Abstract
Natural products with good antioxidative properties have been paid increased attention globally. However, due to its chemical complexity, it is difficult to find out its antioxidative compounds. Herein, the chemical profiling and antioxidant capacity of CiNingJi (CNJ) were analyzed, as an example. By using UHPLC-Q-TOF/MS, a total of 82 compounds were tentatively deduced. Furthermore, its free radical scavenging capacity was assessed by different in vitro spectrophotometric-based assays. The result showed that one ingredient, Rosa roxburghii, plays a critical role in its antioxidant activity. In addition, 18 potential antioxidants were screened out in CNJ by comparing the difference of it with and without DPPH reaction. They were identified mainly as catechin, ellagic acid, kajiichigoside F1, and their derivatives or isomers. With the further quantification of major found antioxidants, our results may provide some knowledge on predicting the antioxidative compounds of natural products.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10068-022-01049-4.
Keywords: CiNingJi beverage, Rosa roxburghii, LC-MS/MS, Antioxidant activity, Quantification
Introduction
As more and more people realize that foods and their derived products containing bioactive compounds can bring benefits to human health recently, the interest of health-conscious consumers towards them is growing considerably. These products, which were known as nutraceuticals and functional foods containing enriched phytochemical extracts from herbs or traditional Chinese medicines (TCMs), are now widely available on the market (Espin et al., 2007; Gonzalez-Sarrias et al., 2013). With their bioactive components, there are many healthy properties were reported about them, including antioxidant, cardioprotective, anticancer, and hypolipidemic (Chang et al., 2020; Chen et al., 2019; Kalhori et al., 2021; Parham et al., 2020). Among them, antioxidant protection effects were reported against various diseases or illnesses relevant to oxidative stress, such as aging, cardiovascular diseases, cancer, Alzheimer’s disease, hypertension, and inflammation (Ayvaz et al., 2018; Grzesik et al., 2018; Jiang et al., 2018; Mansoori et al., 2021; Zhou et al., 2020). Therefore, foods, fruits, medicinal plants, or nutraceuticals and pharmaceutical agents that contained natural antioxidants have drawn increased attention globally. However, due to the chemical complexities of natural products, their antioxidant activities are difficult to acquire, as well as the “real” antioxidative compounds that might contribute to their properties. Herein, the chemical profiling and antioxidative capacity of CiNingJi beverage (CNJ) were analyzed, as an example.
CNJ is a newly developed healthy product, which mainly consisted of Rosa roxburghii, Citrus limon, and Malus domestica (apple). As the main and principal material in CNJ, R. roxburghii is an excellent plant resource that has the concomitant function of both medicine and foodstuff. For now, phytochemical research have shown that it has lots of bioactive compounds, such as phenolics, flavonoids, organic acids, triterpenes, amino acids, etc. (Hou et al., 2020; Liu et al., 2016). However, finding out the compounds with good antioxidative properties in R. roxburghii is still not available, neither is CNJ.
Therefore, in order to acquire the antioxidative capacity of natural products and antioxidative compounds, we used CNJ as an example. First, the chemical profile of CNJ was elucidated by using ultrahigh performance liquid chromatography quadrupole time-of-flight mass spectrometry (UHPLC-Q-TOF/MS). Then, the antioxidant activities of CNJ and its ingredients were evaluated with different methods, such as 1,1-Diphenyl-2-picrylhydrazyl (DPPH), 2,2′-azinobis-(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) scavenging abilities, and ferric-reducing antioxidant power (FRAP). And the potential antioxidant compounds were found based on LC-MS coupled with pre-column DPPH reaction. Finally, major antioxidants in CNJ were also quantified. For the first time, the chemical components of CNJ were illuminated, and the antioxidant profiles were screened and quantified. With more and more attention on the natural antioxidants, it might be useful for predicting the antioxidative compounds of other nutraceuticals or pharmaceutical agents, which contain medicinal plants/food.
Materials and methods
Materials and reagents
Methanol and acetonitrile of LC-MS grade were purchased from Anaqua Chemicals Supply (Houston, TX, USA), and formic acid (LC-MS grade) was provided by Fisher Scientific Laboratories, Inc. (Pittsburgh, PA, USA). Ultrapure water (18.2 MΩ) was produced by a Direct-Q® 8 UV–R Milli-Q purification system (Millipore, MA, USA).
Reference substances, (+)-catechin (25), isoquercitrin (51), rutin (57), quercetin (64), and kajiichigoside F1 (71) were obtained from Shenzhen hiboled century Biotechnology Co. Ltd. (Shenzhen, China), while L-ascorbic acid (1), procyanidin B1 (23), ellagic acid (48) and hyperoside (49) were supplied by Guangzhou zhuanyan Biological Technology Co. Ltd. (Guangzhou, China). Chemical structures of these standards were shown in Figure S1, and their purities were found more than 98.0% by HPLC analysis. 2, 4, 6-tripyridyl-s-triazine (TPTZ), DPPH, potassium persulfate, and ABTS were purchased from Sigma Aldrich (Shanghai, China). Internal standard (IS) substance, 2-chloro-phenylalanine, was purchased from Bide Pharmatech Ltd. (Shanghai, China).
The extracts of R. roxburghii, C. limon, M. domestica and CNJ beverage (batch number: ZX 20210120-010) were provided by Wanlaoji Pharmaceutical Co., Ltd. (Guangzhou, China). The proportion of each raw material in CNJ is R. roxburghii: C. limon: M. domestica = 1.7: 2.3: 1.5 (by weight).
Preparation of solutions
Standard solutions for calibration All 9 reference standards were accurately weighted and prepared in methanol-water (5:5, v/v) as stock solutions (mg/mL): L-ascorbic acid (6.07), procyanidin B1 (10.10), (+)-catechin (10.03), ellagic acid (9.95), hyperoside (10.27), isoquercitrin (10.21), rutin (10.12), quercetin (10.03), kajiichigoside F1 (10.97). An appropriate volume of each stock solution was mixed to yield the mixed stock solution. IS working solution was also obtained with a concentration of 10 µg/mL. Then, working standard solutions were obtained by diluting of mixed stock solution to provide a series of working standard solutions for calibration (Table 1). Working solutions with low, middle, and high concentrations were used as quality controls (QCs) for the investigation of accuracy, precision, and stability. All standard solutions were stored at 4 °C until used, and filtered through a nylon membrane filter (0.22 µm, Phenomenex, CA, USA) before UHPLC-Q-TOF/MS analysis.
Table 1.
Regression equations, linear ranges of nine antioxidants and their contents in CNJ and its ingredients (mean ± SD) (µg/mL)
| No. | Analyte | Measured value [ion form] | Regression equation | r | Linear range (µg/mL) | AP | NM | CL | CNJ |
|---|---|---|---|---|---|---|---|---|---|
| 1 | L-ascorbic acid | 175.0248 [M − H]− | y = 0.0115x + 0.0328 | 0.988 | 12.00-2000 | 23.87 ± 4.35 | 24.81 ± 5.71 | 1819.63 ± 463.46 | 329.44 ± 86.24 |
| 23 | Procyanidin B1 | 577.1351 [M − H]− | y = 0.1077x + 0.5244 | 0.991 | 0.20-200 | ND | ND | 164.81 ± 67.84 | 5.64 ± 3.14 |
| 25 | (+)-Catechin | 289.0717 [M − H]− | y = 0.1761x + 0.2162 | 0.999 | 0.20-40 | ND | ND | 77.06 ± 22.19 | 8.21 ± 3.71 |
| 48 | Ellagic acid | 300.9990 [M − H]− | y = 0.0378x + 0.5683 | 0.996 | 0.40-200 | 0.09 ± 0.04 | ND | 75.41 ± 8.20 | 4.48 ± 1.07 |
| 49 | Hyperoside | 463.0882 [M − H]− | y = 0.0935x + 1.2593 | 0.997 | 0.05-40 | ND | 0.37 ± 0.18 | 1.02 ± 0.17 | 0.75 ± 0.26 |
| 51 | Isoquercitrin | 463.0882 [M − H]− | y = 0.1176x + 1.2437 | 0.992 | 0.05-40 | ND | 0.41 ± 0.07 | 1.56 ± 0.43 | 0.94 ± 0.27 |
| 57 | Rutin | 609.1461 [M − H]− | y = 0.1409x + 1.1948 | 0.994 | 0.20-200 | ND | 0.08 ± 0.01 | 0.10 ± 0.04 | 0.06 ± 0.03 |
| 64 | Quercerin | 301.0354 [M − H]− | y = 0.144x + 1.9332 | 0.990 | 0.40-200 | 0.09 ± 0.02 | ND | ND | ND |
| 71 | Kajiichigoside F1 | 695.4012 [M + COOH]− | y = 0.3951x + 0.5426 | 0.999 | 0.20-200 | ND | ND | 156.87 ± 20.85 | 5.19 ± 2.71 |
CL: R. roxburghii; NM: C. limon; AP: M. domestica. ND, not detected
Sample solutions After samples warmed to room temperature, an appropriate amount of IS was added to obtain a certain concentration of IS (10 µg/mL) (Lin et al., 2018). And then, they were filtered through membrane filters for analysis. As for antioxidant assays, the extract of all ingredients and CNJ were concentrated to same dry weight (88 mg/mL) and diluted to appropriate concentration with distilled water.
