86 3.97 3.05 4.13 3.87 |
|
| Arctiin | −8.5658 | 1.5173 | O6 | Met 165 | H-donor | 3.31 | |
| Quercitrin | −8.4104 | 1.2280 | O5 O11 O3 6-ring |
Cys 145 Thr 26 His 163 Gln 189 |
H-donor H-donor H-acceptor Pi-H |
3.57 3.28 3.01 4.09 |
|
| Luteolin-7-glucoside | −8.0322 | 1.8745 | O3 O5 O28 O3 O4 O8 6-ring |
Leu 141 Cys 145 Met 165 Ser 144 His 163 His 41 Gln 189 |
H-donor H-donor H-donor H-acceptor H-acceptor H-pi pi-H |
2.67 3.13 3.89 2.91 3.14 3.28 3.48 |
|
| Quercetin-3-glucoside | −7.9070 | 1.5491 | O4 O8 O13 O3 O4 O6 6-ring 6-ring 6-ring |
Cys 145 Cys 145 Cys 145 Gly 143 Ser 144 His 163 Gln 189 Gln 189 Gln 189 |
H-donor H-donor H-donor H-acceptor H-acceptor H-acceptor pi-H pi-H pi-H |
3.54 3.63 4.10 3.37 3.08 3.12 4.79 3.71 4.24 |
|
| Daucosterol | −7.8411 | 1.3721 | O1 | Glu 166 | H-acceptor | 3.21 | |
| Hispidulin-7-glucoside | −7.7856 | 1.9530 | O4 C12 O3 O6 |
Cys 145 Cys 145 Gly 143 His 163 |
H-donor H-donor H-acceptor H-acceptor |
3.44 3.55 3.09 3.05 |
|
| β-sitosterol | −7.5755 | 1.9469 | O1 | Thr 26 | H-donor | 2.81 | |
| 7ORB | Rutin | −9.0665 | 1.7144 | O4 O15 O4 |
Phe 347 Ser 399 Ser 349 |
H-donor H-donor H-acceptor |
2.91 2.82 2.83 |
| Tangshenoside III | −7.7140 | 1.9781 | O11 O12 |
Lys 356 Tyr 351 Ala 348 |
H-acceptor H-acceptor pi-H |
3.11 3.27 3.95 |
|
| Hesperidin | −7.5719 | 1.7773 | O6 O8 O9 O10 |
Arg 346 Glu 340 Lys 356 Lys 356 |
H-donor H-donor H-acceptor H-acceptor |
2.76 2.92 3.25 3.14 |
|
| Arctiin | −7.2114 | 1.4840 | O4 O6 O7 |
Ser 399 Arg 346 Arg 346 |
H-donor H-donor H-acceptor |
2.83 3.10 2.94 |
|
| Daucosterol | −7.1701 | 1.9301 | O5 O3 |
Glu 340 Asn 354 |
H-donor H-acceptor |
2.81 3.11 |
|
| Epicatechin gallate | −6.9823 | 1.7679 | O4 O6 O9 O11 |
Ser 349 Ala 352 Ser 399 Ser 399 |
H-donor H-donor H-donor H-donor |
2.77 2.87 2.92 3.04 |
|
| Quercitrin | −6.8851 | 1.8457 | O8 | Ser 399 | H-donor | 2.86 | |
| Quercetin-3-glucoside | −6.694 | 1.5691 | O10 O1 O6 |
Ser 399 Arg 346 Arg 346 |
H-donor H-acceptor H-acceptor |
2.91 2.93 3.20 |
|
| Hispidulin-7-glucoside | −6.6385 | 1.6345 | O3 O7 6-ring |
Ser 399 Arg 346 Arg 346 |
H-donor H-donor Pi-cation |
2.85 3.09 2.99 |
|
| Edultin | −6.4001 | 1.9467 | O2 O7 |
Asn 354 Arg 346 |
H-acceptor H-acceptor |
2.64 3.18 |
|
| 1R42 | Rutin | −8.9636 | 1.239 | O5 O10 C20 O7 O9 |
Glu 406 Asp 367 Glu 406 Gln 442 Lys 441 |
H-donor H-donor H-donor H-acceptor H-acceptor |
2.80 3.15 3.31 3.27 2.94 |
| Hesperidin | −8.4456 | 1.421 | O4 O8 C21 O1 O13 |
Asp 367 Asp 367 Asp 367 Lys 441 Gln 522 |
H-donor H-donor H-donor H-acceptor H-acceptor |
2.84 2.87 3.56 2.94 3.15 |
|
| Tangshenoside III | −8.1305 | 1.176 | O10 C5 O5 |
Glu 375 Asn 290 Lys 441 |
H-donor H-acceptor H-acceptor |
2.84 3.07 3.12 |
|
3.2.2. Binding energy and molecular interaction studies
The molecular docking has been performed using the Molecular Operating Environment (MOE) 2019.0102 software to evaluate the binding mode of ligand and interactions in the active site. Our screening was performed against three major key target RBD of spike glycoprotein SARS-CoV-2 B1.617.2, the main protease (Mpro) and ACE-2.
Docking analysis of SARS-CoV-2 Mpro
According to the 3D structure of SARS-CoV-2 Mpro, the active Mpro homodimer comprises two protomers, constituting three domains. Amino acid residues 8–101 constitute Domain I, amino acid residues 102–184 constitute Domain II, and amino acid residues 201–306 constitute Domain III. Before analyzing ligand-receptor interactions, we have defined binding site residues of Mpro enzyme. Binding site of Mpro enzyme contains Thr24, Thr25, Thr26, Leu27, His41, Met49, Tyr54, Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, His163, His164, Met165, Glu166, Leu167, Pro168, His172, Asp187, Arg188, Gln189, Thr190, Ala191, and Gln192 residues. The S1 binding site is formed by Phe-140, Asn-142, Ser-144, Cys145, His-163, His-172, and Glu-166 side chains, Leu-141, Gly-143, His-164, and Met-165 backbones. The side chains of His-41, Val 42, Asn-119, Thr-25, Cys-145, Gly-143 together with the backbone of Thr-26 define S1’ site. His41 and Cys145 form the catalytic dyad in the active site [67] and His164 is essential for enzyme activity. His163, His172 and Glu166 are believed to provide the opening gate for the substrate in the active state of the protomer [68], and Thr24, Thr26 and Asn119 are predicted to play roles in drug interactions [69,70].
The formation of hydrogen bonds defines the integrity and stable nature of each protein–ligand complex. Table 2 demonstrates the binding energies and different bonds of the best top ten complexes. Most of the docked compounds (1–55) formed relatively stable complexes within the active site of Mpro with Hydrogen bonds; showing moderate to promising energy scores in the range of −4.61 to −9.34 kcal/mol as compared to the inhibitor N3 (−7.44 kcal/mol). Most phytochemicals exhibited higher binding score than the standard. On the competition of docking, total of seven metabolites, namely Tangshenoside III, Rutin, Hesperidin, Quercitrin, Luteolin-7-glucoside, Quercetin-3-glucoside and hispidulin-7-glucoside showed effective binding with HIS41 or CYS145 of catalytic dyad along with multiple interactions with other amino acid residues in active site of SARS CoV2-Mpro.
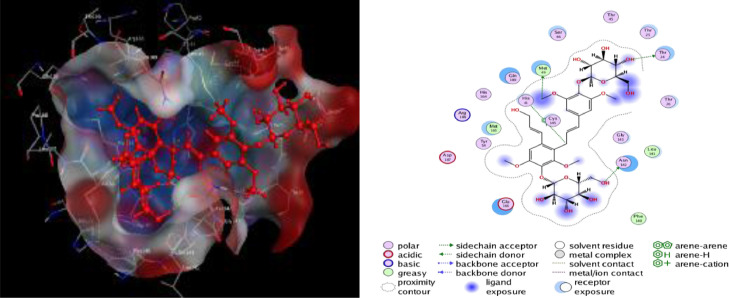
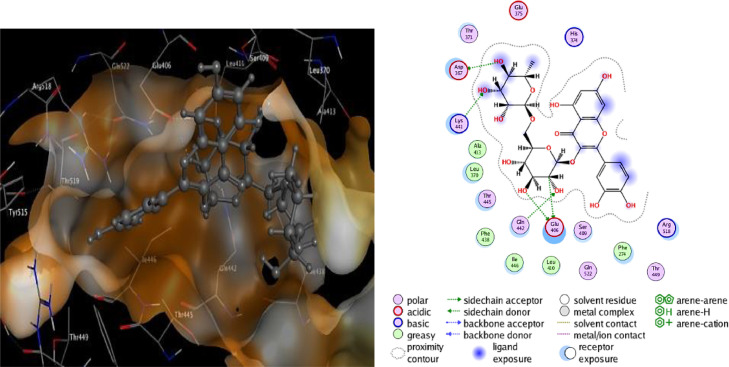
The binding interactions of the top two docked complexes demonstrated that Tangshenoside III had three hydrogen bonds with Mpro, at Thr 24, Asn 142, Met 49 and one H-pi bond were observed for His 41. Rutin exhibited a higher number of hydrogen bonds, forming seven hydrogen bonds three at Cys 145, and four at Thr 26, Gly 143, His 163, Thr 26. This complex also formed a pi-H bond at Gln 189. These amino acids are predicted to play a major role during chemical interactions with these compounds and the inhibition of Mpro. The detail of interaction and visualization of the docking results of Tangshenoside III and Rutin with Mpro are provided in Figs. 1 and 2 . Ghosh et al., [71] reported that polyphenols like broussochalcone A, papyriflavonol A, 3′-(3-methylbut-2-enyl)−3′,4′,7-trihydroxyflavane, broussoflavan A, kazinol F, and kazinol J had good interaction with the catalytic residues of His41 and Cys145 of Mpro. These amino acids interactions with the SARS-CoV-2 main protease Mpro are similar to our findings. Previous studies reported that the inhibitor N3 docks in the active binding site of 6LU7 and forms hydrogen bonds with PHE 140, GLY 143, THR190, HIS 164, GLU166 and GLN189 [72] and these results similar to our finding. A recent study performed by Cherrak et al. showed that Rutin possesses high docking scores of – 9.2 kcal/mol and interacts with Mpro via conventional H-bonds at Thr26, Tyr54, Leu141, and Glu166, π-cation interactions at His41, π-alkyl interactions at Met49, π-sulfur interactions at Cys145, and C-H bond interactions at Met165 [73]. Another investigation reported that Rutin was found to have, with Mpro, a docking score of −9.16 kcal/mol, it was considered to be a potential Mpro inhibitor. Rutin was predicted to form hydrogen bonds involving Cys145, Asn142, Gly143, and Thr190, with additionally the possible formation of σ-π stacking interaction with Gln189. Notably, the major binding affinity was based on the presence of a hydroxyl group, which presented the key to anchoring and blocking the substrate into the active pocket of the catalytic center [74].
Fig. 1.
2D and 3D structure of the Tangshenoside III docking results with the target Main protease Mpro of SARS-CoV-2.
Fig. 2.
2D and 3D structure of the Rutin docking results with the target Main protease Mpro of SARS-CoV-2.
