Abstract
The newly identified coronavirus SARS-CoV-2 is responsible for the worldwide pandemic COVID-19. Considerable efforts have been devoted for the development of effective vaccine strategies against COVID-19. The SARS-CoV-2 spike protein has been identified as the major antigen candidate for the development of COVID-19 vaccines. The Pfizer-BioNTech COVID-19 vaccine comirnaty is a lipid nanoparticle-encapsulated mRNA encoding a full-length and prefusion-stabilized SARS-CoV-2 spike protein. In the present study, synthetic peptide-based ELISA assays were performed to identify linear B-cell epitopes into the spike protein that contribute to elicitation of antibody response in comirnaty-vaccinated individuals. The synthetic S2P6 peptide containing the spike residues 1138/1169 and to a lesser extent, the synthetic S1P4 peptide containing the spike residues 616/644 were recognized by the immune sera from comirnaty vaccine recipients but not COVID-19 recovered patients. We assume that the synthetic S2P6 peptide and to a lesser extent the synthetic S1P4 peptide, could be of interest to measure the dynamic of antibody response to COVID-19 mRNA vaccines. The S2P6 peptide has been identified as immunogenic in adult BALB/c mice that received protein-peptide conjugates in a prime-boost schedule. This raises the question on the role of the B-cell epitope peptide containing the SARS-CoV-2 spike residues 1138/1169 in protective efficacy of the Pfizer-BioNTech COVID-19 vaccine comirnaty.
Subject terms: Immunology, Microbiology
Introduction
SARS-CoV-2 virus is an enveloped RNA virus belonging to betacoronavirus genus of Coronoviridae family1. The COVID-19 disease associated with the emerging SARS-CoV-2 infection is a pandemic public health threat since early 2020 with millions of deaths to date1. The main route for SARS-CoV-2 transmission is through respiratory droplets necessitating the implementation of effective control measures2–6. Convalescent COVID-19 patients develop neutralizing anti-SARS-CoV-2 antibodies produced by the antiviral immune response7–9. The structural spike S protein playing a crucial role in eliciting the immune response during COVID-19 disease has been considered as the predominant viral antigen target for vaccine development10–13. The transmembrane homo-trimer spike protein at the virus surface mediates receptor binding through interaction with cell entry receptor ACE214–18. The spike protein is composed of the S1 and S2 subunits10,11,13. The N-terminal S1 subunit which comprises the Receptor Binding Domain (RBD) triggers binding to ACE2 receptor and anti-RBD antibodies were shown to exert potent neutralizing activity against SARS-CoV-210–12. The C-terminal S2 subunit containing the fusion elements driving viral and host-cell membrane fusion10–12. A polybasic insertion RRAR that can be cleaved by furin has been identified at the S1/S2 boundary playing a major role in the pathogenesis of SARS-CoV-2 infection14–17. The virus binding to ACE2 receptor via the RBD elicits the proteolytic cleavage of pre-protein S into S1 and S2 subunits depending of cell surface protease TMPRSS218–20. Such processing results in large conformational changes causing S1 shedding and fusion elements exposure in S2 leads to a fusion process between viral and host-cell membranes in an endosome-independent pathway10,14,21,22.
Accelerated vaccine programs for COVID-19 prevention have led to the development of commercially available vaccines targeting the SARS-CoV-2 spike protein13,23–25. To date, the most performing current vaccines are based on nucleoside-modified mRNA strategy26–29. The COVID-19 mRNA BNT162b2 vaccine which has been known as the Pfizer-BioNTech COVID-19 vaccine (brand name comirnaty) is a lipid nanoparticle-encapsulated mRNA encoding a full-length and prefusion-stabilized S protein13,26–29. The comirnaty vaccine was initially administered in two doses, 21 days apart, and now in three and even four doses. The approval of this candidate vaccine has been based on a large group of individuals who had been immunized with BNT162b2 in an international and placebo-controlled phase III trial28,29. Vaccinated individuals developed high neutralizing SARS-CoV-2 antibody titers which are indicators of COVID-19 protection28,29. The efficacy study demonstrated that COVID-19 mRNA BNT162b2 vaccine candidate induced 95% protection against COVID-19 in a 2-dose regimen28,29.
To our knowledge, there has been little information on specificities of antibodies induced by comirnaty vaccine. With the aim to design peptide-based vaccine candidates for COVID-19 prevention, we were interested in identifying linear antibody epitopes which are potentially targets of anti-spike antibodies in individuals who have been immunized with COVID-19 mRNA comirnaty vaccine. Neutralization linear antibody epitopes for SARS-CoV-2 have been identified in COVID-19 patients30–33. In the present study, we examined the antibody response post-vaccination with the comirnaty vaccine. Synthetic peptides representing potential B-cell epitopes in the SARS-CoV-2 spike protein were assessed for their antigenic reactivity with immune serum from comirnaty-vaccinated individuals. We showed that administration of comirnaty vaccine to an infection-naïve individual or COVID-19 immune subject elicits a production of antibodies capable of binding to two linear B-cell epitope peptides which are identified in the S1 and S2 subunits of the spike protein.
Results
Synthetic peptides representing potential linear B-cell epitopes in the spike protein
To search for possible linear B-cell epitopes in SARS-CoV-2 spike protein, we used the predictor tool BepiPred-2.0 available in IEDB (for The Immune Epitope Database and Analysis Resource) website. We submitted the spike protein sequence from Wuhan strain of SARS-CoV-2 using 0.50 as cutoff parameter and 23 potential epitope peptides were identified (Table S1). Considering that long peptides would be more appropriate to retain conformational information related to efficiency of antibody binding, we selected a limited number of candidate B-cell epitope peptides around 20 to 30 residues spanning the S1 and S2 subunits of SARS-CoV-2 spike protein (Table 1). Consequently, only the B-cell epitope peptides # 1, 9, 10, 13, 14, 16, and 22 were designed to generate the synthetic peptides S1P1, S1P2, S1P3, S1P4, S1P5 and S2P6 (Table 1). The 29-mer synthetic peptide S1P4 results of the combination of the two adjacent B-cell epitope peptides #13 and #14 whereas 32-mer synthetic peptide S2P6 peptide is based on B-cell epitope peptide # 22.
Table 1.
Sequences of synthetic peptides and their variants.
| Peptide | Amino acid sequence | Spike residues | Length |
|---|---|---|---|
| S1P1 | SQCVNLTTRTQLPPAYTNSFTRGVY | 13–37 | 25-mer |
| S1P1[F,N,s]* | SQCVNFTNRTQLPSAYTNSFTRGVY | 13–37 | 25-mer |
| S1P2 | YNSASFSTFKCYGVSPTKLNDLCF | 369–392 | 24-mer |
| S1P3 | GDEVRQIAPGQTGKIADYNYKL | 404–425 | 22-mer |
| S1P3[N]** | GDEVRQIAPGQTGNIADYNYKL | 404–425 | 22-mer |
| S1P4 | NCTEVPVAIHADQLTPTWRVYSTGSNVFQ | 616–644 | 29-mer |
| S1P5 | ASYQTQTNSPRRARSVASQ | 672–690 | 19-mer |
| S1P5[H]*** | ASYQTQTNSHRRARSVASQ | 672–690 | 19-mer |
| S2P6 | YDPLQPELDSFKEELDKYFKNHTSPDVDLGDI | 1138–1169 | 32-mer |
| S2P6.2.0 | SFKEELDKYFKNHTSPDVD | 1147–1165 | 19-mer |
*Gamma, **Beta/Omicron, and ***Alpha variants of SARS-CoV-2 according to WHO label. The amino-acid substitutions are underlined.