Solutions for antioxidant assays DPPH radical solution was prepared by dissolving an accurately weighed DPPH sample (24.6 mg) into a 100 mL brown volumetric flask right with methanol before the experiments and the whole procedure was protected from light. Meanwhile, 5 mL of ABTS (7 mM) was mixed with 88 µL of potassium persulfate (140 mM) to produce ABTS+ stock solution, and it was incubated in the dark at room temperature for 16 h. And FRAP stock solution contained 2.5 mL of a 10 mM TPTZ solution in 40 mM HCl, 2.5 mL of 20 mM FeCl3 and 25 mL of 0.3 M acetate buffer, pH 3.6. It was prepared daily and kept in the dark at 37 °C before use.
UHPLC-Q-TOF/MS analysis
For the UHPLC-ESI-MS/MS analysis, all samples were analyzed using a Nexera X2 Shimadzu UHPLC system coupled with an AB SCIEX TripleTOF® 5600 mass spectrometer. The chromatographic separation was conducted using Waters ACQUITY UHPLC® BEH C18 column (2.1 × 100 mm, 1.7 µm). The optimal mobile phase consisted of 0.1% formic acid-containing water (A) and 0.1% formic acid-containing acetonitrile (B) at a flow rate of 0.3 mL/min with 40 °C. The solvent gradient was set as follows: 0–0.5 min, 5% B; 0.5–5 min, 5–10% B; 5–10 min, 10–25% B; 10–18 min, 25–35% B; 18–20 min, 35–80% B; 20–20.5 min, 80–95% B; 20.5–28 min, 95% B. The autosampler was set at 6 °C, and the sample injection volume was set at 2 µL.
The mass spectrometer coupled with an electrospray ionization (ESI) source was operated in both positive (POS) and negative (NEG) ion mode. The data was collected using information-dependent acquisition (IDA) mode with mass range m/z 100–1200 for TOF-MS scan, and m/z 50–1200 for TOF-MS/MS scan. After optimization, the nebulizer gas, heater gas, and curtain gas were set at 50, 50, and 35 L/min, respectively. The ion spray voltage and declustering potential in POS mode were set at 5.5 kV and 80 V, while they were at −4.5 kV and −80 V in NEG mode. And for the IDA experiments, the collision energies were set at 35 eV and −35 eV, and collision energy spread was set at 15 eV. The experiments were run with 150 ms accumulation time for TOF-MS and 50 ms accumulation time for TOF-MS/MS. Meanwhile, the accurate MS and MS/MS measurements were acquired with the Automated Calibration Delivery System. As for quantification, the column and conditions were the same with described qualitative method except for acquisition only in NEG mode. And the information for quantified compounds was shown in Table 1.
Antioxidant assays
DPPH free radical scavenging activity DPPH scavenging activity was assessed as described with little revise (Bao et al., 2018). Briefly, different volumes of samples were diluted with methanol to 500 µL, and then mixed with DPPH solution (500 µL). Mixtures were left to react in the dark for 30 min at 25 °C. After incubation, the absorbance (Asample) was measured at 517 nm using Multiskan GO Microplate Reader (Thermo Scientific, USA). L-ascorbic acid was used as the positive control, with different concentrations for standard curve. The DPPH radical scavenging activity of sample was expressed as mg of L-ascorbic acid equivalents (AAE) per gram of dried sample (mg AAE/g). In addition, to determine the IC50 (50% inhibition) of samples on DPPH, five different concentrations were used. And, the distilled water instead of sample was made as blank control (Acontrol). Antioxidant activity of DPPH scavenging was calculated as follow: (%) inhibition = [(Acontrol − Asample) / Acontrol)] ×100%. IC50 value was determined to be the effective concentration at which DPPH radicals were inhibited by 50%.
ABTS free radical scavenging activity ABTS radical scavenging activity was assayed according to the reported method (Sridhar and Charles, 2019). First, ABTS+ stock solution was diluted with ethanol to get a stable absorbance of about 0.700 ± 0.005 at 734 nm. Afterward, 100 µL of appropriately diluted sample was added to 200 µL of ABTS+ solution in a 96-well plate. Then, the mixture was incubated in the dark at room temperature for 30 min and determined the absorbance at 734 nm. Ethanol was selected as blank control, and concentrations of 0 to 50 µg/mL L-ascorbic acid dissolved in water were made to construct standard curve. The scavenging activities of sample against ABTS+ were also expressed as µmol AAE/g. IC50 value was calculated similarly with DPPH scavenging method.
Ferric reducing/antioxidant power assay The FRAP assay was carried out as previously described (Zhu et al., 2015). 50 µL of properly diluted sample was added to 250 µL of freshly prepared FRAP reagent in a 96-well plate and mixed thoroughly. The mixture was incubated at 37 °C for 10 min, and then measured at 593 nm. The FRAP value was calculated as millimoles of Fe2+ equivalents per 100 g of sample (mmol Fe2+ equiv/100 g) based on a calibration curve plotted using FeSO4·7H2O as standard curve.
Screening out of antioxidants by pre-column UHPLC-Q-TOF/MS
DPPH assay combined with UHPLC-Q-TOF/MS was employed to rapidly screen out the antioxidants in extracts. Briefly, after the incubation of sample with DPPH solution, UHPLC-Q-TOF/MS was employed to further screen out the antioxidant profiles and identify the antioxidant components by detecting changes of characteristic peaks compared with the corresponding blank sample (methanol + sample, without DPPH reaction). 2 µL aliquot of the sample with and without DPPH reaction were analyzed under the same conditions as described before. The scavenging percentage of target compound was calculated according to the following formula: [(PAsample − PAsample+ DPPH) / PAsample] × 100%.
Quantitative analysis of major antioxidants
The calibration curves for nine antioxidative compounds was constructed by the plotting peak area ratios (y) of standard/IS versus corresponding concentrations (x). Six replicates of QCs at three different levels (low, medium, high) were analyzed within the same day for intra-day variation, and with three consecutive days for inter-day precision and accuracy. The recovery for these nine compounds were also determined by analyzing them in CNJ beverage spiked with QCs. And, the limit of quantification (LOQ) was estimated as the lowest concentration of the calibration curve with acceptable relative error (≤ 20%) and RSD (≤ 20%), as well as signal-to-noise ratio (≥ 10).
Data analysis
UHPLC-MS data was collected using Analyst TF 1.7.1 software and processed by Peakview (AB Sciex, MA, USA) software. As for antioxidant assays, all the tests were performed in triplicate, and results were analyzed and expressed as mean ± standard deviation using Graph Pad Prism 5.0 (La Jolla, CA, USA).
