Abstract
Purpose
Germline mutations of BRCA1 and BRCA2 are associated with a defined lifetime risk of breast (BC), ovarian (OC) and other cancers. Testing BRCA genes is pivotal to assess individual risk, but also to pursue preventive approaches in healthy carriers and tailored treatments in tumor patients. The prevalence of BRCA1 and BRCA2 alterations varies broadly across different geographic regions and, despite data about BRCA pathogenic variants among Sicilian families exist, studies specifically addressing eastern Sicily population are lacking. The aim of our study was to investigate the incidence and distribution of BRCA pathogenic germline alterations in a cohort of BC patients from eastern Sicily and to evaluate their associations with specific BC features.
Patients and Methods
Mutational status was assessed in a cohort of 389 BC patients, using next generation sequencing. The presence of alterations was correlated with tumor grading and proliferation index.
Results
Overall, 35 patients (9%) harbored a BRCA pathogenic variant, 17 (49%) in BRCA1 and 18 (51%) in BRCA2. BRCA1 alterations were prevalent among triple negative BC patients, whereas BRCA2 mutations were more common in subjects with luminal B BC. Tumor grading and proliferation index were both significantly higher among subjects with BRCA1 variants compared to non-carriers.
Conclusion
Our findings provide an overview about BRCA mutational status among BC patients from eastern Sicily and confirm the role of NGS analysis to identify hereditary BC patients. Overall, these data are consistent with previous evidences supporting BRCA screening to properly prevent and treat cancer among mutation carriers.
Keywords: hereditary breast cancer, BRCA1/2, NGS, tumor grading, Ki-67
Introduction
Breast cancer (BC) represents the most frequent malignancy worldwide and the most lethal in women.1 Over time, the biological features determining BC prognosis and clinical behavior have been extensively investigated and partly elucidated. Indeed, several surrogate markers are currently employed to classify BC into different molecular subtypes. These are estrogen (ER) and/or progesterone receptors (PgR), Human Epidermal growth factor Receptor 2 (HER2) amplification, proliferation index Ki-67 and tumor grading (G).2 The combination of these variables identifies the following BC categories: 1) luminal tumors, which present ER and/or PgR expression and account for up to 75% of BC. These tumors are further classified into luminal A, when Ki-67 is below 20% and HER2 is negative, and luminal B, when Ki-67 is equal or higher than 20% and in case of HER2 amplification, regardless of proliferation index; 2) HER2+ tumors, which are negative for ER and PgR but display HER2 amplification. This group encompasses 10% of all breast tumors; 3) triple negative BC (TNBC), which do not display ER and PgR expression and HER2 amplification and represent about 15% of breast cancer.2–4
Across these BC subtypes, tumor grading and proliferation index represent transversal biomarkers, directly and independently correlating with tumor aggressiveness and prognosis.5,6
Besides the abovementioned biological features, the role of inherited genetic alterations leading to BC development has gained an increasing importance during the last few years.7 Approximately one out of ten breast tumors is hereditary due the presence of germline alterations in specific genes.8 Two large epidemiological studies including more than 180.000 women recently identified a set of 8 genes (ie ATM, BARD1, BRCA1, BRCA2, CHK2, PALB2, RAD51C and RAD51D) mainly responsible for hereditary BC. Among these genes, BRCA1 and BRCA2 (hereafter abbreviated as BRCA1/2) exhibited the strongest correlation with the development of breast tumors.9–12 Indeed, germline BRCA1/2 mutations considerably increase the lifetime risk of BC as well as of other malignancies, including ovarian, prostate, pancreatic, colorectal cancer and melanoma.13 The cumulative incidence of BC by 80 years of age is 72% for women carrying BRCA1 pathogenic variant (PV) and 69% for those with BRCA2 PV.14
Noteworthy, a recent publication showed that BC risk depends on the type of PVs. In fact, photogenic missense variants, especially in BRCA1 gene, are associated with a reduced risk of BC compared with pathogenic truncated mutations, particularly for older women.15
The presence of BRCA1 or BRCA2 PV correlates with distinct biological and clinico-pathological characteristics.16,17 BRCA1-related BC tend to be clinically aggressive, poorly differentiated and highly proliferative. These tumors are frequently triple negative and present an earlier age of onset. Tumors occurring in BRCA2 mutated patients usually display an intermediate to high differentiation grade and a variable proliferation index. These tumors are more often luminal B and usually occur in older individuals.16–18 Of note, mutations in either BRCA1 and BRCA2 confer an increased sensitivity towards specific treatments, including platinum salts and targeted agents, such as poly(ADP-ribose) polymerase inhibitors (PARPi).19,20
In the last few years, the implementation of next generation sequencing (NGS) in the clinical practice gave access to an increasing number of BC patients to molecular tests for cancer predisposition syndromes, including BRCA1/2.21 At the same time, definition of precise criteria based on family history, demographic characteristics and clinicopathological features allowed to better identify individuals who deserve BRCA1/2 testing.22,23 In this context, evidences are accumulating about BRCA1/2 screening in specific populations, highlighting the differences between distinct geographical regions.24–27 While reports exist about BC cohorts from western Sicily, fewer data are available about BRCA1/2 screening among eastern Sicily population.28,29
We describe here the results of germline BRCA1/2 screening in BC patients from eastern Sicily, further correlating the presence of a BRCA1 or BRCA2 mutations with the main clinico-pathological features of these tumors.
Patients and Methods
Study Population
A retrospective study was carried out at the “Center of Experimental Oncology and Hematology” of the Hospital Policlinico “G. Rodolico - San Marco” of Catania. From January 2017 to March 2021, a total of 455 patients with breast and ovarian cancer, melanoma, pancreatic tumour or prostate carcinoma were referred to our Molecular Diagnostics Laboratory for BRCA/2 genetic testing. The present study was conducted in accordance with the Declaration of Helsinki and all participants provided written informed consent before undergoing molecular analysis.
Histological and biological features of BC (ER, PgR, HER2 status, Ki-67 and grading) were evaluated on core biopsies or surgical samples, considering only the invasive tumor component. Based on these features, BC were classified as follow: luminal A (ER+ and/or PgR+, HER2-, Ki-67<20%), luminal B (ER+ and/or PgR+, HER2-, Ki-67≥20%), luminal B-HER2+ (ER and/or PgR+, HER2+), HER2+ (ER and PgR-, HER2+) or triple negative (ER and PgR-, HER2-).