Hesperidin inhibit the cleavage activity of the Mpro in a dose-dependent manner in cell-free and cell-based assays, with an IC50 of 8.3 μM [75]. A previous study also showed that the docked hesperidin compound against Mpro was –13.51 [76], and according to a previous investigation, the best hesperidin position against SARS-CoV-2 Mpro had a score of –10.1 [[77], [78]]. Furthermore, Rameshkumar et al., docked 36 flavonoid to the active site of Mpro and it was observed that the compound agathisflavone had shown the highest binding energy value of −8.4 kcal/mol and interacts with Mpro by t hydrogen bonds at Lys102, Thr111, Ser158, His246 followed by Dracorubin and Cupressuflavone that showed dock score of −8.2 kcal/mol [79]. As presented in a previous study reported that several potentially active compounds have best docking scores which are cannabinoids, rhoifolin, pectolinarin, morin, kaempferol, epigallocatechin gallate, herbacetin, and hesperidin against 3CLpro were –8, –8.2, –8.2, –7.8, –7.8, –7.8, –7.2, and –8.3, respectively [80].
Docking analysis of SARS-CoV-2 B1.617.2 spike glycoprotein
The B.1.617.2 Delta variant is considered to be the most infectious of all variants and as of June 2021 has become one of the most transmissible variants with the highest number of reported cases, followed by the B.1.617.1 [81,82]. Variants are characterized by multiple mutations in the surface spike glycoprotein. Mutations in the RBD can change the ability of the virus spike protein to bind to and enter the host cell. The high rate of mutation and recombination in SARS-CoV2 makes it difficult for scientist to develop specific anti-CoV2 drugs and vaccines.
According to global reports and Health agencies, in comparison with SARS-CoV-2 from Wuhan-2019, the new variants Delta B.1.617.2 present 5 mutations in S protein T19R; L452R; T478K; P681R; D950N [83]. L452R/T478K in B.1.617.2 are assumed to play roles in infectivity, transmissibility of the virus and leads to an increase in virulence by reducing the antibody binding affinity, as well as immune evasion [84].
All the 55 active constituents from S. lappa. and S. involucrata were individually docked to the active site of 7ORB. As compared to the inhibitor, Hydroxychloroquine (binding energy = −7.409), Rutin had shown the highest binding energy value of −9.0665 kcal/mol. it was observed that the key amino acids include Phe 347, Ser 399 and Ser 349 were involved in the hydrogen bond interactions (donor and acceptor). It followed by Tangshenoside III and Hesperidin, that showed binding energy of −7.71 and −7.57 kcal/mol, respectively.
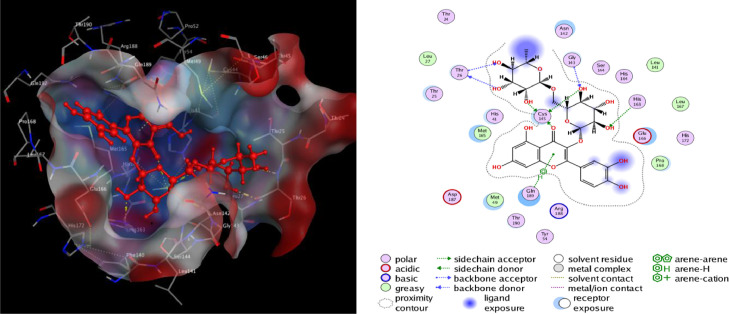
The binding interactions of the top docked complexes demonstrated that Rutin forming three hydrogen bonds at Phe 347, Ser 399 and Ser 349. Tangshenoside III had two hydrogen bonds at Lys 356 and Tyr 351and one pi-H bond were observed for Ala 348. Hesperidin exhibited four hydrogen bonds, two at Arg 346, Glu 340 and two at Lys 356. Arctiin had three hydrogen bonds at Ser 399 and two at Arg 346 (Table 2). These amino acids are predicted to play a major role during chemical interactions with these compounds and the inhibition of RBD of spike glycoprotein. The detail of interaction and visualization of the docking results of Rutin and Tangshenoside III with SARS-CoV-2 B1.617.2 spike glycoprotein is provided in Figs. 3 and 4 . Solo and Doss [85] tries to explore the inhibitory activity of 50 phytochemicals against the spike protein of the Delta SARS-CoV-2. They used Hydroxychloroquine, as a standard in the docking analysis and they reported that this standard had a binding affinity of −6.5 kcal/mol with the delta variant. Solo and Doss identified the 3,5,3′Trimethoxy-6,7:4′,5′-bis(methylenedioxy) flavone as having the highest binding affinity of −8.7 kcal/mol for both the target proteins, in comparison to the other 49 phytochemicals. The major Hydrogen bond interactions were observed with Glu340, Val341, Ala344, Phe347, Ala348, Ser349, Tyr351, Ala352, Lys356, Ser 399 and Tyr451. Except Val341, Ala344 and Ala348 that exhibited a pi-H bond, Amino acids involved in the Hydrogen bond interactions of Delta SARS-CoV-2 spike found in our study were similar with the report of Solo and Doss.
Fig. 3.
2D and 3D structure of the Rutin docking result with the target spike protein (B.1.617.2) of SARS-CoV-2.
Fig. 4.
2D and 3D structure of the Tangshenoside III docking result with the target spike protein (B.1.617.2) of SARS-CoV-2.
Docking analysis of ACE-2
The S-protein is made up of two subunits, the S1 subunit is involved in host cell receptor recognition and is the receptor-binding domain (RBD) and S2 subunit is responsible for fusion of the viral membrane and the host cellular membrane [ 86 ]. Cell entry of SARS CoV-2 depends on two consecutive steps, firstly binding of the viral spike (S-protein) to host cellular receptors followed by priming of S-protein by cell proteases. Recently, researchers showed that SARS CoV-2 uses the ACE-2 receptor for entry [ 87] and the serine protease TMPRSS2 for priming of S-protein. It adheres to the host cell's surface receptor, human angiotensin-converting enzyme 2 (hACE2), allowing viral cellular entry via endosome formation and/or plasma-membrane fusion [ 88,89 ]; ACE-2 is an integral membrane protein type I [ 90 ]. Due to its key role in the process of SARS-CoV-2 infection, this structural component has been confirmed as the entry point for the SARS-CoV-2 virus to infect human cells [ 91]. The SARS CoV-2 virus can colonize any organ that has ACE-2 receptors such as lung endothelial and alveolar type II cells, epithelial cells in the upper esophagus, enterocytes in the ileum and colon, epithelial cells in the bile, heart muscle cells, proximal tubules in the kidneys, and urothelial cells in the bladder [ 92 ].
SARS-CoV-2 binds to ACE2 ten times more strongly than other coronaviruses, making SARS-CoV-2 more infectious than others [ 93]. So the spike protein of SARS-CoV-2 was predicted to have a strong binding affinity to human ACE-2. Mutations in the RBD can help to enable strong affinity and binding capacity to ACE2, leading to higher transmissibility [ [94], [95], [96] ]. Moreover, The Indian (B.1.617.2 or Delta) variant has different mutations in the RBD of SARS-CoV-2: L452R and T478K. Thus, targeting the ACE2-RBD interaction site may be a viable solution that may restrict the coronavirus from entering into the cells by inhibiting the interaction between ACE-2 and the RBD [ 97]. During the COVID-19 pandemic, various molecular docking studies have targeted the interaction regions between the RBD and ACE2, performing docking for drug repositioning or using chemotherapeutic libraries (or using both methods) to search for drugs or compounds that can inhibit the interaction between RBD with ACE-2 [ [98], [99], [100], [101]]. The majority of studies have reported that bioactive molecules, have a higher binding potential than antiviral drugs (hydroxychloroquine and remdesivir) used in SARS-CoV-2 spike protein and ACE-2 [102,103]. In this context, molecular docking was performed to tested the interaction of bioactive compounds from two selected plants, Saussurea costus and Saussurea involucrata with ACE2 to determine their potential to inhibit its binding with RBD of the S-protein.
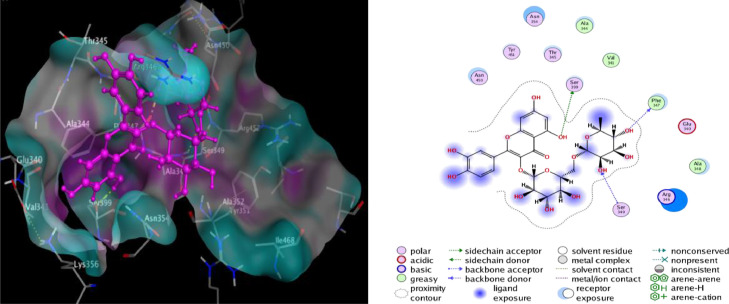
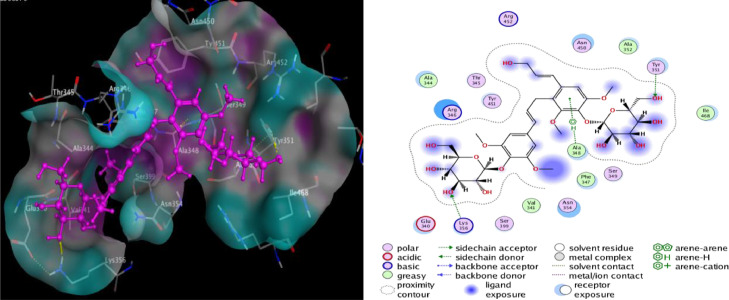
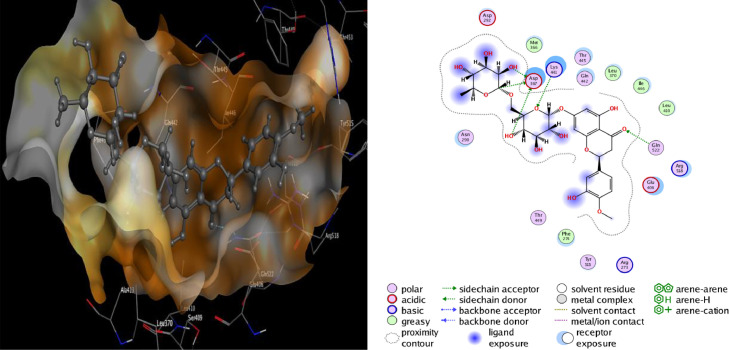
Molecular docking studies with ACE-2 have shown that Rutin, Hesperidin, and Tangshenoside III have the highest binding energy values of −8.9636, −8.4456 and −8.1305 kcal/mol, respectively. Sebsequently, these compounds bind strongly to ACE-2 and may hinder the substrate accessibility and its subsequent inhibition. Rutin involves five hydrogen bonds with Glu 406, one with Asp 367, one with Gln 442 and one with Lys 441. Hesperidin also shows favorable interactions with ACE-2 through five hydrogen bonds three with Asp 367, one with Gln 522 and one with Lys 441 (Table 2). The detail of interaction and visualization of the docking results of these complexes with ACE-2 is provided in Figs. 5 and 6 . Furthermore, another study conducted by Chen and Du (2020) demonstrated that the molecular docking studies of hesperidin with the ACE-2 enzyme showed that hesperidin can bind to ACE-2 with a predicted binding energy of −8.3 kcal/mol and binding sites at Glu-479, Arg-482, Ser-611 and Tyr-613. These results suggest that hesperidin may bind to ACE-2 and thus block 2019-nCoV infection [ 77 ]. Rutin has been identified as one of the three compounds (glycyrrhizin, rutin, and pheophorbide A) with a high binding affinity –6.9 kcal/mol to the ACE2 receptor.166 It created an interaction with the following residues: Phe4, Leu29, Asp30, Asn33, Val93, Ala99,Leu100, Ala387, Asp350, Ala387, Gln388, Pro389, Leu391, and Arg393 [ 104]. The molecular docking studies also showed that baicalin might bind strongly to the ACE-2 enzyme with a predicted binding energy of –8.46 kcal/mol and binding at Asn-149, Arg-273, and His-505. Based on the potential binding to ACE-2, it can be suggested that baicalin is a promising candidate for 2019-nCoV treatment. In addition, Scutellarin has the potential to bind to ACE-2 with a projected binding energy of –14.9 kcal/mol and binding sites at GLU-495, UNK-957, and ARG-482 [77].
