Abstract
Purpose
Merkel cell carcinoma (MCC) is an aggressive cutaneous neuroendocrine carcinoma that can be divided into two classes: virus-positive (VP) MCC, associated with oncogenic Merkel cell polyomavirus (MCPyV); and virus-negative (VN) MCC, associated with photodamage.
Experimental design
We classified 346 MCC tumors from 300 patients for MCPyV using a combination of immunohistochemistry, in situ hybridization, and quantitative PCR assays. In a subset of tumors, we profiled mutation status and expression of cancer-relevant genes. MCPyV and molecular profiling results were correlated with disease-specific outcomes. Potential prognostic biomarkers were further validated by immunohistochemistry.
Results
177 tumors were classified as VP-MCC, 151 tumors were VN-MCC, and 17 tumors were indeterminate. MCPyV positivity in primary tumors was associated with longer disease-specific and recurrence-free survival in univariate analysis, and in multivariate analysis incorporating age, sex, immune status, and stage at presentation. Prioritized oncogene or tumor suppressor mutations were frequent in VN-MCC but rare in VP-MCC. TP53 mutation developed with recurrence in one VP-MCC case. Importantly, for the first time we find that VP-MCC and VN-MCC display distinct sets of prognostic molecular biomarkers. For VP-MCC, shorter survival was associated with decreased expression of immune markers including granzyme and IDO1. For VN-MCC, shorter survival correlated with high expression of several genes including UBE2C.
Conclusion
MCPyV status is an independent prognostic factor for MCC. Features of the tumor genome, transcriptome, and microenvironment may modify prognosis in a manner specific to viral status. MCPyV status has clinicopathologic significance and allows for identification of additional prognostic subgroups.
Keywords: Merkel cell carcinoma, Merkel cell polyomavirus, TP53, IDO-1, biomarker
INTRODUCTION
Merkel cell carcinoma (MCC) is an aggressive cutaneous neuroendocrine malignancy. Clinical outcomes correlate with established staging parameters such as tumor size and regional lymph node involvement. However, a proportion of early-stage patients experience disease progression and mortality, underscoring the need for improved prognostication [1].
MCC can be divided into two molecular subclasses: virus-positive (VP) and virus-negative (VN). Virus-positive tumors are associated with Merkel cell polyomavirus (MCPyV) sequence integrated into the host genome, resulting in expression of viral oncoproteins including large T (LT) and small T (ST) antigens that mediate inactivation of p53 and Rb tumor suppressors, among other functions [2, 3]. Cellular gene mutations are not consistently detected in VP-MCC [1, 4-6] suggesting that viral oncoproteins are sufficient for tumor initiation and maintenance. By contrast, VN-MCC demonstrates genomic evidence of chronic ultraviolet exposure, high tumor mutation burden, and mutational inactivation of tumor suppressor genes, most frequently TP53 and RB1 [1, 4-6].
Viral status in MCC has been associated with differences in prognosis and therapeutic response, although studies have had mixed results [1, 3, 7]. Investigations into the clinical significance of these molecular subclasses have been complicated by debates regarding the relative sensitivity and specificity of MCPyV detection approaches, including immunohistochemical (IHC) detection of viral T-antigens, in situ hybridization (ISH) for detection of viral transcripts or DNA, quantitative polymerase chain reaction (qPCR) for viral DNA, and next-generation sequencing (NGS) [1]. The largest study combined IHC and qPCR approaches to demonstrate improved survival in the VP-MCC cohort relative to VN-MCC [7].
Although TP53/RB1 inactivation is nearly universal in VN-MCC, other genomic changes affecting subgroups of MCC are less well-described. A minority of MCC tumors harbors activating events of the PI3K/AKT pathway [4-6, 8-11]. RAS family mutations are detected at low frequency [4-6]. MYCL amplifications have been described, although reports have varied regarding the incidence of this event in MCC [9, 12]. Evidence supports mutational activation of the beta-catenin pathway in a minority of tumors [13]. Other less frequent oncogene activation events include mutational activation of KNSTRN, RAC1, EZH2, and TERT [4, 8, 9, 14]. In addition to TP53 and RB1, other tumor suppressor inactivation events may be observed in MCC, most frequently inactivation of NOTCH family genes [4-6, 9, 15].
To better understand the clinicopathologic significance of genomic changes in MCC, including MCPyV and cellular gene mutations, we analyzed a large cohort of MCC with disease-specific outcomes.
METHODS
Study Design and Cohort Selection
This study was conducted according to a protocol approved by the University of Michigan Institutional Review Board, in accordance with the U.S. Common Rule, and with informed written consent from each subject upon enrollment in the Michigan Medicine Cutaneous Oncology Database. Patient samples considered for inclusion in the retrospective, single-center analysis were identified by search and review of entries from 1999 through 2019 of patients enrolled in the Cutaneous Oncology Program database, and Pathology laboratory information systems (LIS). 346 samples from 300 unique patients were selected for further analysis based upon adequacy and availability of formalin-fixed, paraffin-embedded (FFPE) tumor material (Table S1). Adequacy was determined by review by a board-certified dermatopathologist (PWH) based upon tumor size adequate for macrodissection, and at least 60% tumor purity. For patients with multiple tumor samples (primary and metastases), the earliest available adequate sample was selected for use, with the exception of 45 cases selected at random for primary-metastasis pair analysis. For one primary-metastasis pair, additional tumor samples were sequenced following identification of an acquired TP53 mutation in the metastasis as described below. Clinicopathologic data was retrieved from the Cutaneous Oncology Program database and the Pathology LIS. Histopathologic parameters were derived from tumor templates, as the original biopsy slides were not available for many patients. Clinical data for some tumors has been previously reported [4, 16, 17].