An oncogenetic counselling was performed for each patient by a multidisciplinary team including an oncologist, a geneticist and a psychologist before assessing BRCA1 and BRCA2 mutational status, in order to identify individuals at high risk of harboring a PV in BRCA1 and/or BRCA2 genes. Patients selection was performed according to the Italian Association of Medical Oncology (AIOM) guidelines and Sicily local recommendations.30,31 These criteria included: (i) family history of known pathogenic variant in a predisposing gene (eg BRCA1, BRCA2, TP53, PTEN); (ii) male with BC; (iii) women with BC and OC; (iv) woman with BC <36 years, with TNBC <60 years or with bilateral BC <50 years; (v) personal history of BC <50 years and at least one first-degree relative with: (a) BC <50 years; (b) non-mucinous and non-borderline OC at any age; (c) bilateral BC; (d) male BC; (e) pancreatic carcinoma; (f) prostate carcinoma; (vi) personal history of BC >50 years and family history of BC, OC or pancreatic carcinoma in two or more relatives who have a first-degree relationship with each other (including one who has a first-degree relationship with her); (vii) personal history of OC and at least one first-degree relative with: (a) BC <50 years; (b) OC; (c) bilateral BC; (d) male BC; (vii) woman with high-grade serous OC.
Molecular Testing
DNA Isolation and Next Generation Sequencing Analysis for BRCA1/2 Genes
Samples of 20 mL of peripheral blood were obtained from each patient and collected into EDTA tubes (BD Biosciences). Genomic DNA was isolated from 0.7 mL of whole blood samples using the QIAsymphony DSP DNA Midi kit Isolation Kit (QIAGEN, Hilden, Italy) and quantified by Qubit®3.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA), according to manufacturer’s instructions. Target enrichment and library preparation was performed by Oncomine™ BRCA Research Assay Chef ready to load into an Ion AmpliSeq™ Chef Reagents DL8 cartridge for automated libraries preparation, according to the manufacturer’s instructions. The kit consists of two multiplex PCR primer pool and allowed to investigate all BRCA1 (NM_007300.3) and BRCA2 (NM_000059.3) genes. Briefly, 15 µL of each diluted sample DNA (10 ng) were added to the barcode plate for library preparation and loaded with all reagents and consumables on the Ion Chef™ Instrument. An automated library preparation and pooling of barcoded sample libraries were then performed on the Ion Chef™ Instrument. The quantity of prepared libraries was then evaluated by Qubit®3.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA), according to manufacturer’s instructions. Finally, libraries were combined in an equimolar ratio in the Ion Chef™ Library Sample Tube (barcoded tube) and loaded onto the Ion Chef™ Instrument. The sequencing was performed with Ion 510 Chip (Thermo Fisher Scientific) using Ion Torrent S5 (Thermo Fisher Scientific) instrument (Thermo Fisher Scientific). Data analysis was done by Amplicon Suite (SmartSeq s.r.l.) and Ion Reporter Software.
Data Analysis, Genetic Classification and Sanger Sequencing
All variants nomenclature followed current guidelines of the Human Genome Variation Society available online (HGVS, http://www.hgvs.org/mutnomen). The clinical significance of BRCA1/2 variants was defined using the classification of the international consortium ENIGMA (evidence-based network for the interpretation of germline mutant alleles, https://enigmaconsortium.org/) and consulting different databases, such as ARUP, BRCAEXCHANGE, ClinVar, IARC_LOVD and UMD. The classification consists of five different classes of risk: benign (class I), likely benign (class II), variant of uncertain significance (VUS, class III), likely pathogenic (class IV) and pathogenic (class V). The impact of mutations on protein structure and function was also analyzed by VarSome, an informatic tool that allows the access to 30 databases.32
To assign a potential clinical significance to each VUS, the following in silico protein prediction algorithms were used: MUTATION TASTER,33 PROVEAN-SIFT (http://provean.jcvi.org/index.php), POLYPHEN-2 (http://genetics.bwh.harvard.edu/pph2/) and Align-GVGD (http://agvgd.hci.utah.edu/agvgd_input.php). Variants classified as class 1 and 2 were considered wild type.
The presence of each pathogenic variants was confirmed by Sanger sequencing. Briefly, a couple of specific primer was designed for each of the detected variants by using the BRCA1 and BRCA2 gene reference sequences (NG_005905.2, NM_007294.3 and NG_012772.3, NM_000059.3, respectively). Therefore, a target- PCR was performed, followed by Sanger sequencing.
CNV Analysis by Multiplex Ligation-Dependent Probe Amplification Analysis (MLPA)
Patients with a negative BRCA1/2 genetic test were tested to evaluate the presence of Large Genomic Rearrangements (LGR) by Multiplex ligation-dependent probe amplification (MLPA), according to the manufacturer’s instructions. Briefly, the DNA samples are denatured and liked up to 60 probes specific for BRCA1 and BRCA2 genes that each detect a specific DNA sequence of approximately 60 nucleotides in length. Probe amplification products, consisting of a set of unique PCR amplicons, are then analyzed by capillary electrophoresis and analyzed by the Coffalyser.Net software, in combination with the appropriate lot-specific Coffalyser sheet (www.mrcholland.com).
Statistical Analysis
Selected clinic-pathological variables (histological grade and Ki-67% proliferation index) were correlated with the presence of BRCA1/2 PVs using the Fisher’s Exact test assuming p-values <0.05 as significant, calculated using Prism software v. 8.4.
