Abstract
Circulating total-tau levels can be used as an endophenotype to identify genetic risk factors for tauopathies and related neurological disorders. Here, we confirmed and better characterized the association of the 17q21 MAPT locus with circulating total-tau in 14,721 European participants and identified three novel loci in 953 African American participants (4q31, 5p13, and 6q25) at P < 5 × 10−8. We additionally detected 14 novel loci at P < 5 × 10−7, specific to either Europeans or African Americans. Using whole-exome sequence data in 2,279 European participants, we identified ten genes associated with circulating total-tau when aggregating rare variants. Our genetic study sheds light on genes reported to be associated with neurological diseases including stroke, Alzheimer’s, and Parkinson’s (F5, MAP1B, and BCAS3), with Alzheimer’s pathological hallmarks (ADAMTS12, IL15, and FHIT), or with an important function in the brain (PARD3, ELFN2, UBASH3B, SLIT3, and NSD3), and suggests that the genetic architecture of circulating total-tau may differ according to ancestry.
Subject terms: Genome-wide association studies, Genetic variation
A meta-analysis of genome-wide association studies of circulating total-tau levels identifies loci specific to European or African American ancestries, providing further insight to the genetic etiology of neurological diseases.
Introduction
The protein tau is an important biomarker of neuronal injury and neurodegeneration. Alzheimer’s disease (AD) and other dementias or related neurological disorders are associated with abnormal intraneuronal tau aggregates (collectively known as tauopathies)1. Newer techniques to diagnose AD now examine CSF biomarkers to improve diagnostic certainty and aid in earlier diagnosis2–4. However, their collection is invasive and user variability can be large in the downstream quantification assays. In addition, CSF tau levels are normal or low in tauopathies like Progressive Supranuclear Palsy (PSP) and in frontotemporal dementia patients with tau mutations5,6.
Using blood biomarkers with high specificity and sensitivity for AD is ideal to lower cost, risk, and burden3. Circulating total-tau (t-tau) levels can be quantified in serum or in plasma7 early in AD due to blood brain barrier breakdown8,9. Particularly, they show promise as a predictive biomarker for dementia and related endophenotypes10, with higher levels in patients with dementia or mild cognitive impairment compared to controls11,12, and higher levels associated with poorer cognitive performance, and smaller hippocampal volumes13,14. However, elevated levels may lack diagnostic specificity for AD, and simply indicate that brain injury is common to several neurological diseases. A recent paper showed for example that higher circulating t-tau predicted a higher risk of incident stroke15. Importantly, circulating biomarkers do not need to mirror their level in CSF to be useful. Altogether, the recent literature suggests that circulating t-tau levels may be a predictive biomarker to improve risk stratification for dementia and assess AD’s progression, help with enrollment of high-risk individuals into dementia prevention trials, be useful in addition to other blood biomarkers of neurodegeneration to determine cognitive improvements in clinical trials, and represent a useful biomarker for AD when added to CSF tau measures10,15–17.
CSF t-tau and phosphorylated tau (p-tau) levels have been used as endophenotypes in genome-wide association studies (GWAS) to detect genetic variants associated with AD risk. Similarly, circulating t-tau levels may be used as an endophenotype to identify genetic risk factors for tauopathies and related neurological disorders. Only two GWAS, conducted in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) study, were published for plasma t-tau or p-tau levels18,19. The modest sample size and the inclusion of only European participants has limited the statistical power to identify potential novel associations (only MAPT and APOE loci associations were statistically significant for t-tau and p-tau respectively), the ability to explore less frequent genetic variation, as well as the generalization of the findings to other ancestries. Therefore, the aim of our study was to perform large-scale meta-analyses of circulating t-tau levels, using 15,674 participants from eight studies representing two ancestries (Europeans and African Americans), to explore genetic variation underlying circulating t-tau levels and assess their overlap with known genetic determinants of neurological diseases. We detected four ancestry-specific loci at the genome-wide significance (17q21 in Europeans, and 4q31, 5p13, and 6q25 in African Americans). We identified pleiotropic associations at 17q21 and 1q24 which, combined with the detection of an enrichment of genes associated with neurological diseases or related traits, suggested that a potential overlap exists between genetic determinants of circulating t-tau levels and several neurological disorders and traits including AD and stroke.
Results
Populations and participants
We included in our meta-analyses 15,674 participants from eight studies representing two major ancestries: Europeans (N = 14,721) and African Americans (N = 953) (Table 1, 2). A description of each study is included in the Supplementary Notes 1–8. Proportion of males varied from 35% (The Cardiovascular Health Study, African Americans) to 100% (The Vietnam Era Twin Study of Aging). Mean age ranged from 49 years (3.5) for The Coronary Artery Risk Development in Young Adults, African Americans to 78 years (4.3) for The Cardiovascular Health Study, African Americans (Table 2).
Table 1.
Description of the European-ancestry participants included in the meta-analysis of circulating total-tau levels.
| FHS (N = 6018) | RSI (N = 2169) | RSII (N = 960) | MEM1 (N = 336) | MEM2 (N = 1738) | CARDIA (N = 315) | CHS (N = 1396) | ARIC (N = 549) | VETSA (N = 754) | ADNI (N = 486) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Men, N (%) | 2790 (46) | 1696 (59) | 951 (44) | 153 (45) | 666 (37) | 149 (47) | 523 (37) | 257 (47) | 754 (100) | 292 (60) |
| Age, mean (SD) | 56.81 (13.98) | 75.30 (6.10) | 67.80 (7.10) | 67.60 (8.58) | 71.47 (8.56) | 51.02 (3.25) | 77.91 (4.3) | 63.52 (4.38) | 67.50 (2.51) | 75.44 (6.71) |
| Age, median [25–75%] | 56.1 [46.4–66.3] | 74.4 [70.5–79.4] | 65.5 [62.8–70.8] | 68.0 [62.9–73.6] | 72.2 [66.1–77.6] | 52.0 [49.0–53.5] | 77.0 [75.0–80.3] | 64.0 [60.0–67.0] | 68.1 [65.4–69.7] | 75.8 [71.5–80.1] |
| Circulating t-tau, mean (SD) | 4.13 (2.12) | 2.60 (0.90) | 2.60 (3.10) | 2.10 (0.88) | 2.23 (1.84) | 0.54 (1.26) | 0.46 (2.18) | 0.41 (0.54) | 2.02 (1.23) | 2.88 (1.64) |
| Circulating t-tau, median [25–75%] | 3.91 [3.24–4.73] | 2.50 [1.90–3.0] | 2.40 [1.90–2.90] | 2.01 [1.55–2.60] | 1.86 [1.41–2.60] | 0.34 [0.20–0.57] | 0.27 [0.17–0.43] | 0.18 [0.09–0.34] | 1.76 [1.29–2.32] | 2.71 [1.93–3.45] |
| Circulating t-tau (log), mean (SD) | 1.97 (0.46) | 1.30 (0.50) | 1.20 (0.50) | 0.91 (0.81) | 0.95 (0.73) | −1.54 (1.14) | −1.31 (0.83) | −1.68 (0.96) | 0.82 (0.72) | 0.92 (0.57) |
| Circulating t-tau (log), median [25–75%] | 1.97 [1.70–2.24] | 1.30 [0.90–1.60] | 1.20 [0.90–1.60] | 1.01 [0.63–1.38] | 0.90 [0.50–1.38] | −1.56 [−2.32, −0.81] | −1.30 [−1.75, −0.85] | −1.71 [−2.34, −1.10] | 0.81 [0.37–1.21] | 1.00 [0.66–1.24] |
FHS Framingham Heart Study, RSI and RSII The Rotterdam Study, MEM1 and MEM2 The MEMENTO Study, CARDIA The Coronary Artery Risk Development in Young Adults Study, CHS The Cardiovascular Health Study, VETSA The Vietnam Era Twin Study of Aging Study, ARIC The Atherosclerosis Risk in Communities Study, ADNI The Alzheimer’s Disease Neuroimaging Initiative Study.