Results and discussion
Identification of Major Constituents in CNJ
Due to the complexity of the natural plant, the structure types and amounts of chemical compounds in CNJ were numerous. Therefore, before data processing, an in-house formula database consisting of name, molecular formula, chemical structure, and accurate mass of the compounds in individual ingredients of CNJ was established by searching from databases, such as TCMSP (https://www.old.tcmsp-e.com/tcmsp.php), PubMed (http://www.ncbi.nlm.nih.gov/pubmed), CNKI (http://www.cnki.net).
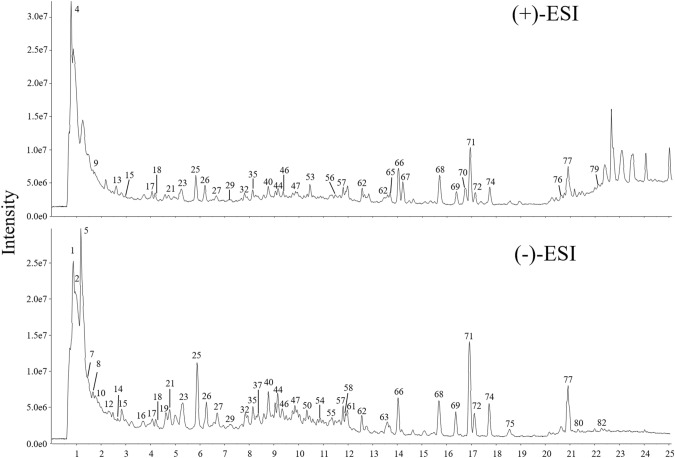
Because of the numerous compounds of natural products and their different chemical properties, both positive and negative ion modes were performed for MS analysis and the typical total ion chromatography (TIC) profiles of CNJ in the positive and negative ion mode were presented in Fig. 1. A total of 82 compounds were discovered, including 26 phenolic acids, 9 organic acids, 26 flavonoids, 11 triterpenoids, 6 other and 4 unknown compounds (Table 2). Among them, 9 compounds were unambiguously characterized by comparing with the reference standards. The other compounds were tentatively identified based on their retention time, mass accuracy of precursor ions, the fragmentation pathways from MS/MS spectra, reported literatures and established in-house database of CNJ. The specific chemical structures of these compounds were illustrated in Figure S1, and the exact ESI-MS/MS mass spectrum of these identified compounds was shown in Supporting Information.
Fig. 1.
Representative total ion chromatograms of the CNJ sample in positive and negative ion mode
Table 2.
Identification of the chemical constituents in CNJ by using UHPLC − ESI-Q-TOF/MS analysis
| No. | Rt (min) | Molecular Formula | [M + H]+ | (+) MS/MS | [M - H]− | (-) MS/MS | Identification | Source |
|---|---|---|---|---|---|---|---|---|
| 1a | 0.74 | C6H8O6 | 175.0231 | 157.0120, 127.0018, 115.0014, 87.0068 | L-ascorbic acid | CL | ||
| 2 | 0.87 | C5H10O6 | 165.0394 | 147.0287, 129.0191, 75.0088 | Ribonic acid or Xylonic acid | CL, AP | ||
| 3 | 0.88 | C7H12O6 | 191.0561 | 173.0446, 127.0381, 85.0724 | Quinic acid | CL, NM | ||
| 4 | 0.93 | C12H22O11 | 365.1023 | 203.0519, 185.0299 | 341.1082 | 179.0567, 119.0354, 89.0249 | 6-O-α-D-Galactopyranosyl-D-glucose or Sucrose or isomer | CL |
| 5 | 0.99 | C4H8O5 | 135.0290 | 117.0194, 89.0246, 75.0092 | Threonic acid | CL, NM, AP | ||
| 6 | 1.33 | C6H8O7 | 191.0173 | 173.0071, 129.0166, 111.0066, 85.0276 | Citric acid | CL, NM, AP | ||
| 7 | 1.33 | C4H6O5 | 133.0132 | 115.0013, 89.0223, 71.0117 | Malic acid | AP, CL | ||
| 8 | 1.33 | C6H8O7 | 191.0176 | 173.0066, 154.9971, 111.0063, 87.0064 | Isocitric acid | CL, NM, AP | ||
| 9 | 1.57 | C9H8O3 | 165.0546 | 147.0389,123.0389,119.0441 | p-Coumaric acid | AP | ||
| 10 | 2.69 | C7H6O4 | 153.0191 | 123.0449, 109.0296, 81.0351, | Protocatechuic acid | AP, CL | ||
| 11 | 1.84 | C13H16O10 | 331.0657 | 169.0127, 125.0225, | Galloyl-glucose | CL | ||
| 12 | 1.88 | C7H6O5 | 169.0145 | 151.0018, 125.0243, 107.0133, 97.0293 | Gallic acid | CL | ||
| 13 | 2.66 | C9H11NO2 | 166.0868 | 120.0813,103.0548 | Phenylalanine | NM, AP | ||
| 14 | 2.71 | C45H38O18 | 867.2125 | 579.1475,289.0709 | 865.2067 | 713.1536, 695.1464, 575.1241, 407.0800, 287.0586 | Procyanidin C1 or isomer | CL |
| 15 | 2.91 | C34H24O22 | 785.0833 | 767.0756, 465.0666, 321.0249, 303.0133, 277.0338 | 783.0701 | 401.0629, 300.9999, 275.0203 | Pedunculaginb or Casuariin | CL |
| 16 | 3.63 | C9H8O5 | 195.0295 | 179.0346, 165.0198, 151.0405, 123.0455 | 3, 4, 5-Trihydroxycinnamic acid | CL | ||
| 17 | 4.13 | C27H22O18 | 635.0535 | 465.0350, 321.0038, 277.0165 | 633.0732 | 481.0625, 300.9987, 169.0138 | Galloyl-HHDP-glucose or Strictinin | CL |
| 18 | 4.24 | C8H12O7 | 221.0538 | 185.0328, 157.0391, 138.9937, 111.0005 | 219.0531 | 157.0509, 111.0089, 87.0088 | 1-Ethyl Citrate | NM |
| 19 | 4.65 | C41H28O27 | 951.0767 | 907.0887, 783.0721, 300.9998 | Geraniin or isomer | CL | ||
| 20 | 4.77 | C34H24O22 | 785.0833 | 767.0756, 465.0666, 321.0249, 303.0133, 277.0338 | 783.0713 | 401.0623, 300.9996, 275.0197 | Pedunculaginb or Casuariin | CL |
| 21 | 4.79 | C41H28O27 | 953.0893 | 935.0157, 633.0293, 615.0203, 470.9874, 427.0006, 277.0165 | Geraniin or isomer | CL | ||
| 22 | 4.85 | C15H18O9 | 341.0879 | 179.04 | Caffeic acid hexoside | CL | ||
| 23a | 5.29 | C30H26O12 | 579.1492 | 427.1009, 409.0905, 287.0540, 247.0593, 163.0375 | 577.1360 | 407.0776, 339.0887, 289.0720, 245.0824, 161.0241, 125.0245 | Procyanidin B1 | CL |
| 24 | 5.77 | C45H38O18 | 867.2125 | 579.1475,289.0709 | 865.2067 | 739.1741, 713.1604, 577.1408, 575.1241, 407.0800, 287.0586 | Procyanidin C1 or isomer | CL |
| 25a | 5.88 | C15H14O6 | 291.0865 | 207.0639, 165.0537, 147.0427, 139.0372, 123.0424 | 289.072, 579.1507 | 245.0834, 203.0724, 151.0408, 123.0456 | (+)-catechin | CL |
| 26 | 6.26 | C21H32O10 | 445.2070 | 265.1419, 247.1320, 229.1204 | 443.1926 | 237.1516, 119.0355, 101.0224 | Penstemide | NM |
| 27 | 6.41 | C27H22O18 | 635.0882 | 465.0350, 321.0038, 277.0165 | 633.0732 | 300.9982, 275.0195 | Galloyl-HHDP-glucose or Strictinin | CL |
| 28 | 6.50 | C30H26O11 | 563.1499 | 291.0865,287.0556,273.0762 | 561.1420 | 289.0727, 245.0841 | Fisetinidol-(4α,8)-catechin or isomer | CL |
| 29 | 6.94 | C30H26O11 | 563.1502 | 393.0964, 291.0865,287.0556,273.0762 | 561.1401 | 435.1067, 407.0756, 289.0712, 273.0760, 125.0244 | Fisetinidol-(4α,8)-catechin or isomer | CL |