Results
Population Characteristics
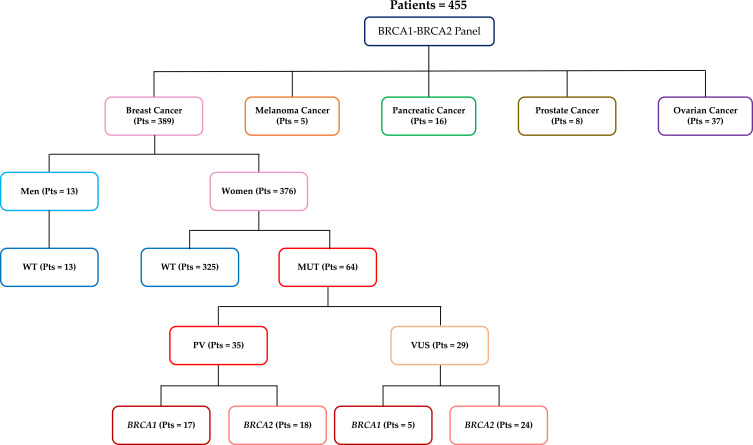
Between January 2017 and March 2021, 455 patients were screened for germline BRCA1/2 mutations. Mutational tests were performed at the Center of Experimental Oncology and Hematology of the Hospital Policlinico “G. Rodolico - San Marco” of Catania, according to Sicily guidelines (http://www.gurs.regione.sicilia.it/Indicep1.htm, N. 02-Venerdì 10 Gennaio 2020) Overall, 389 patients had breast cancer, 37 ovarian cancer, 16 pancreatic cancer, 8 prostate cancer and 5 melanoma. Patients distribution according to cancer type and analysis results is shown in Figure 1.
Figure 1.
Flow chart showing an overview of the study. Patients with breast, melanoma, pancreatic, prostate or ovarian tumors tested for BRCA1 and BRCA2 gene mutation.
Abbreviations: PVs, pathogenic variants; VUS, variants of uncertain significance; WT, wild-type BRCA1/2 sequence.
We selectively focused our study on the breast cancer cohort. These patients displayed a median age of 49 years (range 23–89) and were mainly females (n=376, ie 97%).
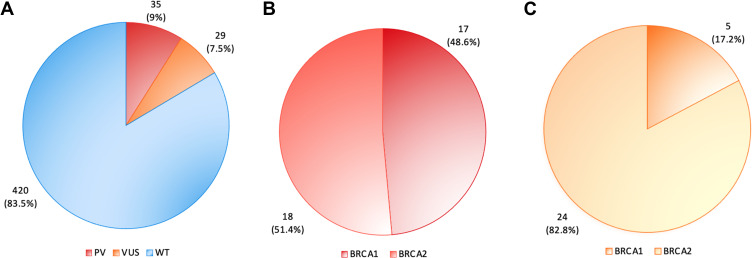
Among these subjects, 64 (17%) carried a BRCA1/2 mutation and all of them were women. Thirty- five (9%) had a PV, while 29 (7.5%) harbored a VUS. Seventeen of the 35 pathogenic variants (48.6%) occurred in BRCA1 and 18 (51.4%) in BRCA2, whereas 5 VUS were in BRCA1 (17.2%) and 24 (82.8%) in BRCA2 (Figures 1 and 2). No LGR emerged from MLPA analysis.
Figure 2.
BRCA1 and BRCA2 mutation analysis of 389 breast cancer patients. (A) Distribution of pathogenic variants (PVs) (red), variants of uncertain significance (VUS) (Orange) and WT (blue) in 389 patients with breast cancer; (B) 35 (9%) out of 389 breast cancer individuals showed BRCA1/2 pathogenic variants (PVs). Among them, 17 (48.6%) were BRCA1 PV carriers (dark red), while 18 (51.4%) were BRCA2 carriers (light red); (C) 29 (7.5%) out 389 subjects harboured VUS, 5 (17.2%) in BRCA1 gene (dark orange) and 24 (82.8%) in BRCA2 gene (light orange).
Abbreviations: PVs, pathogenic variants; VUS, variants of uncertain significance; WT, wild-type BRCA1/2 sequence.
Distribution of Molecular Subtypes
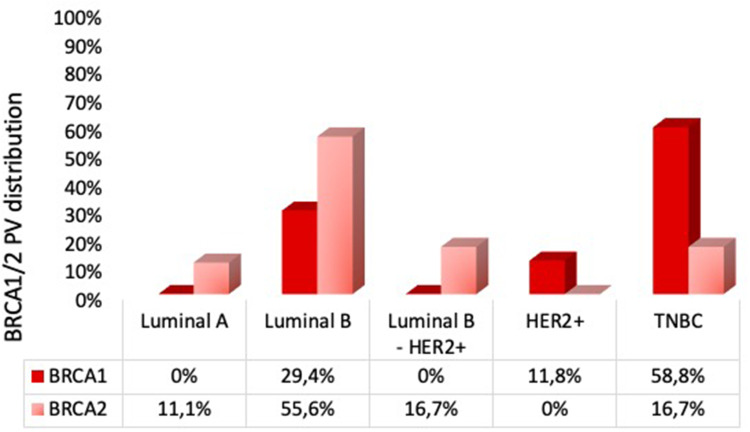
We next investigated the prevalence of BC molecular subtypes in patients carrying BRCA1/2 PVs. The distribution included 2 (5.7%) luminal A, 15 (42.9%) luminal B, 3 (8.6%) luminal B-HER2+, 2 (5.7%) HER2+ and 13 (37.1%) TNBC patients. Among BRCA1 positive patients, 5 (29.4%) presented a luminal B BC, 2 (11.8%) a HER2+ disease and 10 (58.8%) a TNBC. No BRCA1 mutated tumors were luminal A or luminal B-HER2+ (Figure 3). In the BRCA2 positive subgroup, 10 (55.6%) tumors were luminal B, 3 (16.7%) luminal B-HER2+, 3 (16.7%) TNBC and 2 (11.1%) luminal A (Figure 3). No HER2+ tumors were present in this group. Hence, BRCA1 mutations were prevalent in TNBC patients, whereas BRCA2 alterations were predominant in luminal B individuals.
Figure 3.
Prevalence of breast cancer subtypes in patients carrying BRCA1 and BRCA2 pathogenic mutations. The histograms show the distribution of BRCA1- (dark red) and BRCA2- (light red) PVs in molecular subtypes of breast cancer patients. The numbers reported inside each box indicate the percentage of patients with BRCA1 and BRCA2 PVs for each breast cancer subtype.
Abbreviations: PVs, pathogenic variants; HER2 +, human epidermal growth factor receptor 2 positive; TNBC, triple negative breast cancer.