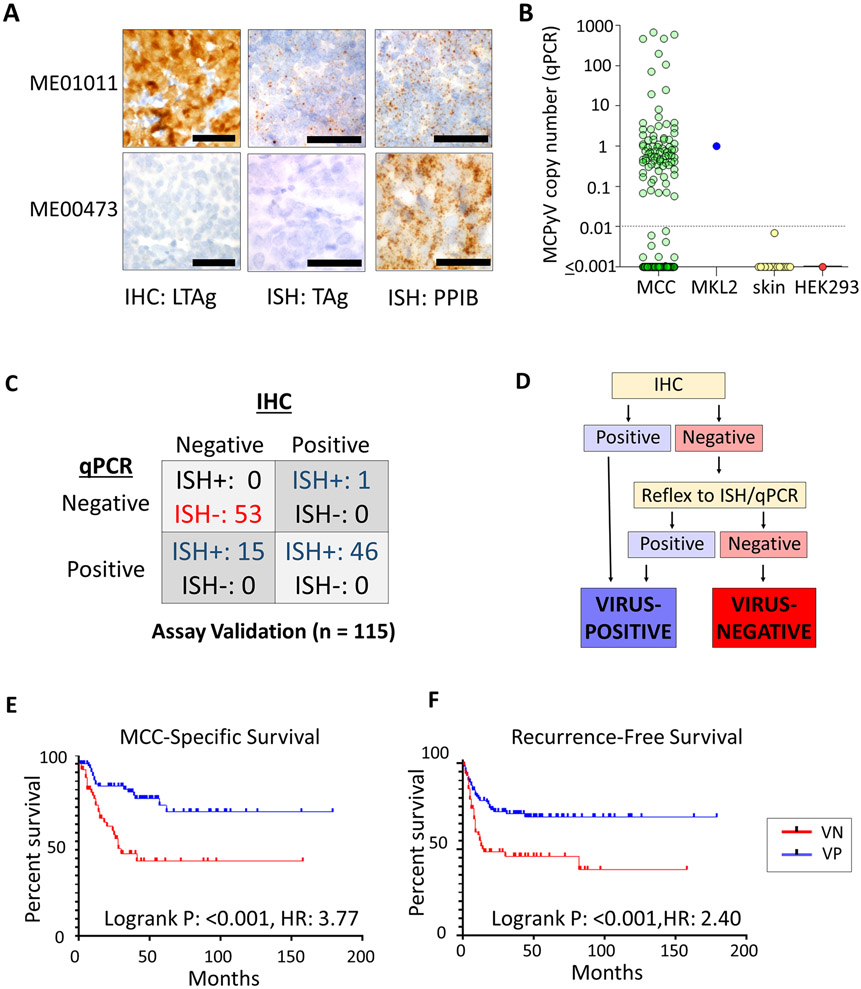
MCC tumors were classified for MCPyV by an approach based upon relative sensitivity of IHC, ISH, and PCR (Figure 1, Table S1) performed according to previously described protocols [4, 16]. Specifically, CM2B4 clone (Santa Cruz) was used for IHC detection of LT at 1:250 dilution on a Ventana automated stainer. IHC was scored as positive (cytoplasmic and/or nuclear staining in >5% of tumor cells), equivocal (blush staining), or negative. Quantitative PCR was performed on 15 ng of tumor DNA using LT2 primer/probes [18] with Taqman detection. Viral copy number was quantitated against the reference cell line MKL-2, with copy number <0.01 considered negative, as well as a negative control cell line (HEK293). Values were also compared against previously reported results in normal skin[16]. In situ hybridization (Advanced Cell Diagnostics RNAScope assay) utilized probes tiling across the common T, ST, and LT transcript regions. Peptidylprolyl isomerase B (PPIB) and bacterial dihydrodipicolinate reductase (DapB) were positive and negative ISH controls, respectively. In situ hybridization was considered positive if at least 5 cells demonstrated positive signal. All assays were evaluated in a blinded manner relative to other modalities and molecular profiling results. We have previously demonstrated that these techniques display comparable performance relative to NGS detection of viral transcripts and mutational analysis of tumor genomes [4, 19]. MCPyV detection results including NGS for some tumors in this study have been previously reported [4, 16, 17, 20].
Figure 1. Merkel cell polyomavirus detection and associated outcomes.
(A) Representative results for detection of Merkel cell polyomavirus (MCPyV) by immunohistochemistry (IHC) for large T antigen (LTAg), and in situ hybridization (ISH) spanning large and small T antigen (TAg) transcripts. PPIB: positive control for RNA integrity. (B) Quantitative PCR evaluation of MCPyV copy number in MCC tumors, as compared to positive control cell line (MKL2, copy number = 1), negative control cell line (HEK293), and negative control formalin-fixed, paraffin-embedded normal skin samples. Normal skin results have been previously reported [16]. Dashed line indicates the threshold of positivity (>0.01 copies). (C) Comparison of MCPyV assay detection methods in initial comparison (subset of total study cases). A subset of tumors with negative IHC have detectable MCPYV RNA and DNA by other approaches, suggesting limited sensitivity for IHC relative to other assays. (D) MCPyV classification approach. Although IHC (Assay 1) was relatively specific, a second assay (ISH or qPCR) is used to confirm negative results. (E) The presence of MCPyV is associated with improved disease-specific survival in MCC. F) The presence of MCPyV is associated with improved recurrence free survival in MCC. HR: Hazard ratio.
For MCPyV-positive MCC with TP53 mutations, whole genome sequencing was performed to further confirm the presence of Merkel cell polyomavirus. DNA was extracted from FFPE sections with a minimal tumor purity of 50%, using the QIAGen AllPrep DNA extraction kit, followed by the NEBNext FFPE DNA repair kit. Bar-coded libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina. Whole Genome Sequencing (WGS) was performed on Novaseq (paired-end 2x151bp) at an average coverage of 45X. Read alignment was performed with BWA-MEM [21] against a customized combination of human reference genome hg19 and Merkel cell polyomavirus isolate 18b genome (GenBank: HM011550.1). Mutations and small indels in the viral genome were called by freebayes [22]. Structural rearrangements, including viral integration sites, were called by LUMPY [23]. Reads predicted to span the viral integration sites were further inspected and confirmed with BLAST. MCPyV genome coverage was visualized by Integrative Genomics Viewer.
Targeted Next Generation Sequencing for Cancer Gene Alterations
DNA profiling was performed using a previously reported version of the StrataNGS laboratory developed test [24, 25], which utilizes multiplex PCR-based Ion Torrent NGS to evaluate mutations and copy number alterations in 63 cancer genes (Table S2). Testing was performed as described, except hematoxylin and eosin–stained sections were used as a guide for dissection from a minimum of 4 FFPE) 10-μm sections by a board-certified dermatopathologist (P. W. H.) to obtain a minimal tumor purity of 60%. DNA extraction prior to specimen transfer for StrataNGS testing. Co-isolation of DNA and RNA, StrataNGS testing, and data analysis were performed as previously described using validated bioinformatics pipelines [24, 25]. Briefly, DNA was extracted using VERSANT Sample Preparation 1.0 Reagents on automated liquid handlers. One aliquot of isolated nucleic acid was treated with DNase I digested to provide an RNA-only sample. Bar-coded libraries were generated from 8 ng of DNA per sample using the Ion Ampliseq library kit 2.0 with the DNA component of the StrataNGS panel targeting 63 cancer-relevant genes. Templates were prepared using the Ion 540 Chef Kit (Thermo Fisher Scientific, Waltham, MA) on the Ion Chef. Sequencing of multiplexed templates was performed using the Ion Torrent S5 on Ion 540 chips using the Ion 540 Chip Kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions. Data analysis was performed using in-house developed, previously validated pipelines employing Torrent Suite 5.8, with alignment by Torrent Mapping Alignment Program and variant calling using the Torrent Variant Caller plugin [24, 25]. For this analysis, significantly decreased copy number was defined as deep deletion (probable homozygous deletion: median copy number <0.5, upper bound of 90% confidence interval <1.0) or copy loss (probable heterozygous copy loss, set to half the magnitude of deep deletion: median copy number <1.25, upper bound of 90% confidence interval <1.5). Mutation and copy number variants were prioritized according to predicted functional consequence per standard StrataNGS pre-specified reporting: amplification or hotspot mutation of oncogenes, or truncating/inactivating mutation of tumor suppressor genes.
We performed targeted, amplicon-based multiplex RNA sequencing to evaluate potential fusions and relative expression in 26 cancer genes (Table S3), using a panel validated against orthogonal truth for the presence of chimeric transcripts (gene fusions) as described [24, 25]. The presence of fusion transcripts was assessed by aligning RNA reads to a custom reference comprised of isoform-specific reference sequences for all measured fusions. Expression of non-chimeric transcripts was measured in normalized reads per million, whereby raw expression target read counts are normalized by a factor that results in the median housekeeping gene expression value matching the same gene's standard reads-per-million (RPM) value in a reference FFPE normal cell line sample (GM24149) run in parallel with all clinically tested samples. Subsequent independent validation of this targeted multiplex RNAseq for relative gene expression was performed vs. gold standard RT-PCR, as well as IHC for PD-L1 expression, and both gene fusions and non-chimeric expression are included in the current clinically validated version of the StrataNGS test [24, 25] and SA Tomlins et al., manuscript submitted).
