Abstract
High plasma lipid/lipoprotein levels are risk factors for various metabolic diseases. We previously showed that circadian rhythms regulate plasma lipids, and deregulation of these rhythms cause hyperlipidemia and atherosclerosis in mice. Here, we show that global and liver-specific Bmal1-deficient mice maintained on a chow or a Western diet developed hyperlipidemia, denoted by the presence of higher amounts of triglyceride- and ApoAIV-rich larger chylomicron and very-low-density lipoprotein, due to overproduction. Bmal1 deficiency decreased Shp and increased MTP, a key protein that facilitates primordial lipoprotein assembly and secretion. Moreover, we show that Bmal1 regulates Crebh to modulate ApoAIV expression and the assembly of larger lipoproteins. This is supported by the observation that Crebh- and ApoAIV-deficient mice, along with Bmal1-deficient mice with knockdown of Crebh, had smaller lipoproteins. Further, overexpression of Bmal1 in Crebh-deficient mice had no effect on ApoAIV expression and lipoprotein size. These studies ind15icate that regulation of ApoAIV and assembly of larger lipoproteins by Bmal1 requires Crebh. Mechanistic studies showed that Bmal1 regulates Crebh expression by two mechanisms. First, Bmal1 interacts with the Crebh promoter to control circadian regulation. Second, Bmal1 increases Rev-erbα expression, and Rev-erbα interacts with the Crebh promoter to repress expression. In short, Bmal1 modulates both the synthesis of primordial lipoproteins and their subsequent expansion into larger lipoproteins by regulating two different proteins, MTP and ApoAIV, via two different transcription factors, Shp and Crebh. It is likely that disruptions in circadian mechanisms contribute to hyperlipidemia, and avoiding disruptions in circadian rhythms may limit/prevent hyperlipidemia and atherosclerosis.
Keywords: Circadian rhythms, Rev-erbα, Lipid metabolism, Lipoprotein assembly, Transcriptional regulation
Graphical Abstract
Introduction
Early studies established that an elevated level of low-density lipoprotein (LDL) cholesterol is a risk factor for atherosclerosis. Therefore, myriad of successful therapeutic interventions have reduced plasma LDL and incidence of atherosclerosis worldwide. Despite these innovations, there are significant residual risks (1–3). Recent studies identified high triglyceride and very-low-density lipoprotein (VLDL) as risk factor for myocardial infarction. In an eleven-year Copenhagen General Population study, VLDL cholesterol explained 50% of the myocardial infarction risk (4). Prenner et al. showed that VLDL cholesterol also associates with coronary artery calcification in type 2 diabetes (5). Thus, mechanisms that regulate plasma triglyceride-rich lipoproteins are of significant interest.
Circadian rhythms are orchestrated by enhancers and repressors called “core clock genes” that exhibit autoregulatory mechanisms. Circadian locomotor output cycles kaput (Clock) and brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1 (Bmal1) heterodimers (Clock:Bmal1) bind to enhancer-boxes (E-boxes) in the promoters of Per and Cry genes to increase expression of period (Per) and cryptochrome (Cry) repressors, respectively. When concentrations of Per and Cry proteins increase, they form heterodimers to repress Clock:Bmal1 activity. Besides this major autoregulatory loop, circadian rhythms are also regulated by an auxiliary loop consisting of a repressor, Rev-erbα (gene Nr1d1), and an activator, RORα that regulates Bmal1 expression (6). Clock and Bmal1 also regulate several physiological processes by modulating the expression of “clock-controlled” genes (7, 8). We previously identified short heterodimer partner (Shp), USF2, and GATA4 as clock-controlled transcription factors that regulate primordial lipoprotein assembly, macrophage cholesterol efflux, and hepatic cholesterol excretion to bile, respectively (9–11).
Lipoprotein assembly likely occurs in two steps. In the first step, the newly-translated apolipoprotein B (apoB) interacts with the endoplasmic reticulum (ER) membrane. ApoB is then co-translationally assembled into smaller high-density lipoprotein (HDL)-size primordial lipoproteins with the assistance of microsomal triglyceride transfer protein (MTP). The primordial lipoproteins undergo a second step of “core expansion,” resulting in the synthesis of chylomicrons (CM) and VLDL by the intestine and liver, respectively. Very little is known about core expansion and its regulation. Because each lipoprotein contains one apoB molecule, the lipoprotein surface is believed to be stabilized by other apolipoproteins during core expansion. Apolipoprotein AIV (ApoAIV, gene Apoa4) may play a role in this process (12, 13). ApoAIV production is significantly induced during the postprandial state and is thought to stabilize CM during core expansion (13, 14). ApoAIV is less abundant in the liver compared to the intestine (13, 15); nevertheless, its role in VLDL assembly in human hepatoma cells and in mice has been noted (15, 16). Increased hepatic ApoAIV is thought to mitigate steatosis by enhancing assembly of larger (≥120 nm) VLDL particles (15, 16).
The assembly of apoB-containing lipoproteins is regulated at the post-translational level (17). However, the synthesis of MTP and ApoAIV is regulated at the transcriptional level (18). MTP and ApoAIV mRNA levels show circadian expression in ad libitum-fed mice (9, 19–21). In ClockΔ19/Δ19 mice, MTP and ApoAIV expression does not show diurnal rhythms (9, 19, 22). However, the role of Bmal1 in ApoAIV-mediated lipid homeostasis is unknown. ApoAIV expression is mainly regulated by cAMP-responsive element-binding protein H (Crebh, gene Creb3l3) (23–25) which is expressed in the liver and intestine (26). Crebh interacts with two Crebh-responsive elements in the Apoa4 promoter to increase expression (24). Understanding the circadian regulatory mechanisms of Crebh expression is of great importance.
Crebh (~75 kDa) is an ER membrane-bound transcription factor (Crebh-ER). Its activation involves transport to and proteolytic cleavage in the Golgi, production of a ~50 kDa N-terminal fragment (Crebh-N), and translocation to the nucleus. Crebh-N interacts with Crebh-responsive elements in the promoters to enhance expression of genes in lipid metabolism (27, 28). Steady-state levels of Crebh-ER and Crebh-N proteins are regulated differently. IL-6 and TNFα strongly induce Crebh-ER but have no effect on Crebh-N. Crebh-ER is regulated by ER-associated degradation involving E3 ubiquitin ligase HRD1 and its co-activator Sel1L (29, 30). Ablation of HRD1 increases Crebh-ER and Crebh target gene expression (30), suggesting that increases in Crebh-ER might augment Crebh-N production and enhance gene expression. Production of Crebh-N shows rhythmicity and is regulated by Bmal1 via AKT-GSK3β signaling (31). As opposed to the breadth of knowledge about the production and catabolism of Crebh proteins, very little is known about the transcriptional regulation of Crebh expression. Earlier studies reported no temporal changes in Crebh mRNA (31); however, recent studies describe robust diurnal regulation of Crebh mRNA in wild-type (WT) mice and an absence of diurnal regulation in Bmal1−/− mice (30). How Bmal1 regulates Crebh expression has not been elucidated.
We previously showed that plasma triglyceride and cholesterol, mainly in apoB-containing lipoproteins, exhibit diurnal rhythms (19). These rhythms are not seen in ClkΔ19/Δ19 mice and in Bmal1−/−Apoe−/− mice (9, 22). Here, we show that Bmal1 and Rev-erbα temporally coordinate Crebh regulation to modulate ApoAIV expression and assembly of larger lipoproteins.
Results
Sustained hyperlipidemia in chow-fed Bmal1-deficient mice
Plasma triglyceride levels in total (Supplementary Figure 1A, left) and apoB-containing lipoproteins (Supplementary Figure 1A, right) were higher at 24:00 (midnight) than at 12:00 (noon) in chow-fed Bmal1+/+ mice. In contrast, triglyceride remained >2-fold higher in Bmal1−/− mice throughout the day (Supplementary Figure 1A, left), due to increases in apoB-containing lipoproteins (Supplementary Figure 1A, right). Plasma CM and VLDL particles were larger in Bmal1−/− than in Bmal1+/+ mice, while LDL and HDL sizes were not different (Figure 1A). These studies indicate that hypertriglyceridemia in Bmal1−/− mice is secondary to increases in larger lipoproteins.
Figure 1: Hypertriglyceridemia and larger lipoproteins in chow-fed Bmal1−/− mice.
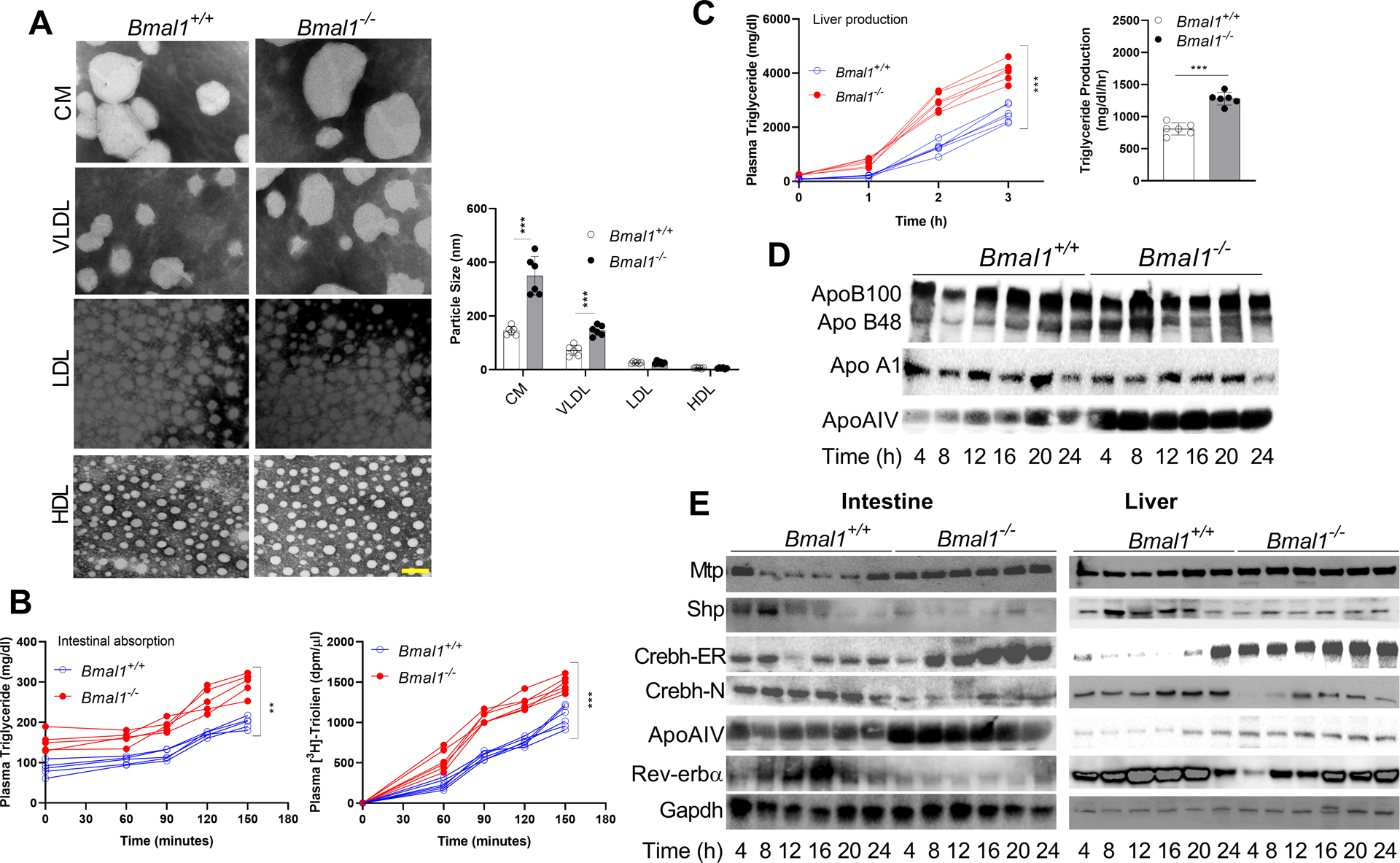
Bmal1−/− and Bmal1+/+ male siblings (12-week-old, ad libitum chow-fed) were sacrificed at 4 h intervals.
