Abstract
A few decades ago, drug discovery and development were limited to a bunch of medicinal chemists working in a lab with enormous amount of testing, validations, and synthetic procedures, all contributing to considerable investments in time and wealth to get one drug out into the clinics. The advancements in computational techniques combined with a boom in multi-omics data led to the development of various bioinformatics/pharmacoinformatics/cheminformatics tools that have helped speed up the drug development process. But with the advent of artificial intelligence (AI), machine learning (ML) and deep learning (DL), the conventional drug discovery process has been further rationalized. Extensive biological data in the form of big data present in various databases across the globe acts as the raw materials for the ML/DL-based approaches and helps in accurate identifications of patterns and models which can be used to identify therapeutically active molecules with much fewer investments on time, workforce and wealth. In this review, we have begun by introducing the general concepts in the drug discovery pipeline, followed by an outline of the fields in the drug discovery process where ML/DL can be utilized. We have also introduced ML and DL along with their applications, various learning methods, and training models used to develop the ML/DL-based algorithms. Furthermore, we have summarized various DL-based tools existing in the public domain with their application in the drug discovery paradigm which includes DL tools for identification of drug targets and drug–target interaction such as DeepCPI, DeepDTA, WideDTA, PADME DeepAffinity, and DeepPocket. Additionally, we have discussed various DL-based models used in protein structure prediction, de novo design of new chemical scaffolds, virtual screening of chemical libraries for hit identification, absorption, distribution, metabolism, excretion, and toxicity (ADMET) prediction, metabolite prediction, clinical trial design, and oral bioavailability prediction. In the end, we have tried to shed light on some of the successful ML/DL-based models used in the drug discovery and development pipeline while also discussing the current challenges and prospects of the application of DL tools in drug discovery and development. We believe that this review will be useful for medicinal and computational chemists searching for DL tools for use in their drug discovery projects.
Keywords: Deep learning, Drug discovery, ADMET, Hit identification, Lead optimization, Virtual screening, Drug development, Property prediction
Introduction
The term artificial intelligence (AI), commonly referred to as the intelligence demonstrated by machines, is used to indicate instances in which a system/machine show cognitive abilities like humans, such as learning and problem solving, have been considered to be a game-changer across all industries, both academic and commercial (Nayak and Dutta 2017). The World Economic Forum stated that the amalgamation of big data and AI would kick start the fourth industrial revolution that can radically alter the practice of scientific discovery (Fouad 2019). Like any other sector, the pharmaceutical sector is considering the unrealized prospects of AI to address key problems influencing drug discovery and productivity (Mak and Pichika 2019). In the pharmaceutical industry, AI started gaining popularity when AI-based models demonstrated biological/chemical property predictions with great accuracy in a short time, especially DL architectures (Chen et al. 2018a). Moreover, both advancements in hardware and the ease of availability of large datasets have contributed to the tremendous growth in the application of AI in pharmaceutical research (Dash et al. 2019).
The majority of the problems arising during the process of drug discovery include unfavorable absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties, which have been known to be a major cause of failure of the potential molecules in the drug development pipeline, contributing to large consumption of time, capital and human resources (Lin et al. 2003). This has increased the interest in the early-stage prediction of ADMET properties of drug candidates so that the success rate of a compound reaching the later stages of drug development can be enhanced (Pathania et al. 2021). AI has been effectively utilized to develop models and prediction tools for ADMET properties. Apart from property predictions, AI has also contributed to early phases of drug discovery like de novo designing of chemical compounds and peptides (Bender and Cortes-Ciriano 2021). Moreover, companies involved in clinical research have ascertained that revising the research strategies by introducing AI-based techniques has resulted in greater success rates in both preclinical and clinical trials (Harrer et al. 2019; Krittanawong et al. 2019; Woo 2019b). Worth mentioning is the contribution of AI to precision medicine, which has been helping researchers maximize patients’ benefit and lower the side effects caused due to the prevalent traditional “one size fits all” approaches. The new revolutionizing AI methodologies have been helping in characterizing different subgroups of diseases, patient stratification, and studying the underlying factor unique to the specific form of the disease (Gardner et al. 2020).
This review mainly focusses on the DL-based tools which are used in different stages of the drug discovery pipeline. We have given a general introduction to drug discovery process, Machine learning and DL techniques, followed by specific examples and discussion on various DL and AI-based tools for drug development. We also pointed out some notable success stories in the use of AI and DL in drug development and medicine.
Introduction to drug discovery and development
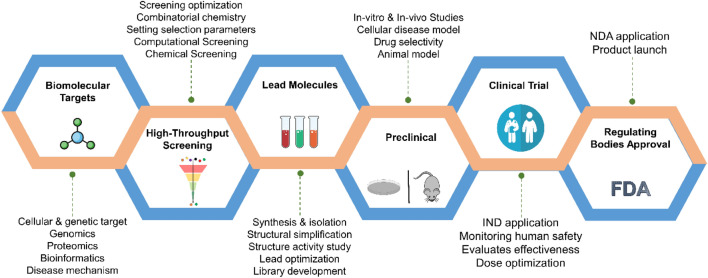
Drug discovery and development is a complex process that aims to identify and develop novel therapeutics against validated biological target intrinsically associated with a particular disease of interest (Mohs and Greig 2017). An overview of drug discovery and development is shown in Fig. 1. It encompasses the whole process of bringing a drug molecule into the market, initially starting with hit identification and ending with clinical trials phases after being approved by regulating bodies such as the Food and Drug Administration FDA (Deore et al. 2019). Traditionally, the drug discovery process starts with identification of bimolecular targets in the body which have been validated to play a vital role in the disease pathology. This is followed by high-throughput screening experiments in which large chemical libraries are screened against the selected target using appropriate assay (Kerns and Di 2003). The main objective of this high-throughput screening is to find promising compounds which have the potential to be developed into a drug candidate to treat the disease under consideration (Wildey et al. 2017). After high-throughput screening, the chosen compounds, often referred to as hit compounds, are analyzed for their biological activity via in vitro studies. The most potent compounds obtained from in vitro activity data are developed into lead compounds through lead optimization process. During the lead optimization phase, the compounds are modified to improve its bioavailability, solubility, partition coefficient, and stability as these factors can have direct impact of the drug therapeutic efficacy and potency. The molecules with optimized ADMET properties are then further evaluated for their effectiveness using suitable animal models (Hooijmans et al. 2018). The optimized compounds are tested in human subjects in order to validate and confirm the potency, therapeutic efficacy, ADMET and possible adverse drug reactions through a four step process called clinical trials, in which each step is carried out in varying number of human subjects in a randomized control manner (Hefti 2008). If the drug candidate yields desired results in the clinical trial phase, then it is approved by the regulating bodies like FDA, after which the drug is released into the market (Fletcher et al. 2022).
Fig. 1.
Overview of the drug discovery and development process. The process includes various stages such as (1) target identification, in which biologically relevant targets for a particular disease condition is selected for drug development, (2) high-throughput screening of compound library to identify hit compounds, (3) lead identification and lead optimization, in which the identified hit molecules are optimizing for their potency, selectivity and ADMET properties, (4) preclinical studies, in which the optimized molecules are tested in animal models to study their pharmacokinetic properties and therapeutic potential, and finally, (5) clinical trials, where the drug candidates are tested on human subjects in four phases to establish their safety and efficacy followed by regulatory approval, molecules which showed good pharmacokinetic properties, potency, therapeutic efficacy and least side effects are approved by regulatory agency for marketing and a drug become available in the clinic
Introduction to machine learning and deep learning
ML is a method of data analysis involving development of new algorithms and models capable of interpreting multitude of data (Elbadawi et al. 2021; Dara et al. 2022). While considering ML, one must not confuse it with AI, as according to the FDA “all ML techniques are AI techniques, but not all AI techniques are ML techniques”. Furthermore, FDA also defines AI as “the science and engineering of making intelligent machines”, and ML as “an AI technique that can be used to design and train software algorithms to learn from and act on data” (Toh et al. 2019).
The algorithms used in recent years have successively improved their performance with the increase in both the quantitative and qualitative aspects of data available for learning (Stephenson et al. 2019; Lavecchia 2015). ML is considered as one of the best options available when applied to solve problems for which a big amount of data and various variables are available to the individual but a model or formula relating these various variables amongst themselves along with the expected result is not known (Musella et al. 2021; Dara et al. 2022; Vamathevan et al. 2019). However, when drug discovery moved into an era of a large amount of data, ML approaches evolved into DL approaches, which are a more powerful and efficient to deal with the massive amounts of data generated from modern drug discovery approaches (Zhang et al. 2017; Jing et al. 2018a; Chen et al. 2018a). ML allows a computer system to make some predictions or decisions, based on its past experience, without being explicitly programmed. In contrast to the traditional physical models that rely on particular physical equations, the ML technique uses several algorithms to create a pattern, leading to the prediction of chemical, biological, and physical properties of the novel compounds. This is mainly done using two learning techniques, supervised learning and unsupervised learning techniques. Supervised ML is the construction of an algorithm capable of generating patterns and hypotheses to predict the fate of future instances, depending on the data provided (Osisanwo et al. 2017). In brief, supervised learning technique uses the input data to train the algorithm and create a decision boundary to classify or predict the outcomes in any similar circumstances. The supervised ML algorithms can be further classified into classification algorithm and regression algorithm (Zhang 2014).
The classification algorithm aims at categorizing the data based on the training dataset. One of the common applications of classification algorithm in bioinformatics is the identification of gene coding regions in a Genome. These tools are considered to be significantly flawless as several classification algorithms are used to train from a given set of datasets and are further used to classify the gene coding regions in a genome (Larranaga et al. 2006). The regression algorithm aims at predicting the fate of future instances depending on the data in the training dataset. It uses various techniques such as rule-based techniques, logic-based techniques, instance-based techniques, stochastic techniques, etc. to make accurate predictions. Recently, the regression algorithms are being widely used to predict the novel targets or structures such as the protein–protein interaction sites. A detailed study about regression algorithms have witnessed some promising results with an accuracy of above 80% to identify the structures in proteomics (Aumentado-Armstrong et al. 2015).
One of the biggest advantages of the supervised learning algorithms is that it can be trained specifically by setting an ideal decision boundary, the user gets the authority to determine the number of classes, and the input data are well-labeled which makes the output of the test algorithm to be more accurate and reliable. While on the other hand, some of the disadvantages of supervised learning techniques include the classification of huge datasets, which can be sometimes be challenging and time consuming, overtraining of decision boundaries due to the unavailability of appropriate examples, which can make the output of the test algorithm to be inaccurate. Moreover, data preparation and pre-processing of the input data can also be a challenging task (Kotsiantis et al. 2007).
The unsupervised ML aims at interpretation and learning an abstract representation of the given data in the absence of any predefined labels, or phenotypes. Unsupervised learning works by clustering data points into patterns to obtain meaningful biological information. Unsupervised learning can be broadly classified into clustering, which relies on the principle of the grouping of unlabeled data based on their similarities and association, which involves discovering some relationships between the attributes of unlabeled data points. Some of the commonly used clustering algorithms include hierarchical or k-means clustering (Parasa et al. 2021). The clustering technique splits the unlabeled dataset into groups based on their similarities. For a huge amount of data, k-means clustering is commonly used to form clusters of small molecule profiles, on the basis of their profile similarity (Tavallali et al. 2021). The k-means clustering and the corresponding heat maps are comparatively simple and require little computational resources. The hierarchical clustering is often used in gene expression profiling or genetic interaction studies which provides a broad visualization of the data, hence helping in better analysis (Shetty and Singh 2021).