Fig. 5.
2D and 3D structure of Rutin docking results with the target ACE-2.
Fig. 6.
2D and 3D structure of Hesperidin docking results with the target ACE-2.
3.3. Screening through pharmacokinetic properties
Drug-like properties and pharmacokinetic properties are intrinsic characteristics of drugs that may need to be optimized independently from pharmacodynamics properties during drug development. It is a balance among molecular properties affecting pharmacodynamics and pharmacokinetics of small molecules.
3.3.1. Chemical descriptors calculation
The drug-likeliness of top ten drug ligands for both SARS-CoV-2 Mpro and spike proteins was evaluated through Lipinski's rule of five parameters and the values are summarised in Table 3 (values of all the molecules are mentioned in supplementary Data (Table S1). This rule is based on physicochemical parameters of the tested ligands, including: Molecular weight (MW) not greater than 500 g/mol; A partition coefficient log P less than or equal five; number of hydrogen bond donors (HBD) (NH and OH groups) not greater than five; and number of hydrogen bond acceptors (HBA) (O and N atoms) not exceeds ten. Drugs having log P ranging from 0 to 5, have high possibility of oral absorption. Not more than one violation of these parameters will reduce the druggability of the molecule [105].
Table 3.
Detailed analyses of parameters of Lipinski's filter for the drug-likeness. The elaborated comparative analysis of all the seven drugs concerning parameters of Lipinski's filter.
| Ligand | MW | Lip acc | lip-Don | Log p (o/w) | Lip drug- like | Violation |
|---|---|---|---|---|---|---|
| Tangshenoside III | 726.72 | 17 | 9 | −1.49 | 0 | 3 |
| Rutin | 610.52 | 16 | 10 | −1.11 | 0 | 3 |
| Hesperidin | 610.56 | 15 | 8 | −0.77 | 0 | 3 |
| Arctiin | 534.6 | 11 | 4 | 0.56 | 0 | 2 |
| Quercitrin | 448.38 | 11 | 7 | 0.80 | 0 | 2 |
| Luteolin-7-glucoside | 448.38 | 11 | 7 | 0 | 0 | 2 |
| Quercetin-3-glucoside | 463.37 | 12 | 7 | −0.11 | 0 | 2 |
| Daucosterol | 576.8 | 6 | 4 | 6.15 | 0 | 2 |
| hispidulin-7-glucoside | 462.4 | 11 | 6 | −0.06 | 0 | 2 |
| β-sitosterol | 414.71 | 1 | 1 | 8.07 | 1 | 1 |
| Edultin | 386.4 | 7 | 0 | 3.53 | 1 | 0 |
| Epicatechin gallate | 442.38 | 10 | 7 | 3.38 | 1 | 1 |
All the screened compounds were found to have little violation and it was believed to effective despite of few violations, except edultin that was fully in agreement to Lipinski's rule of five (without violation). β-sitosterol and Epicatechin gallate also qualified Lipinski's rule of five with one violation concerning Log P and hydrogen donor, respectively. Except for β-sitosterol Daucosterol all other drug molecules showed the value of Log P less than 3.53, which indicates that the molecules are more likely to be in the hydrophilic environment and are favorable for their drug-likeness. Daucosterol has another violation regarding the molecular weight. His molecular weights is slightly more than the recommended values but within the permissible limit. Quercitrin, Luteolin-7-glucoside and Quercetin-3-glucoside exhibited two violation regarding hydrogen acceptor and donor as well as Arctiin showed two violations regarding hydrogen acceptor and molecular weight. Tangshenoside III, Rutin and Hesperidin exhibited three violations concerning hydrogen acceptor, donor and molecular weight.
3.3.2. ADMET parameters screening for drug likeness
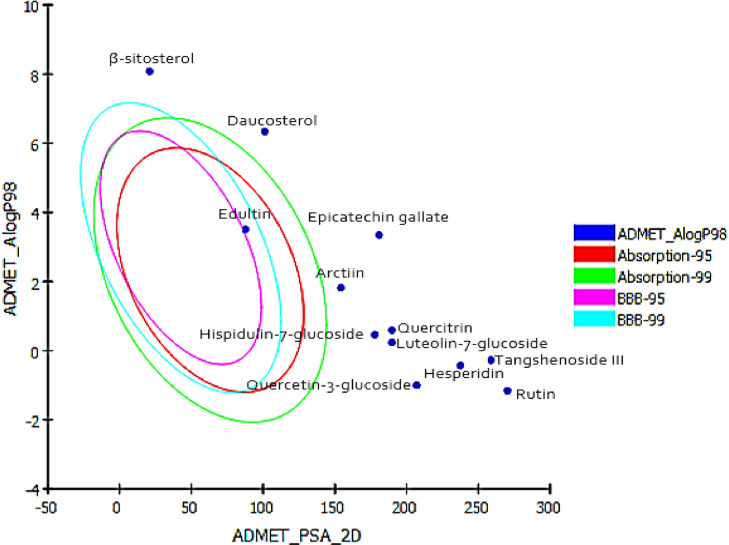
The determination of pharmacokinetic properties of drugs at the preliminary stages of drug discovery is essential to bring the drug up to clinical trial [106]. These properties such as absorption, distribution, metabolism, excretion and hepatotoxicity (ADMET) are important in order to determine the role in drug discovery and success of the development of compound for human therapeutic use. To determine the drug-likeliness of our compounds, we calculated their ADMET properties. ADMET prediction was done using Discovery Studio 2.5 (Accelrys, San Diego, USA) software. Prediction is based on two 2D molecular descriptors, namely AlogP_98 and PSA. As shown in Fig. 7 , Compounds with more than 90% absorbability would lie within the 99% confidence ellipse as the green ellipse Compounds falling out of the ellipse are those with less than 30% absorbability. The results of ADMET were displayed in Table 4 , revealing eight descriptors, such as the absorption of drugs depends on intestinal absorption, polar surface area (PSA) and AlogP98. The distribution depends on factors that include the blood–brain barrier (BBB) and plasma protein binding (PPB). Metabolism is predicted based on the Cytochrome P450 2D6 inhibition and hepatotoxicity. ADMET aqueous solubility predicts the solubility of each compound in water at 25∘C. ADMET solubility descriptors predicts molar solubility of drugs within the ranges: < −8.0 (level 0 = extremely low solubility), −8.0 to −6.0 (level 1 = very low, but possible) −6.0 to −4.0 (level 2 = low), −4.0 to −2.0 (level 3 = good solubility), and −2.0 to 0.0 (level 4 = optimal solubilty). Except β-sitosterol and Rutin ADMET Aqueous solubility logarithmic level of most of the compounds was found to be 2, 3 which indicates low or good aqueous solubility, respectively. ADMET absorption predicts human intestinal absorption (HIA) after oral administration. Intestinal absorption of drug was determined by obtained levels: 0 (good), 1 (moderate), 2 (low), 3 (very low). Daucosterol and Edultin were found to have moderate to good HIA whereas, all other ligands were found to have very low absorption level, therefore are poor in gastro intestinal absorption. High hydrogen-bonding capacity of molecules has ebeen reported as one of the most important factors affecting intestinal absorption of new chemical entities. As the generally respected ‘‘rule of 5′’ suggests, any molecule having more than 5 hydrogen bond donors and 10 hydrogen bond acceptors would normally face poor absorption consequences. Easy distribution of the drug through blood brain barrier (BBB) measured by its AlogP98 value which must be less than 5. The obtained absorption levels determine drug absorption and absorption decreases inversely with the level, i.e., level 0 denotes proper absorption, level 1 denotes moderate absorption and so on. All the compounds were fallen outside the 99% ellipse (undefined). Hence, the compounds may not be able to penetrate the blood brain barrier. ADMET plasma protein binding model predicts whether a compound is likely to be highly bound to carrier proteins in the blood. Predictions are based on AlogP98 and 1D similarities to two sets of “marker” molecules. True symbolizes binding and false symbolizes non-binding. The ADMET plasma protein binding property prediction clearly suggesting that all of 12 compounds with an exception of β-sitosterol and Epicatechin gallate, are not likely to be highly bound to carrier proteins in the blood. ADMET CYP2D6 binding predicts cytochrome P450 2D6 enzyme inhibition using 2D chemical structure as input as well as a probability estimate for the prediction. Predictions are based on a training set of 100 compounds with known CYP2D6 inhibitions. This descriptor determines inhibitory effect by predicted classes: non-inhibitor (false) and inhibitor (true). Except Epicatechin gallate all compounds are predicted as non-inhibitors of CYP2D6 suggesting that these compounds are well metabolized in Phase-I metabolism. Hence, the side effects (i.e., liver dysfunction) are not expected upon administration of these compounds which indicated they did not have hepatotoxicity.
Fig. 7.
Prediction of drug absorption for active phytoconstituents from S. lappa. and S. involucrate considered for anti-SARS-CoV-2.
Table 4.
ADMET of the best ten scoring ligands for both SARS-CoV-2 Mpro and spike glycoprotein.
| Ligand | Solubility | Absorption |
Distribution |
Metabolism | Toxicity | |||
|---|---|---|---|---|---|---|---|---|
| ADMET Aqueous Solubility (Level) | Intestinal Absorption level | ADMET AlogP98 | ADMET PSA-2D | BBB penetration level | Plasma Protein Binding | ADMET CYP2D6 binding | Hepato-toxicity | |
| Tangshenoside III | −4.583 (2) | 3 | −0.274 | 258.78 | 4 | False | False | False |
| Rutin | −6.182 (1) | 3 | −1.158 | 270.1 | 4 | False | False | True |
| Arctiin | −3.057 (3) | 3 | 1.814 | 154.1 | 4 | False | False | False |
| Hesperidin | −4.489 (2) | 3 | −0.431 | 237.4 | 4 | False | False | True |
| Quercitrin | −3.888 (3) | 3 | 0.589 | 189.8 | 4 | False | False | True |
| Luteolin-7-glucoside | −3.328 (3) | 3 | 0.238 | 189.8 | 4 | False | False | True |
| Quercetin-3-glucoside | −2.98 (3) | 3 | −1.003 | 207.1 | 4 | False | False | False |
| Daucosterol | −5.55 (2) | 2 | 6.337 | 101.12 | 4 | False | False | False |
| hispidulin-7-glucoside | −3.426 (3) | 3 | 0.464 | 177.91 | 4 | False | False | True |
| β-sitosterol | −8.256 (0) | 3 | 8.084 | 20.81 | 4 | True | False | False |
| Edultin | −4.856 (2) | 0 | 3.502 | 87.62 | 2 | False | False | True |
| Epicatechin gallate | −5.538 (2) | 3 | 3.339 | 180.87 | 4 | True | True | True |
The common top three ligands, according to the docking results, namely Tangshenoside III, Rutin and Hesperidin pass through the Lipinski's filter with 3 violations regarding molecular weights, hydrogen bond acceptors and donors. They were reported to show low intestinal absorption because of their physicochemical properties. Considering that hydrogen bond acceptor and donor groups influence permeability. In this context, the low oral bioavailability of compounds could be explained in terms of their calculated parameters according to Lipinski's rule, including molecular weights, hydrogen bond acceptors and donors. Thus, these compounds are predicted to be hardly transported, diffused, and absorbed than compared with the small molecules. On the other hand, the most popular and authentic rule for confirming the drug-likeness of the Ligand is Lipinski's rule. However, many drugs that do not pass through the Lipinski's filter but have immense pharmacological properties have been approved by the FDA as potential drug for clinical purposes [107]. Despite violation of some rules, approved anticancer and anti-infective drugs from natural products or their semisynthetic derivatives such as taxol and amphotericin B have also some violations but are biologically effective as drugs. Therefore, these results do not interfere with the development of these compounds as potential SARS-CoV-2 therapeutic agents [108].
