Abstract
The coronavirus disease 2019 (COVID-19) pandemic has posed a threat to public health throughout the world since the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) was discovered in late 2019. Since the beginning of the pandemic, scientists have done a tremendous amount of work in this area. However, among these studies, the investigation of the effect of newly synthesized compounds against coronavirus is rather weak. Examining the newly synthesized compounds with a computer-assisted molecular docking study provides quite an advantage in terms of the estimation and analysis of the biochemical activity and binding affinity of existing synthesized compounds against a biological target in a labor, time, and cost-saving way. In this study, the SNS pincer type 2,6-bis[[(4-methylphenyl)thio]methyl]pyridine ligand(L) (1) and its novel Pd(II) complexes ([Pd(κ2-L)(OAc)2]·3H2O (2) and [Pd(κ2-L)Cl2]·3H2O (3)) were synthesized and characterized by using FT-IR, UV–Vis, NMR, mass and elemental analysis techniques. The synthesized Pd complexes exhibited a square planar structure. The compounds were found to have non-electrolytic behavior. In the meantime, in silico investigations have defined and justified interaction processes between these molecules and Pd(II) at the atomic level. Furthermore, using molecular docking against target proteins of SARS-CoV-2, the efficiency of the SNS pincer type ligand and its Pd (II) complexes produced was studied and discussed for the first time. The experimental data has been supported and illuminated using computational visual methods and molecular docking, and the findings produced indicate compatibility. The binding energy values of the relevant compounds on the four protein model structures of SARS-CoV-2 (Main Protease, Papain-like protease, RdRp without RNA, and RdRp with RNA) are represented. Compound 2 ([Pd(κ2-L)(OAc)2]·3H2O) is the structure that exhibits the highest biochemical activity. According to all of the docking studies, Papain-like protease is the SARS-CoV-2 protein with which the three compounds exhibit mutual interaction. The compound 2 structure, in particular, is the most effective in terms of structural and interaction with the targets, as well as binding orientations.
Keywords: COVID-19, Pincer, Metal complexes, Molecular docking, SARS-CoV-2 proteins
Graphical abstract
1. Introduction
The coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), started in December 2019 in Wuhan, China. From the day it started, it still affects every aspect of life on earth. It is a known fact that SARS-CoV-2 attacks different parts of the human body, such as the lungs, heart, brain, and kidneys, causing multi-organ infections. In addition to vaccines for this situation, studies to find safe, effective, and economical drugs against COVID-19 continue. Based on the literature, focused on the nonstructural protein targets of SARS-CoV-2, Major protease (Mpro), Papain-like protease (PLpro), RdRp-RTR, and RdRp-RNA [[1], [2], [3], [4], [5], [6], [7], [8], [9]]. Because, replication of SARS-CoV-2, a positive-stranded RNA virus, occurs through the multi-subunit replication and transcription complex of viral non-structural proteins (nsps) [10]. Of these, chymotrypsin-like protease (3CLpro, major protease, Mpro, corresponding to nsp5) is a viral cysteine protease responsible for the maturation of the viral polyprotein required for infection. Many drugs are the focus of discovery. Papain-like protease (PLpro, a domain of nsp3) is a viral cysteine protease responsible for the maturation of the viral polyprotein required for infection. It also has a role in suppressing the host immune response. Apart from these, the core component of the replication and transcription complex is the catalytic subunit (nsp12) of an RNA-dependent RNA polymerase (RdRp) [11,12]. Nsp12 is a polymerase that binds to its main cofactors, nsp7 and nsp8 [13,14]. RdRp is also the target of a class of antiviral drugs with nucleotide analogs; this category includes remdesivir [[15], [16], [17]], a prodrug that is converted into the active drug in the form of triphosphate [remdesivir triphosphate (RTP)] within cells [18]. Inhibition of viral replication through inhibition of nsp7, nsp8, and nsp12-nsp7-nsp8 complex [14,[19], [20], [21], [22]] RdRp is a viable pathway to therapy. For the reasons mentioned above, studies on these 4 non-structural protein structures of SARS-CoV-2 were discussed and examined.
The scale of the epidemic all over the world is so worrying that thousands of people die every day. To prevent this, a state of mobilization has been observed all over the world, and working groups have been channeled into this area. Many studies have been done on COVID-19, and drugs that are on the market or newly synthesized substances have been tried in this area. The World Health Organization (WHO) has made a special call to work on this area. Drug or drug active ingredient studies that can be effective in this field have gained great momentum recently. As a result, studies in this field can be a guide for humanity to get rid of this epidemic in a short time. When we look at the literature studies on the thioether compounds used in this study on COVID-19, it can be seen that the compounds have applications in quite different areas. This is the first study to theoretically examine the efficiency of SNS pincer thioether structures against target proteins of SARS-CoV-2.
Thioethers are ether analogs with sulfur replacing oxygen; their general formula is R–S−R′. Recently, a significant increase has been observed in studies on these compounds. This is because the richness in the usage areas of the compound is noticed. Thioethers are a type of compound that attracts attention in terms of catalytic as well as biological importance [[23], [24], [25], [26], [27]]. Moreover, thioether compounds are practically applied in many fields [28,29].
Recently, thioether compounds have gained importance in terms of inorganic chemistry. Due to the donor atom(s) in their structure, thioethers act as ligands and can form complexes. When we look at the literature, various crystal conformation properties and application areas of both thioethers and their complexes attract attention. The properties of thioether compounds include medicinal chemistry applications, catalytic activity, lubricating effect, and biological and photoluminescent properties [[30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46]]. Our aim in this study is to reveal with theoretical calculations whether the SNS-type pincer compound and its metal complexes will be effective against target proteins of SARS-CoV-2.