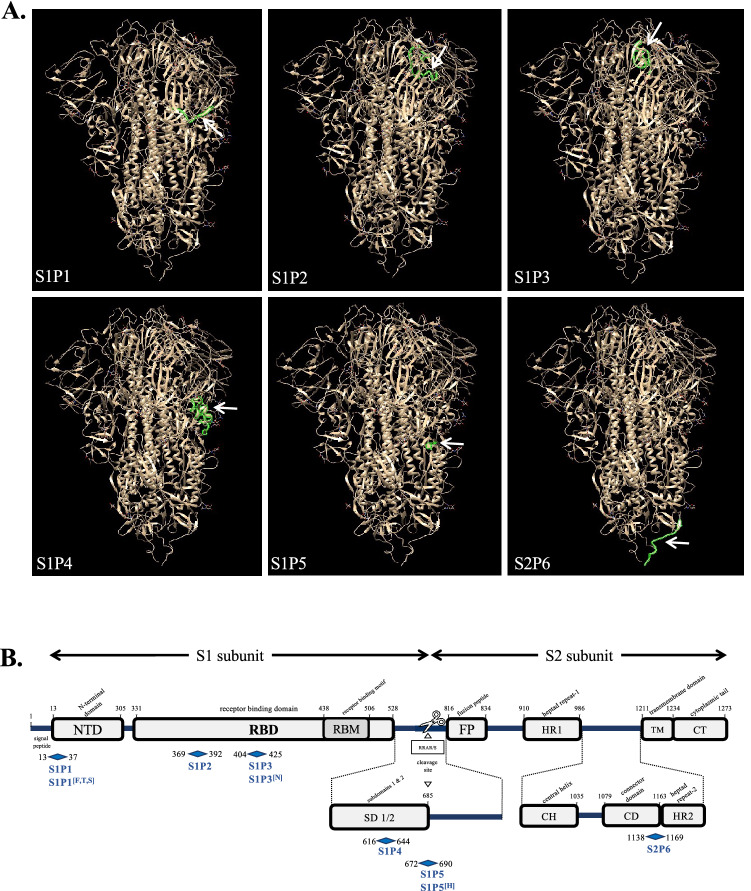
The six identified linear B-cell epitope peptides were mapped on the tridimensional structure prediction of SARS-CoV-2 spike protein trimer (Fig. 1A). The B-cell epitope peptides S1P4 and S2P6 were exposed at the surface of the protomer whereas S1P1, S1P2, S1P3, S1P4 and S1P5 were orientated inside of the structure. In the SARS-CoV-2 spike protein, the S1P1, S1P2, S1P3, S1P4, and S1P5 peptides are identified in the N-terminal domain (NTD), receptor binding domain (RBD), and C-terminal domain 2 (CTD2) sequences of the S1 subunit, respectively (Fig. 1B). The S2P6 peptide spans the connector domain (CD) and heptad repeat-2 (HR2) sequences into the C-terminal region of the S2 subunit (Fig. 1B). To note that the B-cell epitope peptide S2P6 which contains a putative N-glycosylation motif 1158NHT1160 is strictly conserved between SARS-CoV-2 and SARS-CoV spike proteins21.
Figure 1.
Position of B-cell epitope peptides on SARS-CoV-2 spike protein. In (A), positions of the six B-cell epitope peptides colored in green on a single protomer of SARS-CoV-2 spike protein trimer (EMD data resource: EMD-2256). The blank arrow indicates their position on the protomer. In (B), schematic organization of SARS-CoV-2 spike protein with the S1 and S2 subunits and their different domains and motifs is shown. The six synthetic peptides S1P1, S1P2, S2P3, S1P4, S1P5, and S2P6 peptides and their variants S1P1[F,T,S], S1P3[N], and S1P5[H] are mapped on the SARS-CoV-2 spike protein.
The Alpha, Beta, Gamma, Delta, and now Omicron variants of SARS-CoV-2 have been classified as dominant VOC (Variants of Concern) by World Health Organization. Given that SARS-CoV-2 variants display an increased transmissibility and/or change on viral pathogenicity, we developed S1P1[F,N,S], S1P3[N], and S1P5[H] variant peptides bearing amino-acid substitutions that have been identified in VOC (Table 1). We noted that amino-acid changes identified in the spike protein of recent VOC including Delta and Omicron variants have no impact on both S1P4 and S2P6 peptides.
Antigenic reactivity of synthetic peptides with comirnaty vaccine recipient sera
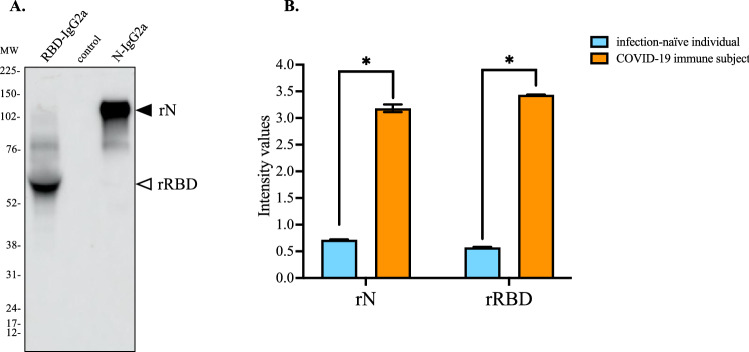
To evaluate the antibody response against SARS-CoV-2 spike protein in comirnaty-vaccinated individuals, we developed an indirect ELISA assay based on soluble recombinant RBD protein (rRBD) produced from transfected CHO cells. A recombinant soluble SARS-CoV-2 nucleoprotein N protein (rN) was also produced as control antigen for natural infection. Both both rN and rRBD were preceded by a heterologous signal peptide and ended by the IgG2A heavy chain region as C-terminal protein tag. The secreted rN and rRBD proteins from transient transfected CHO cells were detected by immunoblot assay (Fig. 2A). Soluble rN and rRBD proteins were quantified by direct ELISA assay using goat anti-mouse IgG heavy chain HRP antibody. Serum samples from COVID-19 immune subjects (Table S2) and infection-naïve individuals (Table S3) were used to validate the antigenic reactivity of rN and rRBD proteins by indirect ELISA (Fig. 2B).
Figure 2.
