Abstract
Porcine hemagglutinating encephalomyelitis virus (PHEV) is a member of the subgenus Embecovirus of the genus Betacoronavirus, and it is ubiquitously distributed in most pig-farming countries worldwide with low clinical incidence. Here, we report the full-length genome sequence and molecular characterization of a novel PHEV strain identified in diarrheic neonates in South Korea. The complete genome of the Korean PHEV strain GNU-2113 was sequenced and analyzed to characterize PHEV circulating in South Korea. The GNU-2113 genome was determined to be 29,982 nucleotides in length, with large unique deletions in the regions encoding nonstructural protein 3 and NS2. It was found to share 95.1–96.9% sequence identity with other global strains. Genetic and phylogenetic analysis indicated that the GNU-2113 strain is distantly related to the existing PHEV genotypes, implying that the virus appears to undergo substantial evolution under endemic pressure. This study provides important information about the genetic diversity of PHEV circulating subclinically in swine herds, which may ensure viral fitness in the enzootic environment.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-022-05414-w.
Porcine hemagglutinating encephalomyelitis virus (PHEV) is a neurological and/or enteric swine coronavirus that belongs to the subgenus Embecovirus of the genus Betacoronavirus within the family Coronaviridae of the order Nidovirales [1]. PHEV is a large enveloped virus with a non-segmented, positive-sense RNA genome of approximately ~ 30 kb containing at least 11 open reading frames (ORFs) [2, 3]. The first two large and partially overlapping ORFs 1a and 1b encode the replicase polyproteins pp1a and pplab, which are cleaved into 16 nonstructural proteins (nsp1–16). The remaining ORFs encode four canonical coronaviral structural proteins – the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins – and the accessory proteins NS2, NS4.9, NS12.7, and N2 [3]. Moreover, similar to other hemagglutinating coronaviruses, PHEV also possesses an envelope-associated glycoprotein, hemagglutinin-esterase (HE), which is encoded by ORF3 [3, 4].
PHEV infection can occur in all age groups, but clinical manifestations are rare and dependent upon age. Only piglets younger than 3–4 weeks old, particularly those born to naïve sows, are highly susceptible to PHEV-related disease, including vomiting and wasting disease (VMD) or encephalomyelitis [1]. Since PHEV is enzootic and circulates subclinically in most pig populations worldwide, most sows that have been asymptomatically infected are immune and provide passive protection to their vulnerable offspring through lactogenic immunity in endemically infected herds [5]. Despite its global distribution, PHEV remains the least studied of the swine enteric coronaviruses because of its low clinical prevalence and impact on the swine industry worldwide. Thus far, a small number of complete nucleotide sequences of PHEV have been reported [3, 6]. In South Korea, the prevalence of PHEV antigens in clinically ill pigs has been reported, indicating the circulation of only one genogroup [7]. However, there has been no investigation yet to characterize the genome of domestic PHEV. In this study, the complete genome sequence of a Korean PHEV strain, GNU-2113, was determined and analyzed.
In early January 2021, an acute outbreak of diarrheic disease in newborn piglets occurred on a commercial farrow-to-finish farm in Gyeongbuk Province in southeastern South Korea. In affected neonates, diarrhea began at 12 hours after birth, and the mortality rate reached 40%. Five fecal specimens collected from diarrheic piglets were submitted to our laboratory for diagnosis on 21 January. Using RT-PCR, the samples were initially tested for the presence of porcine enteric viruses, including porcine epidemic diarrhea virus, porcine deltacoronavirus, transmissible gastroenteritis virus, porcine rotavirus, and porcine torovirus. However, none of these viral pathogens were detected in any of the porcine diarrheic samples. Universal coronavirus RT-PCR assays were conducted to amplify the viral RNA-dependent RNA polymerase gene [8], followed by sequencing and BLAST searches of the PCR amplicons, which revealed the presence of PHEV in the stool samples. This result was confirmed by RT-PCR targeting the HE gene of PHEV [9]. Subsequently, the full-length genome sequence of the Korean PHEV GNU-2113 isolate was determined using the traditional Sanger method and compared with those of other strains available in the GenBank database. To accomplish this, primers were designed based on previously published sequences available in GenBank in addition to the newly amplified GNU-2113 sequences (primer sequences are available upon request). Thirteen overlapping cDNA fragments spanning the entire GNU-2113 genome were amplified by RT-PCR, using a gene-specific primer set. Each PCR amplicon was cloned individually into pGEM-T Easy Vector (Promega, Madison, WI) and sequenced in both directions using commercial vector-specific T7 and SP6 primers and GNU-2113-specific primers as described elsewhere [10]. The 5′ and 3′ ends of the GNU-2113 genome were determined by rapid amplification of cDNA ends as described previously [11]. The GNU-2113 sequence data were deposited in the GenBank database under accession number OL542832.