Quality metrics are shown in Table S4.
Immunohistochemistry for Mutation and Prognostic Markers
Immunohistochemistry for granzyme B (Cell Signaling Technology), IDO-1 (Sigma Aldrich), UBE2C (Abcam), beta catenin (Ventana), and IDH1 R132H (Dianova) expression was performed using the conditions outlined in Table S5. Granzyme B was selected for validation due to common use of this marker in diagnostic laboratories, after confirmation that GZMA and GZMB transcript expression were closely correlated in previously published MCC expression profiles [17]. Potential prognostic markers (granzyme, IDO1, UBE2C) were evaluated using previously described tissue microarrays and whole sections on primary tumors,[26] alongside additional primary tumor cases, to achieve adequate statistical power. Immunostained slides were scanned at 20x magnification on a Vectra Polaris (PerkinElmer). Scoring was performed on representative digitized tumor fields selected by a board-certified dermatopathologist (P.W.H.) using the Positive Cell Detection analysis in QuPATH software to quantitate either density of positive inflammatory cells (granzyme, IDO1) or H-score expression on tumor cells (UBE2C, beta-catenin). Beta-catenin and IDH1 R132H immunohistochemistry were performed on representative mutant and wild type tumors.
Statistical Analysis
Statistical analysis was performed on GraphPad Prism 8.0, R, SAS (version 9.4), or Qlucore software. RNA expression comparisons were performed for each amplicon. Two-sample t tests were used to compare the expression (log transformed) of each gene between negative and positive MCPyV tumors; all p-values were then converted to q-value [27] to control for the false discovery rate. Chi-squared/Fisher’s exact tests were used to assess the association between categorical variables (e.g., stage, ulcer) and MCPyV, and a two-sample t test was used to compare tumor depth (measured by the Breslow method) between positive and negative MCPyV. For survival data, Kaplan-Meier method was used to estimate the survival function for low- and high-expression group (using the median expression value for transcript expression or receiver operator characteristic analysis to identify the most sensitive and specific threshold for protein expression. For outcomes related to molecular markers, primary and secondary (recurrent or metastatic) sample types were included in the analysis (with each patient represented by a single sample) in order to achieve sufficient statistical power, after statistical modeling including regression analysis to confirm that sample type was not a confounder in this group. Cox proportional hazard regression was used to study the effect of a biomarker (e.g., MCPyV) on survival after controlling for patient age, sex, immune status, and stage. Statistical significance was defined as p < 0.05 for clinicopathologic comparisons, and q <0.05 and log2 fold change >1 or <−1 for differential gene expression.
RESULTS
MCPyV Classification, Clinical Associations, and Survival Patterns in MCC
The final study cohort included 346 MCC tumors from 300 unique patients (Table S1), with an average follow-up of 40 months. The majority of tumors (62%) were primary, with the remainder as secondary tumors including regional lymph node/parotid metastases (26%), satellite/in transit metastases (8%), local recurrence (2%), and distant metastases (2%). The most frequent primary site was the extremity (49%), followed by head and neck (43%), trunk/genitalia (6%), and unknown primary (2%). Management included excision (91%), radiotherapy (51%) and chemotherapy (5%). Sentinel lymph node biopsy was performed for clinically node negative cases when surgically feasible, as per standard management guidelines for MCC [1, 28]. There were few tumors treated with immunotherapy (n=2), as our cohort is skewed toward cases with longer follow-up that predate approval of immunotherapy for MCC.
The approach for MCPyV classification was defined by an initial comparison in a subset of our final cohort (117 tumors) evaluated by three modalities (IHC, RNA-ISH, and qPCR)(Figure 1A-D). Of these, 2 were equivocal for IHC and excluded from further comparisons. Of the remaining tumors, 99 (86%) demonstrated concordant results for all three assays. Of 18 tumors with discordance, 15 were negative by IHC and positive by other techniques, and 1 was negative by PCR and positive by other techniques. This data indicated that, by our protocols, IHC has comparable specificity but limited sensitivity relative to ISH and qPCR, suggesting a reflex testing approach to maximize sensitivity while conserving tissue (Figure 1D). Applying this approach to our cohort as a whole, 177 tumors (from 155 unique patients) were classified as VP-MCC, 151 tumors (from 125 unique patients) were VN-MCC, and 17 tumors (from 16 unique patients) had inadequate tissue quantity/quality for definitive classification (Table S1). Of 45 pairs of related tumors (e.g. primary-metastasis pairs), all displayed agreement for MCPyV status by every assay used, with the exception of one pair of tumors that were discordant by IHC staining but were confirmed to be MCPyV positive by other assays (Table S1).
There was no significant association between viral status and tumor classification (primary vs. metastasis [p=0.21]) in this cohort. Significant associations with VP-MCC included female sex, location on extremity, and younger age at diagnosis (Table 1). A greater proportion of VP-MCC tumors were associated with Stage II disease. Histopathologic features significantly associated with VP-MCC included significantly greater tumor depth and size >2 cm. VN-MCC tumors displayed significantly greater frequency of immune compromise, ulceration, coexisting squamous cell carcinoma in situ (SCCIS), and absence of cytokeratin-20 expression (Table S6).
Table 1.
Clinicopathologic features of patient cohort
| VP | VN | ||||
|---|---|---|---|---|---|
| n | 155 | 125 | |||
| Median | SD | Median | SD | p | |
| Age at diagnosis, years | 71.0 | 12.2 | 78.0 | 19.0 | <0.001 |
| n (%) | n (%) | ||||
| Sex | 0.002 | ||||
| Female | 64 (41.3%) | 33 (25.6%) | 0.002 | ||
| Male | 91 (58.7%) | 96 (74.4%) | |||
| Tumor characteristics | |||||
| Primary tumor site | <0.001 | ||||
| Head and neck | 43 (27.7%) | 81 (64.8%) | |||
| Extremity | 103 (66.5%) | 30 (24%) | |||
| Trunk, buttock, genitalia | 8 (5.2%) | 12 (9.6%) | |||
| Unknown primary | 1 (0.6%) | 2 (1.6%) | |||
| Stage at Diagnosis | 0.007 | ||||
| I | 37 (24.5%) | 39 (31.5%) | |||
| II | 27 (17.9%) | 7 (5.6%) | |||
| III | 83 (55.0%) | 73 (58.9%) | |||
| IV | 4 (2.7%) | 5 (4.0%) | |||
| 3 year DSS (primary tumors) | |||||
| DOD | 12 (17.6%) | 25 (33.8%) | 0.04 | ||
| Living | 56 (82.4%) | 49 (66.2%) | |||
| 3 year RFS (primary tumors) | |||||
| Recurrence | 28 (37.8%) | 42 (51.2%) | 0.11 | ||
| No recurrence | 46 (62.2%) | 40 (48.8%) |
P: chi square or Fisher’s exact test. DOD: died of disease. DSS: disease specific survival. RFS: recurrence free survival. SD: standard deviation. VP: Merkel cell polyomavirus positive. VN: Merkel cell polyomavirus negative.