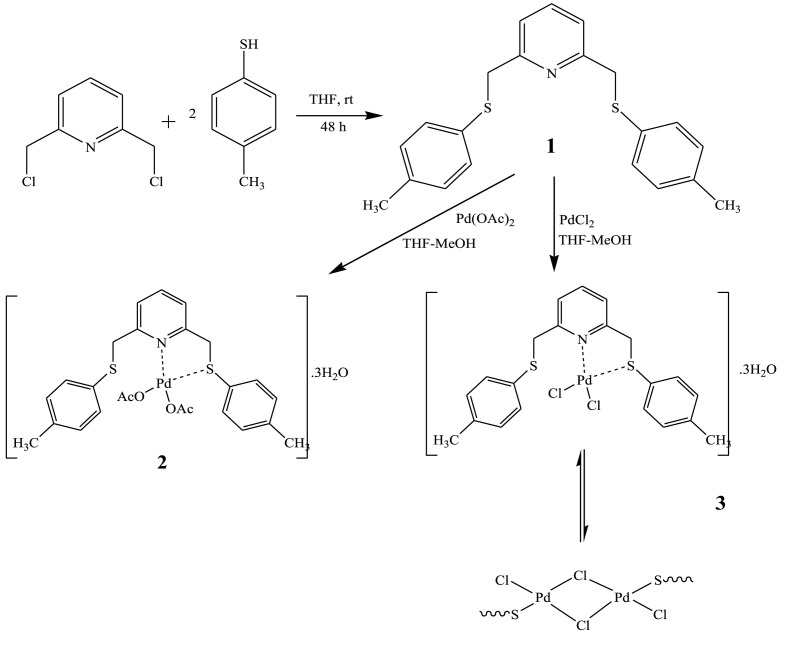
In this study, a thioether-pyridine ligand (L) (1) and novel its Pd(II) complexes ([Pd(κ2-L)(OAc)2]·3H2O (2) and [Pd(κ2-L)Cl2]·3H2O (3)) (Scheme 1 ) were synthesized and characterized by using NMR, UV–Vis, FT-IR, and elemental analysis techniques. In the meantime, interaction mechanisms among these compounds and Pd(II) at the atomic level were described and rationalized by in silico studies. Further, the effectiveness of the pincer ligand and its Pd (II) complexes synthesized was investigated and discussed by molecular docking against COVID-19.
Scheme 1.
Synthesis of the pincer ligand (L) and its metal complexes.
2. Experimental
2.1. General
All the reagents and chemicals (95–99% purity) were purchased from Sigma, Merck, Fluka, and Aldrich and used without further purification. In the literature, the ligand was synthesized for the first time by Parker et al., in 1985 [47]. However, the ligand was obtained by setting up other reaction conditions. The difference between this study from the literature is that it was synthesized using a different starting material (2,6-di(chloromethyl)pyridine) in the presence of a weak base and at room temperature. The reaction parameters in the literature and our study are given comparatively in the supplementary data (Table S1). The FT-IR spectra of the synthesized compounds were recorded using the PerkinElmer Spectrum 100 FT-IR spectrometer system attached to ATR in the range of 4000–400 cm−1. UV–Visible spectra of the synthesized compounds were collected on a PG Instruments T80+ UV/Vis Spectrophotometer. The mass spectra of the compounds were taken on the LCMS-8030 Shimadzu model device. Elemental analyses (C, H, N, and S) were executed using a Thermo Scientific Flash 2000 elemental analyzer. At 25 °C, the molar conductances of the pincer-type metal complexes were measured in DMF using a Thermo Scientific electron corporation model conductivitymeter. The NMR analysis was performed at room temperature on samples dissolved in DMSO-d6 with TMS as an internal standard. 1H and 13C NMR spectra were acquired using a Bruker High-Performance Digital FT-NMR (400 MHz) spectrometer.
2.2. Synthesis
2.2.1. Synthesis of ligand (L) (1)
The ligand 2,6-bis[[(4-methylphenyl)thio]methyl]pyridine (L) (1) was prepared by stiring a solution of 4-methylbenzenethiol (0.248 g, 2 mmol) and 2,6-bis(chloromethyl)pyridine (0.176 g, 1 mmol) in the presence of pyridine (0.158 g, 2 mmol) and potassium iodide (0.166 g, 1 mmol) in THF to room temperature for 48 h. The reaction mixture was then poured into ice water and extracted three times with chloroform. The organic phase was evaporated to dryness. L was obtained as a viscous oil yellow liquid. (74%) (Scheme 1); IR, (ATR) cm-1: 3068, 3051 (C–H)aromatic, 2956, 2920, 2852 (C–H)aliphatic, 1567 (C N)pyridine, 760 (C–S–C); 1H NMR DMSO-d6,400 MHz): δ 7.62 (t, 1H, J = 8 Hz, H-4),7.40 (d, 2H, J = 8 Hz, H-3), 7.23 (t, 4H, J = 8 Hz, H-8), 7.08 (d, 4H, J = 8 Hz, H-7), 4.24 (s, 4H, H-5), 2.24 (s, 6H, H-10); 13C NMR (DMSO-d6,400 MHz): δ 157.55 (C N), 137.84 (C-4), 135.90 (C-9), 132.64 (C-6), 130.06 (C-8), 129.37 (C-7), 121.77 (C-3), 39.55 (C-5), 20.99 (C-10); UV–Vis (DMF) λ max (Abs): 280 (3.47), 271 (3.58) nm; ΛM (10-3 M, in DMF, μS/cm): 8.02. Anal. Calc. For C21H21NS2(351.53): C, 71.75: H, 6.02: N, 3.98: S, 18.24 %. Found: C, 70.93: H, 6.35: N, 3.62: S, 18.67 %.
2.2.2. Synthesis of [Pd(κ2-L)(OAc)2]·3H2O (2)
The 2 complex was synthesized by adding the solution of Pd(OAc)2 (0.168 g, 0.75 mmol) in methanol/THF (1:1) and the ligand 2,6-bis[[(4-methylphenyl)thio]methyl]pyridine (0.263, 0.75 mmol). The obtained product was washed in methanol and purified water. Moreover, the obtained product was gradually purified with the vapor diffusion of Et2O to a THF solution of complex 2, which was a brown-colored compound (Fig. 1 ). Yield: 0.316 g (50%). 175 °C decompose. IR, (ATR) v, cm-1: 3378 (O–H), 3071 (C–H)aromatic, 2990, 2969, 2901 (C–H)aliphatic, 1588 (C N)pyridine, 799 (C–S–C), 598 (M − N), 488 (M − S); 1H NMR DMSO-d6,400 MHz): δ 7.64 (t, 1H, J = 8 Hz, H-4),7.33 (d, 2H, J = 8 Hz, H-3), 7.24 (d, 4H, J = 8 Hz, H-8), 7.08 (d, 4H, J = 8 Hz, H-7), 4.24 (s, 4H, H-5), 2.24 (s, 6H, H-10),1.87 (s, 6H, Hacetate); 13C NMR (DMSO-d6,400 MHz): δ172.64 (C O, C-12),157.98 (C N, C-2), 137.83 (C-4), 135.94 (C-9), 132.62 (C-6), 130.07 (C-8), 129.33 (C-7), 121.82 (C-3), 40.37 (C-5), 21.70 (C-10), 20.98 (C-11); UV–Vis (DMF) λ max (Abs):270 (2.94), 391 (1.27), 485 (0.36) nm; ΛM (10-3 M, in DMF, μS/cm): 1.80. μeff: diamagnetic.ESI-MS, m/z: 578.05 [Pd+L+2(OAc)]]+.Anal.Calc. for C25H33NO7PdS2(630.08); C, 47.66; H, 5.28; N, 2.22; S, 10.18. Found: C, 48.04; H, 4.43; N, 2.84; S, 10.62 %.
