Abstract
COVID-19 is an infectious pneumonia caused by 2019-nCoV. The number of newly confirmed cases and confirmed deaths continues to remain at a high level. RT–PCR is the gold standard for the COVID-19 diagnosis, but the computed tomography (CT) imaging technique is an important auxiliary diagnostic tool. In this paper, a deep learning network mutex attention network (MA-Net) is proposed for COVID-19 auxiliary diagnosis on CT images. Using positive and negative samples as mutex inputs, the proposed network combines mutex attention block (MAB) and fusion attention block (FAB) for the diagnosis of COVID-19. MAB uses the distance between mutex inputs as a weight to make features more distinguishable for preferable diagnostic results. FAB acts to fuse features to obtain more representative features. Particularly, an adaptive weight multiloss function is proposed for better effect. The accuracy, specificity and sensitivity were reported to be as high as 98.17%, 97.25% and 98.79% on the COVID-19 dataset-A provided by the Affiliated Medical College of Qingdao University, respectively. State-of-the-art results have also been achieved on three other public COVID-19 datasets. The results show that compared with other methods, the proposed network can provide effective auxiliary information for the diagnosis of COVID-19 on CT images.
Keywords: Mutex attention network, COVID-19, Deep learning, Attention, Computer-aided diagnosis
Introduction
Coronavirus disease 2019 (COVID-19), named by the World Health Organization, refers to pneumonia caused by the 2019 novel coronavirus (2019-nCoV) [1, 2]. As of August 5, 2021, more than 200 million confirmed cases and 4.2 million confirmed deaths had occurred worldwide. More importantly, the number of newly confirmed cases and new confirmed deaths remains at a high level and some specific variants of SARS-CoV-2 that are more transmissible and possibly more virulent have appeared worldwide [3]. With global vaccine production still insufficient, it is very important to find a fast and effective COVID-19 detection method.
The gold standard for diagnosing COVID-19 is reverse transcription polymerase chain reaction (RT–PCR) [4]. However, existing data show that the sensitivity of RT–PCR to COVID-19 infection is not high [5]. This leads to many patients who are infected with COVID-19 being mistaken as uninfected during rapid screening, which is not conducive to COVID-19 prevention and treatment.
As a common medical imaging tool, computed tomography (CT) imaging technology is a sensitive diagnostic method for COVID-19. Chest CT is an important supplement to RT–PCR in COVID-19 diagnosis[6]. Long et al. [7] reported that CT sensitivity was 97.2%, while the initial rRT-PCR sensitivity was 83.3%. In addition, CT can observe the pulmonary manifestations with different infections in different periods, which can assist doctors in diagnosis and treatment [8].
Although chest CT technology can be used as a diagnostic auxiliary tool for COVID-19, a large number of experienced radiologists are needed for screening. It increases work, and it is also prone to misdiagnosis caused by fatigue and other reasons. With successful deep learning (DL) applications in the image classification [9] and natural language processing fields [10, 11], considerable progress has been achieved in DL-based medical image processing task [12]. Deep learning is widely viewed as a crucial tool in COVID-19 diagnosis[13, 14], as they provide auxiliary information, which is very different from other computer vision tasks, such as head pose estimation [15], intelligent recommendation [16, 17], robot vision [18], and infrared imaging enhancement [19].
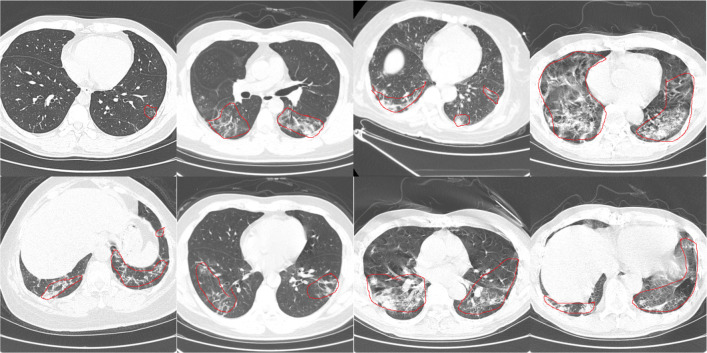
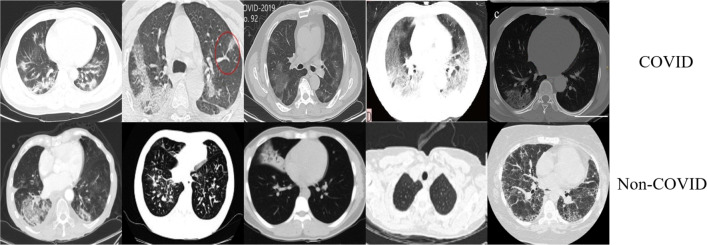
It is difficult to diagnose COVID-19 because the shape, size, and Hounsfield unit (Hu) value of the lesions on the CT images have large variation, as shown in Fig.1. Furthermore, the COVID-19 patients CT images are similar to CT images of other diseases (such as community-acquired pneumonia, H1N1), which also makes the diagnosis of COVID-19 very difficult, as shown in Fig. 2. In response to these problems, we propose a novel network MA-Net including mutex attention blocks and fusion attention blocks to achieve effective feature representation for COVID-19. We also design an adaptive weight loss function for a preferable diagnosis effect. The contributions of this paper are as follows:
Aiming at the problem that the COVID-19 CT images are similar to other diseases, this paper designs a multi-inputs network with shared parameters and a mutex attention block. The mutex input include a pair of positive and negative samples. The purpose of the mutex attention block is to amplify the difference between positive and negative samples, which enables the network to obtain better distinguishable features. It can significantly improve the ability of deep networks to diagnose COVID-19.
In this paper, the fusion attention block is designed to fuse features in the channel direction, which selects features between two inputs to obtain more representative features. It can further strengthen the network COVID-19 diagnostic capabilities.
Joint training of multiple loss functions is used in the work. An adaptive weight adjustment mechanism is proposed to automatically tune the weight of different losses. The experimental results show that the adaptation weight can effectively improve the diagnosis effect of the proposed network.
Fig. 1.
COVID-19 Dataset-A CT images
Fig. 2.
COVID-19 Dataset-C CT images[20]
Related work
Due to the COVID-19 outbreak, many researchers have proposed deep learning methods to analyze COVID-19 CT or X-ray images [21–23]. For COVID-19 diagnosis on X-ray, [24, 25] proposed methods and achieved good results. For COVID-19 diagnosis on CT images [26–28], many methods have been proposed and validated on different COVID-19 datasets.