When evaluating univariate long-term oncologic outcomes in primary tumors, VP-MCC was associated with significantly longer overall survival (OS), recurrence-free survival (RFS) and disease-specific survival (DSS) by Kaplan Meier analysis (Figure 1E, 1F, and S1). At 3 years, disease-specific deaths occurred at nearly twice the rate in VN-MCC relative to VP-MCC (Table 1). In multivariate analyses controlling for age, sex, immune status, and stage at presentation, VP-MCC status in primary tumors was associated with significantly longer DSS (p=0.001) and RFS (p=0.005) (Table 2). Stage, age, and patient immune status were also significantly associated with DSS in multivariate analysis; immune status was also significantly associated with RFS. We also evaluated the performance of individual MCPyV assays in univariate survival analyses, and found significantly improved outcome for the VP-MCC group regardless of assay (Table S7), with the exception of recurrence-free survival for IHC.
Table 2.
Multivariate outcomes analysis for Merkel cell carcinoma primary tumors.
| DSS |
RFS |
|||
|---|---|---|---|---|
| Variable | p | Hazard Ratio | p | Hazard Ratio |
| MCPyV | 0.001 | 2.93 | 0.005 | 2.12 |
| Age | 0.01 | 1.05 | 0.856 | 1.00 |
| Sex | 0.5 | 1.26 | 0.853 | 1.05 |
| Immune Status | <0.001 | 0.08 | 0.016 | 0.039 |
| Stage | <0.001 | multiple | 0.033 | multiple |
Identification of Candidate Driver Genes
247 tumors (131 VP, 111 VN, and 5 equivocal/indeterminate) were successfully further evaluated by targeted genomic profiling (Figure 2A, Tables S1 and S2). As previously described, almost all (97%) VN-MCC tumors were associated with TP53 and/or RB1 mutation or copy loss. TP53 were the most highly recurrent mutations in MCC and were concentrated in VN-MCC (94% of VN-MCC tumors, compared to 8% of VP-MCC tumors) (Table S8). RB1 mutations were the second most frequent (64% of VN-MCC, 2% of VP-MCC). Other tumor suppressor inactivating events involved CDKN2A, ATM, PTEN, BRCA1, and BRCA2. The most frequent oncogene activation events were PIK3CA (12% of VN-MCC, 2% of VP-MCC), followed by TERT promoter mutation and beta-catenin (CTNNB1) activating mutation (Table S8). Of note, CTNNB1 mutations were associated with significantly higher beta-catenin protein expression (Figure 2B). Additional oncogene activation events, to our knowledge not previously described in MCC, included JAK2, IDH1, and IDH2. In the tumor with IDH1 R132H mutation, tumor cell expression of the mutant protein was detectable by immunohistochemistry (Figure 2C). Oncogenic fusions were not detected in any tumor.
Figure 2. Genomic alterations in Merkel cell carcinoma.
(A) Prioritized mutations and copy number variants in MCC. MCPyV-positive tumors (VP-MCC, left) display low frequency of cellular mutations. MCPyV-negative tumors (VN-MCC, right) display near ubiquitous inactivating mutations of TP53 and/or RB1, and prioritized mutations involving additional cellular tumor suppressors or oncogenes in many cases. (B) Pattern of beta-catenin protein expression in a MCC tumor harboring CTNNB1 S33F mutation (bottom panel), compared to a tumor with wild type CTNNB1. Graph: MCC tumors with CTNNB1 mutations (MT) display significantly higher beta catenin protein expression as compared to those with wild-type (WT) CTNNB1. (C) Confirmation of IDH1 R132H mutant protein expression in tumor cells in a representative case of MCC in which this mutation was detected by sequencing (bottom panel). (D) Acquisition of TP53 mutation with progression in VP-MCC. Each oval represents a distinct tumor sample. (E, F) The presence of oncogene activation events (hotspot mutation or amplification) is associated with shorter time to recurrence and disease-specific death for VP-MCC tumors, but not VN-MCC tumors.
Chromosomal copy number losses affecting tumor suppressor genes occurred more frequently in VN-MCC (Table S9). Copy losses in VN-MCC most frequently affected RB1. PTEN loss occurred at a similar frequency in both groups (Table S9). MYC copy gains were detected in 2 VN-MCC. MYCL was not included on the gene panel.
We further evaluated tumors with unexpected molecular results (VP-MCC with TP53 mutation, VN-MCC lacking TP53 mutation). Of 11 VP-MCC tumors with TP53 mutation (from 9 unique patients), the presence of MCPyV was demonstrated by at least one assay in all cases, and could be confirmed by multiple assays in all but one tumor (Table S1, Figure S2A, B), with the remaining tumor being technically inadequate for further testing. 5 VP-MCC tumors with TP53 mutations had sufficient remaining material for whole-genome sequencing; by this approach, MCPyV sequence was detected in all 5 tumors, with each tumor displaying either predicted LT-truncating mutations or predicted integration sites resulting in disruption of the LT 2nd exon distal to the RB-binding motif (Figure S2C-E).
Of 5 VN-MCC tumors (from 5 unique patients) without detected TP53 mutation, all were negative for MCPyV by multiple assays (Table S1, Figure S2A, B). In 2 of these tumors, RB1 truncating mutations were present. None of the 5 tumors had sufficient remaining DNA for whole genome sequencing.
Acquisition of TP53 Mutations in Merkel Cell Carcinoma
Our analysis included 23 sets of related tumors with molecular profiling results on both tumors. Of these, 22 demonstrated concordant mutation calls between related tumors (Table S1). Specifically, 8 pairs had no prioritized mutations in either tumor. 14 pairs had concordant mutation calls including tumor suppressors (TP53, RB1, CDKN2A, PTEN, BRCA2) and oncogenes (PIK3CA, FGFR3, CTNNB1). For one set of related VP-MCC tumors, a TP53 mutation was acquired in a recurrence that was not detected in the primary (Figure 2D, Table S1). Comparison of MCPyV sequences by whole genome sequencing in two tumors of this set (one with TP53 mutation, and one without TP53 mutation) confirmed identical MCPyV sequence variations and predicted integration site (Figure S2C), consistent with clonally identical MCPyV integration.
Prognostic Relevance of Mutations in Merkel Cell Carcinoma
Of tumors with molecular profiling results, 207 had recurrence-free survival data and 173 had MCC-specific survival data. To control for MCPyV, we evaluated VN-MCC and VP-MCC separately. To maximize power, we included secondary tumor samples (recurrent or metastatic) as well as primary tumor samples, with each patient represented by a single sample in the analysis, after first confirming that tumor type did not represent a statistical confounder for outcomes analysis. TP53 mutations did not clearly associate with worse outcome in MCPyV subgroup analysis; specifically, VP-MCC with mutated TP53 were not associated with worse outcomes than VP-MCC tumors with wild type TP53, and VN-MCC with wild type TP53 did not display better outcomes than VN-MCC with mutated TP53 (Figure S3), although some sample groups were small.