One of the major advantages of unsupervised learning techniques over supervised learning algorithms is that it is less complex as compared to supervised learning as training of the dataset is not required and sorting of raw data and understanding the different models of learning makes it useful in real time. Also, it is much easier to get unlabeled data from a computer automatically rather than labeled data which needs human involvement (Brydges et al. 2010). Some of the major disadvantages of unsupervised learning techniques includes data sorting which may not be precise as the data used is not labeled which may lead to less accurate and unpredictable results. Due to the absence of any prior knowledge or training set of data, the spectral classes do not always correspond to the informational classes, hence spectral properties of classes may change with time which makes the class information to vary while moving from one image to another (Dridi 2021). All these learning techniques mentioned above have contributed a lot in making the slow, tedious and expensive process of drug discovery to be efficient, fast and cost effective.
DL is a subset of ML based on artificial neural networks that use multiple layers to progressively extract higher level features from raw input. Due to its ability to learn from data and the environment, DL and neural network (NN), also known as artificial neural networks (ANN) named after its artificial representation of the working of a human nervous system, have become one of the most successful techniques in various AI research areas (LeCun et al. 2015; Min et al. 2017). It has shown superior performance over other ML methods and has recently emerged as one of the most promising tools in the field of pharmaceutical research where its application is not only limited to the bioactivity predictions but has far exceeded in addressing numerous problems in the field of drug design and discovery (Ongsulee 2017). Table 1 enlists some of the extensively used tools in the field of DL (Gupta 2013; Baskin et al. 2016; Abiodun et al. 2018).
Table 1.
Some of the packages that are extensively used for practicing deep learning techniques (Jing et al. 2018b)
| Package | Programming language | Reference link |
|---|---|---|
| Tensorflow | Python | https://www.tensorflow.org/ |
| Torch | Lua | http://torch.ch/ |
| Theano | Python | http://deeplearning.net/software/theano/ |
| Caffe | C++/Python | http://caffe.berkeleyvision.org/ |
| DL4J | Java | https://github.com/deeplearning4j/deeplearning4j |
| Paddle | Python | http://paddlepaddle.org/ |
| Keras | Python | https://keras.io/ |
| CNTK | C++/Python | https://www.microsoft.com/en-us/cognitive-toolkit/ |
| MxNet | R/Python/Julia | http://mxnet.io/ |
| AlexNet | MATLAB | https://www.mathworks.com/products/matlab.html |
| Pytorch | Python | http://pytorch.org/ |
| DeepChem | Python | https://deepchem.io/ |
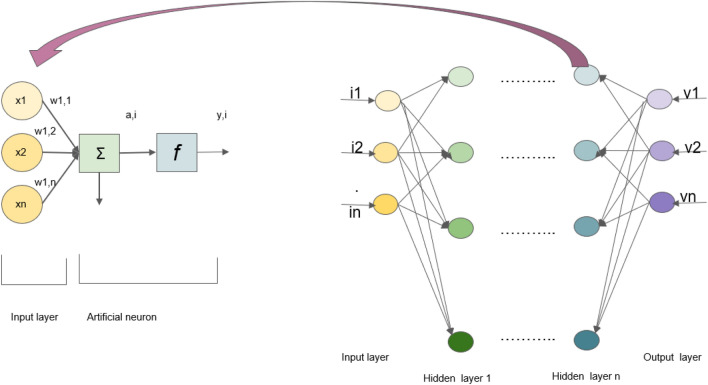
Figure 2 represents a typical NN with the working of a single neuron. Artificial neural network is a structure which has got several input vectors I = [i1, i2, i3, …, in] and an appropriate output vector O = [o1, o2, o3,…, om] along with several connected elementary units, known as neurons. The network initially receives information in the form of input vectors and aims to process or learn from it. From here, the data go through one or more hidden layers which understand the hidden patterns in the data using calculations and carry out transformations accordingly. The activation function or the transfer function also acts upon the processed data to capture the non-linear relationship between the inputs and also convert it to a more usable output (Gurney 2018). The performance of an ANN depends on the number of layers, the number of neurons, transfer function, the presence of a bias, and the way neurons are interconnected (Abraham 2005; Wang 2003; Despotovic 2012; Puri et al. 2016).
Fig. 2.
Schematic diagram of working of a single neuron in artificial neural networks (ANN). It denotes the working performance of an ANN that depends on the number of layers, number of neurons, transfer function, presence of a bias, and the way neurons are interconnected. The figure denotes the various input (i1, i2..) and output vectors (v1, v2..) and their interconnections are depicted as neurons. These interconnected neurons help maintain the architecture of the ANN
Deep learning tools for drug discovery
To realize the enormous potential of DL algorithms in drug discovery and development, computer scientist and medicinal chemist have found a common point to work together to develop DL-based tools, predictive models and algorithms which can be used in the drug discovery and development. Here, we have summarized some of the DL-based tools which are developed to aid the drug discovery and development process.
Deep learning tools for predicting drug–target interactions and binding affinity
Drug–target interaction refers to the interactions between chemical compounds and bimolecular drug targets in the human body and it plays an extremely important role in drug discovery and development as the therapeutic effect is a result of this interaction. The very small and limited knowledge about drug–target interactions based on wet-lab experiments have caused a huge gap between the known and unknown drug–target pairs which has increased the interest in the search for efficient methods of drug–target interactions (DTI) prediction. The traditional practices for DTI prediction have been facing monetary and technical limitations, while computational strategies have been proved to show efficiency in doing the same. At present, the major computational approaches used in DTI prediction includes ligand-based approach, docking simulation, chemo-genomic approach, text mining methods, ML/ DL based methods, and network-based methods, a branch diagram about various tools and approaches is given in Fig. 3 (Luo et al. 2017; Chen et al. 2018b). Some of the DL-based methods for drug–target interaction are discussed below.
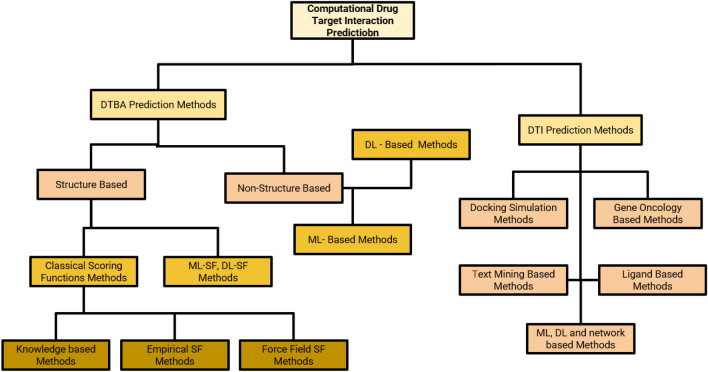
Fig. 3.
Branch Diagram for various computational drug–target interaction predictions. Various DL techniques used for target prediction and identification is depicted in the figure. These techniques are generally classified as either drug target interaction predictions or drug–target binding affinity prediction. DTBA techniques are further classified into structure-based and non-structure-based techniques which makes use of ML and DL, while DTI prediction tools are classified as docking based, ligand based, ML/DL based, gene based and text mining based methods
The computational prediction of drug target in reaction can be largely classified into DTI prediction methods and drug target binding affinity (DTBA) prediction methods. The DTI prediction methods contains docking simulations, gene ontology-based methods, ligand-based methods, text mining-based methods and ML/DL/Network-based methods. DTBA-based methods are further classified into structure based and non-structure based whereas structure based are further classified into classical scoring function methods and ML-SF/DL-SF methods and classical scoring function methods are further classified into knowledge based, empirical SF based and Force-field SF-based methods.
DTI-CNN
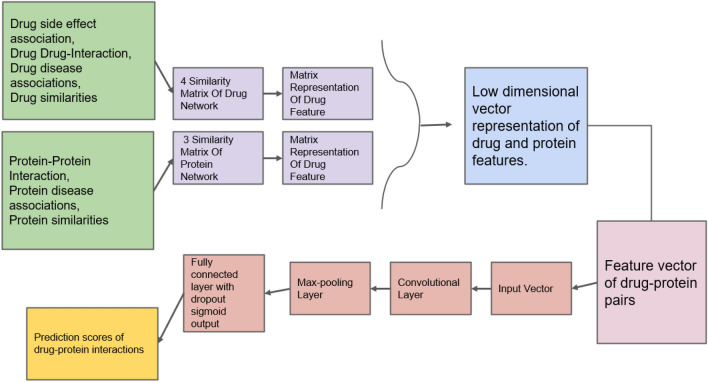
The convolutional neural network (CNN) is a class of NN, commonly used to analyze visual imagery. DTI-CNN (Peng et al. 2020; Li et al. 2022; Ding et al. 2021) is a simple DL-based drug–target interaction prediction tool that is said to outperform the existing state-of-the-art methods by the intelligent interaction of three components namely, (1) heterogeneous-network-based feature extractor, (2) denoising-auto encoder-based feature selector, (3) CNN-based interaction predictor (Peng et al. 2020; Li et al. 2022). As the model is based on random walk with restart (RWR) and denoising auto encoder (DAE) model, it is capable of coping up with low-dimensional feature vectors and noisy incomplete and high-dimensional features from heterogeneous data sources, including drug, proteins, side-effects and diseases information. The general workflow of DTI-CNN-based DTI prediction is shown in Fig. 4.
Fig. 4.
Flowchart for prediction of drug–protein interaction using DTI-CNN. The CNN-based drug–target prediction tool makes use of the available drug–protein interaction data. Using which, the algorithm extracts similarity matrix of the drug and protein network and converts them into drug and protein features. These acts as input for the prediction algorithm, as it uses the convolutional network to make a max pooling layer. The max pooled layer is later used to predict the drug–protein interaction score
The first step in using DTI-CNN for DTI prediction includes the construction of a heterogeneous network by the integration of a large number of drugs and protein related information sources and using the RWR model to obtain the initial drug feature vector and protein feature vector. In the next step with the use of the DAE model, the low-dimensional representations of the high-dimensional features of drugs and proteins are obtained. In the final step, under the known drug–protein interactions, the samples are divided into positive samples and negative samples. The CNN model is then used to predict the association between each pair of drugs and proteins using the feature vector of drug–protein pairs (Fig. 4). Another such DL-based tool called FRnet-DTI, which is a CNN-based classifier for drug target interaction prediction and uses an auto-encoder-based feature manipulation (Rayhan et al. 2020, 2018).
DeepCPI
DeepCPI is a tool based on novel framework that uses DL techniques along with unsupervised representation learning meant for prediction of DTI (Wan et al. 2019). At first, it is said to use latent semantic analysis and Word2vec methods to learn the low-dimensional feature representations of both the compounds and proteins in an unsupervised manner. Then the feature embedding of the compounds and protein from the first step is fed to the DL network for successful prediction of the drug–protein interaction. The development of efficient DTI predicting models is still under progress due to the large number of limitations it faces. For example, in the case of docking simulation, the 3D structure of the target protein is very much required, but is not always available. Moreover, it is evident that in comparison to the DTI, the strength of the binding (binding affinity) between a drug and its target is much more informative for drug development process which helps to gain deeper insights into the intermolecular forces operating between them (Thafar et al. 2019b).
The newly developed DL algorithms to predict DTBA have shown superior performance in comparison to conventional ML algorithms. These DL-based algorithms use simplified molecular input line entry system (SMILES) which is a compact text format of representation of the molecular structure in which each chemical entity is mapped to a single ASCII strings of 20–90 characters, ligand maximum common substructure (LMCS), extended connectivity fingerprint (ECFP) or a mixture of the three as an input data or as drug features. They also use different neural network (NN) types that have their own unique strengths which allow them to be suitable for a varied range of applications (IJzerman and Guo 2019).
DeepDTA
The first DTBA predicting DL-based approach is DeepDTA (Öztürk et al. 2018) which is a non-structure-based method and uses SMILES for input data for drugs. The amino acids and the protein sequences are similarly encoded in SMILES. The CNN used in DeepDTA has three 1D convolutional layers which follow max-pooling functions which are referred to as the first CNN block and are applied on the drug embedding to learn latent features. For the protein embedding, a similar CNN block is constructed and applied to it. DeepDTA is said to tune a large number of hyper-parameters including number of filters, filter length of drug and protein, batch size, optimizer, and learning rate in the validation step. This model aims to minimize the difference in the values of the predicted and real DBTA in the training period. The limitations faced in this model due to the usage of CNN can be overcome by using much more appropriate architectures, which can learn from long protein sequences, like the long-short term memory (Guo et al. 2019).
