Abstract
Aims:
Approximately 30% of the nearly 700,000 Veterans who were deployed to the Gulf War from 1990 to 1991 have reported experiencing a variety of symptoms including difficulties with learning and memory, depression and anxiety, and increased incidence of neurodegenerative diseases. Combined toxicant exposure to acetylcholinesterase (AChE) inhibitors has been studied extensively as a likely risk factor. In this study, we modeled Gulf War exposure in male C57Bl/6J mice with simultaneous administration of three chemicals implicated as exposure hazards for Gulf War Veterans: pyridostigmine bromide, the anti-sarin prophylactic; chlorpyrifos, an organophosphate insecticide; and the repellant N,N-diethyl-m-toluamide (DEET).
Main methods:
Following two weeks of daily exposure, we examined changes in gene expression by whole transcriptome sequencing (RNA-Seq) with hippocampal isolates. Hippocampal-associated spatial memory was assessed with a Y-maze task. We hypothesized that genes important for neuronal health become dysregulated by toxicant-induced damage and that these detrimental inflammatory gene expression profiles could lead to chronic neurodegeneration.
Key findings:
We found dysregulation of genes indicating a pro-inflammatory response and downregulation of genes associated with neuronal health and several important immediate early genes (IEGs), including Arc and Egr1, which were both reduced approximately 1.5-fold. Mice exposed to PB + CPF + DEET displayed a 1.6-fold reduction in preference for the novel arm, indicating impaired spatial memory.
Significance:
Differentially expressed genes observed at an acute timepoint may provide insight into the pathophysiology of Gulf War Illness and further explanations for chronic neurodegeneration after toxicant exposure.
Keywords: Gulf War, RNA-Seq, Gene expression, Pyridostigmine bromide, Chlorpyrifos, DEET, Arc, Immediate early genes, Hippocampus
1. Introduction
Gulf War Illness (GWI) is a chronic multi-system disorder affecting approximately 30% of Veterans deployed during Operations Desert Shield and Desert Storm from August 1990 to February 1991 [1–5]. GWI encompasses a wide spectrum of symptoms which typically include some combination of fatigue/sleep problems, pain, neurological/mood/cognitive impairments, respiratory complaints, gastrointestinal problems, or skin symptoms [6,7]. Of particular interest are neurocognitive impairments and effects on the central nervous system (CNS), as Gulf War Veterans have significantly higher rates of neurological disorders, including amyotrophic lateral sclerosis (ALS), brain cancers, stroke, migraines, neuritis, and neuralgia, than other veteran populations [7]. Research findings in Gulf War animal models have demonstrated that a wide array of physiological alterations including changes in behavior, cognition, neurotransmission, axonal transport, genomic, proteomic, lipidomic, and metabolomic profiling, and mitochondrial dysfunction result from Gulf War exposure [1–5,7,8].
Military personnel deployed during the Gulf War were exposed to an array of chemical exposures in tandem, particularly acetylcholinesterase (AChE) inhibitors. Investigations into the effects of combined Gulf War exposures vary widely but typically include some combination of insecticides, insect repellants, nerve agents, and anti-toxins against nerve agents [1–3,7]. Our Gulf War toxicant mixture includes chemicals from three of the most frequently investigated of these classes: specifically, pyridostigmine bromide (PB, a reversible AChE inhibitor administered as a sarin prophylactic), chlorpyrifos (CPF, an organophosphate pesticide), and N,N-diethyl-m-toluamide (DEET, a common insect repellent).
Significant pathological changes in the hippocampus and corresponding impairments in hippocampal-dependent learning and memory have been observed in several animal models of Gulf War toxicant exposure. Rats exposed to low doses of DEET, permethrin, PB, and restraint stress for four weeks showed significantly reduced hippocampal volume and neuron growth as well as increased occurrence of activated microglia and astrocyte hypertrophy which was accompanied by spatial learning and memory dysfunction [9]. The combination of PB and DEET has been shown to influence cholinesterase activity in the rodent brain and affect seizures [10,11]. Organophosphate exposure has also been shown to impair spatial navigation learning in the Morris Water Maze task [12,13]. Neurotoxicity following administration of PB + CPF + DEET was originally reported by Abou-Donia et al. in hens exposed to 5 mg/kg PB i.o., 10 mg/kg CPF s.c., and 500 mg/kg DEET s.c. 5 days/week for 2 months [14]. Ojo et al. reported significant pathological changes in the hippocampus and cortex of C57Bl/6 mice exposed to PB + CPF + permethrin at an acute timepoint (72 h post-exposure) [15].
Transcriptional changes after Gulf War toxicant exposure in rodent models have mostly focused on epigenetic changes or investigation of specific gene categories of interest at chronic timepoints [8,16–19]. We assessed acute changes in gene expression in mouse hippocampal RNA isolates after exposure to a combined subcutaneous (s.c.) injection of PB, CPF, and DEET for two weeks using whole transcriptome sequencing (RNA-Seq). We focused on genes important for neuronal health, those that could affected by toxicants, and those involved in inflammatory responses. Differentially expressed genes observed at an acute timepoint may set the stage for chronic outcomes and should provide insight into the pathophysiology of Gulf War Illness and help identify potential targets for future treatment.
2. Materials and methods
2.1. Chemicals
HPLC-grade pyridostigmine bromide (PB, P9797) and N,N-diethyl-m-toluamide (DEET, D100951) were obtained from Sigma-Aldrich (St. Louis, MO). Chlorpyrifos (CPF, N-11459) was obtained from Chem-Service, Inc. (West Chester, PA). The toxicant mixture stock was prepared and stored in 500 μL aliquots at −20 °C until use and diluted in PBS immediately prior to injection. Vehicle for injection contained 3.125% dimethyl sulfoxide (DMSO, 99.9%, D2438-5X10ML) obtained from Thermo Fisher Scientific (Waltham, MA) in 1x PBS.
2.2. Subjects
All animal experiments were performed in accordance with the guidelines of the institution, the National Institutes of Health guide for the Care and Use of Laboratory Animals and approved by the East Orange VA and Bay Pines VA Institutional Animal Care and Use Committees. Animals were single-housed in a 22 °C ± 0.5 °C temperature-controlled environment with a 12-hour light/dark cycle. Animals were allowed a 7-day acclimation period before switching to a reverse light cycle (i.e., dark cycle from 10 am-10 pm) for 5 days prior to exposure. Food and water were available ad libitum throughout for all animals.
2.3. Toxicant exposure
Male C57Bl/6 J mice were obtained from Charles River (Wilmington, MA) for RNA-Seq (n = 6/group) and from Jackson Laboratory (Bar Harbor, ME) for behavior (n = 6/group) based on availability. Mice received daily s.c. injections of either the toxicant mixture containing 2.5 mg/kg PB, 12.5 mg/kg CPF, and 7.5 mg/kg DEET with 3.125% DMSO in PBS or vehicle containing 3.125% DMSO in PBS five days a week (M-F) for two weeks beginning at 12 weeks of age. Adverse effects including seizures resulting in removal and euthanasia, were observed at 1.5- and 2.0-fold higher dosages, but this was extremely rare at the dosage used in this study. Experimental cohorts which generated RNA-seq and behavioral data did not display any significant adverse effects. For RNA-Seq, mice were sacrificed 2–4 h after the final exposure via cervical dislocation and decapitation. Whole brains were immediately extracted, and hippocampal tissue from each hemisphere was dissected and snap frozen on dry ice. All fresh frozen tissue samples were stored at −70 °C until use.
2.4. Y-maze task with preference index
To assess hippocampal-dependent memory, subjects underwent a modified Y-maze task 2–4 h after the final exposure. During the training phase, either Arm B or C (novel arm) was blocked off with a barrier. The novel arm was randomly assigned for each trial. Mice were placed in the start arm (Arm A) of the Y-maze facing the wall and allowed to explore the start and familiar arms for 8 min. Mice were then removed from the maze and returned to their home cage for an intertrial interval of 30 min. During the trial phase, the barrier was removed so that all three arms were accessible. Mice were again placed in the start arm and allowed to explore the start and familiar arms for 8 min. Behavior was captured with a video camera (DMK 22AUC03, The Imaging Source, Charlotte, NC) and recorded by ANY-maze (version 6.17, Stoelting, Wood Dale, IL). Time or entry into a zone was scored based on the center point of the animal’s body. All Y-maze trials were performed under red light during the dark cycle.
2.5. RNA isolation
Hippocampal RNA was isolated by TRIzol (Invitrogen, Waltham, MA) extraction followed by cleanup with a RNeasy Mini Kit (QIAGEN, Hilden, Germany). Tissue was resuspended in 0.4 mL TRIzol and homogenized with a Polytron homogenizer (Kinematica USA, Bohemia, NY) on ice for 30–45 s. Samples were incubated at 23 °C for 5 min before adding 80 μL CHCl3 and vortexing for 15 s. Samples were incubated at 23 °C again for 2–3 min. Tubes were centrifuged at 12,000 rcf for 10 min, and the supernatant was transferred into a new tube with an equal volume of 70% EtOH. RNeasy Mini Kit was then used per the manufacturer’s instructions with Tris-EDTA buffer (TE, pH 8.0, AM9858, Invitrogen) for the final elution step. All RNA isolates were stored at −20 °C until use.
2.6. RNA-Seq
RNA isolates were sequenced by the Rutgers-New Jersey Medical School Genomics Center. Total cellular RNA was qualified by confirming integrity with a 2200 TapeStation (Agilent Technologies, Santa Clara, CA). Samples with an RNA integrity number (RIN) > 7.0 were used for subsequent processing. Total RNAs were subjected to two rounds of poly (A) selection using Oligo d(T)25 Magnetic Beads (New England Biolabs, Ipswich, MA). RNA-Seq libraries were prepared using an NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs). cDNA libraries were purified with AMPure XP beads (Beckman Coulter, Brea, CA) and quantified using a Qubit 4 Fluorometer (Thermo Fisher Scientific). Equimolar amounts of barcoded libraries were pooled and sequenced on a NextSeq 500 Sequencing System (Illumina, San Diego, CA) with a 1 × 75 configuration.
2.7. RNA-Seq analysis
RNA-Seq reads were imported into CLC Genomics Workbench (version 20.0.3, QIAGEN) for preliminary analysis using a modified version of the workflow for RNA-Seq analysis with export to IPA (Fig. 1). All reads were batch processed and mapped to the Mus musculus reference genome. Control vs. PB + CPF + DEET samples were quantified using the Differential Analysis for RNA-Seq tool. Differentially expressed genes were considered significant if they met the following criteria: mean reads per kilobase of transcript per million mapped reads (RPKM) > 10.0, fold change in either direction ≥1.2, and p < 0.1. Gene ontology (GO) categories were assigned and analyzed for significance for biological processes, molecular functions, and cellular components using the Gene Set Test tool. GO categories were considered significant if fold change in either direction ≥1.2 and p < 0.05. Significant genes were exported to IPA.
Fig. 1.
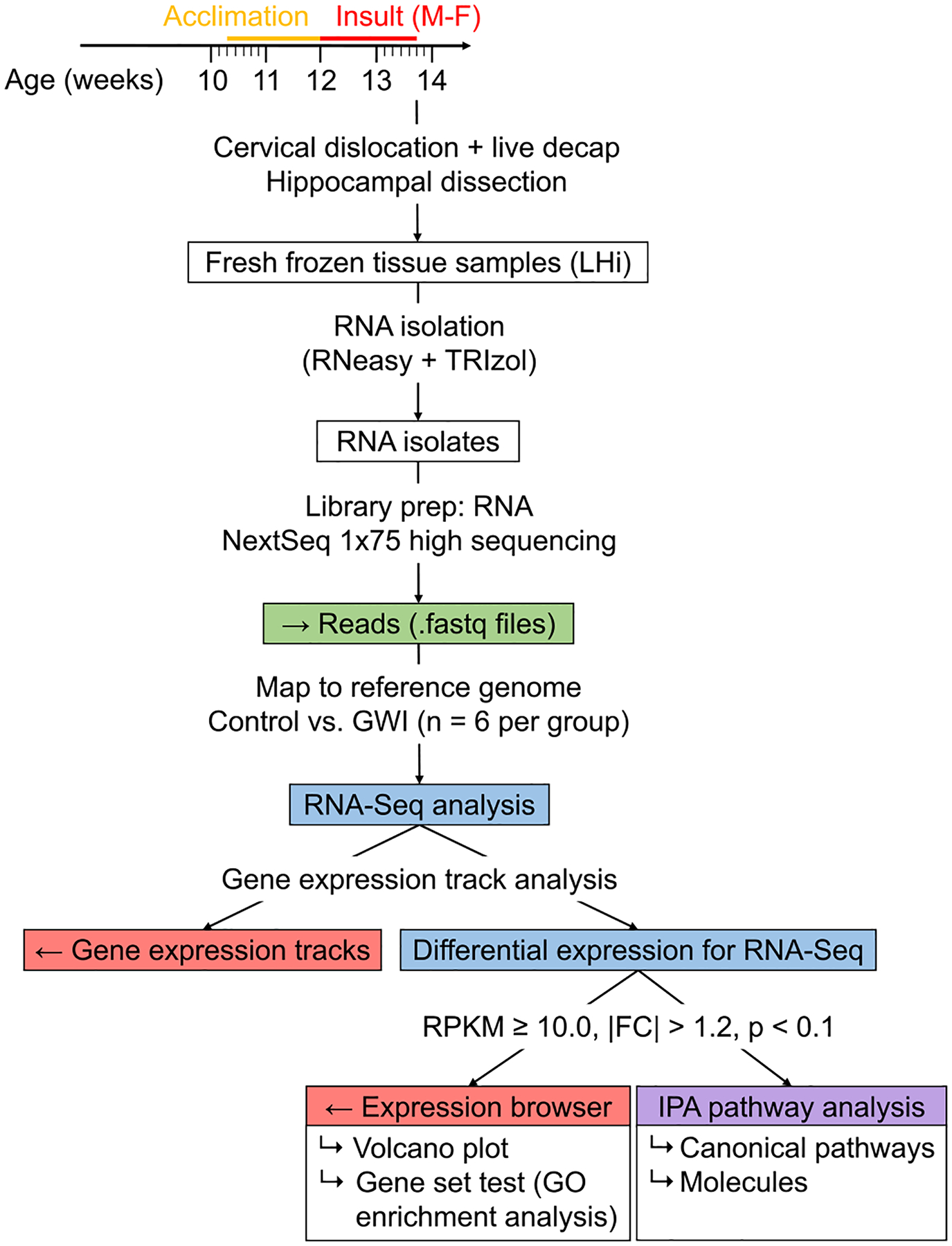
RNA-Seq analysis workflow with CLC Genomics Workbench and Ingenuity Pathway Analysis. Whole transcriptome sequencing was performed using mouse hippocampal RNA isolates collected 2–4 h after final exposure. Gene expression tracks were analyzed using the Differential Expression for RNA-Seq tool with RPKM >10.0, |FC| ≥ 1.2, and p < 0.1 as criteria for significance. GO enrichment analysis was performed on subset of genes that were significantly dysregulated. Data for significant genes was exported to Ingenuity Pathway Analysis to assess canonical pathways, molecules, diseases and functions, and other relevant information.