We then evaluated for the significance of oncogene activation events. Among 6 VP-MCC tumors harboring oncogene activating mutations (specifically, PIK3CA E545K, PIK3CA K111E, PIK3CA C420R, IDH2 R140Q, JAK2 V617F single mutations, or combined BRAF D594N/KRAS A146V mutations), all tumors except the 1 tumor harboring the BRAF/KRAS mutations were associated with recurrence, disease-specific death, or both. Although this was a small set of cases, by Kaplan-Meier analysis, the presence of oncogene hotspot mutations was associated with significantly shorter disease-specific survival (p=0.003) and recurrence-free survival (p=0.03) in VP-MCC (Figure 2E, F). In contrast, oncogene mutations were not associated with outcome in VN-MCC (p=0.82 and 0.70 for DSS and RFS, respectively) (Figure 2E, F). The low frequency of individual mutations precluded further conclusions regarding outcomes. We did not observe differences in outcome associated with tumor suppressor mutations or copy loss for either group (data not shown).
Molecular Biomarker Profiling
118 tumors (49 VN-MCC and 69 VP-MCC) were further evaluated by transcriptional profiling of 26 cancer-relevant genes (Figure S4A). Of these, VP-MCC demonstrated significantly higher expression of certain kinase genes (ALK and NTRK1), tumor suppressors (BRCA1, RB1), and inflammatory genes (CD8A, IFNG, and GZMA), whereas VN-MCC displayed significantly higher expression of the receptor tyrosine kinase MET (Table S3). To control for differences in gene expression related to viral status, we analyzed VP-MCC and VN-MCC separately for prognostically informative transcripts. In VP-MCC, improved disease-specific survival was associated with higher expression of inflammatory transcripts IDO1, IFNG, and GZMA (Figure 3A, 3B, S4A, and Table S10). All 3 transcripts showed significant correlation in expression with each other and CD8A (not shown). For VN-MCC, high expression of certain oncogene (BRAF, RET, UBE2C) and tumor suppressor (BRCA1) transcripts was associated with worse survival (Figure 3C, 3D, and Table S10).
Figure 3. Prognostic biomarkers in Merkel cell carcinoma.
(A) Differentially expressed genes significantly associated with improved disease-specific survival in VP-MCC are immune markers. Hazard ratio is risk for death from disease (high expression over low expression) by gene and MCC subgroup. (B) Representative Kaplan-Meier curve of an inflammatory transcript (IDO1) for which high expression correlates with improved survival in VP-MCC. (C) Differentially expressed genes significantly associated with improved disease-specific survival in VN-MCC are oncogenic and tumor suppressor genes. Hazard ratio is risk for death from disease (high expression over low expression) by gene and MCC subgroup. (D) Representative Kaplan-Meier curve of a transcript (UBE2C) for which high expression correlates with worsened survival in VN-MCC. E) Density of peritumoral granzyme-expressing inflammatory cells (threshold: 75/mm2) by immunohistochemistry defines high and low risk groups for VP-MCC but not VN-MCC. (F) Density of intratumoral granzyme-expressing inflammatory cells (threshold: 5/mm2) by immunohistochemistry defines high and low risk groups for VP-MCC but not VN-MCC. (G) High density of IDO-1 expressing cells at the tumor-inflammatory interface (threshold: 100/mm2) is associated with favorable outcome in VP-MCC, and a trend toward improved outcome in VN-MCC. (H) Higher expression of UBE2C (threshold: h=65) is associated with a strong trend toward favorable outcome in VN-MCC. For RNA transcripts, amplicon details are in Table S10. DSS: disease-specific survival. VP: Virus-positive. VN: virus-negative. IHC: immunohistochemistry. Scale bar: 400 uM.
Of these candidate markers, 3 (GZMA, IDO1, and UBE2C) were selected for orthogonal validation by IHC on 57 VP-MCC and 50 VN-MCC primary tumors. In agreement with transcriptome expression data, higher density of granzyme-positive peritumoral lymphocytes (>75/mm2) and intratumoral lymphocytes (>4/mm2) was associated with improved disease-specific survival in VP-MCC tumors, but not VN-MCC (Figure 3E, F). High granzyme expression was also associated with longer recurrence-free survival (Figure S4B). IDO-1 was expressed predominantly in inflammatory cells at the tumor-stroma interface, with definitive tumor cell expression only identified in two tumors. A higher density of IDO-1-expressing peritumoral inflammatory cells was associated with improved disease-specific survival (Figure 3G). Higher UBE2C expression (H-score >65) correlated with a strong trend toward worsened survival in VN-MCC tumors, similar to transcriptional profiling results (Figure 3H).
DISCUSSION
To our knowledge, this is the largest study to date to combine integrated molecular profiling with disease-specific outcomes in MCC. We found that MCPyV status provides prognostic information independent of tumor stage and patient immune status, with VP-MCC associated with favorable outcome. Additional genomic and transcriptomic changes also have prognostic significance specific to VP-MCC or VN-MCC tumors, supporting roles for oncogene activation (VP-MCC), immune markers (VP-MCC) and oncogene/BRCA1 overexpression (VN-MCC) in modulating tumor aggressiveness.
There is currently no consensus regarding the relative sensitivity and specificity of IHC, RNA-ISH, qPCR, and NGS for detection of MCPyV. IHC was reported in one large study to be sensitive and specific for MCPyV relative to qPCR [7]. However, others have found PCR more sensitive [1, 7, 29, 30]. Multimodal detection of MCPyV may allow for maximal sensitivity and specificity [7, 30]. NGS appears to have high sensitivity and specificity [2, 4, 9, 31, 32]. Of note, while this manuscript was in review, a bait-capture NGS assay effective for demonstrating virus-host junctions in MCC was described [33]. However, MCPyV NGS assays are not standardized, and may not be feasible to implement in most clinical diagnostic laboratories. Our results suggest that assays compatible with common diagnostic laboratory workflows (IHC, ISH, and qPCR) are all prognostically informative when used singly or in combination. For clinical diagnostic testing, maximal sensitivity with tissue conservation may be achieved by a reflex approach (screening by IHC, confirmation of negative results by ISH or qPCR). Alternatively, we found that RNA-ISH and qPCR had high sensitivity and specificity as single assays.
We observed significant clinicopathologic associations with MCPyV status of MCC tumors. Our results clarify the association between virus-positive status and tumor features such as larger tumor size, increased tumor depth, and infiltrative growth, for which previous studies have yielded mixed results [7, 34-36]. In agreement with previous studies, we found that VP tumors are more likely to arise on the extremities and in women, whereas VN tumors predominate on the head and neck, arise more frequently in men, and are associated with SCCIS in a minority of cases [16, 34, 37]. Despite the larger size at presentation, we confirmed the relationship between VP status and improved survival. The association we observed between immune suppression and VN-MCC is in agreement with a smaller study published while this manuscript was in review [33]. Our results augment previous reports on MCPyV and outcome [1, 7, 29, 38, 39], although to our knowledge our study is the first to establish MCPyV as a prognostic factor independent of both immune status and stage.
