Abstract
Introduction
Klebsiella pneumoniae, a multidrug resistant bacterium, that causes nosocomial infections including septicemia, pneumonia etc. Bacteriophages are potential antimicrobial agents for the treatment of antibiotic resistant bacteria.
Methods and Results
In this study, a novel bacteriophage IME268 was isolated from hospital sewage against clinical multi-drug resistant Klebsiella pneumoniae. Transmission electron microscopy and genomic characterization of this phage exhibited it belongs to the Webervirus genus, Drexlerviridae family. Phage IME268 possessed a double-stranded DNA genome composed of 49,552bp with a GC content of 50.5%. The phage genome encodes 77 open reading frames, out of 44 are hypothetical proteins while 33 had assigned putative functions. No tRNA, virulence related or antibiotic resistance genes were found in phage genome. Comparative genomic analysis showed that phage IME268 has 95% identity with 87% query cover with other phages in NCBI database. Multiplicity of infection, one step growth curve and host range of phage were also measured.
Conclusion
According to findings, Phage IME268 is a promising biological agent that infects Klebsiella pneumoniae and can be used in future phage therapies.
Keywords: Klebsiella pneumoniae, bacteriophage, therapy, lytic, drug resistant
Introduction
Klebsiella pneumoniae (K. pneumoniae) is a multidrug resistant bacterium that mostly present in gastrointestinal tract. It is an opportunistic and ubiquitous bacterium that causes nosocomial and community-acquired infections including septicemia, pneumonia and meningitis etc.1 It is one of the bacteria that causes high mortality rate in immuno-compromised patients.2 Its widely spread antibiotic resistance in all over the world pose a major challenge to public health. The insufficient availability of antimicrobials has gained attention to develop alternative therapeutic strategies against infections.3 Multi drug resistant K. Pneumoniae causes severe life threatening nosocomial infections and become resistant against existing antibiotics, recommended to find alternative ways to treat those infections.4,5
Bacteriophages are intracellular parasites of bacteria and have generated interest as an alternative biological agents.6,7 Phages, especially lytic ones, have been used for the treatment against bacterial infections in past. Their bactericidal mechanism have a different behavior towards antibiotics.8 In recent years, phage therapy has gained importance because of antibiotic resistance posed by multidrug resistant bacteria. Traditional antibiotic therapies used before to treat antibiotic resistant bacteria are not working anymore.9,10 In many countries, phage therapy with either modified or natural virulent phages has shown promising results. It reflects the scope of phage therapy in coming times and its effective role as bactericide.11 The availability of phages on wide scale in environment, convenience in isolation, and safety makes them an ideal choice for therapy than other options which lose their importance because of huge risk, long cycle and expensive as far as use is concerned.12 Further, phage therapy along with its uses, it also comes up with few immunological complications and side effects as a drug. Phage therapy has emerged as new approach to treat the MDR K. pneumoniae infections, as it’s treatment has been difficult because of antibiotic resistance to K. pneumoniae.13,14
Emergence of MDR K. Pneumoniae and variations in its genome suggests to isolate and characterize more different lytic phages. Current study is aimed to isolate and characterize a novel lytic phage IME268 from sewage that effectively infect MDR K. pneumoniae. We have analyzed its genome sequence in detail and elucidated its biological properties. This study will serve as the basis for the prevention and control of MDR K. Pneumoniae, provide more knowledge about K. Pneumoniae and facilitate the development of biocontrol technologies.
GenBank Accession Number
Complete phage sequence has been submitted to GenBank under this accession number MZ398242.
Materials and Methods
Bacterial Strains and Culture Conditions
K. Pneumoniae isolate 1733 was used as the host bacterium for bacteriophage IME268. All the tested bacterial strains of K. pneumoniae (74 strains) and Escherichia.coli (10 strains) used in this study for host spectrum assay were clinical isolates (listed in Table S1) and stored at −80° in glycerol (15% v/v). Antibiotic sensitivity test was performed for these clinical isolates and results are listed in Supplementary Table S1. All bacterial strains were cultured in Luria–Bertani (LB) broth at 37°C with shaking at 180 rpm.
Isolation, Propagation and Purification of Phage
Isolation of phage IME268 was done by using K. Pneumoniae isolate 1733 as a host from hospital sewage. Sewage samples were processed in lab followed by centrifugation and then passed from 0.22 μm pore-size membrane (Pall, USA) to wipe out all large fragments and bacteria. Double agar overlay method was used for the isolation and propagation of phage. The phage filtrate about 1 mL was mixed with 5 mL of LB broth and 5 mL culture of overnight grown K. pneumoniae isolate 1733 (1.5 × 108 CFU/mL), and then cultured for 72 h at 37 ◦C with agitation at 100 rpm, followed by centrifugation at 10,000 × g for 10 mins at room temperature. Then, supernatant was passed through a membrane (0.22 µm) to remove residual cells. Further, 100 µL of the phage suspension was added to 0.9 mL of K. Pneumoniae isolate 1733 strain (exponential phase), mixed with 0.7% LB top agar and then mixture was overlaid onto the plate containing solid LB agar. Finally, before incubation plates kept at room temperature to solidify and then incubated at 37 ◦C for overnight to obtain phage plaques. Presence of clear plaques indicated the existence of lytic phage. Sterile toothpicks were used to collect the single plaques and transferred into culture tubes containing 5 mL of SM buffer with 10uL of K. Pneumoniae 1733 strain as indicated bacteria. Phage purification experiment was performed for three times.
Bacteriophage IME268 Purification
Phage IME268 purification experiment was performed as previously illustrated with few modifications.15 Briefly, K. Pneumoniae isolate 1733 which was grown to the exponential phase (1.5×108 CFU/mL), infected by phage IME268 at 37 ◦C for overnight with agitation. The cellular material was removed by centrifugation at 10,000 × g for 20 min at 4 ◦C. The supernatant was filtered with a 0.22 μm filter membrane, obtaining a pure phage lysate. Further, we concentrated this lysate with ultracentrifugation and PBS buffer was used to resuspend. For further purification of phage, sample was placed at the top of a discontinuous CsCl gradient (1.32, 1.45, 1.50, and 1.70 g/mL) and centrifuged at 35,000× g for 3h at 4°C. The phage band was collected and dialyzed with a suspension medium buffer (0.01% gelatin, 100 mM/L NaCl, 50 mmol/L tris-HCl, and 10 mM/L MgSO4) at 4◦C. Finally, pure phage was collected and stored at 4°C. The bacteriophage IME268 showed lytic activity against K. Pneumoniae isolate 1733 and further characterized.
