Abstract
Synechococcus cyanobacteria are ubiquitous and abundant in the marine environment and contribute to an estimated 16% of the ocean net primary productivity. Their light-harvesting complexes, called phycobilisomes (PBS), are composed of a conserved allophycocyanin core, from which radiates six to eight rods with variable phycobiliprotein and chromophore content. This variability allows Synechococcus cells to optimally exploit the wide variety of spectral niches existing in marine ecosystems. Seven distinct pigment types or subtypes have been identified so far in this taxon based on the phycobiliprotein composition and/or the proportion of the different chromophores in PBS rods. Most genes involved in their biosynthesis and regulation are located in a dedicated genomic region called the PBS rod region. Here, we examine the variability of gene content and organization of this genomic region in a large set of sequenced isolates and natural populations of Synechococcus representative of all known pigment types. All regions start with a tRNA-PheGAA and some possess mobile elements for DNA integration and site-specific recombination, suggesting that their genomic variability relies in part on a “tycheposon”-like mechanism. Comparison of the phylogenies obtained for PBS and core genes revealed that the evolutionary history of PBS rod genes differs from the core genome and is characterized by the co-existence of different alleles and frequent allelic exchange. We propose a scenario for the evolution of the different pigment types and highlight the importance of incomplete lineage sorting in maintaining a wide diversity of pigment types in different Synechococcus lineages despite multiple speciation events.
Keywords: cyanobacteria, genomic island, lateral gene transfer, phycobiliprotein, phycobilisome, tycheposon
Significance.
The cyanobacterium Synechococcus, the second most abundant photosynthetic organism of the ocean, has colonized all spectral niches available in this environment, thanks to its sophisticated and diversified light-harvesting complexes. These complexes are encoded in a specialized region of the genome, the gene order and content of which are directly related to the pigment type of the corresponding cells. Here, by looking at a large set of Synechococcus genomes from strains and field populations, we highlight the extent of the genomic variability of these regions within each pigment type and unveil evolutionary mechanisms that led to the growing complexity of light-harvesting antennae. Strikingly, many regions include recombination and transposition genes that could have played a key role in Synechococcus pigment diversification.
Introduction
As the second most abundant phytoplanktonic organism of the ocean, the picocyanobacterium Synechococcus plays a crucial role in the carbon cycle, accounting for about 16% of the ocean net primary productivity (Flombaum et al. 2013; Guidi et al. 2016). Members of this group are found from the equator to subpolar waters and from particle-rich river mouths to optically clear open ocean waters, environments displaying a wide range of nutrient concentrations, temperatures, light regimes, and spectral niches (Olson et al. 1990; Wood et al. 1998; Farrant et al. 2016; Paulsen et al. 2016; Sohm et al. 2016; Grébert et al. 2018; Holtrop et al. 2021). This ecological success is tied to the remarkably large genetic (Ahlgren and Rocap 2012; Huang et al. 2012; Mazard et al. 2012; Farrant et al. 2016) and pigment diversity exhibited by these cells (Six et al. 2007; Humily et al. 2013; Grébert et al. 2018; Xia et al. 2018). The pigment diversity arises from wide variations in the composition of their light-harvesting antennae, called phycobilisomes (PBS).
The building blocks of PBS are phycobiliproteins, which consist of two subunits (α and β). Phycobiliproteins are assembled into trimers of heterodimers (αβ)3 and then hexamers [(αβ)3]2 that are stacked into rod-like structures with the help of linker proteins (Yu and Glazer 1982; Adir 2005). Before PBS can be assembled into their typical fan-like structure, the α and β subunits must be modified by lyases that covalently attach chromophores called phycobilins, at one, two, or three conserved cysteine positions (Scheer and Zhao 2008; Schluchter et al. 2010; Bretaudeau et al. 2013). The PBS core is always made of allophycocyanin (APC), from which radiate six to eight rods (Wilbanks and Glazer 1993; Sidler 1994). Three major pigment types have been defined thus far based on the phycobiliprotein composition of PBS rods (Six et al. 2007; Humily et al. 2013). The simplest rods are found in pigment type 1 (PT 1) and contain only phycocyanin (PC), which binds the red-light absorbing phycocyanobilin (PCB, Amax = 620–650 nm; Six et al. 2007). The rods of pigment type 2 (PT 2) contain PC and phycoerythrin-I (PE-I), which binds the green-light (GL) absorbing phycoerythrobilin (PEB; Amax = 545–560 nm). For pigment type 3 (PT 3), the rods contain the three types of phycobiliproteins, PC, PE-I, and phycoerythrin-II (PE-II), and bind PCB, PEB, and the blue-light (BL) absorbing phycourobilin (PUB, Amax = 495 nm; Ong et al. 1984; Six et al. 2007). Although these three phycobilins are isomers, PCB and PEB are created via oxidation/reduction reactions while PUB is generated by the isomerization of PEB during its covalent binding to a phycobiliprotein. This process is performed by dual-function enzymes called phycobilin lyase-isomerases (Grébert et al. 2021).
Five pigment subtypes have been further defined within PT 3, depending on their PUB:PEB ratio. This ratio is often approximated for living cells by the ratio of the PUB and PEB fluorescence excitation peaks at 495 and 545 nm (Exc495:545), with the emission measured at 585 nm. Subtype 3a strains have a fixed low Exc495:545 ratio (<0.6) and are often called “GL specialists,” 3b strains have a fixed intermediate ratio (0.6 ≤ Exc495:545 < 1.6), while 3c strains display a fixed high Exc495:545 ratio (≥1.6) and are often called “BL specialists” (Six et al. 2007; Sanfilippo et al. 2016). Strains belonging to subtype 3d dynamically tune their PUB:PEB ratio to the ambient GL:BL ratio, a reversible physiological process known as type IV chromatic acclimation (CA4; Palenik 2001; Everroad et al. 2006; Shukla et al. 2012; Humily et al. 2013; Sanfilippo et al. 2016; Sanfilippo, Garczarek, et al. 2019; Sanfilippo, Nguyen, et al. 2019). As a result, the Exc495:545 of these strains varies from 0.6 in GL to 1.6 in BL. Finally, the rare subtype 3e shows only faint changes in Exc495:545 when shifted between GL and BL (Humily et al. 2013).
Comparative genomics analysis of the first 11 marine Synechococcus sequenced genomes (Six et al. 2007) revealed that most genes encoding proteins involved in the biosynthesis and regulation of PBS rods are grouped into a single genomic location called the “PBS rod region.” These authors suggested that the gene content and organization of this region was specific to the different pigment types or subtypes, but they were unable to examine the degree of genomic and genetic variability for strains within each pigment type. Further sequencing of additional PT 3d strain genomes revealed that CA4 capability is correlated with the presence of a small genomic island that exists in one of two configurations, CA4-A and CA4-B, defining the two pigment genotypes 3dA and 3dB (Humily et al. 2013). A novel organization of the PBS rod region was also discovered, first from metagenomes from the Baltic Sea (Larsson et al. 2014) and then from strains isolated from the Black Sea (Sánchez-Baracaldo et al. 2019). Gene content analysis identified these as a new PT 2 genotype named PT 2B, while the original PT 2 was renamed PT 2A. Finally, the genome sequencing of the high-PUB-containing strains KORDI-100 and CC9616 showed that although they display a high, PT 3c-like Exc495:545 ratio, the gene complement, order, and alleles of their PBS rod region differ from PT 3c, establishing an additional pigment subtype called PT 3f (Mahmoud et al. 2017; Grébert et al. 2018; Xia et al. 2018). An interesting feature of the genes within the PBS rod region is that their evolutionary history apparently differs from that of the core genome (Six et al. 2007; Everroad and Wood 2012; Humily et al. 2014; Grébert et al. 2018; Carrigee et al. 2020), but the reason(s) for this remains unclear.
Here, we perform a comprehensive analysis of the phylogenetic and genomic diversity of Synechococcus pigment types by leveraging a large number of recently available genomes of marine Synechococcus and Cyanobium isolates for further comparative genomic analysis. We also use a targeted metagenomics approach to directly retrieve and analyze PBS rod regions from natural populations as well as single-cell amplified genomes (SAGs). This broadened exploration leads us to propose hypotheses for the evolution of the PBS rod regions as well as for the maintenance of the wide pigment diversity found in most lineages, despite multiple speciation events. This study highlights the importance of population-scale mechanisms such as lateral transfers and incomplete lineage sorting (ILS) in shaping the distribution of pigment types among Synechococcus lineages.
Materials and Methods
Genome Information
Genomic regions used in this study were obtained from 69 public complete or draft genomes (Dufresne et al. 2008; Cubillos-Ruiz et al. 2017; Lee et al. 2019; Doré et al. 2020). Information about these genomes can be found in supplementary table S1, Supplementary Material online.
Fosmid Libraries
Samples for the construction of the fosmid library were collected during oceanographic cruises CEFAS (North Sea), BOUM (Mediterranean Sea), and the RRS Discovery cruise 368 (northeastern Atlantic Ocean) as well as from three long-term observatory sites. Two belong to the “Service d'Observation en Milieu Littoral” (SOMLIT), Astan located 2.8 miles off Roscoff and “Point B” at the entrance of the Villefranche-sur-mer Bay, while the “Boussole” station is located 32 miles off Nice in the Ligurian current (Antoine et al. 2008). Details on the sampling conditions, dates, and locations are provided in supplementary table S4, Supplementary Material online. Pyrosequencing of the petB gene, cell sorting, DNA extraction, whole genome amplification, fosmid library construction, screening, and sequencing were performed as previously described (Humily et al. 2014), and the fosmid libraries previously obtained from the Astan station were re-assembled using a different approach, as described below.
Sequencing reads were processed using BioPython v.1.65 (Cock et al. 2009) to trim bases with a quality score below 20, after which reads shorter than 240 nt or with a mean quality score below 27 were discarded. Reads corresponding to the fosmid vector, the E. coli host, or contaminants were removed using a BioPython implementation of NCBI VecScreen (https://www.ncbi.nlm.nih.gov/tools/vecscreen/about/). Paired reads were merged using FLASH v1.2.11 (Magoč and Salzberg 2011), and merged and non-merged remaining reads were assembled using the CLC AssemblyCell software (CLCBio, Prismet, Denmark). The resulting contigs were scaffolded using SSPACE v3.0 (Boetzer et al. 2011), and scaffolds shorter than 500 bp or with a sequencing coverage below 100× were removed. To reduce the number of contigs while preserving the genetic diversity, the second round of scaffolding was done using Geneious v6.1.8 (Biomatters, Auckland, New Zealand). Assembly statistics are reported in supplementary table S5, Supplementary Material online. Assembled scaffolds were manually examined to control for obvious whole genome amplification (WGA)-induced as well as assembly chimeras. Annotation of PBS genes was performed manually using Geneious and the Cyanorak v2.0 information system (http://www.sb-roscoff.fr/cyanorak/). Plotting of regions was conducted using BioPython (Cock et al. 2009).
