Abstract
The cluster of differentiation 36 (CD36) is one of the main receptors implicated in the pathogenesis of the cardiovascular disease. This study aimed to assess the association between CD36 rs1761667 polymorphism and cardiometabolic risk factors including body mass index (BMI), waist circumference (WC), total cholesterol (TC), triglyceride, HDL-C, LDL-C, blood pressure and fasting blood glucose (FBG). PubMed, EMBASE, Scopus, web of science, and Google Scholar were searched up to December 2021. Subgroup and meta-regression analyses were conducted to explore sources of heterogeneity. Eighteen eligible studies (6317 participants) were included in the study. In the overall analysis, a significant association was found between rs1761667 polymorphism of CD36 and TG in allelic (p < 0.001), recessive (p = 0.001) and homozygous (p = 0.006) models. A relationship between this polymorphism and HDL-C and FBG level was observed in the recessive genetic model. In the subgroup analysis, the A allele was associated with impaired lipid profiles (TC, LDL-C and HDL-C) in the Asian population. The influences of health status, design of the study, confounders, and other sources of heterogeneity should be considered when interpreting present findings. Cohort studies with large sample size and in different ethnicities are needed to confirm the relationship between rs1761667 SNP and cardiometabolic risk factors.
Subject terms: Genetics, Cardiology
Introduction
Cardiovascular diseases (CVD) are the most life-threatening conditions which have negative impacts on development and economic growth. Cardiometabolic risk factors including obesity, dyslipidemia, dysglycemia, and elevated blood pressure, increase the risk of cardiometabolic diseases. Impaired cardiometabolic risk factors can be present for years before the clinical symptoms become apparent; therefore, their management is difficult for physicians1. It has been demonstrated that genetic factors play a role in the development of cardiometabolic risk independent of environmental factors2. The cluster of differentiation 36 (CD36), also known as platelet glycoprotein IV or IIIb, is one of the most important membrane proteins presents on the surface of a wide variety of cell types including adipocytes, macrophages, skeletal and cardiac myocytes, hepatocytes, microvascular endothelial cells, breast, kidney, platelets, microvascular endothelial cells, and epithelial cells in the gut3. It has been shown that CD36 plays a role in inflammatory reactions, angiogenesis, orosensory perception of dietary lipid and fat preference, regulating the metabolic pathways of insulin-resistance, transporting long-chain fatty acids (LCFA) into adipose and muscle tissues, chylomicron synthesis, and energy metabolism4. As a consequence of its functions, CD36 might be associated with a wide range of disorders such as CVD, dyslipidemia, hypertension, diabetes, metabolic syndrome, and cancer5,6. A genome-wide association study (GWAS) of the four large cohorts (19,602 white people in whom 1544 cases of stroke) showed that CD36 rs3211928 was significantly associated with stroke. Several studies have been conducted on the association between single-nucleotide polymorphisms (SNPs) in the CD36 gene including rs1761667 (A/G substitution) with CVD7,8, type 2 diabetes mellitus (T2DM)9, consumption of total fat and fat taste perception, obesity10, and metabolic syndrome11. These studies have been performed in various ethnic populations around the world. However, ambivalent results were obtained regarding the association between genotype distribution of rs1761667 and cardiometabolic risk factors. For instance, Bayoumy et al.11 demonstrated that individuals with AA genotype of the CD36 rs1761667 had a significantly lower degree of dyslipidemia, systolic blood pressure (SBP), and waist circumstance (WC) compared to individuals with AG and GG genotype. Boghdady et al.8 also showed that the AG genotype may be involved in the pathogenesis of coronary artery disease, raised body mass index (BMI), metabolic syndrome, and T2DM. On the other hand, Pioltine et al.12 reported that the SNP rs1761667 in the CD36 gene was not associated with obesity risk.
To the best of our knowledge, there is no systematic review and meta-analysis trying to examine the possible relationship between CD36 rs1761667 polymorphism and cardiometabolic risk factors. Thus, the current study aimed to examine the association between this polymorphism and cardiometabolic risk factors including BMI, WC, total cholesterol (TC), triglyceride (TG), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), systolic and diastolic blood pressure, and fasting blood glucose (FBG).
Results
Literature search and study characteristics
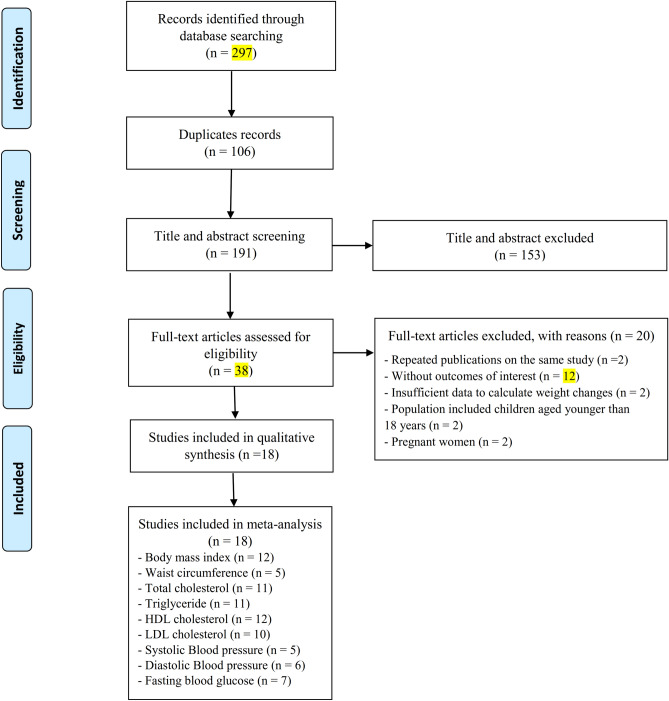
In total, 297 publications were identified in the initial search; from which 106 studies were duplicates, and 153 articles did not meet the eligibility criteria after screening titles/abstracts. The full texts of thirty-eight articles were assessed for further consideration and twenty articles were excluded for the following reasons: reported duplicate data (n = 2), had no data on the outcome variables (n = 12), conducted on pregnant women (n = 2), children and adolescents aged below 18 years (n = 2) and did not provide the sufficient data (n = 2) (Supplementary Table S1). The article selection procedure is illustrated in Fig. 1. Eventually, 18 studies with 6317 participants were included in the systematic review7,8,11,13–26. One study reported BMI before pregnancy; thus, it was included for this variable and this article was not used for other risk factors27.
Figure 1.
Flow diagram for the study selection process.
General characteristics of the 18 eligible studies are provided in Table 1. Six studies were from Asia7,15,19,25–27, five from African8,11,20–22, and six from Europe13,14,17,18,23,24,and one study was from the USA/Italy16. The majority of eligible studies included both sexes and only two20,27 and one17 studies were performed on women and men, respectively. The participants were aged 18–75.73 years and the mean body mass index was ranged from 21.66 to 34.60 kg/m2. Seven studies were conducted on healthy individuals16–18,20,21,23,27, three studies targeted patients with heart diseases7,8,19, one study included individuals with T2DM25, metabolic syndrome11, and the six remaining studies included adults with other health status13–15,22,24,26. In the majority of studies, genotype distributions were in HWE7,11,13,15,16,21,22,24–27. HWE was not reported in six studies8,14,17,18,20,23 and in one publication which reported the results in four different groups, HWE were not in equilibrium in three groups19. Of the total 18 studies that assessed the relationship between CD36 rs1761667 genotypes and cardiovascular risk factors, nine studies were case–control (five studies reported only the data of the case group7,8,11,25,27, two studies combined the data of two groups24,26, the rest of the studies revealed the result separately in the case and control groups18,19), nine studies were cross-sectional, 13–17,20–23 of which 2 were baseline assessment of clinical trials14,17 (Table 1).
Table 1.