Transmission Electron Microscopy (TEM)
For transmission electron microscopy, purified phage lysate was applied to a carbon-coated copper grid and negatively stained with 2% phosphotungstic acid (pH 6.8) for 20s. The morphology of the phage IME268 was inspected using TEM (JEM-1200EX, Japan) at 80 kV.16
An Assay for Host Range
A total of 73 strains of K. pneumoniae and 10 strains of Escherichia. coli were tested to determined the host range of phage. These bacterial strains were isolated from clinical samples and stored at −80 (Table S1). In brief, A 200 µL of bacterial culture of each strain was poured in the plates containing solid agar (0.8%). Then, 5 µL of the purified phage suspension was spotted at the center of the plates containing double-layer agar medium and incubated for 24 hours at 37 ◦C. By observing the plate, a clearing spot showed the phage activity against tested bacteria. The small rounded plaques formation on plates showed bacterial cell lysis. The experiment was performed in triplicate.17
Optimal Multiplicity of Infection (MOI)
Optimal multiplicity of infection (MOI) for phage IME268 was determined by growing serial dilutions of K. Pneumoniae isolate 1733 to its early exponential phase (1×107 CFU/mL) at the optical density of 600 nm. Stock solution of bacteria (1 mL) were inoculated with phage at different MOIs (0.001, 0.01, 0.1, 1, 10, and 100). Then, the mixed solution was incubated for 1 day at 37°C. The phage titers were ascertained via Soft agar overlay method.18 The one which attained the highest phage titer was the optimal MOI. For testing the MOI, experiment was repeated for three times.
Adsorption and One-Step Growth Curve
For phage adsorption assay,19 suspension of bacteriophage IME268 (107 PFU/mL) was mixed with bacterial solution (108 PFU/mL) at a MOI of 0.001 before incubated at 37°C. While during incubation 200 uL aliquots were obtained at 0, 2, 4, 6, 8, 10, 15, 20, 25, 30 mins, centrifuged and then filtered with 0.22 um filter membrane. Number of free phages were determined with the help of soft agar overlay method. Depletion in phage titers exhibited the absorbance rate of phage particles to bacterial cells.
One-step growth curve of bacteriophage IME268 was determined as previously described by yu et al with several minor modifications.20 In brief, exponentially growing K. Pneumoniae 1733 cells (OD600 = 0.5) were mixed with phage IME268 suspension at MOI of 0.001, then mixture was allowed to adsorb for 10 min at 37◦C. Then the suspension was centrifuged and re-suspended in LB broth, followed by incubation at 37 ◦C with agitation at 200 rpm. Samples were taken after every 10 min up to 240 min and phage titers were calculated via soft agar overlay method. The experiment was repeated for three times. The latent period, burst time period and burst size were ascertained through one-step growth curve. The burst size was the ratio of the final phage titer to the number of initial infected bacterial cells.21
Stability Assays
To find out the effect of different temperatures on phage IME268 viability, purified phage samples were incubated at various temperatures (40, 50, 60, 70 and 80 ◦C) for an hour. For the evaluation of phage stability at different pH levels, purified samples of phage lysates were incubated at different levels of pH (range 3 to 14) for 1 hour in LB broth. In addition, for ultraviolet (UV) sensitivity of IME268, phage suspensions were exposed to UV light, and aliquots were taken after every 10 min until 60 min. Further, to find out chloroform sensitivity, an equal volume of pure phage suspension with different concentrations (0, 20%, 50%, and 100%) were mixed with chloroform (v/v) and intensely shaken the solution for 1 min. Then mixture was incubated at 37°C for 24 h followed by centrifugation. Soft agar overlay method was used to tittered the phage samples. All of these experiments were carried out in triplicate.
DNA Extraction of IME268
Phenol-chloroform method was used for the DNA extraction of phage IME268 as previously described.22 Briefly, phage suspension was treated with nucleases DNase (1 mg/mL) and RNase (1mg/mL) for overnight at 37°C. After that Zncl2 was served for phage precipitation and centrifuged. Then, 750 uL of TES buffer was added and subjected to dry bath at 65°C. Then, Purified phage lysate was served with proteinase K and potassium acetate to degrade and precipitate the proteins. Further, centrifugation was done and supernatant was treated with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) followed by centrifugation at 13,000 × g for 5 min at 4°C. After centrifugation, an equal volume of isopropanol was added to supernatant and kept it for 1 h at −20°C. Centrifuged again and final wash was accomplished by adding 70% ethanol and sat down for 1 minute at room temperature. Finally, pellets were air dried and DNA was dissolved in 30 uL of TE solution and stored at −20°C. DNA concentration was confirmed by nanodrop (ThermoFisher, China).
Sequencing and in silico Analysis
Genomic DNA was extracted through Zncl2 precipitation and quantity of DNA was quantified using nanodrop. Sequencing library was prepared through NEBNext Ultra II FS DNA Library Prep Kit (NEB) as manufacturer’s instructions. Sequencing was done by Illumina NovaSeq, with paired-end 2×300 bp reads. FastQC v0.11.5 software was used for analyzing the data quality and filtered pair end reads was assembled by using SPAdes v3.13.023 with default parameters. The complete genome of phage IME268 was annotated by online server RAST (https://rast.nmpdr.org/).24 The protein functions were cross checked by local alignment search tool (BLASTp) of NCBI server (E.value=0.0001) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).25 The circular map of phage genome was generated by using a script which is built in laboratory and further amended by Inkscape 0.92.3.0. tRNAscan-SE v.2.0 (http://lowelab.ucsc.edu/cgi-bin/tRNAscan-SE2.cgi)26 was employed to check the presence of genes encoding tRNAs. ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) and VirulenceFinder (https://cge.cbs.dtu.dk/services/VirulenceFinder/)27 both platforms were used to find out the existence of resistance genes and virulence factors, respectively. Further, for comparative genomic analysis, MEGA5 was used in order to construct phylogenetic trees with neighbor joining method based on terminase large subunit and capsid proteins.28
Statistical Analysis
In this study, we used GraphPad Prism 8.0.1 for analysis and data represented as means and standard error of the mean (± SEM).