| 30 | 7.24 | C13H8O8 | 291.0149 | 247.0240, 219.0294, 191.0344, 175.0407 | Brevifolincarboxylic acid | CL | ||
| 31 | 7.86 | C35H30O15 | 691.1658 | 539.0836, 521.0745, 359.0530, 301.0502, 239.0398 | 689.1522 | 537.1070, 519.0976, 399.0742, 289.0733, 237.0422 | Unknown | CL |
| 32 | 7.89 | C21H10O13 | 469.0050 | 299.9910, 270.9899, | Flavogallonic acid | CL | ||
| 33 | 7.97 | C41H28O27 | 953.0893 | 917.0072, 615.0208, 452.9780, 277.0163 | Geraniin or isomer | CL | ||
| 34 | 8.02 | C21H10O13 | 469.0050 | 299.9910, 270.9899, | Flavogallonic acid | CL | ||
| 35 | 8.19 | C20H18O9 | 403.1029 | 285.0755, 263.0543, 251.0535, 151.0373, 139.0374 | 401.0878 | 301.0696, 289.0699, 203.0703, 151.0394, 137.0236, 109.029 | (Epi)catechin derivative | CL |
| 36 | 8.23 | C30H26O12 | 579.1495 | 561.1374,453.1176,409.0909,291.0861,287.0546 | Procyanidin B2 | CL | ||
| 37 | 8.30 | C20H14O14 | 479.0459 | 302.9928 | 477.0311 | 301.0007 | Ellagic acid glucuronide | CL |
| 38 | 8.38 | C27H22O18 | 633.0737 | 481.0609, 463.0512, 343.0098, 300.9990, 275.0192, 169.0136, 125.0237 | Galloyl-HHDP-glucose or Strictinin | CL | ||
| 39 | 8.63 | C14H24O10 | 353.1251 | 177.1010, 159.0909, 141.0084, 85.0586 | 351.1299 | 249.0619, 175.0266, 113.0245, 101.0610 | 3-Digitalose-quinic acid | AP |
| 40 | 8.83 | C19H30O8 | 387.2012 | 207.1366, 189.1269, 161.1315, 149.0948, 123.0793 | 385.1886 | 223.1355, 205.1253, 153.0932 | Roseoside | CL |
| 41 | 8.91 | C27H30O15 | 595.1659 | 379.0800, 271.0621 | 593.1510 | 473.1084, 383.0796, 353.0671 | Apigenin-6,8- di-C-glucoside | NM |
| 42 | 9.11 | C37H30O16 | 731.1193 | 579.0702, 441.0537, 411.0808, 271.0430, 153.0082 | 729.1458 | 407.0780, 289.0729, 169.0149, 125.0251 | Gallocatechin-(4a, 8)-catechin | CL |
| 43 | 9.21 | C27H30O15 | 593.1551 | 285.04 | Luteolin-7-O-rutinoside | NM | ||
| 44 | 9.22 | C41H28O26 | 937.0403 | 767.0235, 465.0373, 447.0262, 277.0162 | 935.0804, 467.0360 | 633.0739, 300.9988 | Galloylpedunculagin | CL |
| 45 | 9.30 | C30H26O12 | 579.1161 | 561.1400,427.1025,409.0918,291.0861,289.0703 | Procyanidin B3 | CL | ||
| 46 | 9.43 | C28H32O16 | 625.1772 | 607.1631,589.1553,571.1418 | 623.1618 | 503.1187, 413.0869, 383.0763 | Chrysoeriol 6,8-di-C-glucoside (stellarin-2) | NM |
| 47 | 9.82 | C20H16O12 | 449.0467 | 302.99 | 447.0573, 895.1224 | 299.99 | Ellagic acid-4-O-rhamnoside | CL |
| 48a | 9.91 | C14H6O8 | 300.9997 | 283.9963, 229.0142, 201.0192, 173.0245, 145.0296 | Ellagic acid | CL | ||
| 49a | 10.36 | C21H20O12 | 465.1027 | 303.0501 | 463.0907 | 300.0294, 271.0269, 255.0313 | Que-3-gal* (Hyperoside) | CL |
| 50 | 10.40 | C27H32O15 | 597.1812 | 451.1236,435.1288,289.0704 | 595.1667 | 287.0562, 151.0039 | Eriodictyol 7-O-rutinoside (Eriocitrin) | NM |
| 51a | 10.44 | C21H20O12 | 465.1027 | 303.0494 | 463.0912 | 300.0298, 271.0272, 255.0312 | Isoquercitrin | CL |
| 52 | 10.49 | C30H26O13 | 593.1303 | 447.0952, 285.0328, 255.0298, 145.0302 | Tiliroside | NM | ||
| 53 | 10.51 | C23H36O2 | 345.2813 | 155.1330, 137.1232, 81.0645 | Unknown | NM | ||
| 54 | 10.99 | C27H28O16 | 607.1309 | 463.0876, 300.0274, 271.0246, 151.0040 | Quercetin 3-O-HMGlc | CL | ||
| 55 | 11.51 | C27H28O15 | 591.1358 | 529.1361, 489.1044, 447.0942, 285.0418 | Kaempferol 3-O-HMGal | CL | ||
| 56 | 11.70 | C28H32O15 | 609.1814 | 463.1224,301.0705 | Diosmin | NM | ||
| 57a | 11.78 | C27H30O16 | 611.1620 | 303.07 | 609.1825 | 301.07 | Rutin | NM |
| 58 | 11.88 | C29H32O17 | 653.1338 | 347.05 | 651.1572 | 589.1613, 549.1284, 507.1178, 345.0637 | Limocitrin-HMG-glucoside | NM |
| 59 | 11.91 | C27H28O15 | 591.1358 | 529.1361, 489.1044, 447.0942, 285.0418 | Kaempferol 3-O-HMGlc | CL | ||
| 60 | 11.98 | C16H18O12 | 401.0778 | 357.0866, 313.0975 | Carboxyl derivative | CL, NM, AP | ||
| 61 | 12.04 | C30H34O18 | 683.1819 | 377.0870 | 681.1727 | 579.1342, 537.1236, 375.0715, 360.0479 | Limocitrol 3-O-HMGlc | NM |
| 62 | 12.57 | C21H28O9 | 425.1808 | 389.1586, 371.1487, 245.1171, 233.1172, 227.1053, 215.1062 | 423.1689 | 279.1258, 205.1257, 139.0778 | Grandidentatine | CL |
| 63 | 13.56 | C16H24O7 | 346.1671 | 311.1295,177.0285, 165.1166, 135.1170 | 327.1450 | 267.1264, 237.1147, 207.1049, 117.0206 | Rhododendrin or Perilloside B or isomer | NM |
| 64a | 13.60 | C15H10O7 | 303.0456 | 287.0735, 269.0628, 203.0204, 147.0344 | 301.0355 | 257.1551, 203.0169, 178.9988, 151.0036, 121.0295 | Quercetin | CL |
| 65 | 13.85 | C16H16O6 | 305.1010 | 203.0337, 159.0415, 147.0440 | Oxypeucedanin hydrate | NM | ||
| 66 | 13.99 | C16H24O7 | 346.1648 | 311.1487, 265.1263, 135.1170 | 327.1452 | 267.1250, 249.1136, 207.1035, 165.0931, 147.0809 | Rhododendrin or Perilloside B or isomer | NM |
| 67 | 14.28 | C22H21NO5 | 380.1500 | 317.0815, 299.0727, 233.0296, 231.0140, 175.0273 | Unknown | NM | ||
| 68 | 15.61 | C16H24O7 | 346.1648 | 135.11 | 327.1449 | 267.1262, 249.1150, 207.1044, 165.0938, 117.0204 | Rhododendrin or Perilloside B or isomer | NM |
| 69 | 16.27 | C36H58O10 | 651.3716, 673.3924 | 489.3475, 471.3373,453.3276, 407.3232, 247.1645, 223.1646, 205.1546 | 695.4028 | 649.4010, 487.3446 | Kajiichigoside F1 isomer | CL |
| 70 | 16.81 | C13H6N2O | 207.0455 | 192.0281, 164.0351, 149.0125, 121.0556 | Unknown | NM | ||
| 71a | 16.81 | C36H58O10 | 673.3924, 651.4158 | 489.3566, 471.3455, 453.3352, 425.3403, 407.3297, 205.1579 | 695.4026 | 649.3975, 487.3436, 207.0524 | Kajiichigoside F1 | CL |
| 72 | 17.01 | C36H58O10 | 673.3913 | 511.3363,185.0419 | 695.4029 | 649.3975, 487.3447 | Kajiichigoside F1 isomer | CL |