Characteristics of studies that were included in this systematic review and meta-analysis.
| Author (year) | Country | Study design | Number and sex (F/M) | BMI (Kg/cm2) | Genotypes frequencies | Mean age (years) | Notes about participants | Ethnicity | Method of genotype measured |
Reported data | Hardy–Weinberg |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bayoumy et al.11 | Egypt | Case–control, case assessment included | 33 F/67 M | 33.40 ± 3.20 |
GG: 5 AG: 70 AA: 25 |
GG: 45.00 AG: 44.00 AA: 47.00 |
Metabolic syndrome | Arabs | Real-time PCR | WC, TC, TG, HDL-C, LDL-C, SBP, DBP | Equilibrium |
| Boghdady et al.8 | Egypt | Case–control, case assessment included | 16 F/31 M | 29.40 ± 3.40 |
GG: 9 AG: 30 AA: 8 |
GG: 54.80 AG: 53.80 AA: 54.60 |
Coronary artery disease (CAD) | African Americans | Real-time PCR | BMI, WC, TC, TG, HDL-C, LDL-C | NM |
| Dalton et al.13 | UK | Cross-sectional | 85 F&M |
GG: 23.51 ± 4.18 AG: 25.02 ± 4.30 AA: 24.31 ± 3.69 |
GG: 28 AG: 39 AA: 18 |
26.10 | Individuals susceptible to overeating | Caucasian | Sequenom Mass Array system | BMI | Equilibrium |
| Dawczynski et al.14 | Germany | Clinical trial, baseline assessment included | 45 F&M | 26.31 ± 3.77 |
GG: 8 AG: 24 AA: 13 |
60.00 | Mildly hypertriacylglycerolemia | NM | Sequencing | TG, HDL-C | NM |
| Fujii et al.15 | Japan | Cross-sectional | 267 F/228 M |
GG: 23.60 ± 3.40 AG: 23.70 ± 3.40 AA: 23.40 ± 2.80 |
GG: 268 AG: 190 AA: 37 |
GG: 62.90 AG: 64.00 AA: 64.90 |
Community-dwelling individuals (hypertension, dyslipidemia, diabetes, obesity) | Asians | PCR-CTPP | BMI, WC, TG, HDL-C, SBP, DBP, FBG | Equilibrium |
| Ma et al.16 | USA/ Italy | Cross-sectional | 328 F/214 M | 30.67 ± 5.54 |
GG: 115 AG: 273 AA: 154 |
GG: 35.00 AG: 36.00 AA: 38.00 |
Healthy individuals of Caucasian origin | Caucasian | AcycloPrime-FP SNP Detection System | TC, TG, HDL-C, SBP, DBP, FBG | Equilibrium |
| Madden et al.17 | UK | Clinical trial, baseline assessment included | 108 M |
GG: 27.54 ± 4.20 AG: 27.75 ± 3.91 AA:26.90 ± 4.02 |
GG: 27 AG: 51 AA: 30 |
GG: 57.00 AG: 55.80 AA: 54.00 |
Healthy middle-aged men | Caucasian | Real-Time PCR | BMI, TG, HDL-C, LDL-C, FBG | NM |
| Melis et al.18 | Italy | Case–control | 37 F/25 M | 28.56 ± 7.52 |
Case GG: 16/ AG: 31/ AA: 15 |
NM | Healthy obesity | Caucasian | PCR–RFLP | BMI | NM |
| 43 F/21 M |
Control GG: 18/ AG: 39/ AA:7 |
Healthy normal weight | |||||||||
| Momeni-Moghaddam et al.19 | Iran | Case–control | 25 F&27 M |
GG: 25.54 ± 5.26 AG: 25.35 ± 4.88 AA: 24.10 ± 4.35 |
Case GG: 6/ AG: 40/ AA: 6 |
GG: 55.50 AG: 54.90 AA: 52.50 |
HTN without CAD | Asians | PCR–RFLP | BMI, TC, TG, HDL-C, LDL-C, SBP, DBP, FBG | Disequilibrium |
| 13 F&44 M |
Case GG: 17/ AG: 38/ AA: 2 |
GG: 57.47 AG: 56.43 AA: 50.00 |
CAD&HTN | Disequilibrium | |||||||
| 11 F&54 M |
Case GG: 12/ AG: 45/ AA: 8 |
GG: 56.18 AG: 56.69 AA: 54.13 |
CAD | Disequilibrium | |||||||
| 26 F&39 M |
Control GG: 26/ AG: 30/ AA: 9 |
GG: 52.65 AG: 55.93 AA: 50.71 |
Coronary angiography (without HTN and CAD) | Equilibrium | |||||||
| Mrizak et al.20 | Tunisia | Cross-sectional | 203 F | 34.60 ± 4.20 | GG: 42/ AG: 102/ AA: 59 | 38–43 | Healthy obesity women | African | PCR–RFLP | TC, LDL-C | NM |
| Ramos-Lopez et al.21 | Mexico | Cross-sectional | 157 F/75 | 21.66 ± 2.26 |
GG: 46 AG: 104 AA: 82 |
18–25 | Healthy normal-weight | Admixed population | PCR–RFLP | TC, TG, HDL-C, LDL-C | Equilibrium |
| Ramos-Lopez et al.22 | Mexico | Cross-sectional | 40 F/33 M |
GG: 24.40 ± 3.10 AG: 24.90 ± 4.20 AA: 26.60 ± 4.10 |
GG: 11 AG: 40 AA: 22 |
GG: 53.70 AG: 51.40 AA: 48.10 |
Chronic hepatitis C virus infection | Amerindian, Caucasian and African | Real-Time PCR | BMI, TC, TG, HDL-C, LDL-C, FBG | Equilibrium |
| Shen et al.23 | UK | Cross-sectional | 95 F/41 M | 22.9 ± 3.96 |
GG: 37 AG: 66 AA: 33 |
18–55 | Healthy adults | Caucasian, African, West Asian and East Asian | Fluorogenic 5' nuclease assay (Taq-Man) | BMI | NM |
| Solakivi et al.24 | Finland | Case–control |
736 F&M (Case: 314 + Control: 422) |
Case: 28.80 ± 5.20 Control: 25.50 ± 3.60 |
GG: 158 AG: 376 AA: 202 |
50.00 | Hypertension & non-hypertensive healthy subjects | Caucasian | Competitive Allele Specific PCR (KASP) technique | BMI, TC, SBP, DBP, FBG | Equilibrium |
| Yang et al.27 | China | Case–control, case assessment included | 209 F | 21.95 ± 2.76 |
GG: 88 AG: 99 AA: 22 |
32.99 | Healthy women with GDMa | Asians | Taq-man allelic discrimination assay | BMI | Equilibrium |
| Yuan et al.**25 | China | Case–control, case assessment included | 42 F/70 M |
GG: 23.80 ± 2.70 AG: 24.40 ± 3.20 AA: 23.50 ± 2.60 |
GG: 41 AG: 49 AA: 22 |
57.10 | T2DM | Asians | PCR–RFLP | BMI, TC, TG, HDL-C, LDL-C, DBP, FBG | Equilibrium |
| Zhang et al.7 | China | Case–control, case assessment included | 43 F/69 M | 23.78 ± 2.85 |
GG: 43 AG: 60 AA: 9 |
64.04 | Coronary artery heart disease | Asians | PCR–RFLP | BMI, TC, TG, HDL-C, LDL-C | Equilibrium |
| Zhang et al.26 | China | Case–control |
1359 F&M (Case: 367 + Control: 992) |
Case: 25.52 ± 3.42 Control: 25.29 ± 3.48 |
GG: 543 AG: 631 AA: 185 |
75.73 | Atherothrombotic stroke & without stroke | Asians | ligase detection reaction (LDR) probe sequences | TC, HDL-C, LDL-C | Equilibrium |
F female, M male, NM HNW, Healthy normal weight, HO Healthy obesity, not mention, HTN hypertension, T2DM type 2 diabetes mellitus, GDM gestational diabetes mellitus, PCR polymerase chain reaction, PCR-CTPP Polymerase chain reaction with confronting two-pair primers, PCR–RFLP polymerase chain reaction-restriction fragment length polymorphism, BMI body mass index, WC waist circumference, TC total cholesterol, TG triglyceride, HDL-C high-density lipoproteins cholesterol, LDL-C low-density lipoprotein cholesterol, SBP systolic blood pressure, DBP diastolic blood pressure, FBG fasting blood glucose.
aBMI was calculated before gestation.
bInsufficient data to calculate changes in SBP.
Study quality assessment
Based on the Newcastle–Ottawa Scale, four8,18,24,26, and five7,11,19,25,27 case–control studies had a high and moderate methodological quality, respectively. All cross-sectional studies were categorized to be very good or good regarding their quality13–16,20,22,23, except for two studies17,21 (Supplementary Table S2).
The results of quantitative analysis
In this meta-analysis, five common genotype models of CD36 (rs1761667) were considered: allelic model (A vs. G), dominant model (AA + GA vs. GG), recessive model (AA vs. GA + GG), homozygous model (AA vs. GG) and heterozygous model (GA vs. GG).
Anthropometric measures
Twelve articles7,8,13,15,17–19,22–25,27 (n = 2478) reported the data on BMI and five articles8,11,15,16,24 with 1859 subjects were included in quantitative analysis for WC. As shown in Table 2, there was no significant association between CD36 rs1761667 genotypes and BMI in allelic (WMD = 0.029 kg/m2, 95%, CI: − 0.25, 0.31, p = 0.84, I2 = 31.20%), dominant (WMD = 0.23 kg/m2, 95%, CI: − 0.14, 0.61, p = 0.23, I2 = 17.90%), recessive (WMD = − 0.29 kg/m2, 95%, CI: − 0.84, 0.25, p = 0.29, I2 = 41.20%), homozygous (WMD = − 0.10 kg/m2, 95%, CI: − 0.72, 0.51, p = 0.73, I2 = 35.00%), and heterozygous (WMD = 0.35 kg/m2, 95%, CI: − 0.04, 0.75, p = 0.07, I2 = 15.70%) models (Supplementary Figure S5). According to the subgroup analysis, the association was significant for BMI in patients with heart disease (WMD = − 1.36 kg/m2, 95% CI: − 2.53, − 0.20, p = 0.02, I2 = 0.00%) and in the heterozygous model, in studies with medium quality (WMD = 0.58 kg/m2, 95% CI: 0.07, 1.09, p = 0.02, I2 = 0.00%) (Supplementary Table S3).
