Abstract
Background
Chronic postsurgical pain (CPSP) is common among patients receiving major surgeries. CPSP produces suffering in patients, both physically and mentally. However, the mechanisms underlying CPSP remain elusive. Here, a genome-wide expression profiling of ipsilateral spinal cord dorsal horn (SCDH) was performed to identify potential genes related with CPSP.
Methods
A rat skin/muscle incision and retraction (SMIR) model was established to induce CPSP. Immunostaining was used to study glial cell and neuron activation in ipsilateral SCDH of SMIR model rats. RNA sequencing (RNA-Seq), combined with bioinformatics analysis, was undertaken to explore gene expression profiles. qPCR was applied to validate the expression of some representative genes.
Results
The SMIR model rats developed persistent mechanical allodynia in ipsilateral hindpaw for up to 14 days. Ipsilateral SCDH of SMIR rats showed remarkable glial cell and neuron activation. A number of differentially expressed genes (DEGs) were identified in ipsilateral SCDH of SMIR rats by RNA-Seq. qPCR confirmed expression of some representative DEGs. Bioinformatics indicated that chemical synaptic transmission, sensory perception of pain and neuroactive ligand-receptor interaction were predominant functions. We compared our dataset with human pain-related genes and found that several genes exclusively participate in pain modulation and mechanisms.
Conclusion
Our study provided novel understandings of the molecular mechanisms possibly contributing to CPSP. These findings may offer new targets for future treatment of CPSP.
Keywords: postsurgical pain, spinal cord dorsal horn, RNA-Seq, microglia, genome
Introduction
Surgery oftentimes causes persistent pain, a term called chronic postsurgical pain (CPSP).1 It represents a persistent pain condition lasting over 2 months after the surgery that was not induced by other factors, eg apparent inflammation, disease recurrence or nonsurgical-related reasons.1,2 Epidemiological studies estimate that 10–30% of surgical patients will report a certain degree of persistent pain 1 year after the surgery.1,3 For patients receiving major surgeries, such as thoracic surgery and amputation, the occurrence rate of CPSP could reach over 40%, and severe and debilitating CPSP may develop.1,4,5 CPSP can produce unnecessary suffering and discomfort among patients and even result in psychological burdens.6 Unfortunately, there is still no effective treatment for CPSP.
The mechanisms underlying CPSP are still not fully understood. To study its mechanisms, the skin/muscle incision and retraction (SMIR) rat model was developed to mimic CPSP conditions.7 This animal model exhibits long-lasting mechanical allodynia in affected hind limb after the surgery, mimicking human CPSP conditions. The rat SMIR model has become a popular preclinical animal model for mechanistic studies of CPSP. In recent years, much effort has been made to elucidate CPSP mechanisms using the SMIR model. For instance, excessive expression of pain-inducing substances, eg substances P, IL-1β and IL-6, has been observed in dorsal root ganglion (DRG) in model animals.8 SMIR model animals showed activation of P2X7R in DRG satellite glial cell, which triggers ERK activation and TNF-α production and contributes to postsurgical pain.9 SMIR animals also showed increased membrane expression of the nociceptive Nav1.7 channel, which was under regulation by NF-κB signaling. The inhibition of Nav1.7 channel as well as NF-κB phosphorylation attenuated SMIR-induced chronic pain.10 In addition to peripheral mechanisms, central mechanisms have been proposed for SMIR-induced pain as well. Spinal microglia and astrocytes have been observed to be overactivated in SMIR model rats.11,12 Blocking spinal P2X7R attenuates SMIR-induced postsurgical pain via reducing glial cell overactivation and decreasing TNF-α expression.12 TLR4 mediated overproduction of TNF-α in the spinal cord also makes contributions to the chronic pain condition in SMIR model animals.13
Spinal cord dorsal horn (SCDH) receives and integrates nociceptive inputs from the periphery and relays nociceptive signal to higher brain regions. SCDH plays a pivotal role in the integration of nociception signaling and plays an important role in central sensitization. To further study central mechanisms of CPSP, in this study we carried out a genome-wide exploration using RNA-Seq technique to investigate the expression profiles of ipsilateral SCDH from SMIR model rats and sham rats. Our study found a number of differentially expressed genes (DEGs) in ipsilateral side of SCDH from SMIR model animals. We then investigated the major molecular/cellular functions of these DEGs that may participate. We further examined potential genes which could possibly contribute to the mechanisms of SMIR-induced pain. Our work provides some novel understandings of the molecular mechanisms possibly contributing to CPSP, which may offer new targets for CPSP treatment in the future.
Materials and Methods
Experimental Animals
Sprague–Dawley rats (male, 3–4 months of age) were bought from Shanghai Laboratory Animal Center of China. All animals were housed in Zhejiang Chinese Medical University Laboratory Animal Center (5 animals/cage, 12 h dark–light cycle, 24 ± 2 °C). Animals were provided with free access to water and food. The animals were given > 1 w to accommodate the new breeding facility before any test. Animals were randomly allocated using random number tables.
Skin/Muscle Incision and Retraction (SMIR) Operation
The SMIR operations were conducted according to methods previously described.7,11 Sodium pentobarbital (i.p. 50 mg/kg) was used to induce anesthesia in rats. After anesthesia, a 1.5–2 cm cut, approximately 4 mm medial to the saphenous vein, was made in the skin of the thigh. Then the superficial muscle layers of the animal’s thigh were exposed and a 7–10 mm incision to the gracilis muscle was made. The incision was then retracted using a micro-adjustable retractor (Medical Apparatus & Instruments, China). The retraction was extended to 20 mm and lasted for 1 h. During the operation, a heating platform (37°C) was applied to maintain the whole body temperature warmth. In addition, an absorbent bench underpad was applied to cover the animal, which helped to prevent drying of the wound and the loss of heat. After the operation, 3.0 and 4.0 Vicryl sutures were used to seal the muscles and skin. Sham animals were treated with the same skin incision procedure but without the retraction procedure.
Determination of Mechanical Allodynia
Mechanical allodynia was measured by methods as described before.14,15 Briefly, each rat was kept under the testing environment for 30 min beforehand. Mechanical allodynia was measured by applying von Frey filaments to the mid-plantar surface of the rat’s hind paw. The “Up and Down” method was used for measurement and calculation of 50% paw withdrawal threshold.16,17 The experimenter for behavioral test was blinded during the allocation, the conduct of the experiment, the outcome assessment, and the data analysis procedures.
Tissue Collection and RNA Extraction
Methods were described in our previous study.18 Briefly, 14 days after the model establishment, both model and sham groups of rats were anesthetized deeply using sodium pentobarbital at a dosage of 40 mg/kg (i.p.). The animals were then perfused with 0.9% saline with 4 °C. Then the ipsilateral SCDH tissues were harvested and immediately preserved in RNAlater solution (#AM7020, Invitrogen, USA). Trizol reagent (#15596018, Thermo Fisher, USA) was used for total RNA extraction. The concentrations and the purities of tissue samples was examined using Nanodrop Spectrophotometer (NanoDrop One, Thermo Fisher, USA). RNA integrity number (RIN) was then determined by Agilent TapeStation System (Agilent Technologies, USA).
Preparation of the Library Used for RNA-Seq
The total mRNAs from 4 rats of SMIR and 4 rats of sham group were extracted and used to establish the libraries for RNA-Seq. The detailed methods have been described in previous literature19 and are available in Supplementary Materials accompanying this manuscript.