COVID-9 diagnosis on CT
For private datasets, Ma et al. [29] proposed a new COVID-19 diagnosis network based on the multireceptive field attention module. This attention module includes pyramid convolution module (PCM), spatial attention block (SAB) and channel attention block (CAB). PCM is used to obtain multi-receptive field feature sets and send the multi-receiving field feature sets to SAB and CAB to enhance the feature. The proposed method was trained and verified on the DTDB provided by the Beijing Ditan Hospital Capital Medical University, which includes 40 patients infected with COVID-19 in different periods and 40 people without COVID-19 infection. 97.12% accuracy, 96.89% specificity and 97.21% sensitivity are obtained. Li et al. [30] developed a 3D deep learning framework for COVID-19 diagnosis (COVID-19, community-acquired pneumonia and non-pneumonia), referred to as COVNet. It consists of a sequence of shared parameters ResNet50. Experiments on collected dataset containing 4352 chest CT scans from 3322 patients, the per-scan sensitivity and specificity in the independent test set were 90% and 96% , respectively. Harmon et al. [31] proposed 3D model for differentiation of COVID-19 from other clinical entities. Training in a diverse multinational cohort of 1280 patients to localize parietal pleura/lung parenchyma followed by classification of COVID-19 pneumonia, it can achieve up to 90.8% accuracy, with 84% sensitivity and 93% specificity, as evaluated in an independent test set (not included in training and validation) of 1337 patients. For public datasets, Zhao et al. [20] provided a collated COVID-19 CT dataset containing 349 CT images believed to be positive for COVID-19. [20] also proposed a DensNet169 method combining contrastive self-supervised learning (CSSL) [33] and transfer learning (TL) for COVID-19 diagnosis and used CT images with a lung mask as input. Performing COVID-19 diagnosis on the proposed COVID-19 CT dataset, the accuracy, F1-Score and AUC were 85.0%, 85.9% and 92.8% respectively, although the number of CT images for training was only a few hundred. The COVID-19 CT dataset [20] provided by Zhao et al. has been widely used. Due to the limited size of the COVID-19 dataset, Mittal et al. [32] proposed a new clustering method for COVID-19 diagnosis. A novel variant of a gravitational search algorithm is employed to obtain optimal clusters. To validate the performance on the proposed variant, a comparative analysis of recent metaheuristic algorithms is conducted. The proposed method was verified on the COVID-19 CT dataset [20] and an accuracy of 0.6441 was obtained. Underperforming with traditional methods, many researchers focus on using deep learning for COVID-19 diagnosis. He et al. [34] proposed a self-transformation method that integrated comparative self-monitoring learning and transfer learning to learn strong and unbiased feature representations on the limited size dataset [20] to reduce the risk of overfitting. 86% accuracy, 85% F1-Score and 94% AUC were obtained, respectively. Wang et al. [35] proposed a new joint learning framework to achieve accurate diagnosis of COVID-19 by effectively learning heterogeneous datasets with distributed differences. A powerful backbone was built by redesigning the recently proposed COVID-Net [36] in terms of network architecture and learning strategy. Additionally, a contrastive training objective was applied to enhance the domain invariance of semantic embedding to boost the classification performance on each dataset. On the COVID-19 CT dataset [20], the proposed method achieved 78.69% accuracy, 78.83% F1-score and 85.32% AUC.
Ma et al. [29] used the pyramid convolution module to solve the problem of different sizes and shapes of COVID-19 lesions. Zhao et al. [20], Mittal et al. [32] and He et al. [34] focused on solving the problem of limited COVID-19 dataset. Li et al. [30] and Harmon et al. [31] focused on the use of large datasets for the diagnosis of COVID-19 and other lung diseases. These works have achieved excellent results. However, most of them used large-scale private datasets or were evaluated directly using commonly used deep learning network models.In this paper, we propose a COVID-19 diagnostic method based on mutex attention block to distinguish COVID-19 from other pulmonary diseases with limited data.
Image attention mechanism
The attention mechanism was first used in the field of natural language processing(NLP) and achieved state-of-the-art results.Then, using attention mechanism in the field of computer vision has recently received more research. Many outstanding attention modules have been proposed [37, 38]. The attention mechanism can be simply divided into channel attention, spatial attention, mixed attention and special attention models.
SE-Net [39] was proposed in 2017, which is a typical channel attention model and can be embedded in any basic network. SE-Net proposed a novel attention unit, the squeeze-and-excitation module (SE module), which adaptively recalibrates the channel characteristic response by explicitly modeling the interdependence between channels. Experiments show that by embedding the SE module into existing basic models (such as ResNet and VGG), it can bring significant performance improvements to the most advanced deep architecture at a small computational cost. Different from the SE module which only focuses on the feature correction of the channel dimension, CBAM [40] introduces an attention mechanism in both the spatial and channel dimensions. A set of feature maps is input, and the CBAM module sequentially infers the attention map along two independent dimensions (channel and spatial), and then multiplies the attention map with the input feature map to perform adaptive feature refinement.
Datasets and methods
Datasets
We conduct experiments on different COVID-19 datasets to verify our method. All experiments are based on 2D slices. Each dataset is tested separately. The details of the four datasets are shown in Table 1.
Table 1.
The number of COVID and nonCOVID CT images of the three COVID-19 datasets
| Datasets | COVID | NonCOVID | Train | Test |
|---|---|---|---|---|
| COVID-19 | 2499 | 1908 | 2000 COVID | 499 COVID |
| dataset-A | 1527 NonCOVID | 381 NonCOVID | ||
| COVID-19 [41] | 1844 | 1676 | 1107 COVID | 369 COVID |
| dataset-B | 1006 NonCOVID | 335 NonCOVID | ||
| COVID-19 [20] | 349 | 397 | 280 COVID | 70 COVID |
| dataset-C | 317 NonCOVID | 78 NonCOVID | ||
| COVID-19 [42] | 1252 | 1230 | 1001 COVID | 251 COVID |
| dataset-D | 984 NonCOVID | 246 NonCOVID |
The COVID-19 Dataset-A was provided by The Affiliated Hospital of Qingdao University Medical College, including 21 CT scans with COVID-19 and 18 CT scans without COVID-19, which were labeled by experienced doctors. We extracted 2499 slices of positive samples containing COVID-19 infection from CT scans with COVID-19 and randomly selected 1908 slices of negative samples from 18 CT scans without COVID-19 as “NonCOVID” . We conducted slice-level and patient-level experiments. At the slice-level, the dataset was divided into 3527 images for training, and 880 images for verification. At the patient-level, slices from 16 CT scans with COVID-19 and 14 CT scans without COVID-19 were used for training and the rest were used for verification.
The COVID-19 Dataset-B [41] was provided by Ma et al., and it includes CT scans of 20 patients. The infection area was marked by two radiologists and verified by an experienced radiologist. The slices containing COVID-19 infection were extracted as COVID, for a total of 1844. The remaining slices that did not contain COVID-19 were regarded as “NonCOVID”, for a total of 1676. The dataset was divided into 2816 images for training, and 704 images for verification.
The COVID-19 Dataset-C [20] was provided by Zhao et al. The practicability of the dataset was confirmed by a senior radiologist at Tongji Hospital in Wuhan, China, who diagnosed and treated a large number of patients with COVID-19 during the outbreak from January to April. The dataset provides 8-bit CT images of png instead of DICM with the Hu value, which results in resolution loss. Second, the original CT scan contained a series of CT slices, but only a few key slices were selected in the dataset, which also had a negative impact on the diagnosis. The COVID-19 Dataset-C is shown in Fig. 2. This dataset contains 349 COVID-19 CT images and 397 other CT images without COVID-19. The dataset was divided into 597 images for training and 148 images for verification. Because the number of images in the dataset was too small, we enhanced the training data by flipping and rotating.