Table 2.
The association between CD36 rs1761667 polymorphism and anthropometric indices. All analyses were conducted using a random-effects modela.
| No. of data-sets | No. of subjects | Meta-analysis | Heterogeneity | Publication bias | |||||
|---|---|---|---|---|---|---|---|---|---|
| WMD2 (95%CI) | P effect | Q statistic | P within | I2 (%) | Begg's tests | Egger's tests | |||
| Body mass index (kg/m2) | |||||||||
| Allelic model (A vs. G) | 12 | 4956 | 0.029 (− 0.25, 0.31) | 0.84 | 15.99 | 0.14 | 31.20 | 0.11 | 0.07 |
| Dominant model (AA + GA vs. GG) | 2478 | 0.23 (− 0.14, 0.61) | 0.23 | 13.40 | 0.26 | 17.90 | 0.15 | 0.18 | |
| Recessive model (AA vs. GA + GG) | 2478 | − 0.29 (− 0.84, 0.25) | 0.29 | 18.71 | 0.06 | 41.20 | 0.53 | 0.26 | |
| Homozygous model (AA vs. GG) | 1255 | − 0.10 (− 0.72, 0.51) | 0.73 | 16.93 | 0.11 | 35.00 | 0.15 | 0.09 | |
| Heterozygous model (GA vs. GG) | 2028 | 0.35 (− 0.04, 0.75) | 0.07 | 13.04 | 0.29 | 15.70 | 0.45 | 0.30 | |
| Waist circumference (cm) b | |||||||||
| Allelic model (A vs. G) | 5 | 3718 | − 0.92 (− 2.20, 0.35) | 0.15 | 9.45 | 0.05 | 57.70 | – | – |
| Dominant model (AA + GA vs. GG) | 1859 | − 0.70 (− 2.76,1.36) | 0.50 | 8.38 | 0.07 | 52.30 | – | – | |
| Recessive model (AA vs. GA + GG) | 1859 | − 2.32 (− 4.71, 0.06) | 0.056 | 11.87 | 0.01 | 66.30 | – | – | |
| Homozygous model (AA vs. GG) | 949 | − 2.25 (− 5.48, 0.98) | 0.17 | 11.66 | 0.02 | 65.70 | – | – | |
| Heterozygous model (GA vs. GG) | 1452 | − 0.29 (− 2.04, 1.45) | 0.74 | 5.92 | 0.20 | 32.40 | – | – | |
aAll analyses were done using the random-effects model.
bThere was no evidence of publication bias by observing the funnel plots.
WMD, weighted mean difference; 95% CI, 95% confidence interval.
Also, no association was found for WC in allelic (WMD = − 0.92 cm, 95%, CI: − 2.20, 0.35, p = 0.15, I2 = 57.70%), dominant (WMD = -0.70 cm, 95%, CI: − 2.76,1.36, p = 0.50, I2 = 52.30%), recessive (WMD = − 2.32 cm, 95%, CI: − 4.71, 0.06, p = 0.056, I2 = 66.30%), homozygous (WMD = − 2.25 cm, 95%, CI: − 5.48, 0.98, p = 0.17, I2 = 65.70%), heterozygous (WMD = − 0.29 cm, 95%, CI: − 2.04, 1.45, p = 0.74, I2 = 32.40%) models (Table 2 and Supplementary Figure S6).
Lipid profile
Total cholesterol (TC)
The overall meta-analysis of eleven studies with 3755 participants7,8,11,16,19–22,24–26 showed no significant association between CD36 rs1761667 polymorphism and serum TC levels (allelic: WMD = 0.41 mg/dl, 95% CI: − 1.60, 2.44, p = 0.68; dominant: WMD = 0.66 mg/dl, 95% CI: − 2.96, 4.30, p = 0.71; recessive: WMD = − 0.83 mg/dl, 95% CI: − 5.61, 3.95, p = 0.73; homozygous: WMD = 2.18 mg/dl, 95% CI: − 1.24, 5.61, p = 0.21; heterozygous: WMD = − 1.37 mg/dl, 95% CI: − 6.53, 3.79, p = 0.60) models and heterogeneity between included studies was moderate to high (Table 3 and Supplementary Figure S7). In the subgroup analysis, the association was significant in studies conducted in the Asian population in allelic (WMD = 1.04 mg/dl, 95% CI: 0.88, 1.20, p < 0.001, I2 = 0.00%) and homozygous (WMD = 1.17 mg/dl, 95% CI: 0.76, 1.58, p < 0.001, I2 = 0.00%) models and healthy individuals (AA vs. GG: WMD = 5.52 mg/dl, 95% CI: 1.60, 9.43, p = 0.006, I2 = 18.50%). Furthermore, serum TC level was significantly higher in adjusted studies under allelic, dominant and heterozygous models (Supplementary Table S4).
Table 3.
The association between CD36 rs1761667 polymorphism and lipid profile. All analyses were conducted using a random-effects modela.
| No. of data-sets | No. of subjects | Meta-analysis | Heterogeneity | Publication bias | |||||
|---|---|---|---|---|---|---|---|---|---|
| WMD2 (95%CI) | Peffect | Q statistic | Pwithin | I2 (%) | Begg's tests | Egger's tests | |||
| Total cholesterol (mg/dl) | |||||||||
| Allelic model (A vs. G) | 11 | 7510 | 0.41 (− 1.60, 2.44) | 0.68 | 28.23 | 0.002 | 64.60 | 0.35 | 0.33 |
| Dominant model (AA + GA vs. GG) | 3755 | 0.66 (− 2.96, 4.30) | 0.71 | 29.09 | 0.001 | 65.60 | 0.87 | 0.46 | |
| Recessive model (AA vs. GA + GG) | 3755 | − 0.83 (− 5.61, 3.95) | 0.73 | 91.91 | < 0.001 | 89.10 | 0.35 | 0.77 | |
| Homozygous model (AA vs. GG) | 1867 | 2.18 (− 1.24, 5.61) | 0.21 | 20.59 | 0.02 | 51.40 | 0.21 | 0.82 | |
| Heterozygous model (GA vs. GG) | 2962 | − 1.37 (− 6.53, 3.79) | 0.60 | 59.76 | < 0.001 | 83.30 | 0.53 | 0.57 | |
| Triglyceride (mg/dl) | |||||||||
| Allelic model (A vs. G) | 11 | 4210 | − 7.11 (− 11.06, − 3.16) | < 0.001 | 18.97 | 0.04 | 47.30 | 0.062 | 0.069 |
| Dominant model (AA + GA vs. GG) | 2105 | − 7.25 (− 14.64, 0.12) | 0.054 | 21.96 | 0.01 | 54.50 | 0.35 | 0.002 b | |
| Recessive model (AA vs. GA + GG) | 2105 | − 14.54 (− 22.74, − 6.35) | 0.001 | 32.33 | < 0.001 | 69.10 | 0.75 | 0.55 | |
| Homozygous model (AA vs. GG) | 1061 | − 13.94 (− 23.82, − 4.06) | 0.006 | 24.12 | 0.007 | 58.50 | 0.08 | 0.051 | |
| Heterozygous model (GA vs. GG) | 1678 | − 6.22 (− 15.40, 2.96) | 0.18 | 30.66 | 0.001 | 67.40 | 0.75 | 0.004 b | |
| HDL-C(mg/dl) | |||||||||
| Allelic model (A vs. G) | 12 | 6928 | 0.58 (− 0.29, 1.45) | 0.19 | 39.64 | < 0.001 | 72.20 | 0.73 | 0.29 |
| Dominant model (AA + GA vs. GG) | 3464 | 0.21 (− 1.16, 1.58) | 0.76 | 28.30 | 0.003 | 61.10 | 1.00 | 0.54 | |
| Recessive model (AA vs. GA + GG) | 3464 | 1.36 (0.08, 2.64) | 0.03 | 29.03 | 0.002 | 62.10 | 0.73 | 0.14 | |
| Homozygous model (AA vs. GG) | 1789 | 1.64 (− 0.41, 3.70) | 0.11 | 36.79 | < 0.001 | 70.10 | 0.94 | 0.26 | |
| Heterozygous model (GA vs. GG) | 2852 | − 0.31 (− 1.62, 0.99) | 0.64 | 24.45 | 0.01 | 55.00 | 0.73 | 0.87 | |
| LDL-C (mg/dl) | |||||||||
| Allelic model (A vs. G) | 10 | 5170 | − 0.62 (− 4.40, 3.14) | 0.74 | 55.82 | < 0.001 | 83.90 | 0.15 | 0.75 |
| Dominant model (AA + GA vs. GG) | 2585 | − 0.36 (− 3.80, 3.07) | 0.83 | 15.54 | 0.07 | 42.10 | 0.15 | 0.02 b | |
| Recessive model (AA vs. GA + GG) | 2585 | − 2.68 (− 11.53, 6.15) | 0.55 | 138.25 | < 0.001 | 93.50 | 0.72 | 0.90 | |
| Homozygous model (AA vs. GG) | 1295 | − 2.07 (− 9.79, 5.64) | 0.59 | 51.61 | < 0.001 | 82.60 | 0.10 | 0.64 | |
| Heterozygous model (GA vs. GG) | 2118 | − 3.98 (− 8.77, 0.80) | 0.10 | 27.70 | 0.001 | 67.50 | 0.37 | 0.01 b | |
Significant values are in bold.
a All analyses were done using the random-effects model.
b These values were unchanged using the trim and fill method.