Immunofluorescence Staining
After being anesthetized by sodium pentobarbital (40 mg/kg, i.p.), cold 0.9% saline was used to perfuse the animals, followed with perfusion with 4% paraformaldehyde. Ipsilateral spinal cord dorsal horn was then isolated and fixed using paraformaldehyde (4%) for a period of 4 h at 4 °C. Then all specimens were transferred to sucrose gradients for 3 days to dehydrate. After dehydration, the specimens were cut into sections with a thickness of 15 μm with cryostat (CryoStar NX50, Thermo Fisher, USA). The cut sections were then mounted to glass slides pre-coated with gelatin. 1% BSA +10% donkey serum were used to block the sections for 2 h at room temperature. Then the sections were treated overnight at 4 °C using primary antibodies as follows: mouse anti-GFAP (#c9205, Sigma), mouse anti-OX42 (#ab1211, Abcam), rabbit anti-c-Fos (#ab2250, CST), rabbit anti-ATF3 (#HPA001562, Sigma) and mouse anti-NeuN (#ab104224, Abcam). On the second day, the sections were rinsed with PBS and incubated for 1 h with a mixture of corresponding secondary antibodies at room temperature. A Nikon A1R laser scanning confocal microscope (Nikon, Japan) was used to capture fluorescence images. Three to five images were randomly selected per rat tissue, averaged, and then compared according to methods described in our previous studies.20,21 The fluorescent images of SCDH we captured mainly covered from laminar I to III (see Supplementary Figure 1). Therefore, all fluorescent image quantifications were done on laminar I to III of SCDH, accordingly. For image quantification, uniform microscope settings were maintained throughout all image capture sessions and experimenters were blinded to treatment groups.
Clustering Analysis and Screening of DEGs
The analyses of gene expression were carried out with “DESeq2” package of the R software using Bioconductor. The Pheatmap package of R software (Version R 3.5.1) was used for clustering analysis. The screening of DEGs and illustrations were conducted via scatter plot, as reported before.22
qPCR Analysis
The total RNAs from the ipsilateral SCDH tissue samples were extracted using the TRIzol reagent (#15596018, Thermo Fisher, USA). 1,000 ng of total RNA from SCDH tissues was reversely transcribed using Takara PrimeScriptTM Master Mix Kit (#RR047, Takara Bio Inc., China) to generate corresponding cDNAs. qPCR reactions were performed using the LightCycler® 480 SYBR® Green I Master (#04887352001, Roche, Switzerland). Three replicates were performed for each sample. The gene expression level was normalized to β-actin, the housekeeping control gene. LightCycler480 system (Roche, Switzerland) was used for PCR, CT value determination and analysis. Relative mRNA expression levels were determined by the well-established 2ΔΔCT methodology as described before.23,24 For detailed information regarding the sequences for the primers, please see Table 1.
Table 1.
Sequences of the Primers Used for qPCR Validation of RNA-Seq Data
| Gene Name | Gene ID | Primer Sequence (5’-3′) | Amplicon Size (bp) |
|---|---|---|---|
| Nlrp12 | 292541 | 5’-ACCTCAGTGGCAACAGCATTGG-3’ | 112 |
| 5’-GCCTCCAGCAAACACTTCCTCAG-3’ | |||
| Hif3a | 64345 | 5’-CCGATGACCTGATTGGCTGTTCTG-3’ | 123 |
| 5’-GGAAGCGATACTGCCCTGTTACTG-3’ | |||
| Sftpd | 25350 | 5’-CGCCTGGTTATCTGTGAGTTCTGAG-3’ | 111 |
| 5’-TCGGATGGTGGCAGCATAGAGG-3’ | |||
| Serpina3n | 24795 | 5’-CATCAGGAGTCAGCAATCACAGAGG-3’ | 90 |
| 5’-GAGTCACAAGGCGGGTCATCTTC-3’ | |||
| Pcdh20 | 306081 | 5’-CGCTAACGGACCTGGCTGTATTC-3’ | 122 |
| 5’-ACACGACACCATCCATCTCATTTGC-3’ | |||
| Sgk1 | 29517 | 5’-TCTATGGCCTGCCTCCGTTCTAC-3’ | 110 |
| 5’-TCTATGGCCTGCCTCCGTTCTAC-3’ | |||
| Pdk4 | 89813 | 5’-TCTGAGGCTGATGACTGGTGTATCC-3’ | 123 |
| 5’-GCACTGCCGTAGACCCACTTTG-3’ | |||
| Mlf1 | 310483 | 5’-AGCGTGTCTGATGTGGTGAAAGATG-3’ | 112 |
| 5’-AGCGTGTCTGATGTGGTGAAAGATG-3’ | |||
| Aox1 | 54349 | 5’-GCAACACCAGGACCAGACTTCATC-3’ | 110 |
| 5’-GAGCAGAGCATACATGGACATCACC-3’ | |||
| Slc25a48 | 361206 | 5’-ACTGGTATGGTCTCTGTTGGTCTGG-3’ | 102 |
| 5’-CTTCAATCCGTGGCTGGCTTCC-3’ | |||
| Zfp939 | 690136 | 5’-CTGTTGGGTAAGGCAGTAGGTTCAC-3’ | 110 |
| 5’-CGGTGAGAGGGAAAGGGAGAGAG-3’ |
Statistical Analysis
SPSS 19.0 (IBM Corp., USA) was used for all statistical analysis throughout the study. All data were included in data analysis and presentation. To compare data between 2 groups, the Student’s t-test was applied for statistical analysis. To compare data among 3 or more groups, one-way or two-way analysis of variance (ANOVA) with Tukey’s post hoc test was applied for statistical analysis. Statistical significance was accepted at a level of p < 0.05.
The details of some other methods and materials in this study are further summarized in the Supplementary Methods of this article.
Results
The Establishment of the Rat SMIR Model to Mimic Chronic Postsurgical Pain
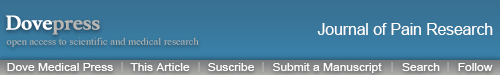
The rat skin/muscle incision and retraction (SMIR) model was established in accordance with procedures previously described.7,11 The SMIR procedures were conducted as illustrated in Figure 1A. The animals recovered well after the surgery and the wound completely healed after the 14 days of observation (Figure 1A). We then measured the mechanical allodynia of SMIR model rats according to time points shown in Figure 1B. The SMIR model rats showed a significantly reduced paw withdrawal threshold in affected hind paw as compared with control (sham rats) (Figure 1C). The mechanical allodynia persisted over 14 days after model establishment. As shown in Figure 1D, the analysis of normalized areas under the curve (AUC) further demonstrated an overall reduction of the paw withdrawal threshold in SMIR model rats. The above results are in line with a previous report,7 which indicates the successful establishment of the rat SMIR model.
Figure 1.
Establishment of SMIR-induced chronic postoperative pain model and nocifensive behavioral evaluation. (A) Photo showing the surgery site and manipulations of the SMIR model (Left). Photos showing the recovery of the surgery site on days 7 and 14 (right). (B) Protocol for model establishment, behavioral assay and tissue collection. (C) 50% paw withdraw threshold (PWT) of ipsilateral hind paw of SMIR model rats vs sham group of rats. (D) Summary of normalized area under the curve (AUC) of time courses shown in panel (C). *p < 0.05 and **p<0.01 vs sham group. n = 6–7 rats/group.