Phylogenetic Analyses
Sequences were aligned using MAFFT v7.299b with the G-INS-i algorithm (default parameters; Katoh and Standley 2013). ML phylogenies were reconstructed using PhyML v20120412 using both SPR and NNI moves (Guindon and Gascuel 2003). Phylogenetic trees were plotted using Python and the ETE Toolkit (Huerta-Cepas et al. 2016).
Inference of Evolutionary Events
Species (vertical) phylogeny was inferred from a set of 73 conserved marker genes (supplementary table S6, Supplementary Material online; Wu et al. 2013). For each marker gene, protein sequences were extracted from 69 isolate genomes and 33 Synechococcus SAGs (supplementary table S1, Supplementary Material online; Berube et al. 2018), aligned using MAFFT, and the alignment trimmed using trimAl (Capella-Gutiérrez et al. 2009). Alignments were concatenated into a multiple alignment, which was used for the reconstruction of the species tree. Phylogenetic reconstruction was done using RAxML 8.2.9, with 100 searches starting from randomized maximum parsimony trees and 100 searches from fully random trees (Stamatakis 2014). The best tree was selected, and 200 bootstraps computed. Next, gene trees were inferred for every gene present in more than half of the considered genomes. For each gene, protein sequences were extracted from genomes and aligned using MAFFT. The resulting alignment was used for phylogenetic reconstruction with RAxML, and 100 bootstraps computed. Evolutionary events (gene duplication, transfer, loss, or speciation) were inferred for each gene from these bootstraps using ALE v0.4, which uses a maximum-likelihood framework to reconcile gene trees with the species tree (Szöllősi et al. 2013).
Results
The 5′ Ends of Many PBS Rod Regions Display Hallmarks of “Tycheposons” Mobile Genetic Elements
We analyzed the PBS rod region from 69 Synechococcus and Cyanobium strains (supplementary table S1, Supplementary Material online), which includes all PTs except 2B, which were extensively described elsewhere (Larsson et al. 2014; Callieri et al. 2019; Sánchez-Baracaldo et al. 2019). This data set contains every sequenced PT 3 strain and covers a very wide range of phylogenetic diversity, with representatives of all three deep branches within Cyanobacteria Cluster 5 sensuHerdman et al. (2001), called subclusters (SCs) 5.1–5.3 (Dufresne et al. 2008; Doré et al. 2020).
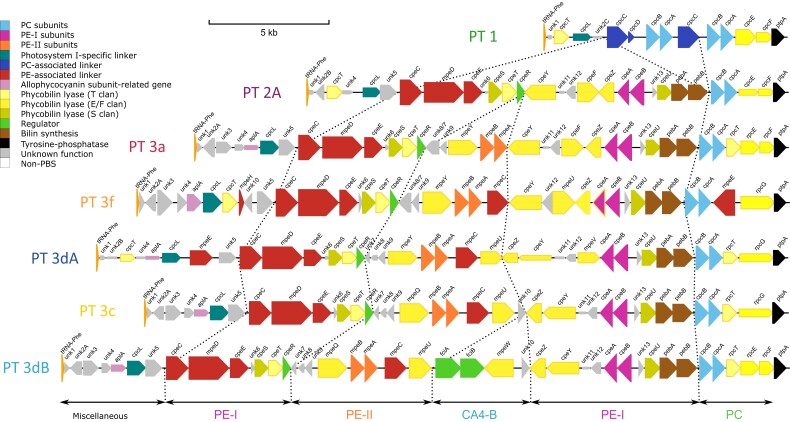
The PBS rod region is always situated between a phenylalanine-tRNA (tRNA-PheGAA) at the 5′-end and the ptpA gene, which encodes a putative tyrosine phosphatase, at the 3′-end (fig. 1 and supplementary figs. S1–S7, Supplementary Material online). Globally, the gene content and synteny of the PBS rod region per se (i.e., from unk1 to ptpA) is remarkably conserved among cultured representatives of a given PT (supplementary figs. S1–S7, Supplementary Material online). PT 1 strains have the simplest rods and the shortest PBS rod region (the smallest is about 8 kb in Cyanobium gracile PCC 6307, supplementary fig. S1, Supplementary Material online), notably containing one to four copies of the cpcBA operon encoding α- and β-PC subunits, two to four rod linker genes (one cpcD and up to three copies of cpcC; supplementary table S2, Supplementary Material online), and three phycobilin lyase genes (supplementary table S3, Supplementary Material online). All other PTs possess a single cpcBA copy and no PC rod linker genes. In PT 2A, these PC genes are replaced by a set of 16–18 genes necessary for the synthesis and regulation of PE-I hexamers (fig. 1 and supplementary fig. S2, Supplementary Material online; supplementary tables S2 and S3, Supplementary Material online), as previously described for strain WH7805 (Six et al. 2007). The main difference between the PBS rod regions of PT 2A and 3a is the presence in the latter of a small cluster of five genes between cpeR and cpeY (fig. 1 and supplementary figs. S2–S7, Supplementary Material online). Differences between PT 3a and other PT 3 subtypes 3c, 3dA, 3dB, and 3f (Mahmoud et al. 2017; Xia, Guo, et al. 2017) are mainly located in the subregion between the PE-II (mpeBA) and PE-I (cpeBA) operons (fig. 1). All PT 3 subtypes other than 3a possess mpeC, encoding a PE-II associated PUB-binding linker (Six et al. 2005), inserted downstream of mpeBA, as well as mpeU encoding a partially characterized phycobilin lyase-isomerase (Mahmoud et al. 2017). Moreover, the conserved hypothetical gene unk10, which is absent from all PT 3a’s, is present in the middle of the PBS rod region of all 3c and 3dB PTs, while in PT 3dA strains it is always located in the CA4-A island, thus outside the PBS rod region. Finally, the lyase gene mpeY is replaced by the lyase-isomerase gene mpeQ in PTs 3c and 3dB (Grébert et al. 2021).
Fig. 1.
PBS rod region for strains of different pigment types. Regions are oriented from the phenylalanine tRNA (left) to the conserved low molecular weight tyrosine phosphatase ptpA (right). Genes are colored according to their inferred function (as indicated in the inset). Their length is proportional to the gene size and their thickness to the protein identity between strains of the same pigment type. The strains represented here are WH5701 (PT 1), WH7805 (PT 2A), RS9907 (PT 3a), KORDI-100 (PT 3f), RS9916 (PT 3dA), WH8102 (PT 3c), and A15-62 (PT 3dB).
A number of differences between PBS rod regions of various strains are more difficult to link with a specific PT. This includes the putative PE-II linker gene mpeE present in all PT 3 strains except SYN20 (PT 3a) but at a highly variable position (supplementary figs. S2–S7, Supplementary Material online and supplementary table S2, Supplementary Material online). Similarly, the distribution of the putative PE-II linker genes mpeG and mpeH (the latter is a truncated version of the former) cannot be linked to either a PT or a clade (supplementary table S2, Supplementary Material online). It is also worth noting that while all PT 3a strains contain the rpcEF operon, encoding the two subunits of a C84 α-PC PEB lyase (Swanson et al. 1992; Zhou et al. 1992), other PT 3 subtypes may have either the rpcEF operon or rpcG, a fusion gene that encodes a C84 α-PC PEB lyase-isomerase and was thought to confer Synechococcus cells a fitness advantage in BL environments (Blot et al. 2009). This interchangeability is also found between closely related strains of the same PT and clade (supplementary figs. S5–S7, Supplementary Material online), and strain MINOS11 (SC 5.3/PT 3dB) even possesses both genes (supplementary fig. S7, Supplementary Material online). Some additional variations of “typical” PBS rod regions are also worth noting. Out of five PT 2A strains, only CB0205 and A15-44 possess the APC-like gene aplA (Montgomery et al. 2004) which, as in all PT 3 strains, is located between unk4 and cpcL (supplementary fig. S2, Supplementary Material online and supplementary table S2, Supplementary Material online). CB0205 also has a unique insertion of nine genes of unknown function, including unk3, in the middle of its PBS rod region between unk12 and cpeF, as observed in some natural PT 2A populations from the Baltic Sea (Larsson et al. 2014).
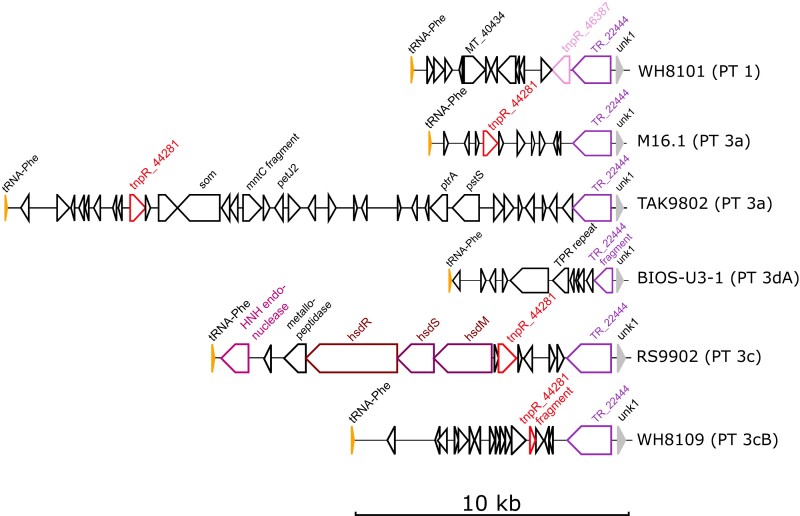
A most striking and previously unreported difference between PBS regions in a number of genomes is the presence of a DNA insertion of variable size between the tRNA-PheGAA and unk1 (fig. 2 and supplementary figs. S1, S3, S5–S7, Supplementary Material online). In TAK9802, the 22.8 kb DNA insertion is almost as large as the 24.5 kb PBS rod region. These insertions have striking similarities to “tycheposons,” a novel type of mobile genetic element that have been found to be responsible for the translocation of 2–10 kbp fragments of heterologous DNA in Prochlorococcus (Hackl et al. 2020). Indeed, the hallmarks of tycheposons include the systematic presence of a tRNA at the 5′-end of the insertion, the localization of this insertion upstream a genomic island important for niche adaptation—in the present case, adaptation to light color—, and the presence of a variety of mobile elements in the DNA insertion. These notably include putative tyrosine recombinases, putative transposon resolvases (TnpR family), and even a complete restriction-modification system (encoded by the hsdMSR operon; fig. 2 and supplementary figs. S1, S3, S6, and S7, Supplementary Material online).
Fig. 2.