WMD, weighted mean difference; 95% CI, 95% confidence interval.
Triglyceride (TG)
The analysis of eleven studies7,8,11,14–17,19,21,22,25 (n = 2105) revealed that in overall, there was a significant association between rs1761667 polymorphism and TG values in allelic (WMD = − 7.11 mg/dl, 95% CI: − 11.06, − 3.16, p < 0.001, I2 = 47.30%), recessive (WMD = − 14.54 mg/dl, 95% CI: − 22.74, − 6.35, p = 0.001, I2 = 69.10%) and homozygous (WMD = − 13.94 mg/dl, 95% CI: − 23.82, − 4.06, p = 0.006, I2 = 58.50%) models, the TG level in the AA genotype group was significantly lower than that in G allele carriers, and the same difference was also observed in the above-mentioned models (Table 3 and Supplementary Figure S8). Subgroup analysis revealed that rs1761667 polymorphism was associated with decreased TG in the healthy population and HWE studies with AA genotype and A allele group, compared to the GG genotype and G allele group in homozygous (healthy participants: WMD = − 5.46 mg/dl, 95% CI: − 8.92, − 2.00, p = 0.002, I2 = 0.00%; equilibrium subgroup: WMD = − 11.85 mg/dl, 95% CI: − 23.02, − 0.68, p = 0.03, I2 = 67.60%) and allelic models (healthy participants: WMD = − 4.26 mg/dl, 95% CI: − 6.12, − 2.40, p < 0.001, I2 = 0.00%; equilibrium subgroup: WMD = − 6.65 mg/dl, 95% CI: − 11.20, − 2.11, p = 0.004, I2 = 58.80%), respectively. Further details about the subgroup analysis are provided in Supplementary Table S5.
High-density lipoprotein cholesterol (HDL-C)
Based on the results of twelve datasets with 3464 individuals7,8,11,14–17,19,21,22,25,26, AA genotype population had a significantly higher HDL-C level (WMD = 1.36 mg/dl, 95% CI: 0.08, 2.64, p = 0.03, I2 = 62.10%) than G allele carriers in the recessive model (Table 3 and Supplementary Figure S9), with regard to subgroup analysis, this relationship was seen in the studies with HWE (WMD = 1.62 mg/dl, 95% CI: 0.23, 3.01, p = 0.02, I2 = 68.60%). Lower serum HDL-C levels were observed in the studies with the Asian population in allelic (WMD = − 0.09 mg/dl, 95% CI: − 0.18, − 0.01, p = 0.02, I2 = 0.00%), dominant (WMD = -0.29 mg/dl, 95% CI: − 0.43, − 0.14, p < 0.001, I2 = 0.00%) and heterozygous (WMD = − 0.38 mg/dl, 95% CI: − 0.52, − 0.23, p < 0.001, I2 = 0.00%) models (Supplementary Table S6).
Low-density lipoprotein cholesterol (LDL-C)
Meta-analysis of ten studies7,8,11,17,19–22,25,26 (n = 2585) revealed, no significant association between CD36 rs1761667 polymorphism and LDL-C levels in different genetic models (allelic model: WMD = − 0.62 mg/dl, 95% CI: − 4.40, 3.14, p = 0.74; dominant model: WMD = -0.36 mg/dl, 95% CI: − 3.80, 3.07, p = 0.83; recessive model: WMD = − 2.68 mg/dl, 95% CI: − 11.53, 6.15, p = 0.55; homozygous model: WMD = − 2.07 mg/dl, 95% CI: − 9.79, 5.64, p = 0.59; heterozygous model: WMD = − 3.98 mg/dl, 95% CI: − 8.77, 0.80, p = 0.10) (Table 3 and Supplementary Figure S10). Allelic (WMD = 5.43 mg/dl, 95% CI: 0.16, 10.70, p = 0.04, I2 = 69.90%), recessive (WMD = 12.77 mg/dl, 95% CI: 1.99, 23.55, p = 0.02, I2 = 84.30%) and homozygous (WMD = 9.10 mg/dl, 95% CI: 0.29, 17.91, p = 0.04, I2 = 54.60%) models were associated with an increase in LDL-C levels in healthy participants and there was moderate to high heterogeneity between the included studies. Under the allelic (WMD = 1.32 mg/dl, 95% CI: 1.18, 1.45, p < 0.001, I2 = 0.00%) and homozygous (WMD = 1.92 mg/dl, 95% CI: 1.57, 2.28, p < 0.001, I2 = 0.00%) genetic models, rs1761667 had a significant association with the LDL-C levels in Asian population and there was no heterogeneity between the included studies. The results of subgroup analysis based on the study design and quality indicated that, the concentration of LDL in moderate quality and cross-sectional studies was lower in heterozygous model, whereas this variable was significantly higher in studies that adjusted the association for the potential confounders (WMD = 2.33 mg/dl, 95% CI: 2.17, 2.48, p < 0.001, I2 = 0.00%) than studies with no adjustment (WMD = − 5.14 mg/dl, 95% CI: − 8.43, − 1.84, p = 0.002, I2 = 1.70%) (Supplementary Table S7).
Blood pressure
The analysis results for five studies (n = 2112)11,15,16,19,24 for systolic (SBP) and six studies with 2224 participants11,15,16,19,24,25 for diastolic blood pressure (DBP) are illustrated in Table 4. No significant association was detected between genetic models of CD36 rs1761667 polymorphism and blood pressure (Table 4 and Supplementary Figure S11–S12). In the recessive model, the average DBP in the AA genotype group was significantly lower than that of G allele carriers in studies which did not adjust the association for confounders (Supplementary Table S8). It should be noted that no relationship was observed regarding this factor in other subgroups.
Table 4.
The association between CD36 rs1761667 polymorphism and blood pressure and fasting blood glucose. All analyses were conducted using a random-effects modela.
| No. of data-sets | No. of subjects | Meta-analysis | Heterogeneity | ||||
|---|---|---|---|---|---|---|---|
| WMD2 (95%CI) | Peffect | Q statistic | Pwithin | I2 (%) | |||
| Systolic Blood pressure (mmHg) b | |||||||
| Allelic model (A vs. G) | 5 | 4224 | − 2.01 (− 4.92, 0.90) | 0.17 | 36.92 | < 0.001 | 89.20 |
| Dominant model (AA + GA vs. GG) | 2112 | − 4.17 (− 9.22, 0.88) | 0.10 | 38.98 | < 0.001 | 89.70 | |
| Recessive model (AA vs. GA + GG) | 2112 | − 4.83 (− 12.95, 3.28) | 0.24 | 126.85 | < 0.001 | 96.80 | |
| Homozygous model (AA vs. GG) | 1050 | − 8.42 (− 18.09, 1.24) | 0.08 | 85.03 | < 0.001 | 95.30 | |
| Heterozygous model (GA vs. GG) | 1669 | − 3.30 (− 7.84, 1.23) | 0.15 | 28.79 | < 0.001 | 86.10 | |
| Diastolic Blood pressure (mmHg) b | |||||||
| Allelic model (A vs. G) | 6 | 4448 | − 0.14 (− 0.75, 0.46) | 0.63 | 4.89 | 0.43 | 0.00 |
| Dominant model (AA + GA vs. GG) | 2224 | − 0.14 (− 1.20, 0.92) | 0.79 | 0.87 | 0.97 | 0.00 | |
| Recessive model (AA vs. GA + GG) | 2224 | − 0.85 (− 2.56, 0.85) | 0.32 | 9.50 | 0.08 | 47.70 | |
| Homozygous model (AA vs. GG) | 1113 | − 0.57 (− 2.14, 1.00) | 0.47 | 5.87 | 0.31 | 14.80 | |
| Heterozygous model (GA vs. GG) | 1759 | 0.001 (− 1.11, 1.11) | 0.99 | 0.33 | 0.99 | 0.00 | |
| Fasting blood glucose (mg/dl) b | |||||||
| Allelic model (A vs. G) | 7 | 4610 | 2.07 (− 0.11, 4.25) | 0.06 | 23.76 | 0.001 | 74.70 |
| Dominant model (AA + GA vs. GG) | 2305 | 3.64 (− 0.89, 8.17) | 0.11 | 49.96 | < 0.001 | 0.88 | |
| Recessive model (AA vs. GA + GG) | 2305 | 2.33 (0.92, 3.74) | 0.001 | 6.03 | 0.42 | 0.40 | |
| Homozygous model (AA vs. GG) | 1173 | 4.28 (− 1.02, 9.60) | 0.11 | 30.42 | < 0.001 | 80.30 | |
| Heterozygous model (GA vs. GG) | 1813 | 3.07 (− 1.17, 7.33) | 0.15 | 39.95 | < 0.001 | 85.00 | |
Significant values are in bold.
aAll analyses were done using the random-effects model.
bThere was no evidence of publication bias by observing the funnel plots.