Results and Discussion
Biological Characterization of Phage IME268
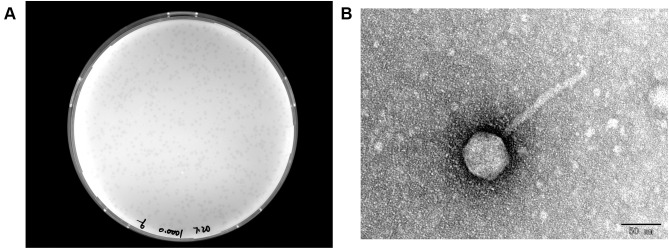
A novel lytic bacteriophage IME268 was isolated from hospital sewage using K. pneumoniae isolate 1733 as a host strain. After incubation of phage IME268 with its host K. pneumoniae isolate 1733 at 37°C for 24 hours, forms a clear and round plaques of about 0.2 mm in diameter (Figure 1A). According to morphology difference tailed bacteriophages classified into three classes including Myoviridae, siphoviridae and podoviridae. Transmission electron microscopy analysis exhibited that phage IME268 possessed icosahedral head (60.6 ± 5 nm, n=3) and long, non contractile tail (132.52 ± 5 nm, n=3) as shown in Figure 1B. Morphological and genomic comparisons revealed it belongs to Webervirus genus, Drexlerviridae family (Siphovirus).
Figure 1.
(A) Morphology of IME268 plaques. Phages were plated in LB agar and overlaid with a liquid culture of K. pneumoniae 1733. The plates were incubated at 37°C. Clear, well-defined IME268 plaques were observed and photographed. (B) The morphology of phage IME268. IME268 was negatively stained with 2% phosphotungstic acid (PTA) and examined by transmission electron microscopy (TEM) at an accelerating voltage of 80 kV. The scale bar represents 50 nm.
Host Range
Lytic spectrum of phage IME268 was determined by using double-layer agar method. A total number of 84 clinical strains (Table S1) were applied in which 74 strains belongs to K. Pneumoniae and 10 strains of E.coli. Results showed Phage IME268 could infect 19 clinical strains of K. Pneumoniae and could not infect any strain of E.coli, suggesting that phage IME268 has narrow host range.
MOI, Adsorption and Single Step Growth Curve
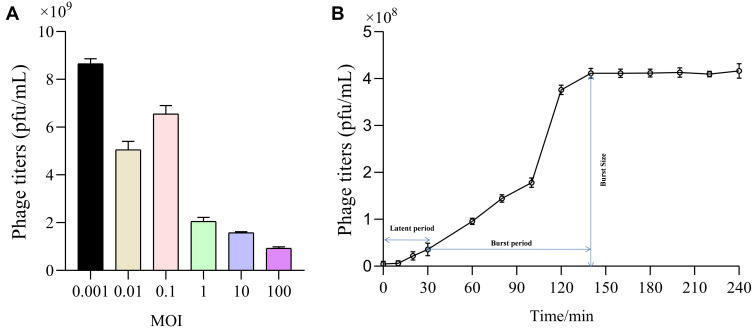
For MOI, we observed that production of phage progeny was at its peak at MOI of 0.001, representing optimal value of MOI for phage IME268 is 0.001 (Figure 2A). After 10 min of phage infection, adsorption of phage IME268 occurred to its host K. Pneumoniae isolate 1733. Almost 90% of phage adsorbed to its bacterial cells and then stabilized. Latent period and burst size of phage IME268 was calculated with the help of one step growth curve. Some factors like incubation temperature, media composition and bacterial growth rate can effect on latent period and phage burst size. As shown in Figure 2B latent period of phage IME268 was almost 30 min. Then, rise phase of phage starts which showed burst size of phage is almost 90 min before it enters into plateau phase.
Figure 2.
Biological characterization of bacteriophage IME268. The optimal MOI (A) and one-step growth curve of phage IME268 (B); Results are presented as mean values ± SD. Strains at the optimal MOI. The supernatants were harvested at 10 min intervals post-infection, and titers were determined using the double-layer method.
Stability Assays
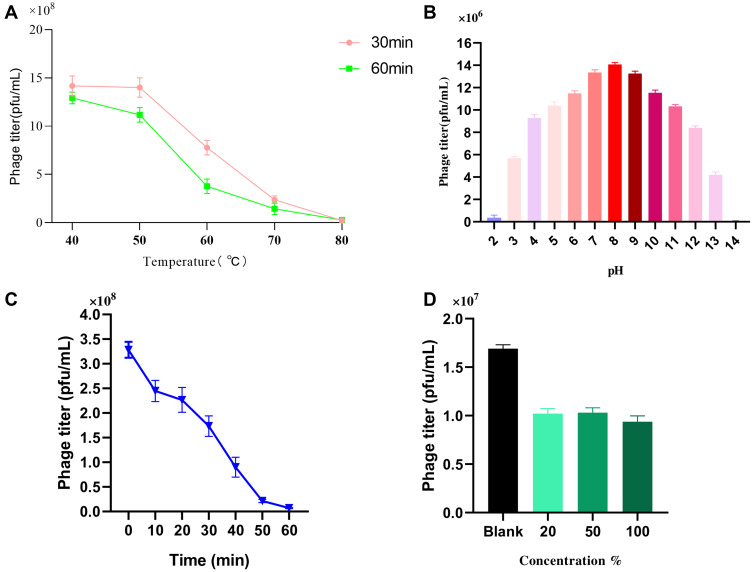
Isolated phages used for therapeutic purposes should be stable during various overstretch conditions. To understand more about phage IME268 characteristics, we have checked its stability at different temperatures and pH values. Thermal stability was checked by measuring the phage titer after incubation of 60 min and 30 min at different temperatures (Figure 3A). Phage IM268 retained its thermo stability at 40°C and 50°C when incubated for one hour and 30 min. No phage titer reduction was observed. However, it is drastically reduced when temperature increased more than 50°C and almost inactivated at 80°C. Phage stability was measured at different pH levels from 2 to 14 (Figure 3B). Results exhibited phage was stable at 4 to 10 value and phage titer was not reduced significantly. Phage titer was declined at 11 to 14 and no viable phages was observed when pH level above 11. Furthermore, results of UV test (Figure 3C) showed that phage is sensitive to UV light as phage growth decreased after exposure. Chloroform sensitivity test of phage IME268 showed phage sensitivity to chloroform (Figure 3D), no plaque formation was observed on plates after treating with chloroform. This is opposite to the results of untreated phage suspension which produced clear plaques with its bacterial host. Results suggests lipid content is not present in phage particles and phage is chloroform sensitive as one-third of tailed bacteriophages.29,30 Phage IME268 has good stability at different stressful conditions and it can be used in different phage cocktail therapies to treat the bacterial infections.
Figure 3.
Stability of IME268 under various conditions. (A) Temperature. (B) pH stability. (C) UV radiation stability. (D) Chloroform sensitivity. Survived phage particles were determined by double-layer tests. Error bars show the SEM among triplicate samples.