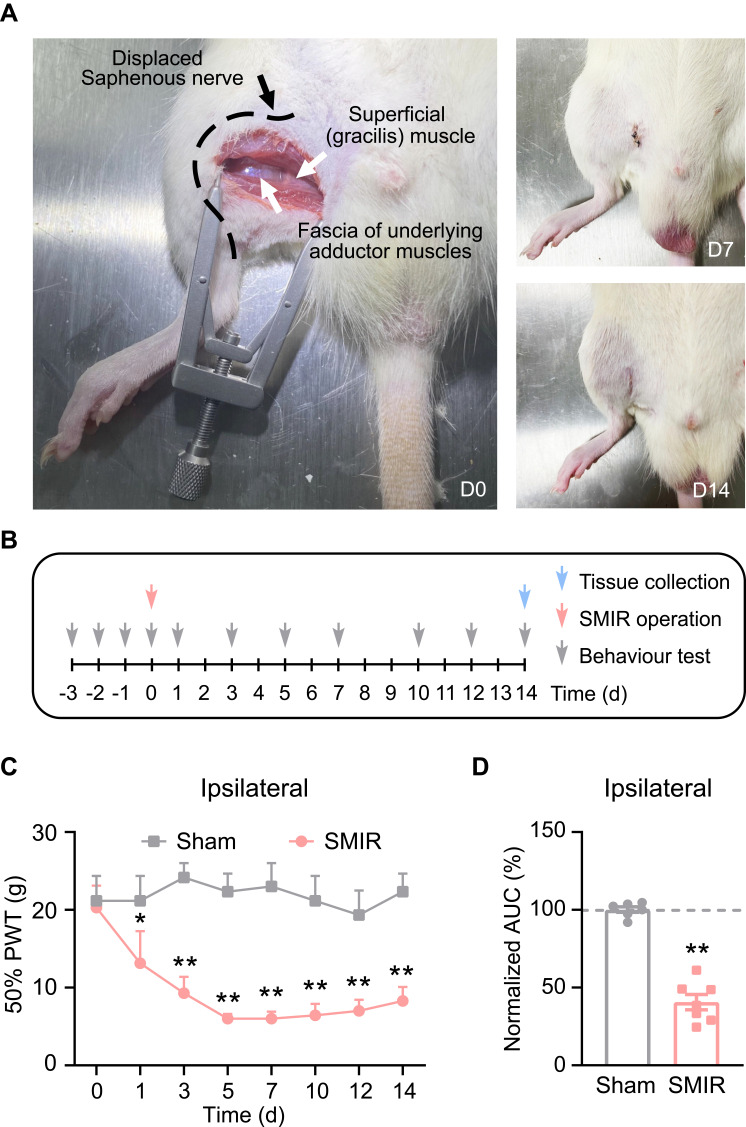
SMIR Model Rats Exhibited Increased c-Fos Immunoreactivity and Glial Cell Overactivation in SCDH
SCDH is responsible for gathering and integrating pain signals from periphery sensory nerves.25,26 We then examined the neuronal activity of SCDH in SMIR model rats. The rats were euthanized 14 days after the surgery, as shown in Figure 1B. The SCDH tissues from the ipsilateral side were harvested and processed for immunofluorescence staining. The expression of c-Fos, a marker indicating neuronal activity following noxious stimulation, was higher in ipsilateral side of SCDH of SMIR group compared with sham group (Figures 2A and B). Spinal glial cells, including astrocytes and microglia, are considered as important players involved in neuron–glia crosstalk and central pain sensitization, which contribute to chronic pain maintaining mechanisms. Next, the status of spinal glia cells of SCDH from SMIR rats was investigated. We observed that the expression of GFAP (an astrocytic marker) was obviously up-regulated in ipsilateral SCDH of SMIR rats (Figures 2C and D). In addition, the expression of OX42, a microglial marker, was remarkably increased in ipsilateral SCDH of SMIR rats as well (Figures 2E and F). Chronic pain, especially neuropathic pain, is usually accompanied by neuronal damage in sensory neurons. We proceeded to check if there was any neuronal damage in ipsilateral SCDH. Figures 2G and H show that the expression of neuronal damage marker ATF3 was not significantly changed in SMIR rats, ruling out the participation of spinal neuronal damage in SMIR-induced chronic pain condition.
Figure 2.
Ipsilateral SCDH of SMIR model rats showed remarkable glial cell overactivation and c-Fos immunoreactivity, but no neuronal damage. (A) Representative immunofluorescence pictures showing c-Fos staining of ipsilateral SCDH from SMIR and sham group of rats. (B) Summary of c-Fos positively stained cell numbers in SMIR and sham groups. (C) Representative immunofluorescence pictures showing GFAP (an astrocytic marker) antibody staining of ipsilateral SCDH from SMIR and sham groups. (D) Normalized percentage increase in GFAP immunostaining intensity in SMIR and sham groups. (E) Representative immunofluorescence pictures showing OX42 (a microglia marker) antibody staining of ipsilateral SCDH from SMIR and sham groups. (F) Normalized percentage increase in OX42 immunostaining intensity in SMIR and sham groups. (G) Representative immunofluorescence images showing ATF3 (a neuronal damage marker) antibody staining of ipsilateral SCDH from SMIR and sham groups. NeuN is used for the labelling of neurons. (H) Summarized data showing ATF3 positively stained cell numbers in SMIR and sham groups. Scale bar indicates 50 μm. **p < 0.01 vs sham group. n = 5 rats/group.
Abbreviation: NS, no significance.
Genome-Wide Expression Profiling of SCDH from SMIR Rats Through RNA-Seq
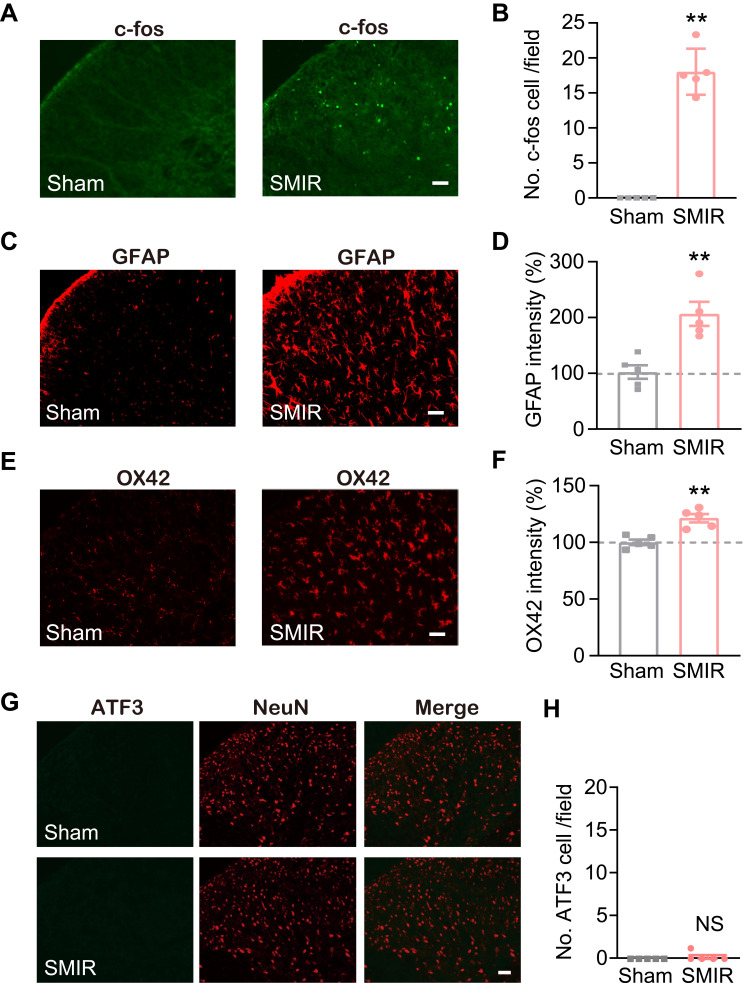
To gain more mechanistic insights into SMIR-induced chronic pain, ipsilateral SCDH (from L4-6 lumbar segment) was collected on day 14 from sham group and SMIR group of rats. Four rats were included in each group. We then employed RNA-Seq technique to examine the global gene expression changes. Supplementary Figure 2 shows the quality check of the RNAs that we managed to extract. The RNA displayed decent qualities (including both purity and integrity) that could be subjected to RNA-Seq processing. After sequencing, each sample generated around 23.92M (million) raw reads (Table 2). Sequencing further showed that clean reads ratio was over 97.0%. Approximately 93% total reads showed clean reads of Q30. Around 94% of the total reads were matched to the genome of rat (Table 2), which identified a total of 22,853 genes together. Supplementary Table 1 further illustrates the overall genes matched to the genome and their corresponding expression changes in each sample. Next, the differentially expressed genes (DEGs) were selected based upon criteria as follows: fold change ≥ 1.25 or ≤ −1.25 with FDR q-value ≤ 0.001 (Supplementary Table 2), as in our recent study.27 A total number of 727 DEGs (401 up-regulated and 326 down-regulated) was obtained from SMIR vs sham rats. The filtered out DEGs were then illustrated in the volcano plot, together with all other non-DEGs (Figure 3A). These DEGs were further illustrated in a heatmap, as shown in Figure 3B. We then performed hierarchical clustering analysis and found a high consistency of the samples within the same group whereas there was distinct segregation between groups. The results demonstrated that we successfully obtained RNA-Seq dataset of high quality that could be further analyzed by bioinformatics.