Type and Localization of BRCA1 and BRCA2 Pathogenic Variants
Subsequently, we evaluated the type and gene location of BRCA1 and BRCA2 PVs. Among BRCA1 PVs we observed 7 single nucleotide variants (SNVs), 6 deletions, 3 duplications and 1 insertion. Only one mutation (c.5522delG) represents a novel finding. The most frequent BRCA1 PV, detected in two subjects, was c.5035_5039delCTAAT. This alteration involves the deletion of five nucleotides (CTAAT) in BRCA1 exon 15, resulting in the substitution of the amino acid leucine with tyrosine at codon 1679 and causing a premature truncation of the protein due to a translational frameshift with a predicted alternative stop codon. All the other alterations were detected in one case only. Of note, one of the reported PV is located in a splice site consensus region (c.4357+1G>T) (Table 1).
Table 1.
BRCA1/2 Pathogenic Variants Identified in Breast Cancer Patients
| Gene | Type of PV | HGVS Nomenclature | Protein Change | No. Cases | |
|---|---|---|---|---|---|
| BRCA1 | Duplication | c.66_67insA | p.(Glu23Argfs) | 1 | |
| BRCA1 | Deletion | c.117_118delTG | p.(Cys39Ter) | 1 | |
| BRCA1 | SNV | c.181T>G | p.(Cys61Gly) | 1 | |
| BRCA1 | Deletion | c.514del | p.(Gln172Asnfs) | 1 | |
| BRCA1 | SNV | c.1204G>T | p.(Glu402Ter) | 1 | |
| BRCA1 | Deletion | c.1360_1361delAG | p.(Ser454Ter) | 1 | |
| BRCA1 | Deletion | c.2350_2351delTC | p.(Ser784Valfs) | 1 | |
| BRCA1 | SNV | c.2536G>T | p.(Glu846Ter) | 1 | |
| BRCA1 | Insertion | c.2835_2836insCC | p.(Ile946Profs) | 1 | |
| BRCA1 | Duplication | c.3253dup | p.(Arg1085Lysfs) | 1 | |
| BRCA1 | SNV | c.4117G>T | p.(Glu1373Ter) | 1 | |
| BRCA1 | SNV | c.4357+1G>T | / | 1 | |
| BRCA1 | SNV | c.4484G>T | p.(Arg1495Met) | 1 | |
| BRCA1 | Deletion | c.5035_5039delCTAAT | p.(Leu1679Tyrfs) | 2 | |
| BRCA1 | Duplication | c.5266dup | p.(Gln1756Profs) | 1 | |
| BRCA1 | SNV | c.5509T>C | p.(Trp1837Arg) | 1 | |
| BRCA1 | Deletion | c.5522delG | p. (Ser1841fs*) | 1 | |
| BRCA2 | Duplication | c.428dup | p.(Val144Cysfs) | 3 | |
| BRCA2 | SNV | c.631G>A | p.(Val211Ile) | 1 | |
| SNV | c.7008–2A>T | / | |||
| BRCA2 | Duplication | c.5073_5074insA | p.(Trp1692Metfs) | 1 | |
| BRCA2 | Deletion | c.5603_5606delACAG | p.(Asp1868Valfs) | 1 | |
| BRCA2 | Deletion | c.5851_5854delAGTT | p.(Ser1951TrpfsTer) | 2 | |
| BRCA2 | Deletion | c.6082_6086 delGAAGA | p.(Glu2028Lysfs) | 1 | |
| BRCA2 | Deletion | c.6486_6489delACAA | p.(Lys2162Asnfs) | 1 | |
| BRCA2 | SNV | c.8331+2T>C | / | 1 | |
| BRCA2 | SNV | c.8487+1G>A | / | 3 | |
| BRCA2 | SNV | c.8754+4A>G | / | 1 | |
| BRCA2 | SNV | c.9004G>A | p.(Glu3002Lys) | 1 | |
| BRCA2 | Deletion | c.9026_9030delATCAT | p.(Tyr3009Serfs) | 1 | |
| BRCA2 | Deletion | c.9455_9456delAG | p.(Glu3152Glyfs) | 1 | |
Note: *Novel alteration.
Abbreviations: HGVS, Human Genome Variation Society; PV, pathogenic variant; SNV, single nucleotide variant; c, chromosome site; p, protein site; del, deletion; fs, frame-shift mutation.
Concerning BRCA2 PVs, we observed 6 deletions, 6 SNVs and 2 duplications. None of the identified alterations was novel. Three mutations were recurrent in our population, namely c.428dup and c.8487+1G>A observed in 3 subjects followed by c.5851_5854delAGTT which has been retrieved in two cases. The c.428dup alteration involves the duplication of C in BRCA2 exon 5 predicting to encode a truncated non-functional protein. The c.8487+1G>A mutation occurs in the intronic region (± 1,2) of BRCA2 intron 19 and affects the splice consensus sequence causing altered splicing leading to an abnormal or absent protein. The c.5851_5854delAGTT pathogenic mutation, results from a deletion of 4 nucleotides at nucleotide positions 5851 to 5854 in the coding exon 10 of the BRCA2 gene and results in a translational frameshift with a predicted alternative stop codon (p.S1951WfsTer). Of note, the two alterations, c.631G>A and c.7008-2A>T, were detected in the same patient, as previously reported.34 The first mutation involves the substitution of one nucleotide containing a guanine (G) with an adenosine (A) in BRCA2 exon 7 causing the change of valine amino acid with an isoleucine, an amino acid with highly similar properties, at codon 211. This change affects the normal mRNA splicing. The second variant is located in an intronic region and causes a double A to thymine (T) substitution before coding exon 13 of the BRCA2 gene. The c.7008-2A>T alteration may produce multiple transcripts of different lengths. Moreover, in the BRCA2 PVs group, 4 (22.2%) of 18 alterations were intronic.