Genomic Characteristics of Phage IME268
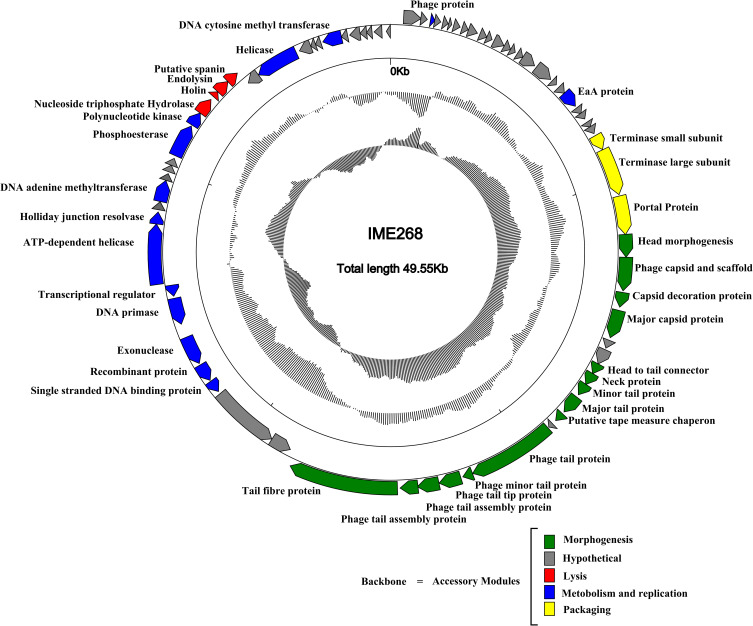
International Committee on the Taxonomy of Viruses (ICTV) has released the main species demarcation criterion for defining new species of bacterial and archaeal viruses is currently set at a genome sequence identity of 95%. This specifies that by using different analytical tools like BLASTn, two viruses of same specie should be vary from each other by 5% at nucleotide level.31,32 The genome of phage IME268 is contiguous, double stranded DNA of 49552bp with a GC content of 50.5%. It showed 95% homology with 87% query cover with other phages in databases. No tRNA was found in the genome of IME268 as it checked by tRNA scanner.33 Phage genome encodes a total of 77 putative open reading frames (ORFs). Mostly ORFs transcribed on positive strand (58/77) while 19 ORFs are transcribed on negative strand. The complete circular map of genome has shown in Table 1 and Figure 4. A total number of 33 gene products showed similarity with proteins of known function while others are hypothetical proteins. According to putative functions of genes encoding by phage, its genome has been divided into four functional modules: Morphogenesis, metabolism and replication, lysis and packaging.
Table 1.
Predicted ORFs in the genome of phage IME268
| ORFs | Strand | Start | Stop | Function | Best-match BLASTp Result | Query Cover | E-values | Identities | GenBank Acc.No |
| ORF1 | + | 444 | 1031 | hypothetical protein | Klebsiella phage KP1801 | 100 | 1e-136 | 98 | YP_009903534.1 |
| ORF2 | + | 1021 | 1242 | hypothetical protein | Klebsiella phage vB-KPNS | 100 | 7e-46 | 98 | QZE51017.1 |
| ORF3 | + | 1347 | 1499 | Phage protein | Klebsiella phage KpKT21phi1 | 100 | 1e-28 | 100 | YP_009818808.1 |
| ORF4 | + | 1489 | 1698 | hypothetical protein | Klebsiella phage TAH8 | 100 | 2e-43 | 97 | YP_009808608.1 |
| ORF5 | + | 1771 | 1950 | hypothetical protein | Klebsiella phage 1513 | 100 | 3e-33 | 100 | YP_009197845.1 |
| ORF6 | + | 1968 | 2075 | hypothetical protein | Klebsiella phage NJR15 | 100 | 6e-17 | 100 | YP_009808765.1 |
| ORF7 | + | 2145 | 2369 | hypothetical protein | Klebsiella phage vB_KpnS-VAC8 | 100 | 4e-47 | 97 | QZE50855.1 |
| ORF8 | + | 2439 | 2651 | hypothetical protein | Gordonia sp. YY1] | 100 | 6e-44 | 100 | WP_159406304.1 |
| ORF9 | + | 2662 | 2982 | hypothetical protein | Klebsiella phage vB_KpnS-VAC5 | 100 | 3e-62 | 90 | QZE50602.1 |
| ORF10 | + | 3069 | 3323 | hypothetical protein | Klebsiella phage ABTNL-2 | 100 | 2e-54 | 97 | QWT56454.1 |
| ORF11 | + | 3320 | 3517 | hypothetical protein | Klebsiella phage KPN N141 | 100 | 3e-37 | 96 | YP_009791610.1 |
| ORF12 | + | 3591 | 3962 | hypothetical protein | Klebsiella phage PhiKpNIH-10 | 100 | 4e-83 | 95 | QHB49652.1 |
| ORF13 | + | 3974 | 4219 | hypothetical protein | Klebsiella phage ABTNL-2 | 100 | 2e-54 | 97 | QWT56454.1 |
| ORF14 | + | 4216 | 4401 | hypothetical protein | Klebsiella phage vB_KpnS_IMGroot | 100 | 2e-50 | 92 | YP_009902547.1 |
| ORF15 | + | 4482 | 4700 | hypothetical protein | Klebsiella phage Sweeny | 100 | 2e-43 | 100 | YP_009902903.1 |
| ORF16 | + | 4700 | 5188 | hypothetical protein | Klebsiella phage PhiKpNIH-10 | 100 | 1e-114 | 98 | QHB49657.1 |
| ORF17 | + | 5264 | 5893 | hypothetical protein | Klebsiella phage GH-K3 | 100 | 5e-146 | 95 | YP_009820134.1 |
| ORF18 | + | 5963 | 6154 | hypothetical protein | Klebsiella phage 13 | 100 | 2e-29 | 77 | YP_009903238.1 |
| ORF19 | + | 6271 | 6543 | hypothetical protein | Klebsiella phage KP36 | 100 | 3e-58 | 97 | YP_009225984.1 |
| ORF20 | + | 6540 | 7115 | EaA protein | Klebsiella phage B1 | 100 | 3e-137 | 97 | QTP96015.1 |
| ORF21 | + | 7185 | 7421 | hypothetical protein | Klebsiella phage 1513 | 100 | 2e-50 | 100 | YP_009197831.1 |
| ORF22 | + | 7409 | 7642 | hypothetical protein | Klebsiella phage KLPN1 | 100 | 8e-49 | 100 | YP_009195358.1 |
| ORF23 | + | 7774 | 7965 | hypothetical protein | Klebsiella phage GH-K3 | 100 | 1e-83 | 100 | YP_009820128.1 |
| ORF24 | + | 7959 | 8222 | hypothetical protein | Klebsiella phage GH-K3 | 100 | 9e-58 | 100 | YP_009820127.1 |
| ORF25 | + | 8302 | 8826 | Phage terminase, small subunit | Klebsiella phage NJS2 | 100 | 3e-126 | 100 | YP_009808708.1 |
| ORF26 | + | 8836 | 10437 | terminase large subunit | Klebsiella phage GML-KpCol1 | 100 | 0.0 | 99 | YP_009796931.1 |
| ORF27 | + | 10483 | 11793 | Phage portal (connector) protein | Klebsiella phage Sweeny | 99 | 0.0 | 93 | YP_009902915.1 |
| ORF28 | + | 11783 | 12550 | head morphogenesis protein | Klebsiella phage vB_KpnS-VAC11 | 100 | 0.0 | 99 | QZE51042.1 |
| ORF29 | + | 12550 | 13680 | Phage capsid and scaffold | Klebsiella phage B1 | 100 | 0.0 | 99 | QTP96001.1 |
| ORF30 | + | 13732 | 14235 | Capsid decoration protein | Klebsiella phage ABTNL-2 | 100 | 1e-114 | 99 | QWT56398.1 |
| ORF31 | + | 14347 | 15306 | MCP | Klebsiella phage vB_KpnS_FZ10 | 100 | 0.0 | 98 | YP_009902976.1 |
| ORF32 | + | 15399 | 15686 | Hypothetical protein | Klebsiella phage Shelby | 100 | 6e-61 | 98 | YP_009903391.1 |