Table 2.
Information on Total Reads and Mapping Ratio for Sham and SMIR Groups in RNA-Seq
| Sample | Total Raw Reads (M) | Total Clean Reads (M) | Total Mapping (%) | Clean Reads Q20 (%) | Clean Reads Q30 (%) | Clean Reads Ratio (%) | Total Mapping (%) |
|---|---|---|---|---|---|---|---|
| SMIR1 | 23.92 | 23.37 | 1.17 | 97.66 | 92.71 | 97.68 | 94.09 |
| SMIR2 | 23.92 | 23.46 | 1.17 | 97.73 | 92.63 | 98.05 | 94.51 |
| SMIR3 | 23.92 | 23.47 | 1.17 | 97.71 | 92.59 | 98.13 | 94.4 |
| SMIR4 | 23.92 | 23.46 | 1.17 | 97.81 | 92.93 | 98.05 | 94.44 |
| Sham1 | 23.92 | 23.39 | 1.17 | 97.77 | 93.1 | 97.79 | 94.02 |
| Sham2 | 23.92 | 23.37 | 1.17 | 97.72 | 92.91 | 97.69 | 93.95 |
| Sham3 | 23.92 | 23.37 | 1.17 | 97.84 | 93.28 | 97.69 | 94.03 |
| Sham4 | 23.92 | 23.35 | 1.17 | 97.73 | 92.9 | 97.61 | 93.47 |
Figure 3.
Genome-wide expression profiling through RNA-Seq of ipsilateral SCDH from SMIR model and sham group of rats. (A) Gene expression profiles of SMIR model group vs sham group shown by volcano graphs. Red and blue spots indicate up- and down-regulated DEGs. Grey spots indicate non-DEGs. (B) Heatmap showing DEGs and hierarchical clustering analysis of the DEGs. n = 4 rats/group.
Examination of Gene Profiles of DEGs from Ipsilateral SCDH of SMIR Rats
Some DEGs were documented to be relevant with inflammation or nociception processing. These genes included Scgb1a1 (secretoglobin family 1A member 1, fold change = 41.4), Pdk4 (Pyruvate dehydrogenase lipoamide kinase isozyme 4, fold change = 2.8) and Serpina3n (serine protease inhibitor A3N, fold change = 2.7), etc.28–30 In total, RNA-Seq identified 63 genes (40 up- and 23 down-regulated), which showed over 10-fold expression changes over 10 fold; 15 genes (4 up- and 11 down-regulated) had expression changes of 5–10 fold. We further summarize the detailed information regarding the top 20 up- or down-regulated genes in Tables 3 and 4, respectively.
Table 3.
Detailed Information on the Top 20 Up-Regulated DEGs
| Up-Regulated Gene | Gene ID | Location | Log2 Fold Change (SMIR/Sham) | Q value | Official Gene Name (NCBI) |
|---|---|---|---|---|---|
| Heatr1 | 361262 | chr17:66446135–66495506 | 10.97753865 | 5.68E-09 | HEAT repeat containing 1 |
| LOC103694869 | 103694869 | NW_007906808.1:134857–155285 | 9.99681143 | 2.19E-83 | Isochorismatase domain-containing protein 1 |
| NEWGENE_1310680 | 108348045 | chr1:220462462–220467269 | 8.387705661 | 9.66E-34 | Transmembrane protein 151A |
| LOC100911730 | 100911730 | chr1:214009687–214013762 | 7.853231583 | 1.01E-24 | CD151 antigen-like |
| LOC100912475 | 100912475 | chr1:77829695–77830416 | 7.579331227 | 3.50E-07 | Consortin-like |
| LOC103690175 | 103690175 | chrX:158265270–158288228 | 7.508770847 | 3.70E-20 | PHD finger protein 6 |
| RGD1566138 | 498167 | chr12:21950846–21980699 | 7.336970389 | 3.39E-18 | Similar to Zinc finger, CW type with PWWP domain 1 |
| LOC102550456 | 102550456 | chr12:21748608–21761500 | 7.111172475 | 6.68E-16 | TSC22 domain family protein 4-like |
| LOC103692570 | 103692570 | chr6:26423841–26446080 | 7.008208521 | 6.11E-15 | Dihydropyrimidinase-related protein 5-like |
| Dgcr2 | 360742 | chr11:87242441–87290806 | 6.683737062 | 2.92E-12 | DiGeorge syndrome critical region gene 2 |
| LOC108348079 | 108348079 | chrX:124516931–124518080 | 6.624716548 | 7.96E-12 | RING finger protein 113A-like |
| LOC100911725 | 100911725 | chr8:118390023–118416270 | 6.478945409 | 8.32E-11 | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4-like |
| LOC100364457 | 100364457 | chr1:77829695–77830416 | 6.361739792 | 4.73E-10 | Ribosomal protein L9-lik |
| Slc40a1 | 170840 | chr9:52819451–5283046 | 6.315701772 | 9.01E-10 | Solute carrier family 40 member 1 |
| LOC100909913 | 100909913 | chrX:6258264–6282760 | 6.275508647 | 1.56E-09 | Norrin-like |
| LOC100911168 | 100911168 | chrX:63392209–63397468 | 6.191793172 | 9.96E-11 | Kelch-like protein 15-like |
| LOC108348080 | 108348080 | chr5:137670134–137674730 | 6.116511993 | 1.22E-08 | Probable rRNA-processing protein EBP2 |
| LOC103689983 | 103689983 | chrX:158623231–158655277 | 5.976710792 | 6.33E-08 | Hypoxanthine-guanine phosphoribosyltransferase |
| LOC100911585 | 100911585 | chr9:20241067–20251271 | 5.887066102 | 1.76E-35 | Leucine-rich repeat-containing protein 23-like |
| Potef | 684969 | chr18:24780521–24783147 | 5.742829959 | 7.42E-07 | POTE ankyrin domain family, member F |
Table 4.