WideDTA
WideDTA is a CNN DL model that uses four text-based sources of information as input which include, (1) ligand SMILES (LS), (2) protein sequences (PS), (3) ligand maximum common substructure (LMCS), (4) protein domains and motifs (PDM). WideDTA differs from DeepDTA as it represents LS and PS as a set of words instead of full-length sequences. In PS, a word is three-residues in the sequences, while in LS it is an eight-residue. The makers claim that the features of the protein that are represented by shorter lengths of residues are not detected in full-length sequences due to the low signal-to-noise ratio and hence the WideDTA is a word-based model instead of a character-based one (Öztürk et al. 2019; Thafar et al. 2019a).
PADME
The DL-based DTBA predicting method which uses drug–target features and fingerprints to different DNN, is called protein and drug molecule interaction prediction (PADME). When extended-connectivity fingerprint is used as the input for the representation of drugs, the tool is known as PADME-ECFP. The other PADME version integrates molecular graph convolution into the model to learn the latent features of drugs from SMILES by adding one more graph convolution neural network and is represented as PADME-GraphConv. Protein sequence composition descriptors are used by both versions which contain rich information for representing target proteins. Once the feature vectors are generated, it is fed into the simple feedforward neural network for a particular drug–target pair for predicting the DTBA (Feng et al. 2018).
DeepAffinity
The DL DTBA predicting model which only relies on SMILES for representing drugs is known as DeepAffinity. To represent the proteins, DeepAffinity relies on the structural property sequence representation that annotates the sequence with structural information, which are shorter than the other representations, provides structural details efficiently and gives higher resolution of the sequences, also the regression task is highly benefited. Using a famous RNN model, seq2seq, the representations are encoded into an embedding form. At later stages of data processing, the RNN encoders are coupled with a CNN model whose output representations for the drug and target are concatenated and to get the final DTBA output values are fed into the FC layers. The complete model which includes data representation, embedding learning, and joint supervised learning trained from end to end. Basically, the RNN-CNN pipeline has been found to produce high accuracy results when compared to other ML-based methods used on the same dataset (Karimi et al. 2019).
Deep docking
Virtual prediction of protein ligand interaction by the help of docking can tremendously reduce the time required for the process of new drug discovery and development (Morris and Lim-Wilby 2008; Meng et al. 2011), but the speed of virtual screening-based DTI prediction is offered a limitation due to the large chemical library with billions of compounds. Thus, a much faster screening technique is required to tackle and filter such huge amount of data, deep docking is a new and novel method that works on the principles of DL platform which is capable of docking billions of molecular structures in a quick and precise manner (Gentile et al. 2020). The main objective of deep docking is to reduce the database of billions of compounds into a few millions of subset of data while retaining the extensive majority of the possible virtual hits which can then be carried forward for the actual docking studies or can be further processed or refined with the combination of other methods of virtual screening to further narrow down the data containing more concentrated virtual hits. Deep docking relies on the deep neural network (DNN) training where the training set is expanded along with the predicted hit molecules form each previous steps of interpretation gradually making more rigorous cutoff toward the end of the calculation. It employs the use of quantitative structure activity relationship (QSAR) models trained on the basis of docking scores obtained for the subsets of a chemical library to predict the docking results for yet to be docked compounds and thus eliminate the molecules not showing promise to yield a good docking score in a constant manner. It utilizes the use of quick computing and large independent QSAR descriptors such as 2D molecular fingerprints (Gao et al. 2020), as a result of using DL to quickly determine the docking scores for a large dataset of compounds for which actual docking is yet to be performed. Deep Docking can attain up to 100 folds improvement in virtual screening speed and about 6000 folds improvement for identification of top ranked compounds thereby avoiding the loss of favorable virtual hits (Pereira et al. 2016; Gentile et al. 2020).
DeepBAR
DeepBAR is a DL-based binding affinity predicting tool developed by the scientists at MIT by combining chemistry and ML (Ding and Zhang 2021). The binding free energy measures the affinity between a drug molecule and its target. To obtain the best drug amongst a given number of potential ones, the drug with the lowest binding free energy value should be chosen as it will be able to disrupt the protein’s normal function effectively.
Therefore, it can be said that fast and accurate calculation of standard binding free energy has many important applications in this drug discovery process. The BAR in DeepBAR refers to the Bennett acceptance ratio method. BAR is an outdated algorithm that is used to calculate binding free power. DeepBAR utilizes the Bennet acceptance ratio along with the information from different endpoints and other intermediate circumstances.
Protein structure prediction
Proteins are the one of the most important biomolecules which plays a wide variety of roles in an organism, like enzymatic activity, receptor activity, cell signaling, hormonal activities, intracellular transport, etc. Most of the currently identified drug targets are also protein molecules, whose malfunction leads to pathological states. To study the function of proteins, it is important to know the structure of the proteins, as the protein structure usually determines its function, activity and also pathological conditions. But the process of identifying the structure is not very straight forward and it requires experimental procedures like X-ray crystallography, NMR spectroscopy, cryo electron microscopy, etc., which are time consuming and in most cases, very difficult to perform. The traditional method of experimental data threw light on the structures of around 100,000 unique proteins, which is just a fraction of the known protein sequences. To address the gap of such a large number of unknown protein structures, accurate computational approaches are needed, which enables large-scale structural bioinformatics (Batool et al. 2019). To make protein structure determination, a much simpler and less tedious procedure, scientist took help of various DL techniques which can predict protein structure with great degree of confidence, some of which are discussed below.
Alphafold
AlphaFold (Wei 2019) is a new computational method that includes analysis of the covariation in the homologous sequences, which are in contact and helps in the prediction of protein structures. The initial step of this modern method includes training a neural network to make precise predictions of the distances between residue pairs, inferring a better idea about the structure. This information can be used to construct a potential of mean force that can precisely mark out the shape of the protein. Considering the chances of having sequences with fewer homologous sequences, the potential of mean force can be optimized by a simple gradient descent algorithm, named AlphaFold, which helps in achieving higher accuracy without complex sampling procedures (Jumper et al. 2021).
AlphaFold generally assembles the best probable fragments based on the analysis of the multiple sequence alignment. It interprets the spatial proximity by detecting the mutations that have occurred throughout the evolutionary timeframes in response to the other mutations. For such a task, it utilizes a huge amount of computational power in order to manage the truly DNN that identify the evolutionary patterns within the protein structure sequences with respect to the contact distributions and angular restraints. Additionally, with the help of DL algorithms it can produce a protein specific statistical potential using a ‘learned reference state’, instead of a physical-based reference state. Thus, AlphaFold has given the researchers a tool that has great potential in protein structure prediction (Ruff and Pappu 2021; Batool et al. 2019).
CASP
The objective of critical assessment of protein structure prediction (CASP) (Kinch et al. 2021; Deng et al. 2018; Kryshtafovych et al. 2019) is to develop a technology that can recognize and construct the three dimensional structure of the protein from the protein sequences. It can be achieved primarily by two ways on the basis of the presence of any template structure previously available or not, namely (1) template-based models and (2) template-free models, amongst which the template-based modeling is the more preferable one if a good template is available, as it utilizes the available protein structure as the base for prediction, thereby more matured technique among the two and can be readily approached by the researchers who have less experience. On the other hand, if there is no template present for the protein structure one can employ template-free modeling to build the structure. Template-free modeling has two kinds of approaches, fragment-based assembly and de novo folding, where the de novo folding approach aims to build the three dimensional structures from the scratch using the fundamentals of physics, where the secret to its success lies within the use of an accurate energy function to efficiently search for the lowest energy state conformation and also to discriminate native like structures from decoys. Yet, the fragment-based assembly still dominates because of its accuracy and better capability in protein structure prediction when there is no good template available.
DL tools for compound de novo design
De novo design is defined as a process to generate novel molecules according to DTBA / DTI data or pharmacophore data, where a pharmacophore is the minimum structural requirements needed for showing biological activity (Schneider and Fechner 2005). The de novo design aims at discovering novel drug-like compounds. In contrast to the early software tools, which have a tendency to discover new compounds of some limited chemical attraction and diversity, the modern de novo design algorithms put focus on the synthesizability and drug-likeness properties of the compounds (Schneider and Schneider 2016). The previous de novo algorithms utilized structure-based approaches to grow ligands that will be able to fit the binding site of the target of interest both sterically and electronically. One of the major drawbacks of these approaches is that the molecules created often possess poor drug metabolism and pharmacokinetic properties and are synthetically intractable (Olivecrona et al. 2017). Apart from the structure-based approach, the ligand-based approaches were also widely used. Although these methods are said to generate novel structures effectively based on the transformation or reaction rules, the inherent rigidness and scope of the synthesizability pose restrictions. The approach usually involves the generation of a large virtual library of compounds, usually a chemical space which is searched using a function that takes several parameters like the drug metabolism and pharmacokinetics profiles into account. These virtual libraries are created either using chemical reactions along with a group of available chemical building blocks or using transformational rules based on the expertise of medicinal chemists (Mauser and Guba 2008; Perron et al. 2022).
Now with the big data revolution, and the development of DL algorithms, de novo methodologies based on deep reinforcement learning (RL) have come into action which helps in generating compounds with the desired physical, chemical, and bioactivity properties (Zhou et al. 2019; Popova et al. 2018). These deep reinforcement algorithms involve the analysis of possible actions and estimation of the statistical relationship between the actions and their possible outcomes, which is followed by the determination of a treatment system that attempts to find the most desirable outcome. Let us look at some DL models for the de novo design.
ReLeaSE
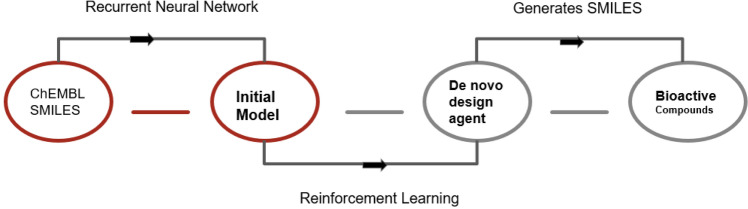
One of the deep RL approaches used here which enables the design of chemical libraries with desired properties is reinforcement learning for structural evolution (ReLeaSE). The most distinct aspect of this approach is the use of SMILES to represent the molecules (Popova et al. 2018). Figure 5 shows the classical representation of reinforcement learning for structural evolution.
Fig. 5.
ReLeaSE (reinforcement learning for structural evolution). General pipeline of reinforce learning system for the generation of novel compounds. This technique uses database such as chEMBL, having chemical and biological information regarding available drug molecules, and uses these data to identify the initial model of a new compound which is likely to have a pharmacological affect. Later through a process of reinforcement learning, the initial model is converted into a de novo design, and later converted into the smiles of the bioactive molecule
The ReLeaSE method includes two DNN, namely the generative and the predictive which are trained in two stages. In stage I, the models are trained separately using different algorithms, while in stage II, the models are trained together using the RL approach. The generative model in this system produces chemically feasible novel molecules, while the predictive model, estimates the generative model’s conduct by allocating a numerical reward or penalty to every molecule that has been generated. The generative model is trained to maximize the expected reward.
Generative artificial intelligence (AI)-based model
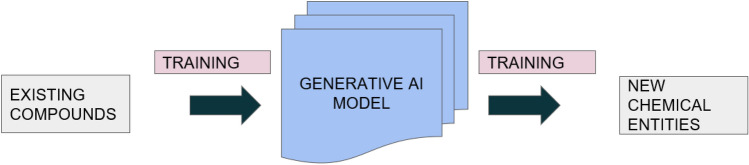
This model is based on generative AI which claims that it autonomously designs novel chemical compounds using the knowledge of known bioactive compounds along with inherited bioactivity and synthesizability (Walters and Murcko 2020). A general overview about generative model is given in Fig. 6
Fig. 6.