Table 2.
Description of the African American participants included in the meta-analysis of circulating total-tau levels.
| CARDIA (N = 111) | CHS (N = 273) | ARIC (N = 569) | |
|---|---|---|---|
| Men, N (%) | 51 (46) | 96 (35) | 218 (38) |
| Age, mean (SD) | 48.78 (3.46) | 76.32 (4.93) | 61.68 (4.47) |
| Age, median [25–75%] | 48.0 [46.0–51.0] | 75.0 [72.0–80.0] | 61.0 [58.0–65.0] |
| Circulating t-tau, mean (SD) | 0.57 (1.41) | 0.46 (0.94) | 0.50 (0.77) |
| Circulating t-tau, median [25–75%] | 0.37 [0.22–0.60] | 0.28 [0.18–0.46] | 0.31 [0.20–0.50] |
| Circulating t-tau (log), mean (SD) | −1.54 (1.03) | −1.24 (0.85) | −1.57 (1.10) |
| Circulating t-tau (log), median [25–75%] | −1.43 [−2.24, −0.78] | −1.27 [−1.73, −0.78] | −1.69 [−2.36, −0.99] |
CARDIA The Coronary Artery Risk Development in Young Adults Study, CHS The Cardiovascular Health Study, ARIC The Atherosclerosis Risk in Communities Study.
Main meta-analysis results
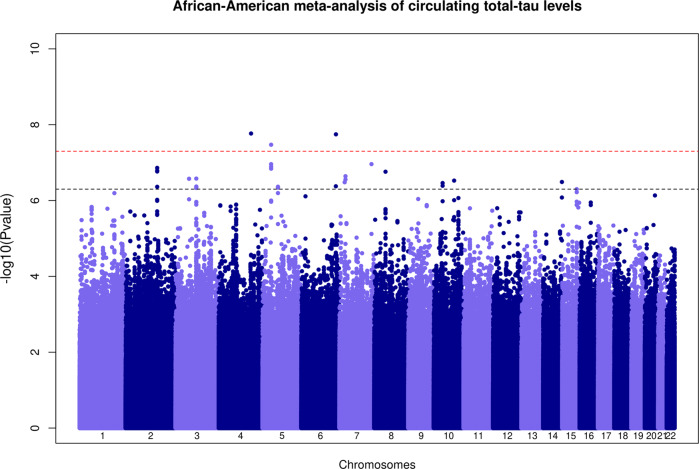
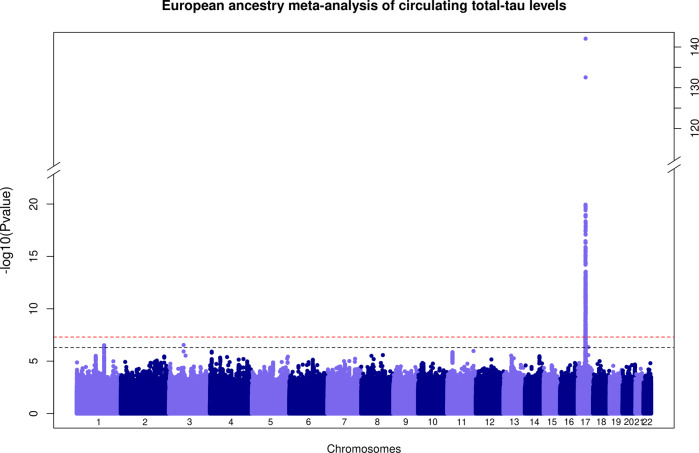
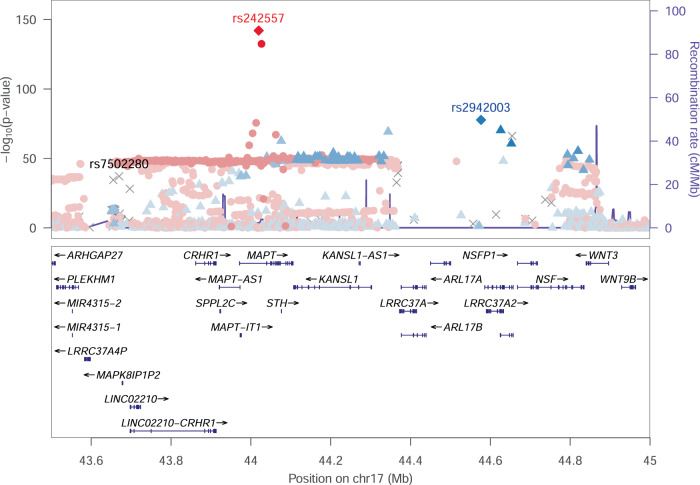
Quantile–Quantile and Manhattan plots for each ancestry-specific meta-analysis of circulating t-tau levels are presented in Figs. 1, 2 and Supplementary Fig. 1. We confirmed and better characterized the strong association of genetic variants at the microtubule associated protein tau (MAPT) 17q21 locus in European participants (lead genetic variant rs242557-A, effect allele frequency = 0.38, beta = 0.20, P = 8.9 × 10−143, proportion of variance explained (PVE) ~4.2%, Table 3). Stepwise model selection procedure at the 17q21 locus identified three distinct signals (rs242557-A, beta = 0.20, P = 2.3 × 10−143; rs7502280-T, beta = 0.17, P = 5.7 × 10−38, PVE ~ 1.1%, and rs2942003-T, beta = 0.16, P = 3.9 × 10−78, PVE ~ 2.3%), Table 3, Supplementary Table 1 and Fig. 3. We also detected, at the genome-wide threshold (P < 5 × 10−8), three potential novel loci in African American participants at 4q31 (lead genetic variant rs111836296-T, effect allele frequency = 0.06, beta = −0.54, P = 1.7 × 10−8), 5p13 (rs74710969-T, effect allele frequency = 0.11, beta = −0.53, P = 3.4 × 10−8, PVE ~ 3%) and 6q25 (rs674432-C, effect allele frequency = 0.97, beta = 0.68, P = 1.8 × 10−8, Table 3). The variants rs111836296 and rs74710969 are extremely rare or monomorphic in European populations. We detected additional associations at P < 5 × 10−7 at eleven loci (2q23, 3p14, 3q11, 5q13, 7p21, 7p15, 7q36, 8p11, 10p11, 10q23, and 14q32) in African American participants and three loci (1q24, 3p14, and 17q23) in European participants (Supplementary Tables 2, 3). All the identified hits were ancestry specific (absence of association or monomorphic variant in one ancestry) as indicated by the high heterogeneity (Table 3 and Supplementary Table 3). Forest plots are presented in Supplementary Figs. 2–11 and indicated consistent direction of effects across studies. Regional association plots for all loci are presented in Supplementary Figs. 12–27.
Fig. 1. Manhattan plot of association P values for the African American specific meta-analysis of GWAS of circulating total-tau levels.
The –log10(P)-value for each single nucleotide variant on the y axis is plotted against the build 37 genomic position on the x axis (chromosomal coordinate). The dashed horizontal red line indicates the genome-wide significance threshold of P = 5 × 10−8 and the dashed horizontal black line indicates the threshold of P = 5 × 10−7.
Fig. 2. Manhattan plot of association P values for the European ancestry specific meta-analysis of GWAS of circulating total-tau levels.
The –log10(P)-value for each single nucleotide variant on the y axis is plotted against the build 37 genomic position on the x axis (chromosomal coordinate). The dashed horizontal red line indicates the genome-wide significance threshold of P = 5 × 10−8 and the dashed horizontal black line indicates the threshold of P = 5 × 10−7. The y axis was truncated for ease of interpretation.