(A) Plasma collected at 12:00 was subjected to sequential density gradient ultracentrifugation. Isolated lipoproteins were negatively stained for electron microscopy (left). Scale bar = 50 nm. Diameters of different particles were quantified (right). Values are mean ± SD. ** p<0.01 and *** p<0.001, t-test.
(B) Mice fasted overnight were injected intraperitoneally with P407. After 1 h, they were gavaged with 3H-triolein in olive oil (50 μL) at 12:00. Blood was collected at the indicated times to measure triglyceride (left) and radioactivity (right). Two-way ANOVA, ** p<0.05, *** p<0.001.
(C) Mice fasted for 5 h were injected with P407 at 12:00, and plasma triglycerides were determined at different times. Time-dependent increases in plasma triglycerides (left) and triglyceride production rates (right) were significantly higher in Bmal1−/− than in Bmal1+/+ mice. Mean ± SD, n = 5–6 for each time point. Two-way ANOVA and t-test *** p<0.001.
(D) Plasma (1 μL) from different times was subjected to Western blotting to measure different apolipoproteins.
(E) Equal amounts of liver protein (25 μg) were used to detect proteins.
We then measured hepatic triglyceride and cholesterol in the livers of these mice (Supplementary Figure 1B, 1C). Bmal1−/− mice had significantly higher amounts of these lipids and showed increased Oil Red O staining. Thus, Bmal1 deficiency leads to hepatosteatosis.
Hypertriglyceridemia can be due to reduced catabolism or overproduction of lipoproteins. Lipoprotein catabolism begins with the hydrolysis of triglyceride by lipases. Post-heparin plasma lipase activity in Bmal1+/+ mice was high at 24:00 and low at 12:00 (Supplementary Figure 1D), consistent with (32). This activity did not change in Bmal1−/− mice and was high at both times (Supplementary Figure 1D). Hepatic lipoprotein lipase expression peaked during the day and was lower at night in Bmal1+/+ mice, but in Bmal1−/− mice expression was high throughout the day (Supplementary Figure 1E). These studies suggest that hyperlipidemia in Bmal1−/− mice may not be due to decreases in lipase activity.
Next, we asked whether hyperlipidemia is secondary to lipoprotein overproduction. Intestinal triglyceride and 3H-triolein absorption (Figure 1B), as well as hepatic triglyceride production (Figure 1C), were significantly increased in Bmal1−/− mice. This indicates that Bmal1 deficiency augments lipoprotein production.
We previously showed that Clock regulates MTP and Shp, and their dysregulation in ClockΔ19/Δ19 mice leads to hypertriglyceridemia (9). Similarly, MTP and Shp mRNA showed diurnal variations in Bmal1+/+, but not in Bmal1−/−, mice (Supplementary Figure 2A). Intestinal and hepatic apoB and apoAI mRNA levels in Bmal1+/+ and Bmal1−/− mice did not change (Supplementary Figure 2B). Plasma ApoAIV protein levels in Bmal1+/+, but not in Bmal1−/−, mice, showed diurnal changes (Figure 1D). Bmal1−/− plasma contained significantly higher amounts of ApoAIV at all times. Consistent with protein levels, intestinal and hepatic ApoAIV mRNA levels show diurnal variations in Bmal1+/+, but not in Bmal1−/− mice (Supplementary Figure 2C). ApoAIV mRNA levels were higher throughout the day in Bmal1−/− mice. These studies show that Bmal1 deficiency abrogates diurnal expression of MTP, Shp, and ApoAIV, and Bmal1−/− mice assimilate larger triglyceride- and ApoAIV-rich lipoproteins in the plasma.
A major regulator of ApoAIV is Crebh (24). In Bmal1+/+ mice, intestinal and hepatic Crebh mRNA levels showed diurnal variations (Supplementary Figure 2C). In contrast, Crebh mRNA levels were significantly increased in the intestine and liver of Bmal1−/− mice and did not show temporal changes. Besides ApoAIV, other targets of Crebh were also increased in Bmal1−/− mice (Supplementary Figure 2D).
Bmal1 regulates Rev-erbα expression (33). We also observed that intestinal and hepatic Rev-erbα mRNA levels peak in the daytime in Bmal1+/+ mice (Supplementary Figure 2C). In Bmal1−/− mice, Rev-erbα expression was significantly reduced, and did not change within a day.
Next, we studied changes in protein levels (Figure 1E). Consistent with mRNA levels, intestinal and hepatic protein levels of MTP, Shp, Crebh-ER, ApoAIV, and Rev-erbα showed diurnal variations in Bmal1+/+ mice. In Bmal1−/− mice, these protein levels did not show temporal changes and remained either high (MTP, Crebh-ER, ApoAIV) or low (Shp, Crebh-N, Rev-erbα).
In summary, chow-fed Bmal1−/− mice accumulate ApoAIV- and triglyceride-rich larger CM- and VLDL-size particles. Additionally, Bmal1−/− mice show increased expression of hepatic and intestinal MTP, ApoAIV, and Crebh; and reduced expression of Shp and Rev-erbα.
A Western diet exacerbates hyperlipidemia in Bmal1−/− mice
Western diet-fed Bmal1−/− mice had higher plasma total and non-HDL triglyceride and cholesterol (Figure 2A), larger CM and VLDL (Figure 2B & Supplementary Figure 3A), and higher apoB100 and ApoAIV protein levels (Figure 2C). Since each lipoprotein contains one apoB, increases in apoB suggest more particles. Intestinal triglyceride and 3H-triolein absorption (Supplementary Figure 3B) and hepatic triglyceride production rates (Supplementary Figure 3C) were significantly higher in Bmal1−/− mice. These mice had significantly higher hepatic lipids and showed enhanced Oil Red O staining (Supplementary Figure 3D, 3E). These studies indicate that Western diet enhances plasma levels of larger apoB100- and ApoAIV-containing triglyceride- and cholesterol-rich particles in Bmal1−/− mice by increasing intestinal and hepatic lipoprotein production.
Figure 2: Hyperlipidemia in Western diet-fed Bmal1−/− mice.
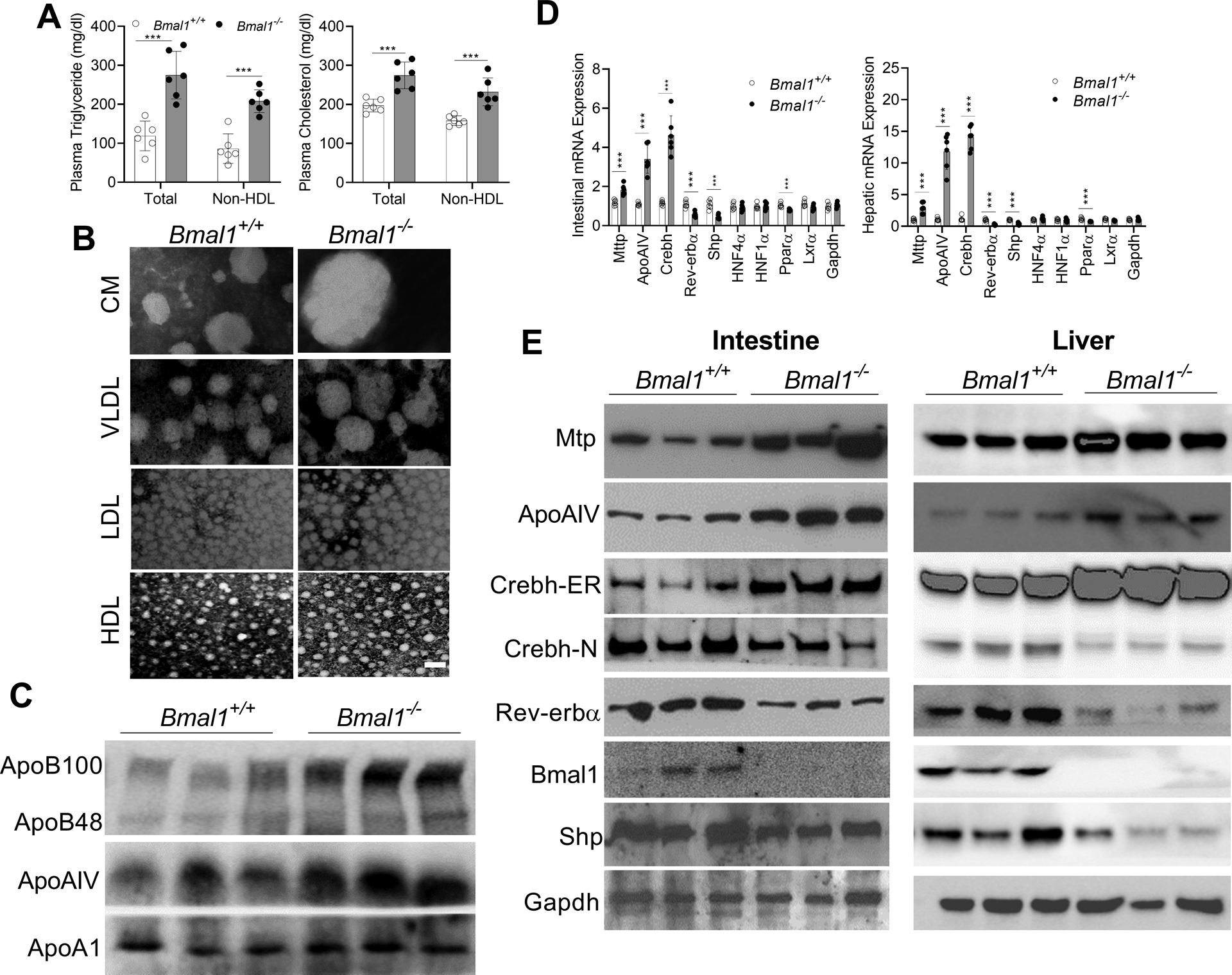
Male, 12-week-old Bmal1+/+ and Bmal1−/− mice were fed a Western diet for 2 months. Plasma and tissues were collected at 12:00.