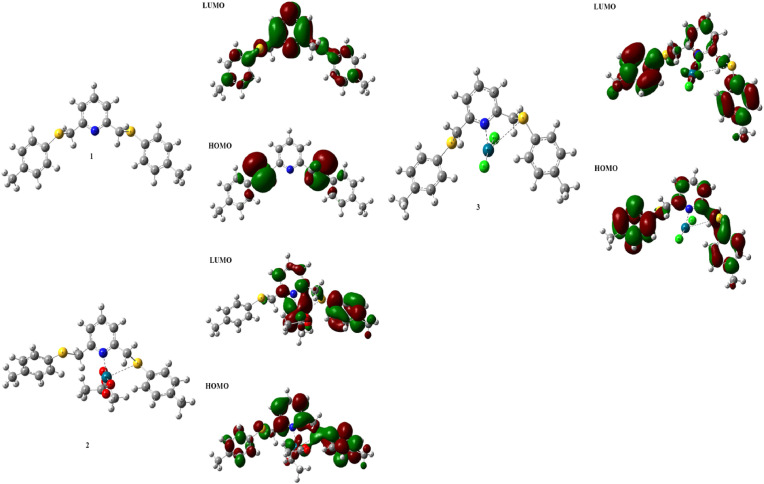
Fig. 1.
The geometry optimized form, HOMO, and LUMO molecular orbitals of the synthesized compounds (1–3) are computed at DFT/B3LYP/LanL2DZ basis set in G09.
2.2.3. Synthesis of [Pd(κ2-L)Cl2]·3H2O (3)
The 3 complex was synthesized by the similar route given as compound 2 using PdCl2 (0.069 g, 0.392 mmol) and 2,6-bis[[(4-methylphenyl)thio]methyl]pyridine L (0.138 g, 0.392 mmol). The obtained product was washed in methanol and purified water. The obtained product was gradually purified with the vapor diffusion of Et2O to a THF solution of complex 3, which was a yellowish brown colored compound (Fig. 1). Yield: 0.139 g (59%). 136 °C decompose. IR, (ATR) v, cm-1: 3381 (O–H), 3058 (C–H)aromatic, 2987, 2970, 2901 (C–H)aliphatic, 1568 (C N)pyridine, 804 (C–S–C), 597 (M − N), 492 (M − S); 1H NMR DMSO-d6,400 MHz): δ 8.20 (t, 1H, J = 8 Hz, H-4),7.81 (d, 4H, J = 8 Hz, H-7), 7.77 (d, 2H, J = 8 Hz, H-3), 7.70 (t, 1H, J = 8 Hz, H-4′), 7.36 (d, 4H, H-8), 7.27–7.22 (m, 6H, H-3' - H-7′), 7.09 (d, 4H, J = 8 Hz, H-8′), 5.38 (s, 4H, H-5 - H-5′), 4.27 (s, 4H, H-5 - H-5′), 2.34 (s, 6H, H-10), 2.25 (s, 6H, H-10′); 13C NMR (DMSO-d6,400 MHz): δ 163.12 (C N), 156.86 (C N)', 141.86 (C-4), 136.30 (C-4)', 131.93 (C-9), 131.50 (C-9)', 131.31 (C-6), 130.52 (C-6)', 130.13 (C-8), 129.80 (C-8)', 128.58 (C-7), 126.16 (C-7)', 123.65 (C-3), 122.55 (C-3)', 50.53 (CH2 and (CH2)'), 38.66 (CH2 and (CH2)'), 21.29 (CH3), 21.02 (CH3)'; UV–Vis (DMF) λ max (Abs):271 (1.428), 328 (0.624), 414 (0.344) nm; ΛM (10-3 M, in DMF, μS/cm): 5.36. μeff: diamagnetic. ESI-MS, m/z: 583.95 [Pd+L+2Cl+3H2O]]+, 493.95 [Pd+L+Cl]]+.Anal.Calc. for C21H23ClNOPdS2 (582.98); C, 43.27; H, 4.67; N, 2.40; S, 11.00. Found: C, 43.90; H, 4.47; N, 2.84; S, 11.45 %.
2.3. Computational part
The related compounds (1–3) underwent geometry optimization at the DFT/B3LYP/LanL2DZ level in Gaussian09 (G09) [48]. Their chemical activity behaviors were estimated and elucidated by the quantum chemical descriptors obtained from in silico studies [49]. In addition, molecular electrostatic potential surface (MEPS) and frontier molecular orbitals (FMOs) were also created to obtain important insights into the Pd(II) complexes. Afterward, molecular docking studies were performed between the aforementioned compounds and different SARS-CoV2 target proteins via Auto Dock 4.2 [50]. Firstly, X-ray structures of the selected SARS-CoV2 targets, as described above part, were uploaded from PDB [51] and given in supplementary data. Then the compounds and targets were prepared by using the sub-protocol of Discovery Studio (DS) 3.5 [52] as described in the previous study [53]. For the related proteins, the active region size was determined as 30 × 30 × 30 with 0.375 Å grid spacing. The other docking parameters were set to the software's default values. To fully search for the possible conformations and binding sites, a Lamarckian genetic algorithm (LGA) was further employed. The DS 3.5 program [52] was utilized for the assessment of the docking results and visualization of the possible protein-ligand interactions.
3. Results
3.1. Synthesis and characterization of compounds
Ligand (1) was obtained from the reaction of 2,6-bis(chloromethyl)pyridine and 4-methylbenzenethiol (1:2) in THF solvent at room temperature. Complexes 2 and 3 were formed by reacting 2,6-bis[[(4-methylphenyl)thio]methyl]pyridine with Pd(OAc)2 and PdCl2 (1:1) in THF/methanol solvent and at 65 °C, respectively. At room conditions, the Pd complexes are solid. In addition, these complexes are dissolved in common organic solvents such as DMSO, DMF and THF. It was observed that the electrolytic conductivity measurements were between 1.80 and 8.02 μS/cm. NMR spectra of the compounds were recorded in DMSO-d6. When the 1H NMR spectrum of the ligand is examined, the H-4 proton in the para position concerning the pyridine nitrogen is seen as a triplet at 7.62 ppm (1H), and the H-3 proton in the meta position for the pyridine nitrogen is seen as a doublet at 7.40 ppm (2H). H-5 Protons belonging to the –CH2-S- groups were observed as a singlet at 4.24 ppm (4H). Methyl group protons (-CH3) (H-10) attached to the phenyl ring were seen singlet at 2.24 ppm (6H) [47]. Complex 2 is partially soluble in NMR-active solvents compared to other compounds. When the 1H NMR spectrum of complex 2 is examined, aromatic protons (11H) as multiplet at 7.64–7.08 ppm, –CH2-S- protons (H-5) (4H) as a singlet at 4.24 ppm, -CH3 protons (H-10) (6H) as a singlet at 2.24 ppm, and -CH3 protons (H-11) (6H) belonging to the acetate group as a singlet at 1.87 ppm were recorded. In the 1H NMR spectrum of complex 3, H-4 (1H) and H-4' (1H) protons at 8.20 and 7.70 ppm and as triplets, H-7 (4H) and H-3 (2H) protons at 7.81 and 7.77 ppm as a doublet, H-3′ and H-7′ as a multiplet at 7.27–7.22 ppm (6H), H-8 (4H) and H-8′(4H) as a doublet at 7.36 and 7.09 ppm, H-5 (4H) and H-5' (4H) protons at 5.38 and 4.27 ppm as a singlet, and finally H-10 (6H) and H-10' (6H) protons were observed as a singlet at 2.34 and 2.25 ppm [32,54]. Details are given in Fig. S1- S6 in the supplementary data.