The COVID-19 Dataset-D [42] is a publicly available COVID-19 CT dataset, containing 1252 CT scans that are positive for SARS-CoV-2 infection (COVID-19) and 1230 CT scans for patients with other pulmonary diseases or normal, 2482 CT scans in total. The dataset was divided into 1984 images for training, 498 images for validation.
Methodology
The network we proposed is based on the ResNet50 [43]. The architecture of the proposed network is shown in Fig. 3. Similar to ResNet, the feature extraction layers of this network consist of 5 mutex attention Res-Layers followed by the global pooling layer and the fully connected layer. The inputs of this network are designed specifically with a pair of mutex CT images that contain opposite categories. In the training process, the pair of mutex inputs are randomly selected from the training data. Since in the forward process, nothing has to do with mutex input, we do not need to enter mutex input in the testing process.
Fig. 3.
Architecture of the proposed network
The architecture of the mutex attention Res-Layer is shown in the blue dashed box in Fig. 3. Similar to ResNet, the Res-Layer in mutex attention Res-Layer0 is a 7 × 7 convolution layer with a stride of 2 and a max pool layer with a stride of 2. The Res-Layers in mutex attention Res-Layer1 to mutex attention Res-Layer4 are made up of different numbers of blocks connected in series. In mutex attention Res-Layer0, the inputs are a pair of mutex CT images. The inputs of mutex attention Res-Layer1 to mutex attention Res-Layer4 are the output feature maps of the previous mutex attention Res-Layer. Assume that the input of the mutex attention Res-Layers is Fi and Fm. Frei and Frem are first obtained through the same Res-Layer, and the two Res-Layers share parameters, as shown in the blue dotted frame of Fig. 3. Frei directly obtainsFo. Fam is obtained by putting Frei andFrem into the mutex attention block (MAB). Then, the obtained Fam and Frem are passed through the fusion attention block (FAB) to obtain the output mutex feature maps Fom. The above process can be described by (1) and (2), as follows:
| 1 |
| 2 |
where Wrl is the Res-Layer parameter. FAB is the fusion attention block. MAB is the mutex attention block. We introduce them in detail as follows.
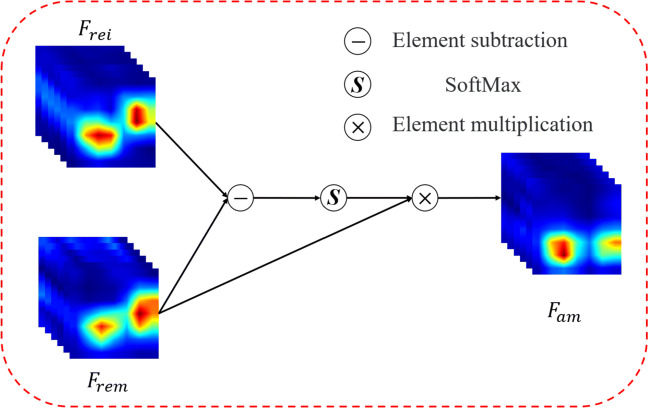
Mutex attention block (MAB)
The MAB inputs are the feature maps Frei and Frem (RC×H×W) after the Res-Layer, as shown in Fig. 4. First, perform elementwise subtraction on Frei and Frem to obtain the distance matrix D(RC×H×W), as equation:
| 3 |
where dcij represents the value of (i, j) of D on c channels, and represent the value of (i, j) of Frei and Frem on c channels. c ∈ [1, C], i ∈ [1, H], j ∈ [1, W]. Then D(RC×H×W) is reshaped to C × HW and softmax is performed in the spatial dimension (HW). The attention map AD is reshaped to (RC×H×W), as shown in (4).
| 4 |
Fig. 4.
Mutex attention block architecture
σ is the softmax function.
Further perform element-wise multiplication between the obtained attention map AD and the input Frem to obtain Fam, as shown in (5).
| 5 |
The purpose of MAB is to reduce the similarity between input feature maps and mutex feature maps. Using the distance between mutex feature maps as an attention map can effectively increase the discrimination of the two inputs. Therefore, the network achieves a better discrimination effect. In the experiments, we visualize the changes in the feature maps to verify the effect of the mutex attention block.
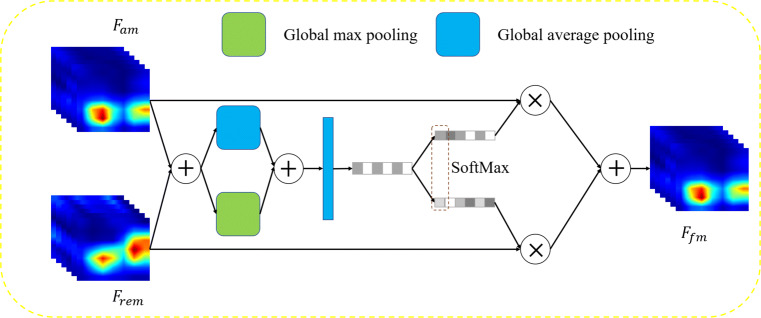
Fusion attention block (FAB)
The structure of FAB is shown in Fig. 5. The inputs of FAB are Fam obtained by MAB and Frem. First, we use elementwise summation on two inputs to obtain mixed feature maps Fmix.
| 6 |
Fig. 5.
Fusion attention block architecture
Then, global average pooling and max pooling are performed on the obtained mixed feature maps, and element addition is used to obtain the channel feature vector V (RC):
| 7 |
where Vc is the value on the c-th channel of the channel feature vector V. Furthermore,two fully connected layers are used to perform feature fitting and channel dimension reduction to obtain the channel feature vector Z:
| 8 |
where C in represents the number of neurons in the fully connected layer, and BR in represents the batch normalization [44] and ReLU [45] layers. The obtained Z is passed through two independent fully connected layers to obtain two weight vectors M and N:
| 9 |
where C is the number of neurons in the fully connected layer, and the obtained M and N all belong to RC. Softmax is used for the corresponding channels of M and N to obtain the attention weights A of Fam for each channel. In addition, the attention weights of Frem is 1 − A. Channelwise multiplication and addition are performed together to obtain the output fusion feature maps Ffm, as following:
| 10 |
| 11 |
where ac is the value of channel weight A on the c-th channel , and is the feature map on the c-th channel of Ffm, c ∈ (0, C − 1).
The FAB function is to perform feature fusion on two sets of feature maps. Through self-learning parameters, the two sets of input feature maps are weighted in the channel dimension.
Loss functions
As shown in Fig. 3, our network contains two losses. LCE is the classification loss corresponding to the input image and the mutex input image, and LCS is a cosine similarity loss between the pair of inputs.
| 12 |
| 13 |
| 14 |
where L1 and L2 are cross-entropy loss, yi and ym are opponent labels of the input and mutex input. , are the predicted values. LCS uses cosine distance to minimize the similarity between two feature maps. In (14), and (RCHW) are the vectors obtained by reshaping the feature maps of the input and the mutex input.