Detailed Information on the Top 20 Down-Regulated DEGs
| Down-Regulated Gene | Gene ID | Location | Log2 Fold Change (SMIR/Sham) | Q value | Official Gene Name (NCBI) |
|---|---|---|---|---|---|
| LOC100912571 | 100912571 | chr11:82886428–82913153 | −8.771523031 | 1.65269E-41 | Eukaryotic translation initiation factor 4 gamma 1-like |
| LOC103693608 | 103693608 | chr11:87765024–87775583 | −8.443487358 | 2.29265E-34 | Integral membrane protein DGCR2/IDD-like |
| LOC100911186 | 100911186 | chr1:141821909–141830770 | −8.016970362 | 7.83502E-27 | Enoyl-CoA hydratase, mitochondrial-like |
| LOC103693640 | 103693640 | chr12:30425332–30427226 | −7.423305636 | 1.05279E-27 | MLV-related proviral Env polyprotein-like |
| LOC100361543 | 100361543 | chrX:37261428–37262470 | −7.270216573 | 3.04548E-17 | RhoA activator C11orf59-like |
| LOC100910207 | 100910207 | chr14:1621021–1632432 | −6.72157611 | 2.23063E-12 | Protein Dr1-like |
| Pirb | 690955 | chr1:64074097–64081568 | −6.705961168 | 2.92195E-12 | Paired Ig-like receptor B |
| LOC103692976 | 103692976 | chr7:140149991–140172615 | −6.579461469 | 2.41046E-11 | Cyclin-T1-like |
| LOC100912259 | 100912259 | chr4:170079196–170083804 | −6.357579032 | 6.82669E-10 | Uncharacterized LOC100912259 |
| LOC100912034 | 100912034 | chr4:31387262–31392791 | −6.347550327 | 7.83703E-10 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-11-like |
| LOC688583 | 688583 | chr14:51458436–51462755 | −6.310416081 | 1.29914E-09 | Similar to high mobility group protein 4 (HMG-4) |
| LOC100912590 | 100912590 | chr12:52670809–52674768 | −6.184080348 | 6.82958E-09 | Putative GTP-binding protein 6-like |
| Retreg1 | 619558 | chr2:78391921–78401569 | −6.113374706 | 4.00516E-47 | Reticulophagy regulator 1 |
| LOC103690002 | 103690002 | chr4:170148955–170149479 | −5.593471284 | 3.55564E-06 | Histone H2A.J |
| LOC100910528 | 100910528 | chr1:32569844–32573188 | −5.573942119 | 4.22151E-06 | 39S ribosomal protein L36, mitochondrial-like |
| LOC100362967 | 100362967 | chr2:238839535–238839941 | −5.065849831 | 0.000182453 | Ribosomal protein 10-like |
| Rcor2 | 305811 | chr1:222513970–222525356 | −4.559740427 | 1.08733E-41 | REST corepressor 2 |
| Kb23 | 407759 | chr7:143134980–143141659 | −4.281431774 | 1.04844E-27 | Type II keratin 23 |
| Krt83 | 681126 | chr7:143078996–143085833 | −4.146112498 | 4.22656E-07 | Keratin 83 |
| LOC103689978 | 103689978 | chr11:72035540–72070679 | −3.957936403 | 3.78505E-92 | Sentrin-specific protease 5 |
Functional Analysis of DEGs from Ipsilateral SCDH of SMIR Rats by Bioinformatics
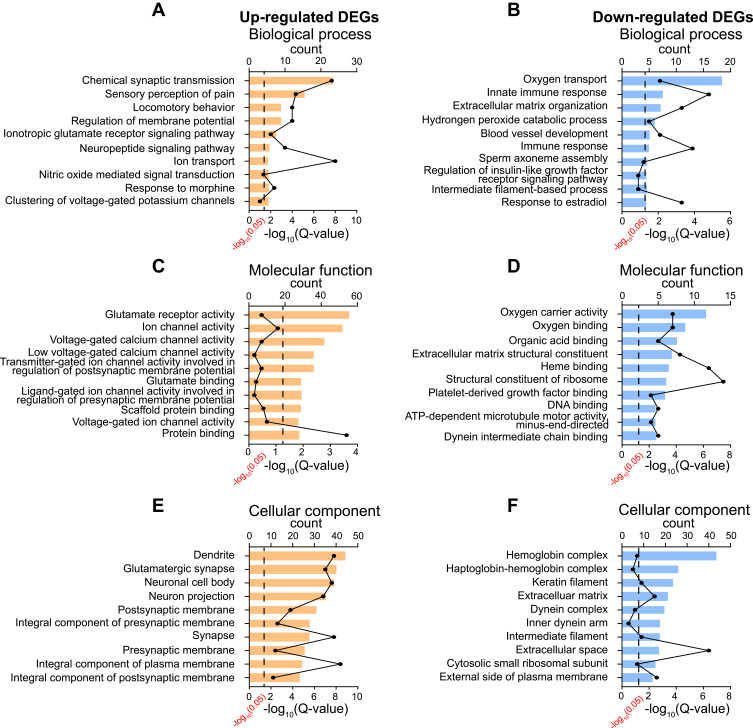
We then utilized gene ontology (GO) analysis to further study the DEGs we had identified. GO analysis showed that the top-ranked biological process of up-regulated or down-regulated genes included chemical synaptic transmission, sensory perception of pain and locomotor behavior or oxygen transport, innate immune response and extracellular matrix organization, respectively (Figures 4A and B). The top-ranked molecular function of up-regulated or down-regulated genes included glutamate receptor activity, ion channel activity, voltage-gated calcium channel activity or oxygen carrier activity, oxygen binding, organic acid binding, respectively (Figures 4C and D). The top-ranked cellular function of up-regulated or down-regulated genes included dendrite, glutamatergic synapse, neuronal cell body or hemoglobin complex, keratin filament, extracellular matrix, respectively (Figures 4E and F). The specific genes belonged to specific GO categories mentioned above are further illustrated in Supplementary Table 3.
Figure 4.
GO enrichment analysis of DEGs identified from ipsilateral SCDH of SMIR model rats. (A, C–E) Top 10 mostly enriched biological processes, molecular functions and cellular components of up-regulated DEGs. (B, D–F) Top 10 mostly enriched biological processes, molecular functions and cellular components of down-regulated DEGs.
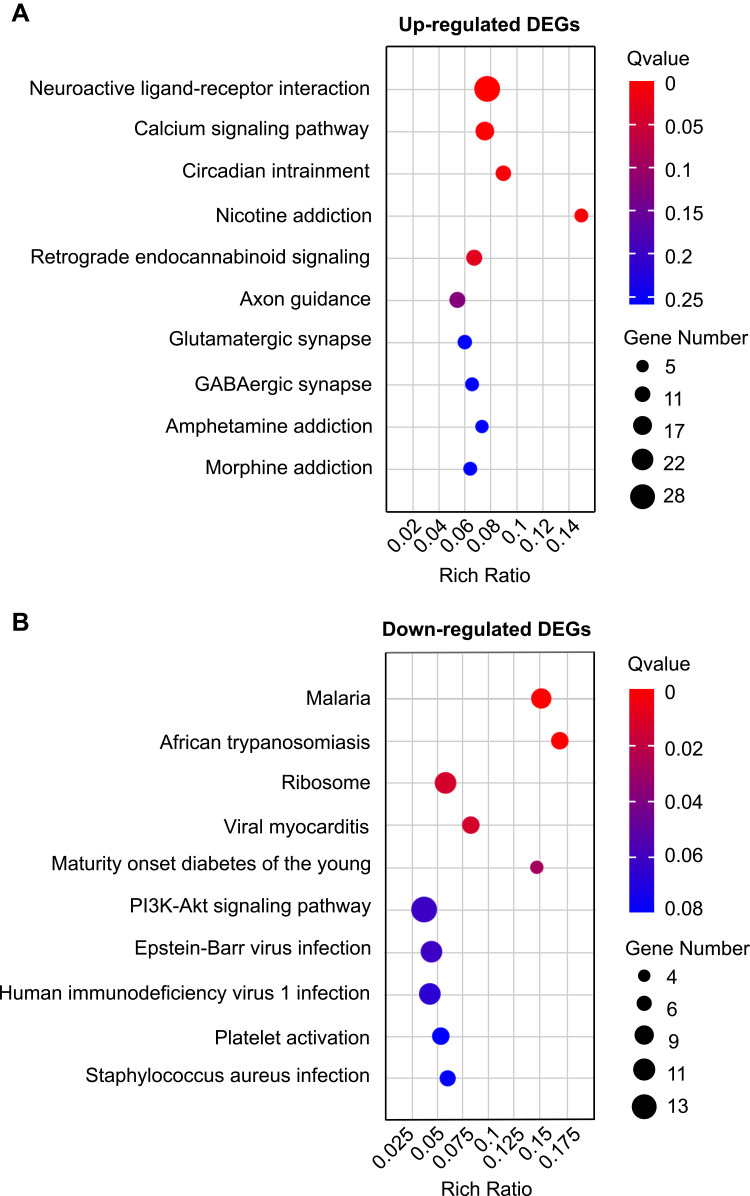
Next, the DEGs were subjected to KEGG pathway enrichment analysis to explore pathways potentially involved in the pathogenesis of CPSP. According to the results of KEGG pathway analysis, the up-regulated genes were mainly involved in neuroactive ligand-–receptor interaction, calcium signaling pathway, circadian entrainment, retrograde endocannabinoid signaling and glutamatergic synapse, etc. (Figure 5A and Supplementary Table 4). Conversely, the down-regulated genes were mainly involved in processes including malaria, African trypanosomiasis and ribosome, etc. (Figure 5B).