Table 3.
Lead genetic variants passing the genome-wide significance threshold (P < 5 × 10−8) in the meta-analysis of GWAS of circulating total-tau levels in European-ancestry participants (N = 14,721) or African American participants (N = 953).
| Europeans (N = 14,721) | African Americans (N = 953) | Multi-ancestry | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsid | Chr:b37Pos | Alleles | EAF | Beta | SE | P | I2 | PQ | EAF | Beta | SE | P | I2 | PQ | PRE2 | I2 | PQ | Gene | |
| rs111836296* | 4:142551581 | T/C | -- | -- | -- | -- | -- | -- | 0.06 | −0.54 | 0.10 | 1.7E-08 | 0 | 0.37 | -- | -- | -- | intergenic | |
| rs74710969* | 5:33618142 | T/G | -- | -- | -- | -- | -- | -- | 0.11 | −0.53 | 0.10 | 3.4E-08 | 0 | 0.88 | -- | -- | -- | ADAMTS12 | |
| rs674432 | 6:158101153 | C/G | 0.83 | −0.001 | 0.01 | 0.90 | 0 | 0.45 | 0.97 | 0.68 | 0.12 | 1.8E-08 | 0 | 0.32 | 7.6E-06 | 96.84 | 1.8E-08 | intergenic | |
| rs7502280* | 17:43670221 | T/G | 0.88 | 0.17 | 0.01 | 7.8E-38 | 0 | 0.53 | -- | -- | -- | -- | -- | -- | -- | -- | -- | intergenic | |
| rs242557 | 17:44019712 | A/G | 0.38 | 0.20 | 0.01 | 8.9E-143 | 42.5 | 0.07 | 0.28 | 0.05 | 0.07 | 0.50 | 76.2 | 0.04 | 8.5E-142 | 76.46 | 0.04 | MAPT | |
| rs2942003 | 17:44576704 | T/G | 0.34 | 0.16 | 0.01 | 1.9E-78 | 44.1 | 0.07 | 0.66 | 0.09 | 0.13 | 0.51 | 0 | 1.00 | 1.5E-77 | 0.00 | 0.57 | intergenic | |
EAF effect allele frequency, Alleles: effect (Alternate) allele/non-effect (Reference) allele.
I2: I-square heterogeneity statistic; PQ: Cochran’s Q statistic’s P value.
P values in bold pass the genome-wide significance threshold of P < 5 × 10−8.
*rs111836296 on chr4 and rs74710969 on chr5 are extremely rare in Europeans and thus they were not included in the multi-ancestry meta-analysis; rs7502280 is not present in the 1000 Genomes reference panel and thus was not present in the African American GWAS results.
PRE2: Han and Eskin’s random effects model to detect associations under heterogeneity59.
Fig. 3. Regional association plot at the MAPT 17q21 locus.
Genetic variants are plotted with their P values (-log10 values, left y axis) as a function of the build 37 genomic position (x axis). Estimated recombination rates (right y axis) reflect the local linkage disequilibrium (LD) structure around the top distinct associated genetic variants (red and blue diamonds, denoting rs242557 and rs2942003, respectively), identified using stepwise model selection procedure, and their correlated proxies. Genetic variants in LD with rs242557 are indicated with circles according to a light to dark red scale, from r² = 0 to 1. Genetic variants in LD with rs2942003 are indicated with triangles according to a light to dark blue scale, from r² = 0 to 1. Gray crosses (X) represent SNPs with missing LD. The third distinct genetic variant (rs7502280), indicated in black on the plot, was not present in the reference panel to calculate the LD with the other variants in the region. Reference panel used was the 1000 Genomes November 2014 European population.
We did not observe an association of the two SNPs defining APOE in the main circulating t-tau European meta-analysis, consistent with previous finding.18 In the additional APOE4-stratified analyses, we observed similar magnitude and consistent direction of effects for the main associations identified in Europeans (Table 4).
Table 4.
Results of the APOE4-stratified analyses for the lead genetic variants in each locus passing the threshold of P < 5 × 10−7 in the European meta-analysis of GWAS of circulating total-tau levels.
| Main analysis (N = 14,721) | APOE4 carriers (N = 3640) | APOE4 Non-carriers (N = 10,574) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsid | Chr | Build 37 Pos (bp) | Eff | NEff | EAF | Beta | P | Beta | P | Beta | P | Gene |
| rs6686805 | 1 | 169,512,643 | A | C | 0.67 | −0.04 | 3.4E-07 | −0.02 | 0.21 | −0.04 | 5.1E-07 | F5 |
| rs139843727 | 3 | 66,316,022 | A | C | 0.99 | 0.22 | 2.9E-07 | 0.08 | 0.44 | 0.24 | 6.5E-07 | SLC25A26 |
| rs7502280 | 17 | 43,670,221 | T | G | 0.88 | 0.17 | 8.0E-38 | 0.16 | 2.2E-09 | 0.18 | 1.4E-30 | intergenic |
| rs242557 | 17 | 44,019,712 | A | G | 0.38 | 0.20 | 8.9E-143 | 0.19 | 1.3E-37 | 0.20 | 1.4E-105 | MAPT |
| rs2942003 | 17 | 44,576,704 | T | G | 0.34 | 0.16 | 1.9E-78 | 0.17 | 3.5E-22 | 0.16 | 3.4E-58 | intergenic |
| rs4968553 | 17 | 59,428,962 | C | G | 0.16 | −0.05 | 4.6E-07 | −0.05 | 0.02 | −0.06 | 1.7E-07 | BCAS3 |
EAF effect allele frequency, Eff effect (alternate) allele, Neff non-effect (reference) allele.
The association of the two SNPs defining APOE in the main European meta-analysis were: rs429358-T (Beta = −0.02, P = 0.10) and rs7412-T (Beta = 0.01, P = 0.49).
Overlap of circulating t-tau genetic determinants with neurological diseases and traits
In addition to the strong associations at the MAPT locus that is known to be pleiotropic, we identified association at 1q24, a locus previously reported for stroke, Supplementary Table 4.20 The lead genetic variant in our analysis (rs6686805) was in linkage disequilibrium with rs1800594, a GWAS hit for ischemic stroke. Analyses conducted with FUMA, based on the main results from the European circulating t-tau meta-analysis, identified significantly differentially expressed genes in brain cerebellar hemisphere and brain cerebellum (Supplementary Fig. 28) and enrichment of genes in gene sets reported by GWAS of neurological diseases or traits including Parkinson Disease (PD), craniofacial microsomia, intracranial volume, cognitive function, subcortical brain region volumes, and AD in APOE E4- carriers, as well as risk factors such as body mass index (Supplementary Fig. 29). Finally, a genetic risk score (GRS) based on the distinct genome-wide associations (two MAPT genetic variants, rs242557 and rs376284405) from our European meta-analysis of circulating t-tau levels (excluding the Framingham Heart Study, FHS) was strongly associated with circulating t-tau levels (beta = 0.3, P = 4 × 10−97, PVE = 7%) and was associated with intracranial volume (beta = 15.1, P = 3 × 10−4) in FHS. We did not detect significant associations with the other traits tested (Supplementary Table 5). Altogether, these findings suggest an overlap of the genetic associations of circulating t-tau levels with known genetic determinants of neurological disorders and associated traits.
Two sample Mendelian Randomization (MR) analyses
Using two sample MR and large GWAS summary statistics, we did not identify significant causal associations between circulating t-tau levels and AD, PD, stroke, or White Matter Hyperintensities (WMH) (Supplementary Table 6). We also tested the opposite hypothesis and did not find significant causal associations (Supplementary Table 7). Our results did not indicate significant heterogeneity or presence of directional horizontal pleiotropy, except for a few analyses (WMH or PD as exposure; stroke as outcome). We also performed power calculations for the MR where circulating t-tau levels was the exposure variable (Supplementary Table 8) that indicated that our analysis would be underpowered if the instruments had small effects on the neurological outcomes (especially for AD and PD analyses with smaller numbers of cases).