In this paper, the adaptive weight loss is shown in (16).
| 15 |
| 16 |
In this way, the three loss functions can be adjusted adaptively to make the final loss more balanced.
Inference process
In the inference process, since the mutex path has no effect on the main path, only the test images need to be input and mutex input is not needed. The prediction network is a simple ResNet50, which is the same as the backbone. Therefore, the time complexity of the proposed method is the same as ResNet50 and smaller than SE-Net and CBAM based on ResNet50. The model size/Flops/speed are also the same as the backbone and our proposed method does not increase any cost in the test process.
Experiment
We provided experiments with four datasets: COVID-19 dataset-A, COVID-19 dataset-B, COVID-19 dataset-C and COVID-19 dataset-D. For COVID-19 dataset-A and COVID-19 dataset-B, to verify the effectiveness of the attention we proposed, we used the state-of-the-art attention methods in image classification and backbone for comparison. For public COVID-19 dataset-C and COVID-19 dataset-D, we compared the excellent algorithms proposed by other researchers, both of which achieved good results on COVID-19 dataset-C or COVID-19 dataset-D. We show the results from quantitative analysis and qualitative analysis. For qualitative analysis, we used Grad-CAM [47] for visualization.
Experimental details
The proposed method was implemented in PyTorch [46]. All the COVID-19 CT images in our experiments were resized to 224 × 224. Due to the small amount of data on COVID-19 dataset-C, we performed data enhancement including horizontal flip, vertical flip and random rotation. Both inputs and mutex inputs during the training process were randomly selected. We used SGD [47] as the optimization function with an initial learning rate of 0.01. The learning rate decays with the epoch, and the change equation is shown as follows:
| 17 |
where epoch is the number of iterations. Momentum was set to 0.9. The batch size was 16, and 120 epochs were run. An NVIDIA GeForce GTX 1070 GPU with 8 GB memory was used. In the test process, only the test images needed to be input. Since in the forward process, nothing has to do with mutex input, we did not need to enter mutex input in the testing process.
For the training process, COVID-19 dataset-A, COVID-19 dataset-B and COVID-19 dataset-C took 3.1 hours, 2.5 hours and 2.1 hours respectively. For the testing process, it took 0.032 seconds to an image.
Evaluation
The metrics employed to quantitatively evaluate classification were accuracy, sensitivity, F1-score, AUC and specificity. The equation of accuracy is as follows:
| 18 |
The sensitivity and specificity measure the classifier’s ability to identify positive samples and negative samples, respectively, as shown in (19) and (20):
| 19 |
| 20 |
F1-score is the classification problems measurement. In machine learning competitions with multiple classification problems, the F1-score is often used as the final evaluation method. It is a harmonic average of the precision and recall, with a maximum of 1 and a minimum of 0.
| 21 |
| 22 |
| 23 |
Results
COVID-19 dataset-A
COVID-19 Dataset-A was provided by The Affiliated Hospital of Qingdao University Medical College. Detailed information was introduced in the previous section (Section 3.1). We used state-of-the-art attention methods in image classification and backbone for comparison, including ResNet [43], ResNet+CBAM [40] and ResNet+SE [39]. The attention module proposed by Ma et al. [29] for COVID-19 diagnosis is also compared. We conducted slice-level and patient-level experiments to verify the effectiveness of our method. Table 2 and 3 summarize the experimental results.
Table 2.
Slice-level results of different methods on COVID-19 Dataset-A
| Methods | Acc (%) | Sen (%) | Spe (%) | AUC(%) | p |
|---|---|---|---|---|---|
| ResNet [43] | 95.11 | 96.19 | 93.70 | 99.28 | 4.4e-8 |
| ResNet+CBAM [40] | 95.45 | 95.79 | 95.01 | 99.46 | 3.7e-14 |
| SE-Net [39] | 96.02 | 98.00 | 93.44 | 99.53 | 7.3e-8 |
| Ma et al. [29] | 97.12 | 97.21 | 96.89 | 99.34 | 1.9e-5 |
| Ours (ResNet50) | 98.18 | 98.60 | 97.64 | 99.84 |
Acc: accuracy, Spe: specificity, Sen: sensitivity (best in bold)
Table 3.
Patient-level results of different methods on COVID-19 Dataset-A
| Methods | Acc (%) | Sen (%) | Spe (%) | AUC(%) | p |
|---|---|---|---|---|---|
| ResNet [43] | 90.15 | 91.03 | 88.95 | 93.37 | 3.2e-9 |
| ResNet+CBAM [40] | 92.49 | 91.85 | 93.37 | 96.98 | 6.3e-11 |
| SE-Net [39] | 93.90 | 95.92 | 91.16 | 95.01 | 3.3e-10 |
| Ma et al. [29] | 93.23 | 93.63 | 93.24 | 96.03 | 7.5e-7 |
| Ours (ResNet50) | 96.36 | 96.94 | 95.58 | 97.63 |
Acc:accuracy, Spe: specificity, Sen: sensitivity (best in bold)
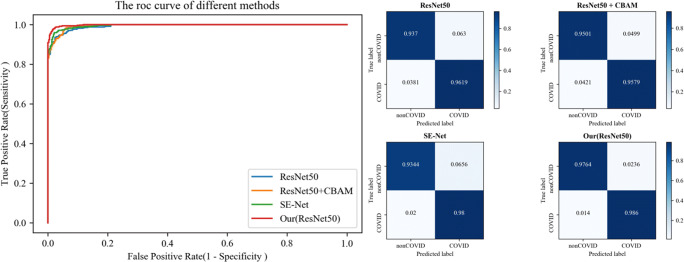
For the slice-level experiment, Table 2 shows that compared with the basic network ResNet, our method improved accuracy, sensitivity and specificity, increasing by 3.07%, 2.41% and 3.96%, respectively. Compared with SE-Net, our method improved accuracy, sensitivity and specificity by 2.16%, 0.6% and 4.20%, respectively. For ResNet + CBAM, 2.67%, 2.81% and 2.63% improvements were obtained. Compared with Ma et al. [29], the proposed attention blocks obtained more satisfactory results in terms of accuracy, sensitivity and specificity. The results illustrate that compared to the widely used classification networks and attention modules, our proposed method had better accuracy and the least false-positives. Fig. 6 shows the ROC curve (left) and confusion matrix (right) of the different methods.
Fig. 6.
The ROC curve (left) and confusion matrix (right) of different methods on COVID-19 dataset-A at the patient-level
As seen in Table 3, first, the proposed method obtained better results at the patient-level compared to state-of-the-art methods, which is similar to the slice-level experiment. Second, there was little difference between patient-level and slice-level experimental results, which verifies the effectiveness of the proposed method.