In the mass spectrum, the molecular ion peak was observed at m/z: 578.05 for complex 2 and 583.95 for complex 3 (see Fig. S7 and Fig. S8 in supplementary data). When the FT-IR spectra are examined, the stretching vibration band of the (C N) group in the pyridine ring is observed at 1567 cm-1 in the ligand, while it is observed at 1588 cm-1 and 1568 cm-1 in the 2 and 3 complexes, respectively. This shift of 1–21 cm-1 in the azomethine group indicates that the complexation may occur over the nitrogen atom of the pyridine ring [55]. However, considering that the shift in complex 3 is as small as 1 cm-1, the structure we have proposed regarding the structure of complex 3 may be in a constant state of transformation. While the M − N interaction was observed at 598 cm-1 in complex 2, it was observed at 597 cm-1 in complex 3 [56,57]. Again, the M − S interaction was recorded at 488 cm-1 in complex 2, while it was recorded at 492 cm-1 in complex 3 [32,38,58] (see Figs. S9- S11 in supplementary data).
UV–Vis measurements of the compounds were taken in DMF solvent and the range of 190–1100 nm. When the UV–Vis spectra were examined, π-π* transitions of phenyl rings at 270 and 280 nm, n- π* transitions of thioether and pyridine rings at 328 and 391 nm, and d-d transitions at 414 and 485 nm were observed [54] (see Fig. S12 in supplementary data).
3.2. Computational part
The DFT/B3LYP/LanL2DZ basis set was used to optimize the geometry of synthesized ligand (1) and complexes (2 and 3). At the same time, the frontier molecular orbitals (HOMO and LUMO) and the molecular electrostatic potential surface of the related compounds were generated with the same basis set in Fig. 1 and Fig. S13, respectively. Then, Fig. 2 displays the biochemical activities of the ligand and the two possible complexes. The electrophilicity ω index has become a useful tool for assessing the reactivity of organic compounds that are engaged in polar reactions [59]. In 2002, a systematic examination of the electrophilicity of several chemicals used in Diels-Alder processes led to the creation of a single electrophilicity ω scale [60]. The electrophilicity ω index was used to study the three-atom components (TACs) involved in [3 + 2] cycloaddition (32CA) reactions in 2003, allowing for rationalization of their reactivity in polar processes [61]. The electrophilicity scale enables organic compounds to be classified as strong electrophiles (ω > 1.5 eV), moderate electrophiles (0.8 < ω < 1.5 eV), and marginal electrophiles (ω < 0.8 eV) [60,62]. Compound 2 is the structure that exhibits the highest biochemical activity (63.231). Compound 3 (48.810) and the last ligand form (9.505) are located, respectively.
Fig. 2.
The biochemical reactivity of the synthesized compounds (1–3).
Besides these, it has been investigated by the molecular docking simulation technique, which is one of the in silico methods, to determine whether the relevant ligand (1) and complexes (2 and 3) have biological activity against proteins of the virus causing the current global problem of COVID-19 disease. The binding energy values of the relevant compounds on the four protein model structures of SARS-CoV-2 (Main Protease, Papain-like protease, RdRp without RNA, and RdRp with RNA) are represented in Fig. 3 . Based on the obtained analysis results, the ligand structure (1) has −8.50 and −8.10 kcal/mol of binding energy values against SARS-CoV-2 proteins, which are especially RdRp with RNA and Papain-like protease target proteins. The first of the complexes of this ligand with Pd metal, compound 2, predominantly demonstrates −9.07 kcal/mol of binding energy with the Main Protease target. It also exhibits the second-best binding tendency with Papain-like protease of SARS-CoV-2. Compound 3, the second possible complex structure of the ligand with Pd metal, forms −8.44 and −8.21 kcal/mol of binding energy values with Papain-like protease and Main Protease, respectively.
Fig. 3.
The binding energy values of the synthesized compounds (1–3) against four different SARS-CoV-2 proteins.
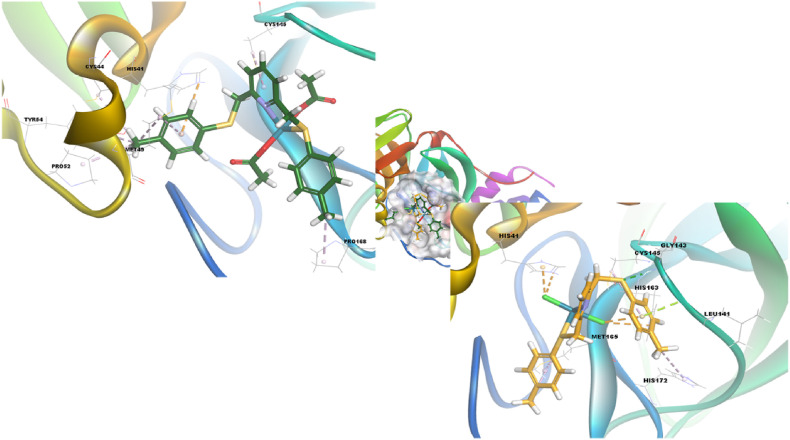
When we consider the interactions of the compounds (1–3) under consideration against different SARS-CoV-2 proteins, first of all, compound 1, namely the ligand structure, tends to bind the best way with the target protein, RdRp with RNA, according to the lowest binding energy values (-8.50 kcal/mol) in Fig. 3. Then, when the interaction of the complex structures formed by the Pd metal and the ligand with different SARS-CoV-2 proteins was investigated, both complexes (2 and 3) showed the best binding affinity with Main Protease and Papain-like protease targets. When we evaluate the details with the visualizations given in Fig. 4 , Compound 2 forms one electrostatic (His41) and seven hydrophobic interactions (Cys44, Met49, Pro52, Pro168, Tyr54, Cys145, and Met49) with Main Protease.
Fig. 4.