Functional analyses were generated using Ingenuity Pathway Analysis (IPA) (QIAGEN). Core analysis was performed on dataset based on RPKM values for genes that met criteria for significance, which generated lists of significant canonical pathways, upstream regulators, associated diseases and functions, and differentially expressed genes. Canonical pathways were based on significant differentially expressed genes, and a pathway itself was considered significant if p < 0.05.
2.8. Statistics
All statistical analyses for behavior were conducted using GraphPad Prism for macOS (version 9.0.0). Mean values for behavioral analyses are depicted ± standard error of the mean (SEM). Data for open field and Y-maze tasks were analyzed using an unpaired t-test with Welch’s correction, and statistical significance was considered when p < 0.05. Entries into each arm during the Y-maze task were analyzed using multiple unpaired t-tests followed by FDR control with the two-stage step-up method of Benjamini, Krieger, and Yekutieli as recommended by GraphPad. Significant fold changes in RNA expression were analyzed by CLC Genomics Workbench using Differential Expression for RNA-Seq as part of the workflow as detailed in Fig. 1.
3. Results
3.1. Effects of Gulf War toxicant exposure on hippocampal-dependent spatial memory in Y-maze task
To assess effects of the exposure on hippocampal-dependent spatial memory, mice underwent a Y-maze task (n = 6/group). Time spent in each arm, number of entries into each arm, and distance travelled were recorded. Preference for the novel arm was significantly lower by 157% in mice exposed to PB + CPF + DEET compared to controls, p = 0.027 (Fig. 2a). The number of entries into the novel arm was also significantly reduced by 33% compared to control mice, p = 0.003 (Fig. 2b). Distance travelled during the test stage was only 12% lower in toxicant-exposed mice compared to controls and therefore did not significantly differ between conditions, p = 0.182 (Fig. 2c).
Fig. 2.
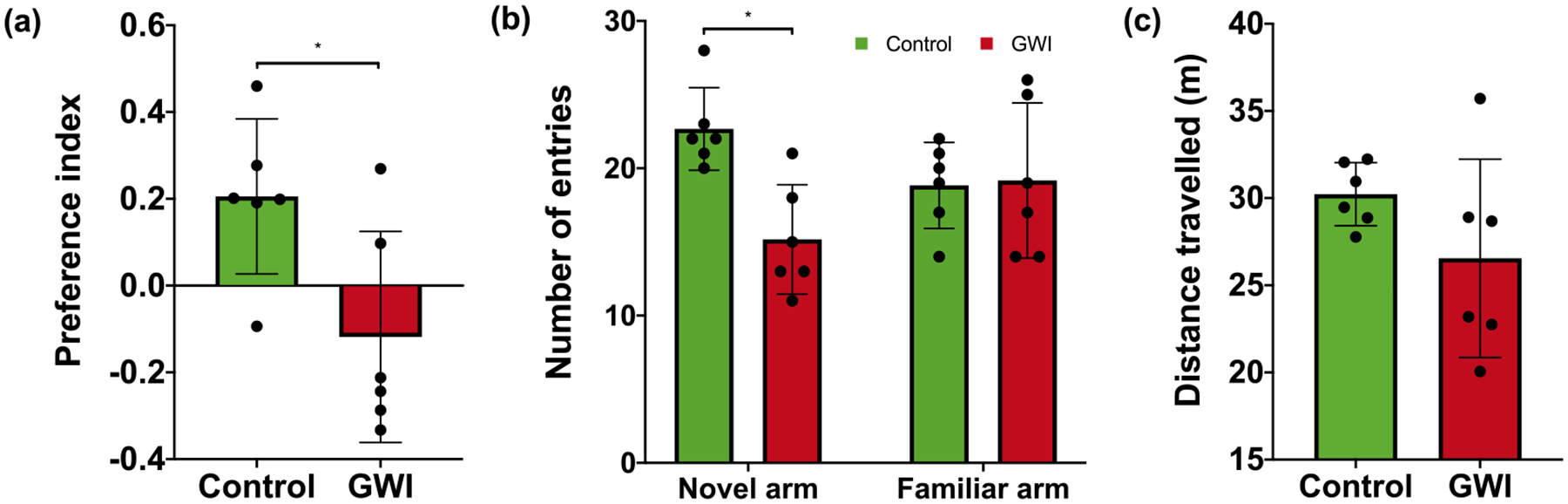
(a) Preference for novel arm, (b) number of entries per arm, and (c) distance travelled during trial phase of Y-maze. Hippocampal-dependent spatial memory was assessed by performance on a Y-maze task 2–4 h after final exposure. (a) Preference for the novel arm was significantly lower in mice receiving PB + CPF + DEET (mean = −0.12 ± 0.099) compared to control mice (mean = 0.21 ± 0.073) (t(9.18) = 2.63, p = 0.027). (b) The number of entries into the novel arm was also significantly lower in mice exposed to PB + CPF + DEET (mean = 15.2 ± 1.15) compared to controls (mean = 22.7 ± 1.52) (t(9.31) = 3.95, p = 0.0031). (c) Distance travelled during the test stage did not significantly differ between conditions (PB + CPF + DEET: mean = 26.6 ± 2.32, control: mean = 30.2 ± 0.74, t(6.00) = 1.51, p = 0.18). All results are graphed as mean ± SEM.
3.2. Gene dysregulation after acute exposure to Gulf War toxicants
In the hippocampus, 158 dysregulated genes were identified with the aid of RNA-Seq analysis which met criteria for differential expression in response to Gulf War toxicant exposure (Fig. 3, Tables 1 and 2). A gene set test (GO enrichment analysis) in CLC Genomics Workbench showed significantly affected gene ontology (GO) categories. Of these categories, 47 were related to biological processes (Table 4a), 138 were related to molecular functions (Table 4b), and 120 were related to cellular components (Table 4c). Pathway analysis in IPA showed 45 significantly affected canonical pathways (Table 3).
Fig. 3.
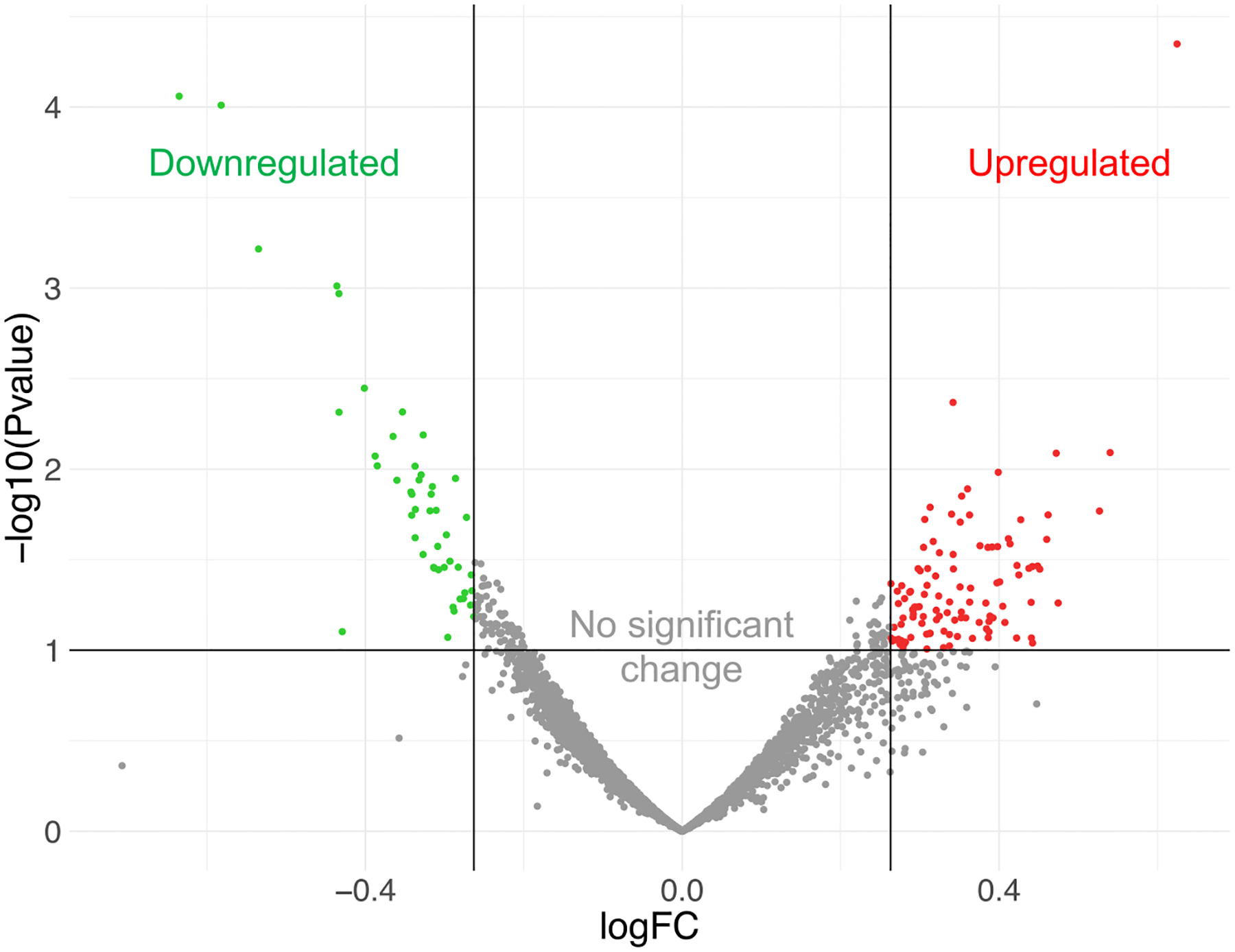
Differentially expressed genes identified by RNA-Seq analysis. Sequence counts from the RNA samples were evaluated with CLC Genomics Workbench and Ingenuity Pathway Analysis software. 158 dysregulated genes were identified in mice exposed to PB + CPF + DEET vs. controls. Genes were considered to be significantly dysregulated if they met the following criteria: RPKM >10.0, |fold change| ≥ 1.2, p < 0.1. Green = downregulated; red = upregulated.
Table 1.
Downregulated genes after exposure to Gulf War insult. Green, negative fold changes indicate downregulation.
| Symbol | Entrez Gene Name | RPKM | FC | P-value |
|---|---|---|---|---|
| Arc | Activity regulated cytoskeleton associated protein | 33.6 | −1.553 | 8.72E-05 |
| Egr1 | Early growth response 1 | 23.3 | −1.497 | 9.77E-05 |
| Nr4a1 | Nuclear receptor subfamily 4 group A member 1 | 16.5 | −1.449 | 0.000608 |
| Apod | Apolipoprotein D | 32.5 | −1.353 | 0.000973 |
| Hba-a2 | Hemoglobin alpha, adult chain 2 | 60.7 | −1.350 | 0.00485 |
| Tmem88b | Transmembrane protein 88B | 16.5 | −1.350 | 0.00107 |
| Wfs1 | Wolframin ER transmembrane glycoprotein | 33.2 | −1.321 | 0.00357 |
| Junb | JunB proto-oncogene, AP-1 transcription factor subunit | 36.6 | −1.308 | 0.00847 |
| Fam163b | Family with sequence similarity 163 member B | 52.3 | −1.306 | 0.00959 |
| Mog | Myelin oligodendrocyte glycoprotein | 22.7 | −1.288 | 0.00659 |
| Mbp | Myelin basic protein | 281.4 | −1.284 | 0.0115 |
| Bcas1 | Breast carcinoma amplified sequence 1 | 41.5 | −1.277 | 0.00483 |
| Cd9 | CD9 molecule | 29.2 | −1.268 | 0.0134 |
| Gsn | Gelsolin | 15.9 | −1.267 | 0.0138 |
| Pllp | Plasmolipin | 21.3 | −1.267 | 0.018 |
| Mag | Myelin associated glycoprotein | 50.6 | −1.263 | 0.00963 |
| Nutf2-ps1 | Nuclear transport factor 2, pseudogene 1 | 19.1 | −1.263 | 0.0167 |
| Pcp4l1 | Purkinje cell protein 4-like 1 | 26.7 | −1.263 | 0.0239 |
| H2-D1 | Histocompatibility 2, D region locus 1 | 11.8 | −1.259 | 0.0115 |
| Trf | Transferrin | 62.8 | −1.257 | 0.0107 |
| Rpl10-ps3 | Ribosomal protein L10, pseudogene 3 | 75.6 | −1.255 | 0.0296 |
| Plekhb1 | Pleckstrin homology domain containing B1 | 65.3 | −1.254 | 0.00647 |
| Srebf1 | Sterol regulatory element binding transcription factor 1 | 11.2 | −1.247 | 0.017 |
| Cnp | 2’,3’-cyclic nucleotide 3’ phosphodiesterase | 105.7 | −1.246 | 0.0137 |
| Septin4 | Septin 4 | 27.6 | −1.244 | 0.0125 |
| Slco1c1 | Solute carrier organic anion transporter family member 1C1 | 11.2 | −1.243 | 0.0349 |
| Pltp | Phospholipid transfer protein | 21.6 | −1.242 | 0.0352 |
| Cldn11 | Claudin 11 | 73.5 | −1.240 | 0.0169 |
| Fa2h | Fatty acid 2-hydroxylase | 11.0 | −1.239 | 0.0267 |
| Rhog | Ras homolog family member G | 12.3 | −1.238 | 0.0359 |
| Prr18 | Proline rich 18 | 17.0 | −1.229 | 0.0231 |
| Egr4 | Early growth response 4 | 16.8 | −1.228 | 0.0849 |
| mt-Atp8 | ATP synthase F0 subunit 8 | 7509.7 | −1.225 | 0.0323 |
| C1ql2 | Complement C1q like 2 | 40.4 | −1.222 | 0.0578 |
| Nfkbia | NFKB inhibitor alpha | 12.4 | −1.221 | 0.0608 |
| Igfbp5 | Insulin like growth factor binding protein 5 | 23.2 | −1.219 | 0.0112 |
| B2m | Beta-2-microglobulin | 57.4 | −1.217 | 0.0348 |
| Hbb-bs | Hemoglobin subunit beta | 45.2 | −1.215 | 0.0521 |
| S100a16 | S100 calcium binding protein A16 | 20.3 | −1.211 | 0.0518 |
| mt-Atp6 | ATP synthase F0 subunit 6 | 8936.0 | −1.210 | 0.0481 |
| Slc6a6 | Solute carrier family 6 member 6 | 15.6 | −1.208 | 0.0185 |
| Ddit4 | DNA damage inducible transcript 4 | 38.8 | −1.204 | 0.0563 |
| Anxa5 | Annexin A5 | 15.5 | −1.203 | 0.0383 |
| S100a1 | S100 calcium binding protein A1 | 35.3 | −1.202 | 0.0469 |
| Chrm3 | Cholinergic receptor muscarinic 3 | 10.5 | −1.200 | 0.0652 |
Table 2.