(A) Cholesterol and triglyceride concentrations were measured in total and non-HDL plasma. *** p<0.001, t-test. Each dot represents individual mouse.
(B) Isolated plasma lipoproteins were subjected to negative staining. Scale bar = 50 nm.
(C) Plasma (1 μL) was used to measure different apolipoproteins by Western blotting.
(D) Intestinal and hepatic mRNA levels of MTP and ApoAIV and the transcription factors that regulate their expression were compared. Values are mean ± SD, n=6. *** p<0.001, t-test.
(E) Intestinal and hepatic proteins were determined using specific antibodies.
To understand mechanisms, we measured mRNA and protein levels of key genes in lipoprotein assembly. Intestines and livers from Bmal1−/− mice had higher mRNA levels of MTP, ApoAIV (7- to 13-fold), and Crebh (~15-fold), while mRNA levels of Shp and Rev-erbα were significantly reduced (Figure 2D). Similar changes in protein levels were observed (Figure 2E). These studies suggest that Bmal1 deficiency significantly alters intestinal and hepatic expression of several lipid metabolism genes in Western diet-fed mice.
Liver-specific ablation of Bmal1 increases Crebh and ApoAIV expression
We generated liver-specific Bmal1-deficient (L-Bmal1−/−) mice by crossing Bmal1fl/fl mice with Alb-Cre mice, and fed a chow diet (11). Plasma triglyceride, cholesterol (Supplementary Figure 4A), apoB100, and ApoAIV (Supplementary Figure 4B) levels were higher in male L-Bmal1−/− mice compared to Bmal1fl/fl mice. L-Bmal1−/− mice had larger plasma VLDL at both 24:00 and 12:00 (Supplementary Figure 4C). Hepatic Crebh, ApoAIV, and Rev-erbα mRNA levels showed diurnal variations in control Bmal1fl/fl mice, but not in L-Bmal1−/−, mice (Supplementary Figure 4D). Hepatic Crebh-ER and ApoAIV protein levels were higher, but Rev-erbα levels were lower, in chow-fed L-Bmal1−/− mice (Supplementary Figure 4E). Diurnal changes in intestinal Crebh, ApoAIV, and Rev-erbα mRNA were not affected (Supplementary Figure 4F), indicating liver-specific changes.
Similar to male mice, chow-fed L-Bmal1−/− female mice had higher plasma triglyceride/cholesterol levels (Supplementary Figure 5A), higher apoB100 and ApoAIV levels (Supplementary Figure 5B), and larger VLDL particles (Supplementary Figure 5C). Hepatic Crebh, ApoAIV, and Rev-erbα mRNA and protein levels showed diurnal variations in Bmal1fl/fl, but not in L-Bmal1−/−, female mice (Supplementary Figure 5D, 5E). These studies show that liver-specific Bmal1 deficiency increases plasma lipids, number of larger VLDL particles, hepatic Crebh and ApoAIV mRNA and protein levels in chow-fed male and female mice.
Total plasma and non-HDL triglyceride and cholesterol levels (Figure 3A), as well as apoB100, apoB48, and ApoAIV protein levels (Figure 3B) were significantly higher in Western diet fed male L-Bmal1−/− than in Bmal1fl/fl mice. VLDL particles were larger in the plasma of L-Bmal1−/− (Figure 3C & Supplementary Figure 6). These studies indicate accretion of greater number of apoB100-containing and ApoAIV-enriched larger VLDL particles.
Figure 3: Hyperlipidemia in L-Bmal1−/− mice fed a Western diet.
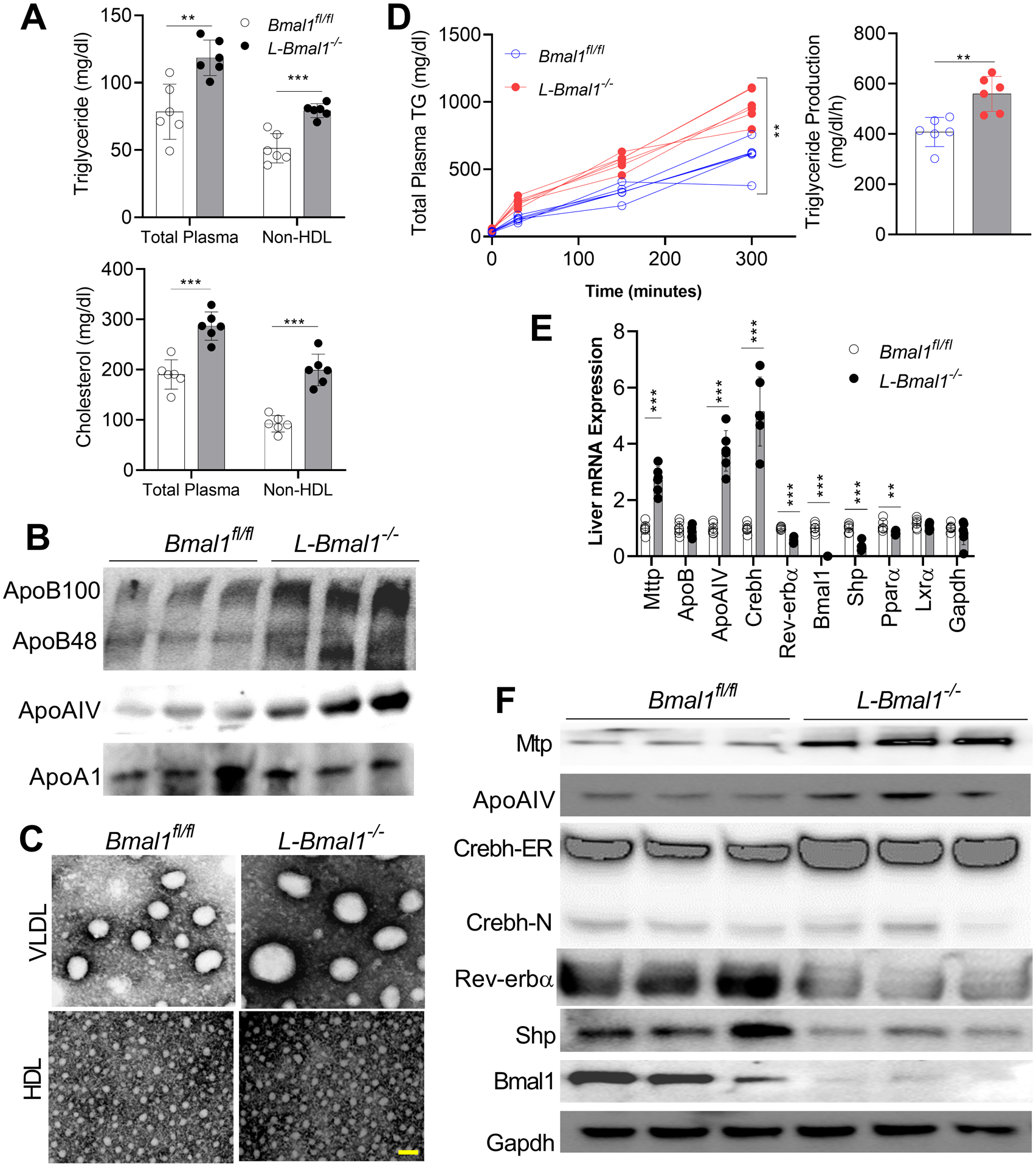
Male 16-week-old Bmal1fl/fl and L-Bmal1−/− mice were fed a Western diet for 2 months. Plasma and tissues were collected at 12:00.
(A) Total and non-HDL plasma cholesterol and triglyceride concentrations. Values are mean ± SD, n=6. ** p<0.01, and *** p<0.001, t-test.
(B) Plasma was used to measure apolipoproteins by western blotting.
(C) Plasma VLDL and HDL particle diameters were measured. Scale bar = 50 nm.
(D) Mice were fasted for 5 h and were injected intraperitoneally with P407 (30 mg/mouse) at 12:00. Triglyceride determinations were done before injection (0 minutes) and at the indicated times (left). The triglyceride production rates were higher in Bmal1−/− than in Bmal1+/+ mice (right). Two-way ANOVA and t-test ** p<0.01.
(E) Expression of MTP and ApoAIV were significantly increased in L-Bmal1−/− livers. Crebh mRNA increased by ~5-fold. Values are mean ± SD. *** p<0.001, t-test.
(F) Hepatic protein expression determined by Western blotting.
We then concentrated on explaining origins of larger VLDL. Liver triglyceride production was significantly higher in Western diet fed L-Bmal1−/− than in Bmal1fl/fl mice (Figure 3D). Expression analysis showed increased expression of MTP (~3-fold) and ApoAIV (~4-fold) with no change in apoB mRNA (Figure 3E). Analysis of the transcription factors that regulate MTP and ApoAIV expression showed significant increases in Crebh (~5-fold) and decreases in Rev-erbα and Shp (Figure 3E). Consistent with changes in mRNA, ApoAIV and Crebh-ER protein levels increased, and Rev-erbα and Shp levels decreased (Figure 3F). Thus, liver-specific Bmal1 deficiency increases plasma apoB100 and ApoAIV levels and hepatic Crebh and ApoAIV expression.
The above studies show that Bmal1 deficiency decreases Rev-erbα and Shp, and augments MTP, Crebh, and ApoAIV protein and mRNA levels. We previously reported that ClockΔ19/Δ19 and Bmal1−/−Apoe−/− mice had reduced hepatic Shp and increased MTP expression (9–11, 19). The several fold increases in Crebh and ApoAIV in Bmal1-deficient livers were, however, novel observations. Therefore, we attempted to establish relationships amongst the expressions of these proteins and their roles in the production of larger lipoproteins.
Nr1d1−/− mice have larger lipoproteins
Nr1d1−/− mice had higher plasma triglyceride and cholesterol levels at 24:00 and 12:00 (Figure 4A & Supplementary Figure 7A). The plasma apoB100, apoB48, and ApoAIV protein levels were elevated, but apoA1 levels were unaffected in Nr1d1−/− mice (Figure 4A, bottom). The Nr1d1−/− mice had larger plasma CM and VLDL compared to controls (Figure 4B & Supplementary Figure 7B), and had higher hepatic mRNA levels of ApoAIV and Crebh (Supplementary Figure 7C). Thus, Rev-erbα deficiency is associated with higher expression of ApoAIV and Crebh, increased plasma apoB and ApoAIV, and larger plasma lipoproteins.
Figure 4: Effects of Rev-erbα, ApoAIV, and Crebh deficiencies on plasma lipoproteins and apolipoproteins.