Examples of tycheposons at 5′-end of PBS regions. Regions are oriented from the phenylalanine tRNA (tRNA-Phe) to unk1, the first coding gene of the PBS region per se. Genes putatively involved in DNA rearrangements (recombination, transposition, etc.) are colored and their orthologs in different regions are shown with the same color. Coding sequences with no gene name are hypothetical, conserved hypothetical, or pseudogenes. Gene length is proportional to the gene size. Abbreviations: TR, tyrosine recombinase; TPR, tetratricopeptide. The number after a gene name corresponds to its CLOG number in the Cyanorak database (Garczarek et al. 2021).
Although the genomic organization of the PBS rod region per se, that is, excluding the tycheposon, is broadly conserved, we found a high level of allelic diversity of the genes of this region and the proteins they encode (fig. 1 and supplementary fig. S8, Supplementary Material online). The most conserved are genes encoding the α- and β-subunits of phycobiliproteins. The sequence of each of these proteins is at most only about 10% different from its closest ortholog. The sequences of the linker proteins show greater variation between strains, with some having less than 70% identity to their closest ortholog. This variability is even greater for phycobilin lyases and uncharacterized conserved proteins, with some showing less than 60% sequence identity to their closest orthologs (supplementary fig. S8, Supplementary Material online). Such highly divergent sequences may in some cases reflect functional differences, as was recently demonstrated for MpeY and MpeQ (Grébert et al. 2021) and for CpeF and MpeV (Carrigee et al. 2020).
Targeted Metagenomics and SAGs Reveal New PBS Rod Region Variants and Natural Deficiency Mutants from Field Populations
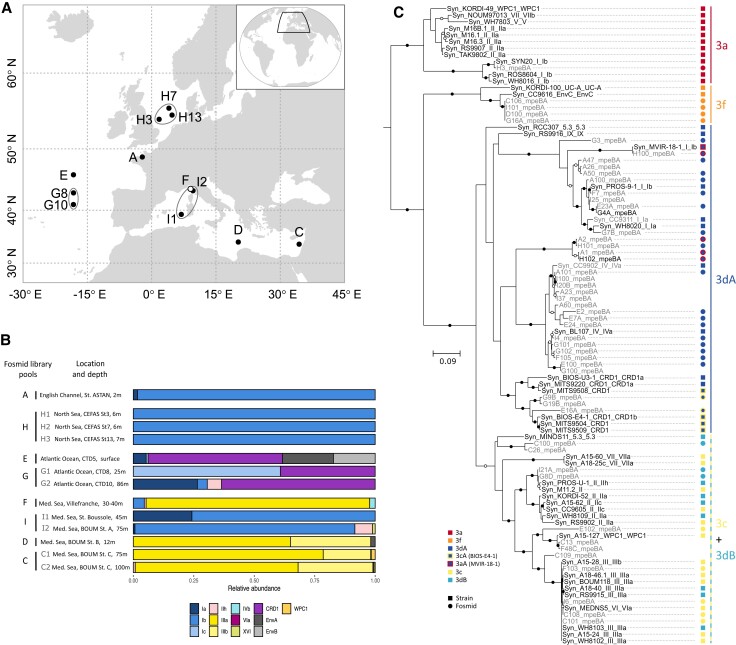
We investigated the genetic variability of PBS rod regions from natural Synechococcus populations using a targeted metagenomic approach that combined flow cytometry, cell sorting, WGA, and fosmid library screening (Humily et al. 2014). This method enabled us to retrieve PBS rod regions from natural populations in the North Sea, northeastern Atlantic Ocean, and various locations within the Mediterranean Sea (fig. 3A and supplementary table S4, Supplementary Material online). In addition, the high-resolution phylogenetic marker petB (Mazard et al. 2012) was sequenced to examine the phylogenetic diversity of these natural Synechococcus populations. Samples collected from the North Sea (fosmid libraries H1-3 in fig. 3B) and English Channel [library A, previously reported by Humily et al. (2014) but re-analyzed here] were exclusively composed of the cold-adapted clade I, mostly subclade Ib. Samples from the northeastern Atlantic Ocean were co-dominated by CRD1 and either clade I (libraries G1 and G2) or the environmental clades EnvA and EnvB (library E; EnvB is sometimes called CRD2; Ahlgren et al. 2019). Synechococcus populations from the western Mediterranean Sea were largely dominated by clade III (exclusively of subclade IIIa) at the coastal “Point B” station located at the entrance of the Bay of Villefranche-sur-Mer (https://www.somlit.fr/villefranche/; library F), while they essentially consisted of clade I (mostly subclade Ib) at station A of the BOUM cruise (library I2; Moutin et al. 2012) and at the long-term monitoring station BOUSSOLE located in the Gulf of Lions (library I1; Antoine et al. 2008). Eastern Mediterranean Sea populations collected at BOUM stations B and C were mainly from clades III, with subclade IIIa dominating. These large differences in clade composition reflect the distinct trophic regimes of the sampled sites and the diversity patterns observed here are globally consistent with previous descriptions of the biogeography of Synechococcus clades (Zwirglmaier et al. 2008; Mella-Flores et al. 2011; Paulsen et al. 2016). In particular, CRD1 and EnvB are known to co-occur in iron-poor areas (Farrant et al. 2016; Sohm et al. 2016) and the northeastern Atlantic Ocean has been reported to be iron-limited (Moore et al. 2013).
Fig. 3.
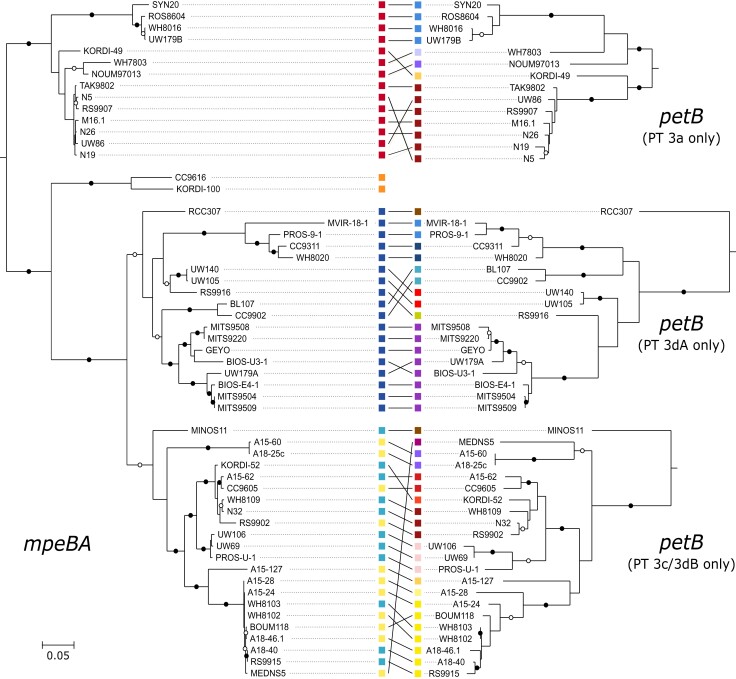
Location and genetic characterization of the samples used to retrieve the PBS rod regions from field populations of Synechococcus shown in figure 4. (A) Location of sampling sites used for fosmid library construction. (B) Synechococcus genetic diversity at each station, as assessed with the phylogenetic marker petB. (C) mpeBA phylogeny for isolates (black) and fosmids (gray). Squares and circles on the right-hand side correspond to reference strains and fosmids, respectively. Within PT 3dA, symbols with a blue center and a red contour correspond to PT 3aA (the reference 3aA strain, MVIR-18-1, exhibits a constitutive low Exc495:545) and those with a blue center and a yellow contour to PT 3cA (the reference 3cA strain, BIOS-E4-1, exhibits a constitutive high Exc495:545; see the text as well as Humily et al. 2013 and Grébert et al. 2018). Bootstrap values higher or equal to 90% are indicated by black circles, those comprised between 70% and <90% by empty circles, and no circles indicate values lower than 70%.
To obtain the largest possible diversity of PBS rod regions from Synechococcus field populations, fosmid libraries generated from similar geographic areas and/or cruises and showing comparable relative clade abundance profiles were pooled (as indicated in figs. 3A and B and supplementary table S4, Supplementary Material online) before screening and sequencing. Assembly of the eight fosmid library pools resulted in the assembly of 230 contigs encompassing either a portion or all of the PBS rod region. These contigs were an average size of approximately 5.5 kb and each library produced at least one contig longer than 10 kb (supplementary table S5, Supplementary Material online). Each contig was assigned to a PT based on its genomic organization and similarity to PBS rod regions of characterized strains (fig. 1 and supplementary figs. S1–S7, Supplementary Material online). Most contigs corresponded to PT 3dA (145 out of 230), but all PT 3 subtypes represented in our reference data set were found. There were 5 PT 3a contigs, 18 PT 3f contigs, and 62 PT 3c/3dB contigs, of which 9 could be unambiguously attributed to PT 3c and 5 to PT 3dB, based on the absence or presence of a CA4-B genomic island between mpeU and unk10, respectively (fig. 1).
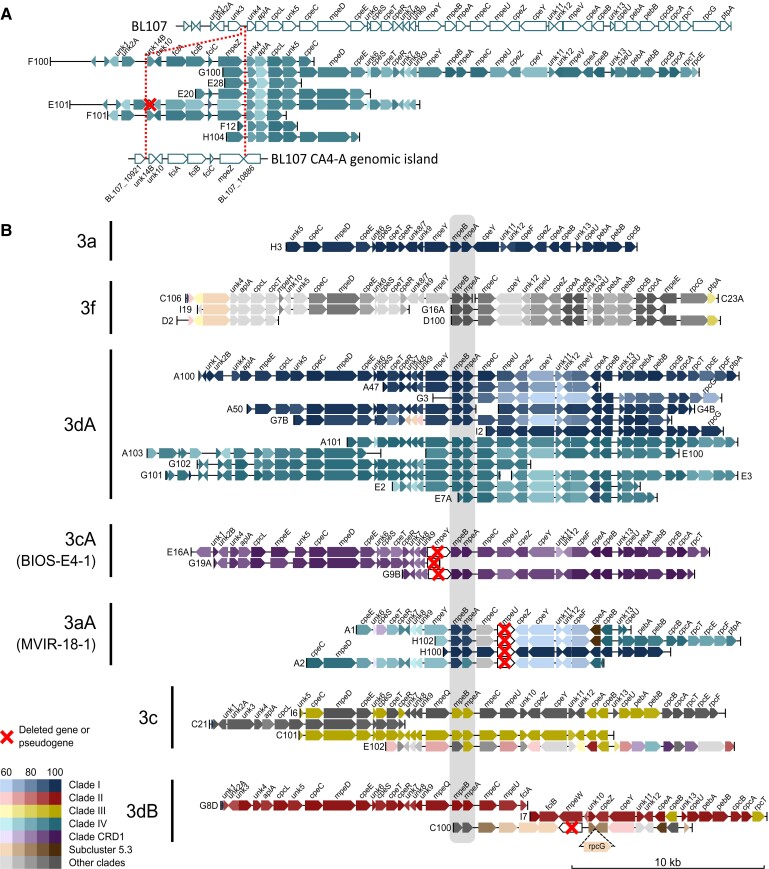
A representative selection of the most complete contigs is provided in figure 4. Most contigs are syntenic with PBS rod regions from characterized strains, notably those assigned to PT 3a retrieved from the North Sea (contig H3), or those assigned to PT 3f either retrieved from the northeastern Atlantic Ocean (contig G16A) or from the Mediterranean Sea (all other contigs assigned to PT 3f; fig. 4B). Yet, a number of contigs assigned to PT 3dA (E101, F100, and F101; fig. 4A) exhibit a novel gene organization compared with reference 3dA strains characterized by the insertion of a complete CA4-A genomic island between unk3 and unk4 at the 5′-end of the PBS rod region (supplementary fig. S5, Supplementary Material online). Five other contigs (G100, E28, E20, F12, and H104) apparently have the same organization because they possess a complete or partial mpeZ gene located immediately upstream of unk4. Despite this novel location, the genes of this CA4-A island are phylogenetically close to those of the PT 3dA/clade IV strains CC9902 or BL107 (fig. 4A). This new arrangement is found in contigs from different sequencing libraries and geographically diverse samples, so it is most likely not artefactual. Contigs corresponding to the canonical 3dA PBS rod region (fig. 1) were also found in most libraries and contain alleles most similar to those of PT 3dA strains from either clade I or IV (fig. 4B).