Antigenic reactivity of recombinant N and RBD proteins with human serum-specific SARS-CoV-2 antibody. In (A), CHO cells were transfected 72 h with plasmids expressing RBD-IgG2A (rN) and N-IgG2A (rRBD) proteins or empty vector plasmid (control). Aliquots of clarified cell culture supernatants (C3S) were analyzed by immunoblot assay using goat anti-mouse IgG heavy chain HRP antibody. The original full-length gel is presented in Supplementary Figure S1. In (B), pools of four COVID-19 donor serum samples (Table S2, serum donors n°16, 18, 19, and 20) (COVID-19 donor serum) and four negative donor serum samples (Table S3) (negative donor serum) at dilution 1:100 were tested for anti-N and anti-rRBD antibodies by indirect ELISA using rN and rRBD for protein-based antibody capture. The intensity values of serum samples were measured at O.D. 450 nm. The results are the mean (± SEM) of three replicates. Unpaired t tests between COVID-19 immune subject and infection-naïve individual were performed and statistically significant comparisons are shown as * p < 10–4.
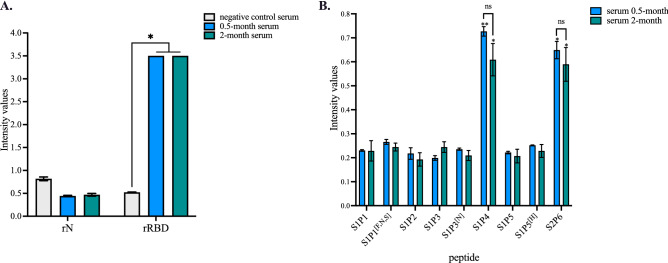
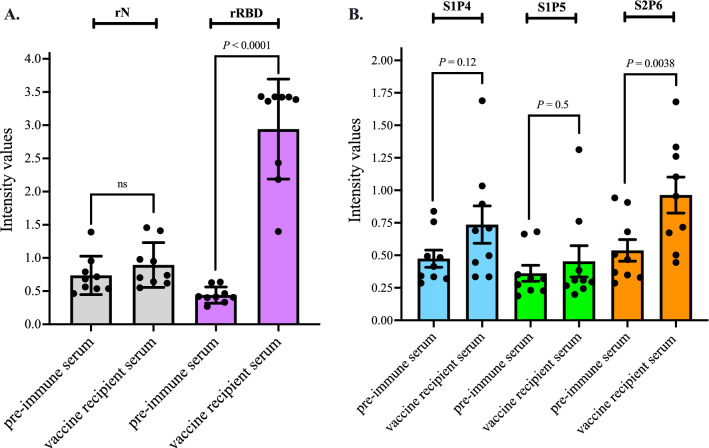
The antigenic reactivity of synthetic peptides was evaluated with the serum sample from an infection-naïve individual who has been fully immunized with comirnaty vaccine in April 2021. The immune reactivity of comirnaty vaccine recipient serum was first evaluated on SARS-CoV-2 spike protein by indirect ELISA using rRBD protein for spike protein-based antibody capture (Fig. 3A). A pool of serum samples from ten infection-naïve subjects (Table S3) served as negative serum control. Administration of comirnaty vaccine resulted in significant production of anti-RBD antibodies in vaccinated subject (Fig. 3A). A high anti-RBD antibody titer was maintained for 2 months after injection of a second vaccine dose (Fig. 3A). As a measure of immunity to natural infection, a lack of antibody response against rN confirmed that individual remained negative for SARS-CoV-2 infection at the time of vaccination (Fig. 3A).
Figure 3.
Antigenic reactivity of synthetic peptides with comirnaty vaccine recipient sera. Serum samples from an infection-naïve individual who received comirnaty vaccine in a 2-dose regimen were collected at 0.5 month or 2 months after the injection of a second dose. A pool of ten infection-naïve individuals collected in 2019 (Table S3) served as negative control serum. In (A), serum samples at dilution 1:100 were assessed for the detection of antibodies against SARS-CoV-2 N and S proteins by indirect ELISA using soluble rN and rRBD proteins for antigen-based antibody capture. The intensity values of serum samples were measured at O.D. 450 nm. The results are the mean (± SEM) of three replicates. Statistical comparisons were performed between serum samples. Statistically significant comparisons are shown as * p < 10–4 (ns: non-statistically significant, p > 0.05). In (B), the serum samples were assayed at dilution 1:50 on synthetic peptides and their variants (200 ng.ml−1) through synthetic peptide-based ELISA. The intensity values of vaccine serum sample were measured at O.D. 450 nm. The results are the mean (± SEM) of three replicates. Pairwise comparisons between peptides showed that the experimental points for S1P4 and S2P6 peptides are significantly different from the other peptides (** p < 10–4, * p < 10–3). The differences between 0.5 month and 2.0 month for the synthetic S1P4 and S2P6 peptides are non-statistically significant (ns, p > 0.05).
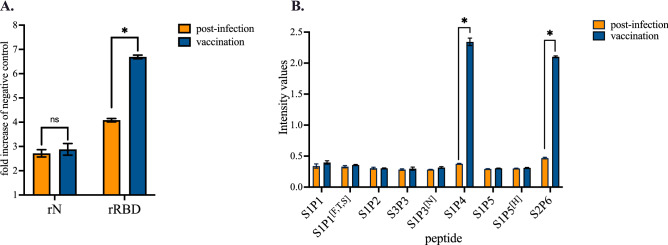
The immune serum from comirnaty-vaccinated individual was assayed for reactivity of antibodies against the synthetic peptides through a peptide-based ELISA (Fig. 3B). Among the six synthetic peptides and their variants, only S1P4 and S2P6 were significantly recognized by the immune serum from the vaccinated subject at serum dilution 1:50. As such, this result is consistent with the accessibility of the two B-cell epitope peptides at the surface of the SARS-CoV-2 spike protein trimer (Fig. 1A). The immune reactivity of comirnaty vaccine recipient serum against the synthetic S1P4 and S2P6 peptides was observed up to two months after the injection of a second vaccine dose (Fig. 3B). We then wondered whether administration of comirnaty vaccine to an individual who had experienced SARS-CoV-2 infection induced production of antibodies targeting the synthetic peptides. Indeed, it has been reported that a single dose of a such COVID-19 mRNA vaccine is sufficient to maximize immune protection in post-infection individuals34. In line with this finding, antibody capture assays were performed on a serum sample from a COVID-19 immune subject who had received a single-dose of comirnaty vaccine three months after recovery from mild disease. The diagnosis of SARS-CoV-2 infection had been performed by PCR during the acute phase of COVID-19. Administration a single dose of vaccine significantly increased the level of anti-RBD antibodies but not anti-N antibodies in COVID-19 immune subject (Fig. 4A). Thus, comirnaty vaccine elicits antibody response against the SARS-CoV-2 spike protein in a COVID-19 immune subject. The reactivity of COVID-19 patient serum was assayed few weeks after recovery through synthetic peptide-based ELISA (Fig. 4B). We observed a lack of antigenic reactivity for all the synthetic peptides tested in relation to COVID-19 donor serum. Thus, the stretches of spike residues 616/644 and 1138/1169 were not or weakly immunogenic in an individual who had experienced a natural infection with SARS-CoV-2. Administration of comirnaty vaccine in COVID-19 recovered individual resulted in production of antibody targeting synthetic S1P4 and S2P6 peptides 0.5 month after injection of a single dose (Fig. 4B). Thus, comirnaty vaccine was efficient to elicit antibody response against the stretches of spike residues 616/644 and 1138/1169 in a COVID-19 patient who previously mounted a humoral immunity against SAR-CoV-2 spike protein in the course of a natural infection. The levels of antibodies directed against the synthetic S1P4 and S2P6 peptides were higher in the vaccinated individual who was previously infected with SARS-CoV-2 than those without prior evidence infection. This is consistent with the finding that vaccination increased anti-SARS-CoV-2 spike antibodies in COVID-19 immune subjects in a greater magnitude than after a prime-boost vaccine strategy in infection-naïve individuals35.