Rare variant analyses using whole-exome sequence data
Using SKAT (variance component test) or CMC (burden test), our rare variant aggregation tests based on whole-exome sequence data identified 10 genes (ELFN2, UBASH3B, RUSF1, ZFP28, LCT, REM1, DELE1, SLIT3, NSD3, and MYO1G) significantly (P ≤ 1.25 × 10−6) associated with circulating t-tau levels when aggregating rare variants (MAF ≤ 5% or MAF ≤ 1%) with high or moderate impact, including some missense and loss of function variants (Supplementary Tables 9, 10 and Supplementary Figs. 30, 31). All except one gene (MYO1G) were detected with SKAT at the gene level significance threshold (P = 1.25 × 10−6), while at least nominally significant associations were observed for most genes with CMC (Supplementary Tables 9, 10). These results indicate that rare variants in those genes were likely to have different magnitudes and directions of effects, including no effect. This is a likely scenario as the number of rare variants aggregated for each gene was somewhat large, ranging from 13 to 64 variants. Similar results were observed for the two sets of annotations tested (missense or loss of function versus high or moderate impact). This observation, combined with the fact that one set of annotations is a subset of the other, and the number of genetic variants contributing to both analyses did not differ drastically, suggested that the same variants were selected to be aggregated in both analyses. Similar results were also observed for the two MAF thresholds tested, except for NSD3 and SLIT3. For these two genes, the addition of a small number of more frequent variants (1% < MAF ≤ 5%) attenuated the association.
Discussion
The goal of our study was to characterize genetic variation underlying circulating t-tau levels and to explore their overlap with known genetic determinants of neurological diseases. By performing large-scale meta-analyses in more than 15,000 participants from two major ancestries, we identified new genetic variants and genes associated with circulating t-tau levels, all associations being observed in only one ancestry (African Americans or Europeans). We identified pleiotropic signals at two regions (17q21 and 1q24) that were previously reported for plasma t-tau, AD, PD, WMH, and PSP (MAPT) or stroke (F5), respectively, and enrichment of genes associated with neurological diseases or related traits. Thus, our analyses highlighted that an overlap may exist between genetic determinants of circulating t-tau levels and several neurological disorders and traits including AD.
Our findings confirmed the importance of genes and pathways already well known to be involved in AD or other tauopathies and neurological diseases. Indeed, we first confirmed the strong association in Europeans of the 17q21 MAPT locus (lead genetic variant rs242557), which has been reported to be associated with circulating t-tau levels18. The MAPT locus has also been associated with AD, PD, and PSP, Supplementary Table 420–22, indicating an important role of MAPT in many neurodegenerative diseases. This locus has also been associated with head size23–25 and notably child head circumference23, which may indicate possible effects of this inversion on brain development very early in life. We found an enrichment for gene sets reported associated with craniofacial microsomia, intracranial volume, and subcortical brain region volumes, which tend to also support this hypothesis (Supplementary Fig. 29). We also identified a significant positive association of a GRS, constructed based on two distinct MAPT genetic variants (rs242557 and rs376284405), with intracranial volume in the FHS, while these variants were distinct from the ones reported by GWAS of intracranial volume at this locus (rs199525, rs8072451, and rs17689882). Despite PD is not a tauopathy, PSP and corticobasal degeneration, two PD subtypes known as Parkinson-plus syndromes, are both associated with the formation of tau deposits26. Here we were also able to identify two additional and distinct signals at 17q21 (rs7502280 and rs2942003). The variant rs7502280 is located at 29 kb of the corticotropin releasing hormone receptor 1 (CRHR1) and at 7.3 kb of the mitogen-activated protein kinase 8 interacting protein 1 (LOC644172), and is a GWAS hit for relative carbohydrate intake27 and sleep duration28. The variant rs2942003 lies at 12 kb and tags the leucine rich repeat containing 37 member A2 (LRRC37A2) gene (Fig. 3). One 17q21 variant (rs439945) reported by the Parkinson Disease GWAS Consortium was found to be significantly associated with nearby gene expression probes targeting LRRC37A and LRRC37A2 by a study investigating the modification of gene expression in prefrontal cortex brain samples of pathologically confirmed PD cases and controls29. Thus, genetic variations in both MAPT and LRRC37A2 appear to be important determinants of tauopathies and neurodegenerative disorders. More research needs to be performed to understand more precisely the mechanisms underlying their contributions to other tauopathies. Particularly, the 17q21 region, a common inversion polymorphism, is complex and may affect the expression of other genes in the region that may also be involved in neurodegenerative disease pathology, possibly in a tissue-specific manner.
In addition to the MAPT 17q21 locus identified in Europeans, we detected three potential novel loci in African American participants (4q31, 5p13, and 6q25) at the genome-wide significance level. Two of the lead genetic variants (rs111836296 and rs74710969) were extremely rare in Europeans and lie in or tag candidate genes (IL15 and ADAMTS12) linked to AD and other neurological disorders. The genetic variant rs111836296 at 4q31 lies at 6 kb and tags the interleukin 15 (IL15) gene (Supplementary Fig. 15). Serum IL15, a pro-inflammatory cytokine, has been studied as a possible marker of AD30. The genetic variant rs74710969 at 5p13 lies in an intron of the ADAM metallopeptidase with thrombospondin type 1 motif 12 (ADAMTS12) gene (Supplementary Fig. 16). Previous studies have associated ADAMTSs family of secreted metalloproteases with the repair of the central nervous system, through its ability to degrade neurocan, a novel component of brain extra-cellular matrix. Alterations in this degradation processes could be associated with the pathogenesis of neurological disorders31. Several studies also suggest a role for ADAMTS12 in stroke32,33.
We also detected 14 loci at P < 5 × 10−7, eleven loci in African American participants and three loci in Europeans.
Three signals identified in African Americans (3p14 and 5q13) or Europeans (17q23) lie in genes related to the tubulin-microtubule system (FHIT, MAP1B, and BCAS3). The signal at 3p14 lies in the fragile histidine triad diadenosine triphosphatase (FHIT) gene (Supplementary Fig. 13) and the encoded protein interacts with tubulin. The signal at 5q13 lies in the microtubule associated protein 1B (MAP1B) gene (Supplementary Fig. 17). Proteins of this family may be involved in microtubule assembly, which is an essential step in neurogenesis. Gene knockout studies of the mouse MAP1B gene suggested an important role in development and function of the nervous system. Several studies are also in favor of a role of MAP1B in AD34,35. MAP1B is also a component of cortical Lewy bodies and binds alpha-synuclein filaments, which suggests that it may be involved in the pathogenesis of Lewy bodies36. The signal at 17q23 lies in the BCAS3 microtubule associated cell migration factor (BCAS3) gene (Supplementary Fig. 27) that is highly expressed in the brain (GTEx). In mice, Rudhira, a murine WD40 domain protein that is 98% identical to BCAS3, has been shown to bind to microtubules and vimentin intermediate filaments to promote cell migration for angiogenic remodeling37.