Fig. 4.
Partial or complete PBS rod region retrieved from natural Synechococcus populations. (A) Description of a new genomic organization related to 3dA pigment type with the CA4-A genomic island inserted at the 5′-end of the PBS rod genomic region. The PBS rod and CA4-A genomic regions of strain BL107 (3dA/clade IV) are shown as a reference. (B) Contigs other than those in (A) and longer than 10 kb sorted according to their organization and inferred corresponding pigment type. Colors represent the clade of the strain giving the best BlastX hit within the given pigment type. The highly conserved mpeBA operon is shaded in gray.
Several contigs from the English Channel and North Sea (A1, A2, H100, and H102) that displayed a similar gene organization to PT 3a strains lacked mpeU (fig. 4B), which encodes a lyase-isomerase that attaches PUB to an as-yet undetermined residue of PEII (Mahmoud et al. 2017). The absence of mpeU was also observed in the genome of strain MVIR-18-1 (clade I/PT 3a; supplementary fig. S5A, Supplementary Material online), which was shown to display a constitutively low Exc495:545 ratio (Humily et al. 2013). MVIR-18-1 was isolated from the North Sea, suggesting that natural populations representative of this mpeU-lacking variant may be common in this area. Similarly, the gene organization of three contigs originating from the northeastern Atlantic Ocean and assigned to CRD1 (E16A, G19A, and G9B; fig. 4B) matched an unusual PBS rod region found in five out of eight reference CRD1/PT 3dA strains (supplementary fig. S5A, Supplementary Material online). In these strains, the mpeY sequence is either incomplete (in MITS9508) or highly degenerate (in BIOS-E4-1, MITS9504, MITS9509, and UW179A) and fciA and fciB, which encode CA4 regulators (Sanfilippo et al. 2016), are missing (supplementary fig. S5B, Supplementary Material online), resulting in a PT 3c phenotype (Humily et al. 2013). This particular genomic organization was recently suggested to predominate in CRD1 populations from warm high nutrient-low chlorophyll areas, in particular in the South Pacific Ocean (Grébert et al. 2018). Thus, these contigs provide additional, more compelling evidence of the occurrence of these natural variants in field populations of CRD1.
A number of contigs corresponding to the phylogenetically indistinguishable PBS rod regions of the 3c and 3dB PTs were assembled from samples from the Mediterranean Sea and the northeastern Atlantic Ocean (fig. 3B). Among these, five (C100, C40, G8D, I7, and I21) could be assigned to PT 3dB due to the presence of a CA4-B genomic island between mpeU and unk10 (fig. 1). The gene organization of contig C100, whose alleles are most similar to the PT 3dB/SC 5.3 strain MINOS11, closely resembles the unique PBS rod region of this strain, which has an additional rpcG gene between unk10 and cpeZ (supplementary fig. S7, Supplementary Material online). Interestingly, the CA4-B genomic island of C100 lacks mpeW (fig. 3B) and thus may represent a novel natural variant. In contrast with the genes of contigs assigned to PT 3dA, which are closely similar to clades I, IV, or CRD1, those assigned to PT 3c and 3dB are most closely related to representatives of different clades, including clades II, III, WPC1, and 5.3. These distinct clade/PT combinations corroborate previous observations made in culture and in the field (Grébert et al. 2018) and are consistent with the population composition observed with petB at the sampling locations of these libraries (fig. 3B). Of note, some contigs from fosmid library E (e.g., contig E102; PT 3c in fig. 4B) possessed alleles that were highly divergent from all reference strains and likely belong to the uncultured clades EnvA or EnvB, which together represented more than 30% of Synechococcus population in sample E (fig. 3B).
We augmented these field observations by examining the PBS rod regions contained within the publicly available SAGs of marine Synechococcus (Berube et al. 2018). Out of 50 Synechococcus SAGs, 11 corresponded to PT 3c, 3 to PT 3dB, 2 to either PT 3c or 3dB, and 17 to PT 3dA (supplementary fig. S9, Supplementary Material online and supplementary table S1, Supplementary Material online). Using a core genome phylogeny based on a set of 73 highly conserved markers (supplementary table S6, Supplementary Material online and supplementary fig. S10, Supplementary Material online), we determined that these SAGs belong to clades I, II, III, IV, and CRD1 as well as some rare clades, including 1 to EnvA, 1 to XV, 3 to SC5.3, and 7 to EnvB. All of the EnvB SAGs contained a PBS rod region, which was very similar to PT 3c except for the insertion of rpcG between unk10 and cpeZ. Due to the absence of any sequenced EnvB isolate in our reference database, genes from these regions appear most similar to genes from a variety of strains and clades. The same is true for SAGs from clades EnvA (AG-676-E04) and XV (AG-670-F04). All SAGs assigned to PT3dB belong to SC5.3 and possess a PBS rod region similar to that of MINOS11 except for a ca. 15 kb insertion between the tRNA-PheGAA and unk2 in AG-450-M17 (supplementary fig. S9B, Supplementary Material online). The ten SAGs within clade CRD1 all belong to PT 3dA, but only one of these contains an intact mpeY gene (supplementary fig. S9C, Supplementary Material online). The three clade I SAGs also belong to PT 3dA. Of these, AG-679-C18 provides the first example of a mpeY deficiency outside of clade CRD1, further highlighting the prevalence of BIOS-E4-1-like populations, which are phenotypically similar to (but genetically distinct from) PT 3c, in the environment (Grébert et al. 2018). Interestingly, all four clade IV SAGs contain the novel PT 3dA arrangement found in several fosmids, where the CA4-A genomic island is located in the PBS rod region between unk3 and unk4. Finally, a number of the SAGs have additional genes between the tRNA-PheGAA gene and unk1, including recombinases and restriction enzymes, sometimes in multiple copies, for example, in the clade II SAGs AG-670-A04 and AG-670-B23 (supplementary fig. S9A, Supplementary Material online).
Altogether, the PBS rod regions retrieved from both fosmids and SAGs were very diverse and some contained alleles whose sequences were highly diverged from those of sequenced isolates. As was found in our comparative analysis of strains, the sequence diversity for lyases and linker proteins was much higher than for phycobiliproteins (supplementary fig. S8, Supplementary Material online).
Genes within the PBS Rod Region Have a Very Different Evolutionary History than the Rest of the Genome
Several phylogenetic analyses based on phycobiliprotein coding genes have shown that strains tend to group together according to their PT rather than their vertical (core or species) phylogeny. The cpcBA operon enables discrimination between PT 1, 2A, 2B, and 3, while the cpeBA operon allows separation of PT 2A, 3a, 3dA, 3f, and the 3c+3dB group and the mpeBA operon is best for distinguishing between the PT 3 subtypes (Everroad and Wood 2012; Humily et al. 2014; Xia, Partensky, et al. 2017; Grébert et al. 2018; Xia et al. 2018). By creating a mpeBA phylogenetic tree using all available genomes from Synechococcus PT 3 strains, we confirmed that alleles within a given PT 3 subtype are more closely related to one other than they are to other PTs from the same clade (fig. 5, left tree). However, we also observed that within each mpeBA clade, the tree topology actually resembles the topology based on the vertically transmitted core gene petB (fig. 5, right tree). The few exceptions to this finding could correspond to inter-clade horizontal gene transfers. The most striking example of this is the clade VI strain MEDNS5, which seemingly possesses a clade III PT 3c/3dB-like mpeBA allele (fig. 5).
Fig. 5.
Correspondence between phylogenies for the mpeBA operon and the marker gene petB, which reproduces the core genome phylogeny. The pigment type for each strain is indicated by a colored square in the mpeBA phylogeny, and its clade is similarly indicated in the petB phylogeny. Bootstrap values higher or equal to 90% are indicated by black circles, those comprised between 70% and <90% by empty circles, and no circles indicate values lower than 70%.
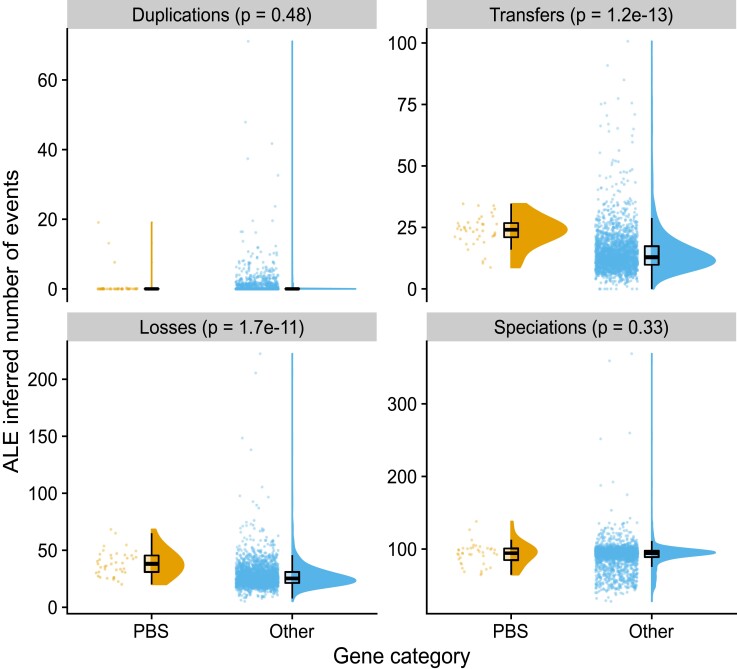
In order to explore the evolution of the PBS genes in greater detail, we used ALE (Szöllősi et al. 2013) to reconcile phylogenetic trees for each gene present in the core genome with the species tree inferred from a set of 73 core genes (supplementary table S6, Supplementary Material online and supplementary fig. S10, Supplementary Material online). This comparison allows the inference of evolutionary events such as duplications, horizontal transfers, losses, and speciations that can best explain the observed gene trees in light of the evolution of the species (fig. 6). Genes from the PBS rod region experienced significantly more transfers (on average 23.5 vs. 14.8 events per gene; Wilcoxon rank sum test P = 1.2 × 10−13) and losses (38.5 vs. 27.5; P = 1.7 × 10−11) than other genes in the genome, with no significant difference in gene duplications and speciations (fig. 6 and table 1). Consistent with the observation that genes within the PBS rod region are single copy except for cpcABC in PT 1, the increase in transfers was very similar to the increase in losses. We conclude that such transfer-and-loss events inferred by ALE actually correspond to allelic exchange, whereby homologous recombination mediates the replacement of one allele by another.