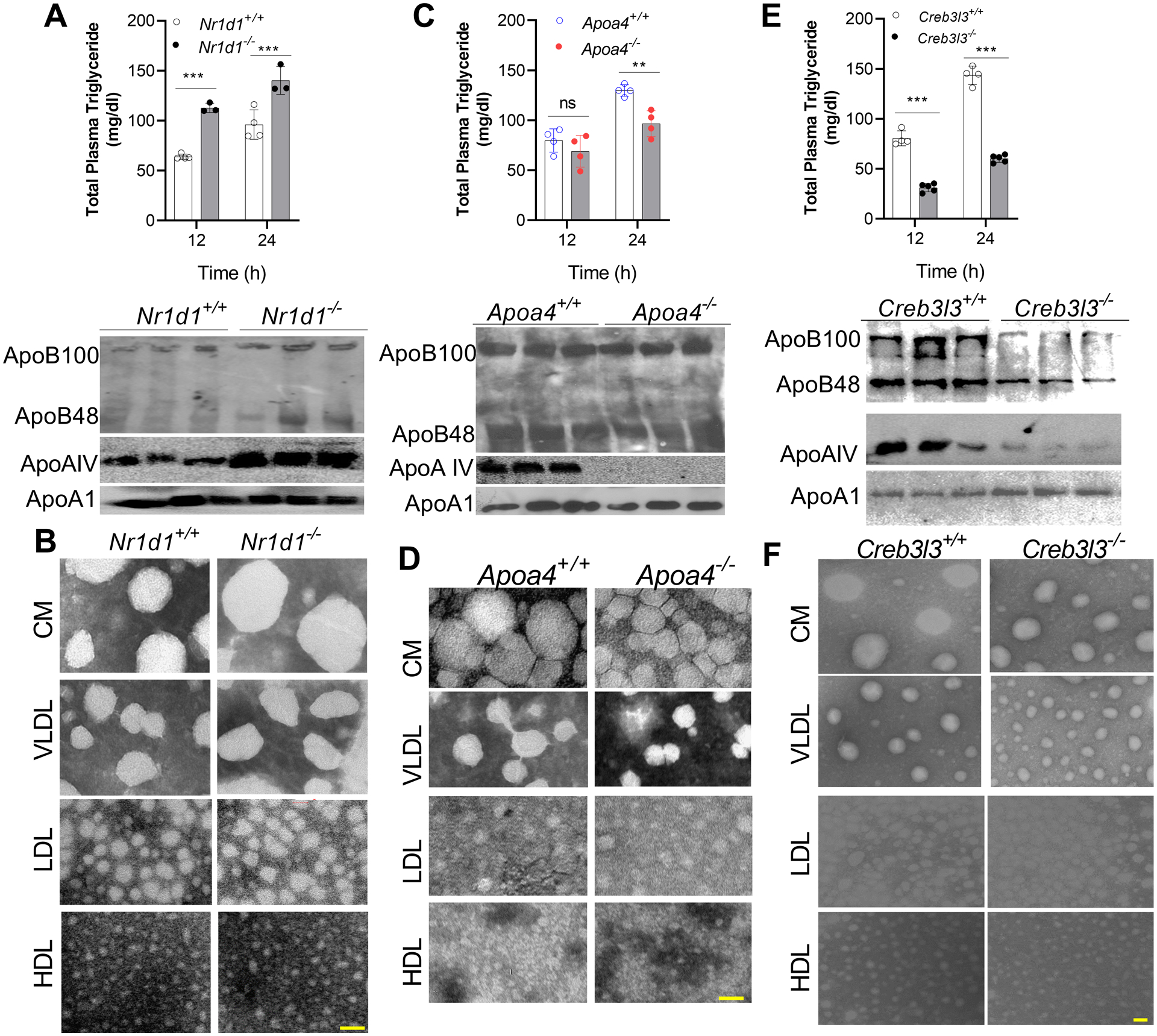
(A-B) Male 12-week-old Nr1d1−/− and Nr1d1+/+ mice were fed a chow diet. (A, top) Plasma triglyceride levels were quantified at 24:00 and 12:00. (A, bottom) Apolipoprotein levels in plasma collected at 12:00 were measured via Western blotting. (B) Isolated lipoproteins were analyzed by electron microscopy. Scale bar = 50 nm.
(C-D) Male 16-week-old Apoa4−/− and Apoa4+/+ mice were fed a chow diet. (C) Triglyceride levels were measured at 24:00 and 12:00 in total plasma (top). Plasma obtained at 12:00 was used to detect apoB and apoA1 levels (bottom), and (D) were used to isolate different lipoproteins for negative staining. Scale bar = 50 nm.
(E-F) Male 16-week-old Creb3l3−/− and Creb3l3+/+ mice on a chow diet were sacrificed at different times. Plasma was used (top) to measure total triglyceride and cholesterol, Values are mean ± SD. *** p<0.001, t-test, and (bottom) to detect apolipoproteins by western blotting. (F) Lipoproteins were isolated at 12:00 and for negative staining. Scale bar = 50 nm.
Apoa4−/− mice have smaller lipoproteins
Apoa4−/− mice had lower plasma levels of total and non-HDL triglyceride at midnight than Apoa4+/+ mice (Figure 4C & Supplementary Figure 8A). ApoAIV deficiency did not significantly affect plasma apoB100, apoB48, and apoAI levels (Figure 4C, bottom). However, Apoa4−/− mice had smaller CM and VLDL particles (Figure 4D & Supplementary Figure 8B). They showed no defect on the diurnal expression of intestinal and hepatic Bmal1, Re-erbα, and Crebh mRNA levels (Supplementary Figure 8C). Thus, Apoa4−/− mice have smaller CM/VLDL and lower triglyceride levels.
Creb3l3−/− mice have lower ApoAIV levels and smaller lipoproteins
Chow-fed nonfasted Creb3l3−/− mice had lower plasma triglyceride and cholesterol levels compared with Creb3l3+/+ mice (Figure 4E & Supplementary Figure 9A). Creb3l3−/− mice had lower plasma apoB100, apoB48, and ApoAIV levels (Figure 4E, bottom) and had smaller CM/VLDL particles (Figure 4F & Supplementary Figure 9B). ApoAIV mRNA levels remained low throughout the day in the livers of Creb3l3−/− mice (Supplementary Figure 9C). The role of Crebh in ApoAIV regulation was further studied in human hepatoma Huh-7 cells (Supplementary Figure 9D) and mouse hepatocytes (Supplementary Figure 9E) after knockdown (KD). SiCreb3l3 significantly reduced Crebh and ApoAIV without affecting Bmal1 expression. These studies support the hypothesis that Crebh regulates ApoAIV expression.
Bmal1 requires Crebh to regulate ApoAIV expression
Next, we asked whether Bmal1 can regulate ApoAIV independently of Crebh. First, we overexpressed Bmal1 in Creb3l3−/− mice. Adv-Bmal1 significantly increased hepatic Bmal1, Rev-erbα, and Shp, and decreased MTP, but had no effect on ApoAIV (Figure 5A). Adv-Bmal1 had no effect on intestinal gene expression (Supplementary Figure 10A), and did not significantly affect plasma apolipoproteins (Figure 5B, top) or lipoprotein sizes (Figure 5B, bottom & Supplementary Figure 10B). These data show that Bmal1 does not regulate ApoAIV or VLDL size in the absence of Crebh.
Figure 5: Overexpression of Bmal1 in the livers of Creb3l3−/− mice and knockdown of Crebh in L-Bmal1−/− mice have no effect on ApoAIV expression.
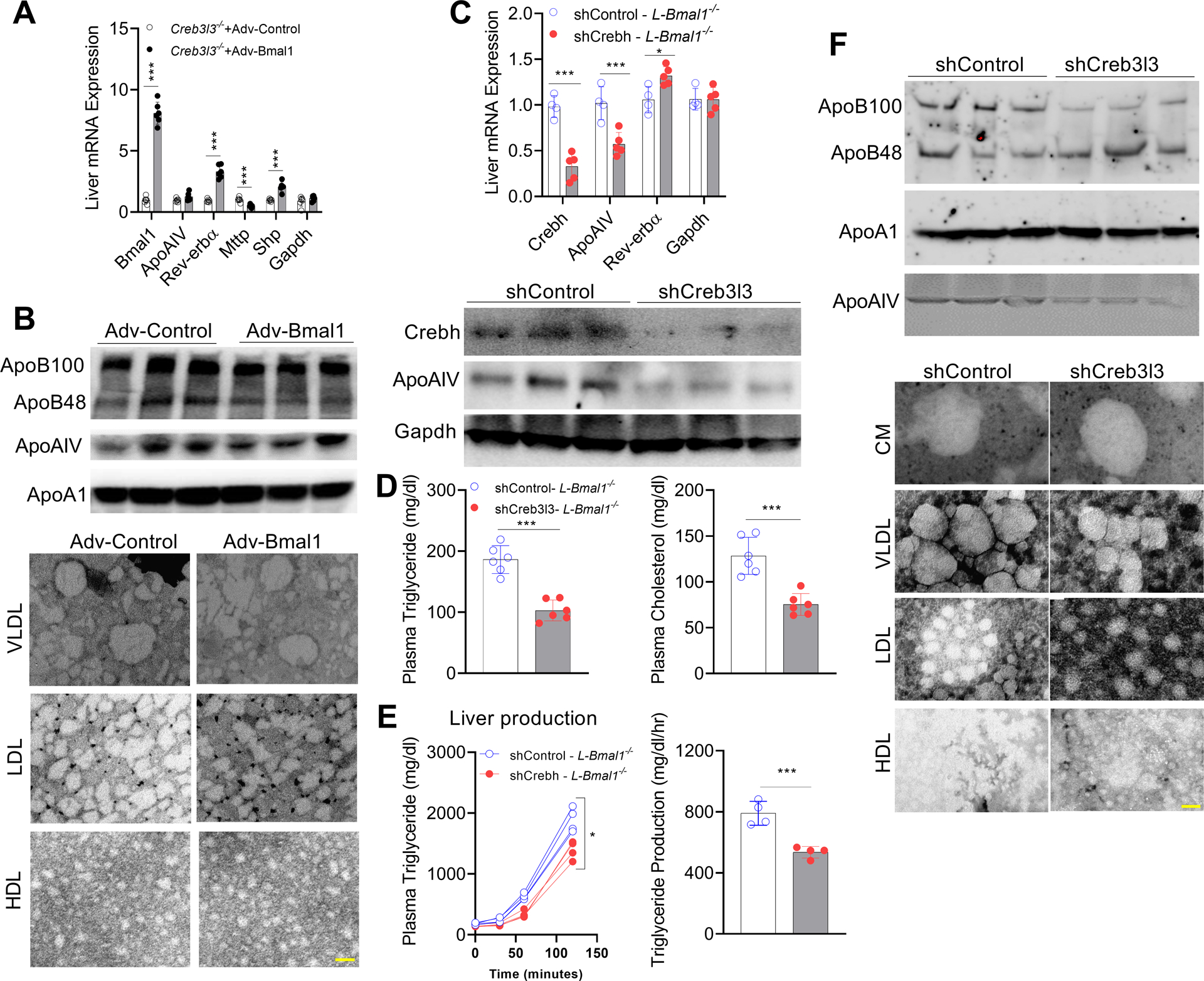
(A-B) Male Creb3l3−/− mice were intravenously injected with adenoviruses expressing GFP or Bmal1 (1.5 × 1011 GC/mouse). After 4 weeks, livers and intestines were used to measure mRNA levels. Mean ± SD. N=5–6, *** p<0.001, t-test. (B) Top, plasma was used to measure apolipoproteins by western blotting. Bottom, isolated lipoproteins were studied by electron microscopy. Scale bar = 50 nm.