Figure 4.
Antigenic reactivity of synthetic peptides with immune sera from a COVID-19 immune subject who received comirnaty vaccine. Serum samples from a COVID-19 patient were collected few weeks after recovery of SARS-CoV-2 infection (post-infection) and then two weeks after the injection of a single dose of comirnaty vaccine (vaccination). Vaccine administration was performed three months after COVID-19 recovery. In (A), serum samples at dilution 1:100 were assayed for the detection of antibodies against SARS-CoV-2 N and spike proteins by indirect ELISA using recombinant rN and rRBD proteins for antigen-based antibody capture. A pool of serum samples from infection-naïve individuals (Table S3) served as negative control serum. The intensity values of serum samples were measured at O.D. 450 nm and their immune reactivity was estimated as a fold increase of intensity values obtained with negative control serum. The results are the mean (± SEM) of three replicates. Statistically significant comparisons are shown as * p < 10–4 (ns: non-statistically significant, p > 0.05). In (B), serum samples at dilution 1:50 were assayed for the detection of specific antibodies through peptide-based ELISA. The intensity values of serum samples were measured at O.D. 450 nm. The results are the mean (± SEM) of three replicates. Paired t tests on experimental points between post-infection and vaccination immune were performed and statistically significant comparisons are shown as * p < 10–4. Differences between experimental points considered as non-statistically significant are not shown.
We next evaluated the antigenic reactivity of the synthetic S1P4 and S2P6 peptides using a group of infection-naïve individuals who had been immunized with comirnaty vaccine in January–February 2021 (n = 9, median age: 50, female to male ratio: 0.9) (Table S4). The serum samples were collected few weeks after injection of a second dose of comirnaty vaccine. The pre-immune serum of each vaccinated subject was used as negative serum control. The comirnaty vaccine recipient sera (serum dilution 1:100) were first assessed for anti-RBD antibodies by indirect ELISA (Fig. 5A). All vaccinated individuals developed anti-RBD antibodies confirming that administration of comirnaty vaccine was effective in eliciting an antibody response against SARS-CoV-2 spike protein. The absence of anti-rN antibodies in comirnaty vaccine recipient sera confirmed that all nine subjects were infection-naïve individuals (Fig. 5A). Synthetic peptide-based ELISA was performed with the serum samples (dilution 1:50) from this group of comirnaty vaccine recipients (Fig. 5B). The peptide S1P5 served as negative peptide control. The synthetic S2P6 peptide but not the synthetic S1P4 peptide showed a significant antigenic reactivity with the comirnaty vaccine recipient sera as compared with pre-immune sera. The fact that comirnaty vaccine recipients had antibodies targeting the synthetic S2P6 peptide highlights for the ability of the stretch of residues 1138/1169 to be recognized as linear B-cell epitope in the recombinant spike protein expressed by comirnaty vaccine. Because the immune reactivity against the synthetic S1P4 peptide was not significantly different between the nine comirnaty vaccine recipient sera and their pre-immune sera, it is likely that individual differences exist in the development of an effective antibody response targeting the stretch of spike residues 616/644.
Figure 5.
Immune reactivity of comirnaty vaccine recipient sera against the synthetic S1P4 and S2P6 peptides. Serum samples from comirnaty vaccine recipients (n = 9) (Table S4) were collected few weeks after the receipt of the second vaccine dose. The pre-immune serum of each individual was collected prior vaccination. In (A), serum samples at dilution 1:100 have been tested for SARS-CoV-2 N and S antibodies by indirect ELISA using soluble rN and rRBD proteins for antigen-based antibody capture. The intensity values of serum samples were measured at O.D. 450 nm. Paired t tests were performed between pre-immune serum and vaccine recipient serum (ns: non-statistically significant, p > 0.05). In (B), the serum samples were assayed on the synthetic S2P4 and S2P6 peptides (200 ng.ml−1) through peptide-based ELISA at serum dilution 1:50. The synthetic S1P5 peptide served as negative control serum. The intensity values of vaccine serum samples were measured at O.D. 450 nm. Paired t tests were performed between pre-immune serum and vaccine recipient serum.
Immune reactivity of COVID-19 patient sera with the synthetic S1P4 and S2P6 peptides
We evaluated antigenic reactivity of the synthetic S1P4 and S2P6 peptides with a group of COVID-19 immune subjects. We selected a panel of thirty COVID-19 donor sera (median age: 52.5 years, female to male ratio: 4) collected in November and December 2020 in France (Table S2). The COVID-19 serum donors developed mild COVID-19 symptoms without requirement for hospital admission. Symptomatic COVID-19 patients with progression to severe disease or admitted in intensive care units in hospital were not included in this study. Serological tests based on semi-quantitative SARS-CoV-2 specific antibody assays indicated that COVID-19 immune subjects developed anti-SARS-CoV-2 IgG and most of them also had specific IgM titers (Table S2). There were no significant differences in humoral immune reactivity among COVID-19 immune subjects related to women/men ratio or different age categories. Although durability of IgM and IgG can vary among COVID-19 patients, the IgM titers are consistent with serum samples collected within the first weeks following the acute infection.
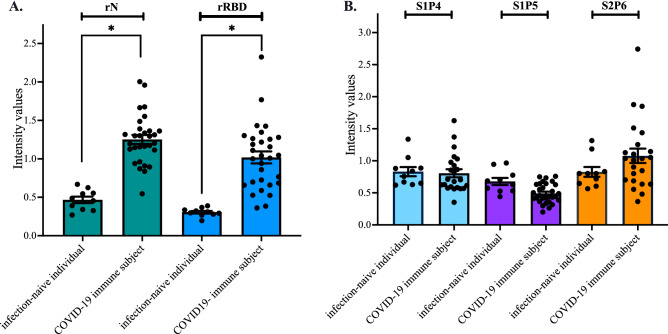
The immune reactivity of COVID-19 donor sera (Table S2) was first assessed for the presence of anti-spike antibodies by indirect ELISA using rRBD for antigen-based antibody capture (Fig. 6A). The rN protein was used as control viral antigen for natural SARS-CoV-2 infection. The serum samples collected before the emergence of COVID-19 in France were used as negative control sera (Table S3). There was a significant immune reactivity of COVID-19 donor sera against both rN and rRBD proteins. The serum samples (n = 23) among the COVID-19 recovered patients who developed higher anti-RBD antibody titers (serum dilution 1:100) were assayed through synthetic peptide-based ELISA (Fig. 6B). A lack of significant antigenic reactivity for S1P4 peptide with COVID-19 donor sera was observed (Fig. 6B). Serum samples from COVID-19 immune subjects showed no significant reactivity with the S2P6 peptide although a great variability in antibody response of relevant specificity suggesting that there might be several populations of responders to natural infection. Taken together, these results suggest that the stretches of spike residues 615/644 and 1138/1169 are not identified as immunodominant B-cell epitope peptides in the COVID-19 immune subjects who had experienced mild SARS-CoV-2 infection.