Overview of generative artificial intelligence (AI)-based model. The generative AI model makes use of the information of known bioactive compounds as input for training the algorithm and use the information to predict new chemical entities which are thought to have similar bioactivity
This approach consists of two steps, one of which includes the creation of a generic model which has learned the constitution of drug like molecules from a set of large unfocussed compounds. In the second step, the already existing model is tuned on more specific molecular aspects from a small target-focused library of actives. A deep recurrent neural network is utilized for training the generic model (Vanhaelen et al. 2020).
DeepScaffold
Drug design aims to find novel compounds which have desirable pharmacological features. One of the efficient methods to develop potential drug candidates is by retaining certain scaffolds as the core structures.
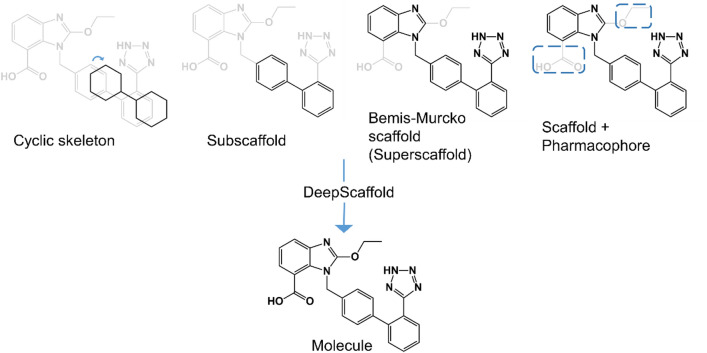
DeepScaffold is a scaffold-based molecular generative model which conducts molecule generation based on scaffold definitions for drug discovery. Scaffold here refers to the core structure of a molecule. These include Bemis–Murcko scaffolds, cyclic skeletons, and scaffolds with specifications on side-chain properties. DeepScaffold is capable of generalizing the learned chemical rules of the addition of atoms and bonds to a given scaffold. The molecules generated by this method were evaluated by molecular docking in the D2 dopamine receptor (DRD2) targets, and the results obtained can then be successfully applied to solve a large number of drug design problems including the generation of compounds containing a given scaffold and de novo drug design of potential drug candidates with specific docking scores. An overview of how DeepScaffold performs the function is shown in Fig. 7.
Fig. 7.
Overview of DeepScaffold. A comprehensive tool for scaffold-directed drug discovery can generate molecules based on CSKs, classical molecular scaffolds, such as Bemis–Murcko scaffolds, and those with additional pharmacophore-based queries for side chains. Reprinted (adapted) with permission from (Li et al. 2019; Rezaei et al. 2020). Copyright (2020) American Chemical Society
AIScaffold
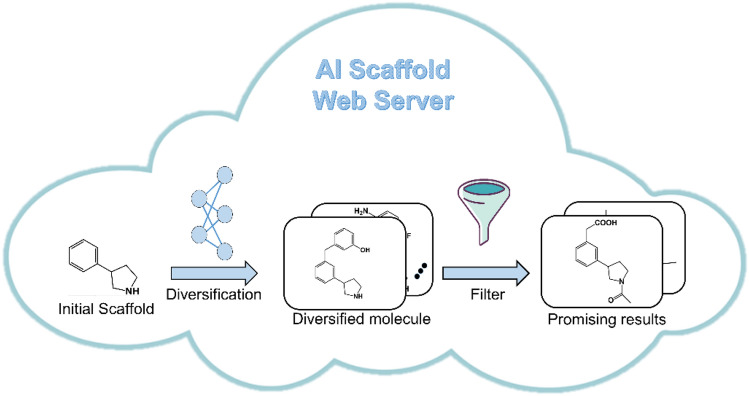
Another such AI-based tool is AIScaffold (https://iaidrug.stonewise.cn), a web-based tool which is mainly utilized for scaffold diversification with the use of a deep generative model (Lai et al. 2020). Scaffold diversification is often used by medicinal chemists for the purpose of lead compound optimization, but the software tools for the same are not readily available. AIScaffold is one such tool that can be utilized for scaffold diversification, unlike other tools which are designed to develop results by utilizing the information in molecular scaffolds. An overview of AIScaffold functioning is shown in Fig. 8
Fig. 8.
Overview of AIScaffold. AI scaffold makes use of an initial input scaffold of a bioactive compound and tries to modify the structural features of the molecule by a diversification process by adding or deleting scaffolds, which is later filtered and predicts a set of chemically diversified molecules with promising bioactivity
This tool is capable of performing large-scale diversification that is up to 500,000 molecules within a matter of several minutes and then as a result recommend the top 500 or the top 0.1% molecules. It also provides additional features such as site-specific diversification. By facilitating the scaffold diversification process, AIScaffold is helping medicinal chemists in accelerating drug design.
DESMILES
DESMILES is based on the principles of DNN model which is a ML approach toward a newer drug design and works on the basis of recurrent neural network (RNN) architecture (Maragakis et al. 2020). DESMILES mainly aims to produce a series of small molecules which could be chemically identical to the given input of small molecule, it utilizes molecular fingerprint as an input and then corresponds it to a matching sequence of SMILES string with an objective of correlating the fingerprint to the SMILES string. DESMILES could be incorporated in the early phase of drug design and discovery process where it could be used in combination with various molecular screening technologies in order to identify some newer scaffolds for possible drug candidates. Extended connectivity fingerprint is another form of representations that are explicitly designed to capture various molecular features that may be relevant to the molecular activity, and is thus advantageous over SMILES form of representation but lacks the structural details of the molecule thereby making it difficult to generate molecular structures from the fingerprints.
DL tools for hit identification using virtual screening
Virtual screening is another in silico technique used in drug discovery to search large libraries of small molecules to identify those hits which have a higher probability to bind to a drug target. The virtual screening technique uses biological, topological, and physicochemical properties of a chemical compound as well as the targets. The virtual screening methods can be mainly divided into two categories: (I) structure-based, which uses the 3D structure of targets and chemical compounds to model and visualize the interactions (Lionta et al. 2014; Li and Shah 2017). The 3D structure can be obtained by X-ray crystallography or by Nuclear magnetic resonance. Once the 3D structural information is collected, docking can be applied to find the interaction between a compound and a particular target. (II) Non-structure based which can be further subdivided into two groups, (a) ligand-based virtual screening (Ripphausen et al. 2011; Chen et al. 2007) which employs the molecular properties of compounds to model and analyzes the interactions with targets and (b) proteo-chemometric modeling which includes combining non-structural descriptors and targets at the input level (Wu et al. 2012).
Many studies have shown that DL algorithms show much better results in comparison with other ML algorithms in the case of virtual screening, especially due to their impactful applications in the de novo molecular design, where molecules with desired properties are generated by the utilization of sequence data (Bahi and Batouche 2018; Liu et al. 2019b). Some of the DL-based tools used in Virtual screening and QSAR are discussed below.
Deep VS
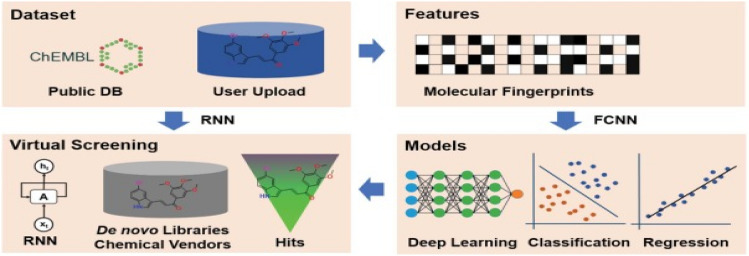
DeepVS is a DL approach to improve docking-based virtual screening. In DeepVS, the output from the docking program is used for the extraction of relevant features from the available basic data from protein ligand complexes. This approach uses atom and amino acid embeddings for implementing an effective way of creating distributed vector representations of protein–ligand complexes by modeling the compound as a set of atom contexts that is further processed by a convolutional layer. It also has an added advantage of having no requirement of feature engineering (Shen et al. 2020). The workflow of Deep VS is represented in the Fig. 9.
Fig. 9.
Representation of deep learning-based virtual screening. It includes (1) dataset preparation: select target of interest, or upload a private dataset for DNN training. (2) Features: selection for molecular vectorization. (3) Parameters: select model parameters for training classification or regression models. (4) Virtual screening: virtual screening against the chemical library or de novo library. Reprinted (adapted) with permission from (Liu et al. 2019a). Copyright (2019) Oxford University Press
SIEVE score
SIEVE score is a newly developed AI-based virtual screening tool, also called Similarity of Interaction Energy Vector Score (Yasuo and Sekijima 2019; Arora and Bist 2020). This Virtual screening tool has been said to be a promising method for hit identification and aims to enrich potentially active compounds from a large library of chemical compounds for the purpose of biological experiments. This tool is said to have a better performance in terms of efficiency as compared to the state-of-the-art virtual screening methods which were based on ML. The screening results obtained from this tool are also human interpretable in the form of important interactions which make it easy to distinguish between active and inactive compounds (Qiao et al. 2020; Yasuo and Sekijima 2017).
Similarity search
Similarity searching is a method that allows fast identification of chemical analogs to biologically active compounds (Cereto-Massagué et al. 2015; Muegge and Mukherjee 2016). It is a deep-learning model which follows the principles of ligand-based drug design, where it utilizes the molecular signatures as vectors to construct a connected neural network. A promising technique to predict various properties/parameters that are essential for hit identification like the bioactivity, aqueous solubility and toxicity based on the structure of the compound under investigation. Such multi-task neural network models can help in predicting the activity for multiple targets for the same hit much more effectively as compared to the single task networks because of the better representation of the data and much accurate recognition of the general patterns within the data.
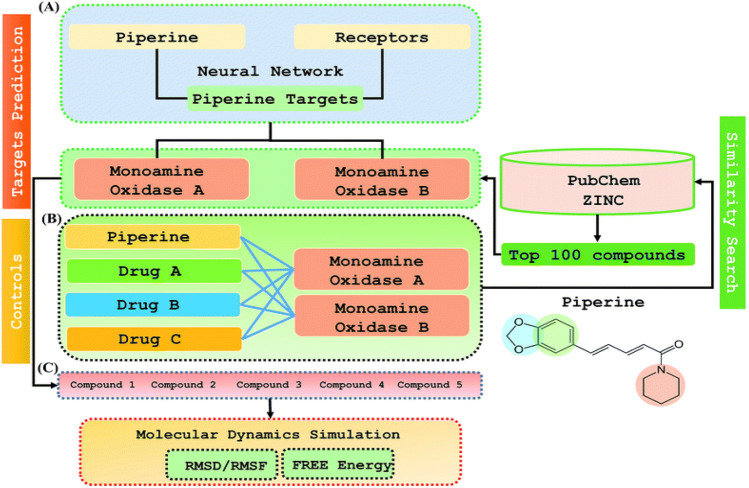
Similarity search is a two-step de novo approach which is based on PERL script, where specific inputs are utilized to improve the efficiency of the resulting outcome. This technique combines the DNN along with the ML approach as a scoring function for the virtual screening or for predicting the affinity of the compound under investigation for the selected target. By this method, one can test numerous compounds against a single target or to test a single target with a single compound (Khan et al. 2019). The model was applied to a single drug, namely piperine, and its experimental targets. A general docking approach and molecular dynamics simulation approaches were used as supplementary validation methods to investigate the potential of the predicted compounds against the prioritized targets. Both structurally close and diverse analogs can be recognized using similarity search based on the applied metrics (Eckert and Bajorath 2007). It is extremely important in the analysis of hits and the schematic representation of Similarity search is shown in Fig. 10.
Fig. 10.