Upregulated genes after exposure to Gulf War insult. Red, positive fold changes indicate upregulation.
| Symbol | Entrez Gene Name | RPKM | FC | P-value |
|---|---|---|---|---|
| Lars2 | Leucyl-tRNA synthetase 2, mitochondrial | 744.328 | 1.542 | 4.48E-05 |
| Gdf1 | Growth differentiation factor 1 | 11.348 | 1.454 | 0.0081 |
| Cdr1 | Cerebellar degeneration related antigen 1 | 23.178 | 1.441 | 0.017 |
| Fam126b | Family with sequence similarity 126 member B | 12.032 | 1.390 | 0.0548 |
| Pak3 | p21 (RAC1) activated kinase 3 | 14.277 | 1.387 | 0.00816 |
| Igip | IgA inducing protein | 31.362 | 1.377 | 0.0179 |
| Pgm2l1 | Phosphoglucomutase 2 like 1 | 45.308 | 1.376 | 0.0244 |
| Smc3 | Structural maintenance of chromosomes 3 | 11.216 | 1.367 | 0.0357 |
| Dgkb | Diacylglycerol kinase beta | 20.952 | 1.365 | 0.0343 |
| Atrx | ATRX chromatin remodeler | 10.147 | 1.359 | 0.0912 |
| Ppp4r2 | Protein phosphatase 4 regulatory subunit 2 | 14.623 | 1.359 | 0.0345 |
| Ankrd12 | Ankyrin repeat domain 12 | 10.712 | 1.357 | 0.0856 |
| Hspa4l | Heat shock protein family A (Hsp70) member 4 like | 12.354 | 1.357 | 0.0544 |
| Ppig | Peptidylprolyl isomerase G | 11.563 | 1.354 | 0.0353 |
| Rabep1 | Rabaptin, RAB GTPase binding effector protein 1 | 16.266 | 1.345 | 0.019 |
| Dnajb4 | DnaJ heat shock protein family (Hsp40) member B4 | 11.871 | 1.343 | 0.0384 |
| Pcmtd1 | Protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 | 16.76 | 1.340 | 0.0856 |
| Reps2 | RALBP1 associated Eps domain containing 2 | 21.405 | 1.340 | 0.034 |
| Ube2q2 | Ubiquitin conjugating enzyme E2 Q2 | 13.79 | 1.332 | 0.0259 |
| Rab3c | RAB3C, member RAS oncogene family | 44.551 | 1.330 | 0.0242 |
| Acbd5 | Acyl-CoA binding domain containing 5 | 11.406 | 1.326 | 0.0703 |
| Fmr1 | FMRP translational regulator 1 | 11.502 | 1.324 | 0.0571 |
| Tax1bp1 | Tax1 binding protein 1 | 22.074 | 1.320 | 0.0419 |
| Nus1 | NUS1 dehydrodolichyl diphosphate synthase subunit | 16.346 | 1.319 | 0.0104 |
| Hsp90aa1 | Heat shock protein 90 alpha family class A member 1 | 213.841 | 1.318 | 0.0268 |
| Gmfb | Glia maturation factor beta | 35.439 | 1.317 | 0.0425 |
| Gpbp1 | GC-rich promoter binding protein 1 | 17.787 | 1.313 | 0.0663 |
| Naa50 | N(alpha)-acetyltransferase 50, NatE catalytic subunit | 16.618 | 1.312 | 0.0269 |
| Gabra2 | Gamma-aminobutyric acid type A receptor alpha2 subunit | 36.465 | 1.309 | 0.0647 |
| Fxr1 | FMR1 autosomal homolog 1 | 11.598 | 1.308 | 0.0792 |
| Kpna3 | Karyopherin subunit alpha 3 | 16.133 | 1.308 | 0.0695 |
| Ipo7 | Importin 7 | 17.7 | 1.307 | 0.0853 |
| Mphosph8 | M-phase phosphoprotein 8 | 15.901 | 1.307 | 0.027 |
| Kif5b | Kinesin family member 5B | 33.739 | 1.305 | 0.0762 |
| Psd3 | Pleckstrin and Sec7 domain containing 3 | 26.285 | 1.304 | 0.0549 |
| Pde1a | Phosphodiesterase 1A | 26.28 | 1.298 | 0.0265 |
| Mob4 | MOB family member 4, phocein | 14.856 | 1.297 | 0.0703 |
| Uba3 | Ubiquitin like modifier activating enzyme 3 | 13.952 | 1.289 | 0.086 |
| Slc8a1 | Solute carrier family 8 member A1 | 11.848 | 1.287 | 0.0454 |
| Ankrd13c | Ankyrin repeat domain 13C | 19.116 | 1.286 | 0.0179 |
| Pten | Phosphatase and tensin homolog | 20.14 | 1.286 | 0.0542 |
| Eif3a | Eukaryotic translation initiation factor 3 subunit A | 27.913 | 1.284 | 0.0128 |
| Gabrb1 | Gamma-aminobutyric acid type A receptor beta1 subunit | 13.845 | 1.282 | 0.0663 |
| Ogfrl1 | Opioid growth factor receptor like 1 | 36.947 | 1.277 | 0.0141 |
| Selenot | Selenoprotein T | 44.851 | 1.277 | 0.0616 |
| Eif5 | Eukaryotic translation initiation factor 5 | 34.859 | 1.276 | 0.0662 |
| Htatsf1 | HIV-1 Tat specific factor 1 | 18.843 | 1.275 | 0.0447 |
| Top1 | DNA topoisomerase I | 22.617 | 1.275 | 0.0196 |
| Slc25a46 | Solute carrier family 25 member 46 | 11.765 | 1.272 | 0.084 |
| Nrxn1 | Neurexin 1 | 30.941 | 1.269 | 0.0682 |
| Gad2 | Glutamate decarboxylase 2 | 17.309 | 1.268 | 0.0356 |
| Fgfr1op2 | FGFR1 oncogene partner 2 | 25.756 | 1.267 | 0.0296 |
| Hspa5 | Heat shock protein family A (Hsp70) member 5 | 46.918 | 1.267 | 0.00428 |
| Zc3h15 | Zinc finger CCCH-type containing 15 | 34.721 | 1.266 | 0.0177 |
| Armcx3 | Armadillo repeat containing X-linked 3 | 22 | 1.264 | 0.0541 |
| Hnrnpa3 | Heterogeneous nuclear ribonucleoprotein A3 | 29.364 | 1.263 | 0.0946 |
| Senp6 | SUMO specific peptidase 6 | 10.207 | 1.263 | 0.0819 |
| Fbxo11 | F-box protein 11 | 23.116 | 1.261 | 0.062 |
| Cert1 | Ceramide transporter 1 | 11.829 | 1.257 | 0.0968 |
| Oxr1 | Oxidation resistance 1 | 23.222 | 1.257 | 0.0785 |
| Impact | Impact RWD domain protein | 38.33 | 1.252 | 0.0648 |
| Psip1 | PC4 and SFRS1 interacting protein 1 | 32.895 | 1.252 | 0.0289 |
| Slmap | Sarcolemma associated protein | 13.2 | 1.252 | 0.0502 |
| Fgf12 | Fibroblast growth factor 12 | 10.635 | 1.249 | 0.0679 |
| Sucla2 | Succinate-CoA ligase ADP-forming beta subunit | 33.008 | 1.249 | 0.0601 |
| Dld | Dihydrolipoamide dehydrogenase | 28.74 | 1.248 | 0.0389 |
| Negr1 | Neuronal growth regulator 1 | 18.551 | 1.246 | 0.0251 |
| Acsl4 | Acyl-CoA synthetase long chain family member 4 | 13.462 | 1.242 | 0.0806 |
| Dnaja1 | DnaJ heat shock protein family (Hsp40) member A1 | 37.903 | 1.242 | 0.0162 |
| Pnrc2 | Proline rich nuclear receptor coactivator 2 | 13.435 | 1.242 | 0.0808 |
| Eif5b | Eukaryotic translation initiation factor 5B | 11.453 | 1.240 | 0.0354 |
| Mib1 | Mindbomb E3 ubiquitin protein ligase 1 | 15.309 | 1.239 | 0.0985 |
| Plcb1 | Phospholipase C beta 1 | 19.494 | 1.239 | 0.0438 |
| Map9 | Microtubule associated protein 9 | 15.383 | 1.238 | 0.0815 |
| Jakmip2 | Janus kinase and microtubule interacting protein 2 | 11.357 | 1.236 | 0.0491 |
| Pura | Purine rich element binding protein A | 19.084 | 1.236 | 0.019 |
| Hsp90b1 | Heat shock protein 90 beta family member 1 | 65.468 | 1.235 | 0.027 |
| Ncl | Nucleolin | 18.087 | 1.235 | 0.0652 |
| Neto1 | Neuropilin and tolloid like 1 | 16.105 | 1.233 | 0.0711 |
| Gda | Guanine deaminase | 30.573 | 1.232 | 0.0364 |
| Cnr1 | Cannabinoid receptor 1 | 25.574 | 1.231 | 0.0575 |
| Bhlhb9 | Basic helix-loop-helix family member b9 | 16.07 | 1.229 | 0.0355 |
| Ythdc1 | YTH domain containing 1 | 13.542 | 1.228 | 0.0578 |
| Golga4 | Golgin A4 | 10.017 | 1.226 | 0.0576 |
| Cir1 | Corepressor interacting with RBPJ, 1 | 10.903 | 1.224 | 0.0657 |
| Mzt1 | Mitotic spindle organizing protein 1 | 27.367 | 1.224 | 0.0631 |
| Rnf6 | Ring finger protein 6 | 10.488 | 1.224 | 0.0599 |
| Gdap1 | Ganglioside induced differentiation associated protein 1 | 20.359 | 1.223 | 0.0596 |
| Lpgat1 | Lysophosphatidylglycerol acyltransferase 1 | 20.943 | 1.221 | 0.0473 |
| Pin4 | Peptidylprolyl cis/trans isomerase, NIMA-interacting 4 | 14.967 | 1.221 | 0.085 |
| Cpne7 | Copine 7 | 93.847 | 1.220 | 0.0478 |
| Ggnbp2 | Gametogenetin binding protein 2 | 18.355 | 1.216 | 0.0902 |
| Etv1 | ETS variant transcription factor 1 | 14.066 | 1.215 | 0.0518 |
| Arl5a | ADP ribosylation factor like GTPase 5A | 15.424 | 1.214 | 0.0924 |
| Pafah1b1 | Platelet activating factor acetylhydrolase 1b regulatory subunit 1 | 59.969 | 1.213 | 0.0964 |
| Tafa1 | TAFA chemokine like family member 1 | 11.298 | 1.213 | 0.0663 |
| Srsf3 | Serine and arginine rich splicing factor 3 | 27.672 | 1.212 | 0.044 |
| Tceal9 | Transcription elongation factor A like 9 | 29.46 | 1.212 | 0.0939 |
| Ccdc47 | Coiled-coil domain containing 47 | 14.385 | 1.211 | 0.0885 |
| Tim2 | Tripartite motif containing 2 | 46.277 | 1.211 | 0.072 |
| Aff4 | AF4/FMR2 family member 4 | 15.746 | 1.210 | 0.093 |
| C5orf24 | Chromosome 5 open reading frame 24 | 18.903 | 1.210 | 0.0917 |
| Msantd4 | Myb/SANT DNA binding domain containing 4 with coiled-coils | 14.084 | 1.208 | 0.0552 |
| Rab39b | RAB39B, member RAS oncogene family | 10.444 | 1.208 | 0.087 |
| Vxn | Vexin | 56.093 | 1.207 | 0.0472 |
| Tmem33 | Transmembrane protein 33 | 10.859 | 1.205 | 0.0876 |
| Slk | STE20 like kinase | 10.826 | 1.204 | 0.0747 |
| Hdgfl3 | HDGF like 3 | 11.325 | 1.202 | 0.0891 |
| Dynlt3 | Dynein light chain Tctex-type 3 | 39.421 | 1.201 | 0.0859 |
| Dyrk2 | Dual specificity tyrosine phosphorylation regulated kinase 2 | 10.698 | 1.200 | 0.0429 |
Table 4a.