Our results expand the spectrum of mutations described in MCC, and identify mutation profiles with prognostic significance in VP-MCC. The high incidence of TP53 and RB1 mutations in VN-MCC, with lower frequency of other mutations in other cellular genes, is consistent with previous reports [4-6, 29]. Although TP53 mutation has been associated with worse prognosis [5], our findings suggest the prognostic differences between VP-MCC and VN-MCC may be related to MCPyV status rather than TP53. In contrast, we found that oncogene mutational activation was associated with a more aggressive course in the small group of VP-MCC tumors harboring these events, but not VN tumors. It is unclear whether the association between oncogene mutations and outcome in VP-MCC is related to direct action of the oncogenes, as investigations into functional consequences of oncogene activation in MCC have been limited. Cultured MCC cell lines demonstrate evidence of PI3K pathway activation and are sensitive to inhibitors of PI3K and PI3K/mTOR [40, 41] suggesting an oncogenic role for aberrant activation of PI3K signaling in MCC. Furthermore, we observed increased beta-catenin expression associated with CTNNB1 mutation in tumors. The contribution of RAS family mutations is less clear, as MCC cell lines are not sensitive to MEK inhibition [6]. BRCA1/2 and CDKN2A mutations may also represent potential therapeutic targets. In addition, we identified rare mutations in oncogenes novel to MCC (IDH1, IDH2, JAK2) representing potentially targetable events. Further evaluation of mutations with prognostic and biological significance are needed to advance targeted therapies. Of note, JAK2, IDH1, and IDH2 mutations have been described as arising in the setting of clonal hematopoiesis, raising potential for false positive tumor mutation calls [42]; we cannot exclude the possibility of a similar phenomenon in our cases, although in one case IHC allowed for direct demonstration of IDH1 R132H protein expression in tumor cells. Finally, given the small number of VP-MCC tumors with oncogene activation in our cohort, continued investigation of this subclass will be helpful in confirming our observations.
Although definitive investigations into heterogeneity and clonal evolution in MCC are lacking, our previous reports support the presence of mutational heterogeneity between primary and metastatic MCC [8, 19]. For the first time, we show that TP53 mutations can be acquired during progression in rare cases of VP-MCC. This observation raises the possibility for acquired resistance to MDM2 inhibitors, that are currently under active investigation for management of advanced MCC [3]. A limitation of our study is that our approach does not allow for evaluation of clonal substructure in tumors.
We evaluated for differential expression of cancer and immune genes based upon outcome and viral status, and observed increased expression of immune markers (IDO1, GZMA, and IFNG) associated with favorable prognosis in VP-MCC. Extensive evidence supports a role for antitumor immunity in the clinical course of MCC, and suggests that MCPyV status may enhance immune infiltration. CD8+ T-cells are more numerous in association with VP-MCC compared to VN-MCC tumors [17, 43, 44] and have been associated with more favorable outcome in MCC [34, 43, 45-48]. Higher transcript expression of GZMA (granzyme A) and IFNG (interferon gamma) has been previously associated with favorable outcome in MCC [45]. However, we found this effect was restricted to VP-MCC by both bulk transcript expression and immunohistochemistry. In contrast to our results, one study found high IDO1 protein expression to be associated with worse prognosis in MCC [49]. Furthermore, as some patients overlapped between the transcriptome and immunohistochemistry cohorts in this study, additional validation in an independent cohort would be optimal to confirm the prognostic utility of these markers. Finally, further investigations beyond the scope of this study are needed to address associations between granzyme and IDO1, and previously described features such as briskness of tumor inflammation or CD8 expression [50].
High expression of transcripts including UBE2C, BRAF, RET, and BRCA1 was associated with poor prognosis for VN-MCC tumors, but not VP-MCC tumors. UBE2C is a ubiquitin conjugating enzyme that promotes cell cycle progression, and has been associated with unfavorable prognosis in many tumor types [51]. As most MCC tumors lack functional mutations in these genes, it is unclear how these gene products might mediate increased tumor aggressiveness. A similar phenomenon has been previously described for the receptor tyrosine kinase c-KIT, in which overexpression has been associated with poor prognosis despite an absence of activating mutations [39, 52]. Although there is benefit to improved prognostication, every prognostic group contained a subset of patients with poor outcome, and therefore effective clinical interventions remain essential for all patients with MCC.
Our study has several important limitations beyond those already mentioned. Our gene panel did not include a subset of recurrently altered genes in MCC including NOTCH genes, MYCL, and KMT2D. Of these, MYCL amplification was initially reported to occur in a substantial minority (36%) of MCC [12]; however, a subsequent large study found a much lower incidence (6%)[9] and therefore the omission of this gene from our panel is of unclear impact. Our approach relies on prioritization of predicted functional variants rather than direct comparison with germline DNA to identify somatic changes. However, as clinical data allows for reasonable exclusion of a background tumor syndrome for most patients, and the size of our cohort allows for effective artifact filtering, we have confidence that most or all of the prioritized variants represent true somatic events. The inclusion requirement for adequate available FFPE material may result in relatively increased representation of tumors with increased size or recurrent/metastatic disease. More significantly, due to the long-term nature of this cohort, we acknowledge that our approach does not account for the impact of immunotherapy on patient survival. Current FDA-approved immune checkpoint inhibitors for management of advanced stage MCC are pembrolizumab and avelumab. Current studies have not shown statistically significant differences in immunotherapy response rates between VP-MCC and VN-MCC [53, 54]. While long-term data is pending, it is likely that survival will improve at a similar rate in both groups, hence preserving the trends reported here. Finally, as treatment with immune checkpoint inhibitors do not impact outcomes until advanced disease, our observations will likely also remain relevant with regard to recurrence/progression of low-stage tumors.
In summary, we confirm and expand previous observations regarding clinicopathologic associations between MCPyV status and MCC. We find that VP-MCC tumors are associated with a more favorable course, and this effect is independent of other prognostic variables including stage. Additional features of the tumor genome, transcriptome, and microenvironment may further modify prognosis in a manner specific to MCC subclass. These observations underscore the prognostic significance of MCPyV, and the necessity of MCPyV classification during investigation of prognostic markers.
Supplementary Material
Statement of Translational Relevance.
Merkel cell carcinoma (MCC) is an aggressive neuroendocrine skin malignancy with high rates of metastasis and mortality, that can be divided into two subclasses: Merkel cell polyomavirus (MCPyV)-positive tumors expressing oncogenic viral T antigens, and MCPyV-negative tumors with mutations of TP53 and RB1. Here, we show that MCPyV-positive tumors display more favorable prognosis in univariate analyses, and in multivariate analysis including stage. Furthermore, we find that the predictive value of prognostic biomarkers is dependent on MCC subclass, with inflammatory markers such as granzyme and IDO1 associated with improved prognosis in MCPyV-positive MCC. Our findings highlight fundamental molecular and prognostic differences between the MCC subclasses, and reveal that incorporating MCPyV status can improve the predictive value of outcome analyses for biomarkers in MCC.
ACKNOWLEDGEMENTS
We thank Nisha Meireles, Sherry Fu, Sharon Kerr, and Threase Nickerson for their efforts to support database review and case retrieval. Tissue processing and immunohistochemical staining was performed in part by the Rogel Cancer Center Tissue and Molecular Shared Resource (supported by NIH P30 CA04659229). Whole-genome sequencing was carried out in the Advanced Genomics Core at the University of Michigan. Funding was provided in part by the Anatomic Pathology Project Funding Committee of the Department of Pathology, Michigan Medicine.
Financial Support:
This project was supported by the Anatomic Pathology Project Funding Committee of the Department of Pathology, Michigan Medicine.