Furthermore, two signals (1q24 and 10p11) lie in genes reported to be associated with AD or stroke or have an important function in the brain (F5, and PARD3). The signal at 1q24 identified in Europeans lies in the coagulation factor 5 (F5) gene (Supplementary Fig. 24). The lead genetic variant rs6686805 is in linkage disequilibrium with rs1800594, a GWAS hit for blood protein levels and ischemic stroke (Supplementary Tables 4, 11),38–40 and with rs6030, a missense variant in F5. A rare protective variant in F5 (rs2027885) has been reported to be associated with AD in African Americans41 and with hippocampal atrophy42. The signal at 10p11 identified in African Americans lies in the par-3 family cell polarity regulator (PARD3) gene (Supplementary Fig. 21) that is required for establishment of neuronal polarity and normal axon formation in cultured hippocampal neurons43,44. Par3 regulates microtubule stability and organization, crucial for neuronal polarization45. Moreover, atypical protein kinase C (aPKC) in complex with PAR-3/PAR-6 negatively regulates microtubule affinity-regulating kinases, which in turn causes dephosphorylation of microtubule-associated proteins, such as tau, leading to the assembly of microtubules and elongation of axons46. Par3 also regulates APP processing and trafficking47, polarized convergence between APP and BACE1 in hippocampal neurons48, and retrograde endosome-to- trans-Golgi network trafficking of BACE1 along with aPKC49. Brain regulatory marks (promoter and enhancer) are reported at the lead variant rs12245909 (HaploReg v4.1), which may suggest a functional role of this variant in the brain.
We looked up the main distinct lead genetic variants from the published ADNI GWAS of circulating tau levels18 in our European meta-analysis (excluding ADNI). We confirmed the strong association of the MAPT rs242557-A genetic variant (Table 5 and Supplementary Table 12). However, we did not find evidence of association for the three other loci that were detected at P < 10−5 in the original ADNI GWAS, suggesting that these signals may have been false positives.
Table 5.
Look-up of the main hits (P < 10−5) from the published ADNI GWAS of circulating tau levels (Chen et al., 2017) in our European meta-analysis of GWAS of circulating total-tau levels (seven studies excluding ADNI).
| Main analysis (N = 14,235) | APOE4 carriers (N = 3401) | APOE4 Non-carriers (N = 10,327) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsid | Chr | Build 37 Pos (bp) | Eff | NEff | EAF | Beta | P | Beta | P | Beta | P | Gene |
| rs2187213 | 6 | 162,634,337 | A | G | 0.35 | 0.0005 | 0.95 | 0.005 | 0.77 | −0.0006 | 0.95 | PARK2 |
| rs7047280 | 9 | 23,297,808 | T | C | 0.60 | −0.0009 | 0.90 | −0.008 | 0.56 | 0.003 | 0.71 | ELAVL2 |
| rs7072793 | 10 | 6,106,266 | T | C | 0.59 | 0.008 | 0.22 | 0.02 | 0.17 | 0.002 | 0.82 | IL2RA |
| rs242557 | 17 | 44,019,712 | A | G | 0.36 | 0.20 | 6.4E-136 | 0.19 | 3.0E-35 | 0.20 | 1.4E-101 | MAPT |
EAF effect allele frequency, Eff effect (alternate) allele, Neff non-effect (reference) allele.
Among the 10 genes identified when leveraging whole exome sequence data and aggregating rare variants with high or moderate impact, four have a function relevant to the brain (ELFN2, UBASH3B, SLIT3, and NSD3). Interestingly, SLIT3 and NSD3 associations were more impacted by the choice of the MAF threshold to select rare variants to aggregate. For both genes, results were only significant with SKAT when using a MAF ≤ 1%, indicating that only rarer variations were contributing to the associations. The extracellular leucine rich repeat and fibronectin type III domain containing 2 gene (ELFN2), is overexpressed in the brain. The encoded protein is a postsynaptic adhesion molecule that selectively binds with group III metabotropic glutamate receptors50,51. ELFN1, a protein of the same family, has been reported to be associated with neuropsychiatric disorders (attention deficit hyperactivity disorder, post-traumatic stress disorder, and epilepsy). Distinct neuronal expression patterns are reported for ELFN1 and ELFN251. The ubiquitin associated and SH3 domain containing B gene (UBASH3B) is overexpressed in the brain. The encoded protein is a phosphatase, and the concerted action of protein kinases and phosphatases represents a critical signaling event controlling synaptic functions and higher-order brain functions, such as learning and memory52. The slit guidance ligand 3 gene (SLIT3) encodes an axon guidance molecule expressed by motor neurons53,54. SLIT3 may also play a role in essential tremor disease pathogenesis55. The nuclear receptor binding SET domain protein 3 gene (NSD3) is highly expressed in the brain. The encoded protein is a SET domain-containing methyltransferase, an epigenetic regulator that is selectively expressed in primary microglia56. Follow-up studies are needed to characterize the potential role of these four genes in tauopathies.
A summary of the neurological traits reported in the GWAS catalog for genetic variants in the main genes identified in the meta-analyses of circulating t-tau levels (IL15, FHIT, ADAMTS12, PARD3, F5, BCAS3, UBASH3B, and SLIT3) and described above is available in Supplementary Table 11.
By performing meta-analyses separately in African Americans and European-ancestry participants, we were able to identify ancestry-specific associations for circulating t-tau levels. The lead variants at the three loci identified at the genome-wide threshold in African American participants were extremely rare in European populations. In addition, most loci identified in African American participants were driven by the largest study, ARIC. Two of the findings identified at the genome-wide threshold in African Americans were low frequency variants, with no linkage disequilibrium support (Supplementary Figs. 15, 18), and with only two of the three cohorts that contributed to the meta-analysis. Caution is thus needed regarding the interpretation of these findings as such results are typically seen in GWAS of admixed populations with small sample sizes and could be driven by a few outliers. We also found that the strong association of the MAPT locus with circulating t-tau levels was specific to European-ancestry participants. This result is consistent with the recent finding from the Florida Consortium for African American Alzheimer’s Disease studies57. The majority of loci identified in European-ancestry participants were driven by the two largest studies, FHS and RSI. The fact that we did not detect an association in the African American participants for the novel loci detected at P < 5 × 10−7 in the European-ancestry participants may be due to a lack of power because of the limited sample size of this subgroup. However, our multi-ancestry meta-analysis showed that the hits identified were ancestry specific (Supplementary Table 3). The high heterogeneity observed across ancestry suggests that the genetic architecture of circulating t-tau levels may differ between European-ancestry and African American populations.
We explored the potential pleiotropy of loci previously reported for several neurological disorders with circulating t-tau levels by performing a look-up of these loci in our European meta-analysis. We also used MR analyses to evaluate the potential causal associations between circulating t-tau levels with several neurological disorders and traits, but we did not identify significant causal associations. Limitations of these analyses are the availability of large, complete, and publicly available GWAS summary statistics, and the strength of the MR instruments due to the limited number of associations in the European circulating t-tau meta-analysis and the limited number of AD and PD cases.
Strengths of our study are the large sample size, with the inclusion of eight population-based cohorts with ancestral diversity, with genotype data and information on circulating t-tau levels measured with ultra-sensitive assays. We leveraged large imputations reference panels (1000 Genomes and the Haplotype Reference Consortium) to study common genetic variations complemented with whole exome sequence data to explore less frequent genetic variations. Some limitations include the modest sample size of the African American sample that has limited our ability to confirm the GWAS findings and perform secondary analyses in this subgroup, such as stratification on APOE4 status, and the fact that the contributed studies only had circulating t-tau measurement available, and not specifically phosphorylated tau. The fact that the APOE locus was not associated with circulating t-tau levels may suggest that circulating t-tau and phosphorylated tau have different genetic architectures. Replication of the African American potential novel loci and the less common variant association identified in Europeans is needed to confirm our findings but the availability of samples with circulating t-tau levels and genetic data is limited.
In conclusion, our large multi-ancestry meta-analysis identified new genetic variants and loci associated with circulating t-tau levels. Notably, our study revealed that the genetic architecture underlying circulating t-tau levels might differs between African American and European-ancestry populations and that genetic variation underlying circulating t-tau levels may overlap with known genetic determinants of neurological disorders. To better understand how these variants may contribute to AD and other tauopathies, further investigations of these findings will be necessary, including cohorts with a broader ancestral diversity, biological experiments, functional and omic studies, and animal models.