Subset of significantly affected gene ontology categories involved in biological processes. Green, negative fold changes indicate downregulation. Red, positive fold changes indicate upregulation.
| GO term | Description | Detected Genes | DE Genes | DE Genes (Names) | P-values |
|---|---|---|---|---|---|
| 0110077 | Vesicle-mediated intercellular transport | 1 | 1 | Arc | 2.89E-4 |
| 0006429 | Leucyl-tRNA aminoacylation | 2 | 1 | Lars2 | 5.78E-4 |
| 0050767 | Regulation of neurogenesis | 934 | 3 | Arc, Gh, Opalin | 1.45E-3 |
| 0006518 | Peptide metabolic process | 262 | 2 | Hmgn5, Lars2 | 2.22E-3 |
| 0090031 | Positive regulation of steroid hormone biosynthetic process | 9 | 1 | Gh | 2.60E-3 |
| 2000969 | Positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity | 9 | 1 | Arc | 2.60E-3 |
| 0032094 | Response to food | 12 | 1 | Gh | 3.47E-3 |
| 0043603 | Cellular amide metabolic process | 392 | 2 | Hmgn5, Lars2 | 4.90E-3 |
| 1900452 | Regulation of long-term synaptic depression | 17 | 1 | Arc | 4.91E-3 |
| 0007405 | Neuroblast proliferation | 19 | 1 | Gh | 5.48E-3 |
| 0099149 | Regulation of postsynaptic neurotransmitter receptor internalization | 23 | 1 | Arc | 6.64E-3 |
| 0032543 | Mitochondrial translation | 31 | 1 | Lars2 | 8.94E-3 |
| 0007616 | Long-term memory | 39 | 1 | Arc | 0.0112 |
| 0007492 | Endoderm development | 40 | 1 | Arc | 0.0115 |
| 0040018 | Positive regulation of multicellular organism growth | 42 | 1 | Gh | 0.0121 |
| 0072089 | Stem cell proliferation | 45 | 1 | Gh | 0.0129 |
| 0048286 | Lung alveolus development | 49 | 1 | Gh | 0.0141 |
| 0048713 | Regulation of oligodendrocyte differentiation | 49 | 1 | Opalin | 0.0141 |
| 0010828 | Positive regulation of glucose transport | 50 | 1 | Gh | 0.0144 |
| 0051028 | mRNA transport | 52 | 1 | Arc | 0.0149 |
| 1900271 | Regulation of long-term synaptic potentiation | 54 | 1 | Arc | 0.0155 |
| 0006749 | Glutathione metabolic process | 55 | 1 | Hmgn5 | 0.0158 |
| 0061001 | Regulation of dendritic spine morphogenesis | 56 | 1 | Arc | 0.0161 |
| 0099601 | Regulation of neurotransmitter receptor activity | 60 | 1 | Arc | 0.0172 |
| 0061351 | Neural precursor cell proliferation | 63 | 1 | Gh | 0.0181 |
| 0048168 | Regulation of neuronal synaptic plasticity | 69 | 1 | Arc | 0.0198 |
| 0045685 | Regulation of glial cell differentiation | 86 | 1 | Opalin | 0.0246 |
| 0032869 | Cellular response to insulin stimulus | 87 | 1 | Gh | 0.0249 |
| 0046889 | Positive regulation of lipid biosynthetic process | 94 | 1 | Gh | 0.0269 |
| 0060998 | Regulation of dendritic spine development | 97 | 1 | Arc | 0.0277 |
| 0032414 | Positive regulation of ion transmembrane transporter activity | 114 | 1 | Arc | 0.0325 |
| 0051260 | Protein homooligomerization | 116 | 1 | Arc | 0.0331 |
| 0071375 | Cellular response to peptide hormone stimulus | 119 | 1 | Gh | 0.0340 |
| 0006575 | Cellular modified amino acid metabolic process | 138 | 1 | Hmgn5 | 0.0393 |
| 0014013 | Regulation of gliogenesis | 148 | 1 | Opalin | 0.0421 |
| 1901564 | Organonitrogen compound metabolic process | 1208 | 2 | Hmgn5, Lars2 | 0.0423 |
| 0010469 | Regulation of receptor activity | 156 | 1 | Arc | 0.0443 |
| 0009952 | Anterior/posterior pattern specification | 170 | 1 | Arc | 0.0482 |
| 0043604 | Amide biosynthetic process | 214 | 1 | Lars2 | 0.0604 |
| 0043933 | Macromolecular complex subunit organization | 1504 | 2 | Arc, Hmgn5 | 0.0633 |
| 1901215 | Negative regulation of neuron death | 244 | 1 | Gh | 0.0686 |
| 0032412 | Regulation of ion transmembrane transporter activity | 249 | 1 | Arc | 0.0700 |
| 0006790 | Sulfur compound metabolic process | 271 | 1 | Hmgn5 | 0.0760 |
| 0009416 | Response to light stimulus | 288 | 1 | Gh | 0.0806 |
| 0050890 | Cognition | 325 | 1 | Arc | 0.0905 |
| 0010769 | Regulation of cell morphogenesis involved in differentiation | 344 | 1 | Arc | 0.0956 |
| 0007005 | Mitochondrion organization | 345 | 1 | Lars2 | 0.0959 |
| 0071417 | 9 Cellular response to organonitrogen compound |
347 | 1 | Gh | 0.0964 |
Table 4b.
Subset of significantly affected gene ontology categories involved in molecular functions. Green, negative fold changes indicate downregulation. Red, positive fold changes indicate upregulation.
| GO term | Description | Detected Genes | DE Genes | DE Genes (Names) | P-values |
|---|---|---|---|---|---|
| 0033592 | RNA strand annealing activity | 3 | 2 | Fmr1, Fxr1 | 2.18E-04 |
| 0097100 | Supercoiled DNA binding | 3 | 2 | Psip1, Top1 | 2.18E-04 |
| 0070840 | Dynein complex binding | 21 | 3 | Fmr1, Pafah1b1, Smc3 | 7.32E-04 |
| 0051082 | Unfolded protein binding | 61 | 4 | Dnajb4, Hsp90aa1, Hsp90b1, Hspa5 | 1.85E-03 |
| 0002151 | G-quadruplex RNA binding | 9 | 2 | Fmr1, Fxr1 | 2.52E-03 |
| 0062061 | TAP complex binding | 9 | 2 | H2-D1, H2-K1 | 2.52E-03 |
| 0031720 | Haptoglobin binding | 9 | 2 | Hba-a2, Hbb-bs | 2.52E-03 |
| 0019911 | Structural constituent of myelin sheath | 10 | 2 | Mbp, Pllp | 3.14E-03 |
| 0030881 | Beta-2-microglobulin binding | 11 | 2 | H2-D1, H2-K1 | 3.81E-03 |
| 0042610 | CD8 receptor binding | 11 | 2 | H2-D1, H2-K1 | 3.81E-03 |
| 0046977 | TAP binding | 11 | 2 | H2-D1, H2-K1 | 3.81E-03 |
| 0003743 | Translation initiation factor activity | 38 | 3 | Eif3a, Eif5, Eif5b | 4.18E-03 |
| 1990825 | Sequence-specific mRNA binding | 13 | 2 | Fmr1, Srsf3 | 5.35E-03 |
| 0022851 | GABA-gated chloride ion channel activity | 13 | 2 | Gabra2, Gabrb1 | 5.35E-03 |
| 0097001 | Ceramide binding | 14 | 2 | Mag, Pltp | 6.20E-03 |
| 0042608 | T cell receptor binding | 15 | 2 | H2-D1, H2-K1 | 7.12E-03 |
| 0004113 | 2’,3’-cyclic-nucleotide 3’-phosphodiesterase activity | 1 | 1 | Cnp | 8.57E-03 |
| 0004148 | Dihydrolipoyl dehydrogenase activity | 1 | 1 | Dld | 8.57E-03 |
| 0043544 | Lipoamide binding | 1 | 1 | Dld | 8.57E-03 |
| 0080132 | Fatty acid alpha-hydroxylase activity | 1 | 1 | Fa2h | 8.57E-03 |
| 0008892 | Guanine deaminase activity | 1 | 1 | Gda | 8.57E-03 |
| 0052858 | Peptidyl-lysine acetyltransferase activity | 1 | 1 | Naa50 | 8.57E-03 |
| 1990631 | ErbB-4 class receptor binding | 1 | 1 | Ncl | 8.57E-03 |
| 0047933 | Glucose-1,6-bisphosphate synthase activity | 1 | 1 | Pgm2l1 | 8.57E-03 |
| 0140339 | Phosphatidylglycerol transfer activity | 1 | 1 | Pltp | 8.57E-03 |
| 0140340 | Cerebroside transfer activity | 1 | 1 | Pltp | 8.57E-03 |
| 0140337 | Diacylglyceride transfer activity | 1 | 1 | Pltp | 8.57E-03 |
| 0140338 | Sphingomyelin transfer activity | 1 | 1 | Pltp | 8.57E-03 |
| 0051717 | Inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity | 1 | 1 | Pten | 8.57E-03 |
| 0051800 | Phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity | 1 | 1 | Pten | 8.57E-03 |
| 0001761 | Beta-alanine transmembrane transporter activity | 1 | 1 | Slc6a6 | 8.57E-03 |
| 0005369 | Taurine:sodium symporter activity | 1 | 1 | Slc6a6 | 8.57E-03 |
| 0004890 | GABA-A receptor activity | 18 | 2 | Gabra2, Gabrb1 | 0.010198 |
| 0019825 | Oxygen binding | 18 | 2 | Hba-a2, Hbb-bs | 0.010198 |
| 0031489 | Myosin V binding | 20 | 2 | Rab39b, Rab3c | 0.012524 |
| 0043022 | Ribosome binding | 57 | 3 | Fmr1, Hspa5, Impact | 0.01287 |
| 0008139 | Nuclear localization sequence binding | 21 | 2 | Kpna3, Nfkbia | 0.013765 |
| 0005104 | Fibroblast growth factor receptor binding | 22 | 2 | Fgf12, Nrxn1 | 0.015057 |
| 0001671 | ATPase activator activity | 23 | 2 | Dnaja1, Dnajb4 | 0.0164 |
| 0004351 | Glutamate decarboxylase activity | 2 | 1 | Gad2 | 0.017065 |
| 0031722 | Hemoglobin beta binding | 2 | 1 | Hbb-bs | 0.017065 |
| 0002135 | CTP binding | 2 | 1 | Hsp90aa1 | 0.017065 |
| 0099609 | Microtubule lateral binding | 2 | 1 | Kif5b | 0.017065 |
| 0004823 | Leucine-tRNA ligase activity | 2 | 1 | Lars2 | 0.017065 |
| 0045547 | Dehydrodolichyl diphosphate synthase activity | 2 | 1 | Nus1 | 0.017065 |
| 0120019 | Phosphatidylcholine transfer activity | 2 | 1 | Pltp | 0.017065 |
| 0030977 | Taurine binding | 2 | 1 | Slc6a6 | 0.017065 |
| 0086038 | Calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential | 2 | 1 | Slc8a1 | 0.017065 |
| 0099580 | Ion antiporter activity involved in regulation of postsynaptic membrane potential | 2 | 1 | Slc8a1 | 0.017065 |
| 0032810 | Sterol response element binding | 2 | 1 | Srebf1 | 0.017065 |
| 0004775 | Succinate-CoA ligase (ADP-forming) activity | 2 | 1 | Sucla2 | 0.017065 |
| 0034986 | Iron chaperone activity | 2 | 1 | Trf | 0.017065 |
| 0019781 | NEDD8 activating enzyme activity | 2 | 1 | Uba3 | 0.017065 |
| 0048027 | mRNA 5’-UTR binding | 24 | 2 | Fmr1, Ncl | 0.017791 |
| 0044183 | Protein folding chaperone | 26 | 2 | Hsp90aa1, Hspa5 | 0.020718 |
| 0035064 | Methylated histone binding | 70 | 3 | Atrx, Fmr1, Mphosph8 | 0.022223 |
| 0048306 | Calcium-dependent protein binding | 70 | 3 | Nrxn1, S100a1, Wfs1 | 0.022223 |
| 0042605 | Peptide antigen binding | 27 | 2 | H2-D1, H2-K1 | 0.022251 |
| 0042165 | Neurotransmitter binding | 28 | 2 | Chrm3, Slc6a6 | 0.02383 |
| 0050750 | Low-density lipoprotein particle receptor binding | 28 | 2 | Dnaja1, Hsp90b1 | 0.02383 |
| 0008081 | Phosphoric diester hydrolase activity | 72 | 3 | Cnp, Pde1a, Plcb1 | 0.023917 |
| 0004949 | Cannabinoid receptor activity | 3 | 1 | Cnr1 | 0.025489 |
| 0044729 | Hemi-methylated DNA-binding | 3 | 1 | Egr1 | 0.025489 |
| 0051033 | RNA transmembrane transporter activity | 3 | 1 | Hnrnpa3 | 0.025489 |
| 0017098 | Sulfonylurea receptor binding | 3 | 1 | Hsp90aa1 | 0.025489 |
| 1905576 | Ganglioside GT1b binding | 3 | 1 | Mag | 0.025489 |
| 0042134 | rRNA primary transcript binding | 3 | 1 | Ncl | 0.025489 |
| 0004719 | Protein-L-isoaspartate (D-aspartate) O-methyltransferase activity | 3 | 1 | Pcmtd1 | 0.025489 |