Schematic representation of similarity search. The figure represents the outline of the workflow, which can be divided into three parts: (A) use of deep-learning methodology for the prediction of piperine targets, (B) similarity search from the ZINC and PubChem databases using a ML approach to get top 100 best hits based on piperine, (C) validation and comparison of how well the compounds predicted the FDA-approved drugs for the selected targets. Reprinted (adapted) with permission from (Khan et al. 2019). Copyright (2019) The Royal Society of Chemistry
DL tools for pharmacokinetic property prediction
Pharmacokinetics is the study of how a drug is being absorbed, distributed, metabolized, and excreted from the body. ADMET includes all the pharmacokinetic parameters as well as the toxicity profile of the xenobiotic (ADME-Tox). Hence, the ADME-Tox profile of a lead compound can impact its efficacy and safety. Drug efficacy and safety are considered to be some of the major causes of clinical failure of drugs in the process of drug discovery. There is an increasing need for an efficient predictive tool of ADMET properties to serve two main purposes—first, at the initial stage of selection of new compounds and compound libraries, to reduce the risk of late-stage attrition, and second, to optimize the screening and testing by predicting the ADMET profile of the lead compound in drug discovery. In this context, ML techniques have been often used in ADME-Tox prediction. These predictions are possible due to the availability of large amount of pharmacokinetic data of compounds to make models with the help of ADMET in-silico modeling, we can predict several properties such as dose size, dose frequency, oral absorption, bioavailability, Blood–brain-barrier (BBB) (Maltarollo et al. 2015; Alqahtani 2017). Table 2 lists few examples of ADMET properties that are being modeled using ML algorithms and Table 3 denotes some of the ML algorithms used in ADMET predictions.
Table 2.
Following are the examples of ADMET properties that are modeled using Machine Learning algorithms:
| Properties | Measurement |
|---|---|
| Solubility | Kinetic solubility |
| Clearance | Rodent in vivo—snapshot PK |
| Permeability | Caco-2, MDCK, PAMPA |
| Transporters such as P-gp | Transporters overexpressing cell lines |
| Metabolic stability | Liver microsomes and hepatocytes |
| Drug–drug interactions | CYP450s, transporters |
| Blood–brain barrier (BBB) | Mouse brain endothelial cell line |
| Cardiotoxicity (hERG) | Binding or flux in different cell types |
Table 3.
Following are some of the examples of common machine learning algorithms which are applied in ADMET prediction
| Algorithm | Summary |
|---|---|
| Random forest (RF) | An ensemble learning method that constructs many decision trees and outputs class or mean prediction |
| Support vector machine (SVM) | A supervised learning method. The examples are mapped in space and classes separated by a hyperplane |
| Neural network | A simple neural network has input, hidden and output layers |
| K nearest neighbors (KNN) | A non-parametric method that uses the K closest training examples in the feature space and classifies objects by a majority vote or in regression uses the average of the values of the nearest members |
| Naive Bayes (NB) | A probabilistic classifier, considers features to contribute independently to the probability |
| Deep learning (DNN) | Uses multiple layers of a neural network, where each layer uses the output from the previous one. It can learn multiple levels of representations at different levels of abstraction |
With the development of different algorithms and detailed analysis over the years, it has been found out that the newly developed DL methodologies when used to predict ADMET, produce efficient results in comparison to the other mentioned model. Researchers all over the world have not only developed specific tools that work on dl techniques for predicting one property at a time but also have been able to make complete in silico ADMET platforms. Some of these tools and platforms have been mentioned below.
Tox_(R)CNN
The cytotoxic changes induced by drugs cause cellular and nuclear morphological changes which are characteristic of the specific cell-death pathway involved (Bácskay et al. 2018). These changes are usually being identified based on the visualization of nuclei using different microscopic approaches for years. Tox_(R)CNN is a modern-day tool that has been designed to detect cytotoxicity from microscopic images of fluorescently stained nuclei, without using specific toxicity labeling. The tool Tox_(R)CNN has been operated using DL technologies, as DL is the most powerful supervised ML methodology available and has the exceptional abilities to solve computer vision tasks (Jimenez-Carretero et al. 2018). This tool uses two convolutional neural networks (CNN):
Tox_CNN—classifies cells based on prior cell segmentation and cropping of nuclei images.
Tox_RCNN—carries out fully automated cell identification and classification.
These networks provide classification outputs that give sensitive screening readouts that detect pre-lethal toxicity and hence make the tool extremely useful and affordable and even applicable to other in vitro toxicity readouts. The model constitutes a robust screening tool for drug discovery.
Metabolite prediction
Studies show that more than about 25% of the compounds are withdrawn from the market or trials due to metabolic, pharmacokinetic, or toxic problems and hence causes a lot of financial loss to the company. The experimental methods developed to identify and study the metabolic processes in drugs are extremely demanding in terms of equipment, expertise, cost, and time and hence researchers are trying to find computational alternatives for the same. The two main research directions which provide necessary support and guidance are metabolic sites (SOMs) and metabolite structure that show extensive help in computer-aided metabolic prediction methods. The model mentioned here performs the function of metabolites prediction in the following steps: it first establishes the database with broad coverage of SMARTS-coded metabolic reaction rules. Then, to construct a DL algorithm-based classification model, the molecular fingerprints of compounds are extracted. The model can identify the reaction types that have a higher probability to occur in comparison to others. According to the researchers, this method is capable of generating higher accuracies than random guess and the rule-based methods used for metabolite prediction (Djoumbou-Feunang et al. 2019; Wang et al. 2020).
Oral bioavailability prediction
Oral bioavailability plays an important role in determining the absorption of a drug in the body. If we are capable of predicting bioavailability, which is difficult to do as it is dependent on highly complex factors and processes, then it would be extremely easy to prioritize drug candidates in the process of drug discovery. This model uses DL and six experimentally determined in vitro and physicochemical endpoints including membrane permeation, free fraction, metabolic stability, solubility, pKa value, and lipophilicity to determine the oral bioavailability in rats. The chemical structure of the drugs is encoded as fingerprints or SMILES. Modeling the available information along with the structural information of the chemical compounds, the model achieves an accuracy of 70%. It might seem that ADMET predicting models or tools should be able to predict direct distribution, absorptions, etc. of drugs directly, but the fact that several parameters govern such pharmacokinetic property and prediction of one such parameter is itself highly significant and serves as an ADMET tool.
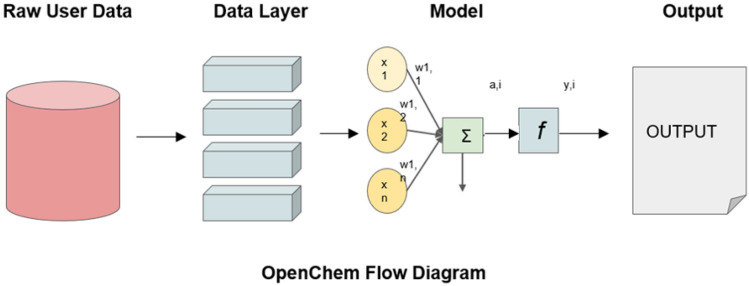
OpenChem
Openchem is a DL toolkit with Pytorch backend which is freely available via the GitHub repository. It aims at making easy-to-use DL models, for computational chemistry and drug design research (Korshunova et al. 2021). This toolkit is very helpful in keeping track of the training set, as well as in visualizing the evaluations, and project embedding in lower dimensional space. The toolkit helps in data preprocessing and has a very fast training process due to the support of multi-GPU. It is user-friendly as new models are built with only configuration files. The above toolkit helps us in the classification of data, regression analysis of data, and helps in generating various models which help in predicting the ADME properties of a lead compound. The workflow diagram of OpenChem is shown in Fig. 11.
Fig. 11.
OpenChem flow diagram. Showing the schematic representation of OpenChem which is PyTorch Based a DL toolkit to carry out the computational chemistry work and drug design
Over the past 2 decades, many in silico fragment-based drug discovery (FBDD) platform has been developed with a goal to generate a variety of models to study the pharmacokinetics and physicochemical for drug discovery and development at its early stage. The main principle for the fragment-based drug discovery includes a screening of low molecular weight compounds against macromolecular targets such as proteins. The screening technology includes differential scanning fluorimetry, surface plasmon resonance, and thermophoresis, followed by structural characterization using nuclear magnetic resonance (NMR) or X-ray crystallography of the individually soaked fragments of the lead compounds.
Tools for drug activity prediction
DL tools are able to identify chemical features in drug molecules and predict the activity of known molecules including their biological activity. One such DL-based tool is discussed below.
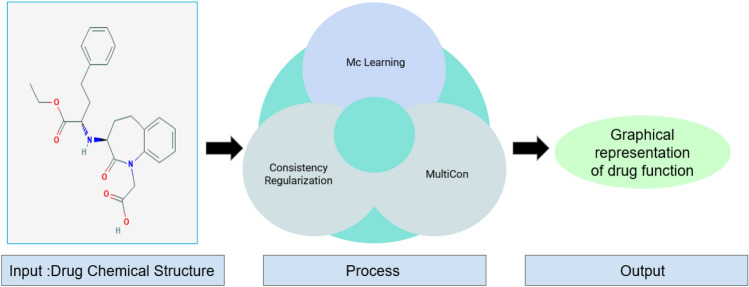
MultiCon
To diminish the workload of a large amount of supervised data and improve the model efficacy, semi-supervised learning algorithms can be used. By predicting the therapeutic application of drugs from their structural formulas using the semi-supervised learning algorithm, the cost, as well as the time consumed, will reduce significantly. Using the MultiCon toolkit (Sahoo et al. 2020), 1 can classify a drug into 12 categories, depending on their therapeutic applications and image analysis of their structural formulas. Studies show that MultiCon has a higher rate of class prediction compared to other pre-existing semi-supervised learning algorithms, because of its rational usage of data balancing, the limited number of labeled data, online argumentations of drug image input during training, along with the combined usage of multi contrastive loss with consistent regularization. The working principle of MultiCon is overviewed in Fig. 12.
Fig. 12.
Overview of the MultiCon toolkit. Working principle of the MultiCon toolkit which predicts the therapeutic application of drugs from their structural formulas using the semi-supervised learning algorithm, where it takes chemical structure as inputs and predicts the drug function
Application of DL in clinical trial design
The clinical study is the next step of the drug development process, which constitute a multi-billion-dollar industry, aiming for a successful evaluation of drug effectiveness by testing the lead molecules on human subjects (Piantadosi 2017). A clinical researcher’s first and foremost objective is to figure out that whether the new treatment, like a new drug or dosage form or medical device (like a pacemaker) is safe and effective for humans to use. Usually, it is designed in a way to understand if the newer treatment under investigation is more efficacious and has less harmful side effects than the conventional treatment available and also design the dosage in such a manner based on these results to attain optimum therapeutic effect where the minimal amount of the drug is needed to elicit a therapeutic response (Friedman et al. 2015).
On an average, it takes 10–15 years with a cost of 1.5–2.0 billion USD for new drug molecule to reach the market. Studies have shown that approximately half of the time, human resource and expenditure during the drug discovery process is spend in the clinical trial phases. The remaining 50% of the time and money covers the preclinical lead compound identification, optimization as well as regulatory processes (Harrer et al. 2019). Records have shown that one of the main obstacles in the drug development cycle is the high failure rate of clinical trials. The factors leading to the high failure rates of the clinical trials are patient cohort selection and recruitment of mechanisms that fail to provide the best suited patients to a trial in time, as well as lack of technical infrastructure to cope with the complexity in the later phases of clinical trials, such as the absence of a reliable and efficient adherence control, patient monitoring, and clinical endpoint detection systems. Past research has witnessed the high potential of AI and other advanced analytics tools to automate clinical trial processes, which makes it a cost-effective approach (Walczak 2018). Some of the currently available AI-based tools used in clinical trial setup is discusses below.
TRIALS.AI
Trials.ai is an AI-based startup which helps in design of clinical trial protocols. It makes use of Natural language processing and other AI techniques. The software collects and analyze data from different source like journals, drug labels, private data from hospitals with which the company have connections. These data are used to study proposed trials, strictness of eligibility criteria, and how it affects the clinical trials outcome like cost, participation retention etc. (Woo 2019a).