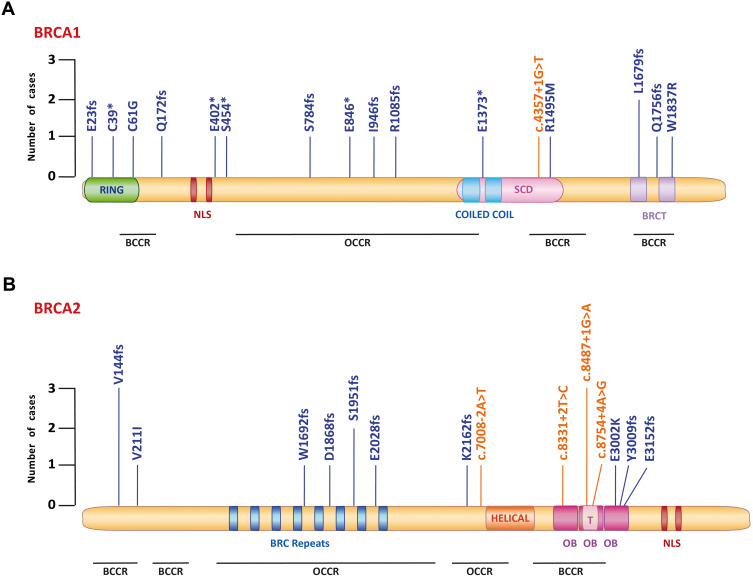
Then, we mapped the BRCA1/2 deleterious mutations across the functional domain and proteins binding regions (Figure 4). In BRCA1 gene, 50% PVs were located in the breast cancer cluster regions (BCCR), while 22% mutations in the ovarian cancer cluster regions (OCCR) (Figure 4A). Among BRCA2 PVs, 35.7% variants were in the BCCR region and 42.8% of mutations in the OCCR (Figure 4B). Next, we evaluated the location of PVs in BRCA1 and BRCA2 protein domains. For BRCA1 protein, we found three PVs placed in both the ring domain and the coiled coil domain, and two mutations in the BRCT domain (Figure 4A). For BRCA2 protein, 4 PVs mapped in the BRC repeats domains, while 3 intronic and 3 exonic alterations were detected in the Oligonucleotide/Oligosaccharide Binding (OB) and Tower (T) domains (Figure 4B).
Figure 4.
Schematic representations of BRCA1 and BRCA2 proteins and localization of pathogenic variants. The diagrams show the distribution of BRCA1 (A) and BRCA2 (B) pathogenic variants in breast cancer patients. The exonic mutations are depicted in blue, while the intronic variants in orange. The bar heights indicate the number of cases. BRCA1 and BRCA2 proteins are reported with their functional domains. (A) The BRCA1 protein contains a RING domain (RING) and a nuclear localization sequence (NLS), a coiled-coil domain, a SQ/TQ cluster domain (SCD) and BRCA1 C terminus domains (BRCT). (B) The BRCA2 protein contains eight BRC repeats, a DNA-binding domain with a helical domain (Helical), three oligonucleotide/oligosaccharide binding (OB) fold, a Tower domain (T) and a NLS at the C-terminus. Regions referred to be breast cancer cluster regions (BCCRs) and ovarian cancer cluster regions (OCCRs) are shown at the bottom. *Represents mutations determining a stop codon.
Correlation Between Clinico-Pathological Characteristics and BRCA Pathogenic Variants
Then we looked at BC clinico-pathological features, which can correlate with the presence of BRCA1/2 PVs. Complete clinical records were available for 181 patients with a negative BRCA1/2 test (non-carriers) and for all the carriers (n=35). A correlation emerged for tumor proliferation rate and grade.
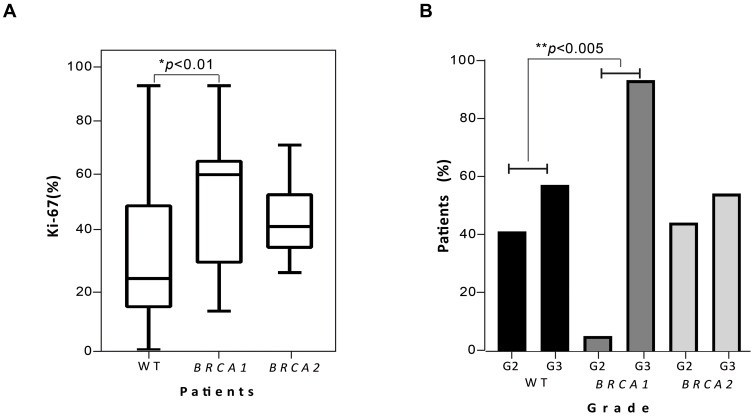
We calculated the distribution of Ki-67 according to median value of our cohort (25%, range <10–90%). Subjects with a Ki-67<25% were defined as “low Ki-67” while individuals with value ≥25% were considered “high Ki-67”. A significant Ki-67 difference was found between non-carriers and BRCA1 PV carriers (p<0.01) (Figure 5A).
Figure 5.
Correlation of Ki-67 and grading distribution in breast cancer women with or without BRCA1 and BRCA2 PVs. (A) Boxplots show comparison of median Ki-67 among 181 non-carriers BC patients and those with BRCA1 (18) or BRCA2 (17) PVs. P value below 0.5 was considered statistically significant. (B) The histograms represent the distribution of BC cancer patients into the histological grade group (G2 and G3) according to the BRCA1 and BRCA2 mutational status (WT subjects, BRCA1 and BRCA2 PVs carriers).
Similarly, we examined whether tumor grading correlates with the presence of BRCA1/2 PVs. Since no G1 BC were present in our population, we stratified patients in two groups (G2 or G3). Consistently with the Ki-67 results, the analysis revealed a statistically significant correlation between tumor grade and BRCA1 mutations, with a higher proportion of G3 tumors among BRCA1 carriers compared to non-carriers (p<0.005) (Figure 5B).
Discussion
The development of DNA sequencing technologies determined an unprecedented progress in BRCA1/2 genetic testing, with crucial consequences for patients presenting a family cancer history. To date, about 20.000 BRCA1/2 variants have been identified and classified according to American College of Medical Genetics35 and ENIGMA system.35,36 It is known that the spectrum of BRCA1/2 mutations varies greatly across different geographic regions.37 Indeed, the rate of BRCA1/2 PVs ranges from 8% to 37% throughout Italian territory, showing a broad intra-national variability.38,39 With a population of almost 5 million people, Sicily represents the fifth Italian region in terms of number of inhabitants. While data exist about BRCA1/2 distribution in western Sicily, no extensive evidences are available for the eastern part of the island.
Our study represents one of the first reports about the incidence of BRCA1/2 PVs in BC patients from eastern Sicily.28 We focused our analysis on BC, since this was by far the most frequent disease in our cohort.