Figure 5.
KEGG pathway analysis of DEGs identified from ipsilateral SCDH of SMIR model rats. (A) Top 10 pathways of up-regulated DEGs by bubble plots. (B) Top 10 pathways of down-regulated DEGs by bubble plots. Larger bubbles indicate higher number of genes and vice versa. The bubble’s color indicates significance.
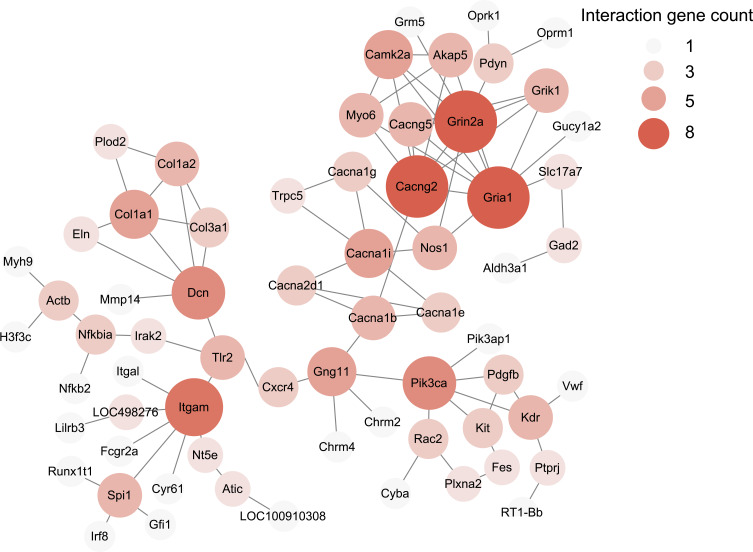
Then PPI network was constructed using “STRING” to explore potential interactions and hub genes of DEGs involved in pathogenesis of CPSP. The analysis revealed that several genes could serves as major hub genes. These genes include Cacng2, Grin2a, Gria1 and Itgam, etc. (Figure 6). To focus more closely on genes related to pain processing and mechanisms, we retrieved a recently published dataset about human pain-related genes.31 We then mapped our DEGs with this database and identified that 58 genes out of 727 DEGs were related to human pain mechanisms (Supplementary Table 5).
Figure 6.
The construction of PPI networks for all DEGs in ipsilateral SCDH of SMIR model rats. Circles represent proteins, whereas lines represent the possible interactions between the protein pairs. Larger circles and deeper color show more protein–protein interactions.
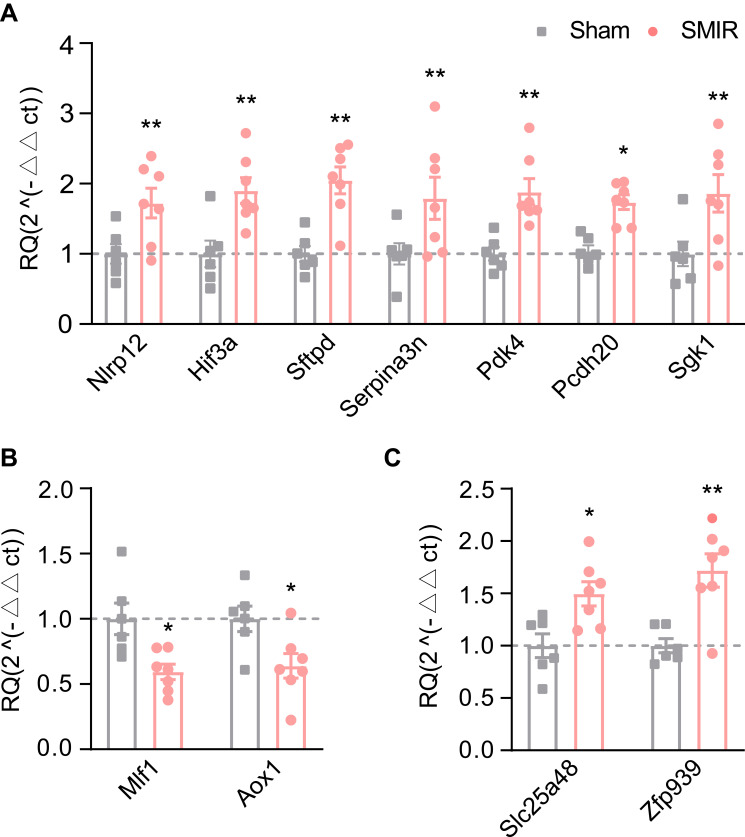
qPCR Validation of the RNA-Seq Dataset
Finally, we set out to validate gene expression changes identified by RNA-Seq using qPCR. We started by evaluating some typical genes known to participate in inflammation or pain modulation processes. These genes include Nlrp12, Hif3a, Sftpd, Serpina3n, Pcdh20 and Sgk1. The data obtained from qPCR indicated that expression of the above mentioned genes increased significantly, showing consistency with RNA-Seq dataset (Figure 7A). Then 2 down-regulated genes (Mlf1 and Aox1) and 2 up-regulated genes (Slc25a48 and Zfp939) were randomly picked up from the DEGs and verified via qPCR. The data showed Mlf1 and Aox1 gene expression was down-regulated, whereas Slc25a48 and Zfp939 gene expression was up-regulated in SMIR model group compared with sham group (Figures 7B and C). These qPCR results demonstrated consistency with the RNA-Seq dataset we obtained.
Figure 7.
Quantitative PCR validation of RNA-Seq data. (A) The expression validations by qPCR of representative genes related to inflammation and pain. (B) Gene expression validations by qPCR of randomly selected up- (B) or down-regulated (C) genes. n = 6–7 rats/group. *p < 0.05, **p < 0.01 vs sham group.
Discussion
In this work, we established a rat SMIR model to replicate the human CPSP condition. The model animals exhibited strong and persistent mechanical hypersensitivities in the affected hind limbs. Ipsilateral side of the SCDH of SMIR rats showed remarkable glial cell activation and elevated neuronal activities, but no neuronal damage occurred. In order to get a systematic view of the possible mechanisms involved, we performed a genome-wide expression profiling of ipsilateral SCDH from SMIR and sham rats. A number of DEGs were identified by RNA-Seq. qPCR confirmed expression of some representative DEGs. Bioinformatics indicated that chemical synaptic transmission, sensory perception of pain and neuroactive ligand–receptor interaction were predominant functions. We further compared our dataset with human pain-related genes and found a number of genes exclusively involved in pain modulation and mechanisms.