3.3.3. Toxicity risks assessment
The toxicity predictions of the compounds were also investigated with Discovery Studio using the TOPKAT protocol. TOPKAT is a computational tool for in-silico quantitatively prediction of toxicity developed by Accelrys [109]. And is used by universities, private companies and government agencies including the US EPA, US FDA, Environment Canada, Health Canada and the Danish EPA for toxicity assessments. It computes and validates assessments of the toxic and environmental effects of chemicals solely from their molecular structure. TOPKAT employs robust and cross-validated Quantitative Structure Toxicity Relationship (QSTR) models to predict a probable value of toxicity . Moreover, with the help of these QSTR models, the product of a structure descriptors and its corresponding coefficient, it calculates probability values and evaluates toxicity through this descriptors contribution. It follows the criterion of checking the components in the optimal predictive space (OPS), and when they lie outside then the results were considered as unreliable.The Optimum Prediction Space (OPS) technology is implemented in TOPKAT as the methodology used to identify model applicability domain, providing a means of cheking whether the compounds under investigation are well represented in the models. The toxicity profiles calculated for all the compounds are tabulated in Table 5 . Our results indicate that all ligands are non-Mutagen, non-carcinogen with Rat female NTP. Therefore, the toxicity of the ligands was found to be suitable for the development into a medical drug. Thus, they experience significant first-pass effect.
Table 5.
Virtual toxicity studies of the best ten scoring ligands for both SARS-CoV-2 Mpro and spike glycoprotein.
| ligand | Ames mutagenicity | TOPKAT Mouse female NTP probability | TOPKAT Mouse male NTP probability | TOPKAT Rat female NTP probability | TOPKAT Rat male NTP probability | Aerobic bio-degradability | Ocular irritancy | Skin irritancy | TOPKAT Daphnia EC50 (mg/L) |
|---|---|---|---|---|---|---|---|---|---|
| Tangshenoside III | Non- Mutagen | 0,597,672 | 0,708,292 | 0,310,728 | 0,578,401 | Degradable | Mild | None | 0.05599 |
| Rutin | Non- Mutagen | 0,208,873 | 0,609,889 | 0,0,827,222 | 0,651,442 | Degradable | Mild | Weak | 1.41631 |
| Hesperidin | Non- Mutagen | 0,368,231 | 0,278,522 | 0,246,047 | 0,576,179 | Degradable | Mild | None | 1.71086 |
| Arctiin | Non- Mutagen | 0,621,887 | 0,614,135 | 0,2685 | 0,667,288 | Degradable | Mild | None | 0.27347 |
| Quercitrin | Non- Mutagen | 0,129,232 | 0,604,399 | 0,0,644,857 | 0,643,589 | Degradable | Mild | None | 2.53329 |
| Luteolin-7-glucoside | Non- Mutagen | 0,203,474 | 0,613,408 | 0,111,993 | 0,586,446 | Degradable | Moderate-severe | Weak | 3.08289 |
| Quercetin-3-glucoside | Non- Mutagen | 0,382,166 | 0,548,789 | 0,16,454 | 0,668,277 | Degradable | Moderate-severe | None | 6.39441 |
| Daucosterol | Non- Mutagen | 0,987,038 (Carcinogen) | 0,99,373 | 0,183,303 | 0,293,199 | Degradable | Moderate-severe | None | 0.152622 |
| hispidulin-7-glucoside | Non- Mutagen | 0,371,303 | 0,578,767 | 0,272,004 | 0,692,262 | Degradable | Moderate-severe | None | 4.27525 |
| β-sitosterol | Non- Mutagen | 0,979,079 | 0,995,694 | 0,280,137 | 0,355,373 | Degradable | None | Weak | 0.217316 |
| Edultin | Non- Mutagen | 0,584,079 | 0,706,606 | 0,364,043 | 686,881 | Degradable | Mild | None | 0.811667 |
| Epicatechin gallate | Non- Mutagen | 0,384,233 | 0,612,907 | 0,281,324 | 0,580,833 | Degradable | None | Strong | 1.61585 |
3.4. Quantum-Chemical Calculations
After sorting the common top three drug ligands (Tangshenoside, Rutin and Hesperidin) based on their highest negative binding energies value. These drug ligands are quite similar with different binding energies towards the three targets. The above-mentioned drug ligands were subjected to Quantum-Chemical Calculations studies.
3.4.1. Geometry optimization
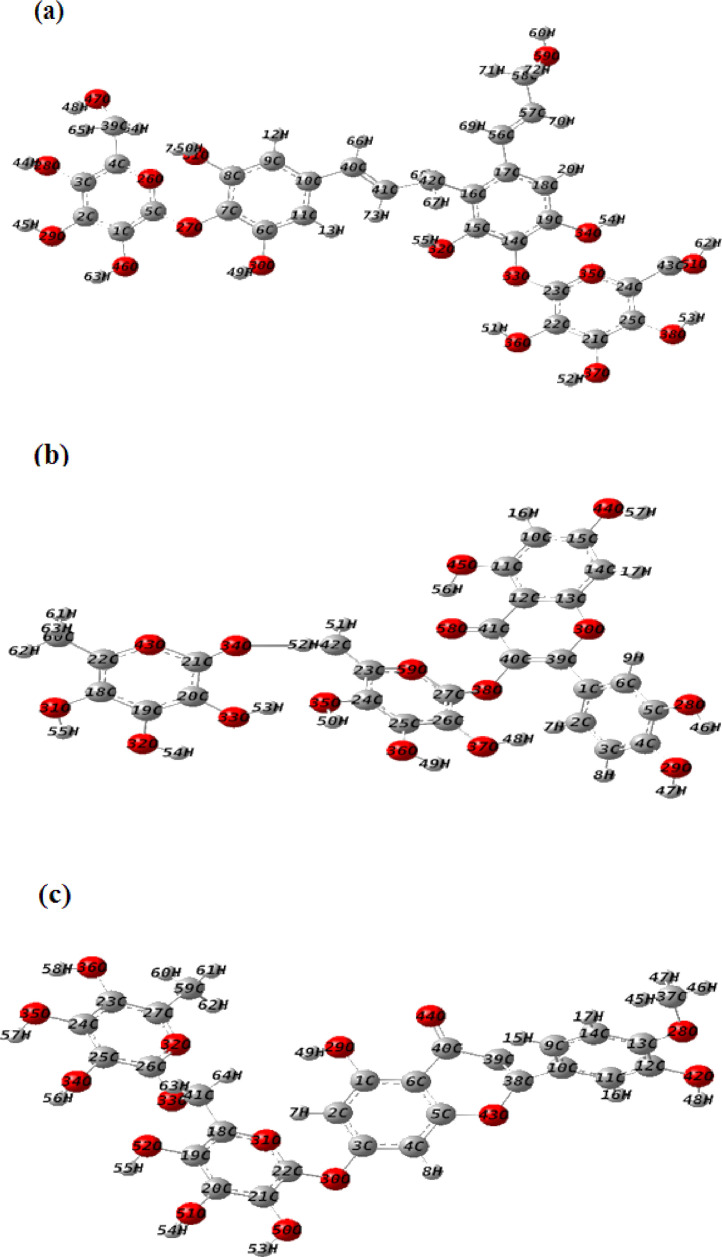
Optimized geometries of the investigated compounds obtained from DFT calculations using B3LYP/6–31G(d,p) method are given in Fig. 8 . The absence of imaginary frequencies confirmed the global minima on the potential energy surface. Calculated geometric parameters of the investigated compounds are given in Supplementary Data (Table S2).
Fig. 8.
Optimized molecular structures of (a) Tangshenoside III, (b) Rutin and (c) Hesperidin calculated with the methodology B3LYP/6–31 G (d,p).
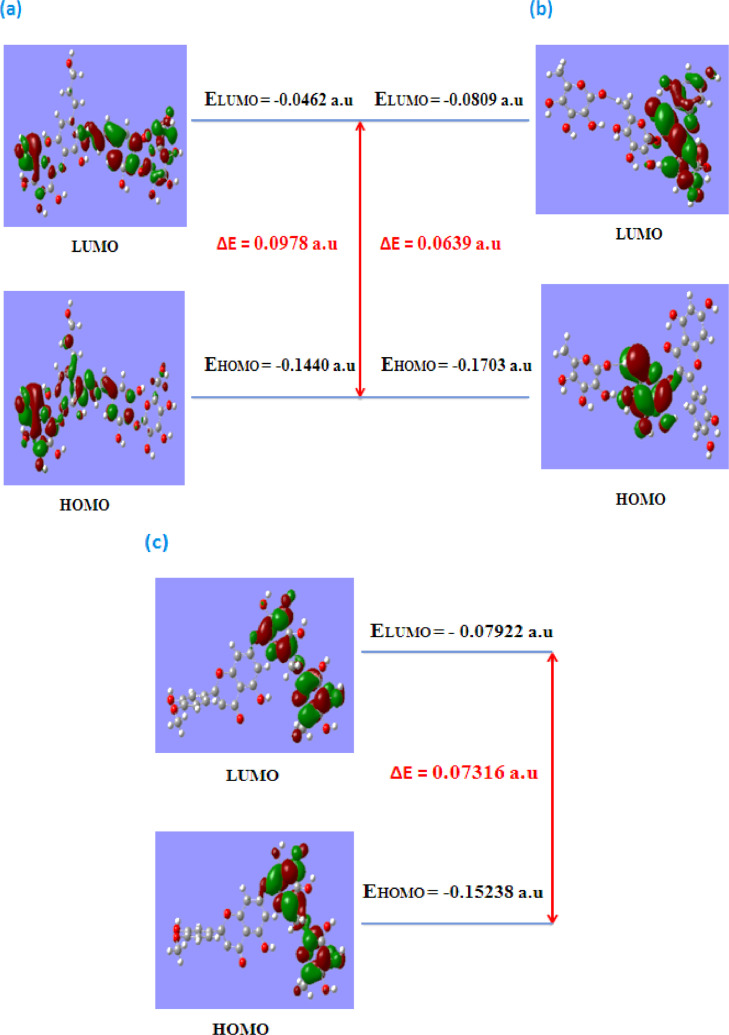
3.4.2. Frontier molecular orbital analysis
The distributions and energy levels of the HOMO and LUMO orbitals, computed with the B3LYP/6–31G(d,p) level for Tangshenoside III, Rutin and Hesperidin are shown in Fig. 2. To understand the relationship between electron delocalization and reactivity of studied compounds, density plots of the HOMO and LUMO composition of studied compounds can be used (Fig. 9 ). As shown in Fig. 9, the positive region is presented in red and the negative one in green to further determine the relationship between delocalization and reactivity, global descriptors were applied. Important factors used to describe inhibition activity of selected phytochemicals plant against two viral proteases, main protease (Mpro) and the Sars-CoV-2 B1.617.2 and ACE-2 are HOMO and LUMO energies and HOMO-LUMO energy gaps (Egap) [110]. The structure with the lower LUMO energy is a weak electron acceptor while the structure with the higher HOMO energy is a good electron donor [111]. The HOMO and LUMO energy values as well as Egap values of the studied compounds are shown in Table 6 .