Upon 389 BC patients tested, 9% harbored a BRCA1/2 PV, evenly distributed between BRCA1 and BRCA2. These results are in line with those previously reported in Italian population.28 Interestingly, our cohort comprised 3% of men (13/389). This rate is higher than expected for male breast cancer (1% of all BC),40 mirroring the selection of our population according to the risk of BRCA1/2 mutations. However, none of the men presented a BRCA1/2 PV, and they are therefore candidate to further molecular analyses to rule out the presence of less frequent mutations (eg PALB2, RAD51C and D, etc.). Variants of uncertain significance were retrieved in 7% of the subjects, with a clear prevalence of BRCA2 VUS. Even this result is consistent with pre-existing evidences.28,41,42
When we analyzed the distribution of BC molecular subtypes among BRCA1/2 mutated women, we confirmed the known association between TNBC and BRCA1 PVs (58.8%) and between luminal B BC and BRCA2 PVs (55.6%).16,43 The scarce number of luminal A and HER2+ tumors among both BRCA1 and BRCA2 PV carriers is in line with the existing literature data.16,43
We then focused on the type and location of BRCA1/2 PVs. In our cohort, the most frequent BRCA1 PV was the c.5035_5039delCTAAT. Although this variant has not been described by Incorvaia et al in their Sicilian cohort,28 it has already been reported as germline BRCA1 PV by other authors.34 Several BRCA1 PVs found in our cohort - such as c.181T>G, c.514del, c.3253dupA and c.5266dupC - have been observed in Sicily.28 Among them, two BRCA1 founder mutations (c.181T>G and c.5266dupC), typically retrieved among Ashkenazi Jews, in Eastern and Central Europe (Polish, Czech, Slovenian, Austrian, Hungarian, Belarusian and German),44,45 but also in United States and Argentina, were recently defined as “recurring germline variants” among Italian BC and OC patients.34 The c.514del variant was previously described in 8 breast cancer patients from Northern Sicily from Palermo and Messina. Interestingly, the c.3253dupA alteration has been found in some families from Catania even by Incorvaia et al.28 The most representative BRCA2 PVs were c.428dup, c.5851_5854delAGTT and the intronic variant c.8487+1G>A, which were already described in a Sicilian population.28 More in detail, the c.428dup was reported in patients from Palermo, the c.5851_5854delAGTT PV was observed among families from north-western Sicily, mainly in the area of Trapani and Palermo, while the c.8487+1G>A variant was more frequently detected in subjects from Messina, Palermo and Caltanissetta.28 The c.5851_5854delAGTT alteration has been previously described by Rebbeck et al in Colombia.37 Another BRCA2 PV, c.631+1G>A, has been identified in BC and OC patients from Sicily (Agrigento, Siracusa and Ragusa).28 Of note, we observed the coexistence of two BRCA2 variants in the same patient (BRCA2 c.631G>A and c.7008-2A>T) that we assume are segregated in cis modality, as previously reported.34,46 These BRCA2 double mutations were frequently observed in Italian regions and were found to introduce a premature stop codons, affecting messenger RNA splicing and leading to a nonfunctioning BRCA2 protein.47,48
We also mapped the BRCA1 and BRCA2 PVs across the protein functional domains and the putative OCCR and BCCR regions of the genes. These regions were described by Rebbeck et al as risk regions for developing ovarian and breast cancer, respectively.49 However, evidences about the correlation between the location of germline variants and the risk of breast or ovarian cancer remain controversial.28,50–52 In our population, BRCA1 PVs were mainly distributed in the BCCR domain, while BRCA2 PVs were mostly located in the OCCR region. However, we were not able to find any association between the putative OCCR and BCCR regions and BC features. This may be due to the limited number BRCA1/2 mutated patients. Looking at the protein domains, BRCA1 PVs were distributed along the entire protein, BRCA2 alterations were preferentially found in the BRC repeats domain.
Lastly, we correlated the BC clinico-pathological features with BRCA1/2 PVs. Because of the limited number of patients included, we only found a significant correlation for Ki-67 and tumor grade. Even though Ki-67 assessment and interpretation still present some controversies, it is certain that a high proliferation rate is associated with an increased risk of disease relapse and worse survival. To date, the cut-off to distinguish between “high” and “low” Ki-67 is 20%. However, this threshold did not apply to our population of BRCA1/2 mutated patients, which presented a Ki-67 median value of 25%. This trend towards a high Ki-67 rate can be explained by the prevalence in our cohort of luminal B and TNBC, with few luminal A tumors. However, several evidences seem to suggest that a higher Ki-67 cut-off (25–30%) could better stratify patients according to their prognosis.53,54 Looking at the results of our analysis, it is not surprising that a significant correlation emerged between high Ki-67 and grade and the presence of BRCA1 PVs. Indeed, BRCA1-associated tumors are classically TNBC and display more aggressive features.16,17
Conclusion
In conclusion, the present study provides a report about BRCA1/2 mutational status in a cohort of BC from eastern Sicily. Overall, our findings are consistent with pre-existing evidences, either in terms of mutations prevalence and clinico-pathological features of BC. Additional studies on a larger population of BRCA1/2 mutated BC patients are warranted, such as the extension of mutational analysis using multigenic panels to assess the presence of PVs different and less frequent than BRCA1/2. This would allow identifying and properly managing a growing number of subjects at increased risk of cancer due to inherited mutations.
Acknowledgments
We acknowledge Prof. Paolo Vigneri for his assistance in breast cancer patients care according to the ethical committee requirements.
Ethics Statement
We confirm that the patients signed an informed consent to release anonymously their tumor samples for research purposes. All patients gave written informed consent in accordance with the Declaration of Helsinki. This study was exempt from ethical review based on the policy of the A.O.U. Policlinico “G.Rodolico - S.Marco” since BRCA1/2 analysis was performed as per clinical practice and all the patients gave their written informed consent. Patients also gave consent for the use of their data for research purposes.
