Figure 3 ∣. Mechanism of GR activation by Hsp90.
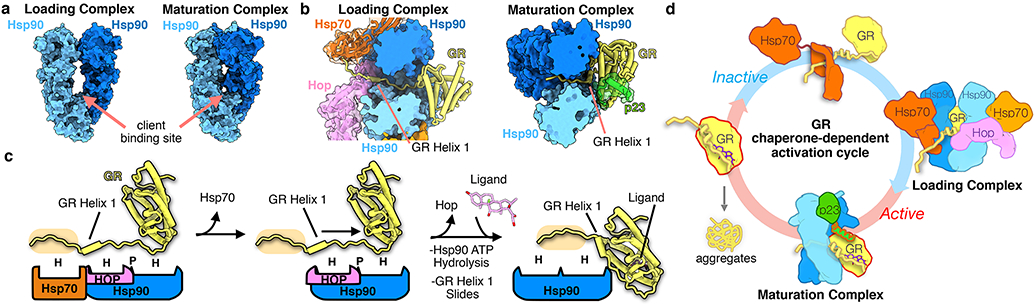
a, Surface representation of the Hsp90 dimer in the GR-loading complex12 versus the GR-maturation complex, showing the Hsp90 conformational change. Hsp90A (dark blue), Hsp90B (light blue). b, Top view of the loading complex (left) with GRHelix 1 extended through the Hsp90 lumen. Top view of the maturation complex (right) with GRHelix 1 docked onto GR. Hsp70 (orange, transparent surface), Hop (pink, transparent surface), Hsp90A (dark blue, surface representation), Hsp90B (light blue, surface representation), GR (yellow), p23 (green, transparent surface). c, Schematic showing the conformational change of GRHelix 1 from the loading complex to the maturation complex. Boxes represent chaperone and co-chaperone binding sites along the GRHelix 1 region (H=hydrophobic interface, P=polar interface). The GRpre-Helix 1 strand is highlighted (tan). Color scheme maintained from (b). d, Schematic of the GR chaperone-dependent activation cycle. Ligand-bound, active GR (left) is aggregation prone under physiological conditions. GR ligand binding is inhibited by Hsp70. Inactive GR is loaded onto Hsp90, with Hop, to form the loading complex, where GRHelix 1 is extended through the Hsp90 lumen. Hsp70/Hop release and p23 binds to form the maturation complex, where GRHelix 1 is redocked back onto the body of GR and ligand binding is restored.