WMD, weighted mean difference; 95% CI, 95% confidence interval.
Fasting blood glucose (FBG)
Seven studies15–17,19,22,24,25 which included a total of 2305 individuals assessed the serum FBG in different genotypes of CD36 rs1761667. According to the pooled analysis, under the recessive genetic model, the homozygous A-allele carriers had a significantly higher FBG concentration compared with G allele carriers (WMD = 2.33 mg/dl, 95% CI: 0.92, 3.74), p = 0.001, I2 = 6.03%) and no heterogeneity was observed between included studies (Table 4 and Supplementary Figure S13). No significant association was observed between the rest of the genotypes with this parameter. Consistent with the results of the overall analysis, a significant relationship of rs1761667 polymorphism with the FBG level was observed under the recessive model in the Caucasian population (WMD = 3.30 mg/dl, 95% CI: 0.66, 5.93, p = 0.01)and the between-study heterogeneity was low (I2 = 39.50% and p = 0.19). Under allelic, recessive and homozygous models, a significant correlation was observed between this SNP and circulating FBG levels in the healthy group and cross-sectional studies. The between-study heterogeneity was not observed in these subgroups (I2 = 0.00%). The findings of other subgroup analyses were reported in Supplementary Table S9.
Meta-regression and cumulative meta-analysis
No significant association was observed between potential modulators (e.g. ethnicity, health status, Hardy–Weinberg equilibrium, quality score, design of the study, adjustment of confounders) and effect sizes for the association between CD36 rs1761667 polymorphism and BMI, HDL-C, LDL-C, SBP and DBP using meta-regression analyses. Ethnicity, quality score, design of the study, adjustment of confounders in the homozygote model and Hardy–Weinberg equilibrium were associated with between-study heterogeneity in the allelic model on the relation of CD36 SNP at rs1761667 on the levels of TC was determined using meta-regression (Supplementary Table S4). Meta-regression based on the mentioned modulators showed that all of them influence the association between rs1761667 polymorphism and the levels of TG in allelic and homozygote models; in addition, ethnicity, health status and Hardy–Weinberg equilibrium were associated with a difference in TG levels in the homozygote model; however, heterogeneity remained unchanged (Supplementary Table S5). Meta-regression according to ethnicity, health status and health status showed that these variables were significantly related to the difference in FBG values in the recessive and allelic models, respectively (Supplementary Table S9). These variables might be the cause of inconsistency in the relationship between SNP rs1761667 and cardiometabolic risk factors and for the sources of the heterogeneity observed in the current study.
The cumulative meta-analysis confirmed the overall estimate was not influenced by the year of study except for FBG in the allelic model (Supplementary Figures S14–S22).
Sensitivity analysis and publication bias
The deletion of each study, individually, from the meta-analysis altered some overall association with BMI (dominant and heterozygous models), WC (dominant model), triglyceride (dominant and heterozygous models), HDL-C (recessive, homozygous, heterozygous models), LDL-C (recessive, heterozygous models), FBG (allelic, homozygous model) which are provided in Supplementary Table S10.
No evidence of publication bias was found for BMI, TC and HDL-C levels. Begg’s and Egger’s asymmetry tests suggested evidence of publication bias for the meta-analysis of serum TG and LDL-C levels in the dominant and heterozygous models. However, the results remained unchanged using the trim and fill analysis, which means it is unlikely that publication bias significantly affected imputes estimates (Tables 2, 3 and 4). There was no evidence of publication bias for studies evaluating the association between CD36 rs1761667 polymorphism and WC, SBP, DBP, and FBG concentrations by visually observing the Begg’s funnel plots (Supplementary Figure S1–S4).
Discussion
The results of the present systematic review and meta-analysis unraveled that CD36 rs1761667 polymorphism had a significant association with cardiometabolic risk factors such as circulating TG, HDL-C, and FBG levels. No significant relationship was observed for other comparisons in the overall analyses. A previous cohort study demonstrated that high triglyceride levels were three times more common in CVD patients than elevated cholesterol levels alone28. The CD36 is a multifunctional receptor that both persistent up-regulation and deficiency of this protein can increase the CVD risk. Abnormally up-regulated CD36 can lead to foam cell formation, endothelial apoptosis, exacerbated inflammation, macrophage trapping, and thrombosis. On the other hand, CD36 deficiency promotes metabolic disorders, dyslipidemia (increasing triglyceride, fatty acid, apoB48, and chylomicron remnants in plasma levels) and sub-clinical inflammation, all of which are cardiometabolic risk factors29. Similarly, regarding higher TG levels in individuals with the G/G genotype, previous studies demonstrated this finding and also showed that improvement in plasma TG levels after a fish oil-rich dietary intervention was lower in these individuals compared with G/A and A/A individuals12,16,17. In addition, a study in North Indian population showed that the presence of the minor rs1761667-allele A was associated with elevating the risk of developing T2DM9. The results of the ethnicity subgroup showed that Caucasians with AA genotype had a higher FBG concentration than G carriers. This discrepancy in this finding might be owing to different ethnicity, sample sizes, gene-environment and gene–gene interactions, publication bias and clinical heterogeneity. Another possible reason for this contrary finding can be a varied selection of the health status in different populations. On the other hand, the majority of eligible studies had evaluated fasting glucose levels. It was better to apply other glucose indicators (glycated hemoglobin A1c) because fasting blood glucose is affected by many factors such as BMI, psychological stress, smoking habits, potassium intake, etc30,31. So, it seems that for a definitive interpretation of this result further studies are needed.
The subgroup analysis demonstrated that the A allele was associated with elevated TC and LDL-C levels and decreased HDL-C levels in Asians. In line with the mentioned conclusion in the present study, some previous studies reported these outcomes and suggested A allele of rs1761667 as a susceptibility factor for high serum cholesterol levels, low HDL-C and atherothrombotic stroke11,26,32. The deficiency of CD36 occurs rarely in Caucasians and is relatively common (3–10%) in the population of Asian and African descent33. As mentioned before, CD36 deficiency is closely related to the elevated prevalence of metabolic abnormalities, including hyperlipidemia, and increased fasting glucose levels. Moreover, clinical investigations have shown that the rs1761667-A allele decreases the CD36 expression and is associated with upper recognition taste thresholds for fat and decreased lipid taste perception34,35. The significant relationship between the CD36 SNP rs1761667 variant and lipid levels were shown in Asians also highlights the interaction between the CD36 SNP rs1761667 variant and ethnicity in modulating the plasma lipids. Bayoumy et al.11 noted in an Egyptian population that the minor allele frequency (MAF) of rs1761667 polymorphism was merely 0.25, Noel et al.36 showed that the MAF was 0.46 in a Hispanic population. Previous studies have been reported the influence of interethnic differences in allele frequencies. These variations can contribute to the differences in gene expression and eventually, disease susceptibility. Therefore, it is important to consider the population variations in the allele frequency when trying to identify an association between a polymorphism and the risk of diseases37–39. Further studies with different ethnicities should be performed to confirm these conclusions.