| 0048101 | Calcium-and calmodulin-regulated 3’,5’-cyclic-GMP phosphodiesterase activity | 3 | 1 | Pde1a | 0.025489 |
| 0004117 | Calmodulin-dependent cyclic-nucleotide phosphodiesterase activity | 3 | 1 | Pde1a | 0.025489 |
| 0003681 | Bent DNA binding | 3 | 1 | Pin4 | 0.025489 |
| 0016314 | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity | 3 | 1 | Pten | 0.025489 |
| 0070139 | SUMO-specific endopeptidase activity | 3 | 1 | Senp6 | 0.025489 |
| 1905060 | Calcium:cation antiporter activity involved in regulation of postsynaptic cytosolic calcium ion concentration | 3 | 1 | Slc8a1 | 0.025489 |
| 0003917 | DNA topoisomerase type I activity | 3 | 1 | Top1 | 0.025489 |
| 0071074 | Eukaryotic initiation factor eIF2 binding | 4 | 1 | Eif5 | 0.033841 |
| 0031721 | Hemoglobin alpha binding | 4 | 1 | Hbb-bs | 0.033841 |
| 0032564 | dATP binding | 4 | 1 | Hsp90aa1 | 0.033841 |
| 0032551 | Pyrimidine ribonucleoside binding | 4 | 1 | Hsp90aa1 | 0.033841 |
| 0002134 | UTP binding | 4 | 1 | Hsp90aa1 | 0.033841 |
| 0044547 | DNA topoisomerase binding | 4 | 1 | Ncl | 0.033841 |
| 0097109 | Neuroligin family protein binding | 4 | 1 | Nrxn1 | 0.033841 |
| 0032422 | Purine-rich negative regulatory element binding | 4 | 1 | Pura | 0.033841 |
| 0015349 | Thyroid hormone transmembrane transporter activity | 4 | 1 | Slco1c1 | 0.033841 |
| 0042162 | Telomeric DNA binding | 34 | 2 | Ncl, Pura | 0.03421 |
| 0031369 | Translation initiation factor binding | 35 | 2 | Eif5, Fmr1 | 0.036084 |
| 0060590 | ATPase regulator activity | 37 | 2 | Dnaja1, Dnajb4 | 0.039946 |
| 0070087 | Chromo shadow domain binding | 5 | 1 | Atrx | 0.042122 |
| 0015616 | DNA translocase activity | 5 | 1 | Atrx | 0.042122 |
| 0055131 | C3HC4-type RING finger domain binding | 5 | 1 | Dnaja1 | 0.042122 |
| 0005131 | Growth hormone receptor binding | 5 | 1 | Gh | 0.042122 |
| 0051022 | Rho GDP-dissociation inhibitor binding | 5 | 1 | Hsp90aa1 | 0.042122 |
| 0005105 | Type 1 fibroblast growth factor receptor binding | 5 | 1 | Nrxn1 | 0.042122 |
| 0036033 | Mediator complex binding | 5 | 1 | Smc3 | 0.042122 |
| 0035255 | Ionotropic glutamate receptor binding | 39 | 2 | Neto1, Pten | 0.043957 |
| 0047676 | Arachidonate-CoA ligase activity | 6 | 1 | Acsl4 | 0.050333 |
| 0016907 | G-protein coupled acetylcholine receptor activity | 6 | 1 | Chrm3 | 0.050333 |
| 0034604 | Pyruvate dehydrogenase (NAD+) activity | 6 | 1 | Dld | 0.050333 |
| 0035368 | Selenocysteine insertion sequence binding | 6 | 1 | Ncl | 0.050333 |
| 0019992 | Diacylglycerol binding | 6 | 1 | Pltp | 0.050333 |
| 1990050 | Phosphatidic acid transporter activity | 6 | 1 | Pltp | 0.050333 |
| 1904121 | Phosphatidylethanolamine transporter activity | 6 | 1 | Pltp | 0.050333 |
| 0004791 | Thioredoxin-disulfide reductase activity | 6 | 1 | Selenot | 0.050333 |
| 0005332 | Gamma-aminobutyric acid:sodium symporter activity | 6 | 1 | Slc6a6 | 0.050333 |
| 0004601 | Peroxidase activity | 44 | 2 | Hba-a2, Hbb-bs | 0.054596 |
| 0099635 | Voltage-gated calcium channel activity involved in positive regulation of presynaptic cytosolic calcium levels | 7 | 1 | Cnr1 | 0.058473 |
| 0010385 | Double-stranded methylated DNA binding | 7 | 1 | Egr1 | 0.058473 |
| 0030911 | TPR domain binding | 7 | 1 | Hsp90aa1 | 0.058473 |
| 1905538 | Polysome binding | 7 | 1 | Impact | 0.058473 |
| 1904315 | Transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential | 47 | 2 | Gabra2, Gabrb1 | 0.061369 |
| 0061797 | pH-gated chloride channel activity | 48 | 2 | Gabra2, Gabrb1 | 0.063687 |
| 0030235 | Nitric-oxide synthase regulator activity | 8 | 1 | Hsp90aa1 | 0.066545 |
| 0031995 | Insulin-like growth factor II binding | 8 | 1 | Igfbp5 | 0.066545 |
| 0010997 | Anaphase-promoting complex binding | 8 | 1 | Pten | 0.066545 |
| 1990247 | N6-methyladenosine-containing RNA binding | 8 | 1 | Ythdc1 | 0.066545 |
| 0008028 | Monocarboxylic acid transmembrane transporter activity | 52 | 2 | Slc6a6, Slco1c1 | 0.073247 |
| 0031957 | Very long-chain fatty acid-CoA ligase activity | 9 | 1 | Acsl4 | 0.074547 |
| 0030957 | Tat protein binding | 9 | 1 | Dnaja1 | 0.074547 |
| 0034046 | Poly(G) binding | 9 | 1 | Fmr1 | 0.074547 |
| 0003691 | Double-stranded telomeric DNA binding | 9 | 1 | Pura | 0.074547 |
| 0005544 | Calcium-dependent phospholipid binding | 53 | 2 | Anxa5, Cpne7 | 0.075705 |
| 0035197 | siRNA binding | 10 | 1 | Fmr1 | 0.082482 |
| 0045159 | Myosin II binding | 10 | 1 | Gsn | 0.082482 |
| 0043208 | Glycosphingolipid binding | 10 | 1 | Mag | 0.082482 |
| 0008199 | Ferric iron binding | 10 | 1 | Trf | 0.082482 |
| 0005388 | Calcium-transporting ATPase activity | 11 | 1 | Anxa5 | 0.090349 |
| 0004143 | Diacylglycerol kinase activity | 11 | 1 | Dgkb | 0.090349 |
| 0016274 | Protein-arginine N-methyltransferase activity | 11 | 1 | Fbxo11 | 0.090349 |
| 0008503 | Benzodiazepine receptor activity | 11 | 1 | Gabra2 | 0.090349 |
| 0008429 | Phosphatidylethanolamine binding | 11 | 1 | Pltp | 0.090349 |
| 1901611 | Phosphatidylglycerol binding | 11 | 1 | Pltp | 0.090349 |
| 0005086 | ARF guanyl-nucleotide exchange factor activity | 11 | 1 | Psd3 | 0.090349 |
| 1990459 | Transferrin receptor binding | 11 | 1 | Trf | 0.090349 |
| 0019829 | Inorganic cation-transporting ATPase activity | 59 | 2 | Anxa5, mt-Atp6 | 0.09098 |
| 0042625 | ATPase coupled ion transmembrane transporter activity | 61 | 2 | Anxa5, mt-Atp6 | 0.096257 |
| 0000900 | Translation repressor activity, mRNA regulatory element binding | 12 | 1 | Pura | 0.098149 |
| 0044548 | S100 protein binding | 12 | 1 | S100a1 | 0.098149 |
| 0042910 | Xenobiotic transporter activity | 12 | 1 | Slc6a6 | 0.098149 |
Table 4c.
Subset of significantly affected gene ontology categories forming cellular components. Green, negative fold changes indicate downregulation. Red, positive fold changes indicate upregulation.
| GO term | Description | Total Genes | DE Genes | DE Genes (Names) | P-values |
|---|---|---|---|---|---|
| 0043218 | Compact myelin | 5 | 4 | Mag, Mbp, Pllp, Pmp22 | 2.639E-07 |
| 0043209 | Myelin sheath | 182 | 12 | Cldn11, Cnp, Dld, Gjc2, Gsn, Hsp90aa1, Hspa5, Mag, Mbp, Mog, Plcb1, Sucla2 | 2.427E-05 |
| 0035749 | Myelin sheath adaxonal region | 6 | 3 | Cnp, Mag, Pten | 6.830E-05 |
| 0000235 | Astral microtubule | 8 | 3 | Dynlt3, Map9, Pafah1b1 | 1.869E-04 |
| 0098982 | GABA-ergic synapse | 104 | 8 | Camk4, Cnr1, Gabra2, Gabrb1, Gabrd, Nrxn1, Plcb1, Slc6a6 | 1.951E-04 |
| 1990015 | Ensheathing process | 2 | 2 | Mag, Myoc | 2.329E-04 |
| 0097453 | Mesaxon | 2 | 2 | Mag, Myoc | 2.329E-04 |
| 0043197 | Dendritic spine | 181 | 10 | Akap5, Arc, Fmr1, Fxr1, Homer1, Lpar1, Mob4, Pten, Slc8a1, Syndig1 | 4.823E-04 |
| 0043198 | Dendritic shaft | 69 | 6 | Akap5, Hcn1, Homer1, Lpar1, Slc8a1, Syndig1 | 6.462E-04 |
| 0034663 | Endoplasmic reticulum chaperone complex | 12 | 3 | Hsp90b1, Hspa5, Sdf2l1 | 7.018E-04 |
| 0042824 | MHC class I peptide loading complex | 14 | 3 | B2m, H2-D1, H2-K1 | 1.135E-03 |
| 0005790 | Smooth endoplasmic reticulum | 31 | 4 | Dnajc3, Fmr1, Hsp90b1, Hspa5 | 1.214E-03 |
| 0043220 | Schmidt-Lanterman incisure | 15 | 3 | Mag, Myoc, Pten | 1.403E-03 |
| 1902737 | Dendritic filopodium | 5 | 2 | Fmr1, Fxr1 | 2.259E-03 |
| 0030139 | Endocytic vesicle | 154 | 8 | Gsn, Kif5b, Lpar1, Nrxn1, Rab8b, Rab9b, Rabep1, Trf | 2.573E-03 |
| 1990712 | HFE-transferrin receptor complex | 6 | 2 | B2m, Trf | 3.354E-03 |
| 0031415 | NatA complex | 6 | 2 | Naa15, Naa50 | 3.354E-03 |
| 0001651 | Dense fibrillar component | 6 | 2 | Ncl, Top1 | 3.354E-03 |
| 0042579 | Microbody | 127 | 7 | Acbd5, Acsl4, Crot, Idi1, Pex13, Pnpla8, Rab8b | 3.400E-03 |
| 0030670 | Phagocytic vesicle membrane | 21 | 3 | B2m, H2-D1, H2-K1 | 3.832E-03 |
| 0005797 | Golgi medial cisterna | 23 | 3 | H2-D1, H2-K1, Yipf6 | 4.990E-03 |
| 0060076 | Excitatory synapse | 46 | 4 | Akap5, Homer1, Neto1, Syndig1 | 5.266E-03 |
| 0060077 | Inhibitory synapse | 24 | 3 | Gabra2, Gad2, Nrxn1 | 5.639E-03 |
| 0005844 | Polysome | 47 | 4 | Fmr1, Fxr1, Impact, Upf2 | 5.688E-03 |
| 0030666 | Endocytic vesicle membrane | 26 | 3 | B2m, H2-D1, H2-K1 | 7.083E-03 |
| 0099524 | Postsynaptic cytosol | 26 | 3 | Fmr1, Homer1, Pten | 7.083E-03 |
| 0005876 | Spindle microtubule | 51 | 4 | Bod1l, Dynlt3, Map9, Pafah1b1 | 7.600E-03 |
| 0035748 | Myelin sheath abaxonal region | 9 | 2 | Cnp, Myoc | 7.809E-03 |
| 0044326 | Dendritic spine neck | 9 | 2 | Fmr1, Fxr1 | 7.809E-03 |
| 0005833 | Hemoglobin complex | 9 | 2 | Hba-a2, Hbb-bs | 7.809E-03 |
| 0051286 | Cell tip | 10 | 2 | Rab8b, Trf | 9.663E-03 |
| 0005777 | Peroxisome | 119 | 6 | Acbd5, Acsl4, Crot, Idi1, Pex13, Pnpla8 | 9.958E-03 |
| 0098845 | Postsynaptic endosome | 12 | 2 | Akap5, Arc | 0.0139 |
| 0009898 | Cytoplasmic side of plasma membrane | 61 | 4 | Akap5, G6pdx, Litaf, Pten | 0.0141 |
| 1990707 | Nuclear subtelomeric heterochromatin | 1 | 1 | Atrx | 0.0153 |
| 0030990 | Intraciliary transport particle | 1 | 1 | Dync2li1 | 0.0153 |
| 0005969 | Serine-pyruvate aminotransferase complex | 1 | 1 | Eea1 | 0.0153 |
| 0071540 | Eukaryotic translation initiation factor 3 complex, eIF3e | 1 | 1 | Eif3a | 0.0153 |
| 0016028 | Rhabdomere | 1 | 1 | Mertk | 0.0153 |
| 0034678 | Integrin alpha8-beta1 complex | 1 | 1 | Npnt | 0.0153 |
| 0005943 | Phosphatidylinositol 3-kinase complex, class IA | 1 | 1 | Pik3ca | 0.0153 |
| 0045239 | Tricarboxylic acid cycle enzyme complex | 13 | 2 | Dld, Sucla2 | 0.0163 |
| 1902711 | GABA-A receptor complex | 13 | 2 | Gabra2, Gabrb1 | 0.0163 |
| 0071556 | Integral component of lumenal side of endoplasmic reticulum membrane | 13 | 2 | H2-D1, H2-K1 | 0.0163 |
| 1990124 | Messenger ribonucleoprotein complex | 14 | 2 | Fmr1, Hnrnpa3 | 0.0188 |