To facilitate mechanistic studies of CPSP, Flatters developed the skin/muscle incision and retraction (SMIR)-induced pain model.7 This animal model mimics the prolonged tissue retraction during surgery and exhibits obvious and long-term mechanical pain in the affected hind limb, which resembles CPSP. Here, in this work, we successfully established the SMIR rat model. In our hands, we observed that the SMIR rats developed similar mechanical hypersensitivities in affected hind limbs, which is consistent with previous reports. We observed that SMIR rats showed strong c-Fos immunoreactivity in ipsilateral SCDH 14 days after model establishment, suggesting that ipsilateral SCDH receives persistent nociceptive inputs. In addition, there are strong activations of astrocytes and microglia in ipsilateral SCDH from SMIR rat group. This observation agrees well with some other recent literature using the same SMIR animal model.11,12
It is known that the spinal glial cell activation triggers the production and release of inflammation mediators or cytokines/chemokines into extracellular space, which bind to corresponding receptors expressed in spinal neurons via neuron–glia crosstalk to produce neuroinflammation and central sensitization.32 It is reported in one recent study that pharmacological inhibition of spinal glial cells attenuates SMIR-induced persistent mechanical allodynia in rats.11 These data clearly indicate that SCDH is critically involved in maintaining SMIR-induced chronic pain condition. Based upon these above reasons, we decided to apply RNA-Seq to characterize gene expression profiles in ipsilateral SCDH of SMIR rats to gain a comprehensive overview of possible mechanisms involved.
Here, we found that the expression of Pcdh20 was obviously increased in ipsilateral SCDH of SMIR rats through RNA-Seq and qPCR. Pcdh20 is a gene-encoding protein, Protocadherin20, a protein belonging to the PCDHs family, which are cell adhesion molecules involved in cellular interactions, cell adhesion and development of central nerve system, etc.33,34 In a previous study, it is reported that the protein expression of Protocadherin20 was significantly increased in SCDH from rats of a bone cancer pain model.35 It is found that Protocadherin20 makes important contributions to the neurite growth and the formations of excitatory synapses of spinal neurons. Gene knockdown of Pcdh20 ameliorated bone cancer pain and reduced the formation of excitatory synapses in ipsilateral SCDH in model rats. Thus, Protocadherin20 makes contributions to bone cancer pain mechanisms via promoting excitatory synapse formation in the spinal dorsal horn.35 Our current findings suggest that increased expression of Pcdh20 gene in SCDH may make contributions to SMIR-induced CPSP mechanism via modulating excitatory synapse formations.
Here, we also observed that Pdk4 gene expression is obviously increased in SCDH of SMIR animals through RNA-Seq and qPCR methods. Pdk4 encodes pyruvate dehydrogenase kinase 4, which modulates the activities of pyruvate dehydrogenase. Pyruvate dehydrogenase is an enzyme in charge of transforming pyruvates to acetyl-CoA or lactates, depending on specific aerobic or anaerobic conditions, respectively.30 PDK4 expression is increased in local inflamed tissues and Pdk4 deficiency attenuates local lactate increase and ameliorates both mechanical and heat pain in hind paws produced by injection of complete Freund’s adjuvant.30 Moreover, animals deficient in Pdk2/4 genes showed reduced lactate levels and macrophage infiltration in DRG, as well as reduced central sensitization in streptozotocin-induced diabetes mouse model.36 A recent study demonstrated that intrathecal lactate injection causes obvious mechanical hypersensitivities in naïve mice. Further experiments using pharmacological methods to reduce lactate supplies in spinal cord significantly attenuated neuropathic pain in model animals.37 These results suggest that lactate production either in periphery or spinal cord is an important contributing factor to chronic pain. Thus, these results suggest a possible involvement of Pdk4 in SMIR-induced CPSP via modulating lactate production in SCDH. Further studies will be needed to confirm the protein expression of PDK4 and lactate levels in SCDH of SMIR model animals.
GO analysis from the present study demonstrated that the “sensory perception of pain” constitutes one of the predominant biological processes. This result indicates that pain-related gene expression changes could be a major biological process that takes place in SCDH from SMIR rats. This result is also consistent with the nature of this model as being a CPSP model. In order to gain more understanding of the genes related to pain processing, we continued to compare our dataset with a comprehensive dataset summarizing all human pain-related genes.31 Among these DEGs, several well-established pain-related genes were identified, including Bdkrb2 and Tacr1. These results suggest that a diversity of pain mechanisms may together contribute to overall CPSP, reflecting the complex nature of mechanisms involved in chronic pain.
In addition, a number of certain well-known endogenous analgesia-relevant genes showed up in the DEGs list. These genes include Oprm1, Oprk1, Cnr1 and Cnr2, which encode protein μ-opioid receptor, κ-opioid receptor, cannabinoid receptor 1 and cannabinoid receptor 2, respectively. These receptors operated as endogenous opioid and endocannabinoid analgesic signaling. These results suggest that endogenous analgesic mechanisms may be activated during the pathogenesis of CPSP. The persistent activation of these endogenous analgesic signaling may help to counteract the chronic pain condition to some extent, which is a reflection of the endogenous protection machinery upon stress insult. Further functional validations of these potential pain- or analgesia-related genes are needed to unravel the detailed mechanisms underlying CPSP.
Conclusions
Our study presented a pilot work to explore genome-wide expression profiles of ipsilateral SCDH of a rat model of CPSP. We found several pain-related genes and pathways that could possibly be related to CPSP. These findings may provide us with some mechanistic insights into the mechanisms of CPSP. We hope further functional validations of these identified DEGs or pathways will be performed to reveal the underlying mechanisms of CPSP.
Funding Statement
This project was supported by the National Natural Science Foundation of China (81873365 & 82105014), Zhejiang Provincial Natural Science Funds (LQ21H270004 & LR17H270001) and research funds from ZCMU (2021JKZDZC07, Q2019J01 & KC201943).
Institutional Review Board Statement
The experimental steps were performed according to National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978) and approved by the Animal Ethics Committee of Zhejiang Chinese Medical University (Permission Number: ZSLL-2017- 183).