| 73 | 17.01 | C30H48O5 | 489.3576 | 471.3472,453.3364,425.3404,407.3297,389.3215,201.1639 | Euscaphic acid or tormentic acid or isomer | CL | ||
| 74 | 17.59 | C36H58O10 | 673.3919 | 489.3563, 471.3452, 453.3344, 425.3403, 407.3287, 205.1574 | 695.4026 | 649.3987, 487.3442 | Kajiichigoside F1 isomer | CL |
| 75 | 18.62 | C36H56O10 | 693.3854 | 647.3844, 485.3265 | Periplocoside O | CL | ||
| 76 | 20.48 | C14H14O6 | 279.0787 | 201.0333, 173.0399, 149.0130 | Columbianetin or isomer | CL, NM, AP | ||
| 77 | 20.97 | C36H56O9 | 633.3631 | 471.3156, 453.3058, 425.3134, 407.3035, 247.1528, 205.1458 | 631.3847, 677.3915 | 469.3318, 425.3425 | Glycyrrhetinic acid-3-O-monoglucose | CL |
| 78 | 21.24 | C30H46O5 | 485.3272 | 467.3164, 425.3049, 375.3079 | 2,19-Dihydroxy-3-oxo-urs-12-en-28-oic acid isomer | CL | ||
| 79 | 21.25 | C18H14O6 | 327.0836 | 287.0899,203.0306 | Rugosaflavonoid A | CL, NM, AP | ||
| 80 | 21.37 | C30H48O5 | 489.3581 | 471.3487, 453.3358,425.3417,407.3316,389.3215, 205.1582, 201.1631 | 487.3429 | 469.3320, 443.3518, 407.3309, 371.2963 | Euscaphic acid | CL |
| 81 | 21.86 | C30H46O5 | 485.3272 | 467.3156, 425.3061, 375.3079 | 2,19-Dihydroxy-3-oxo-urs-12-en-28-oic acid | CL | ||
| 82 | 22.17 | C30H46O4 | 469.3310 | 451.3211, 407.3318 | 2a,3b-Dihydroxylup-20(29)-en-28-oic acid | CL |
a Compounds confirmed with references. CL: R. roxburghii; NM: C. limon; AP: M. domestica. HM, 3-hydroxy-3-methylglutaryl-; Rha, rhamnose; Rut, rutinoside; Gal, galactoside
Flavonoids A total of 26 flavonoids were found in CNJ, including 10 flavonones and 16 flavones, which were mainly originating from R. roxburghii, and C. limon. Among them, compounds 23, 25, 51, 57, and 64 were uniquely assigned as procyanidin B1, catechin, isoquercitrin, rutin, and quercetin, respectively, by comparing with reference substances. With the characteristic fragment ions of reference compounds, it can be learned that: Since most flavonones are derivatives of catechin, they can be preliminarily characterized by specific fragmentation ions in MS/MS spectra, such as m/z 291 in positive mode, and m/z 289, 245 in negative mode. Meanwhile, flavones have similar fragmentation pathways by the neutral loss of glucosyl (162 Da), rhamnosyl (146 Da), or rutinosyl (308 Da). Compounds 36 and 45 shared the same pseudo-molecular ion (m/z 579.14), and similar product ions (m/z 561.13, 409.09, 291.08) with reference 23, procyanidin B1. Thus, they were constitutional isomers of procyanidin B1. According to the referring retention time of these isomers found in R. roxburghii (Liu et al., 2016), they were identified as procyanidin B2 and B3.
Besides the neutral loss of sugar from the cleavage of glycosidic bond, dehydration, successive losses of CO from phenolic hydroxyl groups and ketone group, and Retro-Diels-Alder (RDA) fragmentation are also the most possible fragmentation pathways for flavones (Zhang et al., 2015). For example, compounds 49, 51, 54, and 57 all showed fragment ion at m/z 301.04 in negative mode, which is the same as deprotonated ion of quercetin (65). Thus, they might all share the same aglycone. Compound 49 gave a [M − H]− ion at m/z 463.09 with the molecular formula C21H20O12, indicating the constitutional isomer of isoquercitrin (51). Since isoquercitrin was a glycoside of quercetin with glucose, compound 49 might be glycoside with galactose-hyperoside (Porter et al., 2012). Compound 54 showed [M − H]− ion at m/z 607.13, and fragment ion at m/z 463.09, 301.03, 151.01. Considering the 3-hydroxy-3-methyl-glutaryl (HM) specialized found in Rosa family (Porter et al., 2012), these fragment ions reasonably corresponded to the loss of an HM part ([M + H2O − HM − H]−), further loss of glucose [M + 2H2O − HM − Glc − H]−, and left A ring after RDA fragmentation of aglycon (Liu et al., 2016). Similarly, compounds 43, 50, 52, 55, 56, 58, 59 and 61 were annotated as luteolin 7-O-Rut, eriodictyol 7-O-Rut, tiliroside (Wang et al., 2020), kaempferol 3-O-HMGal, diosmin (Zhang et al., 2018), limocitrin-HMGlc, kaempferol 3-O-HMGlc, and limocitrol 3-O-HMGlc (Ledesma-Escobar et al., 2015), respectively. Two flavone C-glycosides, which were tentatively identified as apigenin 6,8- di-C-Glc (41) (Hwang and Ma, 2018) and chrysoeriol 6,8-di-C-Glc (46) (Guccione et al., 2016), were also discovered in C. limon and CNJ.
Phenolic acids 26 phenolic acids were found in CNJ, and primarily (21) originated from R. roxburghii (Table 2). Among them, compound 48 was unambiguously identified as ellagic acid with the authentic standard. Due to the fragment ions at m/z 300.99 ([M − H]− of ellagic acid), compounds 37 and 47 were both proposed as ellagic acid derivatives. Compound 37 ([M−H]−, m/z 477.03) was then characterized as ellagic acid glucuronide as it yielded a fragment ion at m/z 301.00 (ellagic acid) by the loss of glucuronide (176.03). And compound 47 was assigned as ellagic acid-4-O-Rha due to the loss of rhamnose (146.06) (Zeng et al., 2017). Due to the presence of carboxyl, carbonyl, or hydroxyl groups in structures, loss of CO2, CO and H2O could be found in fragment ions of phenolic acids. Compounds 9, 10, 12, 16, and 30 yielded [M−H2O−H]− and [M−CO2−H]− fragment ions in MS2 spectra. Based on their pseudo-molecular ions and literatures, these compounds were identified as p-coumaric acid, protocatechuic acid, gallic acid, 3, 4, 5-trihydroxycinnamic acid (Liu et al., 2016), and brevifolincarboxylic acid (Yisimayili et al., 2019), respectively.