AICURE
Success of well-designed protocols depends on whether the participants follow the instructions or not. A small mistake, like forgetting to take a pill can negatively impact the study results. AICURE is an AI-based platform which helps clinical trials participants use their smartphone to record videos of them having medication. Using computer vision algorithms, AICURE software can predict the person have taken the medication or not. This tool can also analyze the facial expression of person to track how they respond to treatment which can help in development of therapies (Woo 2019a).
Success stories of the use of DL in drug discovery and development
With the development of DL methodologies, we have seen major pharmaceutical companies shifting toward AI, leaving behind the old traditional methods, to maximize patient’s profit along with their own. Astrazeneca is a global, science-led multinational pharmaceutical company that have reached the heights of success by integrating AI in every step of drug discovery going from virtual screening to clinical trials. The way they incorporated AI to redefine medical science has allowed them to gain a better understanding of existing diseases, identify new targets, design better clinical trials and in general speed up the complete process. Astra Zeneca’s growth is itself a successful example of how integrating AI with medical science can help achieve wonders. Their constant efforts on increasing AI use can be easily seen by the collaborations they are doing with other AI-based companies. One such example is their collaboration with Alibaba subsidiary Ali Health which aims to develop AI-assisted screening and diagnostics platforms in China.
The outbreak of the SARS-CoV-2 virus has brought a large number of companies under the pressure to find out the best drug in the shortest time possible. To achieve these companies have resorted to using AI along with the information available. Some examples of such success stories of the companies who have been able to identify potential leads against the COVID-19 virus have been mentioned below.
MT-DTI (Molecule Transformer Drug Target Interaction Model), a deep-learning based drug-protein interaction prediction model, by Deargen, a South Korea-based company. This model is used to predict the interaction strength between a drug and its target protein using simplified chemical sequences instead of 2D or 3D molecular structures. The model was used on the available FDA-approved antiviral drugs, and it found out that Atazanavir, an HIV medication, is highly probable to bind and block a prominent protein on the COVID-19 causing virus SARS-CoV-2. Along with this, it also identified other three antivirals and a not yet approved drug Remdesivir which is now being tested in patients. Deagen being able to identify antivirals using DL techniques is a great step on advancing pharmaceutical research and making the process less tedious and more efficient. If such drugs are properly tested, there is a great probability, we would be able to overcome this pandemic in the shortest time possible (Beck et al. 2020; Scudellari 2020).
Another example is that of the Benevolent AI, a London based Biotechnology Company that uses biomedical data, AI and ML to accelerate research in the health sector. So far, they have been able to identify six drugs and one of them Ruxolitinib is said to be under clinical trial for COVID-19 (Gatti et al. 2021). The company has been using a large repository of medical data, along with information extracted from the scientific literature by ML, and their AI system to identify potential drugs that might block the viral replication process of SARS-CoV-2. They got the Food and Drug Administration (FDA) approval for the use of their proposed Baricitinib drug in combination with Remdesivir where recovery rate increased for the hospitalized COVID-19 patients (Richardson et al. 2020).
One of the most common types of cancers across the world is skin cancer. As its incidence is continuously increasing and it is becoming extremely important to detect skin cancer in its early stages as studies show early detection and treatment is the key to increase the survival rate of skin cancer patients. With increasing growth in both medical science and AI, a no. of Skin cancer smartphone apps has been launched into the market which provides people with a technological approach who have suspicious lesions to decide whether they should seek medical help. Studies show that about 235 dermatology smartphone apps were developed between the years 2014 and 2017 (Flaten et al. 2018). Earlier, they operated by sending the photo of the lesion to a health professional, but now with inbuilt AI algorithms in smartphones, these apps are successfully capable of detecting and classifying images of lesions into high or low risk along with immediate risk assessment and subsequent recommendations to the patient. One such successful app is SkinVison (de Carvalho et al. 2019).
Conclusion
ML has been adapted in the field of drug discovery and development since 1990s with a long and fruitful history of DL, the current extension of ML was established based on the concept of multi-layer neural networks, and some of these techniques have been engrossed only recently in the pharmaceutical research arena, accelerating the drug discovery process. Unmistakably, application of DL needs a cautious analysis and a rigorous explanation of their respective realm of application that it may provide for the chemists in their quest for new drug discovery. Overall, it is a well-appreciated method in the field of modern drug discovery and development and with the recent advancements in the DL tool box, it can yield far reaching results. Taking into consideration of the recent success of such methodologies and its utilization by the pharmaceutical companies to boost its search for new drugs, it is convincing that the modern DL methodologies will be highly appreciated in the coming era of big data search and analysis for drug design and discovery.
Drug discovery and development has always been a challenging process which involves a lot of time and investment to move a drug candidate into the clinical trials. AI has been proven to demonstrate a substantial progress in boosting the success rate of the process, thereby reducing the overall costs of the research (Paul et al. 2021; Chan et al. 2019; Lavecchia 2019). Several companies have been investing time and money to develop their proprietary algorithms such as to tackle the challenges of the modern drug discovery and development process. The major key to the development of these algorithms is to bring together a group of experts from different domains such as data science engineers, chemists, and biologists under the one ecosystem of in silico drug design (Ekins 2016).
Lack of valid information regarding biological systems have made it difficult to label proper descriptors and endpoints, such that AI or DL tools cannot be applied in an effective manner to expect a reproducible outcome. Hence, the inability to properly model the biological system is one of the main limitations of AI in drug design (Bender and Cortés-Ciriano 2021; Bender and Cortes-Ciriano 2021). Another important limitation of AI in drug discovery is the selection of appropriate data sets and over focus on speed and cost-effectiveness in drug discovery. The enormous amount of available proxy chemical data set and advanced computational techniques, combinatorial chemistry, etc., have made it possible to synthesize more and more molecules in very limited time and that too in a cost-effective manner, but despite the efforts to synthesizing more molecules, the actual number of drugs making into the market via clinical trials is still very less considering the efforts done. This is mainly because all the available data and endpoints are generated and used in ligand design and discovery focusing toward the activity against the targeted disease thus the ligand might have good pharmacodynamics properties but fails to attain optimal pharmacokinetic properties thus the primary focus must be on designing a ligand that displays optimum pharmacokinetic and pharmacodynamics properties to become a good clinical candidate. Rather than focusing on creating more ligands, we have to come up with novel data, and meaningful endpoints which help us to utilize the full potential of AI and ML techniques to predict and bring potential drug candidates to the clinic which have good pharmacokinetic properties, safety, and efficacy (Smith et al. 2018; Fleming 2018; Díaz et al. 2019).
It is a great opportunity to make use of AI and DL in the field of drug discovery to develop methods and techniques for proper modeling of biological systems, by generating well-validated data having well-defined labels and endpoints, providing information regarding the targets and their interactions with ligands.
Funding
Funding was provided by Science and Engineering Research Board, SRG/2021/000415, MTR/2021/000317.
Declarations
Conflict of interest
The authors declare no conflict of interest.
References
- Abiodun OI, Jantan A, Omolara AE, Dada KV, Mohamed NA, Arshad H. State-of-the-art in artificial neural network applications: a survey. Heliyon. 2018;4(11):e00938. doi: 10.1016/j.heliyon.2018.e00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham A (2005) Artificial neural networks. In: Sydenham PH, Thorn R (eds) Handbook of measuring system design. Wiley, Hoboken, NJ
- Alqahtani S. In silico ADME-Tox modeling: progress and prospects. Expert Opin Drug Metab Toxicol. 2017;13(11):1147–1158. doi: 10.1080/17425255.2017.1389897. [DOI] [PubMed] [Google Scholar]
- Arora K, Bist AS. Artificial intelligence based drug discovery techniques for covid-19 detection. Aptisi Trans Technopreneurship (ATT) 2020;2(2):120–126. [Google Scholar]
- Aumentado-Armstrong TT, Istrate B, Murgita RA. Algorithmic approaches to protein-protein interaction site prediction. Algorithms Mol Biol. 2015;10(1):1–21. doi: 10.1186/s13015-015-0033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bácskay I, Nemes D, Fenyvesi F, Váradi J, Vasvári G, Fehér P, Vecsernyés M, Ujhelyi Z. Role of cytotoxicity experiments in pharmaceutical development. London: InTech; 2018. [Google Scholar]
- Bahi M, Batouche M Deep learning for ligand-based virtual screening in drug discovery. In: 2018 3rd international conference on pattern analysis and intelligent systems (PAIS), 2018. IEEE, pp 1–5
- Baskin II, Winkler D, Tetko IV. A renaissance of neural networks in drug discovery. Expert Opin Drug Discov. 2016;11(8):785–795. doi: 10.1080/17460441.2016.1201262. [DOI] [PubMed] [Google Scholar]
- Batool M, Ahmad B, Choi S. A structure-based drug discovery paradigm. Int J Mol Sci. 2019;20(11):2783. doi: 10.3390/ijms20112783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Cortes-Ciriano I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 2: a discussion of chemical and biological data used for AI in drug discovery. Drug Discov Today. 2021;26:1040–1052. doi: 10.1016/j.drudis.2020.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Cortés-Ciriano I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 1: ways to make an impact, and why we are not there yet. Drug Discov Today. 2021;26(2):511–524. doi: 10.1016/j.drudis.2020.12.009. [DOI] [PubMed] [Google Scholar]
- Brydges R, Dubrowski A, Regehr G. A new concept of unsupervised learning: directed self-guided learning in the health professions. Acad Med. 2010;85(10):S49–S55. doi: 10.1097/ACM.0b013e3181ed4c96. [DOI] [PubMed] [Google Scholar]
- Cereto-Massagué A, Ojeda MJ, Valls C, Mulero M, Garcia-Vallvé S, Pujadas G. Molecular fingerprint similarity search in virtual screening. Methods. 2015;71:58–63. doi: 10.1016/j.ymeth.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Chan HS, Shan H, Dahoun T, Vogel H, Yuan S. Advancing drug discovery via artificial intelligence. Trends Pharmacol Sci. 2019;40(8):592–604. doi: 10.1016/j.tips.2019.06.004. [DOI] [PubMed] [Google Scholar]