Footnotes
Conflict of Interest Disclosure
D.R.R., S.A.T., and B.J. are employed by, and equity holders in, Strata Oncology. The remaining authors declare no potential conflicts of interest.
REFERENCES
- 1.Harms PW, Harms KL, Moore PS, DeCaprio JA, Nghiem P, Wong MKK, et al. The biology and treatment of Merkel cell carcinoma: current understanding and research priorities. Nat Rev Clin Oncol 2018; 15: 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng H, Shuda M, Chang Y, and Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008; 319: 1096–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park DE, Cheng J, Berrios C, Montero J, Cortes-Cros M, Ferretti S, et al. Dual inhibition of MDM2 and MDM4 in virus-positive Merkel cell carcinoma enhances the p53 response. Proc Natl Acad Sci U S A 2019; 116: 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harms PW, Vats P, Verhaegen ME, Robinson DR, Wu YM, Dhanasekaran SM, et al. The Distinctive Mutational Spectra of Polyomavirus-Negative Merkel Cell Carcinoma. Cancer Res 2015; 75: 3720–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goh G, Walradt T, Markarov V, Blom A, Riaz N, Doumani R, et al. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget 2016; 7: 3403–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong SQ, Waldeck K, Vergara IA, Schroder J, Madore J, Wilmott JS, et al. UV-Associated Mutations Underlie the Etiology of MCV-Negative Merkel Cell Carcinomas. Cancer Res 2015; 75: 5228–34. [DOI] [PubMed] [Google Scholar]
- 7.Moshiri AS, Doumani R, Yelistratova L, Blom A, Lachance K, Shinohara MM, et al. Polyomavirus-Negative Merkel Cell Carcinoma: A More Aggressive Subtype Based on Analysis of 282 Cases Using Multimodal Tumor Virus Detection. J Invest Dermatol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harms PW, Collie AM, Hovelson DH, Cani AK, Verhaegen ME, Patel RM, et al. Next generation sequencing of Cytokeratin 20-negative Merkel cell carcinoma reveals ultraviolet-signature mutations and recurrent TP53 and RB1 inactivation. Mod Pathol 2016; 29: 240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knepper TC, Montesion M, Russell JS, Sokol ES, Frampton GM, Miller VA, et al. The Genomic Landscape of Merkel Cell Carcinoma and Clinicogenomic Biomarkers of Response to Immune Checkpoint Inhibitor Therapy. Clin Cancer Res 2019; 25: 5961–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hafner C, Houben R, Baeurle A, Ritter C, Schrama D, Landthaler M, et al. Activation of the PI3K/AKT pathway in Merkel cell carcinoma. PLoS One 2012; 7: e31255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki T, Matsushita M, Nonaka D, Kuwamoto S, Kato M, Murakami I, et al. Comparison of Akt/mTOR/4E-BP1 pathway signal activation and mutations of PIK3CA in Merkel cell polyomavirus-positive and Merkel cell polyomavirus-negative carcinomas. Hum Pathol 2014; 46: 210–6. [DOI] [PubMed] [Google Scholar]
- 12.Paulson KG, Lemos BD, Feng B, Jaimes N, Penas PF, Bi X, et al. Array-CGH reveals recurrent genomic changes in Merkel cell carcinoma including amplification of L-Myc. J Invest Dermatol 2009; 129: 1547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veija T, Sarhadi VK, Koljonen V, Bohling T, and Knuutila S. Hotspot mutations in polyomavirus positive and negative Merkel cell carcinomas. Cancer Genet 2016; 209: 30–5. [DOI] [PubMed] [Google Scholar]
- 14.Xie H, Liu T, Wang N, Bjornhagen V, Hoog A, Larsson C, et al. TERT promoter mutations and gene amplification: Promoting TERT expression in Merkel cell carcinoma. Oncotarget 2014; 5: 10048–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter MD, Gaston D, Huang WY, Greer WL, Pasternak S, Ly TY, et al. Genetic profiles of different subsets of Merkel cell carcinoma show links between combined and pure MCPyV-negative tumors. Hum Pathol 2018; 71: 117–125. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Harms PW, Palanisamy N, Carskadon S, Cao X, Siddiqui J, et al. Age and Gender Associations of Virus Positivity in Merkel Cell Carcinoma Characterized Using a Novel RNA In Situ Hybridization Assay. Clin Cancer Res 2017; 23: 5622–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harms PW, Patel RM, Verhaegen ME, Giordano TJ, Nash KT, Johnson CN, et al. Distinct gene expression profiles of viral- and nonviral-associated merkel cell carcinoma revealed by transcriptome analysis. J Invest Dermatol 2013; 133: 936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodig SJ, Cheng J, Wardzala J, DoRosario A, Scanlon JJ, Laga AC, et al. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J Clin Invest 2012; 122: 4645–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harms KL, Lazo de la Vega L, Hovelson DH, Rahrig S, Cani AK, Liu CJ, et al. Molecular Profiling of Multiple Primary Merkel Cell Carcinoma to Distinguish Genetically Distinct Tumors From Clonally Related Metastases. JAMA Dermatol 2017; 153: 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arora R, Choi JE, Harms PW, and Chandrani P. Merkel cell polyomavirus in Merkel cell carcinoma: Integration sites and involvement of the KMT2D tumor suppressor gene. Submitted to Viruses. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013; 1303:3997. [Google Scholar]
- 22.Garrison E and Marth G. Haplotype-based variant detection from short-read sequencing. arXiv 2012; 1207: 3907. [Google Scholar]
- 23.Layer RM, Chiang C, Quinlan AR, and Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 2014; 15: R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.File DM, Morgan KP, and Khagi S. Durable Near-Complete Response to Olaparib Plus Temozolomide and Radiation in a Patient With ATM-Mutated Glioblastoma and MSH6-Deficient Lynch Syndrome. JCO Precision Oncology 2020: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller TI, Zoumberos NA, Johnson B, Rhodes DR, Tomlins SA, Chan MP, et al. A genomic survey of sarcomas on sun-exposed skin reveals distinctive candidate drivers and potentially targetable mutations. Hum Pathol 2020; 102: 60–69. [DOI] [PubMed] [Google Scholar]
- 26.Harms KL, Chubb H, Zhao L, Fullen DR, Bichakjian CK, Johnson TM, et al. Increased expression of EZH2 in Merkel cell carcinoma is associated with disease progression and poorer prognosis. Hum Pathol 2017; 67: 78–84. [DOI] [PubMed] [Google Scholar]
- 27.Storey J, Bass A, Dabney A, and Robinson D. qvalue: Q-value estimation for false discovery rate control. R package version 2.18.0. 2019; Available from: http://github.com/jdstorey/qvalue. [Google Scholar]
- 28.Bichakjian CK, Olencki T, Alam M, Andersen JS, Berg D, Bowen GM, et al. Merkel cell carcinoma, version 1.2014. J Natl Compr Canc Netw 2014; 12: 410–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sihto H, Kukko H, Koljonen V, Sankila R, Bohling T, and Joensuu H. Merkel cell polyomavirus infection, large T antigen, retinoblastoma protein and outcome in Merkel cell carcinoma. Clin Cancer Res 2011; 17: 4806–13. [DOI] [PubMed] [Google Scholar]