Data Sharing Statement
The key data are contained in the figures, tables, and additional files. The datasets used and/or analyzed during this study can be further obtained from the corresponding author, Boyi Liu, on reasonable request.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Bruce J, Quinlan J. Chronic post surgical pain. Rev Pain. 2011;5(3):23–29. doi: 10.1177/204946371100500306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Correll D. Chronic postoperative pain: recent findings in understanding and management. F1000Res. 2017;6:1054. doi: 10.12688/f1000research.11101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macrae WA. Chronic post-surgical pain: 10 years on. Br J Anaesth. 2008;101(1):77–86. doi: 10.1093/bja/aen099 [DOI] [PubMed] [Google Scholar]
- 4.Srivastava D. Chronic post-amputation pain: peri-operative management - review. Br J Pain. 2017;11(4):192–202. doi: 10.1177/2049463717736492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chidambaran V, Gang Y, Pilipenko V, Ashton M, Ding L. Systematic review and meta-analysis of genetic risk of developing chronic postsurgical pain. J Pain. 2020;21(1–2):2–24. doi: 10.1016/j.jpain.2019.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinrib AZ, Azam MA, Birnie KA, Burns LC, Clarke H, Katz J. The psychology of chronic post-surgical pain: new frontiers in risk factor identification, prevention and management. Br J Pain. 2017;11(4):169–177. doi: 10.1177/2049463717720636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flatters SJ. Characterization of a model of persistent postoperative pain evoked by skin/muscle incision and retraction (SMIR). Pain. 2008;135(1):119–130. doi: 10.1016/j.pain.2007.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen YW, Tzeng JI, Lin MF, Hung CH, Wang JJ. Forced treadmill running suppresses postincisional pain and inhibits upregulation of substance P and cytokines in rat dorsal root ganglion. J Pain. 2014;15(8):827–834. doi: 10.1016/j.jpain.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 9.Song J, Ying Y, Wang W, et al. The role of P2X7R/ERK signaling in dorsal root ganglia satellite glial cells in the development of chronic postsurgical pain induced by skin/muscle incision and retraction (SMIR). Brain Behav Immun. 2018;69:180–189. doi: 10.1016/j.bbi.2017.11.011 [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Li Y, Cao J, et al. Membrane protein Nav1.7 contributes to the persistent post-surgical pain regulated by p-p65 in dorsal root ganglion (DRG) of SMIR rats model. BMC Anesthesiol. 2017;17(1):150. doi: 10.1186/s12871-017-0438-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T, Liu T, Chen X, et al. Microglia induce the transformation of A1/A2 reactive astrocytes via the CXCR7/PI3K/Akt pathway in chronic post-surgical pain. J Neuroinflammation. 2020;17(1):211. doi: 10.1186/s12974-020-01891-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying YL, Wei XH, Xu XB, et al. Over-expression of P2X7 receptors in spinal glial cells contributes to the development of chronic postsurgical pain induced by skin/muscle incision and retraction (SMIR) in rats. Exp Neurol. 2014;261:836–843. doi: 10.1016/j.expneurol.2014.09.007 [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Yang M, Tang H, Ma Z, Liang Y, Li Z. The over-production of TNF-alpha via Toll-like receptor 4 in spinal dorsal horn contributes to the chronic postsurgical pain in rat. J Anesth. 2015;29(5):734–740. doi: 10.1007/s00540-015-2011-2 [DOI] [PubMed] [Google Scholar]
- 14.Chai W, Tai Y, Shao X, et al. Electroacupuncture alleviates pain responses and inflammation in a rat model of acute gout arthritis. Evid Based Complement Alternat Med. 2018;2018:2598975. doi: 10.1155/2018/2598975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Yin C, Liu B, et al. Transcriptome profiling of long noncoding RNAs and mRNAs in spinal cord of a rat model of paclitaxel-induced peripheral neuropathy identifies potential mechanisms mediating neuroinflammation and pain. J Neuroinflammation. 2021;18(1):48. doi: 10.1186/s12974-021-02098-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon WJ. Efficient analysis of experimental observations. Annu Rev Pharmacol Toxicol. 1980;20(1):441–462. doi: 10.1146/annurev.pa.20.040180.002301 [DOI] [PubMed] [Google Scholar]
- 17.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53(1):55–63. doi: 10.1016/0165-0270(94)90144-9 [DOI] [PubMed] [Google Scholar]
- 18.Li X, Yin C, Hu Q, et al. Nrf2 activation mediates antiallodynic effect of electroacupuncture on a rat model of complex regional pain syndrome type-I through reducing local oxidative stress and inflammation. Oxid Med Cell Longev. 2022. doi: 10.1155/2022/8035109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin C, Hu Q, Liu B, et al. Transcriptome profiling of dorsal root ganglia in a rat model of complex regional pain syndrome type-I reveals potential mechanisms involved in pain. J Pain Res. 2019;12:1201–1216. doi: 10.2147/JPR.S188758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Q, Zheng X, Li X, et al. Electroacupuncture alleviates mechanical allodynia in a rat model of complex regional pain syndrome type-I via suppressing spinal CXCL12/CXCR4 signaling. J Pain. 2020;21(9–10):1060–1074. doi: 10.1016/j.jpain.2020.01.007 [DOI] [PubMed] [Google Scholar]
- 21.Chen R, Yin C, Hu Q, et al. Expression profiling of spinal cord dorsal horn in a rat model of complex regional pain syndrome type-I uncovers potential mechanisms mediating pain and neuroinflammation responses. J Neuroinflammation. 2020;17(1):162. doi: 10.1186/s12974-020-01834-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nie H, Liu B, Yin C, et al. Gene expression profiling of contralateral dorsal root ganglia associated with mirror-image pain in a rat model of complex regional pain syndrome type-I. J Pain Res. 2021;14:2739–2756. doi: 10.2147/JPR.S322372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin C, Liu B, Wang P, et al. Eucalyptol alleviates inflammation and pain responses in a mouse model of gout arthritis. Br J Pharmacol. 2020;177(9):2042–2057. doi: 10.1111/bph.14967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 25.Chen R, Yin C, Fang J, Liu B. The NLRP3 inflammasome: an emerging therapeutic target for chronic pain. J Neuroinflammation. 2021;18(1):84. doi: 10.1186/s12974-021-02131-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peirs C, Williams SP, Zhao X, et al. Dorsal horn circuits for persistent mechanical pain. Neuron. 2015;87(4):797–812. doi: 10.1016/j.neuron.2015.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B, Chen R, Wang J, et al. Exploring neuronal mechanisms involved in the scratching behavior of a mouse model of allergic contact dermatitis by transcriptomics. Cell Mol Biol Lett. 2022;27(1):16. doi: 10.1186/s11658-022-00316-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu M, Yang W, Wang X, Nayak DK. Lung secretoglobin Scgb1a1 influences alveolar macrophage-mediated inflammation and immunity. Front Immunol. 2020;11:584310. doi: 10.3389/fimmu.2020.584310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vicuna L, Strochlic DE, Latremoliere A, et al. The serine protease inhibitor SerpinA3N attenuates neuropathic pain by inhibiting T cell-derived leukocyte elastase. Nat Med. 2015;21(5):518–523. doi: 10.1038/nm.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jha MK, Song GJ, Lee MG, et al. Metabolic connection of inflammatory pain: pivotal role of a pyruvate dehydrogenase kinase-pyruvate dehydrogenase-lactic acid axis. J Neuro sci. 2015;35(42):14353–14369. doi: 10.1523/JNEUROSCI.1910-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung MK, Park J, Asgar J, Ro JY. Transcriptome analysis of trigeminal ganglia following masseter muscle inflammation in rats. Mol Pain. 2016;12:1744806916668526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donnelly CR, Andriessen AS, Chen G, et al. Central nervous system targets: glial cell mechanisms in chronic pain. Neurotherapeutics. 2020;17(3):846–860. doi: 10.1007/s13311-020-00905-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pancho A, Aerts T, Mitsogiannis MD, Seuntjens E. Protocadherins at the Crossroad of signaling pathways. Front Mol Neurosci. 2020;13:117. doi: 10.3389/fnmol.2020.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirabayashi T, Yagi T. Protocadherins in neurological diseases. Adv Neurobiol. 2014;8:293–314. [DOI] [PubMed] [Google Scholar]
- 35.Ke C, Li C, Huang X, et al. Protocadherin20 promotes excitatory synaptogenesis in dorsal horn and contributes to bone cancer pain. Neuropharmacology. 2013;75:181–190. doi: 10.1016/j.neuropharm.2013.07.010 [DOI] [PubMed] [Google Scholar]
- 36.Rahman MH, Jha MK, Kim JH, et al. Pyruvate dehydrogenase kinase-mediated glycolytic metabolic shift in the dorsal root ganglion drives painful diabetic neuropathy. J Biol Chem. 2016;291(11):6011–6025. doi: 10.1074/jbc.M115.699215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyamoto K, Ishikura KI, Kume K, Ohsawa M. Astrocyte-neuron lactate shuttle sensitizes nociceptive transmission in the spinal cord. Glia. 2019;67(1):27–36. doi: 10.1002/glia.23474 [DOI] [PubMed] [Google Scholar]