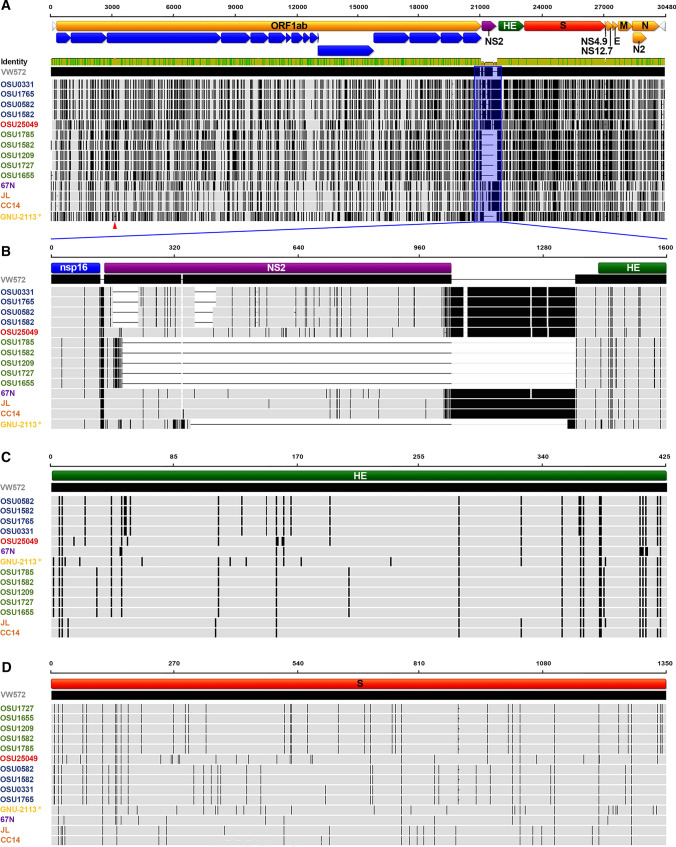
The genome of GNU-2113 is comprised of 29,982 nucleotides, excluding the 3′ poly(A) tail, and is arranged with a gene order typical of members of the subgenus Embecovirus: 5′ untranslated region (UTR)-replicase-HE-S-E-M-N-3′ UTR. The GNU-2113 genome sequence shares 95.1–96.9% identity with the published PHEV sequences (Supplementary Table S1). The percentage of sequence identity and the number of nucleotide (nt)/amino acid (aa) differences between GNU-2113 and other strains are summarized in Supplementary Table S1. In comparison with the reference PHEV strain VW572, GNU-2113 contains large deletions (DELs) in nsp3 and NS2 (Fig. 1A). The former includes a novel 57-nt DEL in ORF1a at positions 3,157–3,213, which leads to a 19-aa DEL in nsp3. Notably, the NS2 gene of GNU-2113 is 126 nucleotides in length, encoding a 42-aa protein, which is 153-aa shorter than that of the VW572 strain. Moreover, the DEL signature in NS2 of GNU-2113 is unique and unprecedented in other isolates, highlighting the diversity of the NS2 DEL (Fig. 1B). Subsequent genome sequence comparisons revealed that the genetic drift position (i.e., a barcode pattern) of GNU-2113 was clearly distinguishable from that of other global strains at the genomic level (Fig. 1A) and in two major embecoviral envelope proteins, HE (Fig. 1C) and S (Fig. 1D).
Fig. 1.