| 0005778 | Peroxisomal membrane | 38 | 3 | Pex13, Pnpla8, Rab8b | 0.0201 |
| 0032590 | Dendrite membrane | 39 | 3 | Akap5, Gabra2, Hcn1 | 0.0215 |
| 0098839 | Postsynaptic density membrane | 39 | 3 | Arc, Neto1, Syndig1 | 0.0215 |
| 0099522 | Region of cytosol | 40 | 3 | Fmr1, Homer1, Pten | 0.0230 |
| 0005753 | Mitochondrial proton-transporting ATP synthase complex | 16 | 2 | mt-Atp6, mt-Atp8 | 0.0243 |
| 0099634 | Postsynaptic specialization membrane | 41 | 3 | Arc, Neto1, Syndig1 | 0.0246 |
| 0045178 | Basal part of cell | 17 | 2 | Cldn11, Trf | 0.0272 |
| 0033270 | Paranode region of axon | 17 | 2 | Gjc2, Mag | 0.0272 |
| 0055037 | Recycling endosome | 113 | 5 | Akap5, Avl9, Eea1, Mctp1, Trf | 0.0298 |
| 0032433 | Filopodium tip | 18 | 2 | Fmr1, Fzd3 | 0.0303 |
| 0030140 | Trans-Golgi network transport vesicle | 18 | 2 | Gopc, Rab8b | 0.0303 |
| 0072563 | Endothelial microparticle | 2 | 1 | Anxa5 | 0.0303 |
| 0043614 | Multi-eIF complex | 2 | 1 | Eif3a | 0.0303 |
| 0032998 | Fc-epsilon receptor I complex | 2 | 1 | Fcer1g | 0.0303 |
| 0061202 | Clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane | 2 | 1 | Gad2 | 0.0303 |
| 0097226 | Sperm mitochondrial sheath | 2 | 1 | Hsp90aa1 | 0.0303 |
| 0098560 | Cytoplasmic side of late endosome membrane | 2 | 1 | Litaf | 0.0303 |
| 0005818 | Aster | 2 | 1 | Map9 | 0.0303 |
| 1904423 | Dehydrodolichyl diphosphate synthase complex | 2 | 1 | Nus1 | 0.0303 |
| 0030426 | Growth cone | 197 | 7 | Cnr1, Fmr1, Fxr1, Hsp90aa1, Kif5b, Nrxn1, Pafah1b1 | 0.0321 |
| 0044449 | Contractile fiber part | 198 | 7 | Anxa5, Fxr1, Homer1, Jph1, Npnt, S100a1, Slc8a1 | 0.0328 |
| 0044295 | Axonal growth cone | 46 | 3 | Hsp90aa1, Kif5b, Nrxn1 | 0.0331 |
| 0090723 | Growth cone part | 19 | 2 | Fmr1, Pafah1b1 | 0.0336 |
| 0043034 | Costamere | 19 | 2 | Fxr1, Homer1 | 0.0336 |
| 0005922 | Connexin complex | 19 | 2 | Gjb1, Gjc2 | 0.0336 |
| 0043679 | Axon terminus | 121 | 5 | Anxa5, Chrm3, Fmr1, Hcn1, Slc8a1 | 0.0383 |
| 0045335 | Phagocytic vesicle | 83 | 4 | Gsn, Kif5b, Rab8b, Rab9b | 0.0384 |
| 0030018 | Z disc | 124 | 5 | Anxa5, Homer1, Jph1, S100a1, Slc8a1 | 0.0418 |
| 0099055 | Integral component of postsynaptic membrane | 167 | 6 | Chrm3, Gabra2, Gabrd, Neto1, Slc6a6, Slc8a1 | 0.0435 |
| 0005921 | Gap junction | 22 | 2 | Gjb1, Gjc2 | 0.0440 |
| 0098855 | HCN channel complex | 3 | 1 | Hcn1 | 0.0452 |
| 0097524 | Sperm plasma membrane | 3 | 1 | Hsp90aa1 | 0.0452 |
| 0014701 | Junctional sarcoplasmic reticulum membrane | 3 | 1 | Jph1 | 0.0452 |
| 0098559 | Cytoplasmic side of early endosome membrane | 3 | 1 | Litaf | 0.0452 |
| 0034457 | Mpp10 complex | 3 | 1 | Mphosph10 | 0.0452 |
| 1990415 | Pex17p-Pex14p docking complex | 3 | 1 | Pex13 | 0.0452 |
| 0042709 | Succinate-CoA ligase complex | 3 | 1 | Sucla2 | 0.0452 |
| 0035327 | Transcriptionally active chromatin | 23 | 2 | Aff4, Psip1 | 0.0477 |
| 0031307 | Integral component of mitochondrial outer membrane | 24 | 2 | Armcx3, Gdap1 | 0.0515 |
| 0032279 | Asymmetric synapse | 25 | 2 | Akap5, Chrm3 | 0.0555 |
| 0005868 | Cytoplasmic dynein complex | 25 | 2 | Dync2li1, Dynlt3 | 0.0555 |
| 0032783 | ELL-EAF complex | 4 | 1 | Aff4 | 0.0598 |
| 0043159 | Acrosomal matrix | 4 | 1 | Dld | 0.0598 |
| 0044308 | Axonal spine | 4 | 1 | Eea1 | 0.0598 |
| 1990812 | Growth cone filopodium | 4 | 1 | Fmr1 | 0.0598 |
| 0097444 | Spine apparatus | 4 | 1 | Fmr1 | 0.0598 |
| 0019034 | Viral replication complex | 4 | 1 | Fmr1 | 0.0598 |
| 0030478 | Actin cap | 4 | 1 | Gsn | 0.0598 |
| 0042567 | Insulin-like growth factor ternary complex | 4 | 1 | Igfbp5 | 0.0598 |
| 0035976 | Transcription factor AP-1 complex | 4 | 1 | Junb | 0.0598 |
| 0098574 | Cytoplasmic side of lysosomal membrane | 4 | 1 | Litaf | 0.0598 |
| 0033269 | Internode region of axon | 4 | 1 | Mbp | 0.0598 |
| 0031021 | Interphase microtubule organizing center | 4 | 1 | Mzt1 | 0.0598 |
| 0030289 | Protein phosphatase 4 complex | 4 | 1 | Ppp4r2 | 0.0598 |
| 0008305 | Integrin complex | 28 | 2 | Npnt, Pmp22 | 0.0679 |
| 0098563 | Intrinsic component of synaptic vesicle membrane | 63 | 3 | Gabra2, Rab3c, Wfs1 | 0.0719 |
| 0070971 | Endoplasmic reticulum exit site | 29 | 2 | H2-D1, H2-K1 | 0.0722 |
| 0031256 | Leading edge membrane | 146 | 5 | Akap5, Gabra2, Hcn1, Hsp90aa1, Psd3 | 0.0737 |
| 0061673 | Mitotic spindle astral microtubule | 5 | 1 | Dynlt3 | 0.0741 |
| 0044094 | Host cell nuclear part | 5 | 1 | Fmr1 | 0.0741 |
| 1990769 | Proximal neuron projection | 5 | 1 | Gjc2 | 0.0741 |
| 0030485 | Smooth muscle contractile fiber | 5 | 1 | Npnt | 0.0741 |
| 0016586 | RSC complex | 5 | 1 | Pbrm1 | 0.0741 |
| 0034991 | Nuclear meiotic cohesin complex | 5 | 1 | Smc3 | 0.0741 |
| 0097433 | Dense body | 5 | 1 | Trf | 0.0741 |
| 0098984 | Neuron to neuron synapse | 30 | 2 | Akap5, Chrm3 | 0.0766 |
| 0030672 | Synaptic vesicle membrane | 66 | 3 | Gad2, Mctp1, Syndig1 | 0.0802 |
| 0005791 | Rough endoplasmic reticulum | 67 | 3 | Ccdc47, Clock, Fmr1 | 0.0830 |
| 0005726 | Perichromatin fibrils | 6 | 1 | Clock | 0.0883 |
| 0031466 | Cul5-RING ubiquitin ligase complex | 6 | 1 | Cul5 | 0.0883 |
| 0071598 | Neuronal ribonucleoprotein granule | 6 | 1 | Fmr1 | 0.0883 |
| 0008274 | Gamma-tubulin ring complex | 6 | 1 | Mzt1 | 0.0883 |
| 0090724 | Central region of growth cone | 6 | 1 | Pafah1b1 | 0.0883 |
| 0000932 | Cytoplasmic mRNA processing body | 72 | 3 | Dcp2, Pnrc2, Top1 | 0.0979 |
| 0032040 | Small-subunit processome | 35 | 2 | Krr1, Mphosph10 | 0.0997 |
Table 3.
Significantly affected canonical pathways after Gulf War insult. Green, negative fold changes indicate downregulation. Red, positive fold changes indicate upregulation.
| Ingenuity Canonical Pathways | −log(p-value) | Ratio | Molecules |
|---|---|---|---|
| Protein Ubiquitination Pathway | 4.08 | 0.033 | B2m, Dnaja1, Dnajb4, Hba-a2, Hsp90aa1, Hsp90b1, Hspa4l, Hspa5, Ube2q2 |
| Aldosterone Signaling in Epithelial Cells | 4.07 | 0.0443 | Dnaja1, Dnajb4, Hsp90aa1, Hsp90b1, Hspa4l, Hspa5, Plcb1 |
| Hypoxia Signaling in the Cardiovascular System | 3.9 | 0.0676 | Hsp90aa1, Hsp90b1, Nfkbia, Pten, Ube2q2 |
| Mitotic Roles of Polo-Like Kinase | 3.03 | 0.0606 | Hsp90aa1, Hsp90b1, Slk, Smc3 |
| Prostate Cancer Signaling | 2.51 | 0.044 | Hsp90aa1, Hsp90b1, Nfkbia, Pten |
| Unfolded protein response | 2.23 | 0.0536 | Hsp90b1, Hspa5, Srebf1 |
| Role of PKR in Interferon Induction and Antiviral Response | 2.13 | 0.0342 | Hsp90aa1, Hsp90b1, Hspa5, Nfkbia |
| Endoplasmic Reticulum Stress Pathway | 2.08 | 0.0952 | Hsp90b1, Hspa5 |
| LXR/RXR Activation | 2.08 | 0.0331 | Apod, Pltp, Srebf1, Trf |
| FXR/RXR Activation | 2.02 | 0.0317 | Apod, Pltp, Srebf1, Trf |
| TCA Cycle II (Eukaryotic) | 1.97 | 0.0833 | Dld, Sucla2 |
| Glutamate Dependent Acid Resistance | 1.88 | 0.5 | Gad2 |
| EIF2 Signaling | 1.79 | 0.0223 | Eif3a, Eif5, Eif5b, Hspa5, Srebf1 |
| Gαq Signaling | 1.69 | 0.0253 | Chrm3, Nfkbia, Plcb1, Rhog |
| Cytotoxic T Lymphocyte-mediated Apoptosis of Target Cells | 1.68 | 0.0588 | B2m, Hba-a2 |
| eNOS Signaling | 1.68 | 0.0252 | Chrm3, Hsp90aa1, Hsp90b1, Hspa5 |
| 0X40 Signaling Pathway | 1.67 | 0.0333 | B2m, Hba-a2, Nfkbia |
| Regulation of Actin-based Motility by Rho | 1.62 | 0.0319 | Gsn, Pak3, Rhog |
| CXCR4 Signaling | 1.61 | 0.024 | Egr1, Pak3, Plcb1, Rhog |
| GABA Receptor Signaling | 1.61 | 0.0316 | Gabra2, Gabrb1, Gad2 |
| Neuregulin Signaling | 1.6 | 0.0312 | Hsp90aa1, Hsp90b1, Pten |
| Branched-chain α-keto acid Dehydrogenase Complex | 1.59 | 0.25 | Dld |
| Antigen Presentation Pathway | 1.57 | 0.0513 | B2m, Hba-a2 |
| Nitric Oxide Signaling in the Cardiovascular System | 1.56 | 0.0303 | Hsp90aa1, Hsp90b1, Pde1A |
| PI3K/AKT Signaling | 1.55 | 0.0229 | Hsp90aa1, Hsp90b1, Nfkbia, Pten |
| Sumoylation Pathway | 1.52 | 0.0291 | Nfkbia, Rhog, Senp6 |
| PPAR Signaling | 1.51 | 0.0288 | Hsp90aa1, Hsp90b1, Nfkbia |
| 2-ketoglutarate Dehydrogenase Complex | 1.49 | 0.2 | Dld |
| 2-oxobutanoate Degradation I | 1.49 | 0.2 | Dld |
| Glutamate Degradation III (via 4-aminobutyrate) | 1.49 | 0.2 | Gad2 |
| BAG2 Signaling Pathway | 1.49 | 0.0465 | Hsp90aa1, Hspa5 |
| Dendritic Cell Maturation | 1.49 | 0.0219 | B2m, Hba-a2, Nfkbia, Plcb1 |
| PD-1, PD-L1 cancer immunotherapy pathway | 1.49 | 0.0283 | B2m, Hba-a2, Pten |
| G-Protein Coupled Receptor Signaling | 1.47 | 0.0184 | Chrm3, Cnr1, Nfkbia, Pde1A, Plcb1 |
| Antioxidant Action of Vitamin C | 1.45 | 0.0275 | Nfkbia, Plcb1, Selenot |
| PPARα/RXRα Activation | 1.44 | 0.0211 | Hsp90aa1, Hsp90b1, Nfkbia, Plcb1 |
| iCOS-iCOSL Signaling in T Helper Cells | 1.44 | 0.027 | Hba-a2, Nfkbia, Pten |
| Type I Diabetes Mellitus Signaling | 1.44 | 0.027 | Gad2, Hba-a2, Nfkbia |
| Glycine Cleavage Complex | 1.41 | 0.167 | Dld |
| Natural Killer Cell Signaling | 1.39 | 0.0203 | B2m, Hba-a2, Hspa5, Pak3 |
| Role of Tissue Factor in Cancer | 1.38 | 0.0256 | Egr1, Plcb1, Pten |
| TNFR1 Signaling | 1.37 | 0.04 | Nfkbia, Pak3 |
| Thioredoxin Pathway | 1.35 | 0.143 | Selenot |
| Acetyl-CoA Biosynthesis I (Pyruvate Dehydrogenase Complex) | 1.35 | 0.143 | Dld |
| Neuroinflammation Signaling Pathway | 1.32 | 0.0167 | B2m, Gabra2, Gabrb1, Gad2, Hba-a2 |
The most significantly affected canonical pathways after exposure included protein ubiquitination (B2m, Dnaja1, Dnajb4, Hba-a2, Hsp90aa1, Hsp90b1, Hspa4l, Hspa5, Ube2q2), aldosterone signaling in epithelial cells (Dnaja1, Dnajb4, Hsp90aa1, Hsp90b1, Hspa4l, Hspa5, Plcb1), hypoxia signaling in the cardiovascular system (Hsp90aa1, Hsp90b1, Nfkbia, Pten, Ube2q2), unfolded protein response (Hsp90b1, Hspa5, Srebf1), endoplasmic reticulum (ER) stress pathway (Hsp90b1, Hspa5), and the neuroinflammation signaling pathway (B2m, Gabra2, Gabrb1, Gad2, Hba-a2).