| ORF33 | + | 15740 | 16273 | Hypothetical protein | Klebsiella phage TSK1 | 100 | 8e-124 | 97 | YP_009817793.1 |
| ORF34 | + | 16270 | 16647 | Head to tail connector | Klebsiella phage ABTNL-2 | 100 | 5e-82 | 99 | QWT56402.1 |
| ORF35 | + | 16640 | 17077 | Neck protein | Klebsiella phage vB_KpnS-VAC5 | 100 | 1e-101 | 97 | QZE50631.1 |
| ORF36 | + | 17067 | 17498 | Minor tail protein | Klebsiella phage Sin4 | 100 | 3e-99 | 97 | YP_009903473.1 |
| ORF37 | + | 17592 | 18248 | major tail protein | Klebsiella phage TSK1 | 100 | 4e-161 | 99 | YP_009817797.1 |
| ORF38 | + | 18325 | 18651 | putative tape measure chaperone | Klebsiella phage ABTNL-2 | 100 | 2e-75 | 100 | QWT56406.1 |
| ORF39 | + | 18798 | 18965 | hypothetical protein | Klebsiella phage TAH8 | 100 | 2e-31 | 100 | YP_009808644.1 |
| ORF40 | + | 19002 | 21968 | Phage tail, tail length tape-measure protein H | Klebsiella phage MezzoGao | 100 | 0.0 | 98 | YP_009792109.1 |
| ORF41 | + | 21971 | 22315 | Phage minor tail protein | Klebsiella phage vB_KpnS-VAC11 | 100 | 1e-77 | 99 | QZE51056.1 |
| ORF42 | + | 22385 | 23137 | Phage tail tip, assembly protein L | Klebsiella phage Sin4 | 100 | 0.0 | 99 | YP_009903479.1 |
| ORF43 | + | 23139 | 23876 | Phage tail assembly protein | Klebsiella phage KpKT21phi1 | 100 | 0.0 | 97 | YP_009818767.1 |
| ORF44 | + | 23851 | 24453 | Phage tail assembly protein | Klebsiella phage KPN N141 | 100 | 6e-141 | 99 | YP_009791644.1 |
| ORF45 | + | 24541 | 28242 | tail fiber protein | Klebsiella phage KP36 | 100 | 0.0 | 96 | YP_009226010.1 |
| ORF46 | - | 29236 | 28481 | hypothetical protein | Klebsiella phage PKP126 | 100 | 1e-174 | 95 | YP_009284924.1 |
| ORF47 | - | 31706 | 29250 | Hypothetical protein | Klebsiella phage PKP126 | 100 | 0.0 | 97 | YP_009284923.1 |
| ORF48 | - | 32261 | 31800 | putative single-stranded DNA binding protein | Klebsiella phage Sweeny | 100 | 2e-109 | 98 | YP_009902938.1 |
| ORF49 | - | 32954 | 32298 | recombination protein | Klebsiella phage KPN N141 | 100 | 2e-160 | 100 | YP_009791650.1 |
| ORF50 | - | 34060 | 33014 | exonuclease | Klebsiella phage Sushi | 100 | 0.0 | 99 | YP_009196679.1 |
| ORF51 | - | 35515 | 34556 | DNA primase/helicase, phage-associated | Klebsiella phage JY917 | 100 | 0.0 | 98 | YP_009903182.1 |
| ORF52 | - | 35992 | 35591 | Transcriptional regulator | Klebsiella phage 1513 | 100 | 1e-92 | 100 | YP_009197873.1 |
| ORF53 | + | 36084 | 38120 | ATP-dependent helicase | Klebsiella phage NJS3 | 99 | 0.0 | 99 | AXQ68106.1 |
| ORF54 | + | 38117 | 38524 | holliday junction resolvase | Klebsiella phage Skenny | 100 | 4e-93 | 97 | YP_009903064.1 |
| ORF55 | + | 38598 | 38876 | Hypothetical protein | Klebsiella phage TSK1 | 100 | 6e-60 | 100 | YP_009817815.1 |
| ORF56 | + | 38879 | 39610 | DNA adenine methyltransferase | Klebsiella phage 1513 | 100 | 8e-180 | 98 | YP_009197869.1 |
| ORF57 | + | 39614 | 39850 | hypothetical protein | Klebsiella phage TSK1 | 100 | 7e-49 | 98 | YP_009817817.1 |
| ORF58 | + | 39915 | 40151 | hypothetical protein | Klebsiella phage vB_KpnS_IMGroot | 100 | 6e-51 | 100 | YP_009902507.1 |
| ORF59 | + | 40148 | 40399 | Hypotheticl protein | Klebsiella phage RAD2 | 100 | 2e-52 | 98 | QUU29426.1 |
| ORF60 | + | 40495 | 41625 | Phosphoesterase | Klebsiella phage KPN N141 | 100 | 0.0 | 100 | YP_009791583.1 |
| ORF61 | + | 41663 | 42151 | polynucleotide kinase | Klebsiella phage Sushi | 100 | 1e-114 | 100 | YP_009196690.1 |
| ORF62 | + | 42148 | 42729 | Nucleoside triphosphate hydrolase | Klebsiella phage vB_KpnS-VAC8 | 100 | 7e-140 | 98 | QZE50879.1 |
| ORF63 | + | 42856 | 43071 | holin | Klebsiella phage 1513 | 100 | 4e-41 | 100 | YP_009197862.1 |
| ORF64 | + | 43073 | 43555 | endolysin | Klebsiella phage Shelby | 100 | 2e-115 | 99 | YP_009903424.1 |
| ORF65 | + | 43552 | 43977 | putative spanin | Klebsiella phage Sushi | 100 | 2e-99 | 99 | YP_009196694.1 |
| ORF66 | - | 44508 | 44047 | Hypothetical protein | Klebsiella phage 13 | 100 | 7e-111 | 99 | YP_009903267.1 |
| ORF67 | - | 46080 | 44512 | helicase | Klebsiella phage MezzoGao | 100 | 0.0 | 99 | YP_009792137.1 |
| ORF68 | - | 46643 | 46206 | Hypothetical protein | Klebsiella phage 1513 | 100 | 2e-100 | 99 | YP_009197857.1 |
| ORF69 | - | 46826 | 46644 | hypothetical protein | Klebsiella phage Sushi | 100 | 9e-35 | 96 | YP_009196698.1 |
| ORF70 | - | 47026 | 46823 | hypothetical protein | Klebsiella phage Sushi | 100 | 1e-38 | 98 | YP_009196699.1 |
| ORF71 | - | 47791 | 47099 | DNA cytosine methyltransferase | Klebsiella phage Sin4 | 100 | 8e-168 | 97 | YP_009903509.1 |
| ORF72 | - | 48015 | 47788 | hypothetical protein | Klebsiella phage Sushi | 100 | 3e-47 | 100 | YP_009196701.1 |
| ORF73 | - | 48465 | 48088 | hypothetical protein | Klebsiella phage KPN N141 | 100 | 3e-87 | 99 | YP_009791596.1 |
| ORF74 | - | 48704 | 48465 | hypothetical protein | Klebsiella phage KP36 | 100 | 5e-51 | 100 | YP_009226038.1 |
| ORF75 | - | 48895 | 48701 | hypothetical protein | Klebsiella phage Sushi | 100 | 2e-36 | 100 | YP_009196704.1 |
| ORF76 | - | 49249 | 48968 | hypothetical protein | Klebsiella phage Sushi | 100 | 3e-61 | 98 | YP_009196705.1 |