Compound 11 was tentatively assigned as galloyl-glucose, because of its characteristic fragment ions (m/z 169.01, 125.02) of gallic acid and neutral loss of glucose (162.05). Compounds 17, 27, and 38 exhibited deprotonated molecules [M−H]− at m/z 633.07 and produced base peak ion at m/z 300.99 in MS2 spectrum, which suggested an ellagic acid unit released. Finally, they were identified as galloyl-HHDP-glucose, or strictinin, or isomers (Xu et al., 2018). And, compounds 32 and 34 produced ions at m/z 300.99 in MS2 spectrum, which was also characterized as an ellagic acid unit and corresponding to neutral loss of 168 Da. Thus, they were identified as flavogallonic acid or isomer (Zeng et al., 2017). Moreover, compound 44 was tentatively identified as galloylpedunculagin according to its [M − H]− at m/z 935.08 and fragment ions at m/z 765.05 [M − H − gallic acid]−, 633.07 [M − H − galloyl dimer]−, 463.05 [M − H − galloyl dimer − gallic acid]−, 300.99 [galloyl dimer]− (Yang et al., 2020).
Triterpenoids The triterpenoids in CNJ were all originated from R. roxburghii, and most of them can be classified as euscaphic acids. Thus, after the prominent loss of H2O or CO2, the cleavage of D ring could be observed in the fragment ions of these compounds. Compounds 69, 71, 72, and 74 all showed the same pseudo-molecular ions [M − H]− at m/z 695.40, indicating their chemical formula C36H58O10. Among them, compound 71 was identified as kajiichigoside F1 by comparison of its retention time and MS/MS spectra with the standard. And the others also generated similar fragment ions with kajiichigoside F1, at m/z 649.39, 487.34 by the neutral losses of HCOOH and glucose groups. Thus, they were identified as kajiichigoside F1 isomers.
Compounds 73 and 80 displayed [M + H]+ ion at m/z 489.35 and [M − H]− ion at m/z 487.34, corresponding to C30H48O5. In MS2 spectra, they generated fragment ions at m/z 471.34/469.33 and 425.34/423.32 by continuous losses of H2O and HCOOH group. Finally, they were identified as euscaphic acid or its isomers by referring to the literature (Liu et al., 2016). With a similar fragmentation pathway, compound 82 was assigned as 2α, 3β-Dihydroxylup-20(29)-en-28-oic acid. Compounds 78 and 81 had the same molecular formula of C30H46O5. Their fragment ions of m/z 467.31 and 425.30 indicated that they were isomers. By comparing their retention time on C18 column, compound 81 was identified as 2α, 19β-Dihydroxy-3-oxo-urs-12-en-28-oic acid, and the other one was its isomer (Liu et al., 2016).
Organic acids 9 organic acids were successfully identified in CNJ using negative ion mode and from three ingredients of CNJ. The characteristic fragment ions of these organic acids were [M − H − H2O]−, [M – H − CO2]−, and [M − HCOOH]−. Compound 1 with a deprotonated molecule at m/z 175.02, which produced ions at m/z 157.01, 129.01, 113.02, by neutral loss of H2O, HCOOH, CO2 groups, was identified as L-ascorbic acid by comparing with standard. According to the exact mass of their deprotonated ions and MS2 fragment ions in previous reports, compounds 2, 3, 5, and 7 were tentatively identified as ribonic acid or xylonic acid, quinic acid, threonic acid, and malic acid, respectively (Liu et al., 2016; Zheng et al., 2020). In addition, compounds 6 and 8, which were isomers of each other, were acknowledged as citric acid, isocitric acid based on their retention time difference.
Others Compound 13 showed quasi-molecular ion at m/z 166.08 with the molecular formula of C9H11NO2. Fragment ions at m/z 149.05, 131.04, 120.08 correspond to the neutral losses of NH3, H2O, HCOOH groups. Finally, it was identified as phenylalanine (Liu et al., 2016). Compound 65 obtained [M + H]+ ion at ions at m/z 305.10. With its biological source, Citrus genus which contained many coumarins, the high-intensity fragment m/z 203.03 indicated a neutral loss of the substituent [M + H − C5H10O2]+, and the sequential cleavage to lose molecules of CO produced the typical fragments m/z 147.04 [203-2CO]+, therefore, it was tentatively identified as oxypeucedanin hydrate (Zhang et al., 2018).
Antioxidant activities of CNJ and its ingredients
Due to the complex nature of phytochemicals and different mechanism-based methods, the antioxidant capacities of CNJ and its ingredients were measured by three commonly used methods: DPPH, ABTS scavenging activity, and FRAP. Results of these evaluations were expressed as L-ascorbic acid equivalents and IC50 values, which are shown in Table 3. The scavenging activity of free radicals determined by DPPH and ABTS+ assays showed that R. roxburghii and CNJ had higher AAEs and lower IC50 values, indicating they had better antioxidant activities. And, the scavenging effects on DPPH radicals (Figure S20) suggested that extracts of C. limon and M. domestica scavenged DPPH radical in a dose-dependent way, while with the same concentration, R. roxburghii and CNJ almost tended to peak value. Therefore, as an important ingredient in CNJ, R. roxburghii might contribute more to the antioxidant activity of CNJ. With regard to the ferric reducing capacities, the trend was almost the same as that of the DPPH and ABTS assay. The result revealed that the order of antioxidant potency was: R. roxburghii > CNJ beverage > C. limon > M. domestica.
Table 3.
DPPH free-radical scavenging capacity of CNJ and its ingredients
| Sample | DPPH | ABTS | FRAP (mmol Fe2+/100 g) | ||
|---|---|---|---|---|---|
| (mg AAE/g) | IC50 (mg/mL) | (mg AAE/g) | IC50 (mg/mL) | ||
| CNJ | 1.35 ± 0.38 | 11.3 ± 0.93 | 1.92 ± 0.93 | 20.5 ± 8.18 | 68.39 ± 12.51 |
| CL | 3.05 ± 0.21 | 4.5 ± 1.42 | 3.84 ± 1.06 | 8.3 ± 2.74 | 103.57 ± 37.26 |
| NM | 0.13 ± 0.06 | 73.5 ± 4.68 | 0.19 ± 0.10 | 79.4 ± 16.22 | 11.05 ± 3.08 |
| AP | 0.08 ± 0.03 | 80.1 ± 1.21 | 0.15 ± 0.07 | 91.3 ± 10.79 | 1.71 ± 0.73 |
| L-Ascorbic acid | nt | 0.026 ± 0.009 | nt | 0.035 ± 0.011 | 1882.72 ± 202.91 |
All values are expressed as mean ± SD of three independent measurements; AAE, L- ascorbic acid equivalents; nt, not tested
Screening potential antioxidants by pre-column UHPLC-DPPH
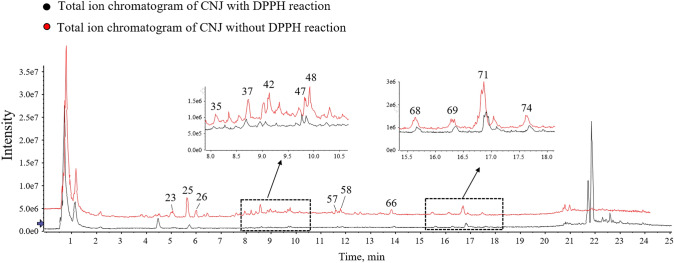
To find out the potential antioxidants in CNJ, the pre-column DPPH reaction coupled with UHPLC was used. After reaction, the antioxidants would be oxidized, leading to a structural change of them. Based on this, the peak intensity of the antioxidants would obviously decrease in the UHPLC chromatogram after with DPPH reaction. The UHPLC chromatograms of CNJ with or without DPPH-treated were shown in Fig. 2. The compounds with peak intensity decreased by more than 50% were considered as potential antioxidants. Finally, 18 potential antioxidant compounds (Table 4) were screened out and identified. Of these, six (procyanidin B1, (+)-catechin, catechin derivative, gallocatechin-(4α, 8)-catechin, rutin, limocitrin-HMGlc) are flavonoids; seven (mainly are ellagic acid and its derivatives, rhododendrin isomers) are phenolic acids; four (kajiichigoside F1 and its isomers) are triterpenoids; and one, penstemide. Moreover, 12 potential antioxidative compounds were originated from R. roxburghii, while 7 were from C. limon. The result further proved that R. roxburghii contributes more to the antioxidant activity of CNJ compared with others, and the selected compounds by pre-column UHPLC-DPPH might play an important role in its antioxidant activity.