(C-F) L-Bmal1−/− (16-weeks-old, male) mice were transduced intravenously with AAV-shCrebh3l3 (2.5 × 1011 GC/mouse). After 3 months, plasma and tissues were collected. (C) Livers were used to measure mRNA (top), and protein (bottom) levels. (D) Plasma was used to measure lipid levels. Values are represented as mean ± SD. * p<0.05, and *** p<0.001, t-test. (E) Mice were injected with P407 to measure plasma triglyceride levels (left) and triglyceride production rates (right). (F) Plasma collected at 12:00 was used to measure apolipoproteins (top) and to isolate lipoproteins for negative staining (bottom). Scale bar = 50 nm.
Second, the role of Crebh was interrogated by KD of Crebh in L-Bmal1−/− mice. ShCreb3l3 reduced hepatic Crebh and ApoAIV mRNA levels without affecting Rev-erbα (Figure 5C, top). It also reduced hepatic Crebh-ER and ApoAIV protein levels (Figure 5C, bottom), and decreased plasma triglyceride and cholesterol (Figure 5D). Furthermore, KD of Creb3l3 in L-Bmal1−/− mice reduced plasma triglyceride production rates (Figure 5E) as well as plasma apoB100 and ApoAIV levels without affecting apoAI levels (Figure 5F, top). KD of hepatic Crebh reduced VLDL size but had no effect on CM size (Figure 5F, bottom & Supplementary Figure 10C). These studies suggest that production of ApoAIV and larger VLDL in L-Bmal1−/− mice is regulated by Crebh.
In short, our studies interrogating the overexpression of Bmal1 in Creb3l3−/− mice and the KD of Crebh in L-Bmal1−/− mice show that Crebh does not regulate Bmal1 or Rev-erbα expression, and Bmal1 regulates ApoAIV expression by modulating Crebh expression. Therefore, we next concentrated on explaining how Bmal1 regulates Crebh.
Bmal1 and Rev-erbα cooperate to regulate Crebh expression
First, we reduced the expression of Bmal1 and Rev-erbα—individually and in combination—using specific siRNAs in mouse primary hepatocytes (Figure 6A) and human hepatoma Huh-7 cells (Supplementary Figure 11). In mouse primary hepatocytes, siBmal1 reduced Bmal1 and Rev-erbα, and increased Crebh and ApoAIV mRNA levels (Figure 6A, top). SiRev-erbα reduced Rev-erbα and increased Bmal1, Crebh, and ApoAIV (Figure 6A, middle). A combination of siBmal1 and siRev-erbα reduced Bmal1 and Rev-erbα and increased ApoAIV and Crebh (Figure 6A, bottom). Protein levels changed similarly to mRNA levels (Figure 6B). Similar observations were made in Huh-7 cells (Supplementary Figure 11). These studies indicate that both Bmal1 and Rev-erbα act as repressors of Crebh and ApoAIV.
Figure 6: Regulation of Crebh expression by Bmal1 and Rev-erbα.
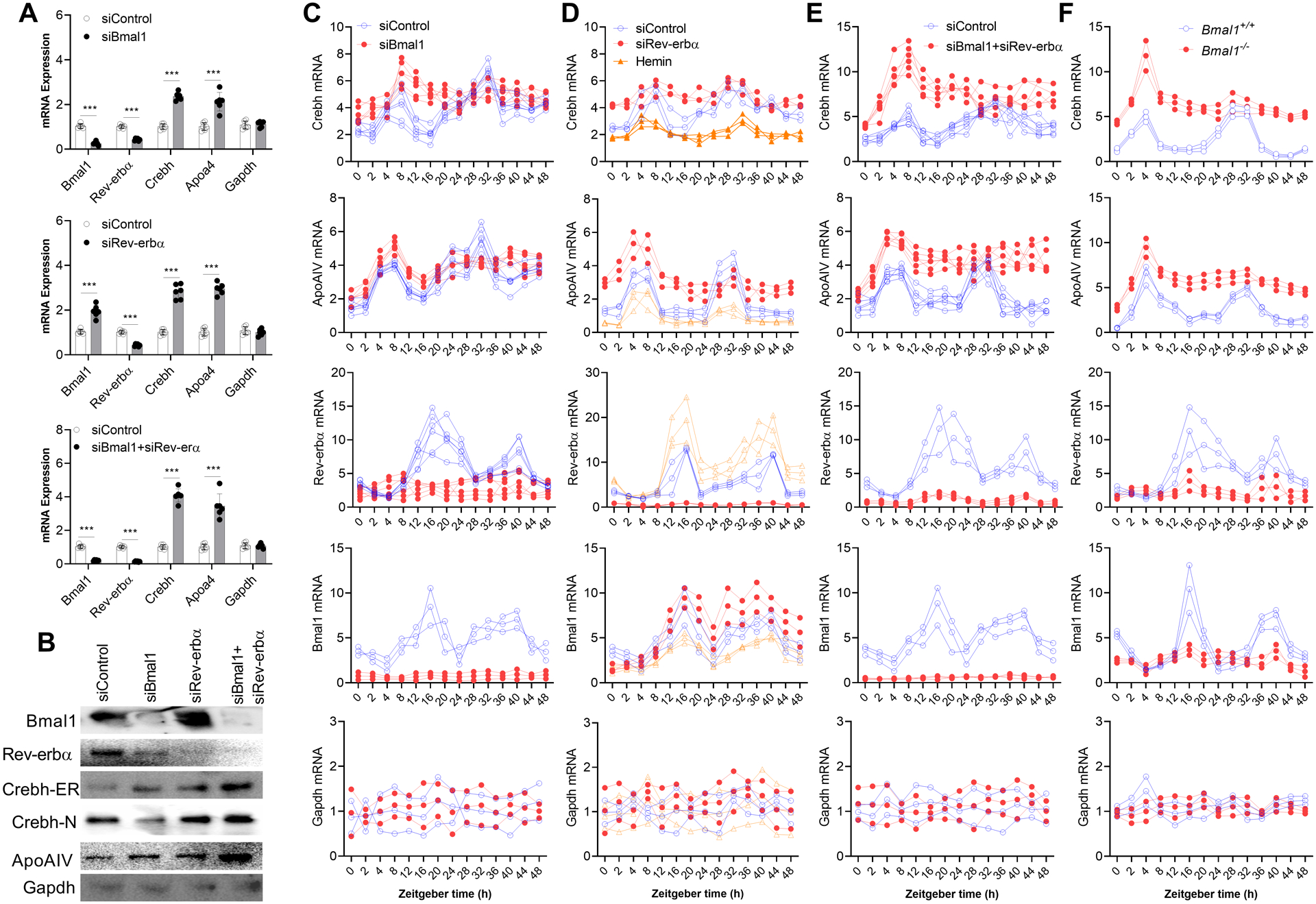
(A-B) (A) Primary hepatocytes from 10 week-old male WT mice were transfected with different indicated siRNAs (pool of four siRNAs) and mRNA levels were quantified after 48 h. (B) Cell lysates were used for Western blotting. Values are represented as mean ± SD. *** p<0.001, t-test.
(C-F) (C) SiBmal1 or siControl were introduced in WT mouse hepatocytes. After 48 h, cells were cultured in serum-free media for 16 h, exposed to media containing 50% FBS for 2 h, and then cultured in serum-free media. At different times, cells were collected to measure mRNA levels. (D) Rev-erbα was knocked down in mouse primary hepatocytes using a pool of four siRNA duplexes, or its expression was increased by treating cells with hemin (25 μM) for 48 h. Cells were then subjected to serum shock as above. In cells treated with hemin, hemin was present at all times. Different wells were collected at indicated times to measure mRNA levels. (E) WT hepatocytes were transfected with siBmal1+SiRev-erbα and subjected to serum shock, and cells collected at different times were used to measure temporal changes in gene expression. (F) Hepatocytes isolated from 12 week-old male Bmal1+/+ and Bmal1−/− mice were subjected to serum shock and used to quantify temporal changes in gene expression.
There are two things worth noting. SiBmal1 reduced both Bmal1 and Rev-erbα, and these reductions were associated with increases in Crebh and ApoAIV. However, siRev-erbα reduced Rev-erbα and increased Bmal1. Despite increases in Bmal1, siRev-erbα increased Crebh and ApoAIV levels. These studies indicate that Rev-erbα is a major regulator of Crebh and ApoAIV. This is consistent with a recent report that Rev-erbα plays an important role in lipid metabolism under conditions of metabolic perturbations (34). Second, increases in Crebh and ApoAIV in siBmal1+siRev-erbα-treated cells were higher than those seen in cells individually treated with these siRNAs, indicating that these transcription factors might additively regulate Crebh and ApoAIV expression. Thus, there might be two mechanisms controlling Crebh expression by Bmal1 and Rev-erbα.
To understand the roles of Bmal1 and Rev-erbα in the temporal expression of Crebh and its target gene ApoAIV, mouse primary hepatocytes were transfected with different siRNAs and then subjected to serum shock (Figure 6C–E). In siControl-treated cells, expression of Crebh and ApoAIV increased soon after serum treatment (peak expression at 4–8 h), followed by reduced expression and a second peak expression at 28–32 h (Figure 6C–E). Expression of Rev-erbα and Bmal1 peaked at 16 h and at 36–40 h (Figure 6C–E). Rev-erbα and Bmal1 mRNA peak expressions were coincident with nadirs in Crebh and ApoAIV mRNA levels. SiBmal1-treated cells showed early peak expression of Crebh and ApoAIV but did not show the second peak (Figure 6C), indicating that Bmal1 plays a role in their cyclic expression. SiRev-erbα significantly increased the Crebh and ApoAIV mRNA levels at all times, and the expression levels showed cyclic changes similar to siControl-treated cells (Figure 6D). Treatment of these cells with hemin, which increases Rev-erbα expression (35), significantly reduced the expression of Crebh, ApoAIV, and Bmal1 mRNA (Figure 6D). These data indicate that Rev-erbα acts as a repressor. A combination of siBmal1 and siRev-erbα increased the expression of Crebh and ApoAIV mRNA levels, and expression levels of these genes did not have second peak (Figure 6E). Bmal1+/+ hepatocytes subjected to serum shock showed two peaks of Crebh and ApoAIV expression (Figure 6F). The Bmal1−/− hepatocytes only showed the first peak. Based on these data, we propose that Rev-erbα regulates basal expression, and that Bmal1 modulates the diurnal expression of Crebh.