- 30.Matsushita M, Nonaka D, Iwasaki T, Kuwamoto S, Murakami I, Kato M, et al. A new in situ hybridization and immunohistochemistry with a novel antibody to detect small T-antigen expressions of Merkel cell polyomavirus (MCPyV). Diagn Pathol 2014; 9: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duncavage EJ, Magrini V, Becker N, Armstrong JR, Demeter RT, Wylie T, et al. Hybrid capture and next-generation sequencing identify viral integration sites from formalin-fixed, paraffin-embedded tissue. J Mol Diagn 2011; 13: 325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starrett GJ, Marcelus C, Cantalupo PG, Katz JP, Cheng J, Akagi K, et al. Merkel Cell Polyomavirus Exhibits Dominant Control of the Tumor Genome and Transcriptome in Virus-Associated Merkel Cell Carcinoma. MBio 2017; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starrett GJ, Thakuria M, Chen T, Marcelus C, Cheng J, Nomburg J, et al. Clinical and molecular characterization of virus-positive and virus-negative Merkel cell carcinoma. Genome Med 2020; 12: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldmeyer L, Hudgens CW, Ray-Lyons G, Nagarajan P, Aung PP, Curry JL, et al. Density, Distribution, and Composition of Immune Infiltrates Correlate with Survival in Merkel Cell Carcinoma. Clin Cancer Res 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higaki-Mori H, Kuwamoto S, Iwasaki T, Kato M, Murakami I, Nagata K, et al. Association of Merkel cell polyomavirus infection with clinicopathological differences in Merkel cell carcinoma. Hum Pathol 2012; 43: 2282–91. [DOI] [PubMed] [Google Scholar]
- 36.Leroux-Kozal V, Leveque N, Brodard V, Lesage C, Dudez O, Makeieff M, et al. Merkel cell carcinoma: histopathologic and prognostic features according to the immunohistochemical expression of Merkel cell polyomavirus large T antigen correlated with viral load. Hum Pathol 2015; 46: 443–53. [DOI] [PubMed] [Google Scholar]
- 37.Haymerle G, Janik S, Fochtmann A, Pammer J, Schachner H, Nemec L, et al. Expression of Merkelcell polyomavirus (MCPyV) large T-antigen in Merkel cell carcinoma lymph node metastases predicts poor outcome. PLoS One 2017; 12: e0180426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kervarrec T, Tallet A, Miquelestorena-Standley E, Houben R, Schrama D, Gambichler T, et al. Morphologic and immunophenotypical features distinguishing Merkel cell polyomavirus-positive and negative Merkel cell carcinoma. Mod Pathol 2019; 32: 1605–1616. [DOI] [PubMed] [Google Scholar]
- 39.Waltari M, Sihto H, Kukko H, Koljonen V, Sankila R, Bohling T, et al. Association of Merkel cell polyomavirus infection with tumor p53, KIT, stem cell factor, PDGFR-alpha and survival in Merkel cell carcinoma. Int J Cancer 2011; 129: 619–28. [DOI] [PubMed] [Google Scholar]
- 40.Lin Z, Mei H, Fan J, Yin Z, and Wu G. Effect of the dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor NVP-BEZ235 against human Merkel cell carcinoma MKL-1 cells. Oncol Lett 2015; 10: 3663–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nardi V, Song Y, Santamaria-Barria JA, Cosper AK, Lam Q, Faber AC, et al. Activation of PI3K signaling in Merkel cell carcinoma. Clin Cancer Res 2012; 18: 1227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ptashkin RN, Mandelker DL, Coombs CC, Bolton K, Yelskaya Z, Hyman DM, et al. Prevalence of Clonal Hematopoiesis Mutations in Tumor-Only Clinical Genomic Profiling of Solid Tumors. JAMA Oncol 2018; 4: 1589–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sihto H, Bohling T, Kavola H, Koljonen V, Salmi M, Jalkanen S, et al. Tumor infiltrating immune cells and outcome of Merkel cell carcinoma: a population-based study. Clin Cancer Res 2012; 18: 2872–81. [DOI] [PubMed] [Google Scholar]
- 44.Ricci C, Righi A, Ambrosi F, Gibertoni D, Maletta F, Uccella S, et al. Prognostic Impact of MCPyV and TIL Subtyping in Merkel Cell Carcinoma: Evidence from a Large European Cohort of 95 Patients. Endocr Pathol 2019. [DOI] [PubMed] [Google Scholar]
- 45.Paulson KG, Iyer JG, Tegeder AR, Thibodeau R, Schelter J, Koba S, et al. Transcriptome-wide studies of merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. J Clin Oncol 2011; 29: 1539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kervarrec T, Gaboriaud P, Berthon P, Zaragoza J, Schrama D, Houben R, et al. Merkel cell carcinomas infiltrated with CD33(+) myeloid cells and CD8(+) T cells are associated with improved outcome. J Am Acad Dermatol 2018; 78: 973–982 e8. [DOI] [PubMed] [Google Scholar]
- 47.von der Grun J, Winkelmann R, Meissner M, Wieland U, Silling S, Martin D, et al. Merkel Cell Polyoma Viral Load and Intratumoral CD8+ Lymphocyte Infiltration Predict Overall Survival in Patients With Merkel Cell Carcinoma. Front Oncol 2019; 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulson KG, Iyer JG, Simonson WT, Blom A, Thibodeau RM, Schmidt M, et al. CD8+ lymphocyte intratumoral infiltration as a stage-independent predictor of Merkel cell carcinoma survival: a population-based study. Am J Clin Pathol 2014; 142: 452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wardhani LO, Matsushita M, Iwasaki T, Kuwamoto S, Nonaka D, Nagata K, et al. Expression of the IDO1/TDO2-AhR pathway in tumor cells or the tumor microenvironment is associated with Merkel cell polyomavirus status and prognosis in Merkel cell carcinoma. Hum Pathol 2019; 84: 52–61. [DOI] [PubMed] [Google Scholar]
- 50.Tetzlaff MT and Harms PW. Danger is only skin deep: aggressive epidermal carcinomas. An overview of the diagnosis, demographics, molecular-genetics, staging, prognostic biomarkers, and therapeutic advances in Merkel cell carcinoma. Mod Pathol 2020; 33: 42–55. [DOI] [PubMed] [Google Scholar]
- 51.Hao Z, Zhang H, and Cowell J. Ubiquitin-conjugating enzyme UBE2C: molecular biology, role in tumorigenesis, and potential as a biomarker. Tumour Biol 2012; 33: 723–30. [DOI] [PubMed] [Google Scholar]
- 52.Andea AA, Patel R, Ponnazhagan S, Kumar S, DeVilliers P, Jhala D, et al. Merkel cell carcinoma: correlation of KIT expression with survival and evaluation of KIT gene mutational status. Hum Pathol 2010; 41: 1405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nghiem P, Bhatia S, Lipson EJ, Sharfman WH, Kudchadkar RR, Brohl AS, et al. Durable Tumor Regression and Overall Survival in Patients With Advanced Merkel Cell Carcinoma Receiving Pembrolizumab as First-Line Therapy. J Clin Oncol 2019; 37: 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D'Angelo SP, Bhatia S, Brohl AS, Hamid O, Mehnert JM, Terheyden P, et al. Avelumab in patients with previously treated metastatic Merkel cell carcinoma: long-term data and biomarker analyses from the single-arm phase 2 JAVELIN Merkel 200 trial. J Immunother Cancer 2020; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.