Fig. 9.
Presentation of the energy levels, energy gaps, and frontier molecular orbitals of (a) Tangshenoside III, (b) Rutin and (c) Hesperidin.
Table 6.
Tangshenoside III, Rutin and Hesperidin geometrical parameters calculated in a.u at the B3LYP/6–31G(d,p) level of theory.
| Compound | HOMO | LUMO | Egap | IP | EA | µ | χ | η |
|---|---|---|---|---|---|---|---|---|
| Tangshenoside III | −0.1440 | −0.0462 | 0.0978 | 0.1440 | 0.0462 | −0.0951 | 0.0951 | 0.0489 |
| Rutin | −0.1703 | −0.0809 | 0.0639 | 0.1703 | 0.0809 | −0.1256 | 0.1256 | 0.0395 |
| Hesperidin | −0.15238 | −0.07922 | 0.07316 | 0.15238 | 0.07922 | −0.1158 | 0.1158 | 0.0365 |
ΔE= EHOMO-ELUMO, IP = -EHOMO, EA = -ELUMO, μ = 1/2(EHOMO + ELUMO), χ = - (ELUMO+EHOMO)/2, η = 1/2(ELUMO-EHOMO).
3.4.3. Global chemical reactivity
The optimized geometries obtained with B3LYP/6–31 G (d,p), were used for global chemical reactivity calculations. The reactivity parameters were calculated according to the approximations of energies EHOMO and ELUMO states, based on the optimization of the fundamental state geometry. The calculated parameters were: Energy Gap (Egap), ionization potential (I), electronic affinity (EA), chemical potential (μ), electronegativity (χ), and chemical hardness (η). The obtained results are shown in Table 6, in which is observed that Rutin presents a higher capacity to form anions due to the electronegativity value (χ), which represents a greater tendency to attract electrons. Regarding chemical hardness (η), Rutin shows a lower value compared to Tangshenoside III and Hesperidin; this parameteris indicative of the good interaction of this molecule with the surrounding environment. As the energy gap increases, the molecule becomes harder and more stable/less reactive [112,113]. Thus, there is short gap between HOMO and LUMO in the Rutin as compared to Tangshenoside III and Hesperidin. The values of electronic chemical potential of Tangshenoside III, Rutin and Hesperidin are presented in Table 6, ligands that have the greater electronic chemical potential (absolute values), was the less stable and more reactive. These results showed that the selected phytochemicals have effective reactivity, as they showed lower band gaps. The difference of the ELUMO and EHOMO was low, ranging from 0.0639 to 0.0978 a.u, implying the strong affinity of these inhibitors towards the target proteins. Among the three phytochemicals, Rutin exhibited higher reactivity against two viral proteases, main protease (Mpro) and the Sars-CoV-2 B1.617.2, and ACE-2 as the band energy gap was lowest among all the three phytochemicals, 0.0639 a.u (Table 6).
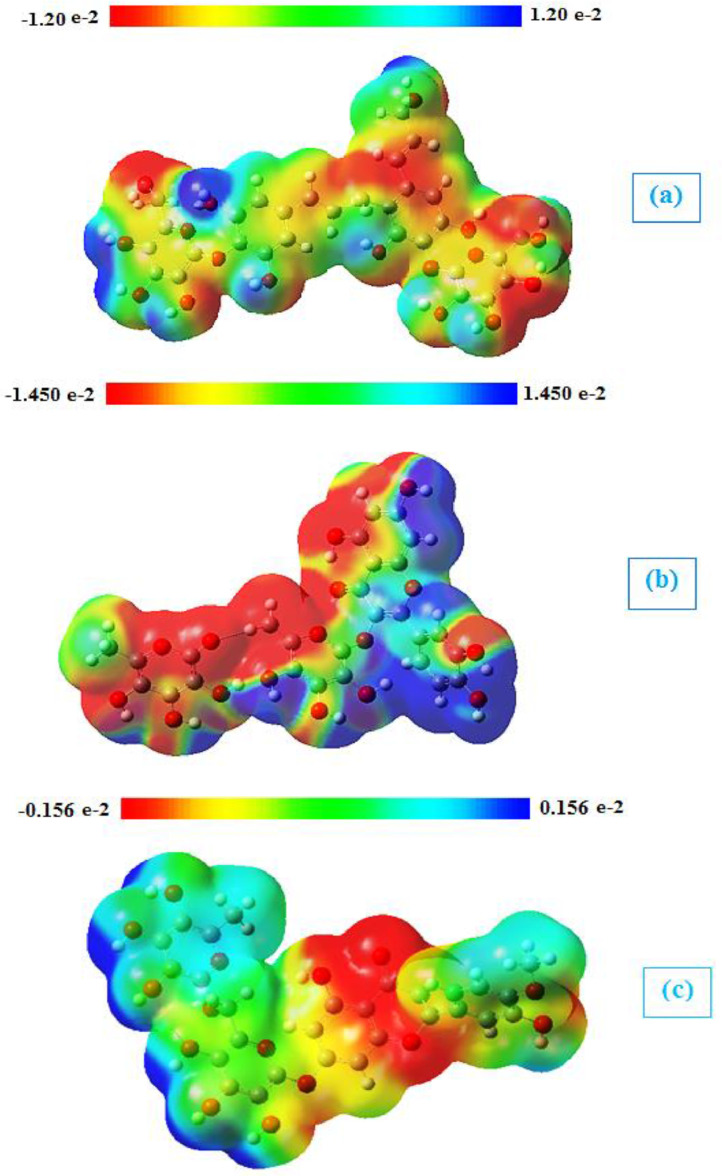
3.4.4. Molecular electrostatic potential (MEP)
The molecular electrostatic potential (MEP) can predict the reactive sites of electrophilic and nucleophilic attack in a molecule. These MEPs were realized to show the electron density within the Tangshenoside III, Rutin and Hesperidin on surfaces that surround the molecules, and the locations of sites for nucleophilic and electrophilic attacks [114,115]. Red indicates negative regions promoting electrophilic attack. Here, the negative potentials are generated over the electronegative oxygen atoms whereas the H-atoms have a positive potential region in the structures and the positive regions represented by the blue color are favorable sites for nucleophilic attack. The yellow regions indicate the slightly rich electron and the green regions indicate neutral. Molecular electrostatic potentials of Tangshenoside III, Rutin and Hesperidin were calculated using B3LYP/6–31 G (d,p) methodology. The surface maps shown in Fig. 10 .
Fig. 10.
Molecular electrostatic potential maps of (a) Tangshenoside III, (b) Rutin and (c) Hesperidin at the B3LYP/6–31G(d,p) level of theory.
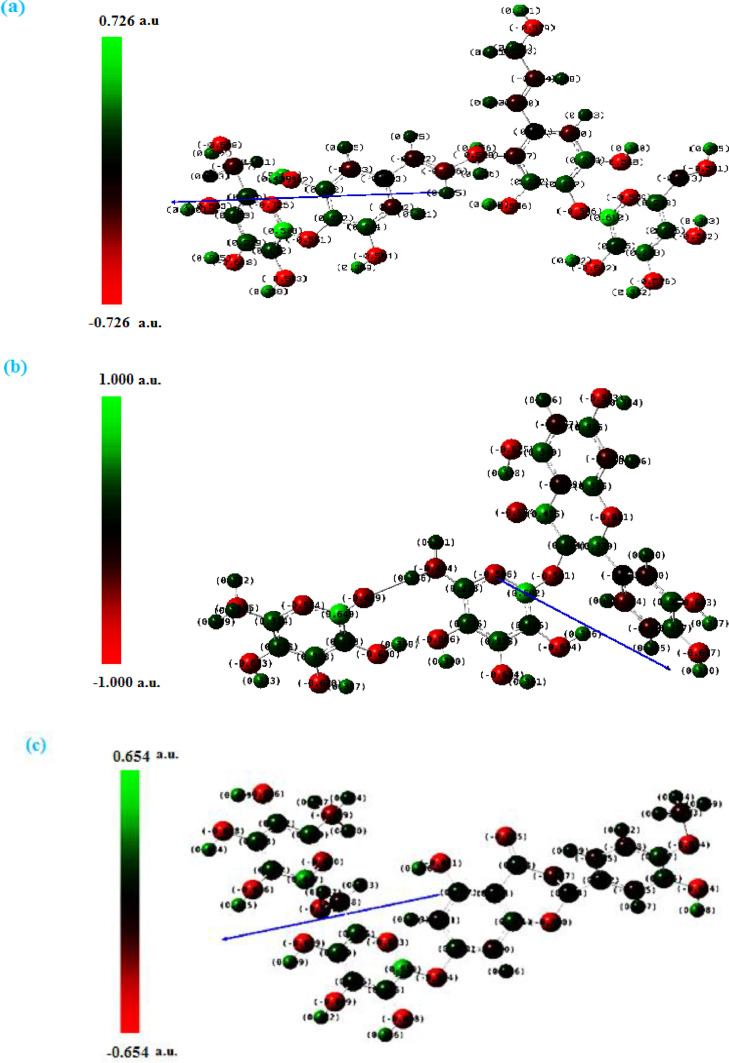
3.4.5. Mulliken atomic charges
The Mulliken atomic charges of Tangshenoside III, Rutin and Hesperidin were calculated by DFT using B3LYP/ 6–31 G (d,p) basis set, the data were tabulated in Table S3. It showed that the C5 is the most positive charge and O26 have the most negative charge for Tangshenoside III. On the other hand, it is observed that the most nucleophilic center of Rutin is O29 which is the most electrophilic susceptibility positions, it is obvious that the nucleophilic susceptibility of the Rutin is recognized on C21 and C27 sites. However, O42 is the most negative charge of Hesperidin and their respective positively charged atom is C22. The positively charged centers are the most susceptible sites for nucleophilic attacks, electron donation. However, the most negatively charged centers are the most susceptible sites for electrophilic one. The graphical representation for Mulliken atomic charges of the Tangshenoside III, Rutin and Hesperidin is shown in Fig. 11 . As it can be observed from this figure, the calculated atomic charges for the more negative atoms are the oxygen atoms while, the opposite case represented by the attached protons, are the most positive atoms. Again, this charge distribution supports the idea that the molecule donates its electrons through these groups.
Fig. 11.
Structures with color range Mulliken atomic charges of Tangshenoside III, Rutin and Hesperidin.
4. Previous studies
To the best of our knowledge, Tangshenoside III was not tested for SARS-CoV-2 inhibition or any other enzyme activity. However, previous studies provide a strong rationale for testing rutin and hesperidin in SARS-CoV-2.