Figure 6.
Antigenic reactivity of the synthetic peptides with human serum-specific SARS-CoV-2 antibody. Serum samples from COVID-19 recovered patients (n = 30) (Table S2) and ten infection- naïve individuals (n = 10) (Table S3) were tested for the antibody reactivity against the N and RBD proteins through indirect ELISA (A), or the S1P4, S1P5, and S2P6 peptides through synthetic peptide-based ELISA (B). The intensity values of serum samples were measured at O.D. 450 nm. In (A), the serum samples at dilution 1:100 were assayed for the detection of antibodies against SARS-CoV-2 N and spike proteins by indirect ELISA using recombinant rN and rRBD proteins (1 µg.ml−1) for antigen-based antibody capture. The intensity values of serum samples were measured at O.D. 450 nm. Unpaired t tests between infection-naïve individuals and COVID-19 immune subjects were performed and statistically significant comparisons are shown as * p < 10–4. In (B), The serum samples (n = 23) among the COVID-19 recovered patients who developed higher anti-RBD antibody titers were assayed for the detection of antibodies against the S1P4, S1P5, and S2P6 peptides (200 ng.ml−1) through synthetic peptide-based ELISA at serum dilution 1:50. The results are the mean (± SEM) of three replicates. The intensity values of serum samples were measured at O.D. 450 nm. Unpaired t tests between infection-naïve individual and COVID-19 immune subject were performed and differences between experimental points considered as non-statistically significant (p > 0.05) are not shown.
Immunogenicity of synthetic S1P4 and S2P6 peptides in mice
To evaluate the in vivo immunogenicity of the two B-cell epitope peptides recognized by the comirnaty vaccine recipient sera, the synthetic S1P4 and S2P6 peptides were N-terminally coupled to Keyhole Limpet Hemocyanin (KLH) protein carrier. The protein-peptide conjugates were assessed with serum sample from a COVID-19 immune subject who received comirnaty vaccine. The KLH-S1P5 served as protein-peptide conjugate control. The antigenic reactivity of KLH-S1P4 and KLH-S2P6 conjugates was verified by indirect ELISA using a comirnaty vaccine recipient serum (Fig. S2A). The antigenic reactivity of protein-peptide conjugates was comparable to that observed with synthetic S1P4 and S2P6 peptides (Fig. S2B), confirming that both KLH-S1P4 and KLH-S2P6 are suitable for further experiments.
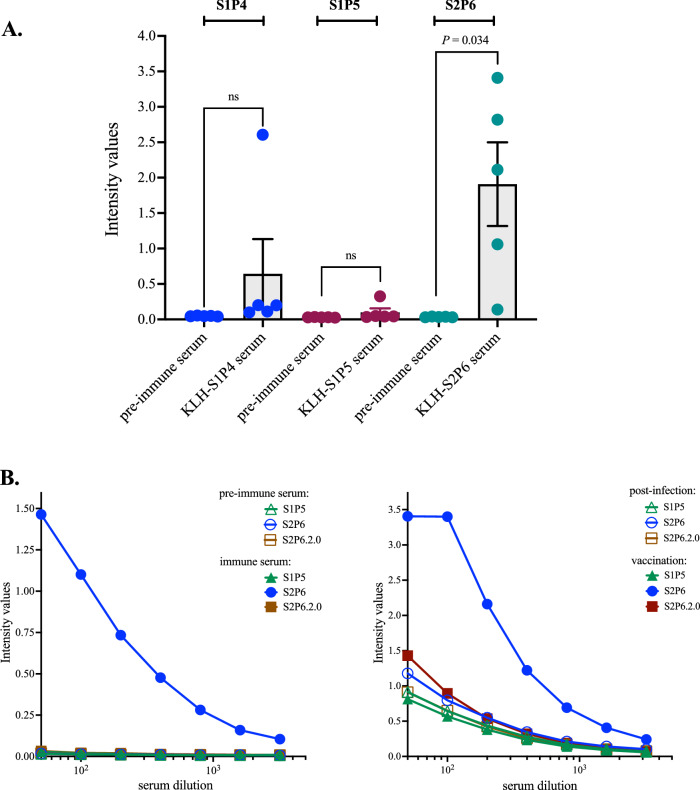
To assess the immunogenicity of KLH-peptide conjugates in inbred laboratory mice, three groups of adult BALB/c mice were subcutaneously (s.c.) inoculated with 20–30 µg of KLH-S1P4, KLH-S1P5 or KLH-S2P6 conjugates in a prime-boost schedule. Immune sera were collected two weeks after the third immunization. The ability of the protein-peptide conjugates to elicit antibody response in mice was assessed by indirect ELISA using protein-peptide conjugates for antigen-mediated antibody capture. Immunization with KLH-S1P4 and KLH-S2P6 conjugates but not KLH-S1P5 conjugate elicited a strong antibody response against protein-peptide conjugates (Fig. S3). To evaluate the ability of the KLH-peptide conjugates to elicit antibody production of relevant specificity, individual mouse immune sera were tested on the synthetic S1P4, S1P5, and S2P6 peptides through a peptide-based ELISA (Fig. 7A). Mouse pre-immune serum served as control serum. Most of BALB/c mice (n = 5) that received KLH-S2P6 conjugates developed S2P6 peptide-reactive antibodies with a median O.D.450 nm value about 2.0 at serum dilution 1:50 (Fig. 7A). In contrast, the KLH-S1P4 conjugates were poorly immunogenic in BALB/c mice whereas KLH-S1P5 conjugates were inefficient to elicit a significant production of specific antibodies. The immunogenicity of the synthetic S2P6 peptide reinforces the notion that the linear B-cell epitope peptide which is delimited by the spike residues 1138/1169 in recombinant SARS-CoV-2 spike protein would be a feature of the Pfizer-BioNTech COVID-19 vaccine comirnaty.
Figure 7.