3D orientations of compounds 2 (green, stick) and 3 (orange, stick) in the active site of Main Protease. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
On the other hand, compound 2 shows predominantly better interactions than compound 3 when looking at their interactions against the Papain-like protease target of compounds 2 and 3. This is illustrated in Fig. 5 where compound 2 makes H-bonding with Gly271, Cys270 residues, electrostatic interaction with Asp164, pi-sulfur bonding with Met208, and Tyr264, and hydrophobic interactions with Leu162, Tyr264, Met208, Leu162, and Pro248 in the binding site of the target model. In contrast, compound 3 exhibited electrostatic interaction with Arg166, hydrogen bonding with Met208, as well as hydrophobic interactions with Tyr264, Tyr207, Met208, and Arg166 of the same target. In addition, it draws attention to the differences in the binding orientations of the related compounds in the binding site of the target.
Fig. 5.
3D orientations of compounds 2 (green, stick) and 3 (orange, stick) in the active site of Papain-like protease. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In line with all the docking results obtained, Papain-like protease is the SARS-CoV-2 protein with which the three compounds exhibited mutual interaction, Fig. 6 A. In addition, compounds 2 and 3, that is, Pd metal complex structures, interact more effectively with the specified targets, and especially Compound 2 structure is the most effective, both with its structural and interaction with the targets and with their binding orientations, Fig. 6B.
Fig. 6.
Superimposed forms of compound 1 (light blue, stick), compound 2 (green, stick) and 3 (orange, stick) in the active site of A) Papain-like protease B) Main Protease. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
SNS pincer-type thioether compounds have been used in many fields. However, this is the first study of such compounds on COVID-19. In this study, SNS pincer-type ligand and two Pd(II) complexes were synthesized and characterized. It is seen that the ligand and its palladium complexes do not have electrolytic conductivity [56,63]. At the same time, the molar conductivity values of the complexes show that the complex structures are neutral and not in the ionic state. All compounds were analyzed by characterization techniques such as NMR, FT-IR, UV–Vis, mass, elemental analysis, and electrolytic conductivity. A surprise was encountered when examining the structure of complex compounds. As expected in complex 2, a 1:1 metal-ligand-containing complex was formed, while in complex 3, we detected a surprisingly different state. When the NMR values of complex 3 were examined, it was seen that the metal-ligand ratio was 2:2 in the structure. However, when the mass results were examined, it was determined that the complex was 1:1. A similar event is found in literature reviews, albeit rarely. Kozlov et al. stated in a study they conducted in 2011 that there is a similar situation. In the study, they explained that the pincer Pd(II) complex actively transforms into each other as 1:1 and 2:2 in the structure [64]. It is thought that this is also the case in Complex 3 and that the formation energies of complexes with metal-ligand ratios of 1:1 and 2:2 are very close to each other. Therefore these structures are converted into each other in the solution environment. Unfortunately, single-crystal X-ray analysis could not be performed because the structures could not be obtained as single crystals despite crystallization efforts. A four-coordinated square-plane structure is proposed for Pd(II) complexes [[65], [66], [67], [68]].
Surprisingly, when the 1H NMR spectrum of Complex 3 was examined, it was seen that the number of protons had doubled. However, it has been observed that the protons, which are described as the same in the structure, are not exactly in the same place, but peak very close to each other. This indicates that the metal-ligand ratio of our structure is likely to be 2:2. When the mass spectra are evaluated, as expected in complex 2 and complex 3, molecular ion peaks of complexes with a 1:1 metal-ligand ratio are observed. Contrary to what is seen in the NMR spectrum of Complex 3, a molecular ion peak of a 2:2 metal-ligand ratio could not be observed in the mass spectrum of this complex. This strengthens our idea that it can be in transformation in both forms in the solution environment [64].
The biochemical activity analysis shows us that the complex state with Pd metal is more reactive than the ligand state, and especially compound 2, containing acetate groups form complexes with Pd metal is more effective than compound 3 with chlorine atoms attached (Figs. 1 and 2).
This is because compound 1 exhibits pi-sulfur bond interactions with the Cys813 residue of the target and the T: A13 nucleotide of DNA, as well as hydrophobic interaction with Leu758, Trp598, Ala688, Val588, Met601, Leu758, Lys593, and Ile589 residues. This situation is given in Figs. S14–A a three-dimensional illustration of the obtained docked complex (see Figs. S14–A in supplementary data). On the other hand, the same compound (1) shows the second-highest binding affinity with the target structure, Papain-like protease. When we focus on the remarked docking here, we see in Figs. S14–B that compound 1 carried out electrostatic interaction (Asp164 and Glu167) and also hydrophobic interaction (Tyr264, Leu185, Leu199, Pro248, Tyr207, Met 208) with Papain-like protease as the target (see Figs. S14–B and Table S2 in supplementary data.).
Compound 3 tends to bind fewer affinity against the target than compound 2, compared to exhibiting three electrostatic interactions, one hydrogen bond, one pi-lone pair interaction, and four hydrophobic interactions with the same target (Fig. 4). This may be due not only to the interaction types, but also to the surface area and functional structure content of the binding compounds, and most importantly, the appropriate orientation of the relevant compounds, especially compound 2, in the binding site of the target protein.
5. Conclusions
In conclusion, the pyridine-thioether-based SNS pincer ligand has been synthesized and metallated with Pd(OAc)2 and PdCl2. In the case of the methylphenyl substituent, the spectroscopic and microanalytical data supported the anticipated four-coordinate geometries of Pd(κ2-L)X2 (X = OAc, Cl). In contrast, when the ligand possessed a 4-methylphenyl substituent, Pd(OAc)2 yielded analogous Pd(κ2-L)(OAc)2 species (2) but surprisingly the PdCl2 metallation yielded analogous, Pd(κ2-L)Cl2/Pd2(κ2-L)2Cl4, (3). It has been reported in the literature that the complex structure can be in constant transformation in the solution medium, albeit rarely. This may be the case within the structure of Complex 3. The computational visual method and molecular docking have been made to support and illuminate the experimental data, and the obtained results show compatibility with each other. Compound 2 is the structure that exhibits the highest biochemical activity. According to all of the docking studies, Papain-like protease is the SARS-CoV-2 protein with which the three compounds exhibit mutual interaction. The structure of compound 2 (Pd(κ2-L)(OAc)2) is very effective, both in terms of structural and interaction with the targets, as well as binding orientations. Moreover, in this study, the computational visual method study of new SNS pincer-type compounds against target proteins of SARS-CoV-2 was performed for the first time, thus expanding the application area of pincer compounds.
Author contributions
Hatice Gamze Sogukomerogullari: Conceptualization; formal analysis; investigation; software; validation; methodology; writing-original draft; writing-review & editing; supervision.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The author thanks Tugba Taskin-Tok, Esin Aki-Yalcin, and the research group for technical assistance. This study received no support from the fund.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105512.
