Extended Data Fig. 6 ∣. The p23tail-helix:GR Interface.
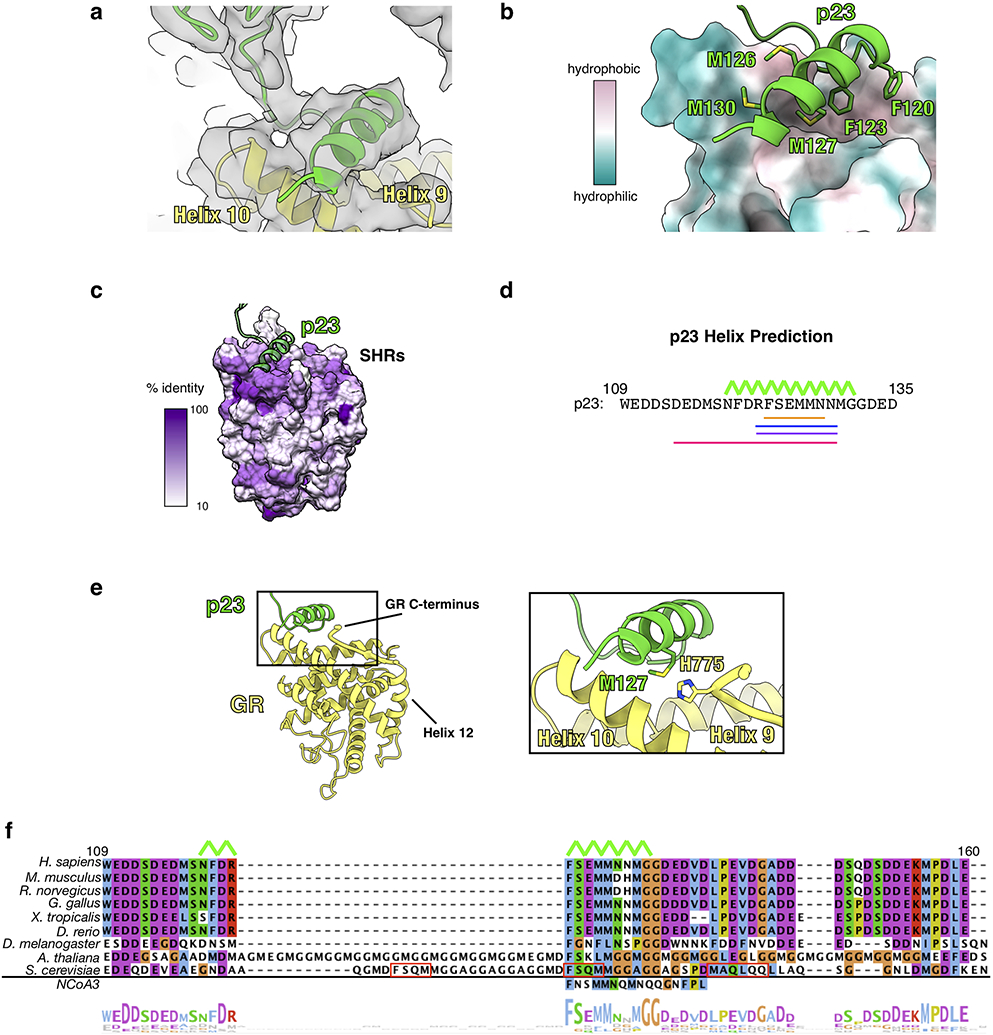
a, Focused map of GR:p23tail-helix showing density for the p23 tail with the atomic model built in. GR (yellow), p23 (green). b, Interface between the p23tail-helix (green) and GR (colored by hydrophobicity, surface representation) showing that the p23tail-helix binds to a hydrophobic patch on GR. p23 side chains interacting with GR are shown. c, Sequence identity across human steroid hormone receptors (GR, mineralocorticoid receptor, androgen receptor, progesterone receptor, estrogen receptor α and β) plotted onto the GR structure. The p23tail-helix (light green) was overlaid to indicate the p23:GR interface. d, Secondary structure predictions for human p23 from three different servers. Porter 4.0 (orange), RaptorX (blue), Psipred (purple), AlphaFold v2.0 (pink). The p23tail-helix from the maturation complex atomic model is shown with the top green line. e, Atomic model of GR (yellow) and p23 (green) from the maturation complex highlighting the interaction between the p23tail-helix and the GR C-terminus, which connects to GRHelix 12. f, Sequence alignment of eukaryotic p23 showing conservation of the p23tail-helix sequence. The p23tail-helix from the maturation complex atomic model is shown with the top green line. The bottom aligned sequence is the p23tail-helix -like motif identified in NCoA3 using the ScanProsite server. Red boxes on the S. cerevisiae p23 sequence indicate predicted helices from the PsiPred server. The alignment is colored according to the ClustalW convention.