Schematic diagram of multiple sequence alignments of the whole genome, NS2, HE, and S relative to the reference strain VW572. PHEV strains that are grouped with the comparable barcode pattern are color-coded. GNU-2113 is shown in yellow and is indicated by an asterisk (*). (A) The top illustration represents the genomic regions, with yellow, purple, green, and red bars representing the identified ORFs and blue arrows indicating the nonstructural proteins produced after translation of ORF1a/1b followed by processing by viral-encoded proteases. Light gray arrows represent the 5′ and 3′ UTRs. The NS2 coding region is shaded and outlined in blue. Lightly shaded areas are those identical to VW572, and each vertical black bar represents a nucleotide sequence that is divergent from that of VW572. Thin horizontal dashed lines indicate DELs. The DEL in nsp3 is indicated by a red arrowhead. (B) The second diagram represents the coding regions for the PHEV NS2 protein (purple) located between nsp16 and HE. Lightly shaded areas are those identical to VW572, and each vertical black bar represents the nucleotide sequence that is different from VW572. Thin horizontal dashed lines indicate DELs. (C) The third diagram represents the coding regions for the PHEV HE protein (green). Lightly shaded areas are those identical to VW572, and each vertical black bar represents the amino acid sequence that is dissimilar to VW572. (D) The bottom illustration represents the coding regions for the PHEV S protein (red). Lightly shaded areas are those identical to VW572, and each vertical black bar represents the amino acid sequence that is divergent from VW572.
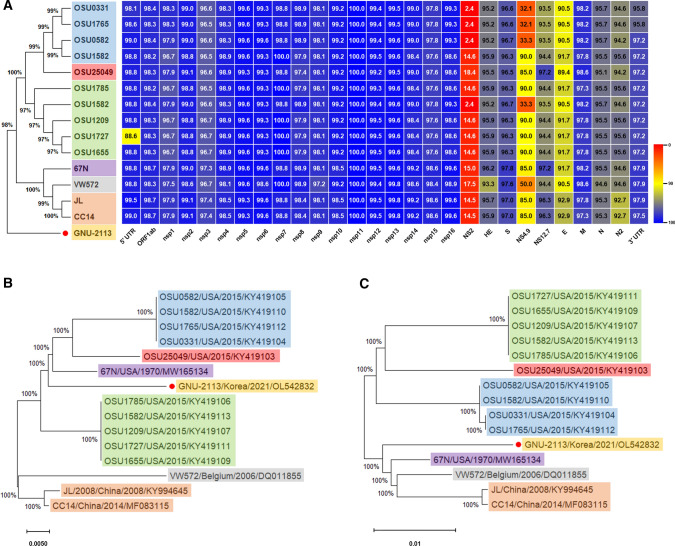
To examine its genetic relationships, phylogenetic analysis was conducted using nt or aa sequences of the full-length genome, HE, and S of the PHEV isolate from this study and those available in the GenBank database (Fig. 2). Analysis based on whole-genome sequences showed the presence of illustrated two different clusters (genotypes 1 and 2), as described previously (Fig. 2A) [3]. However, the Korean isolate GNU-2113 was separate from those two clusters and hence is proposed to represent a new lineage of PHEV (Fig. 2A). In particular, the accessory genes and E protein were divergent and shared a low level of sequence identity with those of other PHEV isolates (Fig. 2A). Phylogenetic analysis was also conducted based on the predicted amino acid sequences of the complete HE (Fig. 2B) and S (Fig. 2C) proteins. The data indicated that the GNU-2113 strain clustered differently depending on the protein sequence that was used to construct the phylogenetic tree.
Fig. 2.
Phylogenetic analysis based on sequences of the PHEV strains. (A) Conservation and variation of the GNU-2113 nonstructural and structural proteins compared with those of global PHEV strains. The whole-genome-based phylogenetic tree of PHEV strains is shown on the left. Heat maps were constructed from the indicated set of PHEV strains, using alignment data paired with neighbor-joining phylogenetic trees, which were built using Geneious prime (v.2022.0.1) and visualized in Prism 8 (v.8.4.3). (B and C) Phylogenetic analysis based on the HE and S amino acid sequences of PHEV strains. Multiple sequence alignments were performed using ClustalX, and phylogenetic trees were constructed from the aligned nucleotide or amino acid sequences using the neighbor-joining method in MEGA11 (v.11.0.8). PHEV strains that are grouped in the same cluster of each phylogenetic tree are indicated in different-colored squares. The novel variant GNU-2113, identified in this study, is indicated in the yellow square with a red circle. The name, country, and date (year) of isolation, and GenBank accession number of each PHEV isolate is shown. Scale bars indicate nucleotide substitutions per site.