In particular, we discovered that SE-Net tended to obtain higher sensitivity, while ResNet + CBAM tended to have higher specificity. Therefore, the probability of missing detection was lower than that of ResNet + CBAM, but ResNet + CBAM had fewer false-positives. From statistical analysis, all p values were less than 0.001, which confirmed that our method was significantly different from other classification methods.
COVID-19 dataset-B
COVID-19 dataset-B [41] was provided by Ma et al. Similar to COVID-19 dataset-A, we also used the state-of-the-art attention modules in image classification and backbone for comparison, including ResNet [43], ResNet+CBAM [40] and ResNet+SE [39]. Table 4 summarizes the experimental results.
Table 4.
Experimental results of different methods on COVID-19 dataset-B
| Methods | Acc (%) | Sen (%) | Spe (%) | AUC (%) | p |
|---|---|---|---|---|---|
| ResNet [43] | 88.90 | 92.53 | 84.39 | 96.49 | 9.3e-18 |
| ResNet+CBAM [40] | 91.62 | 88.08 | 95.52 | 97.84 | 6.4e-12 |
| ResNet+SE [39] | 87.07 | 93.22 | 80.30 | 95.57 | 4.4e-8 |
| Ours (ResNet50) | 95.88 | 95.12 | 96.72 | 98.85 |
Acc: accuracy, Spe: specificity, Sen: sensitivity (best in bold
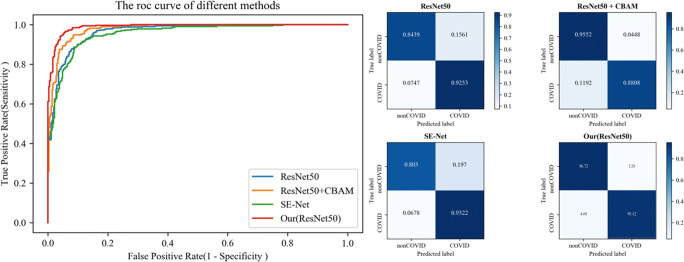
Similar to the results obtained on COVID-19 dataset-A, the proposed method achieved better accuracy, sensitivity and specificity. Comparing with ResNet, ResNet + CBAM and SE-Net, the proposed methods improved accuracy by 6.98%, 4.26% and 8.81%, and improved sensitivity by 2.59%, 7.04% and 1.90%, respectively. The specificity increased by 12.33%, 1.20% and 16.42%, respectively. Fig. 7 shows the ROC curve (left) and confusion matrix (right) of different methods.
Fig. 7.
The ROC curve (left) and confusion matrix (right) of different methods on COVID-19 dataset-B
COVID-19 dataset-C
COVID-19 dataset-C [20] was provided by Zhao et al. It is a public dataset used for COVID-19 diagnosis. The characteristics of COVID-19 Dataset-C were explained in detail in Section 3.1. Due to the small amount of data in it, we choose to compare with the papers that propose different solutions. Yang et al. [20], [32], [34] and [35] were selected for comparison due to their excellent performance. Table 5 summarizes the experimental results.
Table 5.
Experimental results of different methods on COVID-19 dataset-C
| Methods | Acc (%) | F1 (%) | AUC (%) |
| Mittal et al. [32] | 64.41 | 62.25 | - |
| DenseNet-169 [20] | 79.5 | 76.0 | 90.1 |
| DenseNet-169+ [20] | 85.0 | 85.9 | 92.8 |
| ResNet-50 (Self-Trans) [34] | 84 | 83 | 91 |
| DenseNet-169 (Self-Trans) [34] | 86. | 85.0 | 94 |
| Wang et al. [35] | 78.69 | 78.83 | 85.32 |
| Ours (ResNet50) | 84.32 | 83.9 | 91.6 |
| Ours (DenseNet-169) | 87.15 | 87.60 | 95.29 |
+ represents using lung mask as auxiliary input. (Best in bold)
Table 5 reports that compared to the method proposed in [20], our method achieves better accuracy, F1-score and AUC without the assistance of a lung mask. Compared to the method proposed in [34], our method used random initialization, and achieved better results without pretraining on other datasets. Both the F1-score and AUC value were significantly improved. The results confirmed that our method can achieve satisfactory results without using other complex methods when the dataset was small.
COVID-19 dataset-D
COVID-19 dataset-D [42] was provided by Soares et al. It is a public dataset used for COVID-19 diagnosis. The characteristics of COVID-19 Dataset-D were explained in detail in Section 3.1. Some researchers have achieved excellent results on this dataset. we focused on comparing with papers that propose new methods. Wang et al. [35], [29] and [48] were choose for comparison. We also used the state-of-the-art attention modules in image classification for comparing. Table 6 summarizes the experimental results.
Table 6.
Experimental results of different methods on COVID-19 dataset-D
| Methods | Acc (%) | F1 (%) | AUC (%) |
|---|---|---|---|
| Wang et al. [35] | 90.83 | 90.87 | 96.24 |
| ResNet+CBAM [40] | 91.55 | 92.31 | 97.52 |
| ResNet+SE [39] | 91.95 | 91.42 | 96.99 |
| Harsh et al. [48] | 95 | 95 | - |
| Ma et al. [29] | 95.16 | 95.60 | 99.01 |
| Ours (DenseNet-169) | 96.98 | 97.01 | 99.38 |
(Best in bold)
Experimental results illustrated that the proposed network achieved better result than other methods and attention models. Comparing with state-of-the-art methods, our method achieves better accuracy, F1-score and AUC. The experimental results illustrated the effectiveness of the proposed method for COVID-19 discrimination.
Ablation experiments
To verify the effectiveness of our proposed loss and attention blocks, we conducted two ablation experiments. For the loss function, we verified the influence of LCS on the diagnosis results. The results are shown in Table 7.
Table 7.
Diagnosis results using different loss functions (based on COVID-19 dataset-B)
| Loss function | Acc (%) | Sen (%) | Spe (%) |
|---|---|---|---|
| LCE | 94.03 | 93.22 | 94.93 |
| LCE + LCS (1:1) | 94.89 | 96.21 | 93.43 |
| LCE + LCS (Adaptive) | 95.88 | 95.12 | 96.72 |
Adaptive means adaptive weight loss. Acc:accuracy, Spe: specificity, Sen: sensitivity (best in bold)
The experimental results show that the cosine similarity loss LCS can improve the diagnosis result. Especially when combined with the adaptive weighting method, it can significantly improve the accuracy.