Methods
Populations and participants
We included in our multi-ancestry and ancestry-specific meta-analyses of total-tau participants from eight studies: seven cohorts from the Cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium (the Framingham Heart Study (FHS), the Rotterdam Study (RSI and RSII), the MEMENTO Study, the Coronary Artery Risk Development in Young Adults (CARDIA) Study, the Cardiovascular Health Study (CHS), the Vietnam Era Twin Study of Aging (VETSA) Study, and the Atherosclerosis Risk in Communities (ARIC) Study), and the ADNI Study. All participants included in this study provided written informed consent for genetic testing and analyses. Study-specific information including study description, and detailed information about genotyping and imputations and GWAS analysis is included in the Supplementary Notes 1–8.
Tau quantification
Circulating (in plasma or in serum) t-tau levels were quantified using the Human Total Tau kit on the Simoa™ HD-1 analyzer (ADNI, plasma), the Simoa™ Tau 2.0 Kit and the Simoa™ HD-1 analyzer (FHS, plasma), the Simoa™ Tau 2.0 Kit (MEMENTO_1, plasma), the Simoa™ Human Neurology 4-Plex A assay with the Simoa™ HD-X analyzer (the Atherosclerosis Risk in Communities study, MEMENTO_2 and the Coronary Artery Risk Development in Young Adults, plasma and the Cardiovascular Health Study, serum), the Simoa™ Human Neurology 3-Plex A assay with the Simoa™ HD-1 analyzer (the Rotterdam Study, plasma) and high throughput bioassays platforms or single analyte assays using the Simoa™ HD-X or Fujirebio analyzer (the Vietnam Era Twin Study of Aging, plasma).
Genotyping and imputation
Studies used the densest imputation reference panel available to them at the time of analyses, either 1000 Genomes or the Haplotype Reference Consortium. A description of the reference panel used by each study is provided in Supplementary Tables 13–15.
Genome-wide association studies (GWAS) and quality control
Each study evaluated the association of single-nucleotide genetic variants with log2-transformed circulating t-tau levels under an additive model. Analyses were adjusted for age, sex, and additional study specific covariates to control for population structure (including principal components). Studies with both Europeans and African Americans analyzed each ancestry separately. The minimum sample size for each ancestry/phenotype combination for inclusion in this study was fixed to 100. Additional stratified analyses according to APOE4 status (APOE4 carriers vs non APOE4 carriers) were performed in European participants only, given the modest sample size of the African American sample. Study-specific GWAS results were filtered based on an imputation quality score greater or equal to 0.30 and a minor allele count greater or equal to 20. The minor allele frequency threshold to include a genetic variant in the meta-analyses is indicated for each study in Supplementary Tables 13–15.
Multi-ancestry and ancestry-specific meta-analyses
GWAS results across all studies were meta-analyzed by ancestry with METAL using an inverse variance-weighted average method58. For each meta-analysis, we selected results for which two third of the studies contributed. Results from the European and the African American meta-analyses, and for which at least two studies contributed to each meta-analysis, were then meta-analyzed using Metasoft59,60.
Conditional and joint association analysis based on summary statistics
Conditional and joint association analysis in loci associated with circulating t-tau levels at the genome-wide threshold (P < 5 × 10−8) were performed using the Genome-wide Complex Trait Analysis61,62 based on the European meta-analysis results. A stepwise model selection procedure was used to select distinct associated genetic variants (P < 5 × 10−8). The FHS Haplotype Reference Consortium imputed data was used as the reference panel with unrelated participants only.
Overlap of circulating t-tau genetic determinants with neurological diseases and traits
We extracted from the GWAS Catalog20 (https://www.ebi.ac.uk/gwas/downloads/summary-statistics) all reported associations for AD, t-tau (in plasma or CSF), stroke, PSP, Parkinson’s Disease (PD), and White Matter Hyperintensities (WMH), and looked them up in our European meta-analysis results. We also used the FUMA GWAS platform (Functional Mapping and Annotation of Genome-Wide Association Studies, https://fuma.ctglab.nl/) using as input the summary statistics from the European circulating t-tau meta-analysis to leverage functional, biological information to prioritize genes and check expression patterns and shared molecular functions between these genes63,64. Finally, we performed a GRS association analysis in the FHS. We first re-ran the European circulating t-tau levels meta-analysis without FHS. We then used the Genome-wide Complex Trait Analysis to identify distinct genome-wide associations on chromosome 17. We computed the GRS using the distinct variants identified by the Genome-wide Complex Trait Analysis for all FHS participants using the Haplotype Reference Consortium imputed genotypes and weights from the new meta-analysis. We tested the association of the GRS with incident AD (140 cases, 2775 controls) and stroke (149 cases, 3461 controls), and with four brain MRI phenotypes (hippocampal volume, white matter hyperintensities, total brain volume, and intracranial volume, N ranging from 3489 to 4310). Details regarding trait measurements and definitions in the FHS have been published elsewhere65–67. We used logistic or linear mixed-effects model, adjusted for age at baseline or at MRI and sex, while accounting for relatedness. For the brain MRI analyses, we excluded participants with dementia, stroke, large brain infarcts, tumor or any other finding that could have affected the scan and additionally adjusted the hippocampal volume, white matter hyperintensities, and total brain volume analyses for intracranial volume. If a significant association was detected (P = 0.05/6 = 0.008), an additional adjustment for APOE4 was performed.
Two sample mendelian randomization (MR) analyses
We used the TwoSampleMR R package in R68,69 to assess the causal association of circulating t-tau levels with AD, PD, stroke, and WMH using publicly available large European GWAS summary statistics. We selected large GWAS with independent samples from the ones included our meta-analysis. Briefly, we used an arbitrary threshold of P < 5 × 10−6 in the European meta-analysis of circulating t-tau levels to select the genetic variants to be used in the MR and performing clumping to select distinct variants based on 1000 Genomes European linkage disequilibrium reference panel. We then extracted these SNPs from the outcome GWAS based on summary statistics from the GWAS catalog (https://www.ebi.ac.uk/gwas/downloads/summary-statistics) or the IEU GWAS database (IGD, https://gwas.mrcieu.ac.uk/) and performed harmonization of the alleles. As MAPT locus is known to be pleiotropic, we conducted the mendelian randomization using different methods and carefully check for presence of heterogeneity and horizontal pleiotropy. For this analysis, we conducted power calculations for a continuous exposure and a PVE on the exposure of 8%, using the power analysis calculator https://sb452.shinyapps.io/power/. Finally, we tested the opposite hypothesis that AD, PD, stroke, and WMH are causally associated with circulating t-tau levels.
Rare variant analyses using whole exome sequence data
To explore the association of rare variations with circulating t-tau levels, we selected the two largest studies (FHS and RSI) to perform rare-variant aggregation tests based on whole exome sequence data from the Cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium70. A total of 2279 participants were included in the analyses. Information on sequencing and quality control is provided in the Supplementary Notes 9 and 10. To make sure that the same allele was coded as the effect allele for FHS and RSI, we used the effect (alternate) allele from a consensus SNP info file from the Cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium71. Annotations of the exome variants was performed with dbNSFP72. We selected variants with a MAF ≤ 1% or 5%, and (1) missense and loss of function variants only or (2) variants with high or moderate impact from Ensembl Variant Effect Predictor73 including missense and loss of function variants. The analyses were performed using the R package seqMeta (http://cran.r-project.org/web/packages/seqMeta/). Each cohort used the seqMeta prepScores function to generate single variant score statistics and genotype covariance matrices for all variants. Results were then meta-analyzed using the skatMeta and burdenMeta functions. We used a Bonferroni correction for the number of genes included in the analyses and the number of tests (P = 0.05/20,000/2 = 1.25 × 10−6) and filtered the results based on a cumulative minor allele count of 30, that accounts for the number of genetic variants per gene.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank all the participating cohorts from the CHARGE consortium and ADNI participants for contributing to this study. Detailed study-specific acknowledgements are provided in the Supplementary Notes 11 and 12.