- Chen B, Harrison RF, Papadatos G, Willett P, Wood DJ, Lewell XQ, Greenidge P, Stiefl N. Evaluation of machine-learning methods for ligand-based virtual screening. J Comput Aided Mol Des. 2007;21(1):53–62. doi: 10.1007/s10822-006-9096-5. [DOI] [PubMed] [Google Scholar]
- Chen H, Engkvist O, Wang Y, Olivecrona M, Blaschke T. The rise of deep learning in drug discovery. Drug Discov Today. 2018;23(6):1241–1250. doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- Chen R, Liu X, Jin S, Lin J, Liu J. Machine learning for drug–target interaction prediction. Molecules. 2018 doi: 10.3390/molecules23092208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dara S, Dhamercherla S, Jadav SS, Babu C, Ahsan MJ (2022) Machine learning in drug discovery: a review. Artif Intell Rev 55(3):1947–1999 [DOI] [PMC free article] [PubMed]
- Dash S, Shakyawar SK, Sharma M, Kaushik S. Big data in healthcare: management, analysis and future prospects. J Big Data. 2019;6(1):1–25. [Google Scholar]
- de Carvalho TM, Noels E, Wakkee M, Udrea A, Nijsten T. Development of smartphone apps for skin cancer risk assessment: progress and promise. JMIR Dermatol. 2019;2(1):e13376. [Google Scholar]
- Deng H, Jia Y, Zhang Y. Protein structure prediction. Int J Mod Phys B. 2018;32(18):1840009. doi: 10.1142/S021797921840009X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deore AB, Dhumane JR, Wagh R, Sonawane R. The stages of drug discovery and development process. Asian J Pharm Res Dev. 2019;7(6):62–67. [Google Scholar]
- Despotovic DTaV (2012) In: Metallurgy—advances in materials and processes, Yogiraj Pardhi. IntechOpen. 10.5772/47850
- Díaz Ó, Dalton JA, Giraldo J. Artificial intelligence: a novel approach for drug discovery. Trends Pharmacol Sci. 2019;40(8):550–551. doi: 10.1016/j.tips.2019.06.005. [DOI] [PubMed] [Google Scholar]
- Ding X, Zhang B. DeepBAR: a fast and exact method for binding free energy computation. J Phys Chem Lett. 2021;12(10):2509–2515. doi: 10.1021/acs.jpclett.1c00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang J, Guo F. Identification of drug-target interactions via multi-view graph regularized link propagation model. Neurocomputing. 2021;461:618–631. [Google Scholar]
- Djoumbou-Feunang Y, Fiamoncini J, Gil-de-la-Fuente A, Greiner R, Manach C, Wishart DS. BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J Cheminform. 2019;11(1):1–25. doi: 10.1186/s13321-018-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi S (2021) Unsupervised Learning - A Systematic Literature Review. 10.13140/RG.2.2.16963.12323
- Eckert H, Bajorath J. Molecular similarity analysis in virtual screeni ng: foundations, limitations and novel approaches. Drug Discov Today. 2007;12(5–6):225–233. doi: 10.1016/j.drudis.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Ekins S. The next era: deep learning in pharmaceutical research. Pharm Res. 2016;33(11):2594–2603. doi: 10.1007/s11095-016-2029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbadawi M, Gaisford S, Basit AW. Advanced machine-learning techniques in drug discovery. Drug Discov Today. 2021;26(3):769–777. doi: 10.1016/j.drudis.2020.12.003. [DOI] [PubMed] [Google Scholar]
- Feng Q, Dueva E, Cherkasov A, Ester M (2018) Padme: A deep learning-based framework for drug-target interaction prediction. arXiv preprint arXiv:180709741
- Flaten HK, St Claire C, Schlager E, Dunnick CA, Dellavalle RP. Growth of mobile applications in dermatology—2017 update. Dermatol Online J. 2018;24(2):13–16. [PubMed] [Google Scholar]
- Fleming N. How artificial intelligence is changing drug discovery. Nature. 2018;557(7706):S55–S55. doi: 10.1038/d41586-018-05267-x. [DOI] [PubMed] [Google Scholar]
- Fletcher EP, Madabushi R, Sahajwalla CG, Lesko LJ, Huang S-M (2022) The role of the FDA in guiding drug development. In: Huang S-M, Lertora J, Vicini P, Atkinson, A Jr (eds) Atkinson’s principles of clinical pharmacology. Elsevier, pp 681–690
- Fouad F. The fourth industrial revolution is the AI revolution an academy prospective. Int J Inf. 2019;8(5):155–167. [Google Scholar]
- Friedman LM, Furberg CD, DeMets DL, Reboussin DM, Granger CB. Fundamentals of clinical trials. Cham: Springer; 2015. [Google Scholar]
- Gao K, Nguyen DD, Sresht V, Mathiowetz AM, Tu M, Wei G-W. Are 2D fingerprints still valuable for drug discovery? Phys Chem Chem Phys. 2020;22(16):8373–8390. doi: 10.1039/d0cp00305k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S, Das S, Taylor K (2020) AI enabled precision medicine: patient stratification, drug repurposing and combination therapies. In: Cassidy J, Taylor B (eds) Artificial intelligence in oncology drug discovery and development. IntechOpen
- Gatti M, Turrini E, Raschi E, Sestili P, Fimognari C. Janus kinase inhibitors and coronavirus disease (COVID)-19: rationale, clinical evidence and safety issues. Pharmaceuticals. 2021;14(8):738. doi: 10.3390/ph14080738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile F, Agrawal V, Hsing M, Ton A-T, Ban F, Norinder U, Gleave ME, Cherkasov A. Deep docking: a deep learning platform for augmentation of structure based drug discovery. ACS Cent Sci. 2020;6(6):939–949. doi: 10.1021/acscentsci.0c00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li W, Wang B, Liu H, Zhou D. DeepACLSTM: deep asymmetric convolutional long short-term memory neural models for protein secondary structure prediction. BMC Bioinform. 2019;20(1):1–12. doi: 10.1186/s12859-019-2940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N. Artificial neural network. Netw Complex Syst. 2013;3(1):24–28. [Google Scholar]
- Gurney K. An introduction to neural networks. Boca Raton: CRC Press; 2018. [Google Scholar]
- Harrer S, Shah P, Antony B, Hu J. Artificial intelligence for clinical trial design. Trends Pharmacol Sci. 2019;40(8):577–591. doi: 10.1016/j.tips.2019.05.005. [DOI] [PubMed] [Google Scholar]
- Hefti FF. Requirements for a lead compound to become a clinical candidate. BMC Neurosci. 2008;9(3):1–7. doi: 10.1186/1471-2202-9-S3-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooijmans CR, De Vries RB, Ritskes-Hoitinga M, Rovers MM, Leeflang MM, IntHout J, Wever KE, Hooft L, De Beer H, Kuijpers T. Facilitating healthcare decisions by assessing the certainty in the evidence from preclinical animal studies. PLoS ONE. 2018;13(1):e0187271. doi: 10.1371/journal.pone.0187271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJzerman AP, Guo D. Drug–target association kinetics in drug discovery. Trends Biochem Sci. 2019;44(10):861–871. doi: 10.1016/j.tibs.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Jimenez-Carretero D, Abrishami V, Fernandez-de-Manuel L, Palacios I, Quílez-Álvarez A, Díez-Sánchez A, Del Pozo MA, Montoya MC. Tox_ (R) CNN: deep learning-based nuclei profiling tool for drug toxicity screening. PLoS Comput Biol. 2018;14(11):e1006238. doi: 10.1371/journal.pcbi.1006238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Bian Y, Hu Z, Wang L, Xie X-QS. Deep learning for drug design: an artificial intelligence paradigm for drug discovery in the big data era. AAPS J. 2018;20(3):1–10. doi: 10.1208/s12248-018-0210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Bian Y, Hu Z, Wang L, Xie XQ. Deep learning for drug design: an artificial intelligence paradigm for drug discovery in the big data era. Aaps j. 2018;20(3):58. doi: 10.1208/s12248-018-0210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Wu D, Wang Z, Shen Y. DeepAffinity: interpretable deep learning of compound–protein affinity through unified recurrent and convolutional neural networks. Bioinformatics. 2019;35(18):3329–3338. doi: 10.1093/bioinformatics/btz111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns EH, Di L. Pharmaceutical profiling in drug discovery. Drug Discov Today. 2003;8(7):316–323. doi: 10.1016/s1359-6446(03)02649-7. [DOI] [PubMed] [Google Scholar]
- Khan A, Kaushik AC, Ali SS, Ahmad N, Wei D-Q. Deep-learning-based target screening and similarity search for the predicted inhibitors of the pathways in Parkinson’s disease. RSC Adv. 2019;9(18):10326–10339. doi: 10.1039/c9ra01007f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinch LN, Pei J, Kryshtafovych A, Schaeffer RD, Grishin NV. Topology evaluation of models for difficult targets in the 14th round of the critical assessment of protein structure prediction (CASP14) Proteins Struct Funct Bioinform. 2021;89(12):1673–1686. doi: 10.1002/prot.26172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova M, Ginsburg B, Tropsha A, Isayev O. OpenChem: a deep learning toolkit for computational chemistry and drug design. J Chem Inf Model. 2021;61(1):7–13. doi: 10.1021/acs.jcim.0c00971. [DOI] [PubMed] [Google Scholar]
- Kotsiantis SB, Zaharakis I, Pintelas P. Supervised machine learning: a review of classification techniques. Emerg Artif Intell Appl Comput Eng. 2007;160(1):3–24. [Google Scholar]
- Krittanawong C, Johnson KW, Tang WW. How artificial intelligence could redefine clinical trials in cardiovascular medicine: lessons learned from oncology. Future Med. 2019;16:87–92. doi: 10.2217/pme-2018-0130. [DOI] [PubMed] [Google Scholar]
- Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. Critical assessment of methods of protein structure prediction (CASP)—round XIII. Proteins Struct Funct Bioinform. 2019;87(12):1011–1020. doi: 10.1002/prot.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Li X, Wang Y, Yin S, Zhou J, Liu Z. AIScaffold: a web-based tool for scaffold diversification using deep learning. J Chem Inf Model. 2020;61(1):1–6. doi: 10.1021/acs.jcim.0c00867. [DOI] [PubMed] [Google Scholar]
- Larranaga P, Calvo B, Santana R, Bielza C, Galdiano J, Inza I, Lozano JA, Armananzas R, Santafé G, Pérez A. Machine learning in bioinformatics. Brief Bioinform. 2006;7(1):86–112. doi: 10.1093/bib/bbk007. [DOI] [PubMed] [Google Scholar]
- Lavecchia A. Machine-learning approaches in drug discovery: methods and applications. Drug Discov Today. 2015;20(3):318–331. doi: 10.1016/j.drudis.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Lavecchia A. Deep learning in drug discovery: opportunities, challenges and future prospects. Drug Discov Today. 2019;24(10):2017–2032. doi: 10.1016/j.drudis.2019.07.006. [DOI] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Li Y, Hu J, Wang Y, Zhou J, Zhang L, Liu Z. Deepscaffold: a comprehensive tool for scaffold-based de novo drug discovery using deep learning. J Chem Inf Model. 2019;60(1):77–91. doi: 10.1021/acs.jcim.9b00727. [DOI] [PubMed] [Google Scholar]
- Li Q, Shah S (2017) Structure-based virtual screening. In: Wu CH, Arighi CN, Ross KE (eds) Protein bioinformatics. Springer, pp 111–124
- Li Y, Qiao G, Wang K, Wang G (2022) Drug–target interaction predication via multi-channel graph neural networks. Brief Bioinform 23(1):bbab346 [DOI] [PubMed]
- Lin J, Sahakian DC, De Morais S, Xu JJ, Polzer RJ, Winter SM. The role of absorption, distribution, metabolism, excretion and toxicity in drug discovery. Curr Top Med Chem. 2003;3(10):1125–1154. doi: 10.2174/1568026033452096. [DOI] [PubMed] [Google Scholar]
- Lionta E, Spyrou G, Vassilatis D, Cournia Z. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr Top Med Chem. 2014;14(16):1923–1938. doi: 10.2174/1568026614666140929124445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Du J, Fang J, Yin Y, Xu G, Xie L. DeepScreening: a deep learning-based screening web server for accelerating drug discovery. Database. 2019 doi: 10.1093/database/baz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Du J, Fang J, Yin Y, Xu G, Xie L. DeepScreening: a deep learning-based screening web server for accelerating drug discovery. Database J Biol Databases Cur. 2019 doi: 10.1093/database/baz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Zhao X, Zhou J, Yang J, Zhang Y, Kuang W, Peng J, Chen L, Zeng J. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat Commun. 2017;8(1):1–13. doi: 10.1038/s41467-017-00680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak K-K, Pichika MR. Artificial intelligence in drug development: present status and future prospects. Drug Discov Today. 2019;24(3):773–780. doi: 10.1016/j.drudis.2018.11.014. [DOI] [PubMed] [Google Scholar]