Disclosure
Federica Martorana reports honoraria from Istituto Gentili, Lilly, Novartis, Pfizer. The other authors declare no conflicts of interest in this work.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howlader N, Altekruse SF, Li CI, et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst. 2014;106(5). doi: 10.1093/jnci/dju055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curigliano G, Burstein HJ, Winer EP, et al. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol. 2017;28(8):1700–1712. doi: 10.1093/annonc/mdx308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakha EA, Reis-Filho JS, Baehner F, et al. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res. 2010;12(4):207. doi: 10.1186/bcr2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luporsi E, Andre F, Spyratos F, et al. Ki-67: level of evidence and methodological considerations for its role in the clinical management of breast cancer: analytical and critical review. Breast Cancer Res Treat. 2012;132(3):895–915. doi: 10.1007/s10549-011-1837-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiovitz S, Korde LA. Genetics of breast cancer: a topic in evolution. Ann Oncol. 2015;26(7):1291–1299. doi: 10.1093/annonc/mdv022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahdavi M, Nassiri M, Kooshyar MM, et al. Hereditary breast cancer; genetic penetrance and current status with BRCA. J Cell Physiol. 2019;234(5):5741–5750. doi: 10.1002/jcp.27464 [DOI] [PubMed] [Google Scholar]
- 9.Dorling L, Carvalho S, Allen J, et al.; Breast Cancer Association Consortium. Breast cancer risk genes - association analysis in more than 113,000 women. N Engl J Med. 2021;384(5):428–439. doi: 10.1056/NEJMoa1913948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu C, Hart SN, Gnanaolivu R, et al. A population-based study of genes previously implicated in breast cancer. N Engl J Med. 2021;384(5):440–451. doi: 10.1056/NEJMoa2005936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stella S, Martorana F, Manzella L, Vigneri P. The other side of the coin: dissecting molecular mechanisms behind hereditary breast cancer in search of therapeutic opportunities. Transl Oncol. 2021;14(8):101104. doi: 10.1016/j.tranon.2021.101104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kin G, Marks JH, Mandell JB; New York Breast Cancer Study Group. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–646. [DOI] [PubMed] [Google Scholar]
- 13.Pilarski R. The role of BRCA testing in hereditary pancreatic and prostate cancer families. Am Soc Clin Oncol Educ Book. 2019;39:79–86. doi: 10.1200/EDBK_238977 [DOI] [PubMed] [Google Scholar]
- 14.Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317(23):2402–2416. doi: 10.1001/jama.2017.7112 [DOI] [PubMed] [Google Scholar]
- 15.Li H, Engel C, de la Hoya M, et al. Risks of breast and ovarian cancer for women harboring pathogenic missense variants in BRCA1 and BRCA2 compared with those harboring protein truncating variants. Genet Med. 2022;24(1):119–129. doi: 10.1016/j.gim.2021.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen MJ, Kruse TA, Tan Q, et al. Classifications within molecular subtypes enables identification of BRCA1/BRCA2 mutation carriers by RNA tumor profiling. PLoS One. 2013;8(5):e64268. doi: 10.1371/journal.pone.0064268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah PD, Patil S, Dickler MN, Offit K, Hudis CA, Robson ME. Twenty-one-gene recurrence score assay in BRCA-associated versus sporadic breast cancers: differences based on germline mutation status. Cancer. 2016;122(8):1178–1184. doi: 10.1002/cncr.29903 [DOI] [PubMed] [Google Scholar]
- 18.Mavaddat N, Barrowdale D, Andrulis IL, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol Biomark Prev. 2012;21(1):134–147. doi: 10.1158/1055-9965.EPI-11-0775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torrisi R, Zuradelli M, Agostinetto E, et al. Platinum salts in the treatment of BRCA-associated breast cancer: a true targeted chemotherapy? Crit Rev Oncol Hematol. 2019;135:66–75. doi: 10.1016/j.critrevonc.2019.01.016 [DOI] [PubMed] [Google Scholar]
- 20.Tutt A, Robson M, Garber JE, et al. Oral poly (ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376(9737):235–244. doi: 10.1016/S0140-6736(10)60892-6 [DOI] [PubMed] [Google Scholar]
- 21.Pinto P, Paulo P, Santos C, et al. Implementation of next-generation sequencing for molecular diagnosis of hereditary breast and ovarian cancer highlights its genetic heterogeneity. Breast Cancer Res Treat. 2016;159(2):245–256. doi: 10.1007/s10549-016-3948-z [DOI] [PubMed] [Google Scholar]
- 22.Forbes C, Fayter D, de Kock S, Quek RG. A systematic review of international guidelines and recommendations for the genetic screening, diagnosis, genetic counseling, and treatment of BRCA-mutated breast cancer. Cancer Manag Res. 2019;11:2321–2337. doi: 10.2147/CMAR.S189627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pujol P, Barberis M, Beer P, et al. Clinical practice guidelines for BRCA1 and BRCA2 genetic testing. Eur J Cancer. 2021;146:30–47. doi: 10.1016/j.ejca.2020.12.023 [DOI] [PubMed] [Google Scholar]
- 24.Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7(12):937–948. doi: 10.1038/nrc2054 [DOI] [PubMed] [Google Scholar]
- 25.Kwon BS, Byun JM, Lee HJ, et al. Clinical and genetic characteristics of BRCA1/2 mutation in Korean ovarian cancer patients: a multicenter study and literature review. Cancer Res Treat. 2019;51(3):941–950. doi: 10.4143/crt.2018.312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gervas P, Klyuch B, Denisov E, et al. New germline BRCA2 gene variant in the Tuvinian Mongol breast cancer patients. Mol Biol Rep. 2019;46(5):5537–5541. doi: 10.1007/s11033-019-04928-y [DOI] [PubMed] [Google Scholar]
- 27.Cheng J, Peng J, Fu J, et al. Identification of a novel germline BRCA2 variant in a Chinese breast cancer family. J Cell Mol Med. 2020;24(2):1676–1683. doi: 10.1111/jcmm.14861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Incorvaia L, Fanale D, Badalamenti G, et al. Hereditary breast and ovarian cancer in families from Southern Italy (Sicily)-prevalence and geographic distribution of pathogenic variants in BRCA1/2 genes. Cancers. 2020;12(5):1158. doi: 10.3390/cancers12051158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Incorvaia L, Fanale D, Bono M, et al. BRCA1/2 pathogenic variants in triple-negative versus luminal-like breast cancers: genotype-phenotype correlation in a cohort of 531 patients. Ther Adv Med Oncol. 2020;12:1758835920975326. doi: 10.1177/1758835920975326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Italian Association of Medical Oncology (AIOM). Guidelines: breast cancer. Available from: https://www.aiom.it/wp-content/uploads/2021/11/2021_LG_AIOM_Neoplasie_Mammella_11112021.pdf.pdf. Accessed January 17, 2022.