Immune reactivity of mouse antibodies raised against B-cell epitope peptides. In (A), serum samples from mice (n = 5) that received the KLH-peptide conjugates were assessed for peptide-reactive antibodies through peptide-based ELISA using the synthetic S1P4, S1P5, and S2P6 peptides (200 ng.mL−1) for antigen-based antibody capture. The pre-immune serum of each individual that received the KLH-peptide conjugates was tested. The intensity values of serum samples at dilution 1:50 were measured at O.D. 450 nm. Paired t tests between pre-immune serum and KLH-peptide serum were performed (ns: non-statistically significant, p > 0.05). The results are representative of two independent experiments. In (B), synthetic peptide-based ELISA using the S2P6 and S2P6.2.0 peptides (200 ng.mL−1) for peptide-based antibody capture. At the left, pre-immune and immune serum samples of an immunized mouse with KLH-S2P6 conjugates. At the right, serum samples from a COVID-19 immune subject collected after recovery (post-infection) and after injection of a single dose of comirnaty vaccine (vaccination). Synthetic S1P5 peptide served as negative peptide control. The intensity values of serum samples in a dose-curve response were measured at O.D. 450 nm.
A tridimensional structure prediction of the synthetic S2P6 peptide suggests that the amino-acids 10 to 21 (spike residues 1147/1157) could form α-helix (Fig. S4). The identified α-helix might act as potential binding site which enables the antibody recognition of synthetic S2P6 peptide. This prompted us to evaluate whether anti-S2P6 antibodies could target a truncated form of the synthetic S2P6 peptide. Consequently, we developed a 19-mer synthetic S2P6.2.0. peptide containing the amino-acids 10 to 28 of the B-cell epitope S2P6 peptide (Table 1). By peptide-based ELISA, we observed a lack of antigenic reactivity of synthetic S2P6.2.0 peptide with a mouse immune serum wherein the S2P6 peptide-reactive antibody concentration was high and also serum sample from COVID-19 immune subject who received a single dose of comirnaty vaccine after recovery (Fig. 7B). Thus, the motif 1147SFKEELDKYFKNHTSPDVD1165 is not sufficient for the antibody recognition. Given that binding capability of the linear B-cell epitope S2P6 peptide was completely lost when the nine N-terminal and four C-terminal amino-acids were deleted, it may be inferred that motifs 1138YDPLQPELD1146 and 1162LGDI1165 are potentially critical residues for epitope-antibody interaction.
Discussion
The Pfizer/BioNTech COVID-19 vaccine comirnaty, a mRNA-based vaccine against SARS-CoV-2 infection, is highly effective at preventing serious illness and death among individuals of diverse ages. With the aim to identify specific antibody epitopes in comirnaty vaccine, a set of potential linear B-cell epitope peptides in the SARS-CoV-2 spike protein was assayed for its antigenic reactivity against serum samples from comirnaty vaccine recipients and COVID-19 recovered patients.
In the present study, we showed that administration of comirnaty vaccine to infection-naïve individuals elicits production of antibodies capable of binding to the synthetic S2P6 peptide and to a lesser extent the synthetic S1P4 peptide, targeting the SARS-CoV-2 spike residues 1138/1169 and 616/644, respectively. The spike residues 616/644 are located at the C-terminal domain 2 (CTD2) of S1 subunit upstream of the spike furin-cleavage site RRAR at the junction of S1 and S2 subunits whereas the spike residues 1138/1169 are positioned at the junction of the CD and the HR2 in the C-terminal region of S2 subunit (Fig. 1B). In contrast to what it has been observed with comirnaty-vaccinated individuals, serological analysis of COVID-19 patients revealed a lack of reactivity of the S1P4 and S2P6 peptides with antibodies raised against SARS-CoV-2. We showed that administration of comirnaty vaccine to a COVID-19 recovered patient elicits a production of antibodies capable of binding to synthetic S1P4 and S2P6 peptides. Thus, the comirnaty vaccine is effective to elicit an antibody response targeting the SARS-CoV-2 spike residues 1138/1169 and to a lesser extent, the residues 616/644, in COVID-19 immune subjects. The recombinant full-length SARS-CoV-2 spike protein expressed by a such COVID-19 mRNA vaccine is stabilized in a prefusion state due to amino-acid substitutions K986P and V987P13,22,24. One privileged hypothesis is that the immune recognition of spike residues 616/644 and 1138/1169 as linear B-cell epitope peptides would be an antigenic specificity of the pre-fusion stabilized spike protein. Whether the two B-cell epitope peptides representing the SARS-COV-2 spike residues 616/644 and 1138/1169 are a feature of comirnaty vaccine is an important issue that remains to be investigated. This could be achieved by analyzing a larger cohort of comirnaty-vaccinated individuals with or without prior SARS-CoV-2 infection35. It would also be of priority to evaluate whether the other commercially available COVID-19 mRNA vaccine mRNA-1273 licensed as spikevax by Moderna/NIAID also elicits an antibody response targeting the same stretches of spike residues 616/644 and 1138/116913,25.
Phylogenetic analysis of SARS-CoV-2 variants indicated that the spike residues 616/644 and 1138/1169 are strictly conserved among identified VOCs including the Delta and Omicron variants. Also, the residues 1138/1169 mapped in the S2 subunit of spike protein are strictly conserved between SARS-CoV-2 and SARS-CoV. Taking into consideration with the singularity of the synthetic S1P4 and S2P6 peptides, it could be interesting to design peptide-based diagnosis assays to monitor antibody responses in comirnaty-vaccinated individuals without prior evidence of infection or with confirmed prior COVID-19 infection36. Indeed, the design of a such serological assay would be of interest to test for anti-spike antibody levels before administration of vaccine booster doses.
To our knowledge, it is not yet known whether the SARS-CoV-2 spike residues 616/644 and 1138/1169 contribute to the protective efficacy of Pfizer-BioNTech COVID-19 vaccine comirnaty23,31–34,37,38. There is some evidence that potent neutralization of SARS-CoV-2 requires both spike conformational changes and receptor blockade elicited by antibody binding10,13–18,38. The antibody recognition of the spike residues 616/644 into the C-terminal domain 2 of S1 subunit might have an impact on the efficiency of S1/S2 processing at the spike residue 685 by the cell surface protease TMPRSS215–20. The antibody recognition of spike residues 1138/1169 might have an effect on fusion process between viral and host-cell membranes by trapping the prefusion state of spike protein timer21,22. Immunization of adult BALB/c mice with the synthetic S2P6 peptide conjugated to a protein carrier identified the SARS-CoV-2 spike residues 1138/1169 as immunogenic peptide. The SARS-CoV-2 spike protein was recognized by the S2P6 peptide-reactive antibodies suggesting that the synthetic S2P6 peptide could induce antibody response against SARS-CoV-2. Although truncation of the synthetic S2P6 peptide has not allowed to identify a minimal motif for antibody recognition, we assume that the N-terminal part of the B-cell epitope containing the spike residues 1138/1169 might play a key role in the epitope-antibody interaction. It is of priority to generate mouse hybridoma cell lines stably secreting monoclonal antibodies (mAbs) raised against the B-cell epitope peptide S2P6. Such anti-S2P6 mAbs will be assayed for their immune reactivity with spike protein trimer and neutralizing activity against live SARS-CoV-2.
Despite the lack of information on the protection conferred by the S2P6 peptide-reactive antibodies against SARS-CoV-2 infection, the data obtained with the B-cell epitope S2P6 peptide broadens our understanding on the efficacy of COVID-19 mRNA comirnaty vaccine. Finally, our study raises the question on the medical applicability of a vaccine formulation based on linear B-cell epitope peptides, e.g., the S2P6 peptide containing the residues 1138/1169 located in the C-terminal region of S2 subunit from SARS-CoV-2 spike protein37–39.