Various pharmaceutical properties associated with rutin are anti-inflammatory [116], antitumor [117], antiulcer [118], antimalarial [119], antimicrobial [120], anticoagulant [121], nephroprotective [122], hepatoprotective [123]. Rutin was also found to decrease the infectivity of enteroviruses 19. It also showed effects on hepatic monooxygenase activities in experimental influenza virus infection (EIVI) [124]. Molecular docking studies suggested that rutin can act as a novel hepatitis B virus inhibitor [125]. This flavonoid has also been reported to inhibit a broad spectrum of viral proteases such as human norovirus protease 23 and NS3 serine protease, in vitro. A recent analysis, through in silico methods, suggested that the binding and therapeutic property makes the rutin a prominent lead to develop a potential inhibitor of SARS-CoV-2 Mpro, RdRp, PLpro, and S-protein to combat against COVID-19 pandemic [126].
Hesperidin is claimed to possess anti-oxidant as well as anti-inflammatory properties and has the ability to hinder SARS-CoV-2 entry and replication. It exhibited anti-viral activity against the influenza virus through a significant reduction of virus replication [127]. In addition, various studies on the inhibition effects of this compounds have reported. Sakata et al. (2003) indicated that hesperidin inhibits cyclooxygenase-2 (COX-2), inducible nitric oxide synthase (iNOS) proteins, which might be related to the anti-inflammatory and anti-tumorigenic efficacies [128]. It comes out as a multi-potent phytochemical agent in Alzheimer's disease therapeutics exhibiting strong proteases ßsecretase-1 binding ability (BACE1), high amyloid-ß peptide (Aß) aggregation inhibition, from a phytochemical library of 200 phytochemicals through the multi-target screening to identify multi-potent [129]. Furthermore, can block coronavirus from entering host cells through ACE2 receptors which can prevent the infection. Anti-viral activity of hesperidin might constitute a treatment option for COVID-19 through improving host cellular immunity against infection and its good anti-inflammatory activity may help in controlling cytokine storm [130].
5. Conclusion
SARS-CoV-2 spread rapidly, causing the disease known as COVID-19. Variants are characterized by multiple mutations in the surface spike (S) glycoprotein. These mutations in the RBD can change the ability of the virus spike protein to bind to and enter the host cell. The high rate of mutation and recombination in SARS-CoV2 makes it difficult for scientist to develop specific anti-CoV2 drugs and vaccines. In this workflow, we first short-listed 55 Saussurea costus and Saussurea involucrata -based phytochemicals through literature mining. The compounds were screened against the RBD of the SGP of SARS-CoV-2 Delta (B.1.617.2) Variant and the main protease (Mpro) to identify potent inhibitors that might be able to interfere with their catalytic function. The molecular docking studies identified the three ligands namely Tangshenoside III, Rutin and Hesperidin as potent inhibitors. Rutin performed very well when docked with the Sars-CoV-2 proteins producing the highest score with the spike B.1.617.2 and the second highest score with the Mpro Sars-CoV-2.
With these encouraging results, Tangshenoside III, Rutin and Hesperidin can be further explored for structural modification and detailed investigations to arrive at possibly newer potent agents with better anti SARS-CoV-2 Delta (B.1.617.2) Variant therapeutic activity. Accordingly, we recommend further fast in-vivo and in-vitro evaluations, preclinical and clinical studies at least for the aforementioned promising compound.
CRediT authorship contribution statement
Selma Houchi: Methodology, Software, Investigation, Supervision, Formal analysis, Writing – review & editing. Zakia Messasma: Conceptualization, Formal analysis, Writing – original draft.
Declaration of Competing Interest
The authors declare that they have no known competing finan- cial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors convey their hearty thanks to the Algerian Ministry of High Education for providing facilities to carry out the research work.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2022.133032.
Appendix. Supplementary materials
References
- 1.Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Who . WHO; 2021. WHO Coronavirus Disease (COVID-19) Dashboard.https://covid19.who.int/table Accessed 03.18.2021. [Google Scholar]
- 3.Worldometers.info. Available from: (https://www.worldometers.info/coronavirus) COVID-19 Coronavirus pandemic [Internet]. August 1st, 2021.
- 4.Wall E.C., et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet. 2021;397(10292):2331–2333. doi: 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 6.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An Overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy. J. Med. Chem. 2016;59(14):6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis fordesign of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey A., Nitin A., Basavaraj A., Mutalik S.P., Prassl R. Potential therapeutic targets for combating SARS-CoV-2: drug repurposing, clinical trials and recent advancements. Life Sci. 2020;256 doi: 10.1016/j.lfs.2020.117883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C., Abiona O., Graham B.S., Mclellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;1263:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schachter S.C. Complementary and alternative medical therapies. Curr. Opin. Neurol. 2008;21(2):184–189. doi: 10.1097/WCO.0b013e3282f47918. [DOI] [PubMed] [Google Scholar]
- 12.Sundriyal A., Bijjem K.R., Kalia A.N. Antiepileptic potential of Anisomeles indica (Linn.) Kuntze aerial parts in pentylenetetrazole-induced experimental convulsions in Wistar rats. Indian J. Exp. Biol. 2013;51(9):715–720. [PubMed] [Google Scholar]
- 13.Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T., Rizzuti B., Velazquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prateeksha G., Rana T.S., Asthana A.K., Singh B.N., Barik S.K. Screening of cryptogamic secondary metabolites as putative inhibitors of SARS-CoV-2 main protease and ribosomal binding domain of spike glycoprotein by molecular docking and molecular dynamics approaches. J. Mol. Struc. 2021;1240 doi: 10.1016/j.molstruc.2021.130506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu R., Chen L., Lan R., Shen R., Li P. International Journal of Antimicro- bial agents computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int. J. Antimicrob. Agents. 2020;56(2) doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwari G., Bhandari U., Andola H.C., Lohani H., Chauhan N. Volatile constituents of Saussurea costus roots cultivated in Uttarakhand Himalayas, India. Pharmacognosy Res. 2013;5:3. doi: 10.4103/0974-8490.112424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X.Y., Zhao D.X., Fu C.X., Cheng L.Q., Wang N.F., Han L.J., Ma F.S. Determination of flavonoid compounds from Saussurea involucrata by liquid chromatography electrospray ionization mass spectrometry. Nat. Prod. Res. 2009;23:1689–1698. doi: 10.1080/14786410802187742. [DOI] [PubMed] [Google Scholar]
- 18.Chik W.I., Zhu L., Fan L.L, Yi T., Zhu G.Y., Gou X.J., Tang Y.N., J.Xu W.P.Y., Zhao Z.Z., Yu Z.L., Chen H.B. Saussurea involucrata: a review of the botany, phytochemistry and ethnopharmacology of a rare traditional herbal medicine. J. Ethnopharmacol. 2015;172:44–60. doi: 10.1016/j.jep.2015.06.033. [DOI] [PubMed] [Google Scholar]
- 19.Shih InC., Jin S.Y. Flora of China. FRPS. 1999;78(2):57–66. [Google Scholar]
- 20.Cho J.Y., Baik K.U., Jung J.H., Park M.H. In vitro anti-inflammatory effects of cynaropicrin, a sesquiterpene lactone, from Saussurea lappa. Eur. J. Pharmacol. 2000;398:399–407. doi: 10.1016/s0014-2999(00)00337-x. [DOI] [PubMed] [Google Scholar]
- 21.Yaeesh S., Jamal Q., Shah A.J., Gilani A.H. Antihepatotoxic activity of Saussurea lappa extract on d-galactosamine and lipopolysaccharide-induced hepatitis in mice. Phytother. Res. 2010;24:S229–S232. doi: 10.1002/ptr.3089. [DOI] [PubMed] [Google Scholar]
- 22.Li Y., Xu C., Zhang Q., Liu J.Y., Tan R.X. In vitro anti-helicobacter pylori action of 30 Chinese herbal medicines used to treat ulcer diseases. J. Ethnopharmacol. 2005;98:329–333. doi: 10.1016/j.jep.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 23.Kim E.J., Hong J.E., Lim S.S., Kwon G.T., Kim J., Kim J.S., Lee K.W., Park J.H.Y. The hexane extract of Saussurea lappa and its active principle, dehydrocostus lactone, inhibit prostate cancer cell migration. J. Med. Food. 2012;15:24–32. doi: 10.1089/jmf.2011.1735. [DOI] [PubMed] [Google Scholar]
- 24.Patil S.M., Sapkale G.N., Umbrae R.P., Patil M.B. Evaluation of wound healing properties of Saussurea lappa Clarke root extracts. Indian J. Pharm. Sci. 2009;71:210. [Google Scholar]
- 25.Rao K.S., Babu G.V., Ramnareddy Y.V. Acylated flavone glycosides from the roots of Saussurea lappa and their antifungal activity. Molecules. 2007;12:328–344. doi: 10.3390/12030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambavade S.D., Mhetre N.A., Muthal A.P., Bodhankar S.L. Pharmacological evaluation of anticonvulsant activity of root extract of Saussurea lappa in mice. Eur. J. Integr. Med. 2009;1:131–137. [Google Scholar]
- 27.Liu Z.L., He Q., Chu S.S., Wang C.F., Du S.S., Deng Z.W. Essential oil composition and larvicidal activity of Saussurea lappa roots against the mosquito Aedes albopictus (Diptera: culicidae) Parasitol. Res. 2012;110:2125–2130. doi: 10.1007/s00436-011-2738-0. [DOI] [PubMed] [Google Scholar]
- 28.Choi J.Y., Na M.K., Hwang I.H., Lee S.H., Bae E.Y., Kim B.Y., Ahn J.S. Isolation of betulinic acid, its methyl ester and guaiane sesquiterpenoids with protein tyrosine phosphatase 1B inhibitory activity from the roots of Saussurea lappa C.B.Clarke. Molecules. 2011;14:266–272. doi: 10.3390/molecules14010266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwashina T., Smirnov S.V., Damdinsuren O., Kondo K. Saussurea species from the Altai mountain sandadja centarea, and their flavonoid diversity. Bull. Natl. Mus. Nat. Sci. Ser. B. 2010;36:141–154. [Google Scholar]
- 30.The plant list, Version 1.1,2013.URL 〈 http://www.theplantlist.org 〉.