Appendix A. Supplementary Data
All additional information about the characterization of the complexes using NMR (Fig. S1-Fig. S6), mass spectrums (Fig. S7-Fig. S8), FT-IR spectrums (Fig. S9-Fig. S11), UV–Vis spectrum (Fig. S12), the molecular electrostatic potential of the synthesized compounds (1–3) (Fig. S13), 3D orientations of compound 1 (light blue, stick) in the active site of A) RdRp with RNA and B) Papain-like protease (Fig. S14), comparative reaction parameters in the literature and in this study (Table S1), the interaction types and distances of compounds 1, 2 and 3 against Main Protease, Papain-like protease, RdRp without RNA and RdRp with RNA (Table S2) are given in the supporting information available at https://doi.org/10.1016/j.compbiomed.2022.105512.
Appendix B. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. https://doi: 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- 2.W. Rut, Z. Lv, M. Zmudzinski, S. Patchett, D. Nayak, S. J. Snipas, F. E. Oualid, T. T. Huang, M. Bekes, M. Drag, S. K. Olsen,Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design, bioRxiv. org. e-Print archive., https://doi:10.1101/2020.04.29.068890. [DOI] [PMC free article] [PubMed]
- 3.Chakrabarti S., Jayachandran U., Bonneau F., Fiorini F., Basquin C., Domcke S., Le Hir H., Conti E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell. 2011;41(6):693–703. doi: 10.1016/j.molcel.2011.02.010. https://doi:10.1016/j.molcel.2011.02.010 [DOI] [PubMed] [Google Scholar]
- 4.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. https://doi:10.1126/science.abc1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W., Feng B., Han S., Wang P., Chen W., Zang Y., Li J., Hu Y. Discovery of highly potent SARS-CoV-2 Mpro inhibitors based on benzoisothiazolone scaffold. Bioorg. Med. Chem. Lett. 2022;58:128526. doi: 10.1016/j.bmcl.2022.128526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang J.J., Pitsillou E., Ververis K., Guallar V., Hung A., Karagiannis T.C. Investigation of small molecule inhibitors of the SARS-CoV-2 papain-like protease by all-atom microsecond modelling, PELE Monte Carlo simulations, and in vitro activity inhibition. Chem. Phys. Lett. 2022;788:139294. doi: 10.1016/j.cplett.2021.139294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J., Qin Z., Liu X., He X., Yao J., Zhou X., Wen K., Yu N., Wu Q., Xiao W., Zhu L., Wan C., Zhang B., Zhao W. High-specificity targets in SARS-CoV-2 N protein for serological detection and distinction from SARS-CoV. Comput. Biol. Med. 2022;143:105272. doi: 10.1016/j.compbiomed.2022.105272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitsillou E., Liang J., Ververis K., Hung A., Karagiannis T.C. Interaction of small molecules with the SARS-CoV-2 papain-like protease: in silico studies and in vitro validation of protease activity inhibition using an enzymatic inhibition assay. J. Mol. Graph. 2021;104:107851. doi: 10.1016/j.jmgm.2021.107851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maehara S., Kobayashi M., Kuwada M., Choshi T., Inoue H., Hieda Y., Nishiyama T., Hata T. In vitro and in silico studies of potential coronavirus-specific 3C-like protease inhibitors. Chem. Pharm. Bull. 2022;70(3):195–198. doi: 10.1248/cpb.c21-01002. [DOI] [PubMed] [Google Scholar]
- 10.Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn D.G., Choi J.K., Taylor D.R., Oh J.W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012;157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.te Velthuis A.J., Arnold J.J., Cameron C.E., van den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U.S.A. 2014;111 doi: 10.1073/pnas.1323705111. E3900–E3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. Washington state 2019-nCoV case investigation team, first case of 2019 novel coronavirus in the United States, N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.-S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 19.Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peti W., Johnson M.A., Herrmann T., Neuman B.W., Buchmeier M.J., Nelson M., Joseph J., Page R., Stevens R.C., Kuhn P., Wüthrich K. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J. Virol. 2005;79:12905–12913. doi: 10.1128/JVI.79.20.12905-12913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson M.A., Jaudzems K., Wüthrich K. Vol. 402. 2010. pp. 619–628. (NMR structure of the SARS-CoV nonstructural protein 7 in solution at pH 6.5, J. Mol. Biol). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGuinness D.S., Wasserscheid P., Keim W., Morgan D., Dixon J.T., Bollmann A., Maumela H., Hess F., Englert U. First Cr(III)−SNS complexes and their use as highly efficient catalysts for the trimerization of ethylene to 1-hexene. J. Am. Chem. Soc. 2003;125(18):5272–5273. doi: 10.1021/ja034752f. [DOI] [PubMed] [Google Scholar]
- 24.Fernandes R.R., Lasri J., da Silva M.F.C.G., da Silva J.A.L., da Silva J.J.R.F., Pombeiro A.J.L. Bis- and tris-pyridyl amino and imino thioether Cu and Fe complexes. Thermaland microwave-assisted peroxidative oxidations of 1phenylethanol and cyclohexane in the presence of various N-based additives. J. Mol. Catal. A: Chem. 2011;351:100–111. doi: 10.1016/j.molcata.2011.09.022. [DOI] [Google Scholar]
- 25.Vernier W., Chong W., Rewolinski D., Greasley S., Pauly T., Shaw M., Dinh D., Ferre R.A., Nukui S., Ornelas M., Reyner E. Thioether benzenesulfonamide inhibitors of carbonic anhydrases II and IV: structure-based drug design, synthesis, and biological evaluation. Bioorg. Med. Chem. 2010;18(9):3307–3319. doi: 10.1016/j.bmc.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Nekola H., Rehder D. Vol. 337. 2002. pp. 467–469. (A penta-coordinated zinc complex containing a biomimetic NS2S′2 (thiolate/thioether) ligand, Inorg. Chim. Acta). [DOI] [Google Scholar]