- 31.Sicilian Diagnostic, Therapeutic and Assistance Pathway (PDTA) for hereditary breast and ovarian syndromes. Available from: http://pti.regione.sicilia.it/portal/page/portal/PIR_PORTALE/PIR_LaStrutturaRegionale/PIR_AssessoratoSalute/PIR_Infoedocumenti/PIR_DecretiAssessratoSalute/PIR_DecretiAssessoriali/PIR_DecretiAssessorialianno2019/Allegato%20al%20D.A.%20n.32.pdf. Accessed January 17, 2022.
- 32.Kopanos C, Tsiolkas V, Kouris A, et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35(11):1978–1980. doi: 10.1093/bioinformatics/bty897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 34.Santonocito C, Rizza R, Paris I, et al. Spectrum of germline BRCA1 and BRCA2 variants identified in 2351 ovarian and breast cancer patients referring to a reference cancer hospital of Rome. Cancers. 2020;12(5):1286. doi: 10.3390/cancers12051286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsons MT, Tudini E, Li H, et al. Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: an ENIGMA resource to support clinical variant classification. Hum Mutat. 2019;40(9):1557–1578. doi: 10.1002/humu.23818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rebbeck TR, Friebel TM, Friedman E, et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat. 2018;39(5):593–620. doi: 10.1002/humu.23406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ottini L, D’Amico C, Noviello C, et al. BRCA1 and BRCA2 mutations in central and southern Italian patients. Breast Cancer Res. 2000;2(4):307–310. doi: 10.1186/bcr72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giannini G, Capalbo C, Ristori E, et al. Novel BRCA1 and BRCA2 germline mutations and assessment of mutation spectrum and prevalence in Italian breast and/or ovarian cancer families. Breast Cancer Res Treat. 2006;100(1):83–91. doi: 10.1007/s10549-006-9225-9 [DOI] [PubMed] [Google Scholar]
- 40.Gucalp A, Traina TA, Eisner JR, et al. Male breast cancer: a disease distinct from female breast cancer. Breast Cancer Res Treat. 2019;173(1):37–48. doi: 10.1007/s10549-018-4921-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meisel C, Sadowski CE, Kohlstedt D, et al. Spectrum of genetic variants of BRCA1 and BRCA2 in a German single center study. Arch Gynecol Obstet. 2017;295(5):1227–1238. doi: 10.1007/s00404-017-4330-z [DOI] [PubMed] [Google Scholar]
- 42.Dines JN, Shirts BH, Slavin TP, et al. Systematic misclassification of missense variants in BRCA1 and BRCA2 “coldspots”. Genet Med. 2020;22(5):825–830. doi: 10.1038/s41436-019-0740-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toss A, Molinaro E, Venturelli M, et al. BRCA detection rate in an Italian cohort of luminal early-onset and triple-negative breast cancer patients without family history: when biology overcomes genealogy. Cancers. 2020;12:5. doi: 10.3390/cancers12051252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaufman B, Laitman Y, Gronwald J, Lubinski J, Friedman E. Haplotype of the C61G BRCA1 mutation in Polish and Jewish individuals. Genet Test Mol Biomark. 2009;13(4):465–469. doi: 10.1089/gtmb.2009.0001 [DOI] [PubMed] [Google Scholar]
- 45.Janavicius R. Founder BRCA1/2 mutations in the Europe: implications for hereditary breast-ovarian cancer prevention and control. EPMA J. 2010;1(3):397–412. doi: 10.1007/s13167-010-0037-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vietri MT, Caliendo G, D’Elia G, et al. Five Italian families with two mutations in BRCA genes. Genes. 2020;11(12):1451. doi: 10.3390/genes11121451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colombo M, Ripamonti CB, Pensotti V, et al. An unusual BRCA2 allele carrying two splice site mutations. Ann Oncol. 2009;20(6):1143–1144. doi: 10.1093/annonc/mdp241 [DOI] [PubMed] [Google Scholar]
- 48.Pensabene M, Spagnoletti I, Capuano I, et al. Two mutations of BRCA2 gene at exon and splicing site in a woman who underwent oncogenetic counseling. Ann Oncol. 2009;20(5):874–878. doi: 10.1093/annonc/mdn724 [DOI] [PubMed] [Google Scholar]
- 49.Rebbeck TR, Mitra N, Wan F, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA. 2015;313(13):1347–1361. doi: 10.1001/jama.2014.5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park JS, Lee ST, Han JW, Kim TI, Nam EJ, Park HS. Difference in risk of breast and ovarian cancer according to putative functional domain regions in Korean BRCA1/2 mutation carriers. Clin Breast Cancer. 2018;18(5):362–373 e361. doi: 10.1016/j.clbc.2018.02.007 [DOI] [PubMed] [Google Scholar]
- 51.Yoshihara K, Enomoto T, Aoki D, et al. Association of gBRCA1/2 mutation locations with ovarian cancer risk in Japanese patients from the Charlotte study. Cancer Sci. 2020;111(9):3350–3358. doi: 10.1111/cas.14513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallardo-Rincon D, Alvarez-Gomez RM, Montes-Servin E, et al. Clinical evaluation of BRCA1/2 mutation in Mexican ovarian cancer patients. Transl Oncol. 2020;13(2):212–220. doi: 10.1016/j.tranon.2019.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petrelli F, Viale G, Cabiddu M, Barni S. Prognostic value of different cut-off levels of Ki-67 in breast cancer: a systematic review and meta-analysis of 64,196 patients. Breast Cancer Res Treat. 2015;153(3):477–491. doi: 10.1007/s10549-015-3559-0 [DOI] [PubMed] [Google Scholar]
- 54.Nielsen TO, Leung SCY, Rimm DL, et al. Assessment of Ki67 in breast cancer: updated recommendations from the International Ki67 in Breast Cancer Working Group. J Natl Cancer Inst. 2021;113(7):808–819. doi: 10.1093/jnci/djaa201 [DOI] [PMC free article] [PubMed] [Google Scholar]