Fig. 6.
Evolutionary events affecting genes present in more than half of the analyzed genomes inferred by reconciling gene trees with the species tree. Genes were classified either as belonging to the PBS rod region (“PBS genes”) or as other genes (“Other”). P values for the Wilcoxon rank sum exact test are shown.
Table 1.
Frequency of Transfer Events Inferred by ALE for Genes of the PBS Rod Region
| Gene Category | Evolutionary Event | Min. | Median | Mean | Max. |
|---|---|---|---|---|---|
| PBS | Duplications | 0.00 | 0.00 | 1.019 | 19.09 |
| PBS | Transfers | 8.72 | 24.05 | 23.52 | 34.57 |
| PBS | Losses | 20.10 | 37.92 | 38.50 | 68.31 |
| PBS | Speciations | 64.70 | 94.36 | 93.08 | 138.01 |
| Other | Duplications | 0.00 | 0.00 | 0.56 | 71.11 |
| Other | Transfers | 0.00 | 12.91 | 14.80 | 100.68 |
| Other | Losses | 5.22 | 25.38 | 27.53 | 222.40 |
| Other | Speciations | 28.01 | 94.19 | 91.32 | 369.02 |
Finer-grained analysis of transfer events showed that they are slightly more frequent within clades for PBS genes than for other genes (9.7 vs. 8.4, P = 1.1 × 10−3; table 2 and supplementary fig. S11, Supplementary Material online), and more than twice as frequent between clades (13.9 vs. 6.1, P = 10−15; table 2 and supplementary fig. S11, Supplementary Material online). As a result, most transfer events identified for PBS genes occurred between clades, whereas other genes were primarily transferred within the same clade (supplementary fig. S11, Supplementary Material online). We then analyzed transfer events inferred for genes within the PBS rod region by their direction (supplementary table S6, Supplementary Material online). The most frequent recipient of transfer (frequency of 29.07) was strain N32, which can be explained by the fact that this PT 3dB strain is within a lineage of clade II that is otherwise solely made up of PT 3a strains (node 177; supplementary fig. S10, Supplementary Material online). The second most frequent transfer recipient was strain MEDNS5 (clade VI; frequency = 23.71), which possesses the allele of a PT 3c/3dB strain of clade III, as exemplified with mpeBA (fig. 5). Strains RS9902 and A15-44, both within clade II, were the third (21.69) and fourth (18.79) most frequent recipient strains. A15-44 belongs to PT 2, a quite rare PT among strictly marine SC 5.1 Synechococcus strains (Grébert et al. 2018), and indeed groups in the tree with the PT 3c strain RS9902 (supplementary fig. S10, Supplementary Material online). The apparent high transfer frequency to RS9902 could thus represent transfers of PT 2 to the ancestor of RS9902 and A15-44 (node 103 in supplementary fig. S10, Supplementary Material online and supplementary table S7, Supplementary Material online), followed by transfer of PT 3c to RS9902. Other strains for which a high frequency of transfers was inferred are KORDI-49 (WPC1/3aA), RCC307 (SC 5.3/3eA), WH7803 (V/3a), and CB0205 (SC 5.2/2), all of which represent rare combinations of clade and PT (supplementary fig. S10, Supplementary Material online and supplementary table S7, Supplementary Material online). Internal nodes with a high frequency of transfers were mostly deep-branching, representing the ancestor of SC 5.3, clades I, V, VI, VII and CRD1, 5.1B, and 5.1A (nodes 171, 189, 191, and 197, with frequencies of 22.2, 14.5, 13.9, and 12.9, respectively). Taken together, these results indicate that genes of the PBS rod region have a very ancient evolutionary history marked by frequent recombination events both within and between Synechococcus clades.
Table 2.
Frequency of Transfer Events Inferred by ALE for Genes of the PBS Rod Region
| Gene Category | Transfer Event | Min. | Median | Mean | Max. |
|---|---|---|---|---|---|
| PBS | Intra-clade | 5.26 | 9.66 | 9.66 | 13.41 |
| PBS | Extra-clade | 2.08 | 14.57 | 13.86 | 21.29 |
| PBS | Intra-PT | 3.91 | 11.69 | 11.41 | 19.77 |
| PBS | Extra-PT | 4.51 | 12.74 | 12.11 | 17.42 |
| Other | Intra-clade | 0.00 | 8.31 | 8.42 | 40.07 |
| Other | Extra-clade | 0.00 | 4.33 | 6.14 | 81.73 |
| Other | Intra-PT | 0.00 | 5.97 | 6.29 | 40.57 |
| Other | Extra-PT | 0.00 | 6.94 | 8.27 | 68.06 |
Discussion
The Variable Gene Content of the PBS Rod Region Might Partly Rely on a Tycheposon-Like Mechanism
Most of the current knowledge about the genomic organization and genetic diversity of the Synechococcus PBS rod region has relied on analysis of the first 11 sequenced genomes (Six et al. 2007) and later analyses of metagenomic assemblages or strains retrieved from a few specific locations. These include the SOMLIT-Astan station in the English Channel (Humily et al. 2014), the Baltic and Black Seas, where a new organization of the PBS rod region (PT 2B) was found associated with SC 5.2 populations (Larsson et al. 2014; Callieri et al. 2019), and freshwater reservoirs dominated by PT 2A/SC 5.3 populations (Cabello-Yeves et al. 2017). Here, we analyzed a wide set of Synechococcus and Cyanobium genomes (69 genomes and 33 SAGs; supplementary table S1, Supplementary Material online) as well as PBS rod regions directly retrieved from a variety of trophic and light environments (229 contigs; supplementary table S4, Supplementary Material online). Together, these cover all of the genetic and PT diversity currently known for this group (except PT 2B), enabling us to much better assess the extent of the diversity within each PBS rod region type.
Although our data confirmed that the gene content and organization of this region are highly conserved within a given PT, independent of its phylogenetic position (Six et al. 2007), they also highlighted some significant variability within each PT. We notably unveiled a novel and evolutionary important trait of PBS rod regions, namely, the frequent presence of DNA insertions between the tRNA-PheGAA and unk1 in both strains or SAGs (fig. 2 and supplementary figs. S1–S7 and S9, Supplementary Material online). These insertions share striking similarities to “tycheposons,” a novel type of mobile genetic elements recently discovered in Prochlorococcus (Hackl et al. 2020). This notably includes the hallmark presence of a tRNA and, in many of them, of the complete or partial tyrosine recombinase gene. However, Prochlorococcus tycheposons have been associated with seven possible tRNA types (Hackl et al. 2020), but never with a tRNA-PheGAA, which is the tRNA type systematically found upstream of PBS rod regions. Also, while in Prochlorococcus the tyrosine recombinase is most often located immediately downstream the tRNA at the 5′-end of the tycheposon, in Synechococcus it is found at the distal end of the tycheposon. Closer examination of this distal region in several Synechococcus genomes (notably in TAK9802) revealed a remnant of tRNA-PheGAA located between the tyrosine recombinase gene and unk1 in genomes where the former gene was complete. This observation strongly suggests that insertion of DNA material occurred by homologous recombination via a site-specific integrase at the level of this tRNA because this process often leaves the integrated elements flanked on both sides with the attachment site motif (Grindley et al. 2006). The presence of other recombinases in the DNA insertion in a few Synechococcus strains, such as putative site-specific, gamma-delta resolvases (tnpR-like genes in fig. 2 and supplementary figs. S1, S3A, S6 and S7, Supplementary Material online), may have resulted from iterative integrations at the same tRNA site, a phenomenon also reported for Prochlorococcus tycheposons (Hackl et al. 2020). We hypothesize that some of the specific genes present in PBS rod regions, notably the 13 unknown genes (unk1-13; fig. 1 and supplementary figs. S1–S7, Supplementary Material online), could have been acquired by lateral transfer into this specific tycheposon then transposition into the PBS rod region. These genes could have been retained by natural selection because of their yet unknown role in PBS biosynthesis or regulation. Additionally, by facilitating the import of foreign DNA as flanking material to mobile genetic elements (Hackl et al. 2020), tycheposons could also have played a key role in the conservation of the wide genomic diversity of PBS rod regions, and thus pigment type diversity, despite the multiple speciation events that occurred in the Synechococcus lineage, generating the three SCs and many clades observed nowadays in this radiation.
Novel Insights into the Evolution of CA4 Islands and Chromatic Acclimation
The characterization of many PBS rod regions from the environment that were retrieved from SAGs or fosmids led us to the discovery of a new location for the CA4-A genomic island near the 5′-end of the PBS rod region (fig. 4A and supplementary fig. S9C, Supplementary Material online) rather than elsewhere, as found in the genomes of all 3dA strains sequenced thus far (supplementary fig. S5B, Supplementary Material online). All of the genes of the PBS rod region containing these atypical CA4-A islands have the strongest similarity to the corresponding genes in canonical PT 3dA strains. Therefore, this new organization is unlikely to correspond to a new PT phenotype/genotype but rather to a previously unidentified PT 3dA variant. The localization of the CA4-A region at the 5′-end of the PBS rod region strongly supports the abovementioned hypothesis that the increases in the complexity of Synechococcus pigmentation by progressive extension of the PBS rod region primarily occurred via a tycheposon mechanism. This finding also provides a simple solution to the paradox of how two physically separate genomic regions encoding related and interacting components, with evolutionary histories that differ from the rest of the genome, were still able to co-evolve.