We then investigated how Bmal1 and Rev-erbα differentially regulate basal and cyclic expression of Crebh. We found that the human CREB3L3 1kb promoter has one E-box and one RORE (Supplementary Figure 12A). The mouse Creb3l3 promoter contains three E-boxes and one RORE close to the transcription start site (Supplementary Figure 12B). Chromatin immunoprecipitation (ChIP) analysis in mouse hepatocytes showed that Bmal1 binds to all three E-boxes; however, the binding of Clock was obvious at E1 and E2 (Supplementary Figure 12C). Further, Rev-erbα interacted with RORE. Thus, both Bmal1 and Rev-erbα interact with the Creb3l3 promoter at different sites.
Next, we studied temporal binding of Bmal1:Clock and Rev-erbα to E1 and RORE, respectively. Bmal1 and Clock interacted with box E1 at night in WT mice (Figure 7A), whereas, Rev-erbα mainly bound to the RORE in the daytime. Binding of Rev-erbα was significantly lower in Bmal1−/− mice (Figure 7A). We then studied the binding of different transcription factors to the Creb3l3 promoter at noon in chow- or Western diet-fed mice (Figure 7B). The binding of Bmal1 and Clock to the Creb3l3 promoter was low and the binding of Rev-erbα was high in chow-fed mice. Western diet reduced the binding of Bmal1 and Rev-erbα to the Creb3l3 promoter (Figure 7B). These results were further confirmed by qPCR (Figure 7C–F & Supplementary Figure 13). These studies suggest that Bmal1:Clock and Rev-erbα interact with the Crebh promoter at different times. In the absence of Bmal1, the binding of Rev-erbα to the Crebh promoter is significantly reduced and may lead to increased expression of Crebh.
Figure 7: Binding of Bmal1 and Rev-erbα to the Crebh promoter.
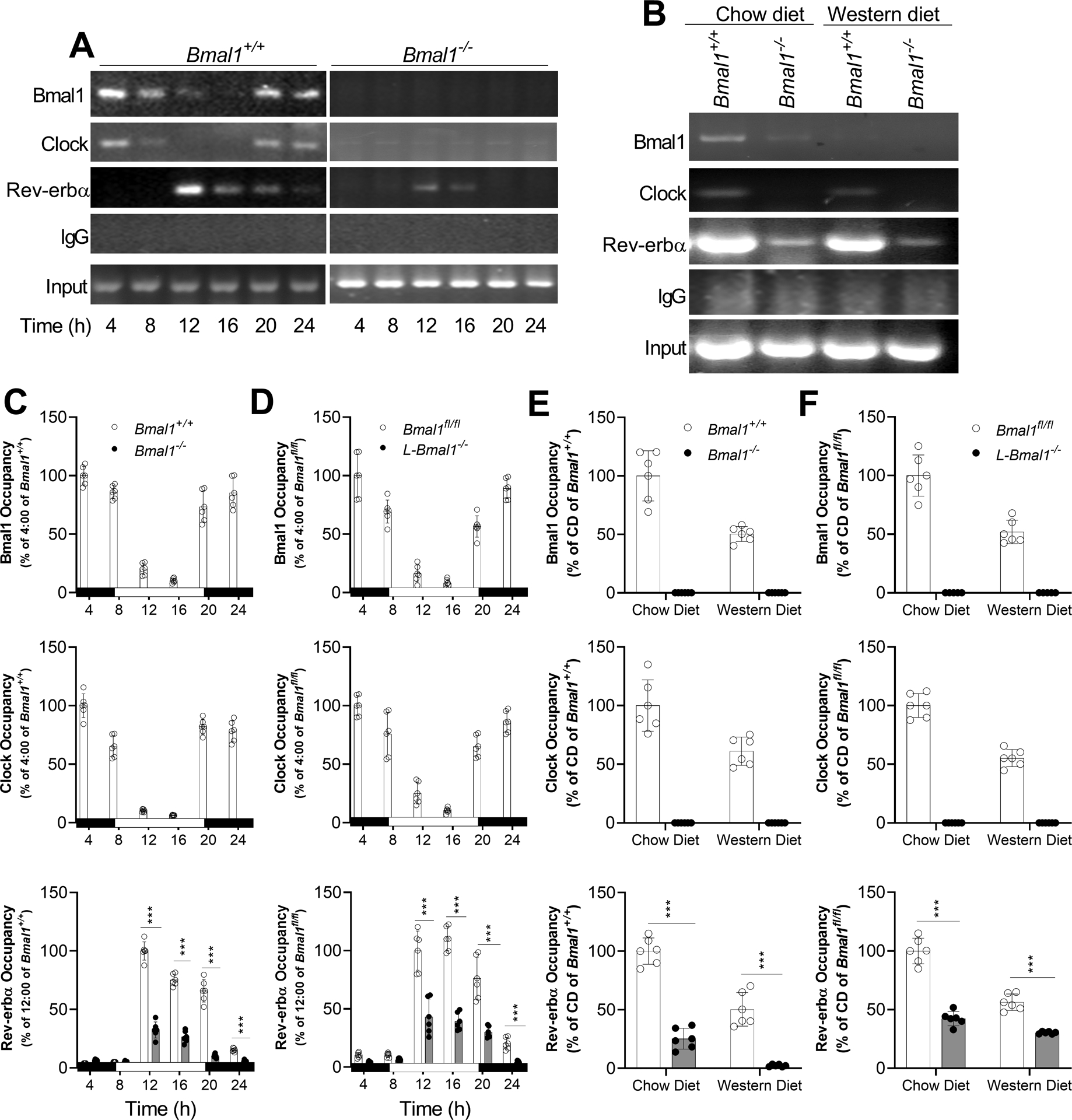
(A-B) Livers from (A) chow-fed or (B) chow- and Western diet-fed Bmal1+/+ and Bmal1−/− mice were collected to study the binding of Bmal1, Clock, or Rev-erbα to E-Boxes or RORE in the Creb3l3 promoter by ChIP.
(C-D) Temporal changes in the occupancy of Bmal1, Clock, and Rev-erbα on the Creb3l3 promoter was studied by ChIP and then quantified by qRT-PCR in the livers of (C) Bmal1−/− and Bmal1+/+ and (D) Bmal1fl/fl and L-Bmal1−/− mice.
(E-F) Livers from chow- and Western diet-fed (E) Bmal1+/+ and Bmal1−/− and (F) Bmal1fl/fl and L-Bmal1−/− mice were collected at 12:00 to study the binding of Bmal1, Clock, or Rev-erbα to the Creb3l3 promoter by ChIP and qPCR. Values are represented as mean ± SD. n=6, *** p<0.001, t-test.
Regulation of Crebh by Bmal1 and Rev-erbα
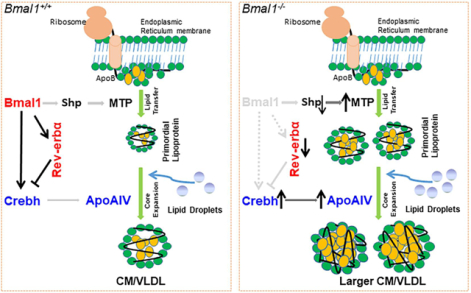
We propose that the regulation of Crebh involves two mechanisms (Figure 8). In Bmal1+/+ mice, Rev-erbα levels (Supplementary Figure 2C) and binding (Figure 7A) to the RORE in the Creb3l3 promoter are high in the day (Figure 8A, 12:00). This reduces Crebh and ApoAIV expression contributing to production of smaller-size CM and VLDL particles in the daytime. At midnight, Bmal1:Clock binding to the Creb3l3 promoter is increased (Figure 7A), resulting in enhanced Crebh and ApoAIV expression and production of larger CM and VLDL particles (Figure 8A, 24:00). These changes are associated with low triglyceride in the daytime and high triglyceride in the nighttime (Figure 8A). In Bmal1−/− mice, Rev-erbα levels are low at all times (Supplementary Figure 2C), resulting in increased expression of Crebh and ApoAIV and more number of larger CM and VLDL particles (Figure 8B). Our studies also suggest that Bmal1 regulates Shp to regulate MTP (Supplementary Figure 2A, Figure 2D, 2E). Thus, Bmal1 regulates two repressors: Shp and Rev-erbα (Figure 8C). Shp regulates MTP to affect primordial lipoprotein assembly. Bmal1 and Rev-erbα regulate Crebh, and this might regulate the second step of the core expansion of apoB-containing lipoproteins by increasing ApoAIV levels in the nighttime (Figure 8C). In Bmal1−/− mice, expression of both repressors is reduced. Repression of Shp increases MTP expression, leading to more synthesis of primordial lipoprotein particles. Repression of Rev-erbα increases Crebh expression and ApoAIV. These changes may lead to production of more number of larger CM and VLDL particles (Figure 8D).
Figure 8: Bmal1 regulates both steps involved in lipoprotein biosynthesis by regulating different transcription factors and proteins.