- 27.Piccirilli L., Pinheiro D.L.J., Nielsen M. Recent progress with pincer transition metal catalysts for sustainability. Catalysts. 2020;10:773. [Google Scholar]
- 28.Tochrop G.P., Sadhukhan S., Koner R.R., Ghosh S. The syntheses and applications of β-benzylmercaptoethylamine derivative. Tetrahedron. 2009;65:10515–10534. doi: 10.1016/j.tet.2009.10.041. [DOI] [Google Scholar]
- 29.Ali Ahmed A.B., Taha R.M. In: Marine Cosmeceuticals Trends and Prospects. Kim Se-Kwon., editor. eBook CRC Press; Florida: 2012. Marine phytochemical compounds and their cosmeceutical applications; pp. 51–61. [Google Scholar]
- 30.Zhang J.-A., Pan M., Jiang J.-J., She Z.-G., Fan Z.-J., Su C.-Y. Syntheses, crystal structures and antimicrobial activities of thioether ligands containing quinoline and pyridine terminal groups and their transition metal complexes. Inorg. Chim. Acta. 2011;374:269–277. doi: 10.1016/j.ica.2011.02.073. [DOI] [Google Scholar]
- 31.Guiu E., Claver C., Castillòn S. Ir(I) complexes with oxazoline-thioether ligands: nucleophilic attack of pyridine on coordinated 1,5-cyclooctadiene and application as catalysts in imine hydrogenati. J. Organomet. Chem. 2004;689:1911–1918. [Google Scholar]
- 32.Sogukomerogullari H.G., Yalçın Ş.P., Ceylan Ü., Aytar E., Aygün M., Richeson D.S., Sönmez M. Synthesis of Fe and Cu metal complexes derived from ‘SNS’ Pincer type ligands and their efficient catalyst precursors for the chemical fixation of CO2. J. Chem. Sci. 2019;131:32–44. doi: 10.1007/s12039-019-1609-6. [DOI] [Google Scholar]
- 33.Soliman S.M., Al-Rasheed H.H., El-Faham A. Synthesis, X-ray structure, hirshfeld analysis of biologically active Mn(II) pincer complexes based on s-triazine ligands. Crystals. 2020;10(10):931. doi: 10.3390/cryst10100931. [DOI] [Google Scholar]
- 34.Zhang J.A., Pan M., Zhang J.Y., Zhang H.K., Fan Z.J., Kang B.S., Su C.Y. Syntheses, structures and bioactivities of silver (I) complexes with a tridentate heterocyclic N- and S-ligand. Polyhedron. 2009;28:145–149. doi: 10.1016/j.poly.2008.09.029. [DOI] [Google Scholar]
- 35.Xu W.-M., Li S.-Z., He M., Yang S., Li X.-Y., Li P. Synthesis and bioactivities of novel thioether/sulfone derivatives containing 1,2,3-thiadiazole and 1,3,4-oxadiazole/thiadiazole moiety. Bioorg. Med. Chem. Lett. 2013;23(21):5821–5824. doi: 10.1016/j.bmcl.2013.08.107. [DOI] [PubMed] [Google Scholar]
- 36.Masdeu-Bultó A.M., Diéguez M., Martin E., Gómez M. Chiral thioether ligands: coordination chemistry and asymmetric catalysis. Coord. Chem. Rev. 2003;242:159–201. [Google Scholar]
- 37.Diéguez M., Pàmies O., Net G., Ruiz A., Claver C. Mixed thioether-phosphite and phosphine-phosphite ligands for copper-catalyzed asymmetric 1,4-addition of organometallic reagents to cyclohexanone. J. Mol. Catal. A: Chem. 2002;185:11–16. doi: 10.1016/S1381-1169(02)00054-7. [DOI] [Google Scholar]
- 38.Sogukomerogullari H.G., Aytar E., Ulusoy M., Demir S., Dege N., Richeson D.S., Sönmez M. Synthesis of complexes Fe, Co and Cu supported by ''SNS" pincer ligands and their ability to catalytically form cyclic carbonates. Inorg. Chim. Acta. 2018;471:290–296. doi: 10.1016/j.ica.2017.11.007. [DOI] [Google Scholar]
- 39.Czerny F., Döhlert P., Weidauer M., Irran E., Enthaler S. Synthesis, characterization and application of nickel(II) complexes modified with N,N′,N′′pincer ligands. Inorg. Chim. Acta. 2015;425:118–123. doi: 10.1016/j.ica.2014.10.003. [DOI] [Google Scholar]
- 40.Muralisankar M., Basheer S.M., Haribabu J., Bhhuvanesh N.S.P., Karvembu R., Sreekanth A. An investigation on the DNA/protein binding, DNA cleavage and in vitro anticancer properties of SNO pincer type palladium(II) complexes with N-substituted isatin thiosemicarbazone ligands. Inorg. Chim. Acta. 2017;466:61–70. doi: 10.1016/j.ica.2017.05.044. [DOI] [Google Scholar]
- 41.Noaimi M.A., Awwadi F.F., Talib W.H., Atia S., Hammud H.H. Cis and trans- palladium (II) complexes derived from SNN amidrazone pincer ligand: synthesis, crystal structures and biological evaluation. J. Mol. Struct. 2019;1197:282–291. doi: 10.1016/j.molstruc.2019.07.062. [DOI] [Google Scholar]
- 42.Naskar R., Ghosh P., Manna C.K., Murmu N., Mondal T.K. Palladium(II) complexes with thioether based ONS donor ligand: synthesis, characterization, X-ray structure, DFT study and anti-cancer activity. Inorg. Chim. Acta. 2022;534:120802. doi: 10.1016/j.ica.2022.120802. [DOI] [Google Scholar]
- 43.Birgül K., Yıldırım Y., Karasulu H.Y., Karasulu E., Uba A.I., Yelekçi K., Bekçi H., Cumaoğlu A., Kabasakal L., Yılmaz Ö., Küçükgüzel Ş.G. Synthesis, molecular modeling, in vivo study and anticancer activity against prostate cancer of (+) (S)-naproxen derivatives. Eur. J. Med. Chem. 2020;208:112841. doi: 10.1016/j.ejmech.2020.112841. [DOI] [PubMed] [Google Scholar]
- 44.Sandoval-Chávez C.I., Velázquez-Jiménez R., Martínez-Otero D., Salazar-Pereda V., Andrade-López N., González-Montiel S. Synthesis and catalytic activity of cationic dinuclear palladium (II) complexes supported by thioether ligands containing two di-(2-picolyl) amine arms. Polyhedron. 2020;182:114489. doi: 10.1016/j.poly.2020.114489. [DOI] [Google Scholar]
- 45.Wang H., Huang Q., Li M.-J. Design and synthesis of cyclometalated Ir(III) complex with thioether groups for highly selective recognition of mercury ions. J. Organomet. Chem. 2021;942:121808. doi: 10.1016/j.jorganchem.2021.121808. [DOI] [Google Scholar]
- 46.Kontham V., Padmaja K.V., Madhu D. Synthesis and tribological investigation of 4-vinyl guaiacol–based thioether derivatives as multifunctional additives and their interactions with the tribo surface using quantum chemical calculations. J. Saudi Chem. Soc. 2020;24(12):942–954. doi: 10.1016/j.jscs.2020.10.002. [DOI] [Google Scholar]