The CA4-A island not only has a highly variable position within the genome, but its gene content is also variable. Indeed, five out of eight CRD1 strains with a 3dA configuration of the PBS rod region possess an incomplete CA4-A island (supplementary fig. S5B, Supplementary Material online). In contrast, the CA4-B island is always found at the same position in the genome and is complete in all 3dB strains sequenced so far (supplementary fig. S7, Supplementary Material online). If the CA4-B island has also been generated in a tycheposon, the mechanism by which it has been transposed in the middle of the PBS rod region remains unknown because there are no known recombination hotspots in this region. Contig C100, which appears to lack the PEB lyase-encoding gene mpeW (fig. 4B; Grébert et al. 2021), is the first documented example of an incomplete CA4-B region. We predict that this new genotype has a constitutively high Exc495:545 ratio because this organism is likely to contain an active lyase-isomerase MpeQ (Grébert et al. 2021). It also has a rpcG gene encoding a PC lyase-isomerase (Blot et al. 2009) located immediately downstream of the CA4-B island and on the same strand as unk10 (fig. 4B and supplementary fig. S7, Supplementary Material online). This arrangement, which has thus far only been observed in MINOS11, suggests that rpcG expression could be controlled by light color and its protein product compete with those encoded by the rpcE-F operon because both act on the same cysteine residue of α-PC (Swanson et al. 1992; Zhou et al. 1992; Blot et al. 2009). This arrangement would be similar to the relationship between mpeZ and mpeY (Sanfilippo, Nguyen, et al. 2019) or between mpeW and mpeQ (Grébert et al. 2021). If confirmed, this would be the first case of chromatic acclimation altering the chromophorylation of PC instead of PE-I and PE-II in marine Synechococcus.
A Hypothesis for the Evolution of the PT 3 PBS Rod Region
The apparent mismatch between PBS pigmentation and vertical phylogeny raises the intriguing question of how different PTs have evolved and been maintained independently from the extensive clade diversification. It is generally agreed that the occurrence of the mpeBA operon in marine Synechococcus spp. PT 3 and the closely related uncultured S. spongiarum group resulted from gene duplication and divergence of a pre-existing cpeBA operon (Apt et al. 1995; Everroad and Wood 2012; Sánchez-Baracaldo et al. 2019). Yet, phycobiliproteins are part of a complex supramolecular structure, interacting with many other proteins such as linkers, phycobilin lyases, and regulators, which all need to co-evolve. Here, we propose an evolutionary scenario of progressively increasing complexity for the diversification of PT 3 from a PT 2/3 precursor (fig. 7). Our model integrates recent advances in our understanding of the functional characterization of PBS gene products, notably phycobilin lyases (Shukla et al. 2012; Sanfilippo et al. 2016; Mahmoud et al. 2017; Kronfel et al. 2019; Sanfilippo, Nguyen, et al. 2019; Carrigee et al. 2020; Grébert et al. 2021). Our proposal does not include PT 2B because strains exhibiting this PT generally possess several PC operons, such as PT 1 (Callieri et al. 2019), and it cannot be established with certainty whether PT 2B or PT 2A occurred first.
Fig. 7.
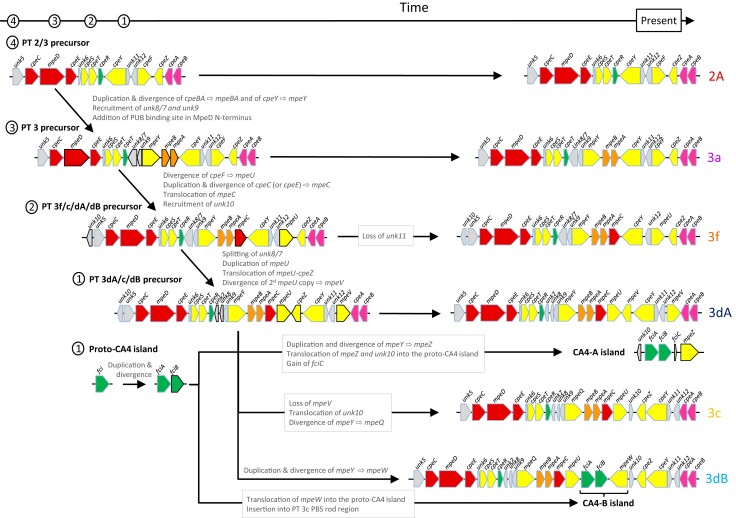
Putative evolutionary scenario for the occurrence of the different Synechococcus PT 3 subtypes. This scenario is globally congruent with individual phylogenies of genes in the PBS rod region. Note that the 5′- and 3′- ends of the PBS rod region are cropped for better visualization of the PE-I/PE-II subregions. Genes that changed between two consecutive PT precursor steps are highlighted by black contours (instead of blue-grey for the other genes).
The first step toward PE-II acquisition by the PT 2/3-like precursor involved the generation of a mpeBA operon precursor by duplication and divergence from an ancestral cpeBA operon. This was accompanied by the concomitant duplication of the ancestral PE-I specific lyase gene cpeY and its divergence to a precursor of the PEII-specific mpeY lyase gene (fig. 7). The origin of the unk8/7 fusion gene and unk9, occurring at the 5′-end of the PE-II subregion in all PT 3 strains (fig. 1 and supplementary fig. S2, Supplementary Material online), is more difficult to assess. However, it is noteworthy that Unk9 and the two moieties of Unk8/7 all belong to the Nif11-related peptide (N11P) family, which shows extensive paralogous expansion in a variety of cyanobacteria (Haft et al. 2010). Although some members of the N11P family have been suggested to be secondary metabolite precursors (Haft et al. 2010; Tang and van der Donk 2012; Cubillos-Ruiz et al. 2017), the functions of the Unk8/7 and Unk9 peptides remain unclear. Yet the localization of their genes in the PE-II subregion of all PT 3 strains strongly suggests a critical role in PE-II biosynthesis or regulation. One possibility is that they modulate the specificity of some PE-I lyases to extend their activity to PE-II subunits. Another, more subtle, change that occurred during the evolution of PT 3 was the change in the N-terminal part of the MpeD linker to include a specific insertion of 17 amino acids near the N-terminal region that is involved in PUB binding, as found in all PEII-specific linkers (Six et al. 2005). Present-day PT 3a would have directly descended from this PT 2/3 last common ancestor (LCA). Accordingly, the PBS rod region from PT 3a is the simplest of all PT 3 strains, and PT 3a sequences form the most basal clade in both mpeBA and mpeWQYZ phylogenies when these are rooted using cpeBA and cpeY sequences, respectively (fig. 3C; Grébert et al. 2021).
The three main differences between the PT 2/3 LCA and other PT 3 LCAs are the acquisition of the linker gene mpeC, which most likely resulted from the duplication and divergence of a pre-existing PE-I linker (either cpeC or cpeE), the acquisition of unk10, encoding an additional member of the N11P family, and the replacement of cpeF by mpeU (fig. 7). The lyase-isomerase MpeU belongs to the same family as the PEB lyase CpeF and is likely to have been derived from it. Even though the CpeF/MpeU phylogeny is unclear due to the deep tree branches having low bootstrap supports (Mahmoud et al. 2017; Carrigee et al. 2020), we hypothesize that the PT 3f/c/dB/dA precursor already had a mpeU-like gene. Phylogenetic trees of mpeBA and of the mpeWQYZ enzyme family place the recently described PT 3f (Xia et al. 2018) in a branch between those formed by PT 3a in one case and PT 3dA and 3c/3dB sequences in the other (fig. 3C). Consistently, the organization of the PT 3f PBS rod region appears to be intermediate between PT 3a and the more complex PT 3c/3dB/3dA regions. The only difference between the PT 3f/c/dB/dA precursor and the present-day PT 3f would be the loss of unk11, a short and highly variable open reading frame (fig. 7).
The PT 3f/c/dB/dA LCA then would have evolved to give the common precursor of PT 3dA/c/dB. This step likely involved four events: (1) the splitting of the unk8/7 gene into two distinct genes, unk8 and unk7, (2) the duplication of mpeU followed by (3) a tandem translocation of one mpeU gene copy and cpeZ between mpeC and cpeY, and (4) the divergence of the second mpeU copy to give mpeV, encoding another recently characterized lyase-isomerase of the CpeF family (Carrigee et al. 2020). Again, the poor bootstrap support of deep branches of the CpeF/MpeU/MpeV phylogeny (Carrigee et al. 2020) makes it difficult to confirm this hypothesis, and we cannot exclude the possibility that mpeV was derived directly from cpeF. Since the 3dA-type PBS rod region does not exist in present-day Synechococcus spp. without co-occurrence of a CA4-A island, the proto-CA4 island must also have evolved concurrently with the PT 3dA precursor. The two regulatory genes it contains, fciA and fciB, likely originate from the duplication and divergence of an ancestral fci precursor gene encoding a member of the AraC family. Both FciA and FciB possess an AraC-type C-terminal helix-turn-helix domain, yet their N-terminal domains have no similarity to any known protein (Humily et al. 2013; Sanfilippo et al. 2016). Generation of a complete CA4-A genomic island required three steps: (1) a translocation of unk10 into the proto-CA4 genomic island, (2) acquisition of fciC, a putative ribbon helix–helix domain-containing regulator that has similarity to bacterial and phage repressors (Humily et al. 2013), and (3) acquisition of mpeZ, possibly by duplication and divergence of mpeY, then translocation into the proto-CA4-A genomic island (fig. 7). It would also require the acquisition of the proper regulatory elements that are still unidentified.
Creation of the PT 3c-type PBS rod region from the same precursor that led to PT 3dA required three events: (1) the loss of mpeV, (2) the translocation of unk10 between mpeU and cpeZ, and (3) the divergence of the pre-existing lyase gene mpeY to make the lyase-isomerase gene mpeQ (Grébert et al. 2021). Then, the development of the PT 3dB from a PT 3c precursor only required the incorporation of a CA4-B genomic island in which the mpeW gene likely originated, like mpeZ, from duplication and divergence of mpeY, then translocation into the genomic island.
As previously noted, PTs 3c and 3dB share the same alleles for all PBS genes. Their only difference is the insertion of the CA4-B genomic island within the PBS rod region. Thus, the conversion between these two PTs appears to be relatively straightforward and may occur frequently. In contrast, the PT 3a and 3dA PBS rod regions differ by a number of genes and often have different alleles for orthologous genes. Thus, although the acquisition of a CA4-A island theoretically should be sufficient to transform a PT 3a-type GL specialist into a chromatic acclimater (Sanfilippo, Garczarek, et al. 2019; Sanfilippo, Nguyen, et al. 2019; Grébert et al. 2021), the divergence between the PT 3a and 3dA PBS components could make this conversion problematic. In accordance with this, although a number of PT 3a strains have naturally acquired either a complete (MVIR-18-1) or a partial (WH8016 and KORDI-49) CA4-A genomic island (so-called PT 3aA strains), none exhibit a functional CA4 phenotype (Choi and Noh 2009; Humily et al. 2013).