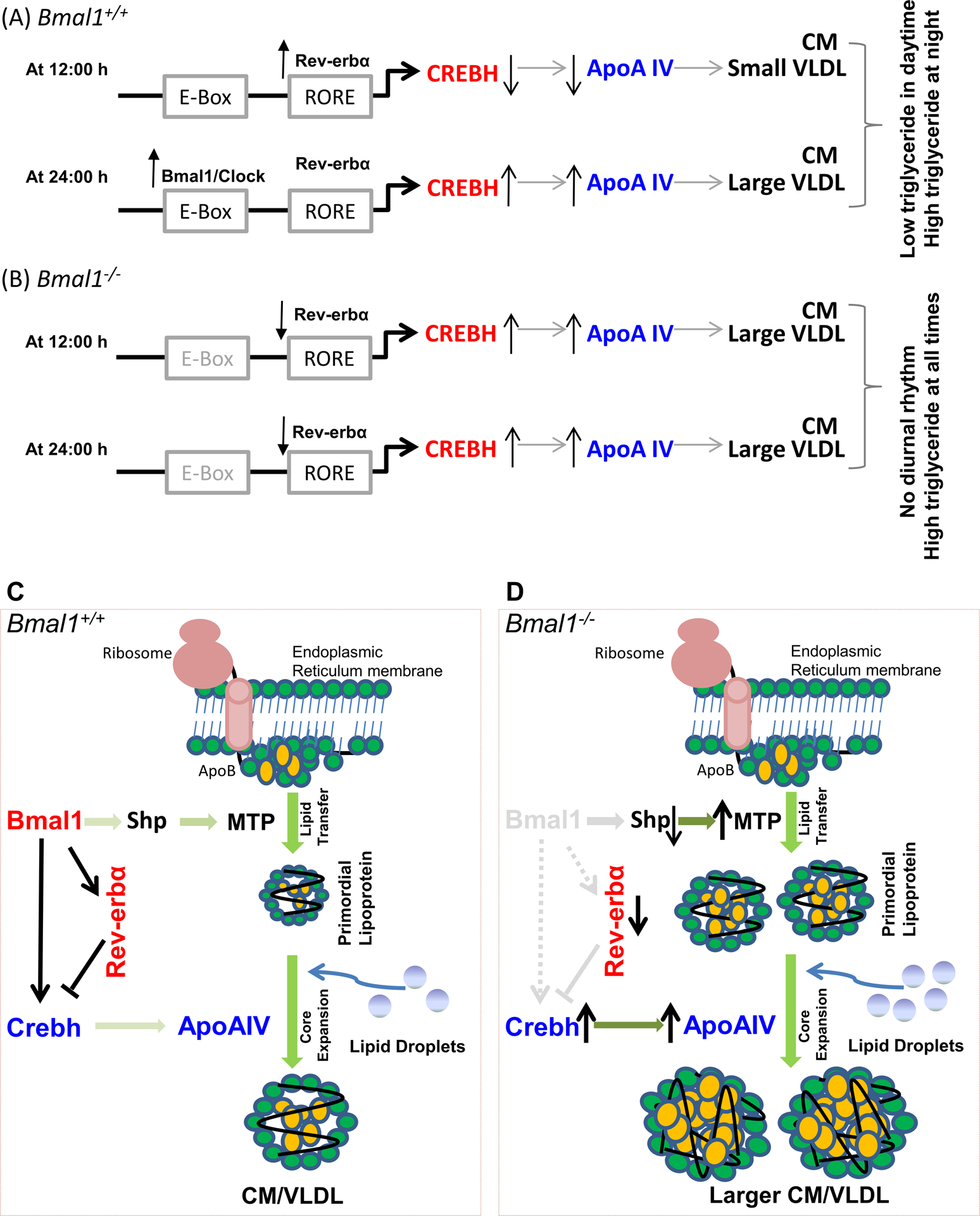
(A-B) Schematic representation of the temporal regulation of Crebh by Bmal1 and Rev-erbα.
(A) Bmal1+/+, 12:00: the binding of Rev-erbα to Creb3l3 promoter is high. This is associated with reduced expression of Crebh in the daytime. Reduced Crebh is associated with reduced apoAIV levels and smaller lipoproteins. Bmal1+/+, 24:00: The Bmal1:Clock heterodimer interacts at night with the E-boxes in the Crebh promoter to enhance expression. This leads to increased apoAIV production and assembly of larger lipoproteins. At this time, binding of Rev-erbα to RORE is low.
(B) Bmal1−/−, 12:00 and 24:00: In Bmal1−/− mice, decreased expression of Rev-erbα results in reduced binding to the RORE in the Crebh promoter, which results in increased gene transcription. In addition, due to the absence of Bmal1, the expression of Crebh does not show daily variations. The sustained increase in Crebh results in higher levels of apoAIV at all times. These higher apoAIV levels may support assembly of larger lipoproteins, contributing to hyperlipidemia in Bmal1−/− mice.
(C) Bmal1+/+: Bmal1 regulates MTP expression involving Shp to control number of primordial lipoprotein assembly. The second step of lipoprotein assembly is “core expansion,” which is less well understood. In this process, primordial lipoproteins increase in size, presumably owing to bulk lipid addition to the core. This is accompanied by surface expansion and requires surface stabilization by the adsorption of exchangeable apolipoproteins, such as apoAIV. Our studies suggest that Bmal1 regulates apoAIV expression by modulating Crebh expression via two mechanisms: (1) Bmal1 interacts with the Crebh promoter to increase expression at night. Increased levels of Crebh enhance apoAIV expression. (2) Bmal1 increases Rev-erbα, which then acts as a repressor of Crebh during the day. These dual mechanisms control the extent and the timing of Crebh and apoAIV expression.
(D) Bmal1−/−: In the absence of Bmal1, there is a decrease in Shp and an increase in MTP expression throughout the day. This leads to increased production of primordial lipoproteins. In addition, direct regulation of Crebh by Bmal1 is abrogated. This results in loss of diurnal regulation of Crebh and apoAIV expression. Bmal1 deficiency also decreases production of Rev-erbα, a repressor of Crebh, contributing to higher expression of Crebh and apoAIV. All these changes may contribute to assembly of greater number of larger VLDL particles and their accretion in the plasma.
Discussion
We provide evidence that Bmal1 deficiency increases number of larger apoB-containing lipoproteins, CM and VLDL, due to their overproduction by the intestine and liver, respectively. We show that Bmal1 regulates the diurnal expression of Crebh and ApoAIV to control the second step of apoB-containing lipoprotein assembly based on several key observations: (1) Mice deficient in Crebh and ApoAIV accumulate smaller lipoproteins; (2) KD of Crebh in Bmal1-deficient mice reduces number and size of apoB-containing lipoproteins; (3) Bmal1 regulates the circadian regulation of Crebh in a tissue- and cell-specific manner; (4) Bmal1 regulates the extent of Crebh expression by modulating Rev-erbα levels. Molecular studies show that Bmal1 and Rev-erbα regulate Crebh expression at the transcriptional level by interacting with the Creb3l3 promoter at different times to impart temporal changes. Bmal1 interacts with the E-boxes in the Creb3l3 promoter at night to increase transcription. Additionally, Bmal1 increases the expression of Rev-erbα, and Rev-erbα binds to the Creb3l3 promoter in the daytime to repress transcription. A combination of these two mechanisms controls the extent and the temporal expression of Crebh. Changes in ApoAIV follow Crebh expression. Increased availability of ApoAIV may support the assembly of larger lipoproteins.
Our observation that MTP expression is high and Shp expression is low in Bmal1−/− and L-Bmal1−/− mice is in concert with previous observations in ClockΔ19/Δ19 and Bmal1−/−Apoe−/− mice (9–11). Thus, Clock and Bmal1 regulate MTP via Shp in mice of different genetic backgrounds, underscoring their dominant role in the regulation of lipoprotein assembly and secretion.
Throughout these studies, we noticed that Bmal1 deficiency increased Crebh mRNA levels and Crebh-ER protein levels but not Crebh-N protein levels. In most instances, Crebh-N levels were low despite increased Crebh mRNA and Crebh-ER protein levels. In a few instances, such as after treatment with siBmal1+siRev-erbα, both Crebh-ER and Crebh-N protein levels were high (Figure 6B, Supplementary Fig 11). These studies indicate differential regulation of Creh-ER and Crebh-N, and are consistent with studies that show that Crebh-N levels are regulated by the AKT-GSK3β pathway (31).
Another discrepancy was that steady state levels of Crebh-N protein were not in concert with expression of Crebh target genes. Even though Crebh-N protein levels did not increase, there was a significant increase in the Crebh target gene, ApoAIV, under various conditions. Changes in ApoAIV correlated with Crebh mRNA and Crebh-ER protein levels. We interpret these results to suggest that Crebh-N protein levels undergo rapid turnover and that steady-state levels are kept low. The rapid turnover is reflected in increased expression of Crebh target genes.
A drawback of our studies is that the observations made in Bmal1−/− and L-Bmal1−/− mice may be either related to a specific role of Bmal1 in lipid metabolism or a reflection of general disruptions in circadian regulatory mechanisms. Our studies in Clock- and Bmal1-deficient mice showed similar phenotypes with regard to Shp and MTP expression. With the use of different mouse models deficient in specific clock genes, it might be possible to identify gene-specific and circadian-related mechanisms of metabolic dysregulations.
It is well established that high plasma lipids and lipoproteins are risk factors for heart disease. Although plasma lipids can be effectively reduced by increasing their removal using drugs such as statins and PCSK inhibitors (36–38), there is still need for new therapeutic modalities, as some people are refractory to these treatments or develop unmanageable side effects (39–41). Therefore, attempts have been made to target lipoprotein assembly and secretion. Very effective drugs targeting apoB and MTP have been discovered but they are associated with steatosis (42). Hence, there is a need for new approaches to modulate lipoprotein assembly and secretion without causing steatosis (43). Our observation that apoAIV could modulate the size of lipoproteins, makes this apolipoprotein a possible target to produce smaller remnant like lipoproteins from the intestine and reduce plasma lipids. This is because apoAIV is mainly expressed in the intestine and its liver expression in humans is low.
In conclusion, we provide evidence that Bmal1 is a key transcription factor that orchestrates the regulation of different steps in the biosynthesis of apoB-containing lipoproteins by modulating the expression of Shp and Crebh. These transcription factors then regulate the expression of MTP and ApoAIV, respectively. Via these mechanisms, Bmal1 appears to regulate both number and size of plasma apoB-containing lipoproteins. Our previous studies showed that ClkΔ19/Δ19 mice also exhibit hyperlipidemia. Thus, chronic disruptions in circadian rhythm genes contribute to dyslipidemia. It is likely that avoidance of circadian disruptions may be beneficial in controlling concentrations of triglyceride-rich larger lipoproteins.
MATERIALS AND METHODS
Cells, animals, and diets:
Huh-7 cells and mouse primary hepatocytes were cultured as before (35). Bmal1−/−, Bmal1+/+, Bmal1fl/fl, L-Bmal1−/−, Apoa4+/+, and Apoa4−/− mice have been described before (11, 21). Nr1d1−/−, Creb3l3−/− mice were from the Jackson Laboratory. All mice were in rooms with a 07:00–19:00 lights-on schedule with unlimited access to chow diet (Lab diet, 5053; % calories: protein 25%, fat 13%, and carbohydrates 62%). Heterozygous pairs were bred to obtain WT and KO siblings for experiments. All experiments were started in 12- to 16-week-old mice. Both male and female mice were sacrificed at different times and plasma and tissues were collected for various analyses. Different viruses were introduced via intravenous tail vein injections. In some experiments, 12- to 16-week-old mice were fed a western diet (Envigo, TD.88137; % calories: protein 15%, fat 43%, and carbohydrates 42%) for 2 months. Procedures were approved by the Animal Care and Use Committees of SUNY Downstate Medical Center and NYU Long Island School of Medicine, conforming to accepted standards of humane animal care. Bmal1−/− and Bmal1+/+ mice received their diets in Petri dishes placed in the bottom. However, water bottles were placed on the top of the cages. Consequently, there were no significant differences in daily food intake and weight gain between Bmal1−/− and Bmal1+/+ mice.
Intestinal lipid absorption and hepatic triglyceride production:
For intestinal absorption, non-fasted mice were injected with 0.5 mL of P407 (1:6, v/v in PBS; 30 mg/mouse). After 1 h, mice were gavaged with 3H-triolein (2 μCi) in olive oil (50 μL). Blood was collected before and after injections at regular intervals to measure triglyceride and radioactivity. For hepatic triglyceride production, mice fasted for 5 h were injected intraperitoneally with 0.5 mL of P407, and blood was collected from the tail vein to measure triglyceride levels.