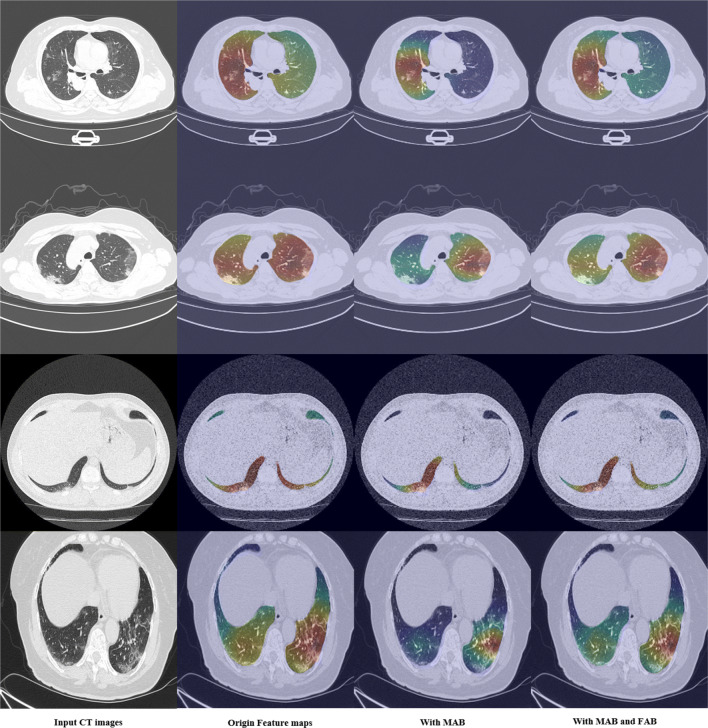
We also performed ablation experiments on the influence of different attention blocks. Table 8 shows that MAB significantly improved the diagnostic effect of the network on COVID-19, and FAB further improved the diagnostic effect of the network on this basis. This result was consistent with the visualized result in Fig. 9.
Table 8.
Diagnosis results using different attention blocks (based on COVID-19 dataset-B)
| MAB | FAB | Acc (%) | Sen (%) | Spe (%) |
|---|---|---|---|---|
| 88.90 | 92.53 | 84.39 | ||
| √ | 93.26 | 92.17 | 94.63 | |
| √ | √ | 95.88 | 95.12 | 96.72 |
Acc: accuracy, Spe: specificity, Sen: sensitivity (best in bold)
Fig. 9.
Visualization results of feature maps after using different attention blocks in mutex attention Res-Layer4. The first column is the input CT images, the second column is the visualization of features after Res-Layer, the third column is the visualization of features after MAB, and the fourth column is the visualization of features after FAB
Discussion
In this work, we proposed a new Res-Layer structure for COVID-19 diagnosis on CT images named mutex attention Res-Layer. It is composed of MAB, FAB and a Res-Layer and can extract more distinguishable features to obtain better COVID-19 diagnosis results with the proposed adaptive weight loss. Experiments on four different COVID-19 datasets verified that our method can achieve better results than other state-of-the-art methods. This reflects the effectiveness and robustness of the proposed method.
For the more regular CT image datasets COVID-19 dataset-A and COVID-19 dataset-B [41], the proposed network obtains 98.18% and 95.88% accuracy, which achieves significant improvement compared to other state-of-the-art attention models. Furthermore, the proposed method achieves higher sensitivity and specificity, which means that our method can better avoid the existence of false negatives (FNs) and false-positives (FPs) than other methods. As shown in Table 2, the experimental results indicate that ResNet+CBAM [40] tends to obtain better specificity. In other words, it will produce many FNs, which is not conducive to COVID-19 diagnosis. Although SE-Net [39] can obtain very good sensitivity, only 0.6% lower than our method, it produces a large number of FPs. The specificity of SE-Net is 4.2% lower than our method. Both experiments demonstrate the superiority of the proposed method.
For the noisy dataset, COVID-19 dataset-C [20], the proposed method also obtains satisfactory results. Compared with the methods proposed by [34], [20], our method can obtain higher accuracy, F1-score and higher AUC with fewer labels than [34] and without transfer learning, as in [20]. This shows that our method can still obtain satisfactory results even when the quantity of data is small and the noise is complex, which fully demonstrates the robustness of our method. For another public COVID-19 dataset-D [42], more satisfactory results are obtained than other state-of-the-art algorithms.
This article proposes two new attention blocks, MAB and FAB. The experimental results indicate the effectiveness of the two attention blocks. In Table 8, compared with the basic ResNet, MAB can significantly improve the accuracy and specificity by 4.36% and 10.24% respectively. MAB can amplify the degree of feature differences in various categories, and tends to obtain fewer FPs. Based on MAB, FAB performs feature fusion to obtain more representative features.With the improvement in accuracy by 1.62% and sensitivity by 3.22%, specificity was nearly unchanged.
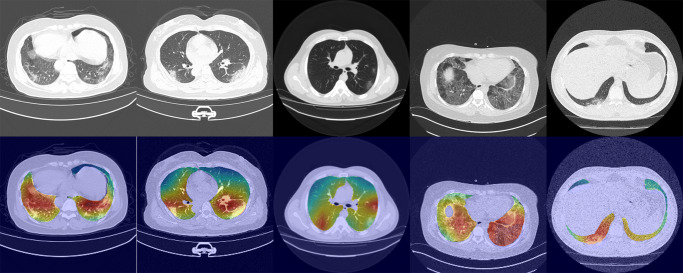
To confirm the results of the experiments, we visualized the feature maps of different categories extracted by the network and their COVID-19 images in Fig. 8. We use Grad-CAM [47] to visualize the feature maps outputted by the mutex attention Res-Layer4.
Fig. 8.
Visualization results of the proposed network using Grad-CAM. The first line is the input CT images; the second line is the visualization results
The visualization results show that our method focuses on the lesion area more accurately. For example, the lesion area in Column 3 was very small, while the lesion area in Column 4 was diffuse in both lungs. From the visualization results of the feature maps in the two columns, we can see that the proposed network is robust to lesions of different sizes. For different types of lesions, our network also achieves satisfactory localization. For example, the first column is mixed ground glass opacity, the third column is ground glass opacity and the second column is solid. The high response areas of these visualizations approach the lesion area. Visualization results confirm that our method can extract more representative features to achieve effective COVID-19 diagnosis, which proves the effectiveness of our method.
We also visualized the influence of different attention blocks on the feature maps. We extracted the feature maps outputted by the mutex attention Res-Layer4 for visualization. Since the output feature map size of mutex attention Res-Layer4 was [2048,7,7], we use the average superposition method to obtain a feature map 7 × 7. The final results were obtained by upsampling to 224 * 224 using bilinear interpolation, as shown in Fig. 9. It shows that MAB focuses the features on the most significant lesion region, but it cannot completely cover some subsidiary lesion areas. For example, in the second row, the original feature map has a diffuse appearance on both sides of the lung, but its coverage area far exceeds the lesion area. As shown in the third column, MAB focuses the feature most on right lung, which is the most distinctive area. The region with the highest response shrank to the lesion area. However, the lesion area in the left lung was ignored. For this problem, the FAB module may provide an explainable improvement. FAB performs feature fusion between the original feature maps and the MAB feature maps. It can also be described as the feature selection of the channel dimension. As the same example, the visualization in the fourth column of the second row shows that FAB focuses on the right lung as well as on the left lung, which can enhance the network’s diagnostic effectiveness. Furthermore, the proposed method will be helpful to precisely localizing and segmenting lesions.