Methods
Antibody epitope mapping
The IEDB (for Immune Epitope Database, https://www.ied.org) as B-cell epitope-prediction tool was applied to the B-cell epitope candidates in the SARS-CoV-2 spike protein (UniprotKB-P0DTC2 (SPIKE_SRAS2)). The BepiPred-2.0 algorithm was used to predict antibody epitopes using 0.50 as the cut-off parameter.
Expression of recombinant SARS-CoV-2 proteins in mammalian cells
Mammalian codon-optimized genes coding either for the SARS-CoV-2 N protein or the RBD domain of spike protein (residues 328/583) (UniProtKB-P0DTC9 (NCAP_SARS2)) were established using Cricetulus griseus codon usage as reference40. The sequence encoding the mouse Ig gamma-2A (IgG2A) chain C region sequence (UniprotKB-P01863 (GCAA_MOUSE) was fused in frame to the carboxy-terminus of N and RBD proteins with a spacer serine-glycine spacer. The protein sequence is preceded by a Secrecon signal peptide. The synthesis of gene sequences and their cloning into Kpn-I and Xho-I restriction sites of the pcDNA3.1 vector plasmid to generate pcDNA3/N-IgG2A and pcDNA3/RBD-IgG2A were performed by Genecust (Boynes, France). The plasmid sequences were verified by Sanger method (Genecust). Chinese Hamster Ovary CHO cells were transiently transfected with pcDNA3/N-IgG2A and pcDNA3/RBD-IgG2A using Lipofectamine 3,000 (Thermo Fisher Scientific, les Ulis, France). After 72 h of transfection, cell supernatants were harvested and clarified cell culture supernatant (C3S) fractions were obtained by centrifugation at 1000×g for 10 min at room temperature. Secreted N-IgG2a (rN) et RBD-IgG2a (rRBD) proteins in C3S fractions were detected by immunoblot assay using goat anti-mouse IgG heavy chain HRP antibody (Abcam, Cambridge, UK). The amounts of soluble rN and rRBD proteins in C3S fractions were estimated at 1.5 and 0.8 µg.mL−1, respectively, by direct ELISA test using goat anti-mouse IgG heavy chain HRP antibody. Standard for ELISA calibration was done using a serial dilution of mouse IgG at 1 mg.mL−1 as coating antigen (Vectors, Clinisciences, France).
Synthetic SARS-CoV-2 spike peptides
All synthetic peptides used in this study were chemically synthesized by Genecust (Boynes, France). Peptides were dissolved in DMSO at concentration of 10 mg.mL−1 and then diluted in sterile H2O at the final concentration 1 mg.mL−1. The stock peptide solutions were stored at − 80 °C. Working peptide solutions at the final concentration 0.2 mg.mL−1 in sterile H2O were stored at − 80 °C.
ELISA methods
For indirect ELISA assay using recombinant soluble rN and rRBD proteins as antibody capture antigens, a 96-well plate was coated with 0.1 ml of CS3 fractions at final concentration of 0.15 µg.mL−1 of rN or 80 ng.mL−1 of rRBD in PBS at 4 °C overnight. For peptide-based ELISA, a 96-well plate was coated with 0.1 ml of peptide at final concentration of 0.2 µg.mL−1 in PBS at 4 °C overnight. For indirect ELISA, a 96-well plate was coated with 0.1 ml of KLH-peptide at final concentration of 1 µg.mL−1 in PBS at 4 °C overnight. At the end of incubation period, the solution was discarded, the wells were washed with PBS supplemented with 0.01% Tween-20 (PBST) and then incubated with a commercial ELISA blocking agent (EBA) at room temperature (RT) for 1 h. After washing with PBST, the wells were incubated with human serum sample at final dilution 1:50 in EBA or mouse serum sample at final dilution 1:100 at 37 °C for 2 h. After washing with PBST, the wells were incubated with goat anti-human IgG-HRP or goat anti-mouse IgG-HRP at final dilution 1:2000 in EBA at room temperature for 1 h. After washing with PBST and PBS, the wells were incubated with TMB substrate solution at RT for 3 min and the reaction was stopped with acidic stopping solution. Absorbance was measured using microplate reader at 450 nm.
Commercial serum samples from COVID-19 immune subjects
Serum samples were collected from de-identified COVID-19 immune subjects (n = 30 total) in 2020 (Table S2) and infection-naïve individuals (donation in 2019; n = 10 total) (Table S3) in France. All serum samples were purchased from Cerba Xpert (Saint-Ouen l’Aumône, France).
Serum samples from comirnaty vaccine recipients
Blood samples were collected from de-identified healthy comirnaty vaccine recipient adults (n = 9) by professional medical caregivers in the Centre Hospitalier Universitaire de La Reunion (CHU de La Reunion, France) (Table S4). Ethical approval for this study was obtained from Research Ethics Committee “Comité de Protection des Personnes, Nord Ouest IV de Lille, France (approval number EudraCT/ID-RCB 2020-A01253-36)”. Written informed consent was obtained from all volunteers before. Age, gender, origin, date of vaccination and date of specimen collection were collected for all volunteers. Study individuals were fasting before venous puncture. Blood was sampled in EDTA tubes and plasma was stored at − 80 °C. All methods were carried out in accordance with relevant guidelines and regulations. Blood samples were collected in infection-naïve individual prior vaccination and few weeks after the receipt of the second dose of comirnaty vaccine.
In this study, two anonymized comirnaty vaccine recipients who have given their consent for the use of their serum samples were also assessed for the reactivity of their antibodies. An infection-naïve individual (Male, 59 years, France) who had been vaccinated with comirnaty vaccine in a two-dose regimen in April 2021. Serum samples were collected 0.5 and 2 months after the receipt of the second dose. A COVID-19 immune subject (Female, 62 years, France) who received the comirnaty vaccine three months after recovery. Serum samples were collected few weeks after COVID-19 recovery in March 2021 and then 0.5 month after the receipt of a single dose of comirnaty vaccine in June 2021.
Mouse experiment and ethical statement
The Animal Ethics Committee of CYROI n°114 approved all the animal experiments with reference APAFIS#32,599-2021060109058958v2 (July 2021). All animal procedures were performed in accordance with the European Union legislation for the protection of animals used for scientific purposes (Directive 2010/63/EU). The study was conducted following the guidelines of the Office Laboratory of Animal Care (agreement n° 974 001 A) at the Cyclotron and Biomedical Research CYROI platform, Sainte-Clotilde, La Reunion, France and in accordance with the ARRIVE guidelines (https://arriveguidelines.org). Three groups of five 6-week-old female BALB/cJRj mice (Janvier Labs, France) were hosted in individually ventilated plastic cages (5 animals per cage) under 50–60% humidity and 22–25 °C temperature in a 12/12-h light dark cycle. Food and drinking were provided ad libitum. The adult BALB/cJRj mice (Janvier Labs, France) were s.c. inoculated with KLH-peptide conjugates in complete Freund’s adjuvant (Sigma, France). The protein-peptide conjugate samples in a final volume of 0.1 ml were distributed over two injection sites. Immunized mice were boosted with the same protein-peptide conjugate in incomplete Freund’s adjuvant at Days 7 and 21 after primary immunization (Day 0). Two weeks after the last immunization, retro-orbital blood sampling was performed in anesthetized mice. All animals were daily observed to detect any stress or suffering.