To our knowledge, this is the first report of the complete genome sequence and molecular characterization of a PHEV isolate from South Korea. Although PHEV is present in most countries, including South Korea [7], it is generally reported to have low clinical prevalence, and its clinical manifestations in swine herds are age-dependent. However, the virus still poses a potential threat to herds of healthy gilts and has caused several outbreaks of VMD and encephalomyelitis affecting entire litters of newborn piglets born to naïve dams [1]. In this study, we identified a novel lineage of PHEV, suggesting continuous viral evolution and diversity, which can affect virulence and clinical manifestations. Therefore, monitoring and surveillance investigations are required to identify circulating PHEV strains and determine their genotypic and phenotypic traits. Our sequence data will provide further insights into the epidemiology and diversity of PHEV and provide information for understanding the molecular biology and pathogenesis of PHEV.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the Animal Disease Management Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (119081-5).
Compliance with ethical standards
Funding
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Animal Disease Management Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (119081-5).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mora-Díaz JC, Piñeyro PE, Houston E, Zimmerman J, Giménez-Lirola LG. Porcine hemagglutinating encephalomyelitis virus: a review. Front Vet Sci. 2019;6:53. doi: 10.3389/fvets.2019.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z, Zhao K, Lan Y, Lv X, Hu S, Guan J, Lu H, Zhang J, Shi J, Yang Y, Song D, Gao F, He W. Porcine hemagglutinating encephalomyelitis virus enters neuro-2a cells via clathrin-mediated endocytosis in a Rab5-, cholesterol-, and pH-dependent manner. J Virol. 2017;91:01083–01017. doi: 10.1128/JVI.01083-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi J, Zhao K, Lu H, Li Z, Lv X, Lan Y, Guan J, He W, Gao F. Genomic characterization and pathogenicity of a porcine hemagglutinating encephalomyelitis virus strain isolated in China. Virus Genes. 2018;54:672–683. doi: 10.1007/s11262-018-1591-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasseville AMJ, Boutin M, Gélinas AM, Dea S. Sequence of the 3'-terminal end (8.1 kb) of the genome of porcine haemagglutinating encephalomyelitis virus: comparison with other haemagglutinating coronaviruses. J Gen Virol. 2002;83:2411–2416. doi: 10.1099/0022-1317-83-10-2411. [DOI] [PubMed] [Google Scholar]
- 5.Saif LJ, Wang Q, Vlasova AN, Jung K, Xiao S. Coronaviruses. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, Zhang J, editors. Diseases of swine. 11. Ames: Blackwell Publishers; 2012. pp. 488–523. [Google Scholar]
- 6.Lorbach JN, Wang L, Nolting JM, Benjamin MG, Killian ML, Zhang Y, Bowman AS. Porcine hemagglutinating encephalomyelitis virus and respiratory disease in exhibition swine, Michigan, USA, 2015. Emerg Infect Dis. 2017;23:1168–1171. doi: 10.3201/eid2307.170019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rho S, Moon HJ, Park SJ, Kim HK, Keum HO, Han JY, Nguyen GV, Park BK. Detection and genetic analysis of porcine hemagglutinating encephalomyelitis virus in South Korea. Virus Genes. 2011;42:90–96. doi: 10.1007/s11262-010-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Souza Luna LK, Heiser V, Regamey N, Panning M, Drexler JF, Mulangu S, Poon L, Baumgarte S, Haijema BJ, Kaiser L, Drosten C. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J Clin Microbiol. 2007;45:1049–1052. doi: 10.1128/JCM.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, He W, Lan Y, Zhao K, Lv X, Lu H, Ding N, Zhang J, Shi J, Shan C, Gao F. The evidence of porcine hemagglutinating encephalomyelitis virus induced nonsuppurative encephalitis as the cause of death in piglets. PeerJ. 2016;4:e2443. doi: 10.7717/peerj.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J, Lee C. Emergence and evolution of novel G2b-like porcine epidemic diarrhea virus inter-subgroup G1b recombinants. Arch Virol. 2020;165:2471–2478. doi: 10.1007/s00705-020-04767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee YN, Lee C. Complete genome sequence of a novel porcine parainfluenza virus 5 isolate in Korea. Arch Virol. 2013;158:1765–1772. doi: 10.1007/s00705-013-1770-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.