| ORF77 | - | 49551 | 49405 | hypothetical protein | Klebsiella phage GML-KpCol1 | 89 | 6e-21 | 93 | YP_009796958.1 |
Figure 4.
Genome map of bacteriophage IME268 and its genetic characteristics. Patterns are divided into four circles. From the outside to the inside: The outermost represents the forward reading frame, followed by the outermost circle represents the reverse reading frame.
Lysis Module
ORF 62 in the genome of phage IME268 encoding nucleoside triphosphate hydrolase that gives rise to bacterial lysis by hydrolysing peptidoglycans.8 In lysis module, holin and endolysin proteins are encoded by putative ORF63 and ORF64, respectively. These two proteins are responsible for host lysis in bacteriophages. Holins are membranous proteins that control the duration of infective cycle to achieve the lysis at an optimal time.34–36 Endolysins rack up in cytosol and causing degradation of bacterial cell wall by folding themselves at the time of vegetative cycle.37 Another protein in phage lysis module encoded by an ORF65 showed 99% homology with putative spanins. Spanin proteins are necessary for the cleavage of bacterial outer membrane as they are phage lysis proteins.38
Metabolism and Replication Module
Metabolism and replication module is located in the downstream region (32,261–42,151 bp) of lysis module. A comparative analysis exhibited that following ORFs are assorted in this module including DNA binding protein ORF48, recombination protein ORF49, exonuclease ORF50, DNA primase ORF51, transcriptional regulator ORF52, helicase ORF53, holliday junction resolvase ORF54, DNA adenine methyltransferase ORF56, Phosphoesterase ORF60 and polynucleotide kinase ORF61.
ORF51 indicated significant amino acid homology (98%) with putative primase/helicase of Klebsiella phage JY917. As replication process of DNA is semi-discontinuous in nature, primases are pre-requisite for DNA replication of lagging strand. They can bind with helicase N-terminus for the formation of replicator, which is crucial for DNA replication and transcription.39
The first part of ORF51 worked as a DNA primase and possessed a conserved DnaG superfamily region while second part of ORF51 belongs to DnaB superfamily of phage and contains ATP binding sites, which act as DNA helicases. This suggests that phage IME268 possessed a bifunctional protein that play a role both as a helicase and primase to complete the unwinding and catalytic synthesis of DNA.40
Further, PSI-BLAST comparison proclaimed that ORF50 and ORF53 of phage genome IME268 encoded putative proteins exonuclease and helicase, respectively. Both these proteins are helping DNA synthesis, they play their role in regulation of DNA replication, repair and recombination.41
Additionally, a transcriptional regulator protein encoded by ORF52, which help in to identify phage-promoter regions and expressing the phage genes instead of host by regulating gene expression.42
Morphogenesis Module
Genes related to morphogenesis are mainly distributed in first half of phage IME268 genome. A comparative analysis revealed ORF29 showed 99% similarity to scaffolding protein of Klebsiella phage B1.
In a comparison with other phages, head structure and assembly of phage IME268 involved ORF28, ORF30 and ORF31. According to BLAST-P analysis ORF28 showed 99% identity with head morphogenesis protein of Klebsiella phage vB_KpnS-VAC11. The deduced products of ORF30 and ORF31exhibited similarity with the capsid protein of Klebsiella phage.
The tail of phage IME268 comprised on many proteins including ORF36, ORF37 and ORF(40–45). The protein products of ORF36 and ORF37 showed homology (97%, 99%) with phage minor and major tail proteins of Klebsiella phage sin4 and Klebsiella phageTSK1, respectively. Further, structural related protein products of ORFs (40–45) have been predicted to encode phage tail proteins including tail fiber protein with almost 98% similarity with Klebsiella phages. Phage tail fibers are protein assemblies that can recognize cell surface of its host and help in attachment and penetrating into the host cells.43
DNA Packaging Module
Gene cluster involved in DNA packaging of phage was observed in the genome of IME268 with following arrangement of proteins: terminase small protein, terminase large protein and portal protein. These three proteins located adjacently and involved in phage DNA packaging and splicing. Terminase protein is mainly involved phage packaging which further divided into small and large subunits.40 The deduced products of ORF25 and ORF26 predicted as terminase small subunit and terminase large subunit, respectively. Large subunit helping in DNA translocation while small one interact with large subunit for the initiation of packaging.44 They are conserved proteins in the genome of phage and help in classification process. Off these packaging proteins, ORF27 encodes a portal protein which showed 93% homology in BLAST-P comparison to YP_009902915.1. Terminase complexes comprised a genome packaging motor along with portal protein.