The analyses also showed a significant association between increased levels of LDL-C, FBG and A allele of CD36 rs1761667 in healthy participants. It is mentioned that the synthesis and translocation of CD36 are influenced by various stimuli. Modifications of CD36 affect cardiac function via altering the cellular uptake of fatty acids in the myocardium. The increased CD36-induced fatty acid uptake could be harmful or beneficial under various pathological conditions40. CD36 is decreased in pathological cardiac hypertrophy and increased in diabetic cardiomyopathy and atherosclerosis. In a healthy heart, insulin promotes CD36 transportation from the endosome to the cell membrane. Simultaneously, the forkhead box O1 (FOXO1) transcription factor promotes CD36 expression41. Long chain fatty acids (LCFAs) absorbed by CD36 are used for oxidation and storage as lipids in mitochondrial. Whereas in diabetes, the enhancement of insulin strongly activates the PI3K-Akt (phosphatidylinositol 3‑kinase-protein kinase B) pathway that leads to a robust CD36 expression. On the other hand, upregulated mir-320 and down-regulated mir-200b-3p accelerate the CD36 transcription and translation, uptake of LCFAs facilitates in cardiomyocytes eventually. Intracellular LCFAs either enter the mitochondria for producing energy and the by-products-reactive oxygen species (ROS) or forms triglycerides. As we know, triglycerides accumulation can result in insulin resistance. The ROS assembly and insulin resistance deteriorate cardiac function and would trigger diabetic cardiomyopathy42. Therefore, CD36 has different functions in diverse conditions and there is a need for more in-depth study with different health status40. Moreover, a significant association with BMI was detected among subjects with heart diseases in the recessive model. The mechanisms underlying how modifications of CD36 affect fat metabolism and cardiometabolic risk factors still remain to be elucidated; however, some evidence has shown that CD36 plays a role in the metabolism of LDL-C and HDL-C and contributes directly to their regulation43,44. A systematic review of studies that investigated the association between CD36 and the metabolic complications of obesity, reported that CD36 may be involved in obesity-related complications in humans. Moreover, CD36 deficiency might affect myocardial uptake of LCFAs, delay clearance of plasma fatty acid after an oral meal, and be associated with abnormalities in chylomicron formation45. The CD36 is a membrane transporter of long-chain polyunsaturated fatty acid in many tissues including skeletal muscles, adipocytes and the heart. Dysfunction of this protein might reduce the intramuscular fatty acid oxidation rate. Therefore the availability of fatty acid enhances their storage in adipocytes46,47. Peroxisome proliferator-activated receptor (PPAR-γ) is a nuclear receptor that regulates adipocyte differentiation and adipogenesis and CD36 is regarded as a key factor in the activation of PPAR-γ and its change might influence PPAR-γ mediated adipocyte differentiation48. It has been suggested that CD36 expression is reduced in circumvallate taste buds among high-fat diet-induced obese rats which leads to a decreased sensitivity to fat taste, as a result the intake of fatty foods increases as a compensatory response 49. Muthuswamy et al.50 reported that a lower CD36 expression (in AA and AG genotype at rs1761667) might be involved in reducing the release of PYY from taste bud cells. The presence of CD36 in gustatory papillae, the main LCFA receptor in taste bud cells51, contributes to dietary fat taste perception and fat preference35. More preference and increased eating of fatty foods have been expressed, which may reflect a decline of oral and gastrointestinal fatty acid sensitivity in obesity52. Based on the results obtained after subgroup analysis according to quality assessment, design of studies, adjustment for confounders, and Hardy–Weinberg equilibrium, high-quality cohort studies considering confounders such as age, sex, BMI, smoking, alcohol consumption and physical activity are needed to confirm the results.
The current evidence showed no significant association of this CD36 SNP with blood pressure; however, subgroup analysis based on adjustment of confounders demonstrated a significant association between decreased DBP and studies which did not consider confounding variables into account. Molecular studies suggested that CD36 contributes to the production of nitric oxide. Since reducing nitric oxide activity in the renal medulla is associated with hypertension, it is proposed that a decreased CD36 in renal cells may be related to hypertension 53. Furthermore, some studies in animals indicated that there is a relationship between CD36 genetic background and regulation of blood pressure54,55. There are some reasons which may explain these inconsistencies. The conflicting results might be due to variations in the health status and the genotyping methods, differences in ethnicity of the populations and their sex, gene–gene interactions in various populations and interactions of rs1761667 polymorphism with other variants in the CD36 gene. Moreover, some environmental factors may affect CD36 expression, as alcohol and fatty acids are recognized to modify the epigenome that includes acetylation of histones, DNA methylation, etc. 56,57. According to the factors mentioned, observations are still controversial and further studies are needed to arrive at a firm conclusion about the association with blood pressure. However, it is worth noting that studies on CD36 suggest that transcriptional activation, post-translational modification and localization alterations in this protein may create new approaches for the treatment of CVD40.
The present meta-analysis has some strengths. To the best of our knowledge, this is the first meta-analysis about the relationship between genetic locus 7q11 rs1761667 polymorphism and cardiometabolic risk factors which tried to include all related publications. Notably, several subgroup analyses and meta-regression were performed to assess the potentially different relation between rs1761667 polymorphism and cardiometabolic risk factors. There are a number of limitations that should be considered when interpreting the current study’s results. First, some information (including sex, lifestyle, other genetic variations, alcohol consumption or smoking, physical activity and so on) was unavailable in studies to perform more detailed subgroup analysis. Age is another risk factor that plays an important role in the enhancement of cardiovascular risk factors and the development of CVD. Unfortunately, conducting subgroup analysis based on age groups was not possible in the present study. In addition, further research considering other environmental and genetic factors is still needed to come up with a more apprehensive estimation of CD36 rs1761667 polymorphism with cardiometabolic risk factors. Second, the number of studies available for some outcome variables and a number of subgroups was fairly small. Furthermore, the total sample size of Asian and European studies are still limited in our research. Another problem is the fact that some of the obtained relations [BMI (dominant and heterozygous models), WC (dominant model), TG (dominant and heterozygous models), HDL-C (recessive, homozygous and heterozygous models), LDL-C (recessive and heterozygous models) and FBG (allelic and homozygous models)] were affected by the removal of one or several studies in the sensitivity analysis. In addition, we could not find any GWAS finding on rs1761667; therefore, it was not possible to compare the results obtained in this study with GWAS result. A high level of heterogeneity was also observed between the studies included in analyses for different genetic models. Thus, the findings should be interpreted with caution.
In summary, the results of the current systematic review and meta-analysis indicated that CD36 rs1761667 polymorphism was significantly associated with decreased TG and elevated HDL-C and FBG concentration in the overall analysis. Asian populations with different genotypes have different levels of lipids (TC, LDL-C and HDL-C), which may affect the susceptibility of the disease in these people. The influence of ethnicity, confounders, and health status on pooled effects should be considered when interpreting the association between rs1761667 polymorphism and elevated cardiometabolic risk factors. Furthermore, cohort studies with a large sample size which take confounders into account are needed to confirm the associations found between rs1761667 SNP and cardiovascular risk factors.
Methods
This systematic review and meta-analysis is reported in accordance with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement58. The study protocol was also registered in the prospective register of systematic reviews (PROSPERO) [protocol code: CRD42021253789].
Search strategy
Relevant articles were identified through online search of the literature in PubMed/MEDLINE, EMBASE, Scopus, ISI (Web of Science) and Google Scholar up to December 2021 without any publication date, language, and any other restrictions. The keywords used to search were: rs1761667 OR “− 31,118 G > A” OR “− 31,118 G > A” OR “− 31,118 G > A” OR “− 31,118 G > A”. The research was also updated by adding the following keywords in the all fields "GWAS", "Genome-wide association studies", "Genome wide association studies", "Genome wide association study", "Genome-wide association study", "GWA study", "whole genome association study", "WGA study", "WGAS", "Whole-genome association study", "whole genome association studies", "whole-genome association studies", "WGA", "Whole Genome Association Analysis", "Whole-Genome Association Analysis", "Genome Wide Association Analysis", "Genome-Wide Association Analysis" to did not lose any article (Supplementary Table S11). The references of the relevant study were also examined manually for any missing related literature.
Eligibility criteria
All published studies (cross-sectional, cohort, case–control designs and baseline of controlled clinical trials) that focused on CD36 rs1761667 polymorphism and cardiometabolic biomarkers such as anthropometric indices (BMI, and WC), lipid profile markers (TC, TG, HDL-C and LDL-C), blood pressure and FBG were included in the present review. Furthermore, articles with or without deviation from the Hardy–Weinberg equilibrium (HWE) were included. If the studies were done in pregnant, lactating women, children, adolescents aged < 18 years, and also if they had no outcomes of interest, were excluded from the review. In the case of repeated publications on the same study, we selected the one which included a higher number of participants.
Data extraction
For each eligible study, data were extracted on the author’s last name, publication year, country, participants’ characteristics (sample size, sex, age, health status and ethnicity), method of genotyping, HWE and mean ± standard deviation (SD) of desired outcomes. Two reviewers (ASA and ZY) independently screened the titles and abstracts, assessed the full text of the relevant articles, and extracted the data. Any possible disagreements or discrepancies were resolved by group discussion.
Risk of bias assessment
Two authors independently evaluated the methodological quality of eligible studies. The quality of each study was assessed by using the Newcastle–Ottawa (NOS) Scale for case–control (eight items)59 and its modified version adapted for cross-sectional studies (seven items)60 with a maximum score of 9 and 10, respectively. The NOS was used for assessing the risk of bias in clinical trials because their baseline assessments were considered in this meta-analysis. According to the obtained NOS scores, case–control studies were classified into three levels: low quality (0–4 points), medium quality (5–6) and high quality (7–9 points) and cross sectional studies were classified into four levels: unsatisfactory (0–4 points), satisfactory (5–6 points), good (7–8 points) and very good (9–10 points) (as previously performed61,62). Any discrepancies were addressed by discussion to reach a consensus.