Author contributions
C.S., E.C.P.F., J.C.B., M.L., M.F., and A.M. conducted the cohort specific GWAS analysis. C.S., S.A., and E.C.P.F. conducted the exome sequence analyses. A.Y., V.C., and V.D. performed secondary analyses. V.B., and R.A.R. were involved in cohort-specific tau quantification. C.S., M.G., J.C.B., M.F., A.S.B., E.B., L.A.C., J.C.L., C.S.D., A.L.D., L.D., T.H.M., K.J.M., M.P.P., C.L.S., R.S.V., S.D., C.E.F., M.J.L., C.D., L.J.L., W.S.K., W.T.L., M.A.I., and S.S. designed and conceived the study. C.S., M.G., M.F., S.A., C.S.D., A.E.F., T.F.K., K.J.M., M.P.P., C.L.S., S.D., W.S.K., W.T.L., M.A.I., and S.S. wrote and edited the manuscript. All authors reviewed and approved the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: George Inglis. Peer reviewer reports are available.
Data availability
All data supporting the findings of this study are available either within the main article or the supplementary information. Summary statistics from the ancestry-specific meta-analyses of circulating levels of total-tau have been deposited and are publicly accessible on the GWAS catalog FTP (study accession numbers GCST90095138, and GCST90095139). Genome-wide summary statistics for complex disorders used in the secondary analyses were downloaded from public repositories (GWAS catalog: https://www.ebi.ac.uk/gwas/downloads/summary-statistics; and IEU GWAS database: https://gwas.mrcieu.ac.uk/).
Competing interests
C.S.D. reports personal fees from Novartis Pharmaceutical outside the submitted work. All the other authors declare no competing interests.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chloé Sarnowski, Mohsen Ghanbari, Joshua C. Bis, Mark Logue, Myriam Fornage, Aniket Mishra.
These authors jointly supervised this work: Stephanie Debette, Carole Dufouil, Lenore J. Launer, William S. Kremen, William T. Longstreth, M Arfan Ikram, Sudha Seshadri.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03287-y.
References
- 1.Schraen-Maschke S, et al. Tau as a biomarker of neurodegenerative diseases. Biomark. Med. 2008;2:363–384. doi: 10.2217/17520363.2.4.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blennow K, Hampel H. CSF markers for incipient Alzheimer’s disease. Lancet Neurol. 2003;2:605–613. doi: 10.1016/s1474-4422(03)00530-1. [DOI] [PubMed] [Google Scholar]
- 3.Olsson B, et al. CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: a systematic review and meta-analysis. Lancet Neurol. 2016;15:673–684. doi: 10.1016/S1474-4422(16)00070-3. [DOI] [PubMed] [Google Scholar]
- 4.Zwan MD, et al. Use of amyloid-PET to determine cutpoints for CSF markers: a multicenter study. Neurology. 2016;86:50–58. doi: 10.1212/WNL.0000000000002081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagshal D, et al. Divergent CSF tau alterations in two common tauopathies: Alzheimer’s disease and progressive supranuclear palsy. J. Neurol. Neurosurg. Psychiatry. 2015;86:244–250. doi: 10.1136/jnnp-2014-308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosso SM, et al. Total tau and phosphorylated tau 181 levels in the cerebrospinal fluid of patients with frontotemporal dementia due to P301L and G272V tau mutations. Arch. Neurol. 2003;60:1209–1213. doi: 10.1001/archneur.60.9.1209. [DOI] [PubMed] [Google Scholar]
- 7.Zetterberg, H. & Burnham, S. C. Blood-based molecular biomarkers for Alzheimer’s disease. Mol. Brain12, 26-019-0448-1 (2019). [DOI] [PMC free article] [PubMed]
- 8.Nation DA, et al. Blood-brain barrier breakdown is an early biomarker of human cognitive dysfunction. Nat. Med. 2019;25:270–276. doi: 10.1038/s41591-018-0297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deane R, Bell RD, Sagare A, Zlokovic BV. Clearance of amyloid-beta peptide across the blood-brain barrier: implication for therapies in Alzheimer’s disease. CNS Neurol. Disord. Drug Targets. 2009;8:16–30. doi: 10.2174/187152709787601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pase, M. P. et al. Assessment of Plasma Total Tau Level as a predictive biomarker for dementia and related endophenotypes. JAMA Neurol. 76, 598–606 (2019). [DOI] [PMC free article] [PubMed]
- 11.Dage JL, et al. Levels of tau protein in plasma are associated with neurodegeneration and cognitive function in a population-based elderly cohort. Alzheimers Dement. 2016;12:1226–1234. doi: 10.1016/j.jalz.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zetterberg H, et al. Plasma tau levels in Alzheimer’s disease. Alzheimers Res. Ther. 2013;5:9. doi: 10.1186/alzrt163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattsson N, et al. Plasma tau in Alzheimer disease. Neurology. 2016;87:1827–1835. doi: 10.1212/WNL.0000000000003246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mielke MM, et al. Association of Plasma Total Tau Level with cognitive decline and risk of mild cognitive impairment or dementia in the mayo clinic study on aging. JAMA Neurol. 2017;74:1073–1080. doi: 10.1001/jamaneurol.2017.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pase MP, et al. Plasma total-tau as a biomarker of stroke risk in the community. Ann. Neurol. 2019;86:463–467. doi: 10.1002/ana.25542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fossati S, et al. Plasma tau complements CSF tau and P-tau in the diagnosis of Alzheimer’s disease. Alzheimers Dement. (Amst.) 2019;11:483–492. doi: 10.1016/j.dadm.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam, E., Lee, Y. B., Moon, C. & Chang, K. A. Serum Tau proteins as potential biomarkers for the assessment of Alzheimer’s disease progression. Int. J. Mol. Sci. 21, 10.3390/ijms21145007 (2020). [DOI] [PMC free article] [PubMed]
- 18.Chen J, et al. Genome-wide association study identifies MAPT locus influencing human plasma tau levels. Neurology. 2017;88:669–676. doi: 10.1212/WNL.0000000000003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lord, J. et al. A genome-wide association study of plasma phosphorylated tau181. Neurobiol. Aging106, 304.e1–304.e3 (2021). [DOI] [PubMed]
- 20.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoglinger GU, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat. Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun G, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol. Psychiatry. 2016;21:108–117. doi: 10.1038/mp.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams HH, et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci. 2016;19:1569–1582. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hibar DP, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin J, et al. Global and regional development of the human cerebral cortex: molecular architecture and occupational aptitudes. Cereb. Cortex. 2020;30:4121–4139. doi: 10.1093/cercor/bhaa035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, et al. Tau pathology in Parkinson’s disease. Front. Neurol. 2018;9:809. doi: 10.3389/fneur.2018.00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meddens, S. F. W. et al. Genomic analysis of diet composition finds novel loci and associations with health and lifestyle. Mol. Psychiatry26, 2056–2069 (2021). [DOI] [PMC free article] [PubMed]
- 28.Doherty, A. et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat. Commun. 9, 5257 (2018). [DOI] [PMC free article] [PubMed]
- 29.Latourelle JC, Dumitriu A, Hadzi TC, Beach TG, Myers RH. Evaluation of Parkinson disease risk variants as expression-QTLs. PLoS ONE. 2012;7:e46199. doi: 10.1371/journal.pone.0046199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bishnoi RJ, Palmer RF, Royall DR. Serum interleukin (IL)-15 as a biomarker of Alzheimer’s disease. PLoS ONE. 2015;10:e0117282. doi: 10.1371/journal.pone.0117282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fontanil T, et al. Neurocan is a new substrate for the ADAMTS12 metalloprotease: potential implications in neuropathies. Cell. Physiol. Biochem. 2019;52:1003–1016. doi: 10.33594/000000069. [DOI] [PubMed] [Google Scholar]