Comparative Genomic Analysis
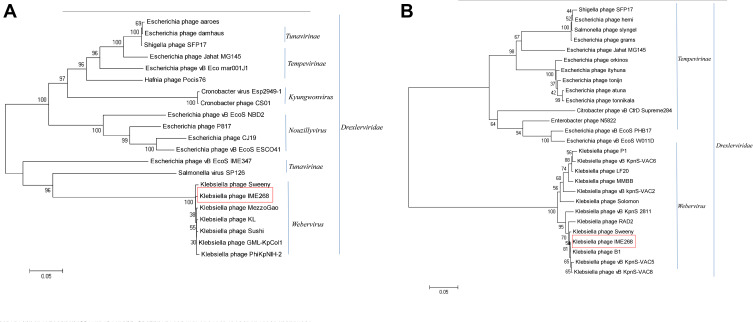
Phylogenetic trees were constructed using terminase large subunit and capsid proteins to infer the evolutionary relationships of IME268 with other bacteriophages as both these proteins are conserved (Figure 5A and B). Results showed, in both trees IME278 clustered with K. pneumoniae phages. Trees represent IME268 is a novel phage that forms a independent branch of Drexlerviridae family, Webervirus genus.
Figure 5.
Phylogenetic analysis of phages belonging to different genera of the family Drexlerviridae based on their terminase large subunit (A) and capsid proteins (B). Phylogenetic trees were constructed using the neighbor-joining method with 1000 bootstrap replicates (red box).
Conclusion
In conclusion, we reported a newly isolated novel phage IME268 that infects K. pneumoniae from hospital sewage samples in China. The already characterized K. pneumoniae phages mostly isolated from hospital sewage, suggesting that hospital sewage is a good source for the isolation of phages against MDR bacterial pathogens. The repeatedly use of a single phage in an antimicrobial treatment may cause phage resistance in hospital settings. We can expand the phage spectrum with the help of phage cocktails to refrain from a single phage resistance. In this perspective, we need to isolate more and more novel lytic phages with unique characteristics, so that phage therapies may highly successful in clinical applications. Genomic and proteomic characteristics presented in detail and evolutionary analysis showed IME268 is a new member of the genus Webervirus, Drexlerviridae family. The current study not only provided new phage resources for phage therapy development against K. pneumoniae but also provided an avenue for the discovery of many unraveled phages.
Funding Statement
This research was supported by Key Project of Beijing University of Chemical Technology (No. XK1803-06), National Key Research and Development Program of China (No. 2018YFA0903000, 2020YFC2005405, 2020YFA0712100, 2020YFC0840805), Funds for First-class Discipline Construction (No. XK1805), Inner Mongolia Key Research and Development Program (No. 2019ZD006), National Natural Science Foundation of China (No. 81672001, 81621005), NSFC-MFST project (China-Mongolia) (No. 31961143024), Fundamental Research Funds for Central Universities (No. BUCTRC201917, BUCTZY2022), and Military Biosecurity Research Program (Grant No. 19SWAQ06).
Ethics Statement
The use of human specimens and all related experimental protocols were approved by the committee on Human Research of Center for Clinical Laboratory, The Affiliated Taian City Central Hospital of Qingdao University, Taian 271000, China Taian City Central Hospital and carried out in accordance with the approved guidelines.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Nazir A, Zhao Y, Li M, et al. Structural genomics of repA, repB1-carrying IncFIB family pA1705-qnrS, P911021-tetA, and P1642-tetA, multidrug-resistant plasmids from Klebsiella pneumoniae. Infect Drug Resist. 2020;13:1889. doi: 10.2147/IDR.S228704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li M, Xiao Y, Li P, et al. Characterization and genome analysis of Klebsiella phage P509, with lytic activity against clinical carbapenem-resistant Klebsiella pneumoniae of the KL64 capsular type. Arch Virol. 2020;165(12):2799–2806. doi: 10.1007/s00705-020-04822-0 [DOI] [PubMed] [Google Scholar]
- 3.Li M, Guo M, Chen L, et al. Isolation and characterization of novel lytic bacteriophages infecting epidemic carbapenem-resistant Klebsiella pneumoniae strains. Front Microbiol. 2020;11:1554. doi: 10.3389/fmicb.2020.01554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):1–12. doi: 10.1186/s12941-017-0191-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu WM, Yuan Z, Zhou HY. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection relative to two types of control patients: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2020;9(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nazir A, Dong Z, Liu J, et al. Genomic analysis of bacteriophage Xoo-sp13 infecting Xanthomonas oryzae pv. oryzae. Arch Virol. 2021;166(4):1263–1265. doi: 10.1007/s00705-021-04985-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nazir A, Ali A, Qing H, Tong Y. Emerging aspects of jumbo bacteriophages. Infect Drug Resist. 2021;14:5041. doi: 10.2147/IDR.S330560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xi H, Dai J, Tong Y, et al. The characteristics and genome analysis of vB_AviM_AVP, the first phage infecting Aerococcus viridans. Viruses. 2019;11(2):104. doi: 10.3390/v11020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y, Tong S, Li P, et al. Characterization and genome sequence of a genetically unique Escherichia Bacteriophage vB_EcoM_IME392. 2021. [DOI] [PubMed]
- 10.Fu P, Zhao Q, Shi L, et al. Identification and characterization of two bacteriophages with lytic activity against multidrug-resistant Escherichia coli. Virus Res. 2021;291:198196. doi: 10.1016/j.virusres.2020.198196 [DOI] [PubMed] [Google Scholar]
- 11.Tian F, Li J, Nazir A, Tong Y. Bacteriophage–a promising alternative measure for bacterial biofilm control. Infect Drug Resist. 2021;14:205. doi: 10.2147/IDR.S290093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Y, Cheng M, Zhai S, et al. Preventive effect of the phage VB-SavM-JYL01 on rabbit necrotizing pneumonia caused by Staphylococcus aureus. Vet Microbiol. 2019;229:72–80. doi: 10.1016/j.vetmic.2018.12.021 [DOI] [PubMed] [Google Scholar]
- 13.Soleimani Sasani M, Eftekhar F. Potential of a bacteriophage isolated from wastewater in treatment of lobar pneumonia infection induced by Klebsiella pneumoniae in mice. Curr Microbiol. 2020;77:2650–2655. doi: 10.1007/s00284-020-02041-z [DOI] [PubMed] [Google Scholar]
- 14.Henry M, Lavigne R, Debarbieux L. Predicting in vivo efficacy of therapeutic bacteriophages used to treat pulmonary infections. Antimicrob Agents Chemother. 2013;57(12):5961–5968. doi: 10.1128/AAC.01596-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q, Xing S, Sun Q, et al. Characterization and complete genome sequence analysis of a novel virulent Siphoviridae phage against Staphylococcus aureus isolated from bovine mastitis in Xinjiang, China. Virus Genes. 2017;53(3):464–476. doi: 10.1007/s11262-017-1445-z [DOI] [PubMed] [Google Scholar]