Statistical analysis
The raw difference in means and its 95% confidence interval (CI) was calculated by five genetic comparison models: allelic model (A vs. G), dominant model (AA + GA vs. GG), recessive model (AA vs. GA + GG), homozygous model (AA vs. GG) and heterozygous model (GA vs. GG) to determine the effect size for the meta-analysis. The fixed-effect model was carried out to combine the subgroup-specific estimates when studies reported separate results across different subgroups and the pooled effect size was used for meta-analysis. A random-effects model which takes the between-study heterogeneity into account was used to derive weighted mean difference (WMD) as the summary estimate and its confidence limit. The heterogeneity between studies was evaluated using Cochran's Q test and the I-squared (I2) statistic (which is an estimate ranging from 0 to 100%). The heterogeneity was regarded as low, moderate, and high when the values of I2 were 25%, 50%, and exceeded 75%, respectively63. Subgroups analyses according to ethnicity, health status (heart disease, healthy and others), Hardy–Weinberg equilibrium, quality score (high or medium quality), design of the study (case–control or cross-sectional) and adjustment of confounders were performed to detect sources of between-study heterogeneity and also meta-regression analysis was used to identify potential sources of heterogeneity. Sensitivity analysis was used to assess whether the overall association depended on a specific study64. The presence of the publication bias was investigated by visual inspection of the funnel plots in case there were < 10 studies in each analysis, and also through statistical asymmetry tests (Begg's adjusted rank correlation and Egger`s tests) for meta-analysis of 10 or more effect sizes65. In the case of asymmetry, Duval and Tweedie’s trim and fill analysis was applied for more adjustment of publication bias66. All statistical analyses were performed by using STATA version 11.2 (StataCorp, College Station, TX). Two-tailed p ≤ 0.05 were considered statistically significant.
Supplementary Information
Acknowledgements
The authors would like to thank the research council of Nutrition and Food Security Research Center, Shahid Sadoughi University of Medical Sciences for the scientific support of the present study.
Abbreviations
- CVD
Cardiovascular disease
- CD36
Cluster of differentiation 36
- LCFA
Long-chain fatty acids
- GWAS
Genome-wide association study
- SNPs
Single-nucleotide polymorphisms
- T2DM
Type 2 diabetes mellitus
- BMI
Body mass index
- HWE
Hardy–Weinberg equilibrium
- SD
Standard deviation
- WMD
Weighted mean difference
- CI
Confidence interval
- WC
Waist circumference
- TC
Total cholesterol
- TG
Triglyceride
- HDL-C
High-density lipoprotein cholesterol
- LDL-C
Low-density lipoprotein cholesterol
- SBP
Systolic blood pressure
- DBP
Diastolic blood pressure
- FBG
Fasting blood glucose
- MAF
Minor allele frequency
- FOXO1
Forkhead box O1
- ROS
Reactive oxygen species
- pi3k-Akt
Phosphatidylinositol 3‑kinase-protein kinase B
- PPAR-γ
Peroxisome proliferator-activated receptor
Author contributions
H.M.-Kh., A.S.A. and Z.Y. designed the research, conducted the electronic searches and study selection; A.S.A. and Z.Y. conducted data extraction and tabulated data; H.M.-Kh. and A.S.A. conducted the data analysis and interpretation of results; Z.Y. wrote the first draft of the manuscript; H.M.-Kh., A.S.A., M.M. and M.H.S. critically revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
The present systematic review was financially supported by the Nutrition and Food Security research center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran.
Data availability
All data analyzed during the current study are available in Supplementary 2.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zeinab Yazdanpanah and Hassan Mozaffari‐Khosravi.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-09908-0.
References
- 1.Roberts LD, Gerszten RE. Toward new biomarkers of cardiometabolic diseases. Cell Metab. 2013;18:43–50. doi: 10.1016/j.cmet.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitfield JB. Genetic insights into cardiometabolic risk factors. Clin. Biochem. Rev. 2014;35:15–36. [PMC free article] [PubMed] [Google Scholar]
- 3.Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Investig. 2001;108:785–791. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao L, et al. CD36 senses dietary lipids and regulates lipids homeostasis in the intestine. Front. Physiol. 2021;12:1–8. doi: 10.3389/fphys.2021.669279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirano K-I, et al. Pathophysiology of human genetic CD36 deficiency. Trends Cardiovasc. Med. 2003;13:136–141. doi: 10.1016/S1050-1738(03)00026-4. [DOI] [PubMed] [Google Scholar]
- 6.Pascual G, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, et al. Associations between CD36 gene polymorphisms and susceptibility to coronary artery heart disease. Braz. J. Med. Biol. Res. 2014;47:895–903. doi: 10.1590/1414-431x20143825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boghdady A, et al. Association between rs1761667 polymorphism of CD36 gene and risk of coronary atherosclerosis in Egyptian population. Cardiovasc. Diagn. Ther. 2016;6:120–130. doi: 10.21037/cdt.2015.12.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee M, Gautam S, Saxena M, Bid HK, Agrawal CG. Association of CD36 gene variants rs1761667 (G > A) and rs1527483 (C > T) with Type 2 diabetes in North Indian population. Int. J. Diabetes Mellit. 2010;2:179–183. doi: 10.1016/j.ijdm.2010.08.002. [DOI] [Google Scholar]
- 10.Sayed A, et al. CD36 AA genotype is associated with decreased lipid taste perception in young obese, but not lean, children. Int. J. Obes. 2015;39:920–924. doi: 10.1038/ijo.2015.20. [DOI] [PubMed] [Google Scholar]
- 11.Bayoumy NM, El-Shabrawi MM, Hassan HH. Association of cluster of differentiation 36 gene variant rs1761667 (G>A) with metabolic syndrome in Egyptian adults. Saudi Med. J. 2012;33:489–494. [PubMed] [Google Scholar]
- 12.Pioltine MB, et al. Genetic variation in CD36 is associated with decreased fat and sugar intake in obese children and adolescents. J. Nutrigenet. Nutrigenomics. 2016;9:300–305. doi: 10.1159/000455915. [DOI] [PubMed] [Google Scholar]
- 13.Dalton, M. Biopsychological investigation of hedonic processes in individuals susceptible to overeating: role of liking and wanting in trait binge eating. (University of Leeds, 2013).
- 14.Dawczynski C, et al. Randomized placebo-controlled intervention with n-3 LC-PUFA-supplemented yoghurt: effects on circulating eicosanoids and cardiovascular risk factors. Clin. Nutr. 2013;32:686–696. doi: 10.1016/j.clnu.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Fujii R, et al. Cluster of differentiation 36 gene polymorphism (rs1761667) is associated with dietary MUFA intake and hypertension in a Japanese population. Br. J. Nutr. 2019;121:1215–1222. doi: 10.1017/s0007114519000679. [DOI] [PubMed] [Google Scholar]
- 16.Ma X, et al. A common haplotype at the CD36 locus is associated with high free fatty acid levels and increased cardiovascular risk in Caucasians. Hum. Mol. Genet. 2004;13:2197–2205. doi: 10.1093/hmg/ddh233. [DOI] [PubMed] [Google Scholar]
- 17.Madden J, et al. Polymorphisms in the CD36 gene modulate the ability of fish oil supplements to lower fasting plasma triacyl glycerol and raise HDL cholesterol concentrations in healthy middle-aged men. Prostaglandins Leukot. Essent. Fatty Acids. 2008;78:327–335. doi: 10.1016/j.plefa.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Melis M, et al. Polymorphism rs1761667 in the CD36 gene is associated to changes in fatty acid metabolism and circulating endocannabinoid levels distinctively in normal weight and obese subjects. Front. Physiol. 2017;8:1006. doi: 10.3389/fphys.2017.01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Momeni-Moghaddam MA, Asadikaram G. CD36 gene polymorphism rs1761667 (G > A) is associated with hypertension and coronary artery disease in an Iranian population. BMC Cardiovasc. Disorders. 2019;19:140. doi: 10.1186/s12872-019-1111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mrizak I, et al. The A allele of cluster of differentiation 36 (CD36) SNP 1761667 associates with decreased lipid taste perception in obese Tunisian women. Br. J. Nutr. 2015;113:1330–1337. doi: 10.1017/s0007114515000343. [DOI] [PubMed] [Google Scholar]
- 21.Ramos-Arellano LE, et al. CD36 haplotypes are associated with lipid profile in normal-weight subjects. Lipids Health Disease. 2013 doi: 10.1186/1476-511x-12-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos-Lopez O, et al. CD36 genetic variation, fat intake and liver fibrosis in chronic hepatitis C virus infection. J. Food Sci. 2016;8:1067–1074. doi: 10.4254/wjh.v8.i25.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen YC, Kennedy OB, Methven L. The effect of genotypical and phenotypical variation in taste sensitivity on liking of ice cream and dietary fat intak. Food Qual. Prefer. 2017;55:79–90. doi: 10.1016/j.foodqual.2016.08.010. [DOI] [Google Scholar]