Statistical Analyses:
All cellular and biochemical experiments were done in triplicate and repeated at least three times. Data are presented as the mean ± standard deviation (SD). Statistical testing was performed using the paired Student’s t-test. Temporal comparisons between 2 groups were performed with 2-way ANOVA followed by Bonferroni post-test (GraphPad Prism 9.0). Comparisons among multiple groups were performed by ANOVA followed by the Student-Newman-Keuls test. A p-value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments:
We are grateful to Wei Quan for performing and helping with negative staining and electron microscopy. This work was supported in part by U.S. National Institutes of Health (NIH) grants DK121490, HL137202, HL158054 and HD094778, and VA Merit Award BX004113, to MMH; and by NIH grant R56 HL137912 and American Heart Association Grant-in-Aid 16GRNT30960027 to XP. The contents of this paper do not represent the views of the Department of Veterans Affairs or the U.S. Government. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations used:
- ApoB
Apolipoprotein B
- ApoAIV
apolipoprotein AIV
- Bmal1
Brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like 1
- ChIP
Chromatin immunoprecipitation
- Clock
Circadian locomotor output cycles kaput
- ClockΔ19/Δ19
Clock mutant mice
- CM
chylomicron
- Crebh
cAMP responsive element-binding protein H
- E-boxes
enhancer boxes
- ER
endoplasmic reticulum
- HDL
high-density lipoproteins
- HNF
hepatocyte nuclear factor
- KD
knockdown
- KO
knockout
- LDL
low-density lipoproteins
- LpL
lipoprotein lipase
- MTP
Microsomal triglyceride transfer protein
- P407
Poloxamer 407
- qPCR
quantitative real-time polymerase chain reaction
- Rev-erbα (Nr1D1)
Nuclear receptor subfamily 1 Group D Member 1
- RORα
RAR related Orphan receptor
- RORE
ROR response elements
- Shp
Small heterodimer partner
- VLDL
very-low-density lipoproteins
- WT
wild-type
References
- 1.Lawler PR, Akinkuolie AO, Chu AY, Shah SH, Kraus WE, Craig D, Padmanabhan L, et al. Atherogenic Lipoprotein Determinants of Cardiovascular Disease and Residual Risk Among Individuals With Low Low-Density Lipoprotein Cholesterol. J Am Heart Assoc 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuura Y, Kanter JE, Bornfeldt KE. Highlighting Residual Atherosclerotic Cardiovascular Disease Risk. Arterioscler Thromb Vasc Biol 2019;39:e1–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Libby P The changing landscape of atherosclerosis. Nature 2021;592:524–533. [DOI] [PubMed] [Google Scholar]
- 4.Balling M, Afzal S, Varbo A, Langsted A, Davey Smith G, Nordestgaard BG. VLDL Cholesterol Accounts for One-Half of the Risk of Myocardial Infarction Associated With apoB-Containing Lipoproteins. J Am Coll Cardiol 2020;76:2725–2735. [DOI] [PubMed] [Google Scholar]
- 5.Prenner SB, Mulvey CK, Ferguson JF, Rickels MR, Bhatt AB, Reilly MP. Very low density lipoprotein cholesterol associates with coronary artery calcification in type 2 diabetes beyond circulating levels of triglycerides. Atherosclerosis 2014;236:244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emery P, Reppert SM. A rhythmic Ror. Neuron 2004;43:443–446. [DOI] [PubMed] [Google Scholar]
- 7.Hussain MM, Pan X. Circadian regulators of intestinal lipid absorption. J. Lipid Res 2014;56:761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain MM. Regulation of intestinal lipid absorption by clock genes. Annu. Rev. Nutr 2014;34:357–375. [DOI] [PubMed] [Google Scholar]
- 9.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab 2010;12:174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan X, Jiang XC, Hussain MM. Impaired Cholesterol Metabolism and Enhanced Atherosclerosis in Clock Mutant Mice. Circulation 2013;128:1758–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan X, Bradfield CA, Hussain MM. Global and hepatocyte-specific ablation of Bmal1 induces hyperlipidemia and enhances atherosclerosis. Nat. Commun 2016;7:13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Kohan AB, Lo CM, Liu M, Howles P, Tso P. Apolipoprotein A-IV: a protein intimately involved in metabolism. J. Lipid Res 2015;56:1403–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinberg RB, Gallagher JW, Fabritius MA, Shelness GS. ApoA-IV modulates the secretory trafficking of apoB and the size of triglyceride-rich lipoproteins. J. Lipid Res 2012;53:736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberg RB, Cook VR, DeLozier JA, Shelness GS. Dynamic interfacial properties of human apolipoproteins A-IV and B-17 at the air/water and oil/water interface. J. Lipid Res 2000;41:1419–1427. [PubMed] [Google Scholar]
- 15.VerHague MA, Cheng D, Weinberg RB, Shelness GS. Apolipoprotein A-IV expression in mouse liver enhances triglyceride secretion and reduces hepatic lipid content by promoting very low density lipoprotein particle expansion. Arterioscler. Thromb. Vasc. Biol 2013;33:2501–2508. [DOI] [PubMed] [Google Scholar]
- 16.Cheng D, Xu X, Simon T, Boudyguina E, Deng Z, VerHague M, Lee AH, et al. Very Low Density Lipoprotein Assembly Is Required for cAMP-responsive Element-binding Protein H Processing and Hepatic Apolipoprotein A-IV Expression. J. Biol. Chem 2016;291:23793–23803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pullinger CR, North JD, Teng BB, Rifici VA, Ronhild de Brito AE, Scott J. The apolipoprotein B gene is constitutively expressed in HepG2 cells: regulation of secretion by oleic acid, albumin, and insulin, and measurement of the mRNA half-life. J. Lipid Res 1989;30:1065–1077. [PubMed] [Google Scholar]
- 18.Hussain MM, Nijstad N, Franceschini L. Regulation of microsomal triglyceride transfer protein. Clin Lipidol 2011;6:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan X, Hussain MM. Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J. Biol. Chem 2007;282:24707–24719. [DOI] [PubMed] [Google Scholar]
- 20.Fukagawa K, Gou HM, Wolf R, Tso P. Circadian rhythm of serum and lymph apolipoprotein AIV in ad libitum-fed and fasted rats. Am. J. Physiol 1994;267:R1385–R1390. [DOI] [PubMed] [Google Scholar]
- 21.Pan X, Munshi MK, Iqbal J, Queiroz J, Sirwi AA, Shah S, Younus A, et al. Circadian regulation of intestinal lipid absorption by apolipoprotein AIV involves forkhead transcription factors A2 and O1 and microsomal triglyceride transfer protein. J. Biol. Chem 2013;288:20464–20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J. Lipid Res 2009;50:1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang M, Kim J, An HT, Ko J. Human leucine zipper protein promotes hepatic steatosis via induction of apolipoprotein A-IV. FASEB J 2017;31:2548–2561. [DOI] [PubMed] [Google Scholar]
- 24.Xu X, Park JG, So JS, Hur KY, Lee AH. Transcriptional regulation of apolipoprotein A-IV by the transcription factor CREBH. J. Lipid Res 2014;55:850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zannis VI, Kan HY, Kritis A, Zanni E, Kardassis D. Transcriptional regulation of the human apolipoprotein genes. Front Biosci 2001;6:D456–D504. [DOI] [PubMed] [Google Scholar]
- 26.Lee JH, Giannikopoulos P, Duncan SA, Wang J, Johansen CT, Brown JD, Plutzky J, et al. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nat. Med 2011;17:812–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M, Zhao S, Tan M. bZIP transmembrane transcription factor CREBH: Potential role in non-alcoholic fatty liver disease (Review). Mol. Med. Rep 2016;13:1455–1462. [DOI] [PubMed] [Google Scholar]
- 28.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 2006;124:587–599. [DOI] [PubMed] [Google Scholar]
- 29.Wei J, Chen L, Li F, Yuan Y, Wang Y, Xia W, Zhang Y, et al. HRD1-ERAD controls production of the hepatokine FGF21 through CREBH polyubiquitination. EMBO J 2018;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H, Wei J, Song Z, Mottillo E, Samavati L, Zhang R, Li L, et al. Regulation of hepatic circadian metabolism by the E3 ubiquitin ligase HRD1-controlled CREBH/PPARalpha transcriptional program. Mol Metab 2021:101192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Z, Kim H, Qiu Y, Chen X, Mendez R, Dandekar A, Zhang X, et al. CREBH Couples Circadian Clock with Hepatic Lipid Metabolism. Diabetes 2016;65:3369–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benavides A, Siches M, Llobera M. Circadian rhythms of lipoprotein lipase and hepatic lipase activities in intermediate metabolism of adult rat. Am J Physiol 1998;275:R811–817. [DOI] [PubMed] [Google Scholar]
- 33.Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, Delaunay F, et al. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. J Mol Endocrinol 2004;33:585–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter AL, Pelekanou CE, Adamson A, Downton P, Barron NJ, Cornfield T, Poolman TM, et al. Nuclear receptor REVERBalpha is a state-dependent regulator of liver energy metabolism. Proc Natl Acad Sci U S A 2020;117:25869–25879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan X, Queiroz J, Hussain MM. Nonalcoholic fatty liver disease in CLOCK mutant mice. J Clin Invest 2020;130:4282–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blumenthal RS. Statins: effective antiatherosclerotic therapy. Am. Heart J 2000;139:577–583. [DOI] [PubMed] [Google Scholar]
- 37.Ehrenstein MR, Jury EC, Mauri C. Statins for atherosclerosis--as good as it gets? N. Engl. J. Med 2005;352:73–75. [DOI] [PubMed] [Google Scholar]
- 38.LaRosa JC, Grundy SM, Waters DD, Shear C, Barter P, Fruchart JC, Gotto AM, et al. Intensive lipid lowering with atorvastatin in patients with stable coronary disease. N. Engl. J. Med 2005;352:1425–1435. [DOI] [PubMed] [Google Scholar]
- 39.Voora D, Shah SH, Reed CR, Zhai J, Crosslin DR, Messer C, Salisbury BA, et al. Pharmacogenetic predictors of statin-mediated low-density lipoprotein cholesterol reduction and dose response. Circ. Cardiovasc. Genet 2008;1:100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmad Z Statin intolerance. Am. J. Cardiol 2014;113:1765–1771. [DOI] [PubMed] [Google Scholar]
- 41.Saxon DR, Eckel RH. Statin Intolerance: A Literature Review and Management Strategies. Prog. Cardiovasc. Dis 2016;59:153–164. [DOI] [PubMed] [Google Scholar]
- 42.Rader DJ, Kastelein JJ. Lomitapide and mipomersen: two first-in-class drugs for reducing low-density lipoprotein cholesterol in patients with homozygous familial hypercholesterolemia. Circulation 2014;129:1022–1032. [DOI] [PubMed] [Google Scholar]
- 43.Hussain MM, Bakillah A. New approaches to target microsomal triglyceride transfer protein. Curr. Opin. Lipidol 2008;19:572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