Conclusion
This paper proposed the COVID-19 diagnosis network MA-Net, which takes a pair of multiple CT images as inputs. In particular, this paper proposed a mutex attention block. The mutex attention block aims to distinguish the features of mutex input pairs. The network can extract distinguishable features to improve the diagnostic effect of the network. Then, the fusion attention block is designed to perform feature fusion further improving the diagnosis accuracy. Regarding the loss function, the proposed network includes three losses, namely the cross-entropy classification loss of the two mutex inputs and the cosine similarity loss of mutex input pairs. Adaptive weight is used to adjust the weights of the three losses. Our method is very robust and achieved satisfactory results on multiple datasets. The accuracy, specificity, sensitivity, and AUC are reported as high as 98.17%, 97.25%, 98.79% and 99.84% on our own COVID- 19 dataset-A and 94.88%, 95.39%, 94.33% and 99.00% on the public COVID-19 dataset-B. The diagnosis result of our method is better than state-of-the-art the attention modules of classification method. For public COVID-19 dataset-C and COVID-19 dataset-D, we achieved better result than other excellent COVID-19 diagnosis methods. The experiments and analysis indicate that the proposed network can provide auxiliary quantitative analysis in COVID-19 diagnosis.
Acknowledgements
This project is supported in part by the Qingdao City Science and Technology Special Fund (20-4-1-5-nsh), Qingdao West Coast New District Science and Technology Project(2019-59), KY-009, Science and Technology Commission of Shanghai Municipality (20DZ2254400), Zhongshan Hospital Clinical Research Foundation (2019ZSGG15) and Shanghai Pujiang Program (20PJ1402400).
Biographies
BingBing Zheng
obtained the B.S. degree in Information Science and Engineering from East China University of Science and Technology in 2015, and is pursuing the Ph.D. degree in East China University of Science and Technology. His main research interests include deep learning for medical image processing, computer vision. His experience includes the identification and detection of pulmonary nodules on CT images, the classification and segmentation of prostate on MRI, and the classification and segmentation of COVID- 19. He has published in journals and conferences in the crossing field of medical and computer vision, and has been involved in publicly and privately funded projects.

Yu Zhu
received the Ph.D. degree from Nanjing University of Science and Technology, China, in 1999. She is currently a professor in the department of electronics and communication engineering of East China University of Science and Technology. Her research interests include image processing, computer vision, multimedia communication and deep learning, especially, for the medical auxiliary diagnosis by artificial intelligence technology. She has published more than 90 papers in journals and conferences.

Qin Shi
was born in Yangzhou, Jiang su, China. She is currently pursuing the B.S. degree in East China University of Science and Technology. She will be a postgraduate at the school of information science and engineering, East China University of Science and Technology in 2021. Her research interests include deep learning, object detection, image processing and pattern recognition.

Dawei Yang
is dedicated in early diagnosis of lung cancer and relevant studies, with special interests on management of pulmonary nodule and validation of diagnostic biomarker panels based on MIOT, CORE and radiomics artificial intelligence (AI) platform. He is the member of IASLC Prevention, Screening & Early Detection Committee. Since 2011, he has published 16 SCI research articles and 9 as first author, including which on Am J Resp Crit Care (2013), Can Lett (2015, 2020) and Cancer (2015 & 2018), etc. As a presenter for oral or poster presentation in ATS, WCLC, APSR, ISRD couple times. He is one of the peer reviewers for international journals, such as J Cell Mol Med, J Transl Med, etc.

Yanmei Shao
was born in 1983 in Shandong Province, China and received her MD degree from Qingdao University in 2013. She is currently an associate chief physician in the department of pulmonary and critical care medicine, the Affiliated Hospital of Qingdao University. She is good at the diagnosis and treatment of chronic obstructive pulmonary disease, bronchial asthma, lung cancer and the other common respiratory diseases, proficiency in various operations such as respiratory endoscopy, internal medicine thoracoscopy and treatment of critically ill patients. She is Youth Member of Respiratory Physician Branch of Shandong Medical Doctor Association, Youth member of Respiratory Rehabilitation Branch of Shandong Rehabilitation Medical Association, Member of Shandong Geriatric Respiratory Intervention Branch.

Tao Xu
was born in 1982 in Shandong Province, China and received his MD degree from Shandong University in 2014. He is good at the standard diagnosis and treatment of lung cancer, especially the diagnosis and treatment of lung nodules, early screening and full management of lung cancer, proficiency in various operations such as respiratory endoscopy, and treatment of critically ill patients. Presided over 2 provincial and ministerial projects published more than 10 SCI papers.

Declarations
Conflict of Interests
The author(s) declared no conflicts of interest with respect to the research, authorship, and publication of this paper.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
BingBing Zheng, Email: bostonkg@outlook.com.
Yu Zhu, Email: zhuyu@ecust.edu.cn.
Qin Shi, Email: sq15052502008@126.com.
Dawei Yang, Email: yang_dw@hotmail.com.
Yanmei Shao, Email: shaoyanmei1983@sina.com.
Tao Xu, Email: xutao1008@163.com.