- 31.Zhai K.F., Duan H., Xing J.G., Huang H. Study on the antiinflammatory and analgesic effects of various purified parts from Saussurea involucrata. Chin. J. Hosp. Pharm. 2010;5:374–377. [Google Scholar]
- 32.Jia J.M., Wu C.F. Effect of Saussurea involucrata culture on mouse immune function. China J. Tradit. Chin. Med. Pharm. 2007;4:238–240. [Google Scholar]
- 33.Qiu J., Xue X., Chen F., Li C., Bolat N., Wang X., Baima Y., Zhao Q., Zhao D., Ma F. Quality evaluation of snow lotus (Saussurea): quantitative chemical analysis and antioxidant activity assessment. PlantCellRep. 2010;29:1325–1337. doi: 10.1007/s00299-010-0919-4. [DOI] [PubMed] [Google Scholar]
- 34.Ma H.P., Fan P.C., Jing L.L., Yao J., He X.R., Yang Y., Chen K.M., Jia Z.P. Anti- hypoxicactivity at simulated high altitude was isolated in petroleum ether extract of Saussurea involucrata. J. Ethnopharmacol. 2012;137:1510–1515. doi: 10.1016/j.jep.2011.08.037. [DOI] [PubMed] [Google Scholar]
- 35.Baima J.C. Tibetan preparatory medicine and preparation method thereof. Chin. Pat. 1997 CN 1161849A. [Google Scholar]
- 36.Hu C.G. Chinese medicinal composition for treatment of trachitis and gastritis. Chin. Pat. 1996 CN 1129134A. [Google Scholar]
- 37.Morgan S., Grootendorst P., Lexchin J., Cunningham C., Greyson D. The cost of drug development: a systematic review. Health Policy. 2011;100:4–17. doi: 10.1016/j.healthpol.2010.12.002. New York. [DOI] [PubMed] [Google Scholar]
- 38.Bai Q., Shao Y., Pan D., Zhang Y., Liu H., Yao X. Search for b2 adrenergic receptor ligands by virtual screening via grid computing and investigation of binding modes by docking and molecular dynamics simulations. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Govindan S.V., Bhattacharaya S.C. Alantolides and cyclocostunolides from Saussurea lappa. Indian J. Chem. 1977;15:956. [Google Scholar]
- 40.Yang J.S., Xie F.Z., Liu Q.H., Wu X. Studies on coumarins of Saussurea involucrata Kar.et. Kir. Chin. Pharm. J. 2006;41:1774–1776. [Google Scholar]
- 41.Yang H.J., Kim M.J., Kang S., Moon N.R., Kim D.S., Leed N.R., Kim K.S., Parkc S. Topical treatments of Saussurea costus root and Thuja orientalis L. synergistically alleviate atopic dermatitis-like skin lesions by inhibiting protease-activated receptor-2 and NF-κB signaling in HaCaT cells and Nc/Nga mice. J. Ethnopharmacol. 2017;199:97–105. doi: 10.1016/j.jep.2017.01.055. [DOI] [PubMed] [Google Scholar]
- 42.T. Julianti, Y. Hata, S. Zimmermann, M. Kaiser, M. Hamburger, M. Adams. Antitrypanosomal sesquiterpene lactones from Saussurea costus Fitoterapia. (2011). Fitoterapia 82 (7), 955-959 Fitoterapia. 82 (2011) 955–959. [DOI] [PubMed]
- 43.Li J.R., Liu L., Liu Y.Q., Sa A.J. Determination of arctiin in seeds of Saussurea involucrata by HPLC. Lishizheng Med. Mater. Med. Res. 2012;23:2119–2120. [Google Scholar]
- 44.Rao A.S., Kelkar G.R., Bhattacharyya S.C. Terpenoids—XXI, the structure of costunolide, a new sesquiterpene lactone from costus root oil. Tetrahedron. 1960;9:275–283. [Google Scholar]
- 45.Yang H., Xie J., Sun H. Study on chemical constituents of Saussurea lappa II. Acta Bot. Yunnanica. 1997;19:92–96. [Google Scholar]
- 46.Jia Z.J., Li Y., Du M., Zhu Z.Q. Studies on the constituents of Saussurea nvolucrata Kar. et. Kir. Chem. J. Chin. Univ. 1983;4:581–584. [Google Scholar]
- 47.Wu W., Qu Y., Gao H.Y., Yang J.Y., Xu J.G., Wu L.J. Novelceramidesfrom aerial partsof Saussurea involucrata Kar.et.Kir.Arch. Pharm. Res. 2009;32:1221–1225. doi: 10.1007/s12272-009-1906-6. [DOI] [PubMed] [Google Scholar]
- 48.Li Y.M., Zhong H. Simultaneous determination of five flavonoidsin Saussurea involucrata by capillaryelectrophoresis. J. Chem. Soc. Pak. 2013;35:1288–1292. [Google Scholar]
- 49.Kusano K., Iwashina T., Kitajima J., Mishio T. Flavonoid diversity of Saussurea and Serratula species in Tien Shan Mountains. Nat. Prod. Commun. 2007;2:1121–1128. [Google Scholar]
- 50.Yi T., Chen H.B., Zhao Z.Z., Jiang Z.H., Cai S.Q., Wang T.M. Identification and determination of the major constituents in the traditional Uighur medicinal plant Saussurea involucrata by LC-DAD-MS. Chromatographia. 2009;69:537–542. [Google Scholar]
- 51.Chen R.D., Zou J.H., Jia J.M., Dai J.G. Chemical constituents from the cell cultures of Saussurea involucrata. J. Asian Nat. Prod. Res. 2010;12:119–123. doi: 10.1080/10286020903482810. [DOI] [PubMed] [Google Scholar]
- 52.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 53.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G., Nakatsuji H., Caricato M., Li X., Hratchian H.P., Izmaylov A.F., Bloino J., Zheng G., Sonnenberg J.L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J.A., Peralta J.E., Ogliaro F., Bearpark M., Heyd J.J., E B., Kudin K.N., Staroverov V.N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Rega N., Millam J.M., Klene M., Knox J.E., Cross J.B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Martin R.L., Morokuma K., Zakrzewski V.J., Voth G.A., Salvador P., Dannenberg J.J., Dapprich S., Daniels A.D., Farkas O., Foresman J.B., Ortiz J.V., Cioslowski J., Fox D.J. Gaussian, Inc.; Wallingford, CT: 2009. D. 0109, Revision D. 01. [Google Scholar]
- 54.Dennington R.D., Keith T.A., Millam J.M. Gaussian Inc; 2008. Gauss View 5.0.8. [Google Scholar]
- 55.Peng C., Ayala P.Y., Schlegel H.B., Frisch M.J. Using redundant internal coordinates to optimize equilibrium geometries and transition states. J. Comput. Chem. 1996;17:49–56. [Google Scholar]
- 56.Stephens P.J., Devlin F.J., Chablowski C.F., Frisch M.J. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 1994;98:11623–11627. [Google Scholar]
- 57.Frisch M.J., Pople J.A., Binkley J.S. J. Chem. Phys. 1984;80:3265–3269. [Google Scholar]
- 58.Kirtikar K.R., Basu B.D. International Book Distributors; Derhadun: 1987. Indian Medicinal Plants; p. 915. [Google Scholar]
- 59.Zahara K., Tabassum S., Sabir S., et al. A review of therapeutic potential of Saussurea lappa-An endangered plant from Himalaya. Asian Pac J. Trop. Med. 2014;7(S1):S60–S69. doi: 10.1016/S1995-7645(14)60204-2. [DOI] [PubMed] [Google Scholar]
- 60.Dhar G.H., Virjee J., Kachroo P., et al. Ethnobotany of Kashmir—I. Sind valley. J. Econ. Taxon. Bot. 1984;5:668–675. [Google Scholar]
- 61.Prawiro S.R., Anam K., Prabowo B., bramanthi R., Fitrianingsih A.A., Hidayati D.Y.N., Imawati S., Fitria E., Winarsih A.S. Generating the responses immune with honey, Saussurea costus, and nigella sativa in cellular and humoral may resolve COVID-19? SRP. 2021;12(1):1588–1593. [Google Scholar]
- 62.Chen H.C., Chou C.K., Lee S.D., Wang J.C., Yeh S.F. Active compounds from Saussurea lappa Clarks that suppress hepatitis B virus surface antigen gene expression in human hepatoma cells. Antivir. Res. 1995;27(1–2):99–109. doi: 10.1016/0166-3542(94)00083-k. [DOI] [PubMed] [Google Scholar]
- 63.Jia Z., He K., Du M., Li Y., Chu T.T. Studies on the constituents of Saussurea involucrata Kar. et Kin. (IV) Chem. J. Chin. Univ. 1988;9:198–200. [Google Scholar]
- 64.Rakib A., Paul A., Chy M., Sami S.A., Baral S.K., Majumder S.M., Tareq A.M., Amin M.N., Shahriar A., Uddin M.Z., Dutta M., Tallei T.E., Emran T.B., Simal-Gandara J. Biochemical and computational approach of selected phytocompounds from Tinospora crispa in the management of COVID-19. Molecules. 2020;25:3936. doi: 10.3390/molecules25173936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jorgensen W.L. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 66.Kontoyianni M., McClellan L.M., Sokol G.S. Evaluation of docking performance: comparative data on docking algorithms. J. Med. Chem. 2004;47:558–565. doi: 10.1021/jm0302997. [DOI] [PubMed] [Google Scholar]
- 67.Tahir M., Qamar U.L., et al. Structural basis of SARS-CoV-2 3CL(pro) and anti- COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020;10:313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang H., et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X., Wang X.J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J. Genet. Genom. 2020;47:119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan S.A., Zia K., Ashraf S., Uddin R., Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2021;39(7):2607–2616. doi: 10.1080/07391102.2020.1751298. [DOI] [PubMed] [Google Scholar]
- 71.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Identification of polyphenols from Broussonetia papyrifera as SARS CoV-2 main protease inhibitors using in silico docking and molecular dynamics simulation approaches. J. Biomol. Struct. Dyn. 2021;39(17):16747–16760. doi: 10.1080/07391102.2020.1802347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choudhary M.I., Shaikh M., Tul-Wahab A., Ur-Rahman A. In silico identification of potential inhibitors of key SARS-CoV-2 3CL hydrolase (Mpro) via molecular docking, MMGBSA predictive binding energy calculations, and molecular dynamics simulation. PLoS One. 2020;15 doi: 10.1371/journal.pone.0235030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cherrak S.A., Merzouk H., Mokhtari-Soulimane N. Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: a molecular docking and simulation studies. PLoS One. 2020;15(10) doi: 10.1371/journal.pone.0240653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu Z., Yang L., Zhang X., et al. Discovery of potential flavonoid inhibitors against COVID-19 3CL proteinase based on virtual screening strategy. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.556481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin C.W., Tsai F.J., Tsai C.H., Lai C.C., Wan L., Ho T.Y., et al. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005;68(1):36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Utomo R.Y., Ikawati M., Meiyanto E. Revealing the potency of citrus andgalangal constituents to halt SARS-CoV-2 infection. Prepr. Org. 2020 doi: 10.20944/preprints202003.0214.v1. [DOI] [Google Scholar]
- 77.Chen H., Du Q. Potential natural compounds for preventing SARS-CoV-2 (2019-nCoV) infection. Preprints. 2020 doi: 10.20944/preprints202001.0358.v3. 2020010358. [DOI] [Google Scholar]
- 78.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rameshkumar M.R., Indu P., Arunagirinathan N., Venkatadri B., El-Serehy H.A., Ahmad A. Computational selection of flavonoid compounds as inhibitors against SARS-CoV-2 main protease, RNA-dependent RNA polymerase and spike proteins: a molecular docking study. Saudi J. Biol. Sci. 2021;28(1):448–458. doi: 10.1016/j.sjbs.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.T.E. Tallei, S.G. Tumilaar, N.J. Niode, B.J. Fatimawali, Kepel, R. Idroes et al., 2020. Potential of Plant Bioactive Compounds as SARS-CoV-2 Main Protease (M) and Spike (S) Glycoprotein Inhibitors: a Molecular Docking Study. Scientifica (Cairo); 2020:6307457. [DOI] [PMC free article] [PubMed]
- 81.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: gISAID's innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2013;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanches P.R.S., Charlie-Silva I., Braz H.L.B., Bittar C., Calmon M.F., Rahal P., Cilli E.M. Recent advances in SARS-CoV-2 Spike protein and RBD mutations comparison between new variants Alpha (B.1.1.7, United Kingdom), Beta (B.1.351, South Africa), Gamma (P.1, Brazil) and Delta (B.1.617.2, India) J. Virus Erad. 2021;7 doi: 10.1016/j.jve.2021.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., et al. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Qand P681R, in the second wave of COVID-19 in Maharashtra, India. BioRxiv. 2021;9(7):1542. doi: 10.3390/microorganisms9071542. 04.22.440932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Solo P., Doss M.A. Potential inhibitors of SARS-CoV-2 (COVID 19) spike protein of the delta and delta plus variant: in silico studies of medicinal plants of North-East India. Curr. Res. Pharmacol. Drug Discov. 2021;2 doi: 10.1016/j.crphar.2021.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bonavia A., Zelus B.D., Wentworth D.E., Talbot P.J., Holmes K.V. Identification ofa receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 2003;77(4):2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bao L., Deng W., Huang B., Gao H., Liu J., Ren L. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 89.Ou X., LiuY Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.J.H. Kuhn, S.R. Radoshitzky, W. Li, S.K. Wong, H. Choe, M. Farzan. The SARS Coronavirus receptor ACE-2 A potential target for antiviral therapy. In: New Concepts of Antiviral Therapy. Springer, (2006): 397–418.