- 16.Nazir A, Dong Z, Liu J, et al. Isolation, characterization, and genome sequence analysis of a novel lytic phage, Xoo-sp15 Infecting Xanthomonas oryzae pv. oryzae. Curr Microbiol. 2021;78:3192–3200. [DOI] [PubMed] [Google Scholar]
- 17.Dong Z, Xing S, Liu J, et al. Isolation and characterization of a novel phage Xoo-sp2 that infects Xanthomonas oryzae pv. oryzae. J Gen Virol. 2018;99(10):1453–1462. doi: 10.1099/jgv.0.001133 [DOI] [PubMed] [Google Scholar]
- 18.Li L, Zhang Z. Isolation and characterization of a virulent bacteriophage SPW specific for Staphylococcus aureus isolated from bovine mastitis of lactating dairy cattle. Mol Biol Rep. 2014;41(9):5829–5838. doi: 10.1007/s11033-014-3457-2 [DOI] [PubMed] [Google Scholar]
- 19.Chang H-C, Chen C-R, Lin J-W, et al. Isolation and characterization of novel giant Stenotrophomonas maltophilia phage φSMA5. Appl Environ Microbiol. 2005;71(3):1387–1393. doi: 10.1128/AEM.71.3.1387-1393.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu X, Xu Y, Gu Y, Zhu Y, Liu X. Characterization and genomic study of “phiKMV-Like” phage PAXYB1 infecting Pseudomonas aeruginosa. Sci Rep. 2017;7(1):1–13. doi: 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, Cong Y, Mi Z, et al. Characterization and complete genome sequence analysis of phage GP4, a novel lytic Bcep22-like podovirus. Arch Virol. 2019;164(9):2339–2343. doi: 10.1007/s00705-019-04309-7 [DOI] [PubMed] [Google Scholar]
- 22.Han P, Hu Y, An X, Song L, Fan H, Tong Y. Biochemical and genomic characterization of a novel bacteriophage BUCT555 lysing Stenotrophomonas maltophilia. Virus Res. 2021;301:198465. doi: 10.1016/j.virusres.2021.198465 [DOI] [PubMed] [Google Scholar]
- 23.Nazir A, Dong Z, Liu J, et al. Sequence analysis of a jumbo bacteriophage, Xoo-sp14, that infects Xanthomonas oryzae pv. oryzae. Microbiol Resour Announc. 2020;9(48):e01072–01020. doi: 10.1128/MRA.01072-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. doi: 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boratyn GM, Camacho C, Cooper PS, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41(W1):W29–W33. doi: 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33(suppl_2):W686–W689. doi: 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinheinz KA, Joensen KG, Larsen MV. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage. 2014;4(2):e27943. doi: 10.4161/bact.27943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kęsik-Szeloch A, Drulis-Kawa Z, Weber-Dąbrowska B, et al. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol J. 2013;10(1):1–12. doi: 10.1186/1743-422X-10-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Msimbira LA, Jaiswal SK, Dakora FD. Identification and characterization of phages parasitic on bradyrhizobia nodulating groundnut (Arachis hypogaea L.) in South Africa. Appl Soil Ecol. 2016;108:334–340. doi: 10.1016/j.apsoil.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adriaenssens E, Brister JR. How to name and classify your phage: an informal guide. Viruses. 2017;9(4):70. doi: 10.3390/v9040070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng H, He W, Li C, et al. Complete genome analysis of a novel phage GW1 lysing Cronobacter. Arch Virol. 2019;164(2):625–628. doi: 10.1007/s00705-018-4084-3 [DOI] [PubMed] [Google Scholar]
- 33.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang Y, Lee J-H, Shin H, Heu S, Ryu S. Characterization and complete genome sequence analysis of Staphylococcus aureus bacteriophage SA12. Virus Genes. 2013;47(2):389–393. doi: 10.1007/s11262-013-0938-7 [DOI] [PubMed] [Google Scholar]
- 35.Bai Q, Zhang W, Yang Y, et al. Characterization and genome sequencing of a novel bacteriophage infecting Streptococcus agalactiae with high similarity to a phage from Streptococcus pyogenes. Arch Virol. 2013;158(8):1733–1741. doi: 10.1007/s00705-013-1667-x [DOI] [PubMed] [Google Scholar]
- 36.Dalmasso M, De Haas E, Neve H, et al. Isolation of a novel phage with activity against Streptococcus mutans biofilms. PLoS One. 2015;10(9):e0138651. doi: 10.1371/journal.pone.0138651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y, Yan Y, Ji W, et al. Characterization and determination of holin protein of Streptococcus suis bacteriophage SMP in heterologous host. Virol J. 2012;9(1):1–11. doi: 10.1186/1743-422X-9-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kongari R, Rajaure M, Cahill J, et al. Phage spanins: diversity, topological dynamics and gene convergence. BMC Bioinform. 2018;19(1):1–26. doi: 10.1186/s12859-018-2342-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guilliam TA, Keen BA, Brissett NC, Doherty AJ. Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes. Nucleic Acids Res. 2015;43(14):6651–6664. doi: 10.1093/nar/gkv625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu H, Yan P, Xiong W, Wang J, Liu X. Genomic characterization of a novel virulent phage infecting Shigella flexneri and isolated from sewage. Virus Res. 2020;283:197983. doi: 10.1016/j.virusres.2020.197983 [DOI] [PubMed] [Google Scholar]
- 41.Guinta D, Lindberg G, Rothman-Denes L. Bacteriophage N4-coded 5’—-3ʹexonuclease. Purification and characterization. J Biol Chem. 1986;261(23):10736–10743. doi: 10.1016/S0021-9258(18)67447-2 [DOI] [PubMed] [Google Scholar]
- 42.O’Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald G, Ross R. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+ C content. J Bacteriol. 2004;186(9):2862–2871. doi: 10.1128/JB.186.9.2862-2871.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.North OI, Sakai K, Yamashita E, et al. Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres. Nature Microbiol. 2019;4(10):1645–1653. doi: 10.1038/s41564-019-0477-7 [DOI] [PubMed] [Google Scholar]
- 44.Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J Biol Chem. 1997;272(6):3495–3501. doi: 10.1074/jbc.272.6.3495 [DOI] [PubMed] [Google Scholar]