ILS Explains PBS Genes Phylogeny
The inconsistency between phylogenies obtained from PBS and core genes has led several authors to suggest that frequent lateral gene transfer (LGT) events of parts of or the whole PBS rod region likely occurred during the evolution of Synechococcus (Six et al. 2007; Dufresne et al. 2008; Haverkamp et al. 2008; Everroad and Wood 2012; Sánchez-Baracaldo et al. 2019). However, by examining additional representatives of each PT/clade combination in this manuscript, we have shown that different alleles of the PBS genes correspond to the different PTs and that the evolutionary history of each of these alleles is finely structured and globally consistent with the core phylogeny (fig. 5). This suggests that LGT events between clades are actually rare. An unambiguous LGT event occurred in strain MEDNS5 (PT 3c, clade VIa) because its mpeBA sequence was clustered with PT 3c/clade III mpeBA sequences (fig. 5 and supplementary table S7, Supplementary Material online). Similar observations were made for other PBS genes such as cpeBA, mpeW/Y/Z, and mpeU (Humily et al. 2013; Mahmoud et al. 2017; Grébert et al. 2018). This suggests that there have been transfers of blocks of co-functioning genes. The match between the evolutionary history of each allele and the corresponding core phylogeny also suggests that most transfer events occurred very early during the diversification of marine Synechococcus. Indeed, the reconciliation analysis using ALE detected a high frequency of transfers to very ancient lineages in Synechococcus phylogeny (supplementary fig. S10, Supplementary Material online and supplementary table S7, Supplementary Material online). However, the reconciliation model implemented in ALE does not account for incongruences between gene trees and species trees arising when an ancestral polymorphism in a population—in the present case, the occurrence of several alleles—is not fully sorted (i.e., resolved into monophyletic lineages) after a speciation event. This is because of the stochastic way in which lineages inherit alleles during speciation (Tajima 1983; Galtier and Daubin 2008; Lassalle et al. 2015), and such incongruences are interpreted by ALE as replacement transfers (i.e., a transfer and a loss). This phenomenon, called “incomplete lineage sorting” (ILS), is predicted to occur for at least some genes in a genome and is expected to be particularly important in prokaryotes with large population sizes (Retchless and Lawrence 2007, 2010; Degnan and Rosenberg 2009). In fact, the coalescent time (i.e., time to the LCA), and hence the frequency of ILS, is predicted to be proportional to the effective population size (Abby and Daubin 2007; Batut et al. 2014). Since Synechococcus is the second most abundant photosynthetic organism in the oceans (Flombaum et al. 2013), we can reasonably expect to observe some ILS in this lineage. Assuming that the Synechococcus effective population size is the same order of magnitude as that estimated for Prochlorococcus (1011 cells; Baumdicker et al. 2012; Batut et al. 2014; Kashtan et al. 2014) and that Synechococcus has a generation time of about 1 day, a tentative allelic fixation time would be of about 280 million years (Myr). This rough estimate is on the same order of timescale as the divergence between SC 5.3 and SC 5.2/SC 5.1 (400–880 Myr ago) or, within SC 5.1, between marine Synechococcus and Prochlorococcus (270–620 Myr ago; Sánchez-Baracaldo et al. 2019). This further supports the possibility of ILS being the major source of the apparent incongruence in PT distribution between clades (Retchless and Lawrence 2007). This new evolutionary scenario would imply that the different PTs appeared before the diversification of SC 5.1 clades and very likely before the divergence of SC 5.1 and 5.3, as was also recently suggested (Sánchez-Baracaldo et al. 2019). The basal position of the two SC 5.3 isolates in the phylogeny of the different mpeBA alleles (fig. 5) and the recent discovery of mpeBA-possessing Prochlorococcus (Ulloa et al. 2021) reinforce this hypothesis. In this view, Synechococcus clades were derived from an ancestral population in which all PT 3 (a through f) co-existed. Some clades seem to have lost some PTs in the course of their separation from other clades such as clade IV or CRD1, in which we only observe isolates of PT 3dA. Others might have conserved most pigment types, such as clade II, which encompasses all PT 3 except 3dA. Thus, recombination would maintain intra-clade diversity (Lassalle et al. 2015), while allowing clades to expand to novel niches defined by environmental parameters such as iron availability or phosphate concentration (Doré et al. 2020). This would have allowed the partial decoupling of adaptation to multiple environmental factors from adaptation to light color (Retchless and Lawrence 2007, 2010). In this regard, the occurrence of tycheposon-like elements at the 5′-end of many PBS rod regions is particularly interesting as it could provide Synechococcus populations with a tool favoring intra-clade recombination.
In conclusion, the analyses of the PBS rod region of newly sequenced Synechococcus isolates and of those retrieved from wild populations allowed us to clarify previous findings regarding the relationships between gene content and organization of this region, allelic variability, and Synechococcus PTs. We proposed a scenario for the evolution of the different PTs and present a new hypothesis based on population genetics to explain the observed discrepancies between PT and core phylogenies. These results demonstrate that analyzing Synechococcus evolution from the perspective of its demographic history provides a promising avenue for future studies.
Supplementary Material
Acknowledgments
This work was supported by the collaborative program METASYN with the Genoscope, the French “Agence Nationale de la Recherche” programs CINNAMON (ANR-17-CE02-0014-01) and EFFICACY (ANR-19-CE02-0019) as well as the European Union program Assemble+ (Horizon 2020, under grant agreement number 287589) to F.P. and L.G. and by National Science Foundation Grants (USA) MCB-1029414 and MCB-1818187 to D.M.K. We thank Fabienne Rigaud-Jalabert for collecting sea water, Thierry Cariou for providing physico-chemical parameters from the SOMLIT-Astan station and Thomas Hackl for useful discussions about tycheposons. We are also most grateful to the Biogenouest genomics core facility in Rennes (France) for petB sequencing and the platform of the Centre National de Ressources Génomiques Végétales in Toulouse (France) for fosmid library screening. We also warmly thank the Roscoff Culture Collection for maintaining and isolating some of the Synechococcus strains used in this study as well as the ABIMS Platform (Station Biologique de Roscoff) for helping in setting up the genome database used in this study and for providing storage and calculation facilities for bioinformatics analyses.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Data Availability
The genomic data underlying this article are available in GenBank and accession numbers are provided in supplementary table S1, Supplementary Material online. Annotated fosmid sequence data are available online at https://figshare.com/collections/Diversity_and_evolution_of_light-harvesting_complexes_in_marine_Synechococcus_cyanobacteria/5607200. Other data are available in the article and in its online Supplementary material.
Literature Cited
- Abby S, Daubin V. 2007. Comparative genomics and the evolution of prokaryotes. Trends Microbiol. 15:135–141. [DOI] [PubMed] [Google Scholar]
- Adir N. 2005. Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth Res. 85:15–32. [DOI] [PubMed] [Google Scholar]
- Ahlgren NA, Belisle BS, Lee MD. 2019. Genomic mosaicism underlies the adaptation of marine Synechococcus ecotypes to distinct oceanic iron niches. Environ Microbiol. 22:1801–1815. [DOI] [PubMed] [Google Scholar]
- Ahlgren NA, Rocap G. 2012. Diversity and distribution of marine Synechococcus: multiple gene phylogenies for consensus classification and development of qPCR assays for sensitive measurement of clades in the ocean. Front Microbiol. 3:213–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine D, et al. 2008. Assessment of uncertainty in the ocean reflectance determined by three satellite ocean color sensors (MERIS, SeaWiFS and MODIS-A) at an offshore site in the Mediterranean Sea (BOUSSOLE project). J Geophys Res. 113:C07013. [Google Scholar]
- Apt KE, Collier JL, Grossman AR. 1995. Evolution of the phycobiliproteins. J Mol Biol. 248:79–96. [DOI] [PubMed] [Google Scholar]
- Batut B, Knibbe C, Marais G, Daubin V. 2014. Reductive genome evolution at both ends of the bacterial population size spectrum. Nat Rev Microbiol. 12:841–850. [DOI] [PubMed] [Google Scholar]
- Baumdicker F, Hess WR, Pfaffelhuber P. 2012. The infinitely many genes model for the distributed genome of bacteria. Genome Biol Evol. 4:443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube PM, et al. 2018. Single cell genomes of Prochlorococcus, Synechococcus, and sympatric microbes from diverse marine environments. Sci Data. 5:180154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blot N, et al. 2009. Phycourobilin in trichromatic phycocyanin from oceanic cyanobacteria is formed post-translationally by a phycoerythrobilin lyase-isomerase. J Biol Chem. 284:9290–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. [DOI] [PubMed] [Google Scholar]
- Bretaudeau A, et al. 2013. CyanoLyase: a database of phycobilin lyase sequences, motifs and functions. Nucleic Acids Res. 41:D396–D401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Yeves PJ, et al. 2017. Novel Synechococcus genomes reconstructed from freshwater reservoirs. Front Microbiol. 8:1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callieri C, et al. 2019. The mesopelagic anoxic Black Sea as an unexpected habitat for Synechococcus challenges our understanding of global “deep red fluorescence”. ISME J. 13:1676–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrigee LA, et al. 2020. MpeV is a lyase isomerase that ligates a doubly-linked phycourobilin on the β-subunit of phycoerythrin I and II in marine Synechococcus. J Biol Chem. 296:100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DH, Noh JH. 2009. Phylogenetic diversity of Synechococcus strains isolated from the East China Sea and the East Sea. FEMS Microbiol Ecol. 69:439–448. [DOI] [PubMed] [Google Scholar]
- Cock PJA, et al. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz A, Berta-Thompson JW, Becker JW, van der Donk WA, Chisholm SW. 2017. Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc Natl Acad Sci USA. 114:E5424–E5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 24:332–340. [DOI] [PubMed] [Google Scholar]
- Doré H, et al. 2020. Evolutionary mechanisms of long-term genome diversification associated with niche partitioning in marine picocyanobacteria. Front Microbiol. 11:567431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, et al. 2008. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everroad C, et al. 2006. Biochemical bases of Type IV chromatic adaptation in marine Synechococcus spp. J Bacteriol. 188:3345–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everroad RC, Wood AM. 2012. Phycoerythrin evolution and diversification of spectral phenotype in marine Synechococcus and related picocyanobacteria. Mol Phylogen Evol. 64:381–392. [DOI] [PubMed] [Google Scholar]