- 24.Solakivi T, Kunnas T, Nikkari ST. Contribution of fatty acid transporter (CD36) genetic variant rs1761667 to body mass index, the TAMRISK study. Scand. J. Clin. Lab. Invest. 2015;75:254–258. doi: 10.3109/00365513.2014.1003596. [DOI] [PubMed] [Google Scholar]
- 25.Yuan Y, et al. Relationship between the polymorphism of rs17154181 and rs1761667 sites of CD36 gene and macroangiopathy in patients with type 2 diabetes mellitus in Guangxi province. Chin. Gen. Pract. 2015;18:2421–2425. [Google Scholar]
- 26.Zhang Y, et al. CD36 genotype associated with ischemic stroke in Chinese Han. Int. J. Clin. Exp. Med. 2015;8:16149–16157. [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Luo BR, Hu M, Zhao DM, Jing WJ. Association of CD36 gene single nucleotide polymorphism with gestational diabetes mellitus in Chinese Han population. Clin. Exp. Obstet. Gynecol. 2018;45:266–271. doi: 10.12891/ceog3844.2018. [DOI] [Google Scholar]
- 28.Goldstein JL, Hazzard WR, Schrott HG, Bierman EL, Motulsky AG. Hyperlipidemia in coronary heart disease I. Lipid levels in 500 survivors of myocardial infarction. J. Clin. Investig. 1973;52:1533–1543. doi: 10.1172/JCI107331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao L, Varghese Z, Moorhead J, Chen Y, Ruan XZ. CD36 and lipid metabolism in the evolution of atherosclerosis. Br. Med. Bull. 2018;126:101–112. doi: 10.1093/bmb/ldy006. [DOI] [PubMed] [Google Scholar]
- 30.Ridwanto M, Indarto D, Hanim D. Factors affecting fasting blood glucose in patients with type 2 diabetes mellitus. Int. J. Nutr. Sci. 2020;5:13–18. doi: 10.30476/IJNS.2020.84492.1048. [DOI] [Google Scholar]
- 31.Yitshak-Sade M, Mendelson N, Novack V, Codish S, Liberty IF. The association between an increase in glucose levels and armed conflict-related stress: a population-based study. Sci. Rep. 2020;10:1–6. doi: 10.1038/s41598-020-58679-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Ramos O, Panduro A, Martinez-Lopez E. Genetic variant in the CD36 gene (rs1761667) is associated with higher fat intake and high serum cholesterol among the population of West Mexico. J. Nutr. Food Sci. 2005;5:1–5. [Google Scholar]
- 33.Aitman TJ, et al. Malaria susceptibility and CD36 mutation. Nature. 2000;405:1015–1016. doi: 10.1038/35016636. [DOI] [PubMed] [Google Scholar]
- 34.Pepino MY, Love-Gregory L, Klein S, Abumrad NA. The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. J. Lipid Res. 2012;53:561–566. doi: 10.1194/jlr.M021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller KL, et al. Common variants in the CD36 gene are associated with oral fat perception, fat preferences, and obesity in African Americans. Obesity. 2012;20:1066–1073. doi: 10.1038/oby.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noel SE, et al. Variants of the CD36 gene and metabolic syndrome in Boston Puerto Rican adults. Atherosclerosis. 2010;211:210–215. doi: 10.1016/j.atherosclerosis.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ioannidis JP, Ntzani EE, Trikalinos TA. 'Racial'differences in genetic effects for complex diseases. Nat. Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 38.Spielman RS, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goddard KA, Hopkins PJ, Hall JM, Witte JS. Linkage disequilibrium and allele-frequency distributions for 114 single-nucleotide polymorphisms in five populations. Am. J. Hum. Genet. 2000;66:216–234. doi: 10.1086/302727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shu H, et al. The role of CD36 in cardiovascular disease. Cardiovasc. Res. 2020 doi: 10.1093/cvr/cvaa319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chistiakov DA, Orekhov AN, Bobryshev YV. The impact of FOXO-1 to cardiac pathology in diabetes mellitus and diabetes-related metabolic abnormalities. Int. J. Cardiol. 2017;245:236–244. doi: 10.1016/j.ijcard.2017.07.096. [DOI] [PubMed] [Google Scholar]
- 42.Li H, et al. Nuclear miR-320 mediates diabetes-induced cardiac dysfunction by activating transcription of fatty acid metabolic genes to cause lipotoxicity in the heart. Circ. Res. 2019;125:1106–1120. doi: 10.1161/CIRCRESAHA.119.314898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luangrath V, Brodeur MR, Rhainds D, Brissette L. Mouse CD36 has opposite effects on LDL and oxidized LDL metabolism in vivo. Arterioscler. Thromb. Vasc. Biol. 2008;28:1290–1295. doi: 10.1161/ATVBAHA.107.161653. [DOI] [PubMed] [Google Scholar]
- 44.Brundert M, et al. Scavenger receptor CD36 mediates uptake of high density lipoproteins in mice and by cultured cells. J. Lipid Res. 2011;52:745–758. doi: 10.1194/jlr.M011981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love-Gregory L, Abumrad NA. CD36 genetics and the metabolic complications of obesity. Curr. Opin. Clin. Nutr. Metab. Care. 2011;14:527–541. doi: 10.1097/MCO.0b013e32834bbac9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ibrahimi A, Abumrad NA. Role of CD36 in membrane transport of long-chain fatty acids. Curr. Opin. Clin. Nutr. Metab. Care. 2002;5:139–145. doi: 10.1097/00075197-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 47.Bonen A, et al. Triacylglycerol accumulation in human obesity and type 2 diabetes is associated with increased rates of skeletal muscle fatty acid transport and increased sarcolemmal FAT/CD36. FASEB J. 2004;18:1144–1146. doi: 10.1096/fj.03-1065fje. [DOI] [PubMed] [Google Scholar]
- 48.Feng J, et al. Induction of CD36 expression by oxidized LDL and IL-4 by a common signaling pathway dependent on protein kinase C and PPAR-γ. J. Lipid Res. 2000;41:688–696. doi: 10.1016/S0022-2275(20)32377-4. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X-J, et al. Decreased expression of CD36 in circumvallate taste buds of high-fat diet induced obese rats. Acta Histochem. 2011;113:663–667. doi: 10.1016/j.acthis.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 50.Muthuswamy K, et al. Single nucleotide polymorphism in CD36: Correlation to peptide YY levels in obese and non-obese adults. Clin. Nutr. 2021;40:2707–2715. doi: 10.1016/j.clnu.2021.02.044. [DOI] [PubMed] [Google Scholar]
- 51.Simons PJ, Kummer JA, Luiken JJ, Boon L. Apical CD36 immunolocalization in human and porcine taste buds from circumvallate and foliate papillae. Acta Histochem. 2011;113:839–843. doi: 10.1016/j.acthis.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 52.Stewart JE, et al. Marked differences in gustatory and gastrointestinal sensitivity to oleic acid between lean and obese men. Am. J. Clin. Nutr. 2011;93:703–711. doi: 10.3945/ajcn.110.007583. [DOI] [PubMed] [Google Scholar]
- 53.Pravenec M, et al. Identification of renal Cd36 as a determinant of blood pressure and risk for hypertension. Nat. Genet. 2008;40:952–954. doi: 10.1038/ng.164. [DOI] [PubMed] [Google Scholar]
- 54.Aitman TJ, et al. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat. Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 55.Pravenec M, et al. Genetics of Cd36 and the clustering of multiple cardiovascular risk factors in spontaneous hypertension. J. Clin. Investig. 1999;103:1651–1657. doi: 10.1172/JCI6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burdge GC, Lillycrop KA. Fatty acids and epigenetics. Curr. Opin. Clin. Nutr. Metab. Care. 2014;17:156–161. doi: 10.1097/MCO.0000000000000023. [DOI] [PubMed] [Google Scholar]
- 57.Choi S-W, Friso S. Epigenetics: a new bridge between nutrition and health. Adv. Nutr. 2010;1:8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shamseer L, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. Br. Med. J. 2015 doi: 10.1136/bmj.g7647. [DOI] [PubMed] [Google Scholar]
- 59.Wells, G. et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Ottawa Hospital Research Institute. Clinical Epidemiology. Available from: http://www.ohri. ca/programs/clinical_epidemiology/oxford. asp (2016).
- 60.Herzog, R. et al. Newcastle-Ottawa Scale adapted for cross-sectional studies. Available from https://eje.bioscientifica.com/supplemental/journals/eje/176/3/R137.xml/supplementary_figure_1.pdf (2013).
- 61.Cinar E, Saxena S, Gagnon I. Differential effects of concurrent tasks on gait in typically developing children: a meta-analysis. J. Mot. Behav. 2021;53:509–522. doi: 10.1080/00222895.2020.1791038. [DOI] [PubMed] [Google Scholar]
- 62.Lopes LB, Machado V, Mascarenhas P, Mendes JJ, Botelho J. The prevalence of molar-incisor hypomineralization: a systematic review and meta-analysis. Sci. Rep. 2021;11:1–20. doi: 10.1038/s41598-020-79139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 64.Egger M, Davey-Smith G, Altman D. Systematic reviews in health care: meta-analysis in context. New Jersey: John Wiley & Sons; 2008. [Google Scholar]
- 65.Higgins J. P., G. S. Cochrane handbook for systematic reviews of interventions. Version 5.1. 0 [updated March 2011]. The Cochrane Collaboration. www. cochrane-handbook. org (2011).
- 66.Duval S, Tweedie R. Trim and fill: a simple funnel-plot–based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341X.2000.00455.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analyzed during the current study are available in Supplementary 2.