Statistical analysis
Unpaired and pairedt t tests were used to compare quantitative data. GraphPad Prism 9 was used for all statistical analysis.
Supplementary Information
Acknowledgements
We thank the MAVIVH team (Tours) for the helpful discussion. We thank the PIMIT and DeTROI teams for their interest in this study. This research was funded by grants from POE FEDER 2014-20 of the Conseil Régional de La Réunion (TFORCE-COVIR and LIPICOR programs, N°20201437-0027601) with additional support of the COVID-19 Emergency Seed from the La Reunion University. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Author contributions
J.A. contributed to formal analysis, investigation, methodology and validation; W.V. contributed to formal analysis, investigation, methodology, supervision, and writing review & editing; C.C. contributed to investigation, methodology, and resources; C.H. contributed to investigation, methodology and validation; C.P. contributed to investigation and resources; B.R. contributed to methodology, resources, and writing review & editing; M.L-X. contributed to methodology and resources; C.E-K. contributed to investigation, methodology and validation; O.M. contributed to methodology, resources and writing review & editing; P.M. contributed to methodology, funding acquisition, project administration and writing review & editing; D.C. contributed to methodology, resources and writing review & editing; G.G. contributed to formal analysis, investigation, methodology, and writing review & editing.; P.D. contributed to conceptualization, formal analysis, investigation, methodology, resources, visualization, validation, writing-original draft and writing review & editing.
Competing interests
All authors declare no conflict of interest.
Footnotes
The original online version of this Article was revised: The original version of this Article contained a repeated error in the Title, in the Results section under the subheading ‘Antigenic reactivity of synthetic peptides with COMIRNATY vaccine recipient sera’ and in the Methods section under the subheading ‘Serum samples from COMIRNATY vaccine recipients’ where “COMINARTY” now reads: “COMIRNATY”.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jessica Andries and Wildriss Viranaicken.
Change history
4/20/2022
A Correction to this paper has been published: 10.1038/s41598-022-10885-7
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-10057-7.
References
- 1.Gralinski LE, Menachery VD. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19: A review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 3.Yuki K, Fujiogi M, Koutsiogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parasher A. COVID-19: Current understanding of its pathophysiology, clinical presentation and treatment. Postgrad. Med. J. 2021;97:312–320. doi: 10.1136/postgradmedj-2020-138577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osuchowski MF, et al. The COVID-19 puzzle: Deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir. Med. 2021;9:622–642. doi: 10.1016/S2213-2600(21)00218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGonagle D, Bridgewood C, Meaney JFM. A tricompartmental model of lung oxygenation disruption to explain pulmonary and systemic pathology in severe COVID-19. Lancet Respir. Med. 2021;9:665–672. doi: 10.1016/S2213-2600(21)00213-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suthar MS, et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell. Rep. Med. 2020;1:100040. doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni L, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;53:971–977. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Beltran, et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184:476–488. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;180:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharm. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asarnow D, et al. Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncitia. Cell. 2021;184:3192–3204. doi: 10.1016/j.cell.2021.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat. Rev. 2020;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu C, He G, Yin Q, Ye X, Shi Y, Xu W. Molecular biology of the SARS-CoV-2 spike protein: a review of current knowledge. J. Med. Virol. 2021 doi: 10.1002/jmv.27132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. (USA) 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson BA, et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peacock TP, et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021;6:899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 21.Kirchdoerfer RN, et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadarangani M, Marchant A, Kollman TR. Immunological mechanisms of vaccine-induced protection against Covid-19 in humans. Nat. Rev. Immunol. 2021;1:10. doi: 10.1038/s41577-021-00578-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinz FX, Stiasny K. Distinguishing features of current COVID-19 vaccines: knows and unknows of antigen presentation and modes of action. NPJ Vacc. 2021;6:104. doi: 10.1038/s41541-021-00369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teo SP. Review of COVID-19 mRNA vaccines: BNT162b2 and mRNA-1273. J. Pharm. Pract. 2021;1:5. doi: 10.1177/08971900211009650. [DOI] [PubMed] [Google Scholar]
- 26.Vitiello A, Ferrara F. Brief review on the mRNA vaccines COVID-19. Inflammopharmacology. 2021;29:645–649. doi: 10.1007/s10787-021-00811-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anand P, Stahel VP. Review the safety of Covid-19 mRNA vaccines: A review. Patient Saf. Surg. 2021;15:20. doi: 10.1186/s13037-021-00291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polack FP, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skowronski DM, De Serres G. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2021;384:1576–1578. doi: 10.1056/NEJMc2036242. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, et al. Linear epitope landscape of the SARS-CoV-2 spike protein constructs from 1,051 COVID-19 patients. Cell Rep. 2021;34:1089015. doi: 10.1016/j.celrep.2021.108915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poh CM, et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralizing antibodies in COVID-19 patients. Nat. Comm. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, et al. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell Mol. Immunol. 2020;17:1095–1097. doi: 10.1038/s41423-020-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu S, et al. The immunodominant and neutralization linear epitopes for SARS-CoV-2. Cell Rep. 2021;34:108666. doi: 10.1016/j.celrep.2020.108666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD 3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44:W449–W454. doi: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anglya A, et al. T-cell and antibody responses to first BNT162b2 vaccine dose in previously infected and SARS-CoV-2 UK health-care workers: A multicentre prospective cohort study. Lancet Microbe. 2022;3:e21–31. doi: 10.1016/S2666-5247(21)00275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manisty C, et al. Antibody response to first BNT162b2 dose in previously SARS-CoV-2-infected individuals. Lancet. 2022;397:1057–1058. doi: 10.1016/S0140-6736(21)00501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang M, et al. Epitope profiling reveals the critical antigenic determinants in SARS-CoV-2 RBD-based antigen. Front. Immunol. 2021;12:707977. doi: 10.3389/fimmu.2021.707977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finkelstein MT, Mermelstein AG, Miller EP, Seth PC, Stancokski EDT, Fera D. Structural analysis of neutralizing epitopes of the SARS-CoV-2 spike to guide therapy and vaccine design strategies. Viruses. 2021;13:134. doi: 10.3390/v13010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrscher C, et al. ELISA-based analysis reveals an anti-SARS-CoV-2 protein immune response profile associated with disease severity. J. Clin. Med. 2022;11:405. doi: 10.3390/jcm11020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frumence E, et al. Robust and low-cost ELISA based on IgG-Fc tagged recombinant proteins to screen for anti-SARS-CoV-2 antibodies. J Virol Methods. 2021;495:113082. doi: 10.1016/j.jim.2021.113082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.