- Farrant GK, et al. 2016. Delineating ecologically significant taxonomic units from global patterns of marine picocyanobacteria. Proc Natl Acad Sci USA. 113:E3365–E3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flombaum P, et al. 2013. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA. 110:9824–9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Daubin V. 2008. Dealing with incongruence in phylogenomic analyses. Phil Trans Roy Soc B Biol Sci. 363:4023–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garczarek L, et al. 2021. Cyanorak v2.1: a scalable information system dedicated to the visualization and expert curation of marine and brackish picocyanobacteria genomes. Nucl Acids Res. 49:D667–D676. 10.1093/nar/gkaa958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grébert T, et al. 2018. Light color acclimation is a key process in the global ocean distribution of Synechococcus cyanobacteria. Proc Natl Acad Sci USA. 115:E2010–E2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grébert T, et al. 2021. Molecular bases of an alternative dual-enzyme system for light color acclimation of marine Synechococcus cyanobacteria. Proc Natl Acad Sci USA. 118:e2019715118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley NDF, Whiteson KL, Rice PA. 2006. Mechanisms of site-specific recombination. Annu Rev Biochem. 75:567–605. [DOI] [PubMed] [Google Scholar]
- Guidi L, et al. 2016. Plankton networks driving carbon export in the oligotrophic ocean. Nature 532:465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Hackl T, et al. 2020. Novel integrative elements and genomic plasticity in ocean ecosystems. BioRxiv Available from: 10.1101/2020.12.28.424599 [DOI] [PubMed] [Google Scholar]
- Haft DH, Basu MK, Mitchell DA. 2010. Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family. BMC Biol. 8:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp T, et al. 2008. Diversity and phylogeny of Baltic Sea picocyanobacteria inferred from their ITS and phycobiliprotein operons. Environ Microbiol. 10:174–188. [DOI] [PubMed] [Google Scholar]
- Herdman M, Castenholz RW, Waterbury JB, Rippka R. 2001. Form-genus XIII. Synechococcus. In: Boone D, Castenholz R, editors. Bergey’s Manual of Systematics of Archaea and Bacteria, Volume 1. 2nd ed. New York: Springer-Verlag. p. 508–512. [Google Scholar]
- Holtrop T, et al. 2021. Vibrational modes of water predict spectral niches for photosynthesis in lakes and oceans. Nat Ecol Evol. 5:55–66. [DOI] [PubMed] [Google Scholar]
- Huang S, et al. 2012. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J. 6:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 33:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humily F, et al. 2013. A gene island with two possible configurations is involved in chromatic acclimation in marine Synechococcus. PLoS One. 8:e84459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humily F, et al. 2014. Development of a targeted metagenomic approach to study in situ diversity of a genomic region involved in light harvesting in marine Synechococcus. FEMS Microbiol Ecol. 88:231–249. [DOI] [PubMed] [Google Scholar]
- Kashtan N, et al. 2014. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344:416–420. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronfel CM, et al. 2019. The roles of the chaperone-like protein CpeZ and the phycoerythrobilin lyase CpeY in phycoerythrin biogenesis. Biochim Biophys Acta Bioenerg. 1860:549–561. [DOI] [PubMed] [Google Scholar]
- Larsson J, et al. 2014. Picocyanobacteria containing a novel pigment gene cluster dominate the brackish water Baltic Sea. ISME J. 8:1892–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle F, et al. 2015. GC-content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLoS Genet. 11:e1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MD, et al. 2019. Marine Synechococcus isolates representing globally abundant genomic lineages demonstrate a unique evolutionary path of genome reduction without a decrease in GC content. Environ Microbiol. 21:1677–1686. [DOI] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud RM, et al. 2017. Adaptation to blue light in marine Synechococcus requires MpeU, an enzyme with similarity to phycoerythrobilin lyase isomerases. Front Microbiol. 8:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazard S, Ostrowski M, Partensky F, Scanlan DJ. 2012. Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus: taxonomic resolution and biogeography of marine Synechococcus. Environ Microbiol. 14:372–386. [DOI] [PubMed] [Google Scholar]
- Mella-Flores D, et al. 2011. Is the distribution of Prochlorococcus and Synechococcus ecotypes in the Mediterranean Sea affected by global warming? Biogeosciences 8:2785–2804. [Google Scholar]
- Montgomery BL, Casey ES, Grossman AR, Kehoe DM. 2004. AplA, a member of a new class of phycobiliproteins lacking a traditional role in photosynthetic light harvesting. J Bacteriol. 186:7420–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CM, et al. 2013. Processes and patterns of oceanic nutrient limitation. Nat Geosci. 6:701–710. [Google Scholar]
- Moutin T, Van Wambeke F, Prieur L. 2012. Introduction to the Biogeochemistry from the Oligotrophic to the Ultraoligotrophic Mediterranean (BOUM) experiment. Biogeosciences 9:3817–3825. [Google Scholar]
- Olson RJ, Chisholm SW, Zettler ER, Armbrust EV. 1990. Pigment, size and distribution of Synechococcus in the North Atlantic and Pacific oceans. Limnol Oceanogr. 35:45–58. [Google Scholar]
- Ong LJ, Glazer AN, Waterbury JB. 1984. An unusual phycoerythrin from a marine cyanobacterium. Science 224:80–83. [DOI] [PubMed] [Google Scholar]
- Palenik B. 2001. Chromatic adaptation in marine Synechococcus strains. Appl Environ Microbiol. 67:991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen ML, et al. 2016. Synechococcus in the Atlantic gateway to the Arctic Ocean. Front Mar Sci. 3:191. [Google Scholar]
- Retchless AC, Lawrence JG. 2007. Temporal fragmentation of speciation in bacteria. Science 317:1093–1096. [DOI] [PubMed] [Google Scholar]
- Retchless AC, Lawrence JG. 2010. Phylogenetic incongruence arising from fragmented speciation in enteric bacteria. Proc Natl Acad Sci USA. 107:11453–11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Baracaldo P, Bianchini G, Di Cesare A, Callieri C, Chrismas NAM. 2019. Insights into the evolution of picocyanobacteria and phycoerythrin genes (mpeBA and cpeBA). Front Microbiol. 10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilippo JE, et al. 2016. Self-regulating genomic island encoding tandem regulators confers chromatic acclimation to marine Synechococcus. Proc Natl Acad Sci USA. 113:6077–6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilippo JE, et al. 2019. Interplay between differentially expressed enzymes contributes to light color acclimation in marine Synechococcus. Proc Natl Acad Sci USA. 116:6457–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilippo JE, Garczarek L, Partensky F, Kehoe DM. 2019. Chromatic acclimation in Cyanobacteria: a diverse and widespread process for optimizing photosynthesis. Annu Rev Microbiol. 73:407–433. [DOI] [PubMed] [Google Scholar]
- Scheer H, Zhao K-H. 2008. Biliprotein maturation: the chromophore attachment: biliprotein chromophore attachment. Mol Microbiol. 68:263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluchter WM, et al. 2010. Phycobiliprotein biosynthesis in cyanobacteria: structure and function of enzymes involved in post-translational modification. In: Hallenbeck PC, editor. Recent advances in phototrophic prokaryotes. Vol. 675. Advances in experimental medicine and biology. New York: Springer. p. 211–228. Available from: http://link.springer.com/10.1007/978-1-4419-1528-3_12 [DOI] [PubMed] [Google Scholar]
- Shukla A, et al. 2012. Phycoerythrin-specific bilin lyase-isomerase controls blue-green chromatic acclimation in marine Synechococcus. Proc Natl Acad Sci USA. 109:20136–20141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler WA. 1994. Phycobilisome and phycobiliprotein structure. In: Bryant DA, editor. The molecular biology of cyanobacteria. Advances in photosynthesis. Dordrecht: Springer Netherlands. p. 139–216. Available from: 10.1007/978-94-011-0227-8_7 [DOI] [Google Scholar]
- Six C, et al. 2005. Two novel phycoerythrin-associated linker proteins in the marine cyanobacterium Synechococcus sp. strain WH8102. J Bacteriol. 187:1685–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six C, et al. 2007. Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study. Genome Biol. 8:R259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohm JA, et al. 2016. Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, macronutrients and iron. ISME J. 10:333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson RV, et al. 1992. Characterization of phycocyanin produced by cpcE and cpcF mutants and identification of an intergenic suppressor of the defect in bilin attachment. J Biol Chem. 267:16146–16154. [PubMed] [Google Scholar]
- Szöllősi GJ, Rosikiewicz W, Boussau B, Tannier E, Daubin V. 2013. Efficient exploration of the space of reconciled gene trees. Syst Biol. 62:901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, van der Donk WA. 2012. Structural characterization of four prochlorosins: a novel class of lantipeptides produced by planktonic marine cyanobacteria. Biochemistry 51:4271–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa O, et al. 2021. The cyanobacterium Prochlorococcus has divergent light-harvesting antennae and may have evolved in a low-oxygen ocean. Proc Natl Acad Sci USA. 118:e2025638118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbanks SM, Glazer AN. 1993. Rod structure of a phycoerythrin II-containing phycobilisome. I. Organization and sequence of the gene cluster encoding the major phycobiliprotein rod components in the genome of marine Synechococcus sp. WH8020. J Biol Chem. 268:1226–1235. [PubMed] [Google Scholar]
- Wood A, Phinney D, Yentsch C. 1998. Water column transparency and the distribution of spectrally distinct forms of phycoerythrin-containing organisms. Mar Ecol Prog Ser. 162:25–31. [Google Scholar]
- Wu D, Jospin G, Eisen JA. 2013. Systematic identification of gene families for use as “markers” for phylogenetic and phylogeny-driven ecological studies of Bacteria and Archaea and their major subgroups. PLoS One. 8:e77033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, et al. 2017. Phylogeography and pigment type diversity of Synechococcus cyanobacteria in surface waters of the northwestern Pacific Ocean. Environ Microbiol. 19:142–158. [DOI] [PubMed] [Google Scholar]
- Xia X, Guo W, Tan S, Liu H. 2017. Synechococcus assemblages across the salinity gradient in a salt wedge estuary. Front Microbiol. 8:1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Liu H, Choi D, Noh JH. 2018. Variation of Synechococcus pigment genetic diversity along two turbidity gradients in the China Seas. Microb Ecol. 75:10–21. [DOI] [PubMed] [Google Scholar]
- Yu MH, Glazer AN. 1982. Cyanobacterial phycobilisomes. Role of the linker polypeptides in the assembly of phycocyanin. J Biol Chem. 257:3429–3433. [PubMed] [Google Scholar]
- Zhou J, Gasparich GE, Stirewalt VL, De Lorimier R, Bryant DA. 1992. The cpcE and cpcF genes of Synechococcus sp. PCC 7002: construction and phenotypic characterization of interposon mutants. J Biol Chem. 267:16138–16145. [PubMed] [Google Scholar]
- Zwirglmaier K, et al. 2008. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol. 10:147–161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic data underlying this article are available in GenBank and accession numbers are provided in supplementary table S1, Supplementary Material online. Annotated fosmid sequence data are available online at https://figshare.com/collections/Diversity_and_evolution_of_light-harvesting_complexes_in_marine_Synechococcus_cyanobacteria/5607200. Other data are available in the article and in its online Supplementary material.