- 47.Parker D., Lehn J.M., Rimmer J. Mono- and di-nuclear rhodium and palladium complexes of macrocyclic ligands containing the 2,6-Di(thiomethyl)pyridine sub-unit. J. Chem. Soc. Dalton Trans. 1985:1517–1521. doi: 10.1039/DT9850001517. [DOI] [Google Scholar]
- 48.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., et al. Gaussian, Inc.; Wallingford CT: 2009. Gaussian 09, Revision E.01. [Google Scholar]
- 49.Taskin T., Sevin F. Theoretical investigation on chemical and biochemical activities of 5,6-dihydro-11H-benzo[α]carbazole and its derivatives. J. Mol. Struct. THEOCHEM. 2007;803(1):61–66. doi: 10.1016/j.theochem.2006.09.028. [DOI] [Google Scholar]
- 50.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose Y., Duarte J.M., Lowe R., Segura J., Bi C., Bhikadiya C., Chen L., Rose A.S., Bittrich S., Burley S.K., Westbrook J.D. RCSB protein data bank: architectural advances towards integrated searching and efficient access to macromolecular structure data from the PDB archive. J. Mol. Biol. 2021;433(11):166704. doi: 10.1016/j.jmb.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Accelrys Software Inc . Release 3.5 Accelrys Software Inc; San Diego: 2013. (Discovery Studio Modeling Environment). [Google Scholar]
- 53.Aki-Yalcin E., Ertan-Bolelli T., Taskin-Tok T., Ozturk O., Ataei S., Ozen C., Yildiz I., Yalcin I. Evaluation of inhibitory effects of benzothiazole and 3-amino-benzothiazolium derivatives on DNA topoisomerase II by molecular modeling studies. SAR QSAR Environ. Res. 2014;25(8):637–649. doi: 10.1080/1062936X.2014.923039. [DOI] [PubMed] [Google Scholar]
- 54.Sogukomerogullari H.G., Taskin Tok T., Uruş S., Sönmez M. Pd (II) complexes bearing “SNS” pincer-type thioether ligands: application as catalysts in the synthesis of vitamin K3. Appl. Organomet. Chem. 2021:e6467. doi: 10.1002/aoc.6467. [DOI] [Google Scholar]
- 55.Dede B., Özen N., Görgülü G. Synthesis, characterization, theoretical calculations and enzymatic activities of novel diimine-dioxime ligand and its homodinuclear Cu(II) complex. J. Mol. Struct. 2018;1163:357–367. doi: 10.1016/j.molstruc.2018.03.015. [DOI] [Google Scholar]
- 56.Sönmez M., Sogukomerogullari H.G., Öztemel F., Berber İ. Synthesis and biological evaluation of a novel ONS tridentate Schiff base bearing pyrimidine ring and some metal complexes. Med. Chem. Res. 2014;23:3451–3457. doi: 10.1007/s00044-014-0925-0. [DOI] [Google Scholar]
- 57.Uçan S.Y., Uçan M., Mercimek B. Synthesis and characterization of new schiff bases and their cobalt(II), nickel(II), copper(II), zinc(II), cadmium(II) and mercury(II) complexes. Syn. React. Inorg. Met. 2005;35:417–421. doi: 10.1081/SIM-200059233. [DOI] [Google Scholar]
- 58.Miecznikowski J.R., Lynn M.A., Jasinski J.P., Reinheimer E., Bak D.W., Pati M., Butrick E.E., Drozdoski A.E.R., Archer K.A., Villa C.E., Lemons E.G., Powers E., Siu M., Gomes C.D., Morio K.N. Synthesis, characterization, and computational study of three-coordinate SNS-copper(I) complexes based on bis-thione precursors. J. Coord. Chem. 2014;67:29–44. doi: 10.1080/00958972.2014.883070. [DOI] [Google Scholar]
- 59.Pérez P., Domingo L.R., Aizman A., Contreras R. Theoretical Aspects of Chemical Reactivity. Elsevier; New York, NY, USA: 2007. The electrophilicity index in organic chemistry. [Google Scholar]
- 60.Domingo L.R., Aurell M.J., Pérez P., Contreras R. Quantitative characterization of the global electrophilicity power of common diene/dienophile pairs in Diels-Alder reactions. Tetrahedron. 2002;58:4417–4423. doi: 10.1016/S0040-4020(02)00410-6. [DOI] [Google Scholar]
- 61.Pérez P., Domingo L.R., Aurell M.J., Contreras Quantitative characterization of the global electrophilicity pattern of some reagents involved in 1,3-dipolar cycloaddition reactions. Tetrahedron. 2003;59:3117–3125. doi: 10.1016/S0040-4020(03)00374-0. [DOI] [Google Scholar]
- 62.Domingo L.R., Ríos-Gutiérrez M., Pérez P. Applications of the conceptual density functional theory indices to organic chemistry reactivity. Molecules. 2016;21:748. doi: 10.3390/molecules21060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maravalli P.B., Dhumwad S.D., Goudar T.R. Synthetic, spectral, thermal and biological studies of lanthanide (III) complexes with a schiff base derived from 3-N-Yetaylpiperidino-4-Amino-5-Yercapto-1,2,4-Triazole. Synth. React. Inorg. Met. Org. Chem. 1999;29:525–540. doi: 10.1080/00945719909349467. [DOI] [Google Scholar]
- 64.Kozlov V.A., Aleksanyan D.V., Korobov M.V., Avramenko N.V., Aysin R.R., Maloshitskaya O.A., Korlyukov A.S., Odinets I.L. The first solid phase synthesis of pincer palladium complexes. Dalton Trans. 2011;40:8768–8772. doi: 10.1039/c1dt10680e. [DOI] [PubMed] [Google Scholar]
- 65.Fiebor A., Tia R., Makhubela B.C.E., Kinfe H.H. Water-soluble SNS cationic palladium(II) complexes and their Suzuki–Miyaura cross-coupling reactions in aqueous medium. Beilstein J. Org. Chem. 2018;14:1859–1870. doi: 10.3762/bjoc.14.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu K., Sommer W., Richardson J.M., Weck M., Jones C.W. Evidence that SCS pincer Pd(II) complexes are only precatalysts in heck catalysis and the implications for catalyst recovery and reuse. Adv. Synth. Catal. 2005;347:161–171. doi: 10.1002/adsc.200404264. [DOI] [Google Scholar]
- 67.Flores-Rojas G.G., González-Sebastián L., Reyes-Martínez R., Aguilar-Castillo B.A., Hernández-Ortega S., Morales-Morales D. Evaluation of Their Microwave Assisted Catalytic Activity in C-C Coupling Reactions, Polyhedron. Vol. 185. 2020. Synthesis and characterization of Pd(II) complexes bearing NS, CS, SNS and SCS ligands; p. 114601. [DOI] [Google Scholar]
- 68.Shimokawa R., Kawada Y., Hayashi M., Kataoka Y., Ura Y. Oxygenation of a benzyl ligand in SNS-palladium complexes with O2: acceleration by anions or Brønsted acids. Dalton Trans. 2016;45:16112. doi: 10.1039/C6DT02948E. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.