- Maltarollo VG, Gertrudes JC, Oliveira PR, Honorio KM. Applying machine learning techniques for ADME-Tox prediction: a review. Expert Opin Drug Metab Toxicol. 2015;11(2):259–271. doi: 10.1517/17425255.2015.980814. [DOI] [PubMed] [Google Scholar]
- Maragakis P, Nisonoff H, Cole B, Shaw DE. A deep-learning view of chemical space designed to facilitate drug discovery. J Chem Inf Model. 2020;60(10):4487–4496. doi: 10.1021/acs.jcim.0c00321. [DOI] [PubMed] [Google Scholar]
- Mauser H, Guba W. Recent developments in de novo design and scaffold hopping. Curr Opin Drug Discov Devel. 2008;11(3):365–374. [PubMed] [Google Scholar]
- Meng X-Y, Zhang H-X, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform. 2017;18(5):851–869. doi: 10.1093/bib/bbw068. [DOI] [PubMed] [Google Scholar]
- Mohs RC, Greig NH. Drug discovery and development: role of basic biological research. Alzheimer’s Dementia Transl Res Clin Interv. 2017;3(4):651–657. doi: 10.1016/j.trci.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Lim-Wilby M (2008) Molecular docking. In: Kukol A (eds) Molecular modeling of proteins. Springer, pp 365–382 [DOI] [PubMed]
- Muegge I, Mukherjee P. An overview of molecular fingerprint similarity search in virtual screening. Expert Opin Drug Discov. 2016;11(2):137–148. doi: 10.1517/17460441.2016.1117070. [DOI] [PubMed] [Google Scholar]
- Musella S, Verna G, Fasano A, Di Micco S. New perspectives on machine learning in drug discovery. Curr Med Chem. 2021;28:6704–6728. doi: 10.2174/0929867327666201111144048. [DOI] [PubMed] [Google Scholar]
- Nayak A, Dutta K Impacts of machine learning and artificial intelligence on mankind. In: 2017 international conference on intelligent computing and control (I2C2), 2017. IEEE, pp 1–3
- Olivecrona M, Blaschke T, Engkvist O, Chen H. Molecular de-novo design through deep reinforcement learning. J Cheminform. 2017;9(1):1–14. doi: 10.1186/s13321-017-0235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongsulee P (2017) Artificial intelligence, machine learning and deep learning. In: 2017 15th international conference on ICT and knowledge engineering (ICT&KE), 2017. IEEE, pp 1–6
- Osisanwo F, Akinsola J, Awodele O, Hinmikaiye J, Olakanmi O, Akinjobi J. Supervised machine learning algorithms: classification and comparison. Int J Comput Trends Technol (IJCTT) 2017;48(3):128–138. [Google Scholar]
- Öztürk H, Özgür A, Ozkirimli E. DeepDTA: deep drug–target binding affinity prediction. Bioinformatics. 2018;34(17):i821–i829. doi: 10.1093/bioinformatics/bty593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öztürk H, Ozkirimli E, Özgür A (2019) WideDTA: prediction of drug-target binding affinity. arXiv preprint arXiv:190204166
- Parasa NA, Namgiri JV, Mohanty SN, Dash JK (2021) Introduction to unsupervised learning in bioinformatics. In: Data analytics in bioinformatics: a machine learning perspective. Wiley-Scrivener, Hoboken, NJ, pp 35–49
- Pathania A, Kumar R, Sandhir R. Hydroxytyrosol as anti-parkinsonian molecule: Assessment using in-silico and MPTP-induced Parkinson’s disease model. Biomed Pharmacother. 2021;139:111525. doi: 10.1016/j.biopha.2021.111525. [DOI] [PubMed] [Google Scholar]
- Paul D, Sanap G, Shenoy S, Kalyane D, Kalia K, Tekade RK. Artificial intelligence in drug discovery and development. Drug Discov Today. 2021;26(1):80. doi: 10.1016/j.drudis.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Li J, Shang X. A learning-based method for drug-target interaction prediction based on feature representation learning and deep neural network. BMC Bioinform. 2020;21(13):1–13. doi: 10.1186/s12859-020-03677-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JC, Caffarena ER, Dos Santos CN. Boosting docking-based virtual screening with deep learning. J Chem Inf Model. 2016;56(12):2495–2506. doi: 10.1021/acs.jcim.6b00355. [DOI] [PubMed] [Google Scholar]
- Perron Q, Mirguet O, Tajmouati H, Skiredj A, Rojas A, Gohier A, Ducrot P, Bourguignon M-P, Sansilvestri-Morel P, Do Huu N, Gellibert F, Gaston-Mathé Y (2022) Deep generative models for ligand-based de novo design applied to multi-parametric optimization. J Comput Chem 43(10):692–703 [DOI] [PubMed]
- Piantadosi S. Clinical trials: a methodologic perspective. New York: Wiley; 2017. [Google Scholar]
- Popova M, Isayev O, Tropsha A. Deep reinforcement learning for de novo drug design. Sci Adv. 2018;4(7):eaap7885. doi: 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri M, Solanki A, Padawer T, Tipparaju SM, Moreno WA, Pathak Y (2016) Introduction to artificial neural network (ANN) as a predictive tool for drug design, discovery, delivery, and disposition: Basic concepts and modeling. In: Puri M, Pathak Y, Sutariya V, Tipparaju S, Moreno W (eds) Artificial neural network for drug design, delivery and disposition. Elsevier, pp 3–13
- Qiao R, Tran NH, Xin L, Chen X, Shan B, Li M (2020) Systems and methods using artificial neural network and de novo peptide sequencing to identify patient-specific neoantigens for personalized immunotherapy. US20200243164
- Rayhan F, Ahmed S, Mousavian Z, Farid DM, Shatabda S. FRnet-DTI: deep convolutional neural network for drug-target interaction prediction. Heliyon. 2020;6(3):e03444. doi: 10.1016/j.heliyon.2020.e03444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayhan F, Ahmed S, Mousavian Z, Farid DM, Shatabda S (2018) FRnet-DTI: convolutional neural networks for drug-target interaction. arXiv preprint arXiv:180607174 7 [DOI] [PMC free article] [PubMed]
- Rezaei MA, Li Y, Wu D, Li X, Li C. Deep learning in drug design: protein–ligand binding affinity prediction. IEEE/ACM Trans Comput Biol Bioinform. 2020 doi: 10.1109/TCBB.2020.3046945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P, Griffin I, Tucker C, Smith D, Oechsle O, Phelan A, Rawling M, Savory E, Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet (london, England) 2020;395(10223):e30. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripphausen P, Nisius B, Bajorath J. State-of-the-art in ligand-based virtual screening. Drug Discov Today. 2011;16(9–10):372–376. doi: 10.1016/j.drudis.2011.02.011. [DOI] [PubMed] [Google Scholar]
- Ruff KM, Pappu RV. AlphaFold and implications for intrinsically disordered proteins. J Mol Biol. 2021;433(20):167208. doi: 10.1016/j.jmb.2021.167208. [DOI] [PubMed] [Google Scholar]
- Sahoo P, Roy I, Wang Z, Mi F, Yu L, Balasubramani P, Khan L, Stoddart JF. MultiCon: a semi-supervised approach for predicting drug function from chemical structure analysis. J Chem Inf Model. 2020;60(12):5995–6006. doi: 10.1021/acs.jcim.0c00801. [DOI] [PubMed] [Google Scholar]
- Schneider G, Fechner U. Computer-based de novo design of drug-like molecules. Nat Rev Drug Discov. 2005;4(8):649–663. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- Schneider P, Schneider G. De novo design at the edge of chaos: miniperspective. J Med Chem. 2016;59(9):4077–4086. doi: 10.1021/acs.jmedchem.5b01849. [DOI] [PubMed] [Google Scholar]
- Scudellari M (2020) Five companies using AI to fight coronavirus. https://spectrum.ieee.org/the-human-os/artificial-intelligence/medical-ai/companies-ai-coronavirus. Accessed 14 Feb 2022
- Shen C, Ding J, Wang Z, Cao D, Ding X, Hou T. From machine learning to deep learning: advances in scoring functions for protein–ligand docking. Wiley Interdiscip Rev Comput Mol Sci. 2020;10(1):e1429. [Google Scholar]
- Shetty P, Singh S. Hierarchical clustering: a survey. IJAR. 2021;7(4):178–181. [Google Scholar]
- Smith JS, Roitberg AE, Isayev O. Transforming computational drug discovery with machine learning and AI. New York: ACS Publications; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson N, Shane E, Chase J, Rowland J, Ries D, Justice N, Zhang J, Chan L, Cao R. Survey of machine learning techniques in drug discovery. Curr Drug Metab. 2019;20(3):185–193. doi: 10.2174/1389200219666180820112457. [DOI] [PubMed] [Google Scholar]
- Tavallali P, Tavallali P, Singhal M. K-means tree: an optimal clustering tree for unsupervised learning. J Supercomput. 2021;77(5):5239–5266. [Google Scholar]
- Thafar M, Raies AB, Albaradei S, Essack M, Bajic VB. Comparison study of computational prediction tools for drug-target binding affinities. Front Chem. 2019;7:782. doi: 10.3389/fchem.2019.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh TS, Dondelinger F, Wang D. Looking beyond the hype: applied AI and machine learning in translational medicine. EBioMedicine. 2019;47:607–615. doi: 10.1016/j.ebiom.2019.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, Li B, Madabhushi A, Shah P, Spitzer M. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18(6):463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaelen Q, Lin Y-C, Zhavoronkov A. The advent of generative chemistry. ACS Med Chem Lett. 2020;11(8):1496–1505. doi: 10.1021/acsmedchemlett.0c00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak S. The role of artificial intelligence in clinical decision support systems and a classification framework. Int J Comput Clin Pract (IJCCP) 2018;3(2):31–47. [Google Scholar]
- Walters WP, Murcko M. Assessing the impact of generative AI on medicinal chemistry. Nat Biotechnol. 2020;38(2):143–145. doi: 10.1038/s41587-020-0418-2. [DOI] [PubMed] [Google Scholar]
- Wan F, Zhu Y, Hu H, Dai A, Cai X, Chen L, Gong H, Xia T, Yang D, Wang M-W. DeepCPI: a deep learning-based framework for large-scale in silico drug screening. Genom Proteom Bioinform. 2019;17(5):478–495. doi: 10.1016/j.gpb.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu W, Shen Z, Jiang L, Wang J, Li S, Li H. Deep learning based drug metabolites prediction. Front Pharmacol. 2020;10:1586. doi: 10.3389/fphar.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-C (2003) Artificial neural network. In: Wang S-C (eds) Interdisciplinary computing in java programming. Springer, pp 81–100
- Wei G-W. Protein structure prediction beyond AlphaFold. Nat Mach Intell. 2019;1(8):336–337. doi: 10.1038/s42256-019-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildey MJ, Haunso A, Tudor M, Webb M, Connick JH. High-throughput screening. Annu Rep Med Chem. 2017;50:149–195. [Google Scholar]
- Woo M. An AI boost for clinical trials. Nature. 2019;573(7775):S100–s102. doi: 10.1038/d41586-019-02871-3. [DOI] [PubMed] [Google Scholar]
- Wu D, Huang Q, Zhang Y, Zhang Q, Liu Q, Gao J, Cao Z, Zhu R. Screening of selective histone deacetylase inhibitors by proteochemometric modeling. BMC Bioinform. 2012;13(1):1–10. doi: 10.1186/1471-2105-13-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuo N, Sekijima M. Development of postprocessing method of protein-ligand docking using interaction fingerprint. Biophys J. 2017;112(3):452a. [Google Scholar]
- Yasuo N, Sekijima M. Improved method of structure-based virtual screening via interaction-energy-based learning. J Chem Inf Model. 2019;59(3):1050–1061. doi: 10.1021/acs.jcim.8b00673. [DOI] [PubMed] [Google Scholar]
- Zhang L, Tan J, Han D, Zhu H. From machine learning to deep learning: progress in machine intelligence for rational drug discovery. Drug Discov Today. 2017;22(11):1680–1685. doi: 10.1016/j.drudis.2017.08.010. [DOI] [PubMed] [Google Scholar]
- Zhang HH (2014) Supervised Learning. In: Wiley StatsRef: Statistics Reference Online. Wiley, pp 1–17
- Zhou Z, Kearnes S, Li L, Zare RN, Riley P. Optimization of molecules via deep reinforcement learning. Sci Rep. 2019;9(1):1–10. doi: 10.1038/s41598-019-47148-x. [DOI] [PMC free article] [PubMed] [Google Scholar]