- 32.Arning A, et al. A genome-wide association study identifies a gene network of ADAMTS genes in the predisposition to pediatric stroke. Blood. 2012;120:5231–5236. doi: 10.1182/blood-2012-07-442038. [DOI] [PubMed] [Google Scholar]
- 33.Witten A, et al. ADAMTS12, a new candidate gene for pediatric stroke. PLoS ONE. 2020;15:e0237928. doi: 10.1371/journal.pone.0237928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gevorkian G, et al. Amyloid-beta peptide binds to microtubule-associated protein 1B (MAP1B) Neurochem. Int. 2008;52:1030–1036. doi: 10.1016/j.neuint.2007.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasegawa M, Arai T, Ihara Y. Immunochemical evidence that fragments of phosphorylated MAP5 (MAP1B) are bound to neurofibrillary tangles in Alzheimer’s disease. Neuron. 1990;4:909–918. doi: 10.1016/0896-6273(90)90144-5. [DOI] [PubMed] [Google Scholar]
- 36.Jensen PH, et al. Microtubule-associated protein 1B is a component of cortical Lewy bodies and binds alpha-synuclein filaments. J. Biol. Chem. 2000;275:21500–21507. doi: 10.1074/jbc.M000099200. [DOI] [PubMed] [Google Scholar]
- 37.Joshi D, Inamdar MS. Rudhira/BCAS3 couples microtubules and intermediate filaments to promote cell migration for angiogenic remodeling. Mol. Biol. Cell. 2019;30:1437–1450. doi: 10.1091/mbc.E18-08-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng YC, et al. Genome-Wide Association analysis of young-onset stroke identifies a locus on chromosome 10q25 Near HABP2. Stroke. 2016;47:307–316. doi: 10.1161/STROKEAHA.115.011328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun BB, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinds DA, et al. Genome-wide association analysis of self-reported events in 6135 individuals and 252 827 controls identifies 8 loci associated with thrombosis. Hum. Mol. Genet. 2016;25:1867–1874. doi: 10.1093/hmg/ddw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Logue MW, et al. Targeted Sequencing of Alzheimer disease genes in African Americans implicates novel risk variants. Front. Neurosci. 2018;12:592. doi: 10.3389/fnins.2018.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melville SA, et al. Multiple loci influencing hippocampal degeneration identified by genome scan. Ann. Neurol. 2012;72:65–75. doi: 10.1002/ana.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khazaei MR, Puschel AW. Phosphorylation of the par polarity complex protein Par3 at serine 962 is mediated by aurora a and regulates its function in neuronal polarity. J. Biol. Chem. 2009;284:33571–33579. doi: 10.1074/jbc.M109.055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, et al. Rare Deleterious PARD3 Variants in the aPKC-Binding Region are implicated in the pathogenesis of human cranial neural tube defects via disrupting apical tight junction formation. Hum. Mutat. 2017;38:378–389. doi: 10.1002/humu.23153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, et al. Regulation of microtubule stability and organization by mammalian Par3 in specifying neuronal polarity. Dev. Cell. 2013;24:26–40. doi: 10.1016/j.devcel.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen YM, et al. Microtubule affinity-regulating kinase 2 functions downstream of the PAR-3/PAR-6/atypical PKC complex in regulating hippocampal neuronal polarity. Proc. Natl Acad. Sci. U. S. A. 2006;103:8534–8539. doi: 10.1073/pnas.0509955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun M, Asghar SZ, Zhang H. The polarity protein Par3 regulates APP trafficking and processing through the endocytic adaptor protein Numb. Neurobiol. Dis. 2016;93:1–11. doi: 10.1016/j.nbd.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun M, Huang C, Wang H, Zhang H. Par3 regulates polarized convergence between APP and BACE1 in hippocampal neurons. Neurobiol. Aging. 2019;77:87–93. doi: 10.1016/j.neurobiolaging.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun M, Zhang H. Par3 and aPKC regulate BACE1 endosome-to-TGN trafficking through PACS1. Neurobiol. Aging. 2017;60:129–140. doi: 10.1016/j.neurobiolaging.2017.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunn HA, Zucca S, Dao M, Orlandi C, Martemyanov KA. ELFN2 is a postsynaptic cell adhesion molecule with essential roles in controlling group III mGluRs in the brain and neuropsychiatric behavior. Mol. Psychiatry. 2019;24:1902–1919. doi: 10.1038/s41380-019-0512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsunaga H, Aruga J. Trans-Synaptic regulation of metabotropic glutamate receptors by Elfn proteins in health and disease. Front. Neural Circuits. 2021;15:634875. doi: 10.3389/fncir.2021.634875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smidak R, et al. Quantitative proteomics reveals protein kinases and phosphatases in the individual phases of contextual fear conditioning in the C57BL/6J mouse. Behav. Brain Res. 2016;303:208–217. doi: 10.1016/j.bbr.2015.12.033. [DOI] [PubMed] [Google Scholar]
- 53.Punnamoottil B, Rinkwitz S, Giacomotto J, Svahn AJ, Becker TS. Motor neuron-expressed microRNAs 218 and their enhancers are nested within introns of Slit2/3 genes. Genesis. 2015;53:321–328. doi: 10.1002/dvg.22852. [DOI] [PubMed] [Google Scholar]
- 54.Brose K, et al. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 55.Odgerel Z, et al. Whole genome sequencing and rare variant analysis in essential tremor families. PLoS ONE. 2019;14:e0220512. doi: 10.1371/journal.pone.0220512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Das, A. et al. Transcriptome sequencing reveals that LPS-triggered transcriptional responses in established microglia BV2 cell lines are poorly representative of primary microglia. J. Neuroinflammation13, 182 (2016). [DOI] [PMC free article] [PubMed]
- 57.Deniz K, et al. Plasma Biomarkers of Alzheimer’s Disease in African Americans. J. Alzheimers Dis. 2021;79:323–334. doi: 10.3233/JAD-200828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang EY, et al. ForestPMPlot: a flexible tool for visualizing heterogeneity between studies in meta-analysis. G3 (Bethesda) 2016;6:1793–1798. doi: 10.1534/g3.116.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed]
- 64.Watanabe, K., Umicevic Mirkov, M., de Leeuw, C. A., van den Heuvel, M. P. & Posthuma, D. Genetic mapping of cell type specificity for complex traits. Nat. Commun. 10, 3222 (2019). [DOI] [PMC free article] [PubMed]
- 65.Malik R, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Satizabal C, Beiser AS, Seshadri S. Incidence of dementia over three decades in the Framingham Heart Study. N. Engl. J. Med. 2016;375:93–94. doi: 10.1056/NEJMc1604823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarnowski C, et al. Whole genome sequence analyses of brain imaging measures in the Framingham Study. Neurology. 2018;90:e188–e196. doi: 10.1212/WNL.0000000000004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife7, 10.7554/eLife.34408 (2018). [DOI] [PMC free article] [PubMed]
- 70.Psaty BM, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grove ML, et al. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS ONE. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McLaren, W. et al. The ensembl variant effect predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available either within the main article or the supplementary information. Summary statistics from the ancestry-specific meta-analyses of circulating levels of total-tau have been deposited and are publicly accessible on the GWAS catalog FTP (study accession numbers GCST90095138, and GCST90095139). Genome-wide summary statistics for complex disorders used in the secondary analyses were downloaded from public repositories (GWAS catalog: https://www.ebi.ac.uk/gwas/downloads/summary-statistics; and IEU GWAS database: https://gwas.mrcieu.ac.uk/).