We observed dysregulation of genes indicative of a pro-inflammatory response, including downregulation of B2m and Hba-a2 and upregulation of Gabra2, Gabrb1, and Gad. There was significant downregulation of several genes associated with neuronal health, particularly genes involved in the integrity of the myelin sheath (Mog, Mbp, Mag, Pllp, Pmp22, Cldn11, Cnp), neurogenesis (Arc, Opalin), dendritic cell maturation (B2m, Hba-a2), NF-κB inhibition (Nfkbia, Plcb1), and learning and memory (Arc). Additionally, we found significant downregulation of mitochondrial genes coding for the F0 subunit of the proton-transporting ATP-synthase complex (mt-Atp6, mt-Atp8). There was significant upregulation of pro-apoptotic genes (Pten), genes involved in ER stress response (Hspa5, Hsp90b1), and genes involved in organonitrogen compound metabolism (Lars2, Hmgn5). There was also upregulation of genes implicated in related neurodegenerative diseases, including Oxr1, Top1, and Cdr1.
We observed dysregulation in GO categories of interest relating to biological processes, molecular functions, and cellular components. Significantly affected biological processes included leucyl-tRNA aminoacylation (Lars2), regulation of neurogenesis (Arc, Opalin), peptide metabolic process (Hmgn5, Lars2), regulation of long-term synaptic depression (Arc), regulation of postsynaptic neurotransmitter receptor internalization (Arc), and mitochondrial translation (Lars2). Notably affected GO categories involved in molecular functions included RNA strand annealing activity (Fmr1, Fxr1), supercoiled DNA binding (Psip1, Top1), and unfolded protein binding (Dnajb4, Hsp90aa1, Hsp90b1, Hspa5). Significantly affected GO categories forming cellular components of interest included the myelin sheath (Cldn11, Cnp, Dld, Gjc2, Gsn, Hsp90aa1, Hspa5, Mag, Mbp, Mog, Plcb1, Sucla2), GABAergic synapses (Camk4, Cnr1, Gabra2, Gabrb1, Gabrd, Nrxn1, Plcb1, Slc6a6), dendritic spines (Akap5, Arc, Fmr1, Fxr1, Homer1, Lpar1, Mob4, Pten, Slc8a1, Syndig1), ER chaperone complex (Hsp90b1, Hspa5, Sdf2l1), MHC class I peptide loading complex (B2m, H2-D1, H2-K1), and endocytic vesicles (Gsn, Kif5b, Lpar1, Nrxn1, Rab8b, Rab9b, Rabep1, Trf), among others.
4. Discussion
Our results showed that subcutaneous administration of PB + CPF + DEET for two weeks induced acute changes in gene expression in mouse hippocampal tissue, including dysregulation of genes indicating a pro-inflammatory response, downregulation of genes associated with neuronal health, and upregulation of pro-apoptotic genes, genes involved in ER stress response, and genes implicated in neurogenerative diseases, among others. We also observed significant effects of our Gulf War exposure on spatial memory.
The three most significantly downregulated genes after exposure were Arc, Egr1, and Nr4a1, all of which are neuronal immediate early genes (IEGs). Arc is predominantly expressed in cortical and hippocampal glutamatergic neurons and is involved in numerous neuronal signaling pathways [20,21]. Arc knockout mice display deficits in long-term memory formation in implicit and explicit learning tasks and impaired long-term potentiation (LTP) and depression (LTD) [22]; similar effects on LTP and spatial learning were shown in rats after chemical inhibition of Arc [23]. Egr1 is required for stabilization of synaptic plasticity in the hippocampus as well as formation of both hippocampal and non-hippocampal-dependent long-term memory [24] and is a direct transcriptional regulator of Arc [25].
Although IEGs are classified as such due to their early and transient response to environmental stimuli, both Arc and Egr1 also play critical roles in mediating the structural changes that underlie neuronal and synaptic plasticity, suggesting that their dysregulation could trigger long-term morphological changes with negative impacts on learning and memory formation. Several mouse models of Alzheimer’s disease (AD) report early dysregulation of IEGs involved in LTP and synaptic plasticity [26]. Dickey et al. observed a significant decrease in basal Arc, Egr1, and Nr4a1 expression in amyloid-containing hippocampus and cortex of APP/PS1 transgenic mice [27]. Levels of basal and exploration-induced Arc expression are significantly reduced in granule cells of the dentate gyrus of hAPPFAD transgenic mice [28]. Induced Arc expression was also dysregulated in the CA3 region and dentate gyrus of rats chronically infused with lipopolysaccharide (LPS) to induce neuroinflammation, suggesting altered patterns of Arc expression may contribute to cognitive and memory impairments in neurodegeneration [29]. IEGs have been investigated as a potential therapeutic target in AD treatment [30,31].
IEGs such as Arc and Egr1 have also been suggested to play a critical role in the interaction between genes and environment to determine the risk of developing psychiatric illness, particularly major depressive disorder (MDD), which is typically comorbid with GWI [1–7,32–35]. Chronic treatment with various antidepressants targeting serotonin and norepinephrine can also restore Arc expression in the hippocampus and prefrontal cortex [36,37].
Additionally, Arc inhibits the binding of heat shock factor 1 (HSF1) to the heat shock element (HSE) in heat shock protein (HSP) gene promoters and prevents activation of HSP genes [38]. Accordingly, we observed upregulation of Hsp genes, including Hsp40s (Dnajb4, Dnaja1), Hsp70s (Hspa4l, Hspa5), and Hsp90s (Hsap90aa1, Hsp90b1) and found that these genes were involved in several significantly affected pathways, including protein ubiquitination, aldosterone signaling, hypoxia signaling, unfolded protein response, interferon induction and antiviral response, and the ER stress pathway, among others. Thus, dysregulation of IEGs may play a role in acute neuroinflammation, leading to chronic neurodegeneration.
Interestingly, several genes encoding proteins that are structural components of myelin were downregulated, including Mbp, Mag, Mog, and Cnp. Myelin basic protein (Mbp) is phosphorylated by MAP kinase in response to action potential firing during LTP in the hippocampus [39,40]. Plasma autoantibodies against Mbp have also been found to be significantly increased in Veterans with symptoms of GWI compared to healthy controls [41,42]. We also observed dysregulation of genes related to the GABAergic synapse, including Camk4, Cnr1, Gabra2, Gabrb1, Gabrd, Nrxn1, Plcb1, and Slc6a6. Chronically, decreased GABA has been reported in hippocampi of mice exposed to PB + permethrin + DEET three months after exposure [43]. Additionally, we found decreased expression of Chrm3, which codes for the M3 muscarinic receptor. Decreased M3 receptor density has been reported in the CA1 region, CA3 region, and molecular layer of the hippocampus in C57Bl/6 mice exposed to PB + stress [44]. This suggests that changes in GABAA and M3 receptor expression may begin during the acute phase of chronic sublethal exposure to our Gulf War toxicants.
Reported dosages and routes of administration of Gulf War toxicants in rodent models have varied widely throughout the literature. The subcutaneous route of administration for exposure to PB + CPF + DEET has several advantages. PB was taken orally by military personnel and is frequently administered via gavage in animal models; however, PB has been shown to have poor bioavailability, suggesting that injection may deliver a more precise dosage [45]. There has also been a significant amount of investigation into the effects of stress in combination with PB and other toxicants, with results that indicate increased BBB permeability to toxicants in stressed animals [46]. Friedman et al. reported significant effects of PB + stress on levels of c-Fos and AChE mRNAs in mouse whole-brain homogenates, indicating that stress can be a confounding variable in gene expression data examining an early transcriptional response [47]. The subcutaneous route would not present potential stress from repeated oral gavage.
Subcutaneous administration also avoids variable absorption via dermal application of CPF and DEET, which would have been in contact with the skin of military personnel. A study by Keil et al. examining the immunotoxicology of DEET in female B6C3F1 mice elaborated on factors which are necessary to accurately compare exposures in animal models but are often not considered [48]. Many human and animal studies refer to dermal penetration rather than absorption into the bloodstream, which is not an equivalent measure due to the variability of absorption levels within and between species. Keil et al. reported that s.c. administration of 7.7 mg/kg/day DEET equates to an estimated mouse blood exposure level that encompasses estimated military exposure levels as well as estimated DEET usage by the general population. Additionally, Keil et al. argue that the emphasis placed on relevant route of exposure in the literature has limited utility, particularly in the case of dermal exposures such as DEET or CPF. CPF, a lipophilic organophosphate, could accumulate within the brain to cause AChE inhibition at our acute timepoint, which could have an effect on behavioral outcomes. There are wide ranges of estimated absorption and metabolic rates between rodents and humans.
It should be noted that military personnel would have been exposed to these compounds at lower dosages, but this exposure occurred over longer periods of time. In rodent models, higher dosages are often used in a shorter time frame due to the lifespan of the animal and the window in which to study effects. Other studies have reported using similar dosages at these intervals: Lamproglou et al. reported i.o. administration of 1.5 mg/kg PB for 12 days (5 days on, 2 days off, 5 days on) in male Wistar rats [49]; Peden-Adams et al. treated female B6C3F1 mice treated with 15.5 mg/kg DEET, 2 mg/kg PB, and 500 mg/kg JP-8 s.c. for 14 days as a “low dose” group [50]; Torres-Altoro et al. reported treatment of female C57Bl/6 mice with 30 mg/kg CPF s.c. for 7 days, male FVB mice with 2.5 mg/kg PB + 5 mg/kg DEET s.c. for 15 days, and male C57Bl/6 treated with 1 mg/kg PB s.c. for 10 days [51]; and Mauck et al. treated male C57Bl/6 mice with 3 or 10 mg/kg PB for 7 days via s.c. ALZET pump [44]. These studies illustrate the similar range of concentrations over shorter time frames, as well as the potential advantages of s.c. administration for certain experiments.
Whole transcriptome sequencing has been used previously in several rodent models of Gulf War exposure. A similar study by Shetty et al. examined changes in gene expression using qRT-PCR after 4 weeks of exposure to PB + DEET + stress in male Sprague-Dawley rats; however, their samples, collected at a longer 6-month time point after the last exposure, presented a gene expression profile indicative of chronic neuroinflammation [16]. Gene expression profiles of GWI patients have also been studied to identify novel treatment strategies by examining the overlap of dysregulated genes with drug targets and in comparison with expression profiles of other diseases [52]. In contrast, our acute Gulf War exposure model shows early effects that do not appear in chronic exposure models, such as dysregulation of IEGs. Xu et al. also recently reported on acute transcriptional changes in BXD mouse strains after exposure to corticosterone + diisopropyl fluorophosphate (DFP) [19]. We believe that acute changes may prime chronic neurodegenerative processes; therefore, further research should investigate mechanistic connections between early responses to toxicant exposure and chronic symptoms, including memory deficits, mood disorders, and neurodegeneration.
5. Conclusion
This study provides an assessment of changes in gene expression in combined exposure to PB, CPF, and DEET and a unique gene expression profile at an acute timepoint. Many of the dysregulated genes involve inflammatory signaling and other pathways that are important for the health of neurons. The neurological effects of toxicants, including memory deficits, may begin soon after exposure, and future research will further define the way these responses increase with time due to aging and other influences.
Acknowledgments
We thank Julia A. Burton and Mihal Grinberg for their expert technical assistance and Joshua Karp, Shannon Clare, Elizabeth Chang, and Gabrielle Gallant for their input. We also thank the Rutgers-NJMS Genomics Center (Drs. Patricia Soteropoulos and Mainul Hoque) for RNA-Seq processing and advice. This study was supported by the Department of Veterans Affairs (Veterans Health Administration, Office of Research and Development, Rehabilitation Research and Development (I01RX001520 and IK2RX003253) and Biomedical Laboratory Research and Development (I21BX003514)), the Assistant Secretary of Defense for Health Affairs through the Congressionally Directed Gulf War Illness Research Program (W81XWH-16-1-0626), the War Related Illness and Injury Study Center (NJ), the Bay Pines Foundation, and the Veterans Bio-Medical Research Institute. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE180786 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE180786).
Footnotes
Declaration of competing interest
No competing financial interests exist. The contents do not represent the views of the Department of Veterans Affairs or the United States Government, and the opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense.