References
- 1.Baker DM, Bhatia S, Brown S, Cambridge W, Kamarajah SK, McLean KA, Xu W. Medical student involvement in the COVID-19 response. The Lancet. 2020;395(10232):1254. doi: 10.1016/S0140-6736(20)30795-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.W. H. Organization (2020) “Novel Coronavirus(2019-nCoV) Situation Report – 22,” Accessed on: February. 11, 2020 [Online]. Available: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---20-april-2021
- 3.Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, Macneil A, Dugan VG. Emergence of SARS-cov-2 b. 1.1. 7 lineage—united states, december 29, 2020–january 12, 2021. Morb Mortal Wkly Rep. 2021;70(3):95. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao AT, Tong YX, Zhang S, False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol (2020) [DOI] [PMC free article] [PubMed]
- 6.Ye Z, Zhang Y, Wang Y, Huang Z, Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur Radiol. 2020;30(8):4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long C, Xu H, Shen Q, Zhang X, Fan B, Wang C, Li H. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol. 2020;126:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mei X, Lee HC, Diao KY, Huang M, Lin B, Liu C, Yang Y. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020;26(8):1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Nie H, Zhang Z, Li YF. Anisotropic angle distribution learning for head pose estimation and attention understanding in human-computer interaction. Neurocomputing. 2021;433:310–322. doi: 10.1016/j.neucom.2020.09.068. [DOI] [Google Scholar]
- 10.Li Z, Liu H, Zhang Z, Liu T, Xiong NN (2021) Learning knowledge graph embedding with heterogeneous relation attention networks. IEEE Transactions on Neural Networks and Learning Systems. 10.1109/TNNLS.2021.3055147 [DOI] [PubMed]
- 11.Zhang Z, Li Z, Liu H, Xiong NN (2020) Multi-scale dynamic convolutional network for knowledge graph embedding. In: IEEE Transactions on Knowledge and Data Engineering. 10.1109/TKDE.2020.3005952
- 12.Liang W, Yao J, Chen A, Lv Q, Zanin M, Liu J, He J. Early triage of critically ill COVID-19 patients using deep learning. Nat Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-17280-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozsahin I, Sekeroglu B, Musa MS, Mustapha MT, Ozsahin DU. Review on diagnosis of covid-19 from chest ct images using artificial intelligence. Computational and Mathematical Methods in Medicine. 2020;2020:1–10. doi: 10.1155/2020/9756518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohamadou Y, Halidou A, Kapen PT. A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of covid-19. Appl Intell. 2020;50(11):3913–3925. doi: 10.1007/s10489-020-01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Fang S, Zhang Z, Li D, Lin K, Wang J (2021) MFDNet: Collaborative Poses Perception and Matrix Fisher Distribution for Head Pose Estimation. IEEE Transactions on Multimedia. 10.1109/TMM.2021.3081873
- 16.Li D, Liu H, Zhang Z, Lin K, Fang S, Li Z, Xiong NN. CARM: Confidence-aware recommender model via review representation learning and historical rating behavior in the online platforms. Neurocomputing. 2021;455:283–296. doi: 10.1016/j.neucom.2021.03.122. [DOI] [Google Scholar]
- 17.Shen X et al (2021) Deep variational matrix factorization with knowledge embedding for recommendation system. In: IEEE Transactions on Knowledge and Data Engineering. 10.1109/TKDE.2019.2952849, vol 33, pp 1906–1918
- 18.Liu T, Liu H, Li Y , Zhang Z, Liu S Fast Blind Reconstruction with Wavelet Transforms Regularization and Total Variation Minimization for FTIR Imaging Spectrometer. IEEE/ASME Transactions on Mechatronics. 10.1109/TMECH.2018.2870056
- 19.Liu T, Liu H, Li Y, Chen Z, Zhang Z, Liu S (Jan. 2020) Flexible FTIR spectral imaging enhancement for industrial robot infrared vision sensing. In: IEEE Transactions on Industrial Informatics. 10.1109/TII.2019.2934728, vol 16, pp 544–554
- 20.Yang X, He X, Zhao J, Zhang Y, Zhang S, Xie P (2020) COVID-CT-dataset: a CT scan dataset about COVID-19. https://github.com/UCSDAI4H/COVID-CT
- 21.Wang G, Liu X, Li C, Xu Z, Ruan J, Zhu H, Zhang S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Transactions on Medical Imaging. 2020;39(8):2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ. Deep-COVID: Predicting COVID-19 from chest x-ray images using deep transfer learning. Med Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahuja S, Panigrahi BK, Dey N, Rajinikanth V, Gandhi TK. Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. Appl Intell. 2021;51(1):571–585. doi: 10.1007/s10489-020-01826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamshidi M, Lalbakhsh A, Talla J, Peroutka Z, Hadjilooei F, Lalbakhsh P, Mohyuddin W. Artificial intelligence and COVID-19: deep learning approaches for diagnosis and treatment. Ieee Access. 2020;8:109581–109595. doi: 10.1109/ACCESS.2020.3001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell. 2021;51(2):854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian X, Fu H, Shi W, Chen T, Fu Y, Shan F, Xue X. M 3 Lung-Sys: a deep learning system for multi-class lung pneumonia screening from CT imaging. IEEE Journal of Biomedical and Health Informatics. 2020;24(12):3539–3550. doi: 10.1109/JBHI.2020.3030853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B, Liu P, Dai L, Yang Y, Xie P, Tan Y, He K. Assisting scalable diagnosis automatically via CT images in the combat against COVID-19. Scientific Reports. 2021;11(1):1–8. doi: 10.1038/s41598-021-83424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao K, Su J, Jiang Z, Zeng LL, Feng Z, Shen H, Hu D. Dual-branch combination network (DCN): Towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med Image Anal. 2021;67:101836. doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma X, Zheng B, Zhu Y, Yu F, Zhang R, Chen B. COVID-19 lesion discrimination and localization network based on multi-receptive field attention module on CT images. Optik. 2021;241:167100. doi: 10.1016/j.ijleo.2021.167100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Xia J. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harmon SA, Sanford TH, Xu S, Turkbey EB, Roth H, Xu Z, Turkbey B. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat Comput. 2020;11(1):1–7. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mittal H, Pandey AC, Pal R, Tripathi A. A new clustering method for the diagnosis of CoVID19 using medical images. Appl Intell. 2021;51(5):2988–3011. doi: 10.1007/s10489-020-02122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He K, Fan H, Wu Y, Xie S, Girshick R (2020) Momentum contrast for unsupervised visual representation learning, pp 9729– 9738
- 34.He X, Yang X , Zhang S, Zhao J, Zhang Y, Xing E, Xie P (2020) Sample-efficient deep learning for COVID-19 diagnosis based on CT scans. medrxiv. 10.1101/2020.04.13.20063941
- 35.Wang Z, Liu Q, Dou Q. Contrastive cross-site learning with redesigned net for covid-19 ct classification. IEEE Journal of Biomedical and Health Informatics. 2020;24(10):2806–2813. doi: 10.1109/JBHI.2020.3023246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, Lin ZQ, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-Ray images. Scientific Reports. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Wang, Lv Y, Lei J, Yu L (2021) Global and local-contrast guides content-aware fusion for RGB-d saliency prediction. In: IEEE Transactions on Systems, Man, and Cybernetics: Systems. 10.1109/TSMC.2019.2957386, vol 51, pp 3641–3649
- 38.Zhou W, Liu J, Lei J, Yu L, Hwang JN. GMNet: graded-feature multilabel-learning network for RGB-thermal urban scene semantic segmentation. IEEE Trans Image Process. 2021;30:7790–7802. doi: 10.1109/TIP.2021.3109518. [DOI] [PubMed] [Google Scholar]
- 39.Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 7132–7141
- 40.Woo S, Park J, Lee JY, Kweon IS (2018) Cbam: Convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV), pp 3–19
- 41.Ma J, Cheng G, Wang YX, An XL, Gao JT, Yu ZQ, Zhu QJ “COVID-19 CT Lung and Infection Segmentation Dataset. 10.5281/zenodo.3757476
- 42.Soares E, Angelov P, Biaso S, Froes MH, Abe DK (2020) “SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification,” medRxiv preprint. 10.1101/2020.04.24.20078584
- 43.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
- 44.Ioffe S, Szegedy C (2015) Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: International conference on machine learning. PMLR, pp 448–456
- 45.Glorot X, Bordes A, Bengio Y (2011) Deep sparse rectifier neural networks. In: Proceedings of the fourteenth international conference on artificial intelligence and statistics, pp 315-323. JMLR Workshop and Conference Proceedings
- 46.Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Chintala S. Pytorch: An imperative style, high-performance deep learning library. Advances in Neural Information Processing Systems. 2019;32:8026–8037. [Google Scholar]
- 47.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision, pp 618–626
- 48.Panwar H, Gupta PK, Siddiqui MK, Morales-Menendez R, Bhardwaj P, Singh V. “A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-scan images,”. Chaos, Solitons Fractals. 2020;140(110190):39. doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]