- 91.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., et al. Evolution of the novelcoronavirus from the ongoing Wuhan outbreak and modeling of its spike protein forrisk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drugrepurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2spike protein in the interaction with the human ACE2 receptor: an in silicoanalysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020;10:20248640. [Google Scholar]
- 95.Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021;236:7045–7057. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains ofSARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2020.26.1.2002106. Bull. Eur. Mal. Transm. Eur. Commun. Dis. Bull. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith M., Smith J.C. Repurposing therapeutics for covid-19: supercomputerbased docking to the sars-cov-2 viral spike protein and viral spike protein-humanace2 interface. ChemRxiv. 2020:1–15. doi: 10.26434/chemrxiv.12725465. [DOI] [Google Scholar]
- 98.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Souza P.F.N., Lopes F.E.S., Amaral J.L., Freitas C.D.T., Oliveira J.T.A. A moleculardocking study revealed that synthetic peptides induced conformational changes inthe structure of SARS-CoV-2 spike glycoprotein, disrupting the interaction withhuman ACE2 receptor. Int. J. Biol. Macromol. 2020;164:66–76. doi: 10.1016/j.ijbiomac.2020.07.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Br B., Damle H., Ganju S., Damle L. In silico screening of known small molecules to bind ACE2 specific RBD on Spike glycoprotein of SARS-CoV-2 for repurposingagainst COVID-19. F1000Res. 2020;9:663. doi: 10.12688/f1000research.24143.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Prajapat M., Shekhar N., Sarma P., Avti P., Singh S., Kaur H., Bhattacharyya A., Kumar S., Sharma S., Prakash A., Medhi B. Virtual screening and moleculardynamics study of approved drugs as inhibitors of spike protein S1 domain andACE2 interaction in SARS-CoV-2. J. Mol. Graph. Model. 2020;101 doi: 10.1016/j.jmgm.2020.107716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mady F.M., Sarhan H.A., Rateb H.S., et al. Optimization and Evaluation of propolis liposomes as a promising therapeutic approach for COVID-19. Int. J. Pharm. 2020;592 doi: 10.1016/j.ijpharm.2020.120028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Güler H.I., Tatar G., Yildiz O., Belduz A.O., Kolayli S. Investigation of potential inhibitor properties of ethanolic propolis extracts against ACE-II receptors for COVID-19 treatment by molecular docking study. Arch. Microbiol. 2021 doi: 10.1007/s00203-021-02351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gupta S., Singh A.K., Kushwaha P.P., et al. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 2021;9(12):4334–4345. doi: 10.1080/07391102.2020.1776157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Prinsa R.N., Normi D.G., Tejas M.D. In search of SARS CoV-2 replication inhibitors: virtual screening,molecular dynamics simulations and ADMET analysis. J. Mol. Struc. 2021;1246 doi: 10.1016/j.molstruc.2021.131190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prateeksha G., Rana T.S., Asthana A.K., Singh B.N., Barik S.K. Screening of cryptogamic secondary metabolites as putative inhibitors of SARS-CoV-2 main protease and ribosomal binding domain of spike glycoprotein by molecular docking and molecular dynamics approaches. J. Mol. Struc. 2021;1240 doi: 10.1016/j.molstruc.2021.130506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Drugbank Database Site: https://go.drugbank.com/
- 108.Sayed A.M., Alhadrami H.A., El-Gendy A.O., Shamikh Y.I., Belbahri L., Hassan H.M., Abdelmohsen U.R., Rateb M.E. Microbial natural products as potential inhibitors of SARS-CoV-2 main protease (Mpro) Microorganisms. 2020;8:970. doi: 10.3390/microorganisms8070970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Topkat Version 6.2. Accelrys. http://www.accelrys.com/products/topkat/
- 110.Rajan V.K., Muraleedharann K. A computational investigationon the structure, global parameters and antioxidant capacity of a polyphenol, Gallic acid. Food Chem. 2017;220:93–99. doi: 10.1016/j.foodchem.2016.09.178. [DOI] [PubMed] [Google Scholar]
- 111.Veselinović J.B., Veselinović A.M., Vitnik Ž.J., et al. Antioxidant properties of selected 4-phenyl hydroxycoumarins: integrated in vitro and computational studies. Chem. Biol. Interact. 2014;214:49–56. doi: 10.1016/j.cbi.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 112.Fukui F.T., Yonezawa F., Nagata C., Shingu H. Molecular orbital theory of orientation inaromatic, heteroaromatic, and other conjugated molecules. J. Chem. Phys. 1954;20:1433–1441. [Google Scholar]
- 113.Zhou Z., Parr R.G. Activation hardness: new index for describing the orientation of electrophilic aromatic substitution. J. Am. Chem. Soc. 1990;112:5720–5724. [Google Scholar]
- 114.Sudhir M., Hiremath S., Chidanandayya S.H, Mahantesha B., et al. Structural, spectroscopic characterization of 2-(5-methyl-1-benzofuran-3-yl) acetic acid in monomer, dimer and identification of specific reactive, drug likeness properties: experimental and computational study. J. Mol. Struc. 2016;1178:1–14. [Google Scholar]
- 115.Rublova L., Zarychta B., Olijnyk V., Mykhalichko B. Synthesis, crystal and molecular-electronic structure, and kinetic investigation of two new sterically hindered isomeric forms of the dimethyl[methyl(phenylsulfonylamino] benzensulfony chloride. J. Mol. Struc. 2017;214:1–8. [Google Scholar]
- 116.Guardia T., Rotelli A.E., Juarez A.O., Pelzer L.E. Antiinflammatory properties of plant flavonoids. Effects of Rutin, quercetin and hesperidin on adjuvant arthritis in rat. Farmaco. 2001;56(9):683–687. doi: 10.1016/s0014-827x(01)01111-9. [DOI] [PubMed] [Google Scholar]
- 117.Alonso-Castro A.J., Domínguez F., García-Carrancá A. Rutin exerts antitumor effects on nude mice bearing SW480 tumor. Arch. Med. Res. 2013;44(5):346–351. doi: 10.1016/j.arcmed.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 118.Dubey S., Ganeshpurkar A., Shrivastava A., Bansal D., Dubey N. Rutin exerts antiulcer effect by inhibiting the gastric proton pump. Indian J. Pharmacol. 2013;45(4):415–417. doi: 10.4103/0253-7613.115011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Silveira P., Vashist U., Cabral A., Amaral K.B., Soares G.L.G., Dagosto M. Effect of Rutin and chloroquine on white leghorn chickens infected with Plasmodium (Bennettinia) juxtanucleare. Trop. Anim. Health Prod. 2009;41(7):1319–1323. doi: 10.1007/s11250-009-9317-8. [DOI] [PubMed] [Google Scholar]
- 120.Soni H., Malik J., Singhai A.K., Sharma S. Antimicrobial and antiinflammatory activity of the hydrogels containing Rutin delivery. Asian J. Chem. 2013;25(15):8371–8373. [Google Scholar]
- 121.Jasuja R., Passam F.H., Kennedy D.R., et al. Protein disulfideisomerase inhibitors constitute a new class of antithrombotic agents. J. Clin. Invest. 2012;122(6):2104–2113. doi: 10.1172/JCI61228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kandemir F.M., Ozkaraca M., Yildirim B.A., et al. Rutin attenuatesgentamicin-induced renal damage by reducing oxidative stress, inflammation, apoptosis, and autophagy in rats. Ren Fail. 2015;37(3):518–525. doi: 10.3109/0886022X.2015.1006100. [DOI] [PubMed] [Google Scholar]
- 123.Janbaz K.H., Saeed S.A., Gilani A.H. Protective effect of Rutin on paracetamol- and CCl4-induced hepatotoxicity in rodents. Fitoterapia. 2002;73(7–8):557–563. doi: 10.1016/s0367-326x(02)00217-4. [DOI] [PubMed] [Google Scholar]
- 124.Savov V.M., Galabov A.S., Tantcheva L.P., et al. Effects of Rutin and quercetin on monooxygenase activities in experimental influenza virus infection. Exp. Toxicol. Pathol. 2006;58(1):59–64. doi: 10.1016/j.etp.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 125.Parvez M.K., Tabish Rehman M., Alam P., Al-Dosari M.S., Alqasoumi S.I., Alajmi M.F. Plant-derived antiviral drugs as novel hepatitis B virus inhibitors: cell culture and molecular docking study. Saudi Pharm. J. 2019;27(3):389–400. doi: 10.1016/j.jsps.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rahman F., Tabrez S., Ali R., Alqahtani A.S., Ahmed M.Z., Rub A. Molecular docking analysis of Rutin reveals possible inhibition ofSARS-CoV-2 vital proteins. J. Tradit. Complement Med. 2021;11:173–179. doi: 10.1016/j.jtcme.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dong W., Wei X., Zhang F., Hao J., Huang F., Zhang C.A. dual character of flavonoids in influenza A virus replication and spread through modulating cell-autonomous immunity by MAPK signaling pathways. Sci. Rep. 2014;4:7237. doi: 10.1038/srep07237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chakraborty J.B, Chakraborty S., Basu S. Multi-target screening mines hesperidin as a multi-potent inhibitor: implication in Alzheimer's disease therapeutics. Eur. J. Med. Chem. 2016;121:810–822. doi: 10.1016/j.ejmech.2016.03.057. European Journal of Medicinal Chemistry. [DOI] [PubMed] [Google Scholar]
- 129.Sakataa, Hirosea Y., Qiaoa Z., Tanakab T., Mori H. Inhibition of inducible isoforms of cyclooxygenase and nitric oxide synthase by flavonoid hesperidin in mouse macrophage cell line. Cancer Lett. 2003;199:139–145. doi: 10.1016/s0304-3835(03)00386-0. [DOI] [PubMed] [Google Scholar]
- 130.Haggag Y.A., El-Ashmawy N.E., Okasha K.M. Is hesperidin essential for prophylaxis and treatment of COVID-19 Infection? Med. Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.109957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.