References
- [1].Institute of Medicine [IOM], Gulf War and Health: Volume 8: Update of Health Effects of Serving in the Gulf War, Washington, D.C, 2010. [PubMed] [Google Scholar]
- [2].Institute of Medicine [IOM], Chronic Multisymptom Illness in Gulf War Veterans: Case Definitions Reexamined, Washington, D.C, 2014. [PubMed] [Google Scholar]
- [3].United States Department of Veterans Affairs, Research Advisory Committee on Gulf War Veterans' Illnesses [RAC-GWI], Gulf War Illness and the Health of Gulf War Veterans: Scientific Findings and Recommendations, Washington, D.C, 2008. [Google Scholar]
- [4].Institute of Medicine [IOM], Gulf War Veterans: Treating Symptoms and Syndromes, Washington, D.C, 2001. [PubMed] [Google Scholar]
- [5].Institute of Medicine [IOM], Gulf War and Health: Treatment for Chronic Multisymptom Illness, Washington, D.C, 2013. [PubMed] [Google Scholar]
- [6].Steele L, Prevalence and patterns of gulf war illness in Kansas veterans: association of symptoms with characteristics of person, place, and time of military service, Am. J. Epidemiol 152 (2000) 992–1002, 10.1093/aje/152.10.992. [DOI] [PubMed] [Google Scholar]
- [7].White RF, Steele L, O’Callaghan JP, Sullivan K, Binns JH, Golomb BA, Bloom FE, Bunker JA, Crawford F, Graves JC, Hardie A, Klimas N, Knox M, Meggs WJ, Melling J, Philbert MA, Grashow R, Recent research on Gulf War illness and other health problems in veterans of the 1991 Gulf War: effects of toxicant exposures during deployment, Cortex 74 (2016) 449–475, 10.1016/j.cortex.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dickey B, Madhu LN, Shetty AK, Gulf war illness: mechanisms underlying brain dysfunction and promising therapeutic strategies, Pharmacol. Ther 107716 (2020), 10.1016/j.pharmthera.2020.107716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Parihar VK, Hattiangady B, Shuai B, Shetty AK, Mood and memory deficits in a model of gulf war illness are linked with reduced neurogenesis, partial neuron loss, and mild inflammation in the hippocampus, Neuropsychopharmacology 38 (2013) 2348–2362, 10.1038/npp.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chaney LA, Rockhold RW, Mozingo JR, Hume AS, Moss JI, Potentiation of pyridostigmine bromide toxicity in mice by selected adrenergic agents and caffeine, Vet. Hum. Toxicol 39 (1997) 214–219. [PubMed] [Google Scholar]
- [11].Chaney LA, Wineman RW, Rockhold RW, Hume AS, Acute effects of an insect repellent, N, N-diethyl-m-toluamide, on cholinesterase inhibition induced by pyridostigmine bromide in rats, Toxicol. Appl. Pharmacol 165 (2000) 107–114, 10.1006/taap.2000.8936. [DOI] [PubMed] [Google Scholar]
- [12].Prendergast MA, Terry AV Jr., Buccafusco JJ, Chronic, low-level exposure to diisopropylfluorophosphate causes protracted impairment of spatial navigation learning, Psychopharmacology 129 (1997) 183–191, 10.1007/s002130050179. [DOI] [PubMed] [Google Scholar]
- [13].Terry AV Jr., Stone JD, Buccafusco JJ, Sickles DW, Sood A, Prendergast MA, Repeated exposures to subthreshold doses of chlorpyrifos in rats: hippocampal damage, impaired axonal transport, and deficits in spatial learning, J. Pharmacol. Exp. Ther 305 (2003) 375–384, 10.1124/jpet.102.041897. [DOI] [PubMed] [Google Scholar]
- [14].Abou-Donia MB, Wilmarth KR, Abdel-Rahman AA, Jensen KF, Oehme FW, Kurt TL, Increased neurotoxicity following concurrent exposure to pyridostigmine bromide, DEET, and chlorpyrifos, Fundam. Appl. Toxicol 34 (1996) 201–222, 10.1006/faat.1996.0190. [DOI] [PubMed] [Google Scholar]
- [15].Ojo JO, Abdullah L, Evans J, Reed JM, Montague H, Mullan MJ, Crawford FC, Exposure to an organophosphate pesticide, individually or in combination with other gulf war agents, impairs synaptic integrity and neuronal differentiation, and is accompanied by subtle microvascular injury in a mouse model of gulf war agent exposure, Neuropathology 34 (2014) 109–127, 10.1111/neup.12061. [DOI] [PubMed] [Google Scholar]
- [16].Shetty GA, Hattiangady B, Upadhya D, Bates A, Attaluri S, Shuai B, Kodali M, Shetty AK, Chronic oxidative stress, mitochondrial dysfunction, Nrf2 activation and inflammation in the hippocampus accompany heightened systemic inflammation and oxidative stress in an animal model of gulf war illness, Front. Mol. Neurosci 10 (2017) 182, 10.3389/fnmol.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pierce LM, Kurata WE, Matsumoto KW, Clark ME, Farmer DM, Long-term epigenetic alterations in a rat model of gulf war illness, Neurotoxicology 55 (2016) 20–32, 10.1016/j.neuro.2016.05.007. [DOI] [PubMed] [Google Scholar]
- [18].Ashbrook DG, Hing B, Michalovicz LT, Kelly KA, Miller JV, de Vega WC, Miller DB, Broderick G, O’Callaghan JP, McGowan PO, Epigenetic impacts of stress priming of the neuroinflammatory response to sarin surrogate in mice: a model of gulf war illness, J. Neuroinflammation 15 (2018) 86, 10.1186/s12974-018-1113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xu F, Ashbrook DG, Gao J, Starlard-Davenport A, Zhao W, Miller DB, O’Callaghan JP, Williams RW, Jones BC, Lu L, Genome-wide transcriptome architecture in a mouse model of gulf war illness, Brain Behav. Immun 89 (2020) 209–223, 10.1016/j.bbi.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Epstein I, Finkbeiner S, The arc of cognition: signaling cascades regulating arc and implications for cognitive function and disease, Semin. Cell Dev. Biol 77 (2018) 63–72, 10.1016/j.semcdb.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Korb E, Finkbeiner S, Arc in synaptic plasticity: from gene to behavior, Trends Neurosci 34 (2011) 591–598, 10.1016/j.tins.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, Bosl MR, Lipp HP, Grant SG, Bliss TV, Wolfer DP, Kuhl D, Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories, Neuron 52 (2006) 437–444, 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- [23].Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA, Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory, J. Neurosci 20 (2000) 3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S, A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories, Nat. Neurosci 4 (2001) 289–296, 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- [25].Li L, Carter J, Gao X, Whitehead J, Tourtellotte WG, The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors, Mol. Cell. Biol 25 (2005) 10286–10300, 10.1128/MCB.25.23.10286-10300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Perusini JN, Cajigas SA, Cohensedgh O, Lim SC, Pavlova IP, Donaldson ZR, Denny CA, Optogenetic stimulation of dentate gyrus engrams restores memory in Alzheimer's disease mice, Hippocampus 27 (2017) 1110–1122, 10.1002/hipo.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dickey CA, Loring JF, Montgomery J, Gordon MN, Eastman PS, Morgan D, Selectively reduced expression of synaptic plasticity-related genes in amyloid precursor protein presenilin-1 transgenic mice, J. Neurosci 23 (2003) 5219–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Palop JJ, Chin J, Bien-Ly N, Massaro C, Yeung BZ, Yu GQ, Mucke L, Vulnerability of dentate granule cells to disruption of arc expression in human amyloid precursor protein transgenic mice, J. Neurosci 25 (2005) 9686–9693, 10.1523/JNEUROSCI.2829-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rosi S, Ramirez-Amaya V, Vazdarjanova A, Worley PF, Barnes CA, Wenk GL, Neuroinflammation alters the hippocampal pattern of behaviorally induced arc expression, J. Neurosci 25 (2005) 723–731, 10.1523/JNEUROSCI.4469-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Perry KW, Nisenbaum LK, George CA, Shannon HE, Felder CC, Bymaster FP, The muscarinic agonist xanomeline increases monoamine release and immediate early gene expression in the rat prefrontal cortex, Biol. Psychiatry 49 (2001) 716–725, 10.1016/s0006-3223(00)01017-9. [DOI] [PubMed] [Google Scholar]
- [31].Tong XK, Lecrux C, Rosa-Neto P, Hamel E, Age-dependent rescue by simvastatin of Alzheimer's disease cerebrovascular and memory deficits, J. Neurosci 32 (2012) 4705–4715, 10.1523/JNEUROSCI.0169-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gallitano AL, Editorial: the role of immediate early genes in neuropsychiatric illness, Front. Behav. Neurosci 14 (2020) 16, 10.3389/fnbeh.2020.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Duclot F, Kabbaj M, The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders, Front. Behav. Neurosci 11 (2017) 35, 10.3389/fnbeh.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xu Y, Pan J, Sun J, Ding L, Ruan L, Reed M, Yu X, Klabnik J, Lin D, Li J, Chen L, Zhang C, Zhang H, O’Donnell JM, Inhibition of phosphodiesterase 2 reverses impaired cognition and neuronal remodeling caused by chronic stress, Neurobiol. Aging 36 (2015) 955–970, 10.1016/j.neurobiolaging.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Covington HE 3rd, Lobo MK, Maze I, Vialou V, Hyman JM, Zaman S, LaPlant Q, Mouzon E, Ghose S, Tamminga CA, Neve RL, Deisseroth K, Nestler EJ, Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex, J. Neurosci 30 (2010) 16082–16090, 10.1523/JNEUROSCI.1731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gallo FT, Katche C, Morici JF, Medina JH, Weisstaub NV, Immediate early genes, memory and psychiatric disorders: focus on c-fos, Egr1 and arc, Front. Behav. Neurosci 12 (2018) 79, 10.3389/fnbeh.2018.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li Y, Pehrson AL, Waller JA, Dale E, Sanchez C, Gulinello M, A critical evaluation of the activity-regulated cytoskeleton-associated protein (Arc/Arg3.1)& apos;s putative role in regulating dendritic plasticity, cognitive processes, and mood in animal models of depression, Front. Neurosci 279 (2015), 10.3389/fnins.2015.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Park AY, Park YS, So D, Song IK, Choi JE, Kim HJ, Lee KJ, Activity-regulated cytoskeleton-associated protein(Arc/Arg3.1) is transiently expressed after heat shock stress and suppresses heat shock factor 1, Sci. Rep 9 (2019) 2592, 10.1038/s41598-019-39292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Atkins CM, Yon M, Groome NP, Sweatt JD, Regulation of myelin basic protein phosphorylation by mitogen-activated protein kinase during increased action potential firing in the hippocampus, J. Neurochem 73 (1999) 1090–1097, 10.1046/j.1471-4159.1999.0731090.x. [DOI] [PubMed] [Google Scholar]
- [40].Lee PR, Fields RD, Regulation of myelin genes implicated in psychiatric disorders by functional activity in axons, Front. Neuroanat 3 (2009) 4, 10.3389/neuro.05.004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Abou-Donia MB, Conboy LA, Kokkotou E, Jacobson E, Elmasry EM, Elkafrawy P, Neely M, Bass CR, Sullivan K, Screening for novel central nervous system biomarkers in veterans with gulf war illness, Neurotoxicol. Teratol 61 (2017) 36–46, 10.1016/j.ntt.2017.03.002. [DOI] [PubMed] [Google Scholar]
- [42].Abou-Donia MB, Lapadula ES, Krengel MH, Quinn E, LeClair J, Massaro J, Conboy LA, Kokkotou E, Abreu M, Klimas NG, Nguyen DD, Sullivan K, Using plasma autoantibodies of central nervous system proteins to distinguish veterans with gulf war illness from healthy and symptomatic controls, Brain Sci 10 (2020), 10.3390/brainsci10090610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [43].Carreras I, Aytan N, Mellott T, Choi JK, Lehar M, Crabtree L, Leite-Morris K, Jenkins BG, Blusztajn JK, Dedeoglu A, Anxiety, neuroinflammation, cholinergic and GABAergic abnormalities are early markers of gulf war illness in a mouse model of the disease, Brain Res 1681 (2018) 34–43, 10.1016/j.brainres.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mauck B, Lucot JB, Paton S, Grubbs RD, Cholinesterase inhibitors and stress: effects on brain muscarinic receptor density in mice, Neurotoxicology 31 (2010) 461–467, 10.1016/j.neuro.2010.06.001. [DOI] [PubMed] [Google Scholar]
- [45].Abdullah L, Crynen G, Reed J, Bishop A, Phillips J, Ferguson S, Mouzon B, Mullan M, Mathura V, Mullan M, Ait-Ghezala G, Crawford F, Proteomic CNS profile of delayed cognitive impairment in mice exposed to gulf war agents, Neuromolecular Med 13 (2011) 275–288, 10.1007/s12017-011-8160-z. [DOI] [PubMed] [Google Scholar]
- [46].Abdel-Rahman A, Shetty AK, Abou-Donia MB, Disruption of the blood-brain barrier and neuronal cell death in cingulate cortex, dentate gyrus, thalamus, and hypothalamus in a rat model of gulf-war syndrome, Neurobiol. Dis 10 (2002) 306–326, 10.1006/nbdi.2002.0524. [DOI] [PubMed] [Google Scholar]
- [47].Friedman A, Kaufer D, Shemer J, Hendler I, Soreq H, Tur-Kaspa I, Pyridostigmine brain penetration under stress enhances neuronal excitability and induces early immediate transcriptional response, Nat. Med 2 (1996) 1382–1385, 10.1038/nm1296-1382. [DOI] [PubMed] [Google Scholar]
- [48].Keil DE, McGuinn WD, Dudley AC, EuDaly JG, Gilkeson GS, Peden-Adams MM, N, N,-diethyl-m-toluamide (DEET) suppresses humoral immunological function in B6C3F1 mice, Toxicol. Sci 108 (2009) 110–123, 10.1093/toxsci/kfp001. [DOI] [PubMed] [Google Scholar]
- [49].Lamproglou I, Barbier L, Diserbo M, Fauvelle F, Fauquette W, Amourette C, Repeated stress in combination with pyridostigmine part I: long-term behavioural consequences, Behav. Brain Res 197 (2009) 301–310, 10.1016/j.bbr.2008.08.031. [DOI] [PubMed] [Google Scholar]
- [50].Peden-Adam MM, Eudaly J, Eudaly E, Dudley A, Zeigler J, Lee A, Robbs J, Gilkeson G, Keil DE, Evaluation of immunotoxicity induced by single or concurrent exposure to N, N-diethyl-m-toluamide (DEET), pyridostigmine bromide (PYR), and JP-8 jet fuel, Toxicol. Ind. Health 17 (2001) 192–209, 10.1191/0748233701th120oa. [DOI] [PubMed] [Google Scholar]
- [51].Torres-Altoro MI, Mathur BN, Drerup JM, Thomas R, Lovinger DM, O’Callaghan JP, Bibb JA, Organophosphates dysregulate dopamine signaling, glutamatergic neurotransmission, and induce neuronal injury markers in striatum, J. Neurochem 119 (2011) 303–313, 10.1111/j.1471-4159.2011.07428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Craddock TJ, Harvey JM, Nathanson L, Barnes ZM, Klimas NG, Fletcher MA, Broderick G, Using gene expression signatures to identify novel treatment strategies in gulf war illness, BMC Med. Genet 8 (2015) 36, 10.1186/s12920-015-0111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