Fig. 2.
Comparison of the chromatograms of the CNJ treated with DPPH-methanol solution and methanol in negative ion mode
Table 4.
Potential antioxidative compounds founded in CNJ
| No. | Name | Intensity reduced (%) |
|---|---|---|
| 23 | Procyanidin B1 | 67 |
| 25 | (+)-Catechin | 52 |
| 26 | Penstemide | 64 |
| 35 | Catechin derivative | 71 |
| 37 | Ellagic acid glucuronide | 74 |
| 42 | Gallocatechin-(4, 8)-catechin | 75 |
| 47 | Ellagic acid 4-O-rhamnoside | 58 |
| 48 | Ellagic acid | 53 |
| 57 | Rutin | 71 |
| 58 | Limocitrin-HMGlc | 78 |
| 60 | Carboxyl derivative | 71 |
| 63 | Rhododendrin or isomer | 79 |
| 66 | Rhododendrin or isomer | 60 |
| 68 | Rhododendrin or isomer | 59 |
| 69 | Kajiichigoside F1 isomer | 64 |
| 71 | Kajiichigoside F1 | 62 |
| 72 | Kajiichigoside F1 isomer | 70 |
| 74 | Kajiichigoside F1 isomer | 68 |
Contents of major antioxidants in CNJ and its ingredients
With the discovery of potential antioxidative compounds in CNJ, they should be quantified as much as possible to make sure its antioxidant activity. In this study, five selected potential antioxidants and four other compounds in CNJ were determined and quantified. The results of intra- and inter-day precision, accuracy, and recoveries Table S1 and S2 suggested this quantification method was validated with good reliability and sensitivity.
The content of these nine compounds in CNJ and its ingredients is shown in Table 1. From the results, the contents of most analytes in R. roxburghii were higher than other ingredients, which further proved its good antioxidant activity. And it was worthy that, comparing with other compounds, procyanidin B1 (23), (+)-catechin (25), ellagic acid (48), and kajiichigoside F1 (71) not only had good antioxidative capacities (decreased more than 50% after DPPH-treated, Table 4), but also had a higher amount in both CNJ (5.64, 8.21, 4.48, and 5.19 µg/mL) and R. roxburghii (164.81, 77.06, 75.41, and 156.87 µg/mL) (Table 1). Thus, these four compounds might contribute more to the antioxidant activity. By contrast, even though rutin also had a large decrease after DPPH reaction (71%), due to its low content (0.06 and 0.10 µg/mL in CNJ and R. roxburghii), it might contribute less. As the commonly used positive control, L-ascorbic acid was also quantified despite no significant decrease after the reaction. And L-ascorbic acid, vitamin C, did have much higher contents in both CNJ and R. roxburghii (329.44 and 1819.63 µg/mL, Table 1).
As we know, the components of natural products are not only highly complex, but also with wide scales of content. Furthermore, there is still a lack of knowledge on the antioxidant capacities of most components, especially those unidentified compounds. Therefore, it is really difficult to find out its “real” antioxidative compounds of natural products or antioxidants. In this study, the chemical profiling and antioxidant capacity of CNJ were analyzed, as an example. Although there is still a lack of information on antioxidative capacities (IC50) of most compounds, the degree to which they react with DPPH (percentage of peak intensity decrease after reaction) can somehow explain it. With the chemical identification, and further quantitation, it may provide some knowledge on predicting the antioxidative compounds of natural products.
Finally, for the first time, the chemical composition profile of CNJ beverage was analyzed, as well as its antioxidant activity. By using UHPLC-Q-TOF/MS analysis, a total of 82 compounds were identified or tentatively deduced, mainly consisting of flavonoids, phenolic acids, triterpenoids, and organic acids. And, the antioxidant capacities of CNJ and its ingredients were assessed by different mechanism-based assays. The result suggested that the ingredient, R. roxburghii, plays a critical role in the antioxidant activity of CNJ. Furthermore, 18 potential antioxidants in CNJ were screened out by pre-column DPPH-UHPLC. Based on the identification, these antioxidants are considered mainly as catechin and its derivatives, ellagic acid and its derivatives, and kajiichigoside F1 or isomers. This study not only provides information on the chemical composition of CNJ and its antioxidant activity but also most importantly, by further quantification of these antioxidants in CNJ, it helps us find out the “real antioxidative compounds” in it. This study may provide a practical strategy for rapid screening out antioxidants in natural plants or foods and a better understanding of their antioxidative activities.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was funded by the Guangzhou Institute of Respiratory Health Open Project (Funds provided by China Evergrande Group) (Project No.:2020GIRHHMS04).
Author contributions
Yida Zhang, Peiyan Zheng and Guanyu Yan contributed equally to this work. Yida Zhang analyzed the data, interpreted the results, and drafted the manuscript. Guanyu Yan performed the experiments. Yue Zhuo provided guidance for experiments. Peiyan Zheng critically revised the manuscript. Jian-lin Wu and Baoqing Sun conceived and designed the study. All authors read and approved the final manuscript.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yida Zhang, Peiyan Zheng and Guanyu Yan are co-first authors and contributed equally to this work.
Contributor Information
Yida Zhang, Email: xiaoda0610@163.com.
Peiyan Zheng, Email: gdmcslxx@126.com.
Guanyu Yan, Email: sharpgyyan@126.com.
Yue Zhuo, Email: zy2008213037@126.com.
Jian-lin Wu, Email: jlwu@must.edu.mo.
Baoqing Sun, Email: sunbaoqing@vip.163.com.
References
- Ayvaz MC, Omur B, Erturk O, Kabakci D. Phenolic profiles, antioxidant, antimicrobial, and DNA damage inhibitory activities of chestnut honeys from Black Sea Region of Turkey. Journal of Food Biochemistry. 42: e12502 (2018)
- Bao YT, Qu Y, Li JH, Li YF, Ren XD, Maffucci KG, Li RP, Wang ZG, Zeng R. In Vitro and In Vivo antioxidant activities of the flowers and leaves from Paeonia rockii and identification of their antioxidant constituents by UHPLC-ESI-HRMSn via pre-column DPPH reaction. Molecules. 23: 392 (2018) [DOI] [PMC free article] [PubMed]
- Chang X, Zhang WJ, Zhao ZY, Ma CX, Zhang T, Meng QY, Yan PZ, Zhang L, Zhao YP. Regulation of mitochondrial quality control by natural drugs in the treatment of cardiovascular diseases: Potential and advantages. Frontiers in Cell and Developmental Biology. 8 (2020) [DOI] [PMC free article] [PubMed]
- Chen GL, Zhu MZ, Guo MQ. Research advances in traditional and modern use of Nelumbo nucifera: phytochemicals, health promoting activities and beyond. Critical Reviews in Food Science and Nutrition. 2019;59:S189–S209. doi: 10.1080/10408398.2018.1553846. [DOI] [PubMed] [Google Scholar]
- Espin JC, Garcia-Conesa MT, Tomas-Barberan FA, Nutraceuticals Facts and fiction. Phytochemistry. 2007;68:2986–3008. doi: 10.1016/j.phytochem.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sarrias A, Larrosa M, Garcia-Conesa MT, Tomas-Barberan FA, Espin JC. Nutraceuticals for older people: Facts, fictions and gaps in knowledge. Maturitas. 2013;75:313–334. doi: 10.1016/j.maturitas.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Grzesik M, Naparlo K, Bartosz G, Sadowska-Bartosz I. Antioxidant properties of catechins: Comparison with other antioxidants. Food Chemistry. 2018;241:480–492. doi: 10.1016/j.foodchem.2017.08.117. [DOI] [PubMed] [Google Scholar]
- Guccione C, Bergonzi MC, Piazzini V, Bilia AR. A Simple and Rapid HPLC-PDA MS Method for the Profiling of Citrus Peels and Traditional Italian Liquors. Planta Medica. 2016;82:1039–1045. doi: 10.1055/s-0042-108735. [DOI] [PubMed] [Google Scholar]
- Hou ZQ, Yang HZ, Zhao Y, Xu L, Zhao L, Wang YT, Liao XJ. Chemical characterization and comparison of two chestnut rose cultivars from different regions. Food Chemistry. 323: 126806 (2020) [DOI] [PubMed]
- Hwang YH, Ma JY. Preventive Effects of an UPLC-DAD-MS/MS Fingerprinted Hydroalcoholic Extract of Citrus aurantium in a Mouse Model of Ulcerative Colitis. Planta Medica. 2018;84:1101–1109. doi: 10.1055/a-0604-2797. [DOI] [PubMed] [Google Scholar]
- Jiang XL, Wang L, Wang EJ, Zhang GL, Chen B, Wang MK, Li F. Flavonoid glycosides and alkaloids from the embryos of Nelumbo nucifera seeds and their antioxidant activity. Fitoterapia. 2018;125:184–190. doi: 10.1016/j.fitote.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Kalhori MR, Khodayari H, Khodayari S, Vesovic M, Jackson G, Farzaei MH, Bishayee A. Regulation of long non-coding RNAs by plant secondary metabolites: a novel anticancer therapeutic approach. Cancers. 13: 1274 (2021) [DOI] [PMC free article] [PubMed]
- Ledesma-Escobar CA, Priego-Capote F, Luque de Castro MD. Characterization of lemon (Citrus limon) polar extract by liquid chromatography-tandem mass spectrometry in high resolution mode. Journal of Mass Spectrometry. 2015;50:1196–205. doi: 10.1002/jms.3637. [DOI] [PubMed] [Google Scholar]
- Lin CZ, Zhang RJ, Yao YF, Huang XD, Zheng RB, Wu BJ, Zhu CC. Qualitative and quantitative analysis of the major constituents in WLJ herbal tea using multiple chromatographic techniques. Molecules. 23: 2623 (2018) [DOI] [PMC free article] [PubMed]
- Liu MH, Zhang Q, Zhang YH, Lu XY, Fu WM, He JY. Chemical analysis of dietary constituents in Rosa roxburghii and Rosa sterilis fruits. Molecules. 21: 1204 (2016) [DOI] [PMC free article] [PubMed]
- Mansoori S, Dini A, Chai SC. Effects of tart cherry and its metabolites on aging and inflammatory conditions: Efficacy and possible mechanisms. Ageing Research Reviews. 66: 101254 (2021) [DOI] [PubMed]
- Parham S, Kharazi AZ, Bakhsheshi-Rad HR, Nur H, Ismail AF, Sharif S, RamaKrishna S, Berto F. Antioxidant, antimicrobial and antiviral properties of herbal materials. Antioxidants. 9 (12): 1309 (2020) [DOI] [PMC free article] [PubMed]
- Porter EA, van den Bos AA, Kite GC, Veitch NC, Simmonds MSJ. Flavonol glycosides acylated with 3-hydroxy-3-methylglutaric acid as systematic characters in Rosa. Phytochemistry. 2012;81:90–96. doi: 10.1016/j.phytochem.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Sridhar K, Charles AL. In vitro antioxidant activity of Kyoho grape extracts in DPPH center dot and ABTS(center dot) assays: Estimation methods for EC50 using advanced statistical programs. Food Chemistry. 2019;275:41–49. doi: 10.1016/j.foodchem.2018.09.040. [DOI] [PubMed] [Google Scholar]
- Wang D, Fu ZF, Xing YC, Tan Y, Han LF, Yu HY, Wang T. Rapid identification of chemical composition and metabolites of Pingxiao Capsulein vivousing molecular networking and untargeted data-dependent tandem mass spectrometry. Biomedical Chromatography. 34: e4882 (2020) [DOI] [PubMed]
- Xu H, Wang Y, Yuan F, Gao Y. Polyphenols In Chinese Kushui Rose (Rosa sertata × Rosa rugosa) leaves. Acta Alimentaria. 2018;47:433–442. doi: 10.1556/066.2018.47.4.6. [DOI] [Google Scholar]
- Yang QQ, Zhang D, Farha AK, Yang X, Li HB, Kong KW, Zhang JR, Chan CL, Lu WY, Corke H, Gan RY. Phytochemicals, essential oils, and bioactivities of an underutilized wild fruit Cili (Rosa roxburghii). Industrial Crops And Products. 143: 111928 (2020)
- Yisimayili Z, Abdulla R, Tian Q, Wang YY, Chen MC, Sun ZL, Li ZX, Liu F, Aisa HA, Huang CG. A comprehensive study of pomegranate flowers polyphenols and metabolites in rat biological samples by high-performance liquid chromatography quadrupole time-of-flight mass spectrometry. Journal of Chromatography A. 1604: 460472 (2019) [DOI] [PubMed]
- Zeng FF, Ge ZW, Limwachiranon J, Li L, Feng SM, Wang YS, Luo ZS. Antioxidant and tyrosinase inhibitory activity of Rosa roxburghii fruit and identification of main bioactive phytochemicals by UPLC-Triple-TOF/MS. International Journal Of Food Science And Technology. 2017;52:897–905. doi: 10.1111/ijfs.13353. [DOI] [Google Scholar]
- Zhang YD, Huang X, Zhao FL, Tang YL, Yin L. Study on the chemical markers of Caulis Lonicerae japonicae for quality control by HPLC-QTOF/MS/MS and chromatographic fingerprints combined with chemometrics methods. Analytical Methods. 2015;7:2064–2076. doi: 10.1039/C4AY02744B. [DOI] [Google Scholar]
- Zhang Q, Tan CN, Cai L, Xia FB, Gao D, Yang FQ, Chen H, Xia ZN. Characterization of active antiplatelet chemical compositions of edible Citrus limon through ultraperformance liquid chromatography single quadrupole mass spectrometry-based chemometrics. Food & Function. 2018;9:2762–2773. doi: 10.1039/C8FO00403J. [DOI] [PubMed] [Google Scholar]
- Zheng YT, Liu S, Xie JH, Chen Y, Dong RH, Zhang XJ, Liu SQ, Xie JY, Hu XB, Yu Q. Antioxidant, alpha-amylase and alpha-glucosidase inhibitory activities of bound polyphenols extracted from mung bean skin dietary fiber. Lwt-Food Science And Technology. 132: 109943 (2020)
- Zhou RR, Liu XH, Chen L, Huang JH, Liang XJ, Wan D, Zhang SH, Huang LQ. Comparison of the antioxidant activities and phenolic content of five Lonicera flowers by HPLC-DAD/MS-DPPH and chemometrics. International Journal Of Analytical Chemistry. 2020: 2348903 (2020) [DOI] [PMC free article] [PubMed]
- Zhu MZ, Wu W, Jiao LL, Yang PF, Guo MQ. Analysis of Flavonoids in Lotus (Nelumbo nucifera) Leaves and Their Antioxidant Activity Using Macroporous Resin Chromatography Coupled with LC-MS/MS and Antioxidant Biochemical Assays. Molecules. 2015;20:10553–10565. doi: 10.3390/molecules200610553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.